Abstract
The mammalian gastrointestinal tract and bloodstream are highly disparate biological niches that differ in concentrations of nutrients such as iron. However, some commensal-pathogenic microorganisms, such as the yeast Candida albicans, thrive in both environments. We report the evolution of a transcription circuit in C. albicans that controls iron uptake and determines its fitness in both niches. Our analysis of DNA-binding proteins that regulate iron uptake by this organism suggests the evolutionary intercalation of a transcriptional activator called Sef1 between two broadly conserved iron-responsive transcriptional repressors, Sfu1 and Hap43. Sef1 activates iron uptake genes and promotes virulence in a mouse model of bloodstream infection, whereas Sfu1 represses iron uptake genes and is dispensable for virulence but promotes gastrointestinal commensalism. Thus, C. albicans can alternate between genetic programs conferring resistance to iron depletion in the bloodstream versus iron toxicity in the gut, and this may represent a fundamental attribute of gastrointestinal commensal-pathogens.
INTRODUCTION
The unique chemical properties of iron underlie its broad utility as a cofactor for essential cellular processes as well as its toxicity (via hydroxyl radicals produced by the Fenton reaction) to proteins, lipids, and nucleic acids (Pierre et al., 2002). Virtually all organisms have evolved mechanisms to precisely regulate the uptake and storage of iron. This task is particularly challenging for commensal-pathogens such as the yeast, Candida albicans, that inhabit the mammalian gastrointestinal tract (Odds, 1988) but also enter the bloodstream (Edmond et al., 1999). Gastrointestinal commensals are bathed in comparatively high levels of iron (~15 mg or 0.27 mmol/day in humans) since the majority of dietary iron is not absorbed (McCance and Widdowson, 1938; Miret et al., 2003). In contrast, bloodstream pathogens face extraordinary iron depletion (~10−24 M free Fe3+) because of active sequestration by the host (Martin et al., 1987). Only a handful of clinically important commensal-pathogenic microorganisms are adapted to both environments.
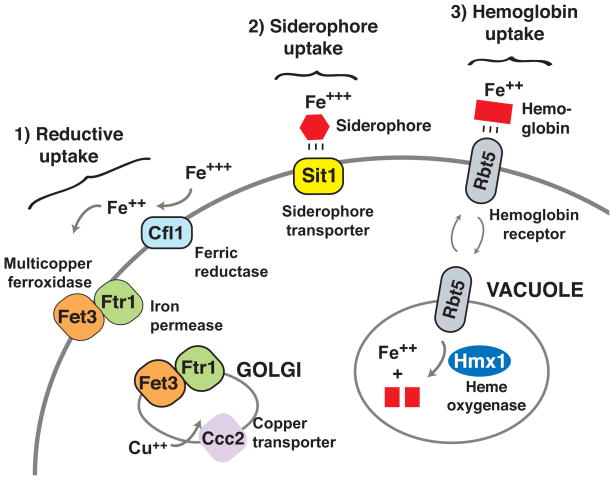
To survive in the bloodstream, C. albicans maintains at least three systems of iron extraction from the host (Figure 1). Iron is recovered from red blood cells by means of secreted hemolysins (Luo et al., 2001; Manns et al., 1994) and the Rbt5 system of hemoglobin utilization (Weissman and Kornitzer, 2004; Weissman et al., 2008). Iron bound to small molecule siderophores is imported via the Sit1 siderophore transporter (Ardon et al., 2001; Heymann et al., 2002; Hu et al., 2002). High-affinity uptake of free or chelated Fe3+ occurs through sequential reduction and oxidation-internalization steps mediated, respectively, by a family of cell surface ferric reductases (Hammacott et al., 2000; Knight et al., 2002) and complexes of the Ftr1 iron permease with any of several multicopper ferrous oxidases (Knight et al., 2002). The reductive system is also used in tissues to extract iron from host ferritin, in a process dependent on the cell surface adhesin Als3 (Almeida et al., 2008). In contrast to this detailed understanding of C. albicans adaptation to iron limitation in the bloodstream, comparatively little is known about how C. albicans defends against iron excess in the gut.
Figure 1. Iron acquisition in C. albicans.
Shown are key factors mediating the three known pathways of high-affinity iron uptake in this organism: reductive iron uptake, siderophore-iron uptake, and hemoglobin-iron uptake. Gene families encode ferric reductases and multicopper oxidases of the reductive pathway, and only a subset are expressed in an iron-dependent fashion. Ccc2 is a copper transporter required for proper assembly and function of Fet3/iron permease complexes. Genes for each depicted protein are directly activated by Sef1, and genes for Ftr1, Sit1, and Rbt5 are directly repressed by Sfu1.
In most ascomycetes and the basidiomycete, Cryptococcus neoformans, a simple switch regulates the expression of iron homeostasis genes (Haas et al., 1999; Hortschansky et al., 2007; Jung et al., 2010; Jung et al., 2006; Mercier et al., 2006; Pelletier et al., 2002). When environmental iron is replete, a GATA family transcription factor directly represses genes for iron acquisition as well as the gene encoding the regulatory component of the CCAAT-binding complex. When environmental iron is low, the CCAAT-binding complex directly represses the GATA factor gene as well as genes for noncritical iron-dependent processes. C. albicans was thought to share this system because its GATA factor ortholog, called Sfu1, can functionally substitute for the GATA factor of S. pombe (Pelletier et al., 2007), and deletion of SFU1 leads to inappropriate expression of iron acquisition genes when environmental iron is replete (Lan et al., 2004). Moreover, mutants affecting the C. albicans regulatory component of the CCAAT-binding complex exhibit multiple abnormalities on iron-depleted media, including defective growth (Baek et al., 2008; Homann et al., 2009; Hsu et al., 2011), failure to upregulate the FRP1 ferric reductase gene (Baek et al., 2008), and inappropriate expression of iron-utilization genes (Hsu et al., 2011). This hap43ΔΔ mutant is also defective for virulence in a mouse bloodstream infection model (Hsu et al., 2011), like mutants affecting orthologs in the fungal pathogens Aspergillus fumigatus (Schrettl et al., 2010) and C. neoformans (Jung et al., 2010).
We identified C. albicans SEF1, which encodes a predicted Zn(2)Cys(6) DNA-binding protein, in a screen for candidate virulence factors (Noble et al., 2010). Our discovery that SEF1 is also required for growth on iron-depleted media (Homann et al., 2009) suggested that it may transcriptionally activate genes involved in iron acquisition—but this hypothesis was at odds with the prevailing model of iron regulation, described above. We therefore determined the gene regulatory activities of Sef1, Sfu1, and Hap43 using whole-genome RNA expression and chromatin immunoprecipitation (ChIP) approaches. The results combined with systematic comparisons of mutants in C. albicans, S. pombe, and Saccharomyces cerevisiae suggest that C. albicans has evolved a unique, feed forward transcriptional circuit in which Sef1 is intercalated into the broadly conserved switch between the GATA factor and the CCAAT-binding complex. In C. albicans, Sfu1 (GATA factor) represses SEF1 and iron uptake genes, Sef1 activates HAP43 (CCAAT-binding complex) and iron uptake genes, and Hap43 represses SFU1 and iron utilization genes (i.e. genes for processes that require iron). We investigated the in vivo roles of SEF1 and SFU1 by profiling the respective deletion mutants in mouse models of virulence and commensalism. Only SEF1 was critical for virulence in the bloodstream, whereas SFU1 was selectively required for persistence in the gastrointestinal tract. These results suggest that the reciprocal abilities of C. albicans to activate iron uptake upon entry into the iron-depleted bloodstream, while efficiently restricting it in the potentially iron toxic environment of the gut, are critical to its success as a commensal-pathogen.
RESULTS
Sef1 activates the machinery for iron uptake in C. albicans
We identified C. albicans sef1ΔΔ in two separate genetic screens (note that C. albicans is an obligate diploid organism, necessitating the disruption of two copies of any gene): (1) an in vivo screen for genes required in competitive bloodstream infections (Noble et al., 2010) and (2) an in vitro screen for genes promoting growth under iron-limiting conditions (Homann et al., 2009). The hap43ΔΔ mutant was also sensitive to iron limitation in the latter study and previously (Baek et al., 2008; Homann et al., 2009; Hsu et al., 2011).
SEF1 encodes a predicted Cys(6)Zn(2) zinc binuclear cluster DNA binding protein without a clear ortholog in S. pombe; its S. cerevisiae ortholog has not been characterized. To test the hypothesis that Sef1 regulates iron homeostasis, we compared global RNA expression profiles of sef1ΔΔ and wild type C. albicans under iron-limiting conditions. Deletion of SEF1 resulted in downregulation of 170 genes and upregulation of 53 genes relative to wild type (minimum 2-fold change relative to wild type, 0.1% false discovery rate; Figure 2a and Table S1a). Morever, GO term analysis supported a significant association between Sef1-activated genes and iron homeostasis (6.5% of Sef1-activated genes vs. 0.5% in the genome, p=1×10−7). Indeed, the Sef1-activated gene set encodes every iron uptake factor depicted in Figure 1, as well as multiple components of the CCAAT-binding complex (Hap43, Hap2, and Hap3; Table 1).
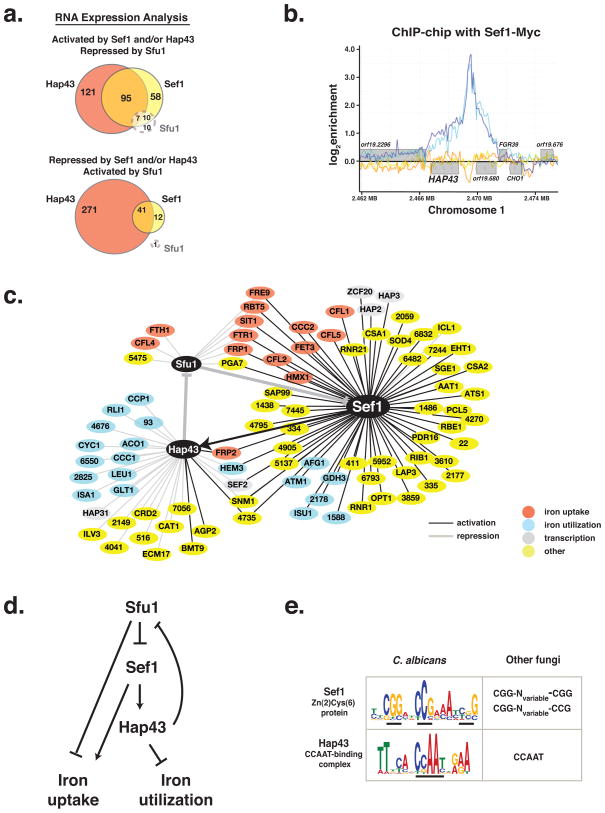
Figure 2. Transcriptional regulatory activities of Sef1, Sfu1, and Hap43.
a) Gene sets controlled by Sef1, Sfu1, and Hap43, based on whole genome ORF microarray analysis of the respective knockout mutants. Activation was defined by a minimum 2-fold decrease of gene expression in the deletion mutant relative to wild type, and repression by a minimum 2-fold increase. sef1ΔΔ and hap43ΔΔ were assessed in low iron medium, and sfu1ΔΔ in iron replete medium. Expression of the lone Sfu1-activated gene (IRO1) was unaffected by mutation of SEF1 or HAP43.
b) Sef1 binds to the HAP43 promoter. ChIP enrichment profiles of duplicate Sef1-Myc extracts are plotted in dark and light blue, and results from untagged controls are in yellow and orange. Genes are transcribed from left to right above the baseline, and from right to left below.
c) Direct gene regulation by Sef1, Sfu1, and Hap43. Gene activation is indicated by a black line and repression by a grey line. Targets involved in iron uptake are shaded red, iron utilization targets are blue, transcription factors are grey, all others are yellow. Targets lacking common names are depicted by the unique numerical components of their standard names; i.e. “123” for orf19.123.
d) Simplified scheme of C. albicans iron homeostasis.
e) Sef1 and Hap43 DNA recognition motifs compared to consensus sequences of orthologs in other species
See also Figure S1 and Tables S1a and S1b.
Table 1. Sef1 activation targets that encode iron uptake factors or components of the CCAAT-binding complex.
Fold expression changes were calculated from microarray analysis of sef1ΔΔ and wild type strains grown in iron-limiting medium.
| Systematic name | Gene name | Predicted protein | Expression sef1ΔΔ vs. WT |
|---|---|---|---|
|
REDUCTIVE IRON UPTAKE
| |||
| orf19.1263 | CFL1 | Putative ferric reductase | −4.8 |
| orf19.1930 | CFL5 | Putative ferric reductase | −32.8 |
| orf19.1264 | CFL2 | Putative ferric reductase | −7.1 |
| orf19.3538 | FRE9 | Putative ferric reductase | −4.4 |
| orf19.5634 | FRP1 | Putative ferric reductase | −5.8 |
| orf19.7112 | FRP2 | Putative ferric reductase | −4.4 |
| orf19.7219 | FTR1 | High-affinity iron permease | −2.9 |
| orf19.7231 | FTR2 | High-affinity iron permease | −2.4 |
| orf19.4211 | FET3 | Multicopper oxidase | −10.5 |
| orf19.4328 | CCC2 | Copper-transporting P-type ATPase of Golgi that is required for wild-type iron assimilation | −3.5 |
|
| |||
|
SIDEROPHORE UPTAKE
| |||
| orf19.2179 | SIT1 | Transporter of ferrichrome siderophores | −3.2 |
|
| |||
|
HEMOGLOBIN UPTAKE AND UTILIZATION
| |||
| orf19.5636 | RBT5 | GPI-anchored cell wall protein involved in hemoglobin utilization | −2.4 |
| orf19.5674 | PGA10 | Plasma membrane protein involved in heme-iron utilization | −3.4 |
| orf19.6073 | HMX1 | Heme oxygenase | −4.5 |
| orf19.2501 | FLC1 | Putative FAD transporter involved in uptake of heme | −2.1 |
|
| |||
|
CCAAT-BINDING COMPLEX
| |||
| orf19.1228 | HAP2 | Component of the CCAAT-binding complex | −3.3 |
| orf19.4647 | HAP3 | Putative component of the CCAAT-binding complex | −23.2 |
| orf19.681 | HAP43 | Component of the CCAAT-binding complex required for iron-limitation response | −3.1 |
Sef1, Sfu1, and Hap43 regulate each other’s expression
Because C. albicans was previously understood to control iron homeostasis similarly to S. pombe and other fungi (Baek et al., 2008; Hsu et al., 2011; Lan et al., 2004; Pelletier et al., 2007), we analyzed RNA expression in mutants affecting C. albicans orthologs of the GATA factor and a CCAAT-binding complex component. Under iron-replete conditions, deletion of SFU1 (GATA factor) resulted in upregulation of SEF1, HAP43 (CCAAT-binding complex), and 25 other genes largely associated with iron homeostasis (37% of Sfu1-repressed genes vs. 0.5% in the genome, p=1.7×10−15; Figure 2a and Table S1a). Only the gene encoding Iro1 was consistently ~2-fold downregulated in the sfu1ΔΔ mutant. Although the molecular function of Iro1 is not known, ectopic expression of IRO1 complements the growth defect of a S. cerevisiae mutant with a defect in iron acquisition (Garcia et al., 2001).
Our results with Sfu1 differed from those of Lan et al. (2004), who previously profiled a different sfu1ΔΔ mutant. Similar to us, these investigators observed upregulation of 22 genes (as currently annotated) in their mutant, including SEF1, HAP43, and multiple genes involved in iron uptake; however, a much larger set of 97 genes was downregulated. To determine whether our microarrays had failed to detect bona fide Sfu1-activation targets, we used RT-PCR to reassess the expression of the 5 genes that were most strongly downregulated (9 to 14-fold) in the Lan et al. mutant. Comparison between RNA levels in our sfu1ΔΔ mutant and wild type confirmed virtually identical expression of four of the genes, whereas the fifth gene (orf19.1774) was 2-fold downregulated in the mutant (Figure S1a). These results validate our microarray studies, which likely identified the vast majority of Sfu1 targets but may have missed some weakly regulated genes, in keeping with the stringent 0.1% false discovery rate. An alternative explanation for the discrepant results may be that the “sfu1ΔΔ” mutant profiled by Lan et al. was actually a sfu1ΔΔiro1ΔΔ double mutant, since the 3′ half of IRO1 is absent from the parental strain (Garcia et al., 2001).
We evaluated the role of HAP43 under iron-limiting conditions, where deletion of this gene has been shown to impair cellular proliferation (Baek et al., 2008; Homann et al., 2009; Hsu et al., 2011), activation of FRP1 (Baek et al., 2008), and repression of 9 genes associated with iron-dependent processes (shown by RT-PCR of selected targets in Hsu et al., 2011). We observed upregulation of 286 genes in hap43ΔΔ relative to wild type (Figure 2a, Table S1a). These included SFU1, FRP1 and other iron homeostasis genes (2.8% of Hap43-repressed genes vs. 0.5% in the genome, p=0.023), as well as genes involved in a variety of iron-dependent processes (including the 9 identified by Hsu et al., 2011) such as aerobic respiration (7.7% vs. 0.9% in the genome, p=1.9×10−13), the respiratory electron transport chain (4.5% vs. 0.4%, p=2.2×10−8), heme biosynthesis (2.8% vs. 0.2%, p=4.6×10−6), and iron-sulfur cluster assembly (2.1% vs. 0.2%, p=0.013). Another 223 genes were downregulated in hap43ΔΔ (Figure 2a, Table S1a), with a slight bias towards those associated with cytokinesis (4.5% vs. 1.0% in the genome, p=0.020). In aggregate, these experiments suggest important roles for all three transcription factors (Sef1, Sfu1, and Hap43) in the regulation of iron homeostasis in this species.
DNA-binding analysis of Sef1, Sfu1, and Hap43 reveals a tightly knit circuit
To dissect the direct versus indirect regulatory roles of Sef1, Sfu1, and Hap43, we performed chromatin immunoprecipitation experiments using Myc epitope-tagged versions of the three transcription factors. Fusion proteins were fully (Sef1-Myc and Sfu1-Myc) or partially (Hap43-Myc) functional when expressed as the only copy in the cell (Figure S1b). Matched pairs of strains differing only by the presence of the epitope tag were grown in iron-depleted (Sef1, Hap43) or iron-replete (Sfu1) medium. Whole cell extracts were incubated with monoclonal antibodies to the Myc epitope, and immunoprecipitated DNAs were amplified, fluorescently labeled, and hybridized to C. albicans genomic tiling microarrays. Shown in Figure 2b is a representative peak of highly specific binding of Sef1-Myc to the HAP43 promoter. Dark and light blue plots depict two independent experiments using Sef1-Myc extracts, and orange and yellow plots depict results with the untagged controls. Similar strong peaks of specific binding were observed across the genome for Sef1-Myc and Hap43-Myc, whereas Sfu1-Myc produced somewhat lower signal to noise (Table S1b). Sfu1 targets were validated with four additional ChIP experiments, using qPCR to quantify levels of the 9 putative direct targets vs. 4 controls; every target but no control was at least 2-fold enriched in the Sfu1-Myc extracts (Figure S1c).
We defined a gene regulatory event to comprise: (1) a significant and specific peak of DNA association by a given transcription factor (Table S1b) and (2) significant dependence on the associated transcription factor for normal expression of the regulated gene (Table S1a). Gene regulatory events mediated by Sef1, Sfu1, and/or Hap43 are depicted in Figure 2c (full dataset in Table 2), where black lines indicate transcriptional activation and grey lines, repression. Interactions among Sef1, Sfu1, and Hap43 themselves are marked with arrows (activation) and bars (repression). Examination of Figure 2c reveals Sef1 to function primarily as a transcriptional activator, with a large direct regulon of 64 genes, whereas Hap43 and Sfu1 are primarily transcriptional repressors, with smaller direct regulons of 25 and 10 genes, respectively. Sef1 and Sfu1 control most of the iron uptake genes (red), whereas Hap43 controls genes involved in iron-utilizing processes (e.g. aerobic respiration, heme biosynthesis, etc.; blue). In addition, Sef1 and Hap43 each regulate at least one other transcription factor (grey), potentially accounting for much larger number of (directly plus indirectly) regulated genes observed in the RNA expression studies (Figure 2a and Table 1).
Table 2. Gene regulatory events mediated by Sef1-, Sfu1-, and Hap43.
Direct gene regulatory targets of Sef1, Sfu1, and/or Hap43 are presented with descriptions, published virulence associations, fold expression changes, and physically-associated transcription factor. sfu1ΔΔ was assessed under iron-replete conditions, and sef1ΔΔ and hap43ΔΔ under iron-depleted conditions. Citations for published virulence associations are: FTR1 (Ramanan and Wang, 2000), CFL2 (Noble et al., 2010), HAP43 (Hsu et al., 2011), orf19.4905 (Noble et al., 2010), SEF1 (Noble et al., 2010), HMX1 (Navarathna and Roberts), HEM3 (Kirsch and Whitney, 1991), CAT1 (Nakagawa et al., 2003), and ICL1 (Barelle et al., 2006; Lorenz and Fink, 2001; Ramirez and Lorenz, 2007).
| REGULATED GENE | Wild type expression (replete vs. low iron) |
C. albicans Transcription Factor | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Systematic name |
Gene name |
Description | Cellular process | Role in virulence? (VIR) |
Sfu1 | Sef1 | Hap43 | ||||
| Expression (sfu1ΔΔ vs. WT) |
Promoter binding (Sfu1) | Expression (sef1ΔΔ vs. WT) |
Promoter binding (Sef1) |
Expression (hap43ΔΔ vs. WT) |
Promoter binding (Hap43) |
||||||
| orf19.539 | LAP3 | Putative ortholog of S.c. Lap3, a cysteine aminopeptidase that protects against homocysteine toxicity | Metabolism | −100.9 | −2.63 | Sef1 | −2.5 | ||||
| orf19.7114 | CSA1 | Surface antigen on elongating hyphae and buds | Cell wall composition: protein | −64.6 | −52.46 | Sef1 | −9.2 | ||||
| orf19.1263 | CFL1 | Putative ferric reductase, with sequence similarity to S.c. Fre2 which with Fre1 is a major cellular ferric reductase | Iron uptake: ferric reductase | −42.9 | 45.6 | −4.81 | Sef1 | 2.3 | |||
| orf19.5636 | RBT5 | GPI-anchored cell wall protein involved in hemoglobin utilization | Iron uptake: heme utilization pathway | −41.1 | 14.1 | Sfu1 | −2.39 | Sef1 | |||
| orf19.3117 | CSA2 | Putative cell surface protein | Cell wall composition: protein | −39.5 | 6.3 | −26.79 | Sef1 | −15.2 | |||
| orf19.5634 | FRP1 | Putative ferric reductase, with sequence similarity to S.c. Fre5 | Iron uptake: ferric reductase | −36.6 | 20.6 | Sfu1 | −5.78 | Sef1 | −3.2 | ||
| orf19.1930 | CFL5 | Putative ferric reductase, with similarity to S.c. Fre3 | Iron uptake: ferric reductase | −34.9 | 22.9 | −32.80 | Sef1 | 4.2 | |||
| orf19.5952 | Conserved hypothetical protein | Unknown | −32.5 | −46.24 | Sef1 | ||||||
| orf19.2179 | SIT1 | Transporter of ferrichrome siderophores, but not ferrioxamine B; putative ortholog of S.c. Arn1 | Iron uptake: siderophore transporter | −27.0 | 12.1 | Sfu1 | −3.16 | Sef1 | −4.1 | ||
| orf19.4802 | FTH1 | Putative ortholog of S.c. Fth1 high affinity iron transporter for intravacuolar iron stores | Iron homeostasis: vacuolar transporter | −22.4 | 22.6 | Sfu1 | |||||
| orf19.3538 | FRE9 | Putative ferric reductase with similarity to S.c. Fre3 | Iron uptake: ferric reductase | −20.0 | 8.9 | Sfu1 | −4.44 | Sef1 | −2.2 | ||
| orf19.7219 | FTR1 | High-affinity iron permease, putative ortholog of S.c. Ftr1 | Iron uptake: reductive pathway | VIR | −17.6 | 14.4 | Sfu1 | −2.94 | Sef1 | ||
| orf19.4647 | HAP3 | Conserved hypothetical protein with similarity to S.c. Hap3, a subunit of the Hap2p/3p/4p/5p CCAAT-binding complex | Iron regulation: transcription factor | −17.5 | −23.25 | Sef1 | −2.7 | ||||
| orf19.1264 | CFL2 | Putative ferric reductase with sequence similarity to S.c. Fre4 | Iron uptake: ferric reductase | VIR | −17.3 | 21.2 | −7.10 | Sef1 | 2.3 | ||
| orf19.5635 | PGA7 | Putative hyphal surface antigen with predicted GPI anchor | Cell wall composition: protein | −12.5 | 5.8 | Sfu1 | −4.70 | Sef1 | −2.9 | ||
| orf19.334 | Hypothetical protein, conserved in C.d., C.t. | Unknown | −9.7 | −2.01 | Sef1 | ||||||
| orf19.4795 | Conserved hypothetical protein | Unknown | −8.4 | −2.85 | Sef1 | −2.3 | |||||
| orf19.7112 | FRP2 | Putative ferric reductase, with sequence similarity to S.c. Fre5 | Iron uptake: ferric reductase | −7.2 | −4.40 | Sef1 | −2.6 | Hap43 | |||
| orf19.4270 | Putative mannosyltransferase | Cell wall composition: mannosylation | −5.2 | −6.17 | Sef1 | −6.4 | |||||
| orf19.5801 | RNR21 | Putative ortholog of S.c. Rnr2, a component of ribonucleotide-diphosphate reductase that catalyzes the rate-limiting step in dNTP synthesis | Metabolism: biosynthesis of dNTPs | −5.0 | −2.58 | Sef1 | −2.2 | ||||
| orf19.6793 | Putative protein conserved in C.d. and C.t. | Unknown | −5.0 | −4.35 | Sef1 | −4.7 | |||||
| orf19.681 | HAP43 | CCAAT-binding factor (CBF)-associated transcription factor required for iron-limitation response, | Iron regulation: transcription factor | −4.6 | 3.8 | −3.06 | Sef1 | (−26.0) | |||
| orf19.1228 | HAP2 | Putative ortholog of S.c. Hap2, a component of CCAAT-binding factor | Transcription: transcription factor | −4.5 | −3.33 | Sef1 | −2.2 | ||||
| orf19.5475 | Hypothetical protein | Unknown | −4.4 | 3.2 | Sfu1 | −2.08 | −2.2 | ||||
| orf19.4905 | Putative MFS transporter | Transporter | VIR | −4.4 | −5.39 | Sef1 | −2.1 | ||||
| orf19.3859 | Putative ortholog of S.c. Ifa38, a microsomal beta-keto-reductase important for VLCFA synthesis | Metabolism: biosynthesis of lipids | −4.1 | −2.43 | Sef1 | −2.1 | |||||
| orf19.7218 | RBE1 | Putative cell wall protein, no predicted GPI anchor | Cell wall composition: protein | −4.1 | −4.03 | Sef1 | −4.0 | ||||
| orf19.4211 | FET3 | Multicopper oxidase with similarity to S.c. Fet3 | Iron uptake: reductive pathway | −4.0 | 3.2 | −10.47 | Sef1 | ||||
| orf19.4735 | Putative ornithine cyclodeaminase, putative ortholog of S.c. YGL159W | Metabolism: interconversion of amino acids | −3.9 | −4.85 | Sef1 | −2.9 | Hap43 | ||||
| orf19.2062 | SOD4 | Member of a family of superoxide dismutases, predicted GPI-anchor | Response to stress: oxidative, cell wall composition: protein | −3.7 | −3.71 | Sef1 | −2.6 | ||||
| orf19.853 | SAP99 | Putative secreted aspartyl protease | Secreted enzyme | −3.7 | −2.38 | Sef1 | −2.1 | ||||
| orf19.5779 | RNR1 | Putative ortholog of S.c. Rnr1, a regulatory component of ribonucleotide reductase | Metabolism: biosynthesis of dNTPs | −3.7 | −5.25 | Sef1 | −3.1 | ||||
| orf19.411 | Conserved hypothetical protein | Unknown | −3.7 | −2.56 | Sef1 | −2.2 | |||||
| orf19.4328 | CCC2 | Copper-transporting P-type ATPase of Golgi that is required for wild-type iron assimilation, putative ortholog of S.c. Ccc2 | Iron uptake: reductive pathway | −3.6 | 3.0 | −3.53 | Sef1 | ||||
| orf19.6482 | Conserved hypothetical protein | Unknown | −3.6 | −2.71 | Sef1 | ||||||
| orf19.2177 | Hypothetical protein | Unknown | −3.6 | −6.84 | Sef1 | −2.3 | |||||
| orf19.1932 | CFL4 | Putative ortholog of S.c. Fre3, a ferric reductase | Iron uptake: ferric reductase | −3.6 | 24.4 | Sfu1 | 19.5 | ||||
| orf19.7445 | Putative ortholog of S.c. Vid24, a peripheral membrane protein located at Vid (vacuole import and degradation) vesicles | Vacuolar function | −3.5 | −2.79 | Sef1 | ||||||
| orf19.6832 | Conserved hypothetical protein with K-Cl cotransporter domain | Transporter: ions | −3.3 | −2.23 | Sef1 | ||||||
| orf19.4145 | ZCF20 | Putative zinc finger transcription factor | Transcription: transcription factor | −3.3 | −2.63 | Sef1 | |||||
| orf19.3753 | SEF1 | Putative transcription factor with zinc cluster DNA-binding motif, ortholog of S.c. Sef1 | Iron regulation: transcription factor | VIR | −3.0 | 5.4 | Sfu1 | −8.89 | |||
| orf19.335 | Conserved hypothetical protein | Unknown | −2.9 | −2.49 | Sef1 | ||||||
| orf19.6073 | HMX1 | Heme oxygenase, acts in utilization of hemin iron, putative ortholog of S.c. Hmx1 | Iron uptake: heme utilization pathway | VIR | −2.9 | −4.52 | Sef1 | −6.8 | |||
| orf19.22 | Conserved hypothetical protein with homology to peroxisomal membrane proteins | Peroxisomal function | −2.7 | −3.16 | Sef1 | ||||||
| orf19.1588 | Putative ortholog of S.c. Fmp21, a mitochondrial protein of unknown function | Mitochondrial function | −2.7 | −2.72 | Sef1 | ||||||
| orf19.2059 | Conserved hypothetical protein with homology to magnesium-dependent endonucleases and phosphatases involved in intracellular signalling | Signaling | −2.6 | −2.70 | Sef1 | ||||||
| orf19.4743 | AFG1 | Conserved hypothetical protein with similarity to S.c. Afg1 mitochondrial ATPase | Mitochondrial function | −2.4 | −3.19 | Sef1 | |||||
| orf19.1942 | SGE1 | Putative MFS-MDR transporter | Transporter | −2.4 | −2.52 | Sef1 | |||||
| orf19.1926 | SEF2 | Putative zinc cluster protein | Transcription: transcription factor | 2.2 | −12.54 | Sef1 | 9.0 | Hap43 | |||
| orf19.4869 | SFU1 | Transcriptional regulator of iron-responsive genes | Iron regulation: transcription factor | 3.5 | −17.1 | 5.9 | Hap43 | ||||
| orf19.6550 | Putative ortholog of S.c. Yor228c, a mitochondrial outer membrane protein | Mitochondrial function | 3.7 | 2.9 | Hap43 | ||||||
| orf19.6948 | CCC1 | Putative vacuolar Fe2+/Mn2+ transporter | Iron storage: vacuolar, Transporter: ions | 3.8 | 2.6 | Hap43 | |||||
| orf19.1742 | HEM3 | Hydroxymethylbilane synthase (uroporphyrinogen I synthase) | Iron utilization: biosynthesis of heme | VIR | 3.8 | −2.67 | Sef1 | 8.5 | Hap43 | ||
| orf19.7056 | Putative amino acid permease | Transporter: amino acids | 4.0 | 4.6 | 5.2 | Hap43 | |||||
| orf19.6229 | CAT1 | Catalase; role in resistance to oxidative stress, neutrophils, peroxide | Resistance to stress: oxidation | VIR | 4.2 | 2.0 | 10.6 | Hap43 | |||
| orf19.3034 | RLI1 | Putative ortholog of S.c. Rli1, an iron-sulfur protein required for ribosome biogenesis and translation initiation | Translation: initiation, ribosome biogenesis; iron utilization: iron-sulfur cluster protein | 4.5 | 3.9 | Hap43 | |||||
| orf19.5521 | ISA1 | Conserved hypothetical protein with strong similarity to S.c. Isa1, a mitochondrial matrix protein involved in biogenesis of iron-sulfur (Fe/S) cluster proteins | Iron utilization: biogenesis of iron-sulfur cluster proteins, mitochondrial function | 4.9 | 10.1 | Hap43 | |||||
| orf19.4099 | ECM17 | Putative ortholog of S.c. Met5, a sulfite reductase beta subunit | Metabolism: biosynthesis of amino acids | 5.1 | 2.01 | 13.4 | Hap43 | ||||
| orf19.6385 | ACO1 | Putative ortholog of S.c. Aco1, an aconitase required for the TCA cycle and maintenance of the mitochondrial genome | Metabolism: TCA cycle, mitochondrial function | 5.4 | 49.9 | Hap43 | |||||
| orf19.6257 | GLT1 | Putative ortholog of S.c. Glt1, an NAD(+)-dependent glutamate synthase | Metabolism: biosynthesis of glutamate | 5.7 | 7.2 | Hap43 | |||||
| orf19.1770 | CYC1 | Cytochrome c | Mitochondrial electron transport chain | 8.4 | 16.4 | Hap43 | |||||
| orf19.7498 | LEU1 | Putative ortholog of S.c. Leu1, an isopropylmalate isomerase that catalyzes the second step in the leucine biosynthesis pathway | Metabolism: biosynthesis of amino acids | 8.7 | 20.4 | Hap43 | |||||
| orf19.238 | CCP1 | Putative ortholog of S.c. Ccp1, a mitochondrial cytochrome-c oxidase | Mitochondrial function | 9.0 | 26.9 | Hap43 | |||||
| orf19.4040 | ILV3 | Putative ortholog of S.c. Ilv3, a dihydroxyacid dehydratase involved in biosynthesis of branched chain amino acids | Metabolism: biosynthesis of amino acids | 9.6 | 6.2 | Hap43 | |||||
| orf19.4674.1 | CRD2 | Metallothionein | Resistance to stress: copper | 24.9 | 3.3 | Hap43 | |||||
| orf19.4716 | GDH3 | Putative ortholog of S.c. Gdh3, a NADP(+)-dependent glutamate dehydrogenase that synthesizes glutamate from ammonia and alpha-ketoglutarate | Metabolism: biosynthesis of glutamate | −6.13 | Sef1 | ||||||
| orf19.1486 | Conserved hypothetical protein | Unknown | −3.48 | Sef1 | |||||||
| orf19.2602 | OPT1 | Oligopeptide transporter; transports 3-to-5-residue peptides | Transporter: peptides | −3.34 | Sef1 | −4.6 | |||||
| orf19.4673 | BMT9 | Putative beta-mannosyltransferase | Cell wall composition: mannosylation | −3.26 | −2.6 | Hap43 | |||||
| orf19.1027 | PDR16 | Phosphatidylinositol transfer protein, putative ortholog of S.c. Pdr16 that controls levels of various lipids | Metabolism: regulation of lipids | −2.98 | Sef1 | ||||||
| orf19.3610 | Conserved hypothetical protein with LgrB domain associated with murein hydrolase activity and penicillin tolerance | Unknown | −2.83 | Sef1 | |||||||
| orf19.2178 | Putative ortholog of S.c. Mrs4, an iron transporter that mediates Fe2+ transport across the inner mitochondrial membrane under low iron conditions | Iron utilization: mitochondrial transporter | −2.73 | Sef1 | |||||||
| orf19.6548 | ISU1 | Putatvie ortholog of S.c. Isu1, mitochondrial matrix protein that performs a scaffolding function during assembly of iron-sulfur clusters | Iron utilization: biogenesis of iron-sulfur cluster proteins, mitochondrial function | −2.57 | Sef1 | 2.0 | |||||
| orf19.3554 | AAT1 | Putative aspartate aminotransferase | Metabolism | −2.54 | Sef1 | −2.3 | |||||
| orf19.5137 | Hypothetical protein | Unknown | −2.54 | Sef1 | −4.1 | ||||||
| orf19.7244 | Conserved hypothetical protein with homology to fumarylacetoacetate (FAA) hydrolases | Unknown | −2.50 | Sef1 | 3.1 | ||||||
| orf19.3040 | EHT1 | Putative ortholog of S.c. Eht1, an Acyl-coenzymeA:ethanol O-acyltransferase that plays a minor role in medium-chain fatty acid ethyl ester biosynthesis | Metabolism: biosynthesis of lipids | −2.50 | Sef1 | ||||||
| orf19.1077 | ATM1 | Putative ortholog of S.c. Atm1, a mitochondrial inner membrane ABC transporter | Mitochondrial function, transporter | −2.30 | Sef1 | 2.9 | |||||
| orf19.1438 | Conserved hypothetical protein with homology to NADH dehydrogenase, FAD-containing subunit | Unknown | −2.27 | Sef1 | |||||||
| orf19.2862 | RIB1 | Putative ortholog of S.c. Rib1, a GTP cyclohydrolase II that catalyzes the first step of the riboflavin biosynthesis | Metabolism: biosynthesis of cofactors | −2.12 | Sef1 | ||||||
| orf19.1927 | SNM1 | Conserved hypothetical protein with Rpr2 motif of Rnase MRP family members | Translation: rRNA processing | −2.04 | Sef1 | 3.2 | Hap43 | ||||
| orf19.4012 | PCL5 | Putative cyclin | Cell cycle | −2.02 | Sef1 | −2.5 | |||||
| orf19.6399 | ATS1 | Putative ortholog of S.c. Ats1, a protein required, with Elongator complex, Kti11p, and Kti12p, for modification of wobble nucleosides in tRNA | Translation: tRNA modification | −2.01 | Sef1 | ||||||
| orf19.6844 | ICL1 | Putative ortholog of S.c. Icl1, a key enzyme of the glyoxylate cycle | Metabolism: glyoxylate cycle | VIR | 15.04 | Sef1 | 32.9 | ||||
| orf19.4679 | AGP2 | Putative ortholog of S.c. Agp2, an amino acid permease | Transporter: polyamines | −2.1 | Hap43 | ||||||
| orf19.516 | Putative ortholog of S.c. Rft1, a protein required for translocation of Man5GlcNac2-PP-Dol from the cytoplasmic side to the lumenal side of the ER membrane | Cell wall composition: N-glycosylation | 2.1 | Hap43 | |||||||
| orf19.4676 | Conserved hypothetical protein with homology to mitochondrial intermembrane space proteins | Mitochondrial function | 2.2 | Hap43 | |||||||
| orf19.93 | Putative ortholog of S.c. Mic17, a mitochondrial intermembrane space protein | Mitochondrial function | 2.2 | Hap43 | |||||||
| orf19.4041 | Putative ortholog of S.c. Pex4, a peroxisomal ubiquitin conjugating enzyme | Peroxisome function | 2.2 | Hap43 | |||||||
| orf19.517 | HAP31 | Putative ortholog of S.c. Hap3, a subunit of the CCAAT-binding complex | Iron regulation: transcription factor | 2.7 | Hap43 | ||||||
| orf19.2825 | Putative ortholog of S.c. Dre2, a protein required for iron-sulfur cluster assembly and sister chromatid cohesion | Iron utilization: iron-sulfur cluster assembly | 3.3 | Hap43 | |||||||
| orf19.2149 | Putative ortholog of S.c. Ypr003c, a putative sulfate permease | Transporter: sulfate | 3.3 | Hap43 | |||||||
A simplified interaction network focused on iron homeostasis is presented in Figure 2d. Under iron-replete conditions, Sfu1 directly represses Sef1 and iron uptake genes. Under iron-limiting conditions, Sef1 directly activates Hap43 and iron uptake genes, and Hap43 directly represses Sfu1 and iron utilization genes. The intercalation of Sef1 into this network reconfigures the reciprocal switch between the GATA factor (Sfu1) and the CCAAT-binding complex (Hap43) that is found in most other ascomycetes; however, the overall logic is unchanged, with iron uptake genes being repressed and iron utilization genes expressed under iron-replete conditions, and the reverse when environmental iron is low.
Putative DNA binding motifs recognized by Sef1 and the Hap43-associated CCAAT-binding complex were determined by MEME analysis of the highest-confidence DNA binding targets. As shown in Figure 2e, the top MEME hits strongly resemble consensus sequences of orthologs in other species. The Sef1 motif contains three CGG repeats (one inverted). Such repeats are characteristic of fungal zinc binuclear finger proteins, with spacing that is specific to individual family members (MacPherson et al., 2006). The Hap43-associated motif contains the signature CCAAT sequence of the CCAAT-binding complex, as defined in multiple eukaryotic species (Chodosh et al., 1988). Although unbiased searches of putative Sfu1 targets did not reveal a unifying motif, the conserved HGATAR motif (H represents A,T, or G; (Scazzocchio, 2000)) appears at least once in 7 of the 9 targets (Table S1b).
Sef1 is uniquely important in C. albicans
We questioned whether the prominence of Sef1 in C. albicans iron homeostasis is unique to this species vs. existing unrecognized among other fungi. Reciprocal amino acid sequence comparisons revealed no clear Sef1 ortholog in the S. pombe fungal lineage (Figure 3a). To exclude the presence of a functional homolog that is divergent in sequence, we tested mutants affecting 29 of 31 predicted zinc binuclear finger proteins for sensitivity to iron depletion (deletions of SPBC1773.12 and SPCC965.10 were not recovered and may be inviable). None of the mutants was highly sensitive to iron depletion (Figure S2a), arguing against the presence of a functional Sef1 homolog.
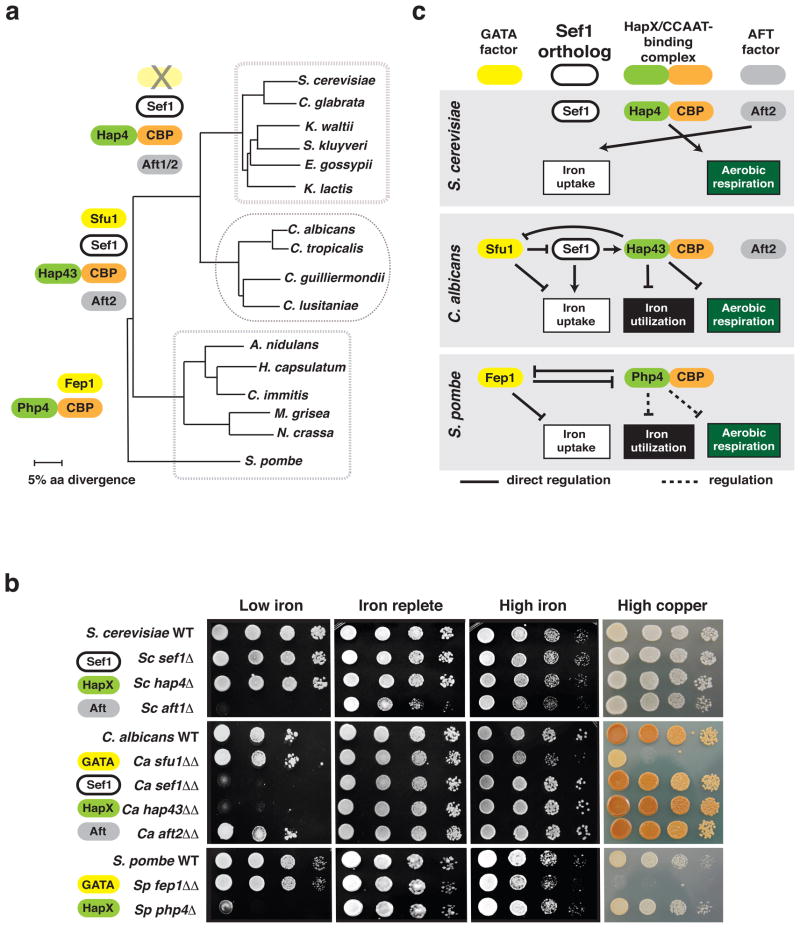
Figure 3. Analysis of transcription factor orthologs in C. albicans, S. cerevisiae, and S. pombe.
a) Phylogeny of iron-related transcription factors, based on amino acid sequence. Transcription factors present in each boxed lineage are shown on the left. Notably, Sef1 and Aft factors were gained by the C. albicans and S. cerevisiae lineages, and the GATA factor was lost by the S. cerevisiae lineage. Although the Hap2, Hap3, and Hap5 components of the CCAAT-binding complex are conserved at the sequence level in all three lineages, orthologs of the “HapX” regulatory component could not be unambiguously identified based on amino acid sequence and were instead defined functionally (Baek et al., 2008; Forsburg and Guarente, 1989; Hortschansky et al., 2007; Mercier et al., 2006).
b) Phenotypic comparison of orthologous mutants in C. albicans, S. cerevisiae, and S. pombe. Strains were plated on low iron medium (with iron chelators), iron replete medium (standard), high iron medium (with ferrichrome), or high copper medium (with copper sulfate) and incubated at 30°C. Note that densely growing C. albicans is darkly pigmented on high copper medium.
c) Updated comparison of iron homeostasis among three model fungi. The schematic integrates published information with our current analysis of C. albicans, S. pombe, and S. cerevisiae. S. pombe Php4/CBP has been implicated in repression of iron utilization genes (Mercier et al., 2006), but direct regulation has not yet been demonstrated.
See also Figure S2. Strains and primers are described in Tables S2a and S2b, respectively.
In contrast to S. pombe, the S. cerevisiae lineage has maintained a Sef1 ortholog that is recognizable at the amino acid level (Figure 3a). However, this lineage (along with the C. albicans lineage, Figure 3a) also acquired Aft family transcription factors that are known to regulate iron uptake in S. cerevisiae (Courel et al., 2005; Yamaguchi-Iwai et al., 1995; Yamaguchi-Iwai et al., 1996).
To clarify the iron-related roles of orthologous transcription factors among C. albicans, S. pombe, and S. cerevisiae, we profiled knockout mutants in each species on media containing low, standard, or elevated levels of iron, as well as high copper medium (that promotes iron uptake through enhanced assembly of iron permease/oxidase complexes; phenotypes are presented in Figure 3b). On standard medium, all mutants grew similarly to wild type except for S. cerevisiae aft1Δ (Aft factor), which exhibited a mild defect. On iron-depleted media, S. cerevisiae aft1Δ, C. albicans sef1ΔΔ and hap43ΔΔ (CCAAT-binding complex), and S. pombe php4Δ (CCAAT-binding complex) exhibited substantial growth defects, indicating roles for the deleted genes in resistance to iron depletion. On ferrichrome-supplemented media (“high iron”), only C. albicans sfu1ΔΔ GATA factor and S. pombe fep1Δ (GATA factor) exhibited growth defects, suggesting roles for these factors in resistance to iron toxicity; these phenotypes were enhanced on copper-supplemented media.
These results emphasize the importance of the GATA factor and CCAAT-binding complex in S. pombe and C. albicans iron homeostasis (whereas Aft1 is the major player in S. cerevisiae). The GATA factor protects against toxicity when environmental iron is abundant, and the CCAAT-binding complex prevents deficiency when environmental iron is scarce. The central role of Sef1 under iron-limiting conditions appears restricted to C. albicans, since this factor is not conserved in S. pombe and since the S. cerevisiae ortholog is not required for growth (nor is S.c.SEF1 expression upregulated, Figure S2b) under such conditions. These observations are integrated into an updated comparison of the three fungal regulatory circuits in Figure 3c.
SEF1 and SFU1 are differentially expressed in the bloodstream and gut
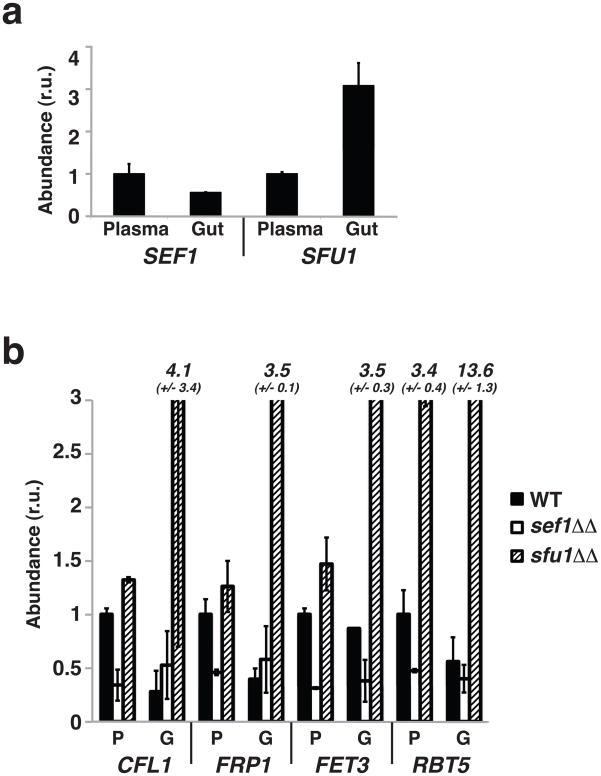
Given the divergent roles of Sef1 and Sfu1 in protection from iron depletion vs. iron toxicity in vitro, we considered whether they might also play specialized roles in the host. We compared SEF1 and SFU1 expression in wild type C. albicans grown for 1 hour at 37°C in human plasma or propagated for 5 days in the murine gastrointestinal tract (see below for description of this model). RT-PCR revealed a 2-fold induction of SEF1 vs. 3-fold repression of SFU1 in the plasma relative to the gut (Figure 4a), with concomitant induction of iron uptake genes (Figure 4b, black bars). Disparate effects on the expression of iron uptake genes were seen in sef1ΔΔ and sfu1ΔΔ mutants grown in the same environments (Figure 4b). Deletion of SEF1 attenuated the expression of iron uptake genes primarily in plasma (white bars), whereas deletion of SFU1 enhanced expression especially in the gut (crosshatched bars). These results suggest that Sef1 activates iron uptake genes in iron poor niches such as the bloodstream, whereas Sfu1 restricts expression of these genes, particularly in iron replete niches such as the gut.
Figure 4. Analysis of C. albicans gene expression in host niches that differ in iron content.
a) SEF1 is induced in the bloodstream, and SFU1 is induced in the gut. RT-PCR was used to analyze RNA extracted from wild type C. albicans grown for 1 hour in human plasma or for 5 days in the mouse gastrointestinal infection model. Transcript levels were normalized to levels of 16S ribosomal RNA. Error bars indicate the standard deviation.
b) Expression of iron uptake genes in wild type, sef1ΔΔ, and sfu1ΔΔ after growth in human plasma or the murine gastrointestinal tract. P stands for plasma and G for gut. Numerical values (with standard deviation) are presented for results that exceed the scale of the chart.
Sef1 but not Sfu1 is required for virulence
To determine whether Sef1 or Sfu1 contributes to virulence, we performed intravenous infections of BALB/c mice with sef1ΔΔ, sfu1ΔΔ, or wild type C. albicans and determined the time to illness (Figure 5a). As shown in Figure 5b, sef1ΔΔ but not sfu1ΔΔ displayed a significant virulence defect (p<0.0001, log-rank test); the defect was complemented by restoration of a copy of wild type SEF1, confirming genetic linkage.
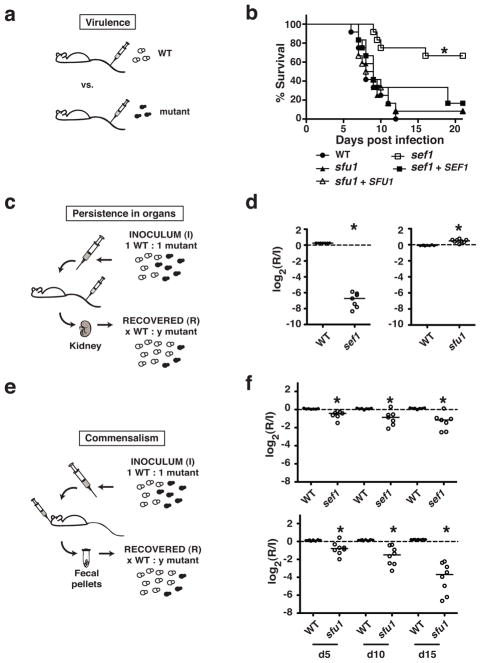
Figure 5. Roles of C. albicans Sef1 and Sfu1 in virulence and commensalism.
a) Virulence experiment. BALB/c mice were infected by tail vein injection with individual C. albicans strains (wild type, sef1ΔΔ, sfu1ΔΔ, or gene addback strains), and time to illness was monitored.
b) Sef1 but not Sfu1 is essential for virulence. Only the sef1ΔΔ mutant exhibited a significant decrease in virulence compared to wild type (asterisk indicates p<0.0001, log rank test).
c) Persistence experiment. BALB/c mice were infected by tail vein injection with 1:1 mixtures of wild type and sef1ΔΔ or sfu1ΔΔ). After mice developed clinical disease, the abundance of each strain in the inoculum (I) vs. mouse kidneys (R) was determined by qPCR.
d) Sef1 but not Sfu1 is required for persistence in host kidneys. Compared to wild type, sef1ΔΔ was significantly depleted from kidneys (p<0.0001, unpaired t-test), whereas sfu1ΔΔ was significantly enriched (p<0.0001).
e) Commensalism experiment. BALB/c mice were infected by gavage with 1:1 mixtures of wild type and sef1ΔΔ or sfu1ΔΔ. The abundance of each strain in the inoculum (I) and after recovery from fecal pellets (R) was determined by qPCR.
f) Sfu1 and Sef1 promote commensalism. sef1ΔΔ and sfu1ΔΔ were progressively depleted from fecal pellets relative to wild type (sef1ΔΔ: p=0.0017 at day 5, p=0.0047 at day 10, p=0.0012 at day 12; sfu1ΔΔ: p<0.002 at day 5, p<0.0003 at day 10, p<0.0001 at day 15; unpaired t-test). Comparison between the competitive indices of each mutant on day 15 indicated a more severe defect for sfu1ΔΔ (p=0.002, unpaired t-test).
We next investigated the abilities of the mutants to compete with wild type C. albicans for persistence in host tissues. BALB/c mice were infected intravenously with 1:1 mixtures of wild type and sef1ΔΔ or sfu1ΔΔ, followed by euthanasia when they developed signs of clinical illness. C. albicans was recovered from kidneys (the primary target organ in this model), and the relative abundance of each strain was determined by qPCR (Figure 5c). As shown in Figure 5d, sef1ΔΔ was significantly outcompeted by wild type C. albicans in mouse kidneys (p<0.0001, unpaired t-test), whereas sfu1ΔΔ displayed a significant competitive advantage (p<0.0001). These experiments indicate that C. albicans Sef1 but not Sfu1 is required for virulence and persistence in a mammalian bloodstream infection model.
Sef1 and Sfu1 promote commensalism
To assess the roles of Sef1 and Sfu1 in commensalism, we utilized a mouse model of gastrointestinal infection. In this model, infected mice remain healthy despite persistent, high-grade colonization with C. albicans (~107 CFUs/g stool; data not shown). BALB/c mice were infected by gavage with 1:1 mixtures of wild type C. albicans and either sef1ΔΔ or sfu1ΔΔ, and the abundance of each strain in fecal pellets over 15 days was monitored by qPCR (Figure 5e). Both mutants exhibited competitive defects compared to wild type throughout the time course (p<0.002, unpaired t-test; Figure 5f). Comparison between the competitive indices of each mutant on Day 15 revealed sfu1ΔΔ to have the more substantial defect (p=0.002, unpaired t-test). These results indicate that both transcription factors contribute to commensalism, with Sfu1 perhaps playing the more prominent role.
DISCUSSION
C. albicans is a ubiquitous commensal of the human microbiome, as well as the most common cause of disseminated fungal infections. Our detailed analysis of the regulatory system governing iron homeostasis in this microorganism underscores its intense adaptation to the mammalian host. Unlike the bipartite system used by most ascomycetes, the C. albicans system hinges on three transcriptional regulators, Sef1, Sfu1, and the Hap43-associated CCAAT-binding complex. These compose a tightly wired network in which each component directly regulates the expression of another transcription factor in the circuit as well as genes for iron uptake (Sfu1 and Sef1) or iron utilization (Hap43). Functional profiling of sef1ΔΔ and sfu1ΔΔ mutants in mouse models of bloodstream and gastrointestinal infection revealed a tradeoff of importance between these regulatory components that depends on the microenvironment of the host.
The C. albicans system for regulating iron homeostasis (Figure 2d) was deduced from global RNA expression and chromatin immunoprecipitation experiments. What distinguishes this system from that of most ascomycetes is the intercalation of Sef1 between the GATA factor (Sfu1) and the CCAAT-binding complex (Hap43). In other fungi, orthologs of Sfu1 and Hap43 create a simple switch, in which each component represses a discrete set of genes as well as the other transcription factor. In C. albicans, Sfu1 regulates Hap43 only indirectly via Sef1, and Sef1 directly activates target genes that are also directly repressed by either Sfu1 or Hap43. Our comprehensive analysis of mutants affecting transcription factor orthologs in C. albicans, S. pombe, and S. cerevisiae supports the idea that Sef1 plays a unique role in C. albicans iron homeostasis and was not simply missed in the other fungi.
Despite substantial rewiring of this transcriptional circuit among the three species, the overall logic has been preserved. That is, high-affinity iron uptake genes are expressed only under circumstances of environmental iron depletion. Nevertheless, specific features of each system likely impart additional properties that may be adaptive for the organism. In engineering parlance, the C. albicans mode of iron homeostasis in which one transcription factor (Sfu1) regulates the expression of a second transcription factor (Sef1)—and both regulate a common target (iron uptake genes)—is known as a feed forward loop (Mangan and Alon, 2003). An emergent feature of such loops is that expression of coregulated genes is buffered against transient perturbations of the activating signal, such that the level of expression of Sef1-Sfu1 coregulated iron uptake genes should be stabilized relative to short-term fluctuations in environmental iron, and therefore guarded against “spurious” activation or repression. The ability to maintain continuous expression of survival factors or continuous repression of toxicity factors in the appropriate setting could conceivably be of selective advantage.
Our findings that Sef1 but not Sfu1 is critical for virulence and competitive infection, whereas Sfu1 has a more important role in commensalism, suggest that extreme contrasts in iron availability within the mammalian host may have helped to shape the C. albicans iron regulatory circuit. A requirement for upregulation of high-affinity iron uptake genes in the host bloodstream is a common and well-recognized feature of many bacterial pathogens, some of which also use iron depletion to cue the expression of virulence genes (Mey et al., 2005). In keeping with this precedent, Sef1 directly activates multiple genes with known or suspected roles in virulence (Table 2).
Far less is known about microbial adaptations to the mammalian gastrointestinal tract, yet the microbiome is a primary source of pathogens causing disseminated disease. Our studies are a starting point for understanding such adaptations in C. albicans. The findings that Sfu1 promotes competitiveness in the gut and resistance to iron toxicity in vitro suggest that iron toxicity is an important selective pressure on gut commensals. Iron depletion is likely also important, at least in certain microniches, since sef1ΔΔ was also defective in the commensal model. In more general terms, our results emphasize that different host niches differ dramatically in ways that must be sensed by microbes. We hypothesize that successful commensals, whether bacterial or fungal, have evolved signaling and regulatory mechanisms to promote success in the commensal habitat, whereas commensal-pathogens that also enter the bloodstream must be capable of rapid cellular reprogramming to survive in this environment. An understanding of these mechanisms will be needed to decipher the transition between the commensal and pathogenic lifestyles.
METHODS
Strains
Strains are described in Table S2a, and primers are listed in Table S2b. C. albicans mutants, complemented strains, and Myc-tagged alleles of Sef1, Sfu1, and Hap43 were created as described (Nobile et al., 2009; Noble et al., 2010; Noble and Johnson, 2005).
S. pombe deletion mutants affecting Fep1, Php4, and zinc binuclear finger proteins were created by homologous recombination in reference strain SP286 using gene disruption fragments containing KanMX6 (G418 resistance) flanked by 700–900 bp of DNA homologous to sequences upstream and downstream of the target ORFs. Colony PCR was used to screen G418-resistant transformants for the expected 5′ and 3′ recombination junctions, and absence of the targets ORFs was confirmed using primers internal to the disrupted ORFs.
Media
Liquid “iron replete” medium was YPD (Guthrie and Fink, 1991), and “low iron” medium was YPD plus 500 μM bathophenanthrolinedisulfonic acid (BPS). Solid “iron replete” medium for C. albicans and S. cerevisiae was SC/2% agar (Guthrie and Fink, 1991); “low iron” was SC/2% agar with 350 μM BPS; “high iron” was SC/2% with 25 μM ferrichrome; and “high copper” was YPD/2% agar with 6 mM (C. albicans) or 800 μM (S. cerevisiae) CuSO4. For S. pombe, solid “iron replete” medium was YES/2% agar (Forsburg, 2003); “low iron” was YES/2% agar with 140μM 2,2′-dipyridyl (DIP); “high iron” was YES/2% agar with 25 μM ferrichrome; and “high copper” was YES/2% agar with 800 μM CuSO4.
Gene Expression Analysis
Saturated overnight cultures of sef1ΔΔ (SN330), hap43ΔΔ (SN694), sfu1ΔΔ (SN515), and isogenic wild type C. albicans (SN250) were inoculated into YPD to OD600=10−4 and incubated with shaking at 30°C. The next morning, logarithmically growing cells were diluted to OD600 0.01 in iron-replete (wild type and sfu1ΔΔ) or low iron (wild type, sef1ΔΔ, and hap43ΔΔ) medium and incubated at 30°C for 5–6 hours before harvesting at OD600=0.5–0.6. 5–6 biological replicates were performed per strain per condition. Total RNA was prepared using a hot phenol method (Miller and Johnson, 2002). 10 μg of each RNA was treated with DNase I (Turbo DNA-free kit, Ambion) and reverse transcribed using aminoallyl-dUTP and Superscript II (Invitrogen) according to manufacturers’ instructions. cDNA was labeled with Cy3 and Cy5 (Amersham), and 0.5 μg of each channel was hybridized to custom Agilent C. albicans ORF arrays (15,000 spots/array, 70-mer probes).
Fluorescently labeled cDNAs from mutants were directly hybridized against those from wild type grown under the same conditions; dye flip controls were included. 6 additional arrays were performed using wild type grown under iron-replete vs. iron-limiting conditions. Arrays were scanned using a Genepix 4000A Axon scanner, and spots were filtered using GenePix Pro software. Data were normalized using Goulphar (LOWESS normalization) and subjected to One-class Significance Analysis of Microarrays (SAM) analysis with a median false discovery rate of 0.1%. Candidates meeting SAM criteria were also required to exhibit median 2-fold changes among 5–6 experiments. Primary data are available at the Geo Expression Omnibus (GEO) website (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE30593.
GO Term analysis was performed using the GO Term Gene Ontology Finder tool on the Candida Genome Database website (Skrzypek et al., 2010).
Whole Genome Chromatin Immunoprecipitation Analysis
Saturated cultures of untagged wild type C. albicans (SN250), Sef1-Myc (SN423), Hap43-Myc (SN840), and Sfu1-Myc (SN646) were inoculated into low iron (untagged, Sef1-Myc, Hap43-Myc) or iron replete (untagged, Sfu1-Myc) liquid medium to OD600=0.05. Cultures were incubated with shaking at 30°C until OD600 0.4, when formaldehyde was added to 1% final (with shaking, room temperature, 15 minutes), followed by glycine to 125mM final (with shaking, room temperature, 5 minutes). Cells were collected by centrifugation at 4°C and washed twice with 20 mM Tris-HCl pH 7.5/150 mM NaCl, followed by freezing in liquid N2 and storage at −80°C. Cell lysis, DNA shearing, and ChIP-Chip were performed as described (Nobile et al., 2009).
12 independent hybridization experiments were performed on 6 biological replicates of the untagged control and 2 biological replicates each of Sef1-Myc, Sfu1-Myc, and Hap43-Myc. Agilent Chip Analytics software v1.2 (Agilent Technologies) was used for initial data normalization and analysis (Tuch et al., 2008), followed by visualization and additional analysis using MochiView v.1.39 (http://johnsonlab.ucsf.edu/). High confidence regulatory events were associated with Agilent segment p-values of ≥4 (−log10 p-value based on the enrichment statistic for each probe in the region) and minimum 2-fold (Sef1-Myc and Hap43-Myc) or 1.5-fold (Sfu1-Myc) enrichment in both biological replicates of the epitope-tagged strains (Table S1b). Genes with enrichment peaks in untagged controls were excluded. Primary data are available at the GEO website (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE30593.
Identification of DNA recognition motifs
MEME v.3.5.7 software (Bailey and Elkan, 1994) was applied to 250 bp sequences centered at the midpoints of the most significant binding peaks of Sef1, Sfu1, and Hap43. Analysis was performed using minw=6, maxw=20, and nmotif=3.
Phylogenetic analysis
Protein sequences of iron homeostasis regulators in C. albicans, S. pombe, and S. cerevisiae were BLASTed (Altschul et al., 1997) against a database of all fungal ORFs. Matching ORFs with E-values <10−5 were extracted from the database and multiply aligned with MUSCLE (Edgar, 2004), followed by inference of an NJ tree using ClustalW (Higgins and Sharp, 1988). The resulting gene tree was inspected and gain/loss/duplication events were mapped to a fungal species tree inferred previously (Tuch et al., 2008). The results of this analysis (summarized in Figure 2a) are consistent with those in the YGOB (Byrne and Wolfe, 2005) and CGOB (Fitzpatrick et al.) databases, which cover a narrower range of species but also account for conservation of gene synteny. Whereas Sfu1 and components of CBP clearly existed prior to divergence of the ascomycetes studied here, evidence for an early origin of Sef1 and Aft1/2 is lacking.
In vitro growth assays
Freshly streaked C. albicans, S. cerevisiae, and S. pombe strains were inoculated into YPD (C. albicans, S. cerevisiae) or YES (S. pombe) and incubated overnight at 30°C. Saturated cultures were diluted with sterile water to A600=0.8, serial 10-fold dilutions were made, and 5–10 μl of each dilution series was applied to solid test media, followed by incubation at 30°C for 2–4 days.
In vivo assays
Procedures involving animals were approved by the UCSF Institutional Animal Care and Use Committee. Virulence analysis was conducted by tail vein injection of groups of 8–10 week old female BALB/c with 5 × 105 CFUs of wild type (SN425), sef1ΔΔ (SN452), sef1ΔΔ/SEF1 (SN436), sfu1ΔΔ (SN668), or sef1ΔΔ/SFU1 (SN664). Mice were monitored twice daily and euthanized when morbidity criteria were met (weight loss>15%, hunched posture, inactivity).
Competitive bloodstream infections were performed with wild type (SN250), sef1ΔΔ (SN330), and sfu1ΔΔ (SN515) as previously described (Noble et al., 2010).
The mouse model of C. albicans commensalism was adapted from published protocols (Koh et al., 2008; White et al., 2007). Groups of 8–10 week female BALB/c mice received penicillin 1500 un/ml and streptomycin 2 mg/ml in their drinking water for 3–5 days prior to gavage with 108 CFUs of a 1:1 mix of wild type (SN250) and sef1ΔΔ (SN330) or sfu1ΔΔ (SN515). Antibiotics were continued, and fecal pellets were collected at specified intervals. C. albicans recovery and quantification were performed as described (Noble et al., 2010).
Supplementary Material
Highlights.
C. albicans uses a transcriptional circuit to regulate iron acquisition from the host.
Sef1 activates iron uptake genes and promotes virulence in the bloodstream.
Sfu1 represses iron uptake genes and promotes commensalism in the gut.
Acknowledgments
We are grateful to C. Nobile and A. Hernday for useful advice and technical assistance with RNA expression and ChIP-Chip experiments, to B. Green and A. Johnson for the Sfu1-Myc strain, to O. Homann for C. albicans mutants and guidance with his excellent MochiView software, to P. Hartley for generating ChIP-chip heatmaps, and to A. Johnson and H. Madhani for use of laboratory equipment. S. Braun and H. Madhani provided unpublished S. pombe strains. D. Breslow and J. Weissman provided S. cerevisiae strains. J. Kronstad generously provided ferrichrome. H. El-Samad provided insights into feed forward networks. H. Madhani, A. Sil, and J. Cox provided helpful comments on the manuscript. This work was supported by NIH KO8AI062800 and awards from the Burroughs Wellcome Foundation, Hellman Family Foundation, and the UCSF Program in Breakthrough Biomedical Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, Edwards JE, Filler SG, Hube B. the hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 2008;4:e1000217. doi: 10.1371/journal.ppat.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardon O, Bussey H, Philpott C, Ward DM, Davis-Kaplan S, Verroneau S, Jiang B, Kaplan J. Identification of a Candida albicans ferrichrome transporter and its characterization by expression in Saccharomyces cerevisiae. J Biol Chem. 2001;276:43049–43055. doi: 10.1074/jbc.M108701200. [DOI] [PubMed] [Google Scholar]
- Baek YU, Li M, Davis DA. Candida albicans ferric reductases are differentially regulated in response to distinct forms of iron limitation by the Rim101 and CBF transcription factors. Eukaryot Cell. 2008;7:1168–1179. doi: 10.1128/EC.00108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Paper presented at: Second International Conference on Intelligent Systems for Molecular Biology (AAAI Press); 1994. [PubMed] [Google Scholar]
- Barelle CJ, Priest CL, Maccallum DM, Gow NA, Odds FC, Brown AJ. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 2006;8:961–971. doi: 10.1111/j.1462-5822.2005.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth LN, Tuch BB, Johnson AD. Intercalation of a new tier of transcription regulation into an ancient circuit. Nature. 2010;468:959–963. doi: 10.1038/nature09560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh LA, Olesen J, Hahn S, Baldwin AS, Guarente L, Sharp PA. A yeast and a human CCAAT-binding protein have heterologous subunits that are functionally interchangeable. Cell. 1988;53:25–35. doi: 10.1016/0092-8674(88)90484-9. [DOI] [PubMed] [Google Scholar]
- Courel M, Lallet S, Camadro JM, Blaiseau PL. Direct activation of genes involved in intracellular iron use by the yeast iron-responsive transcription factor Aft2 without its paralog Aft1. Mol Cell Biol. 2005;25:6760–6771. doi: 10.1128/MCB.25.15.6760-6771.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis. 1999;29:239–244. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick DA, O’Gaora P, Byrne KP, Butler G. Analysis of gene evolution and metabolic pathways using the Candida Gene Order Browser. BMC Genomics. 11:290. doi: 10.1186/1471-2164-11-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL. Current Protocols in Molecular Biology. John Wiley and Sons; 2003. S. pombe Strain Maiintenance and Media; pp. 13.15.11–13.15.15. [DOI] [PubMed] [Google Scholar]
- Forsburg SL, Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- Garcia MG, O’Connor JE, Garcia LL, Martinez SI, Herrero E, del Castillo Agudo L. Isolation of a Candida albicans gene, tightly linked to URA3, coding for a putative transcription factor that suppresses a Saccharomyces cerevisiae aft1 mutation. Yeast. 2001;18:301–311. doi: 10.1002/1097-0061(20010315)18:4<301::AID-YEA672>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR, editors. Guide to Yeast Genetics and Molecular Biology. San Diego, CA, USA: Academic Press; 1991. [Google Scholar]
- Haas H, Zadra I, Stoffler G, Angermayr K. The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J Biol Chem. 1999;274:4613–4619. doi: 10.1074/jbc.274.8.4613. [DOI] [PubMed] [Google Scholar]
- Hammacott JE, Williams PH, Cashmore AM. Candida albicans CFL1 encodes a functional ferric reductase activity that can rescue a Saccharomyces cerevisiae fre1 mutant. Microbiology. 2000;146(Pt 4):869–876. doi: 10.1099/00221287-146-4-869. [DOI] [PubMed] [Google Scholar]
- Heymann P, Gerads M, Schaller M, Dromer F, Winkelmann G, Ernst JF. The siderophore iron transporter of Candida albicans (Sit1p/Arn1p) mediates uptake of ferrichrome-type siderophores and is required for epithelial invasion. Infect Immun. 2002;70:5246–5255. doi: 10.1128/IAI.70.9.5246-5255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;5:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortschansky P, Eisendle M, Al-Abdallah Q, Schmidt AD, Bergmann S, Thon M, Kniemeyer O, Abt B, Seeber B, Werner ER, et al. Interaction of HapX with the CCAAT-binding complex--a novel mechanism of gene regulation by iron. EMBO J. 2007;26:3157–3168. doi: 10.1038/sj.emboj.7601752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PC, Yang CY, Lan CY. Candida albicans Hap43 Is a Low Iron-Induced Repressor Essential for Iron-Responsive Transcriptional Regulation and Virulence. Eukaryot Cell. 2011;10:207–225. doi: 10.1128/EC.00158-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CJ, Bai C, Zheng XD, Wang YM, Wang Y. Characterization and functional analysis of the siderophore-iron transporter CaArn1p in Candida albicans. J Biol Chem. 2002;277:30598–30605. doi: 10.1074/jbc.M204545200. [DOI] [PubMed] [Google Scholar]
- Jung WH, Saikia S, Hu G, Wang J, Fung CK, D’Souza C, White R, Kronstad JW. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog. 2010;6:e1001209. doi: 10.1371/journal.ppat.1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sham A, White R, Kronstad JW. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 2006;4:e410. doi: 10.1371/journal.pbio.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch DR, Whitney RR. Pathogenicity of Candida albicans auxotrophic mutants in experimental infections. Infect Immun. 1991;59:3297–3300. doi: 10.1128/iai.59.9.3297-3300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SA, Lesuisse E, Stearman R, Klausner RD, Dancis A. Reductive iron uptake by Candida albicans: role of copper, iron and the TUP1 regulator. Microbiology. 2002;148:29–40. doi: 10.1099/00221287-148-1-29. [DOI] [PubMed] [Google Scholar]
- Koh AY, Kohler JR, Coggshall KT, Van Rooijen N, Pier GB. Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS Pathog. 2008;4:e35. doi: 10.1371/journal.ppat.0040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan CY, Rodarte G, Murillo LA, Jones T, Davis RW, Dungan J, Newport G, Agabian N. Regulatory networks affected by iron availability in Candida albicans. Mol Microbiol. 2004;53:1451–1469. doi: 10.1111/j.1365-2958.2004.04214.x. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Fink GR. The glyoxylate cycle is required for fungal virulence. Nature. 2001;412:83–86. doi: 10.1038/35083594. [DOI] [PubMed] [Google Scholar]
- Luo G, Samaranayake LP, Yau JY. Candida species exhibit differential in vitro hemolytic activities. J Clin Microbiol. 2001;39:2971–2974. doi: 10.1128/JCM.39.8.2971-2974.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev. 2006;70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns JM, Mosser DM, Buckley HR. Production of a hemolytic factor by Candida albicans. Infect Immun. 1994;62:5154–5156. doi: 10.1128/iai.62.11.5154-5156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RB, Savory J, Brown S, Bertholf RL, Wills MR. Transferrin binding of Al3+ and Fe3+ Clin Chem. 1987;33:405–407. [PubMed] [Google Scholar]
- McCance RA, Widdowson EM. The absorption and excretion of iron following oral and intravenous administration. J Physiol. 1938;94:148–154. doi: 10.1113/jphysiol.1938.sp003669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier A, Pelletier B, Labbe S. A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell. 2006;5:1866–1881. doi: 10.1128/EC.00199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect Immun. 2005;73:8167–8178. doi: 10.1128/IAI.73.12.8167-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- Miret S, Simpson RJ, McKie AT. Physiology and molecular biology of dietary iron absorption. Annu Rev Nutr. 2003;23:283–301. doi: 10.1146/annurev.nutr.23.011702.073139. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Kanbe T, Mizuguchi I. Disruption of the human pathogenic yeast Candida albicans catalase gene decreases survival in mouse-model infection and elevates susceptibility to higher temperature and to detergents. Microbiol Immunol. 2003;47:395–403. doi: 10.1111/j.1348-0421.2003.tb03376.x. [DOI] [PubMed] [Google Scholar]
- Navarathna DH, Roberts DD. Candida albicans heme oxygenase and its product CO contribute to pathogenesis of candidemia and alter systemic chemokine and cytokine expression. Free Radic Biol Med. 49:1561–1573. doi: 10.1016/j.freeradbiomed.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, Nantel A, Andes DR, Johnson AD, Mitchell AP. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 2009;7:e1000133. doi: 10.1371/journal.pbio.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC. Candida and Candidosis, a Review and Bibliography. 2. London: W.B. Saunders; 1988. [Google Scholar]
- Pelletier B, Beaudoin J, Mukai Y, Labbe S. Fep1, an iron sensor regulating iron transporter gene expression in Schizosaccharomyces pombe. J Biol Chem. 2002;277:22950–22958. doi: 10.1074/jbc.M202682200. [DOI] [PubMed] [Google Scholar]
- Pelletier B, Mercier A, Durand M, Peter C, Jbel M, Beaudoin J, Labbe S. Expression of Candida albicans Sfu1 in fission yeast complements the loss of the iron-regulatory transcription factor Fep1 and requires Tup co-repressors. Yeast. 2007;24:883–900. doi: 10.1002/yea.1539. [DOI] [PubMed] [Google Scholar]
- Pierre JL, Fontecave M, Crichton RR. Chemistry for an essential biological process: the reduction of ferric iron. Biometals. 2002;15:341–346. doi: 10.1023/a:1020259021641. [DOI] [PubMed] [Google Scholar]
- Ramanan N, Wang Y. A high-affinity iron permease essential for Candida albicans virulence. Science. 2000;288:1062–1064. doi: 10.1126/science.288.5468.1062. [DOI] [PubMed] [Google Scholar]
- Ramirez MA, Lorenz MC. Mutations in alternative carbon utilization pathways in Candida albicans attenuate virulence and confer pleiotropic phenotypes. Eukaryot Cell. 2007;6:280–290. doi: 10.1128/EC.00372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scazzocchio C. The fungal GATA factors. Curr Opin Microbiol. 2000;3:126–131. doi: 10.1016/s1369-5274(00)00063-1. [DOI] [PubMed] [Google Scholar]
- Schrettl M, Beckmann N, Varga J, Heinekamp T, Jacobsen ID, Jochl C, Moussa TA, Wang S, Gsaller F, Blatzer M, et al. PLoS Pathog. Vol. 6. 2010. HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypek MS, Arnaud MB, Costanzo MC, Inglis DO, Shah P, Binkley G, Miyasato SR, Sherlock G. New tools at the Candida Genome Database: biochemical pathways and full-text literature search. Nucleic Acids Res. 2010;38:D428–432. doi: 10.1093/nar/gkp836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 2008;6:e38. doi: 10.1371/journal.pbio.0060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman Z, Kornitzer D. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol Microbiol. 2004;53:1209–1220. doi: 10.1111/j.1365-2958.2004.04199.x. [DOI] [PubMed] [Google Scholar]
- Weissman Z, Shemer R, Conibear E, Kornitzer D. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol Microbiol. 2008;69:201–217. doi: 10.1111/j.1365-2958.2008.06277.x. [DOI] [PubMed] [Google Scholar]
- White SJ, Rosenbach A, Lephart P, Nguyen D, Benjamin A, Tzipori S, Whiteway M, Mecsas J, Kumamoto CA. Self-regulation of Candida albicans population size during GI colonization. PLoS Pathog. 2007;3:e184. doi: 10.1371/journal.ppat.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Dancis A, Klausner RD. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 1995;14:1231–1239. doi: 10.1002/j.1460-2075.1995.tb07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Stearman R, Dancis A, Klausner RD. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 1996;15:3377–3384. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.