Abstract
The function of the conserved zinc-binding domains in the related Pol II- and Pol III-specific factors TFIIB and Brf was investigated. Three-dimensional structure modeling and mutagenesis studies indicated that for both factors, the functional surface of the zinc ribbon fold consists of a small conserved patch of residues located on one face of the domain comprised mainly of the second and third antiparallel β strands. Previous studies have shown that the TFIIB zinc ribbon is essential for recruitment of Pol II into the preinitiation complex. In contrast, Pol III recruitment assays and in vitro transcription demonstrate that the disruption of the Brf zinc ribbon does not lead to a defect in Pol III recruitment but, rather, a defect in open complex formation. Therefore, the same conserved surface of the zinc ribbon domain has been adapted to serve distinct roles in the Pol II and Pol III transcription machinery.
Keywords: TFIIB, Brf, RNA Pol II, zinc ribbon domain, transcription
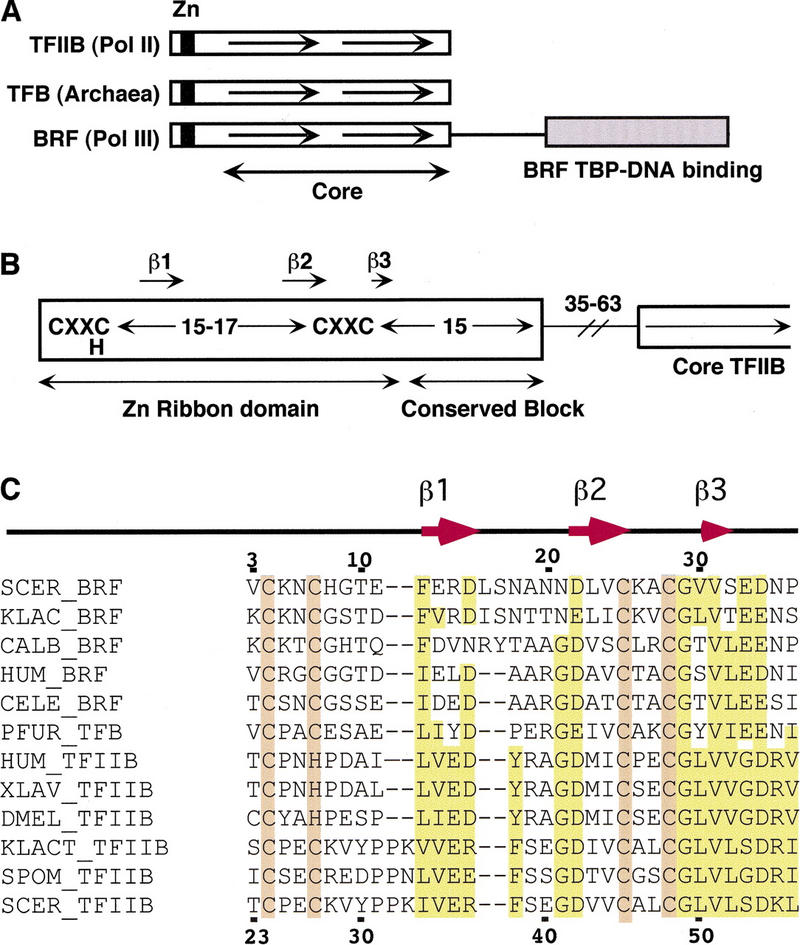
The TFIIB family is comprised of three classes of general transcription factors (Fig. 1A). TFIIB functions specifically in RNA polymerase II (Pol II) transcription, in which it binds the TATA-binding protein (TBP)–DNA complex and recruits RNA Pol II/TFIIF to the promoter (Orphanides et al. 1996; Hampsey 1998). In vivo, TFIIB is a component of the RNA Pol II holoenzyme (Myer and Young 1998) and likely binds the TFIID–TFIIA–DNA complex as a subunit of holoenzyme (Ranish et al. 1999). The Pol III-specific factor Brf, together with TBP and B”, are subunits of TFIIIB, which forms stable promoter complexes and functions to recruit RNA Pol III to promoters (Chedin et al. 1998; Colbert et al. 1998; Kumar et al. 1998; Shen et al. 1998). TFB is an Archaea general factor, and along with Archaea TBP, promotes transcription by Archaea RNA polymerase (Hausner et al. 1996; Qureshi et al. 1997). TFIIB and Brf have been widely conserved among eukaryotes, and TFB has been conserved among all Archaea species examined.
Figure 1.
The TFIIB family and the conserved zinc ribbon fold. (A) The three classes of TFIIB family members; (B) schematic of the TFIIB family amino-terminal domain; (C) sequence alignment of Brf, TFB, and TFIIB family members. Conserved secondary structural elements are indicated. The numbering at the top is for Saccharomyces cerevisiae BRF and numbering at the bottom is for S. cerevisiae TFIIB. The models derived in this study were for S. cerevisiae Brf residues 3–33 and TFIIB residues 23–53.
The amino-terminal region of all three factors is comprised of a zinc-binding region followed by a conserved block of ∼15 residues (Fig. 1B,C). The conserved block forms a domain distinct from the zinc ribbon and in TFIIB plays a role in transcription start site selection (Pinto et al. 1994; Pardee et al. 1998), in polymerase activity after preinitiation complex formation (Ranish et al. 1999), and probably makes an intramolecular interaction with the core domain (Roberts and Green 1994).
The zinc-binding region of the TFIIB family contains the sequence motif CXXC(H)-15/17-CXXC. The structure of this zinc ribbon fold was determined for one TFB factor using NMR (Zhu et al. 1996). To date, the structures of three distinct zinc ribbon domains have been determined by NMR and the presence of this fold in a number of other proteins has been inferred from sequence homology (Qian et al. 1993; Olmsted et al. 1998; Wang et al. 1998). Factors containing this mini domain have very diverse amino acid sequence and function. Examples include the RNA Pol II elongation factor TFIIS, subunits of RNA Pol I, Pol II, and Pol III (Chedin et al. 1998), the general transcription factor TFIIE, DNA polymerase α, and T4 DNA primase. The only conserved sequence features of this fold appear to be the CXXC(H) and CXXC motifs. This fold consists of a rubredoxin knuckle containing the first CXXC followed by a β strand of variable length (see Fig. 2B). This is connected to an antiparallel strand by a flexible loop. The second β strand is followed by a second CXXC containing a rubredoxin knuckle, which connects to a third very short antiparallel β strand. It has been proposed that the great sequence diversity of zinc ribbons is due to the planar nature of this fold, along with zinc coordination, which eliminates the need for a hydrophobic core (Schwabe and Klug 1994; Wang et al. 1998). This great sequence diversity allows the fold to be used in proteins of very different function.
Figure 2.

Structure modeling of the Brf and TFIIB zinc ribbons. (A) Polypeptide backbone of two P. furiosis TFB zinc ribbon domains used as templates for modeling (blue) and the backbone of the three best Brf models (pink). This as well as other data described in this paper can be viewed as three-dimensional models at the WWW site indicated in the text. (B) Cartoon of one Brf model with the three β strands indicated. The side chains of four Cys residues (yellow and green) coordinating the zinc atom (red) are shown. (C) Polypeptide backbone of two P. furiosis TFB zinc ribbon domains used as templates for modeling (blue) and the backbone of the three best TFIIB models (pink).
Disruption of the ribbon domain by mutation of conserved cysteine residues in yeast TFIIB and Brf is lethal (Buratowski and Zhou 1993; this work). Mutation of the TFIIB ribbon severely compromises in vitro transcription under all conditions tested (Buratowski and Zhou 1993; Pardee et al. 1998; Ranish et al. 1999). The zinc ribbon of TFIIB likely interacts with Pol II and/or TFIIF. Disruption of the domain blocks coimmune precipitation of Pol II and TFIIB (Pardee et al. 1998) and prevents recruitment of Pol II/TFIIF to the TBP–TFIIB–DNA complex (Barberis et al. 1993; Buratowski and Zhou 1993; Hisatake et al. 1993; Ranish et al. 1999). The ribbon domain of TFIIB has also been reported to interact with two transcription activators (Colgan et al. 1995; Masuyama et al. 1997).
In a highly purified system, deletion of a large portion of the amino terminus of yeast Brf including the zinc ribbon, the conserved block, and the first repeat of the core domain leads to a strong decrease in initiation but does not completely eliminate transcription (Kassavetis et al. 1997, 1998). The target of the Brf ribbon domain is unknown. It is possible that the Brf ribbon domain contacts Pol III, or alternatively contacts a Pol III general factor.
At least three Pol II general factors function after recruitment of Pol II to the preinitiation complexes (PICs) TFIIB, TFIIF, and TFIIH (Coin and Egly 1998; Lei et al. 1998; Ranish et al. 1999). In the Pol III system, two factors were shown to affect open complex formation by Pol III. Mutations were isolated in the Pol III subunit C34, which affected open complex formation but not Pol III recruitment (Brun et al. 1997). Similar defects were observed with several small internal deletions in the TFIIIB subunit B”, which specifically affected open complex formation (Kassavetis et al. 1998). In addition, these B” deletions were observed to have a more severe transcription defect with supercoiled rather than linear DNA. In contrast, the large Brf amino-terminal deletion mentioned above had a slightly lower defect on supercoiled DNA compared with linear DNA, but was severely compromised for function on both templates (Kassavetis et al. 1998).
As a step in understanding the function and target of the zinc ribbon domains in the TFIIB family, three-dimensional structure modeling and systematic mutagenesis was used to identify the functional surface of this domain in TFIIB and Brf. The resulting mutations in the Brf ribbon domain were used to probe for the function of the ribbon domain. Our results show that the two homologous domains play distinct roles in the Pol II and Pol III systems.
Results
Structure modeling of the Brf zinc ribbon domain
As a first step in identification of the functional surface of the Brf zinc ribbon, the three-dimensional structure of the Brf ribbon fold was modeled. The NMR structure of Pyrococcus furiosis TFB (Zhu et al. 1996) was used as a template for modeling the structure of Brf. An overlay of the 25 NMR structure models for P. furiosis TFB in the protein database showed that the polypeptide backbone of these models was very consistent within the zinc ribbon fold (residues 6–34). However, the residues amino- or carboxy-terminal to this fold had no consistent structure in the models and were not used in Brf modeling. The sequence alignment used for modeling Brf residues 3–33 is shown in Figure 1C. The three yeast Brfs contain an insertion of two residues in the loop between β strands one and two compared with the other TFIIB family members. The position of this insertion in the sequence alignment was arbitrary, as there is no homology between TFB and Brf in this region. Using the program Modeller (Sali and Blundell 1993), three TFB NMR models were used as templates to generate a number of Brf Models (Materials and Methods). Statistically based structural validation methods were used to select the best three models that closely followed the characteristics for proteins of known structure. The three best Brf models were very similar and all scored at least as well or better in these tests than the NMR models used as templates. Figure 2A shows the backbone of two TFB structure models (blue) along with several Brf models (pink). The main difference in the models is the polypeptide backbone of residues within the loop connecting β strands one and two. This occurred because this loop contains the two extra residues mentioned above and also because there is no sequence homology between Brf and TFB in this segment. A cartoon representation of one Brf model is shown in Figure 2B along with the location of the four Cys side chains (green and yellow) coordinating the zinc atom (red).
Interactive three-dimensional models of both the Brf and TFIIB zinc ribbons, along with display of the functionally important residues and coordinates of the models, can be viewed on the World-Wide Web (WWW) at: http://www.fhcrc.org/science/basic/labs/hahn/chime_ pages/3dstruct_index.html.
Radical and alanine scanning mutagenesis
Residues most likely to be surface exposed were identified using the three best structure models for the BRF zinc ribbon. Because the flexible loop region did not give a consistent structure in the three models, surface residues in the loop could not be identified. The ribbon domain is unlike a typical globular protein in that it does not contain a hydrophobic core. Instead, the domain is formed by coordination of the zinc atom as well as several van der Waals interactions and a network of six hydrogen bonds holding the anti-parallel β strands. Thus, there are no truly buried side chains in this domain. Residues targeted for mutagenesis were identified using surface accessibility calculations as well as identification of residues that do not make intramolecular interactions predicted to be important for structure or stability of the domain or disrupt coordination of the zinc atom.
The side chains identified above were targeted for radical mutagenesis, in which amino acid side chains were replaced by either glutamic or aspartic acid. Radical mutagenesis was used because in previous tests in vivo, this type of substitution gave a stronger phenotype and was more likely to identify functional regions of the protein (Bryant et al. 1996; Tang et al. 1996; Chou and Struhl 1997; Lee and Struhl 1997). Radical substitutions that cause a defect in function do not necessarily imply that the wild-type side chain is important for function. Two alternative explanations are that the newly introduced bulky-charged side chain interferes with binding an interacting factor, or that it affects the structural integrity of the domain. To test for the requirement of particular side chains, alanine substitutions were also introduced at all surface-exposed residues causing truncation of the side chain at the β carbon. Finally, to test the in vivo requirement for this domain in cell growth, truncations lacking the first 12 or 24 residues of Brf were generated.
All of the above mutations were made in a low-copy number centromere containing vector that expresses HA epitope-tagged Brf under control of its own promoter (Materials and Methods). The HA epitope did not affect function of Brf in vivo or in vitro. The mutant plasmids were used to replace the wild-type yeast BRF gene by plasmid shuffle. The resulting growth phenotype was measured on synthetic glucose plates at 25, 30, and 35.5°C (Table 1). In contrast to our earlier findings, disruption of the zinc ribbon domain by deletion of the first 12 or 24 amino acids of Brf was lethal and did not cause a cold-sensitive phenotype as reported previously (Colbert and Hahn 1992). The reason for this difference is unknown.
Table 1.
In vivo function of Brf mutations
| Mutation
|
18°C
|
25°C
|
30°C
|
35.5°C
|
|---|---|---|---|---|
| Wild type | +++ | +++ | +++ | +++ |
| V3E | +++ | +++ | +++ | |
| V3A | +++ | +++ | +++ | |
| K5E | +++ | +++ | +++ | |
| K5A | +++ | +++ | +++ | |
| N6E | +++ | +++ | +++ | |
| N6A | +++ | +++ | +++ | |
| H8E | +++ | +++ | +++ | |
| H8A | +++ | +++ | +++ | |
| T10E | +++ | +++ | +++ | |
| T10A | +++ | +++ | +++ | |
| E11R | +++ | +++ | +++ | |
| E11A | +++ | +++ | +++ | |
| E13R | +++ | +++ | +++ | +++ |
| R14E | +++ | +++ | +++ | |
| R14A | +++ | +++ | +++ | |
| D22R | NA | lethal | NA | NA |
| D22A | ++ | ++ | ++ | ++ |
| V24E | NA | lethal | NA | NA |
| V24A | +++ | +++ | +++ | |
| K26E | +++ | +++ | +++ | |
| K26A | +++ | +++ | +++ | |
| V30E | +/− | + | + | + |
| V30A | +++ | +++ | +++ | +++ |
| V31E | +/− | ++ | ++ | + |
| V31A | ++ | ++ | ++ | + |
| E33R | ++ | ++ | ++ | |
| E33A | +++ | +++ | +++ | |
| Δ2-12 | NA | lethal | NA | NA |
| Δ2-26 | NA | lethal | NA | NA |
Yeast growth phenotypes on minimal complete glucose media caused by radical and alanine substitution as well as deletion of the domain are indicated relative to wild-type Brf. Growth phenotypes different from wild type are indicated by shading: dark gray for slow growth or temperature sensitive; light gray for lethal.
Figure 3, A and B (also see the WWW site), shows two faces of the Brf zinc ribbon model with side chains color coded according to the in vivo phenotype caused by radical substitution as follows: (yellow) no growth defect; (blue) lethal; (red) temperature sensitive and/or slow growth phenotype. It is striking that one face of the fold is insensitive to mutagenesis, whereas on the other face, four residues important for function (D22, V24, V30, and V31) are clustered. A fifth residue identified by radical mutagenesis, E33, is near this surface. However, because E33 is the last residue in the model, it is not as certain whether this side chain is likely to be surface exposed in the context of full-length Brf, and whether it functions as part of the ribbon fold or the conserved sequence block domain carboxy-terminal to the ribbon domain. Of the four clustered residues, D22 and V24 are located in the second β strand, V31 is located in the third β strand, and V30 is positioned at the junction between the second knuckle and the third β strand.
Figure 3.

In vivo phenotypes of Brf and TFIIB zinc ribbon mutations and their position on the structure models. (A,B) Two faces of the Brf ribbon structure model are shown with the side chains targeted for mutagenesis color coded by growth phenotype caused by radical substitution. (Yellow) No growth defect; (blue) lethal; (red) temperature sensitive and/or slow growth phenotype. Also indicated are mutations that cause the in vivo phenotypes. (C,D) Two faces of the TFIIB ribbon structure model are shown with the side chains targeted for mutagenesis color coded by in vivo phenotype as was done in A and B. The views of the Brf model in A and B correspond to the same faces of the TFIIB model shown in C and D, respectively. These data can also be viewed three-dimensionally at the web site indicated in the text.
Of the side chains that showed a phenotype when mutated, D22 and V24 are the least accessible. However, in two of the three best Brf structure models, the D22 side chain is not predicted to interact with any other residue. In one model, D22 makes a hydrogen bond with the side chain of residue N20. N20 is not conserved in Brfs, so this interaction is not predicted to be important. Modeling of the Brf structure with the D22R or A mutations gave models that scored well in the statistical tests and had structures essentially identical to the wild-type Brf models (see WWW site). The side chain V24 is predicted to make two van der Waals interactions with V31 and D15. Modeling the V24E mutation gave models that again scored well and in which the E side chain fit nicely into the space occupied by the V side chain with the charged part of the surface exposed. The interaction with D15 is preserved in the modeled structure (see WWW site). Therefore, mutation of D22 and V24, the least exposed side chains in the Brf ribbon domain, is not predicted to be detrimental to the ribbon structure.
As an in vivo test for the structural integrity of the mutagenized ribbon fold, the in vivo stability of the mutant proteins was measured. Cells containing both an HA-tagged mutant and wild-type untagged copy of Brf were grown in minimal medium and cellular-protein extracted. The levels of mutant protein were assayed by Western blot, probing for the HA epitope inserted on the mutant copy of Brf. All of the mutant proteins were stably expressed at least as well as wild-type Brf, with the exception of the truncation Δ2–12, which disrupts the domain and was expressed at threefold-lower steady state levels (not shown).
Modeling and mutagenesis of the TFIIB zinc ribbon
Because it is not clear as to what extent the function of TFIIB and Brf have been conserved in their respective transcription systems, it was of interest to compare the function of the zinc ribbon domains in both factors. Because the precise molecular target of the domain for either factor is unknown, it was impossible to predict whether the location of the functional surfaces in the two factors would be conserved. To answer this question, the structure of the yeast TFIIB zinc ribbon fold was modeled with the same strategy used for Brf. The sequence alignment with TFB used for the modeling is shown in Figure 1C. The yeast TFIIB factors all contain an insertion of two residues between the first rubredoxin knuckle and β strand. The position of this insertion in the alignment was arbitrary, as there is no homology between the family members in this region. With the same structure validation methods used for the Brf models, the best three TFIIB models were selected, were all very similar, and scored well in the statistical tests. The polypeptide backbone of three models along with two P. furiosis NMR models closely align except for the region between the first knuckle and β strand at the location of the two residue insertion (Fig. 2C; see WWW site).
Amino acid side chains in the TFIIB ribbon were targeted for radical mutagenesis in a centromere containing yeast expression vector with TFIIB expression driven by the natural promoter. The mutagenized TFIIB contained a Flag epitope tag at the carboxyl terminus, which had no effect on in vivo function. The mutagenized TFIIB plasmids were used to replace the wild-type copy of the TFIIB gene, SUA7, by plasmid shuffle, and the growth phenotype was measured as was done for Brf mutations (Table 2).
Table 2.
In vivo function of TFIIB mutations
| Mutation
|
18°C
|
25°C
|
30°C
|
35.5°C
|
|---|---|---|---|---|
| Wild type | +++ | +++ | +++ | +++ |
| T23E | +++ | +++ | +++ | +++ |
| P25E | +++ | +++ | +++ | +++ |
| E26R | ++ | ++ | ++ | + |
| E26A | +++ | +++ | +++ | +++ |
| E26N | +++ | +++ | +++ | ++ |
| K28E | +++ | +++ | +++ | +++ |
| Y30E | +++ | +++ | +++ | +++ |
| P32E | +++ | +++ | +++ | +++ |
| K33E | +++ | +++ | +++ | +++ |
| I34E | +++ | +++ | +++ | +++ |
| V35E | +++ | +++ | +++ | +++ |
| E36R | +++ | +++ | +++ | +++ |
| R37E | +++ | +++ | +++ | +++ |
| F38E | +++ | +++ | +++ | +++ |
| S39E | +++ | +++ | +++ | +++ |
| E40R | +++ | +++ | +++ | +++ |
| D42R | N.D. | + | + | + |
| D42A | N.D. | +++ | +++ | +++ |
| D42C | N.D. | +++ | +++ | +++ |
| D42E | +++ | +++ | +++ | +++ |
| V43E | +++ | +++ | +++ | +++ |
| V44E | +++ | ++ | ++ | ++ |
| V44A | +++ | +++ | +++ | ++ |
| L47E | +++ | +++ | +++ | +++ |
| L50E | ++ | ++ | ++ | + |
| L50A | ++ | ++ | ++ | + |
| L50V | +++ | +++ | +++ | +++ |
| V51E | ++ | ++ | ++ | + |
| V51A | +++ | +++ | ++ | + |
| S53E | +++ | +++ | +++ | +++ |
Yeast growth phenotypes on minimal complete glucose media caused by site-directed mutagenesis of the yeast TFIIB zinc ribbon domain are indicated relative to wild-type TFIIB. Growth phenotypes different from wild type are indicated by shading; which indicates slow growth.
Figure 3, C and D, shows two faces of the TFIIB zinc ribbon model with side chains color coded by growth phenotype. Again, it is striking that one face of the TFIIB fold is insensitive to mutagenesis, whereas on the other face, five residues important for function (E26, D42, V44, L50, and V51) are clustered. Of these residues, four are in the analogous position to residues identified by radical mutagenesis in Brf. TFIIB D42 is analogous to Brf D22, V44 is analogous to Brf V24, V51 is analogous to Brf V31, and L50 is analogous to Brf V30. The positions of these five residues are all on the second or third β strands or on the second rubredoxin knuckle (E26). These five residues were also targeted for alanine substitution and assayed for function in vivo. In contrast to Brf, in which only one of these common residues was sensitive to alanine substitution, three common residues in TFIIB were sensitive to alanine substitution (Table 2). Figure 4 compares the molecular surface and electrostatic potential at the functional surface of the Brf and TFIIB ribbon domains. The four functionally conserved residues are predicted to form a nearly identical surface in both factors.
Figure 4.

Molecular surface and electrostatic potential of the Brf and TFIIB zinc ribbon functional surfaces. Surface and potential indicated using Grasp (Nicholls et al. 1991). Positive potential, (blue); negative potential, (red); hydrophobic, (white).
Interestingly, TFIIB residue E26 is important for TFIIB function, as substitution with arginine significantly decreases in vivo function, but the analogous substitution in BRF (N6E) causes no phenotype. Substitution of TFIIB E26 by alanine causes no noticeable effect on in vivo function. However, substitution by asparagine, the same residue as in yeast BRF, surprisingly decreased in vivo function of TFIIB (Table 2), demonstrating that for yeast TFIIB, E26 is the optimal residue compared with the side chain found in BRF.
As an in vivo test for the expression of the TFIIB mutants, the in vivo stability of the mutant proteins was measured. Cells containing both a mutant tagged and wild-type untagged copy of TFIIB were grown in minimal medium, and cellular protein was extracted. The levels of mutant protein were assayed by Western blot, probing for the Flag epitope inserted on the mutant copy of TFIIB. All of the mutant proteins were expressed to within 70% of the wild-type level (not shown).
Transcription defects in Brf zinc ribbon mutants
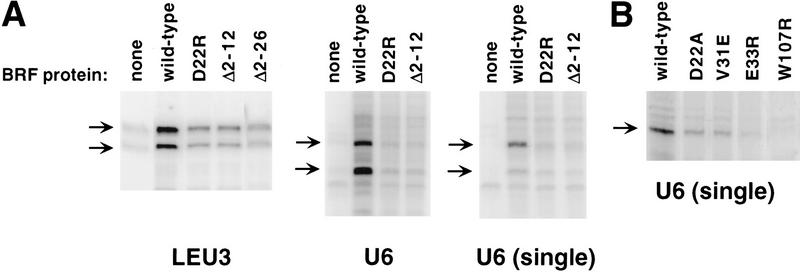
To determine how strongly mutations in the Brf zinc ribbon domain affected Pol III transcription, in vitro transcription was conducted with three defective proteins identified above. These mutant Brf proteins were expressed and purified from Escherichia coli, the radical substitution mutant D22R, and two deletion mutations that disrupt the domain, Δ2–12 and Δ2–24. To assay the transcriptional defects, a whole cell extract was prepared from a strain containing the Brf temperature-sensitive mutation W107R (Colbert 1997). Extracts made from this mutant are severely defective for in vitro transcription and TFIIIB–DNA complex formation, even when assayed at the permissive temperature (S. Roberts and T. Colbert, unpubl.). A total of 10 ng of either wild-type or mutant Brf was added to the W107R extracts and multiround transcription was assayed on supercoiled templates (Fig. 5A). Transcription from the LEU3 tRNA promoter was stimulated 5-fold by wild-type Brf and was only stimulated 2- to 2.5-fold when the extract was supplemented with the Brf ribbon mutants. Transcription from the SNR6 gene (U6) was stimulated 14-fold by wild-type Brf and was only stimulated 4.6-fold by the ribbon mutants. Finally, 5S rRNA transcription was stimulated 3-fold by wild-type Brf and only 1.8- to 1.4-fold by the ribbon mutants (not shown). Brf stimulation of U6 transcription was also decreased from 14- to 3–4-fold by the zinc ribbon mutants when nucleotides were added for 3.5 min to limit transcription reinitiation (Fig. 5A, single). The above results are consistent with previous studies that showed that deletion of a large portion of the Brf amino terminus, including the zinc ribbon and at least two other Brf domains, caused a significant drop in initiation but did not eliminate transcription entirely (Kassavetis et al. 1997, 1998).
Figure 5.
In vitro transcription defects caused by Brf zinc ribbon mutations. (A) In vitro transcription was performed for 30 min as described in Materials and Methods using the indicated promoter contained on a supercoiled plasmid. A total of 10 ng of the indicated Brf was used to supplement the Brf W107R extract as indicated. For the U6 (single) panel, preinitiation complexes were performed in the absence of nucleotides for 30 min followed by addition of nucleotides for 3.5 min to limit transcription to a single round. For LEU3 and U6 products, the top arrow indicates the full-length product; the bottom arrow, a processed product derived from full-length transcript. (B) Whole cell extracts were made from the indicated Brf mutant strains grown at 25°C and used for in vitro transcription at 32°C. Complexes were formed in the absence of NTPs for 30 min followed by NTP addition for 3.5 min.
In vitro transcription defects were also found for several of the nonlethal Brf ribbon mutants, D22A, V31E, and E33R (Fig. 5B). In vitro transcription was performed with extracts made from these mutant strains grown at the permissive temperature. These extracts were most defective for transcription at U6 showing 1.5- to 2.7-fold lower transcription compared with wild-type. The defects seen at the LEU3 promoter were much milder, with defects ranging from 0.2- to 2-fold (not shown).
The Brf zinc ribbon mutants are not defective in polymerase recruitment
On the basis of the above results along with other published studies, the Brf ribbon mutants could be defective in transcription because of a defect in either polymerase recruitment or some later step such as open complex formation or initiation. To assay polymerase recruitment, PICs formed with wild-type or mutant Brfs were directly isolated and analyzed (Fig. 6). Biotinylated ∼340-bp fragments of the wild-type U6 promoter or a U6 promoter containing TATA and B-block mutations (U6 mut) were immobilized to streptavidin-coated magnetic beads. These fragments also contained a BamHI site engineered upstream of the promoter. To assay PIC formation, the W107R extract with or without addition of Brf was incubated with the immobilized templates at 30°C, washed, and then the PIC complex isolated by liberation with BamHI digestion. The liberated protein–DNA complex was then assayed by SDS–PAGE and Western blot for the presence of Pol III factors specifically bound to the U6 but not the U6 mut fragment.
Figure 6.

Isolation of preinitiation complexes using immobilized promoter template. (A) Cartoon of the immobilized U6 and mutant templates used. (B) Western blot of complexes formed on wild-type and mutant U6 templates probed with Tfc4 or Rpc34 antisera. Brf was added to the Brf W107R extract as indicated. (C) in vitro transcription activity of isolated PICs. Complexes were formed as in B, except that after washing, nucleotides were added for 5 min and RNA products analyzed by PAGE.
Figure 6B shows that TFIIIC binds to the immobilized promoter in the absence of added Brf as assayed by the presence of the TFIIIC subunit Tfc4. However, Pol III was not recruited as measured by the absence of the Pol III-specific subunit Rpc34. Brf and B” were not detected under these conditions (not shown). TBP bound nearly as well to both the U6 and U6 mut templates (not shown), probably because of the fairly strong nonspecific-binding capacity of TBP as well as the A T-rich nature of the U6 sequence. When wild-type Brf was added, Pol III was specifically recruited to the U6 but not the U6 mut promoter. Brf and B” were also detected in these complexes, although the available antisera against these proteins was not as sensitive in Western blots as the Tfc4 and Rpc34 antisera and the signals were sometimes difficult to detect quantitatively (not shown). Strikingly, both the D22R and Δ2–12 ribbon mutants recruited Pol III to the U6 promoter as well as wild-type Brf. In contrast, transcription from the washed PICs was drastically different comparing wild-type and mutant Brfs (Fig. 6C) showing that the ribbon domain is critical for initiation in the washed complexes.
The Brf ribbon domain functions in open complex formation
Probing PICs with potassium permanganate was used to determine whether the Brf ribbon mutants were defective in open complex formation. Permanganate preferentially reacts with single-stranded T residues and has been used previously at both tRNA and the U6 promoter to measure the amount of open complex formed (Kassavetis et al. 1992, 1998; Brun et al. 1997). For technical reasons, it was not possible to assay permanganate sensitivity on the same DNA fragment used in the immobilized template assay. First, template utilization of short DNA fragments in the extracts is low compared with plasmid templates, and second, the DNA fragments are either dephosphorylated and/or degraded on prolonged incubation in the extract at 30°C. To avoid these problems, a supercoiled U6 plasmid was used in these assays analogous to the experiments shown in Figure 5. PICs were formed for 30 min in the W107R extract after the addition of wild-type or mutant BRF. Permanganate was added for 1 min, the reaction was stopped, and modification of the nontranscribed strand assayed by primer extension (Materials and Methods). Figure 7 shows that addition of wild-type BRF stimulates KMnO4 sensitivity threefold at position T −5 with respect to the transcription start site. In contrast, addition of either Brf D22R or Δ2–12 did not significantly stimulate reactivity compared with no Brf addition.
Figure 7.
Open complex defect in the Brf zinc ribbon mutants. PICs were formed on 100 ng of supercoiled plasmid DNA as in Fig. 5 (U6 single) using the Brf W107R extract supplemented with 15 ng of the indicated Brf proteins (lanes 1–4). In lane 5, extract made from wild-type yeast was supplemented with 15 ng of wild-type Brf. KMnO4 was added to 10 mm for 1 min, followed by DNA isolation and detection of modified DNAs on the nontranscribed strand by primer extension. The arrow marks the position of base T −5.
Discussion
The zinc ribbon fold has been conserved in the three classes of TFIIB family members, TFIIB, TFB, and Brf. The TFIIB zinc ribbon is known to interact with Pol II and/or TFIIF, although the specific subunits with which this domain interacts is unknown. Structural studies of the TFIIB- and TFB–TBP–DNA complexes as well as cross-linking of TFIIB to promoters suggests that the amino-terminal region of TFIIB is located downstream of the TATA element, where Pol II and TFIIF also interact (Ebright 1998). Mutation of the TFIIB ribbon domain can eliminate recruitment of Pol II to PICs when assembled by either the stepwise assembly pathway or the holoenzyme pathway (Ranish et al. 1999).
In contrast to the function of the TFIIB ribbon, disruption of the Brf ribbon was shown to have no effect on recruitment of Pol III to the PIC. This effect is understandable, as Pol III makes at least three other contacts with subunits of TFIIIB and TFIIIC. First, the Pol III subunit Rpc34 contacts the carboxy-terminal half of Brf and possibly the amino-terminal half as well (Khoo et al. 1994; Brun et al. 1997). Second, the Pol III subunit Rpc17 also appears to contact Brf (Ferri et al. 2000). Finally, the TFIIIC subunit Tfc4 contacts the Pol III subunit Rpc53 (Flores et al. 1999). Despite not having a role in Pol III recruitment, the Brf ribbon domain is important for transcription from both supercoiled and linear DNA templates. In the experiments reported here using Brf D22R and two deletion mutants, in vitro transcription was decreased 2- to 4-fold in the Brf ribbon mutants assayed at the different classes of Pol III promoters and at least 10-fold in the washed PICs. Previously, it has been shown that deletion of a large portion of the amino-terminal half of Brf decreased in vitro transcription, consistent with the results reported here (Kassavetis et al. 1997, 1998), although this and another amino-terminal deletion reported recently (Kassavetis et al. 1999) removed other domains in addition to the zinc ribbon. Finally, a previous report from our laboratory showed that deletion of the zinc ribbon led to a cold-sensitive phenotype and an in vitro transcription defect (Colbert and Hahn 1992). However, this analysis with the zinc ribbon is suspect, because it was found in the present work that the zinc ribbon domain is essential for growth of yeast.
The transcription defect in the Brf ribbon mutants was shown to be due to a defect in open complex formation. When PICs were formed with the ribbon mutants on supercoiled DNA, significantly less permanganate reactivity was seen near the transcription start site compared with wild-type Brf. A similar effect on open complex formation was seen using mutations in both the Pol III subunit Rpc34 as well as in the TFIIIB subunit B” (Brun et al. 1997; Kassavetis et al. 1998). Because B” has been proposed to be a scaffold or structural component of the TFIIIB complex (Kumar et al. 1998), it is possible that the effect of these B” mutations on Pol III is indirect and is the result of an altered structure of the TFIIIB–DNA complex. In contrast, the Brf ribbon domain does not play a role in the structure of TFIIIB–DNA (Colbert et al. 1998; Kumar et al. 1998) and would be more likely to play a direct role in promoting a conformational change in Pol III to the open complex state. However, as the molecular target for the Brf ribbon domain is unknown, it cannot be said whether this involves a direct interaction with Pol III. To solve this problem, it will be necessary to determine which factor interacts with the ribbon domain and how this relates to Rpc34 function. Because the ribbon domain probably does not contact the active site of Pol III, a more general question that needs to be answered is how distant protein–protein interactions can promote a conformational change in the Pol III–DNA complex. Similar questions have been raised for prokaryotic activators that contact sites on Polymerase far from the active site but nevertheless promote open complex formation (Niu et al. 1996). It has been proposed that these prokaryotic activators may cause an allosteric change in polymerase or stabilize the transition state between closed and open complexes.
Strikingly, the functional surface identified in both the Brf and TFIIB ribbon domains has a common core of four identical or conserved residues on one face of the domain. For both factors, this core region is a small hydrophobic surface with one negative charge comprised of β strands two and three. From the location of the functional surface in both factors, an important role of zinc coordination and the first β strand in the ribbon domain is to orient the second and third antiparallel β strands to form the functional surface of the domain. From the mutagenesis results with TFIIB and Brf, it is likely that both the side chains and polypeptide backbone at this surface are involved in interaction with the target of this domain. However, the different sensitivity of these residues to alanine substitution leaves open the possibility that the molecular details of how the two ribbons interact with their targets are different. These surfaces in TFIIB and Brf likely make multiple interactions with their targets, since most single mutations did not completely disrupt the function of either domain compared with mutation of the conserved Cys residues.
Previous mutagenesis of yeast TFIIB has identified several non-cysteine residues important for function in the ribbon domain. Mutation of two glycine residues in the rubredoxin loop, G49S and G41E, both affected function in vivo (Knaus et al. 1996) and likely interfered with zinc coordination by altering the bending of the knuckle. The mutation L50D in yeast TFIIB, one of the residues identified in this study, was found to impair Pol II binding and caused a cold-sensitive growth phenotype (Bangur et al. 1997; Pardee et al. 1998). The mutation L52P was reported to cause either a cold-sensitive or lethal phenotype (Knaus et al. 1996; Bangur et al. 1997). From inspection of the TFIIB ribbon model, residue L52 is not surface exposed and is likely important for folding of the domain. Consistent with this finding, attempts at modeling the TFIIB ribbon with this mutation did not result in any reasonable three-dimensional models. Finally, mutation S53P was found to cause a cold-sensitive phenotype (Knaus et al. 1996) and reduced transcription activation in vivo by two activators (Wu and Hampsey 1999), in contrast to no phenotype observed in this study with the S53E mutation. It should be noted that this is the last residue in the TFIIB model, and it is uncertain whether this residue acts as part of the ribbon domain or the conserved sequence block and whether it is surface exposed.
Among proteins of diverse function, the amino acid sequence of zinc ribbon folds is not conserved outside of the two CXXC(H) motifs. This sequence divergence allows this fold to function in very different contexts in many proteins. Mutagenesis of TFIIS has identified five residues important for function, the location of all but one differing markedly from the functional surface identified in TFIIB and Brf (Cipres-Palacin and Kane 1995; Olmsted et al. 1998; Yoon et al. 1998). Comparison of the important residues for TFIIS and the two TFIIB family members discussed here demonstrates that these two classes of factors interact very differently with their targets.
The structure modeling and functional analysis described here are important for understanding the function of the ribbon domains of the TFIIB family and how these factors interact with their targets. In the future, identification of the target of the TFIIB and Brf ribbon domains will lead to an understanding of how distant protein–protein interactions contribute to conformational changes in Polymerase as well as the evolution and specificity of the machinery for the three nuclear RNA polymerases.
Materials and methods
General methods
General genetic, biochemical, and molecular biology methods used in this study are detailed at the Hahn laboratory WWW site: www.fhcrc.org/science/basic/labs/hahn. For determination of Brf or TFIIB temperature-sensitive mutant levels in vivo, yeast were grown to ∼A600 = 1.0 in synthetic complete glucose medium lacking both uracil and leucine to select for both the wild-type and mutant copy of either BRF or SUA7. Rapid protein extracts were made by boiling cells in SDS buffer and assayed by PAGE and Western blot as described on the above web site. Results were quantitated using ImageQuant (Molecular Dynamics). Polyclonal antisera directed against Rpc34 and amino acids 1–170 of Tfc4 were generated by purification of recombinant protein from E. coli and immunization of rabbits (provided by K. Coachman, Fred Hutchinson Cancer Research Center).
Structure modeling and model evaluation
The program ClustalX (Thompson et al. 1997) was used to align the TFIIB family members for three-dimensional modeling. The Brf family was first aligned and then this Brf profile was aligned with P. furiosis TFB weighting the alignment for secondary structural elements in the TFB NMR structure. The TFIIB family was aligned separately and this profile was aligned with TFB as above.
Of the 25 NMR models for the TFB zinc ribbon listed in the Protein Data Bank, 3 were chosen to use as templates in the structure modeling of Brf (models 1, 15, and 25). Each of the three NMR models was used to generate 20 independent structure models of yeast Brf residues 3–33 using the program Modeller 4 (Sali and Blundell 1993). The Modeller parameters for refinement were set as follows: library schedule 1, iterations 300, molecular dynamics level 1, and repeat optimization 3. The resulting 60 models were evaluated by statistically based structural validation methods to compare the properties of the models with properties of proteins of known structure. The models were first evaluated using the program ERRAT, which analyzes the interaction of nonbonded atoms (Colovos and Yeates 1993). This program was found to be most useful as an initial screening test. The best models were then evaluated using the Procheck suite of programs (Laskowski et al. 1993), which examines the distribution of dihedral angles and deviation of bond length and angles from ideal. The best of this group were evaluated using 3-D Profile (Bowie et al. 1996), which evaluates the fitness of a residue to its environment in the structure. By use of these evaluation methods, one of the original TFB NMR models did not score well (model 1) and all models derived from model 1 as a template also did not score well and were discarded. The final group of the three best models scored at least as well, and in most cases, better than the original NMR models used as templates. The same methods were used to model the yeast TFIIB residues 23–53 and select the best three structure models for further analysis.
Mutagenesis and tests for in vivo function
The residues most likely to be surface exposed were identified by comparing the three best models for surface accessibility using the programs Quanta (Molecular Simulations, Inc.) and naccess (Hubbard and Thornton 2000). No consistent structure of side chains or polypeptide backbone was seen for the loop between β strands one and two in the Brf models, so this region was not targeted for mutagenesis. The same methods were used to identify the likely surface-exposed residues in yeast TFIIB. The polypeptide backbone between the first knuckle and β strand in the TFIIB models was not consistent between the three best models due to reasons described in Results. However, all three models predicted the same residues to lie on the surface in this region, so all of these residues were targeted for mutagenesis.
Brf mutations were made in the vector sBRF–HA–int-2 (C. Landel and G. Schimmack, unpubl.), which contains yeast Brf expressed under control of its own promoter with the coding sequence altered to remove many rare E. coli codons without altering the amino acid sequence, the mutation M166L to reduce internal initiation when expressed in E. coli (Librizzi et al. 1996), and insertion of an alanine residue after the initiation methionine codon. In addition, an HA epitope and 8-His tag were inserted in a nonconserved region near the carboxy-terminal end of BRF (sequence available upon request) cloned in the Ars Cen vector pRS315 (Sikorski and Hieter 1989). None of these changes had any apparent effect on the in vivo or in vitro function of Brf. Zinc ribbon mutations were introduced using site-directed mutagenesis and confirmed by DNA sequencing. Plasmids were transformed to yeast strain SHY285 (matα, ade2Δ::hisG, his3Δ200, leu2Δ0, lys2Δ0, met15Δ0, trp1Δ63, ura3Δ0, brf1Δ::HIS3/pSH524 (Ars Cen URA3, BRF1), and the wild-type BRF1 gene replaced with the mutant gene by plasmid shuffle at 25°C. Cells were tested for growth phenotype on synthetic complete glucose plates (−leucine) at 18°C, 25°C, 30°C, and 35.5°C.
TFIIB mutations were made in pLB2 (L. Boric, unpubl.), which contains the SUA7 gene with two copies of a carboxy-terminal Flag epitope tag (Hopp et al. 1988) expressed under control of the Sua7 promoter cloned in the PRS315 vector. The mutant sua7 genes were introduced by plasmid shuffle and scored as above in strain SHY98 [matα, ade6, leu2, his4, sua7Δ::HIS4/pSH374 (Ars Cen URA3, SUA7)].
In vitro transcription
Yeast whole cell extracts were made from a strain with the Brf temperature-sensitive mutation W107R (Colbert 1997). This mutation disrupts in vitro transcription as well as TFIIIB complex formation (Colbert 1997; S. Roberts, unpubl.). Yeast were grown at the permissive temperature of 26°C and extracts made by a modified method of Schultz et al. (1991) and P. Aprikian and R. Reeder (unpubl.) as detailed on the above web site. In vitro transcription was carried out under the following conditions and as detailed on the web site using 30 μg of whole cell extract: 2% glycerol, 20 mm HEPES (pH 7.9), 80 mm KCl, 5 mm MgCl2, 1 mm EDTA, 1 mm DTT, 200 ng of α-amanitin, 4 units of RNase Inhibitor (Amersham/Pharmacia), 140 ng of plasmid template, 500 μm ATP, UTP, CTP, 50 μm GTP, and 0.5 μl of [α-32P]GTP (10 mCi/ml, 3000 Ci/mmole). For multi-round transcription reactions, proteins and DNA were mixed in 20 μl-reactions on ice. Initiation was started by addition of nucleotides and incubation at 30°C for 30 min and stopped by the addition of 200 μl of 0.1 m Na acetate, 10 mm EDTA, 0.5% SDS, and 5 μg/ml tRNA. For single-round transcription, proteins and DNA were mixed on ice as above except that nucleotides were omitted and the reactions incubated at 30°C for 20 min. Nucleotides were then added and reactions stopped after 3.5 min. After phenol chloroform extraction and ethanol precipitation, RNA products were analyzed on 6% denaturing urea acrylamide gels and quantitated by PhosphorImager analysis.
Open complex assay
Open complex formation was probed by DNA modification with KMnO4 on supercoiled DNA and detected by primer extension (Sasse-Dwight and Gralla 1991). PICs were formed for 30 min as detailed above for single-round transcription, except that plasmid template was 30 or 100 ng in 20 μl, and DTT, α-amanitin, and RNase inhibitor were omitted from the reactions. After 30 min, KMnO4 was added to a final concentration of 5 or 10 mm for 1 min and stopped by addition of 2 μl of 2-mercaptoethanol and 200 μl of transcription stop mix. Reactions were extracted with phenol/chloroform and precipitated with ethanol. Modified DNAs were resuspended in 35 μl of water and purified using MicroSpin G-25 columns (Amersham/Pharmacia). These purified DNAs were then used as templates for linear amplification with Taq polymerase using the 5′ 32P-labeled oligonucleotide: CACAGCCTGGCATGAACAGTGGTA with the following amplification profile: 94°C for 20 sec; 50°C for 30 sec; 72°C for 2 min, and after 18 cycles, followed by 8 min at 72°C. Reaction products were ethanol precipitated and analyzed on 8% urea acrylamide gels and quantitated by PhosphorImager analysis.
Brf purification
Brf and several zinc ribbon mutants from sBRF–HA–int2 were subcloned to the expression vector pet21D (Novagen). Brf was expressed and purified by Ni–NTA chromatography under denaturing conditions and renatured by dialysis as described previously (Colbert et al. 1998). This renatured Brf was further purified by chromatography on Source 15Q (Amersham/Pharmacia) and resulted in nearly homogenous full-length Brf.
Immobilized template assay
DNAs for immobilized templates were prepared and attached to beads essentially as described by Ranish et al. (1999). A 5′ biotin-labeled 344-bp U6 promoter fragment was amplified by PCR using the oligonucleotides 5′ Biotin-TTCCGGAACGGGATCCCACAGCCTGGCATGAACAGTGGTA and 5′-ACCGATAGCAAAGGCTTAGG using pCH6 (Brow and Guthrie 1990) as a template. The U6 mut fragment was amplified from the plasmid U6 mutTATA (S. Roberts, unpubl.), which contains the mutation TAGAGAAA at the U6 TATA box. The mutant biotinylated fragment was synthesized by PCR using the biotin-labeled oligo from above and the oligonucleotide CGCGAGACAATTTTCTATTCGAG, which deletes the TFIIIC box B-binding site. A typical binding experiment contained 20 μg of streptavidin magnetic beads (Dynal) linked to 60 ng of U6 DNA in Pol III transcription buffer from above lacking nucleotides with 0.01% Tween 20 and 30 μg of Brf W107R whole cell extract in a final volume of 50 μl. Twenty nanograms of Brf was added where indicated. Before adding the beads, extracts diluted in transcription buffer were incubated 10 min on ice and then spun 5 min in a microcentrifuge at 4°C to remove any insoluble material. After addition of all components on ice, reactions were incubated for 30 min with shaking at 30°C. Beads were washed three times in the above binding buffer and resuspended in 10 μl of the same buffer including 30 units of BamHI. The restriction digest was allowed to proceed for 25 min at room temperature and then the supernatant was removed and analyzed by SDS-PAGE and Western blot. For in vitro transcription analysis, the washed beads were resuspended in 50 μl of transcription buffer with nucleotides and incubated at 30°C for 5 min.
Acknowledgments
We thank K. Zhang and J. Geiger for generous help and advice on the structural modeling and model evaluation, T. Colbert for additional help with modeling, C. Landel, G. Schimmack, B. Moorefield, and L. Boric for plasmids, K. Coachman for Rpc34 and Tfc4 antisera, P. Aprikian for the modified whole cell extract method, and J. Movius for wild-type recombinant Brf. We thank J. Geiger, R. Reeder, T. Young, N. Yudkovsky, and K. Zhang for comments on the manuscript, and members of the Hahn Laboratory for valuable comments and sharing of reagents throughout the course of this work. This work was funded by grant GM53451 from the NIH. S.H. is an investigator of the HHMI.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL shahn@fred.fhcrc.org; FAX (206) 667-6497.
References
- Bangur CS, Pardee TS, Ponticelli AS. Mutational analysis of the D1/E1 core helices and the conserved N-terminal region of yeast transcription factor IIB (TFIIB): Identification of an N-terminal mutant that stabilizes TATA-binding protein-TFIIB-DNA complexes. Mol Cell Biol. 1997;17:6784–6793. doi: 10.1128/mcb.17.12.6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberis A, Muller CW, Harrison SC, Ptashne M. Delineation of two functional regions of transcription factor TFIIB. Proc Natl Acad Sci. 1993;90:5628–5632. doi: 10.1073/pnas.90.12.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie JU, Zhang K, Wilmanns M, Eisenberg D. Three-dimensional profiles for measuring compatibility of amino acid sequence with three-dimensional structure. Methods Enzymol. 1996;266:598–616. doi: 10.1016/s0076-6879(96)66037-6. [DOI] [PubMed] [Google Scholar]
- Brow DA, Guthrie C. Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes & Dev. 1990;4:1345–1355. doi: 10.1101/gad.4.8.1345. [DOI] [PubMed] [Google Scholar]
- Brun I, Sentenac A, Werner M. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 1997;16:5730–5741. doi: 10.1093/emboj/16.18.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant GO, Martel LS, Burley SK, Berk AJ. Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes & Dev. 1996;10:2491–2504. doi: 10.1101/gad.10.19.2491. [DOI] [PubMed] [Google Scholar]
- Buratowski S, Zhou H. Functional domains of transcription factor TFIIB. Proc Natl Acad Sci. 1993;90:5633–5637. doi: 10.1073/pnas.90.12.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedin S, Ferri ML, Peyroche G, Andrau JC, Jourdain S, Lefebvre O, Werner M, Carles C, Sentenac A. The yeast RNA polymerase III transcription machinery: A paradigm for eukaryotic gene activation. Cold Spring Harb Symp Quant Biol. 1998;63:381–389. doi: 10.1101/sqb.1998.63.381. [DOI] [PubMed] [Google Scholar]
- Chou S, Struhl K. Transcriptional activation by TFIIB mutants that are severely impaired in interaction with promoter DNA and acidic activation domains. Mol Cell Biol. 1997;17:6794–6802. doi: 10.1128/mcb.17.12.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipres-Palacin G, Kane CM. Alanine-scanning mutagenesis of human transcript elongation factor TFIIS. Biochemistry. 1995;34:15375–15380. doi: 10.1021/bi00046a046. [DOI] [PubMed] [Google Scholar]
- Coin F, Egly JM. Ten years of TFIIH. Cold Spring Harb Symp Quant Biol. 1998;63:105–110. doi: 10.1101/sqb.1998.63.105. [DOI] [PubMed] [Google Scholar]
- Colbert T. “Characterization of Brf1, an RNA polymerase III transcription factor,” Ph.D. thesis. Seattle: University of Washington; 1997. [Google Scholar]
- Colbert T, Hahn S. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes & Dev. 1992;6:1940–1949. doi: 10.1101/gad.6.10.1940. [DOI] [PubMed] [Google Scholar]
- Colbert T, Lee S, Schimmack G, Hahn S. Architecture of protein and DNA contacts within the TFIIIB-DNA complex. Mol Cell Biol. 1998;18:1682–1691. doi: 10.1128/mcb.18.3.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan J, Ashali H, Manley JL. A direct interaction between a glutamine-rich activator and the N terminus of TFIIB can mediate transcriptional activation in vivo. Mol Cell Biol. 1995;15:2311–2320. doi: 10.1128/mcb.15.4.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colovos C, Yeates TO. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright RH. RNA Polymerase-DNA interaction: Structure of intermediate, open, and Elongation complexes. Cold Spring Harb Symp Quant Biol. 1998;63:11–20. doi: 10.1101/sqb.1998.63.11. [DOI] [PubMed] [Google Scholar]
- Ferri ML, Peyroche G, Siaut M, Lefebvre O, Carles C, Conesa C, Sentenac A. A novel subunit of yeast RNA polymerase III interacts with the TFIIB-related domain of TFIIIB70. Mol Cell Biol. 2000;20:488–495. doi: 10.1128/mcb.20.2.488-495.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores A, Briand JF, Gadal O, Andrau JC, Rubbi L, Van Mullem V, Boschiero C, Goussot M, Marck C, Carles C, et al. A protein-protein interaction map of yeast RNA polymerase III. Proc Natl Acad Sci. 1999;96:7815–7820. doi: 10.1073/pnas.96.14.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausner W, Wettach J, Hethke C, Thomm M. Two transcription factors related with the eucaryal transcription factors TATA-binding protein and transcription factor IIB direct promoter recognition by an archaeal RNA polymerase. J Biol Chem. 1996;271:30144–30148. doi: 10.1074/jbc.271.47.30144. [DOI] [PubMed] [Google Scholar]
- Hisatake K, Roeder RG, Horikoshi M. Functional dissection of TFIIB domains required for TFIIB-TFIID-promoter complex formation and basal transcription activity. Nature. 1993;363:744–747. doi: 10.1038/363744a0. [DOI] [PubMed] [Google Scholar]
- Hopp TP, Prickett KS, Price VL, Libby RT, March CJ, Cerretti DP, Urdal DL, Conlon PJ. A short polypeptide marker sequence useful for recombinant protein identification and purification. BioTechnology. 1988;6:1204–1210. [Google Scholar]
- Hubbard, S. and J. Thornton. 2000. naccess Home Page: http://sjh.bi.umist.ac.uk/naccess.html.
- Kassavetis GA, Blanco JA, Johnson TE, Geiduschek EP. Formation of open and elongating transcription complexes by RNA polymerase III. J Mol Biol. 1992;226:47–58. doi: 10.1016/0022-2836(92)90123-2. [DOI] [PubMed] [Google Scholar]
- Kassavetis GA, Bardeleben C, Kumar A, Ramirez E, Geiduschek EP. Domains of the Brf component of RNA Polymerase III transcription factor IIIB (TFIIIB): Functions in assembly of TFIIIB-DNA complexes and recruitment of RNA polymerase to the promoter. Mol Cell Biol. 1997;17:5299–5306. doi: 10.1128/mcb.17.9.5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Kumar A, Letts GA, Geiduschek EP. A post-recruitment function for the RNA polymerase III transcription- initiation factor IIIB. Proc Natl Acad Sci. 1998;95:9196–9201. doi: 10.1073/pnas.95.16.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Letts GA, Geiduschek EP. A minimal RNA polymerase III transcription system. EMBO J. 1999;18:5042–5051. doi: 10.1093/emboj/18.18.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo B, Brophy B, Jackson SP. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes & Dev. 1994;8:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- Knaus R, Pollock R, Guarente L. Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. EMBO J. 1996;15:1933–1940. [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Grove A, Kassavetis GA, Geiduschek EP. Transcription factor IIIB: The architecture of its DNA complex, and its roles in initiation of transcription by RNA polymerase III. Cold Spring Harb Symp Quant Biol. 1998;63:121–129. doi: 10.1101/sqb.1998.63.121. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Lee M, Struhl K. A severely defective TATA-binding protein-TFIIB interaction does not preclude transcriptional activation in vivo. Mol Cell Biol. 1997;17:1336–1345. doi: 10.1128/mcb.17.3.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Ren D, Finkelstein A, Burton ZF. Functions of the N- and C-terminal domains of human RAP74 in transcriptional initiation, elongation, and recycling of RNA polymerase II. Mol Cell Biol. 1998;18:2130–2142. doi: 10.1128/mcb.18.4.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librizzi MD, Moir RD, Brenowitz M, Willis IM. Expression and purification of the RNA polymerase III transcription specificity factor IIIB70 from Saccharomyces cerevisiae and its cooperative binding with TATA-binding protein. J Biol Chem. 1996;271:32695–32701. doi: 10.1074/jbc.271.51.32695. [DOI] [PubMed] [Google Scholar]
- Masuyama H, Jefcoat SC, Jr, MacDonald PN. The N-terminal domain of transcription factor IIB is required for direct interaction with the vitamin D receptor and participates in vitamin D-mediated transcription. Mol Endocrinol. 1997;11:218–228. doi: 10.1210/mend.11.2.9879. [DOI] [PubMed] [Google Scholar]
- Myer VE, Young RA. RNA polymerase II holoenzymes and subcomplexes. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Niu W, Kim Y, Tau G, Heyduk T, Ebright RH. Transcription activation at class II CAkP-dependent promoters: Two interactions between CAP and RNA Polymerase. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted VK, Awrey DE, Koth C, Shan X, Morin PE, Kazanis S, Edwards AM, Arrowsmith CH. Yeast transcript elongation factor (TFIIS), structure and function. I: NMR structural analysis of the minimal transcriptionally active region. J Biol Chem. 1998;273:22589–22594. doi: 10.1074/jbc.273.35.22589. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Pardee TS, Bangur CS, Ponticelli AS. The N-terminal region of yeast TFIIB contains two adjacent functional domains involved in stable RNA polymerase II binding and transcription start site selection. J Biol Chem. 1998;273:17859–17864. doi: 10.1074/jbc.273.28.17859. [DOI] [PubMed] [Google Scholar]
- Pinto I, Wu W-H, Na JG, Hampsey M. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J Biol Chem. 1994;269:30569–30573. [PubMed] [Google Scholar]
- Qian X, Gozani SN, Yoon H, Jeon CJ, Agarwal K, Weiss MA. Novel zinc finger motif in the basal transcriptional machinery: Three-dimensional NMR studies of the nucleic acid binding domain of transcriptional elongation factor TFIIS. Biochemistry. 1993;32:9944–9959. doi: 10.1021/bi00089a010. [DOI] [PubMed] [Google Scholar]
- Qureshi SA, Bell SD, Jackson SP. Factor requirements for transcription in the Archaeon Sulfolobus shibatae. EMBO J. 1997;16:2927–2936. doi: 10.1093/emboj/16.10.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: Holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes & Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SGE, Green MR. Activator-induced conformational change in general transcription factor TFIIB. Nature. 1994;371:717–720. doi: 10.1038/371717a0. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S, Gralla JD. Footprinting protein-DNA complexes in vivo. Methods Enzymol. 1991;208:146–168. doi: 10.1016/0076-6879(91)08012-7. [DOI] [PubMed] [Google Scholar]
- Schultz MC, Choe SY, Reeder RH. Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc Natl Acad Sci. 1991;88:1004–1008. doi: 10.1073/pnas.88.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe JW, Klug A. Zinc mining for protein domains. Nature Struct Biol. 1994;1:345–349. doi: 10.1038/nsb0694-345. [DOI] [PubMed] [Google Scholar]
- Shen J, Kassavetis G, Bryant GO, Berk AJ. Pol III TAF Brf binds to a surface on TBP also required for activated Pol II transcription. Mol Cell Biol. 1998;18:1692–1700. doi: 10.1128/mcb.18.3.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Sun X, Reinberg D, Ebright RH. Protein-protein interactions in eukaryotic transcription initiation: Structure of the preinitiation complex. Proc Natl Acad Sci. 1996;93:1119–1124. doi: 10.1073/pnas.93.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Jones DN, Kaine BP, Weiss MA. High-resolution structure of an archaeal zinc ribbon defines a general architectural motif in eukaryotic RNA polymerases. Structure. 1998;6:555–569. doi: 10.1016/s0969-2126(98)00058-6. [DOI] [PubMed] [Google Scholar]
- Wu W-H, Hampey M. An activation-specific role for transcription factor TFIIB in vivo. Proc Natl Acad Sci. 1999;96:2764–2769. doi: 10.1073/pnas.96.6.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H, Sitikov AS, Jeon C, Agarwal K. Preferential interaction of the mRNA proofreading factor TFIIS zinc ribbon with rU.dA base pairs correlates with its function. Biochemistry. 1998;37:12104–12112. doi: 10.1021/bi980924n. [DOI] [PubMed] [Google Scholar]
- Zhu W, Zeng Q, Coangelo CM, Lewis M, Summers MF, Scott RA. The N-terminal domain of TFIIB from Pyrococcus furiosus forms a zinc ribbon. Nature Struct Biol. 1996;3:122–124. doi: 10.1038/nsb0296-122. [DOI] [PubMed] [Google Scholar]