Abstract
The Cdc7p protein kinase plays an essential, but undefined, role promoting S phase in the budding yeast, Saccharomyces cerevisiae. Previous experiments have shown that the essential function of Cdc7 is executed near the G1–S boundary; after Start but before the elongation phase of DNA replication. Origins of DNA replication fire throughout S phase in budding yeast. Therefore, the G1–S transition is a cell-cycle event that precedes, and is distinct from, the activation of individual origins. Consequently, we have asked whether Cdc7 is only required for S-phase entry or if it plays a role during S phase in origin firing. In this article, we show that partial loss of Cdc7 function results in slow progression through S phase rather than slow entry into S phase and that Cdc7 is still required for the timely completion of S phase after a block to elongation with hydroxyurea. This is because Cdc7 is still required for the activation of late-firing origins after the hydroxyurea block. These experiments show that, rather than acting as a global regulator of the G1–S transition, Cdc7 appears to play a more direct role in the firing of replication origins during S phase.
Keywords: Cdc7 protein kinase, S. cerevisiae, origin firing, S phase
Considerable progress has been made in understanding how the initiation of DNA replication is regulated in eukaryotic cells (Diffley 1996; Muzi-Falconi et al. 1996; Nasmyth 1996; Stillman 1996; Wuarin and Nurse 1996). Evidence now supports a model that explains how the genome can be precisely duplicated just once in each cell cycle. At the heart of this model is the notion that cells exist in two mutually exclusive states. In the first state, which occurs during G1, cells are competent to assemble prereplicative complexes (pre-RCs) at their origins (Diffley et al. 1994). Pre-RCs are likely to be bona fide preinitiation complexes because they contain a number of proteins that are essential for initiating DNA replication including the origin recognition complex (ORC) (Diffley and Cocker 1992; Diffley et al. 1994; Santocanale and Diffley 1996; Tanaka et al. 1997), the Cdc6 protein (Cocker et al. 1996; Tanaka et al. 1997), and the Mcm2-7 family of proteins (Aparicio et al. 1997; Donovan et al. 1997; Tanaka et al. 1997). Similar results have been obtained with the Xenopus cell-free replication system, suggesting that this pathway has been conserved in evolution (Chong et al. 1995; Kubota et al. 1995; Madine et al. 1995a,b; Carpenter et al. 1996; Coleman et al. 1996; Romanowski et al. 1996; Rowles et al. 1996). In actively cycling cells, pre-RCs assemble at origins at the end of mitosis (Diffley et al. 1994), however, pre-RCs can also assemble later in G1 (Piatti et al. 1996). This is important because pre-RCs are lost from origins when cells enter stationary phase and must reassemble upon re-entry into the cell cycle (Diffley et al. 1994). Although G1 cells are competent to assemble pre-RCs, they are not competent to fire origins.
At the end of G1, cells enter the second state in which origins containing preformed pre-RCs are now competent to fire, but new pre-RCs can no longer assemble at origins (Dahmann et al. 1995; Piatti et al. 1996). The activation of the major cyclin dependent kinase, Cdc28, together with its regulatory subunits, the B-type cyclins, is essential for entry into this second state; Clb–Cdc28 kinase is required both to initiate DNA replication and to block the formation of new pre-RCs after origins fire (Schwob et al. 1994; Dahmann et al. 1995; Piatti et al. 1996). Thus, origins cannot be reset until the Clbs are degraded at the end of mitosis. The dual requirement for Clb–Cdc28 in activation of origins and inhibition of pre-RC assembly ensures that only a single round of origin firing can occur during the cell cycle.
In addition to Cdc28, a second protein kinase, Cdc7, also plays an essential role in promoting S-phase progression (Hartwell 1974, 1976; Hartwell et al. 1974; Hereford and Hartwell 1974; Patterson et al. 1986; Bahman et al. 1988; Hollingsworth and Sclafani 1990; Buck et al. 1991). Cdc7, like Cdc28, has been conserved in evolution and homologs have been found in fission yeast (Masai et al. 1995) and metazoans including Xenopus and humans (Sato et al. 1997). cdc7 mutants arrest as budded cells with a 1C DNA content and high levels of Clb kinase (Amon et al. 1992). Genetic experiments have shown that Cdc7 is required for DNA replication after Start (α factor mating pheromone) and after the action of the Cdc4 protein. Cdc7, in fact, appears to act quite late in G1 because cycloheximide does not block S-phase progression after release from a cdc7-1 block. Cdc7 is not required, however, to complete the cell cycle after a block to elongation with hydroxyurea (HU) (Hereford and Hartwell 1974; Hartwell 1976). Taken together, these results argue that Cdc7 executes its essential function in a narrow window around the end of G1 or the beginning of S phase.
Cdc7 does not act alone; evidence suggests that the Dbf4 protein is a positive regulator of Cdc7p (Jackson et al. 1993; Kitada et al. 1993). The terminal phenotype of dbf4 mutants is very similar to that of cdc7 mutants (Johnston and Thomas 1982; Solomon et al. 1992); overexpression of Dbf4 can suppress cdc7 mutants (Kitada et al. 1993); dbf4 mutants are synthetically lethal with cdc7 mutants (Kitada et al. 1993); and Dbf4 and Cdc7 interact in two-hybrid experiments (Jackson et al. 1993; Dowell et al. 1994) and in yeast extracts (Dixon and Campbell 1997). The Cdc7p protein kinase activity is dependent on Dbf4 and extracts from cdc7 and dbf4 mutants are both defective in Cdc7 kinase, but can complement each other in vitro (Jackson et al. 1993). In addition to being required for activation of the Cdc7 protein kinase, Dbf4 interacts with replication origins through the ORC-binding site by use of a one-hybrid assay (Dowell et al. 1994). Whether Dbf4 interacts directly with ORC or indirectly with some other ORC-interacting protein has not yet been determined, however, the origin-interacting domain of Dbf4 can be separated from its Cdc7-interacting domain, indicating that origin interaction does not occur through Cdc7 (Dowell et al. 1994). In addition, genetic analysis has shown that Dbf4 and Cdc7 interact with ORC and Mcm proteins (Fox et al. 1995; Loo et al. 1995; Hardy 1996; Hardy et al. 1997; Lei et al. 1997). Both the origin-interacting domain and the Cdc7p-interacting domain of Dbf4 are essential for its function, consistent with the hypothesis that Dbf4 acts to recruit Cdc7 to replication origins (Dowell et al. 1994).
Origins do not normally fire synchronously at the onset of S phase, but instead, some origins fire early in S phase, whereas others fire late (for review, see Ferguson et al. 1991a; Fangman and Brewer 1992; Brewer et al. 1993). Factors regulating this are currently not well understood. Recent experiments have indicated that the timing of origin firing during S phase is established during the preceding mitosis (Raghuraman et al. 1997). An interesting possibility is that some aspect of pre-RC composition may be different between early and late-firing origins.
Because origins fire throughout S phase, the G1–S transition can be considered as a cell-cycle event that is distinct from actual origin firing. We have been interested in understanding whether Cdc7 functions as the global promoter of this G1–S transition or if Cdc7 acts more directly throughout S phase for the firing of replication origins. In this article, we provide evidence that argues strongly against the former possibility.
Results
cdc7 mutants are defective in S-phase progression but not G1–S transit
The first approach we used has been to examine cell-cycle kinetics in cdc7 mutants at semipermissive temperatures. The rationale for this approach is as follows. At fully permissive temperatures, cdc7 temperature-sensitive mutants should proceed normally through S phase after release from a G1 block, whereas at fully restrictive temperatures, cells should arrest in late G1. At semipermissive temperatures, cdc7 mutants should have partial Cdc7 activity and should, therefore, only be partially defective in the essential function of Cdc7. We reasoned that if Cdc7 were only required as a switch for the G1–S transition, then at semipermissive temperatures, cdc7 mutants should show a measurable defect in entry into S phase, but once in S phase, these mutants should show no defect in S-phase progression. This would be manifested as accumulation of cells with a 1C DNA content for an extended period of time after release from a G1 block as cells accumulate at the G1–S boundary. Once cells cross this G1–S boundary, however, they should proceed through S phase at a wild-type rate. Alternatively, if the action of Cdc7 is manifested during S phase rather than at the G1–S transition, then cdc7 mutants should enter S phase with normal kinetics but should show defects in S-phase progression, which would be seen as a measurably slower passage of cells from 1C to 2C DNA content.
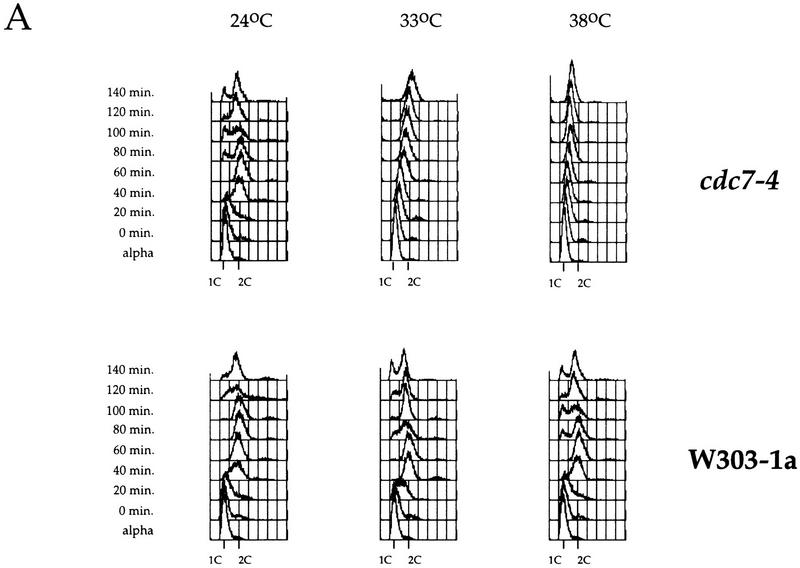
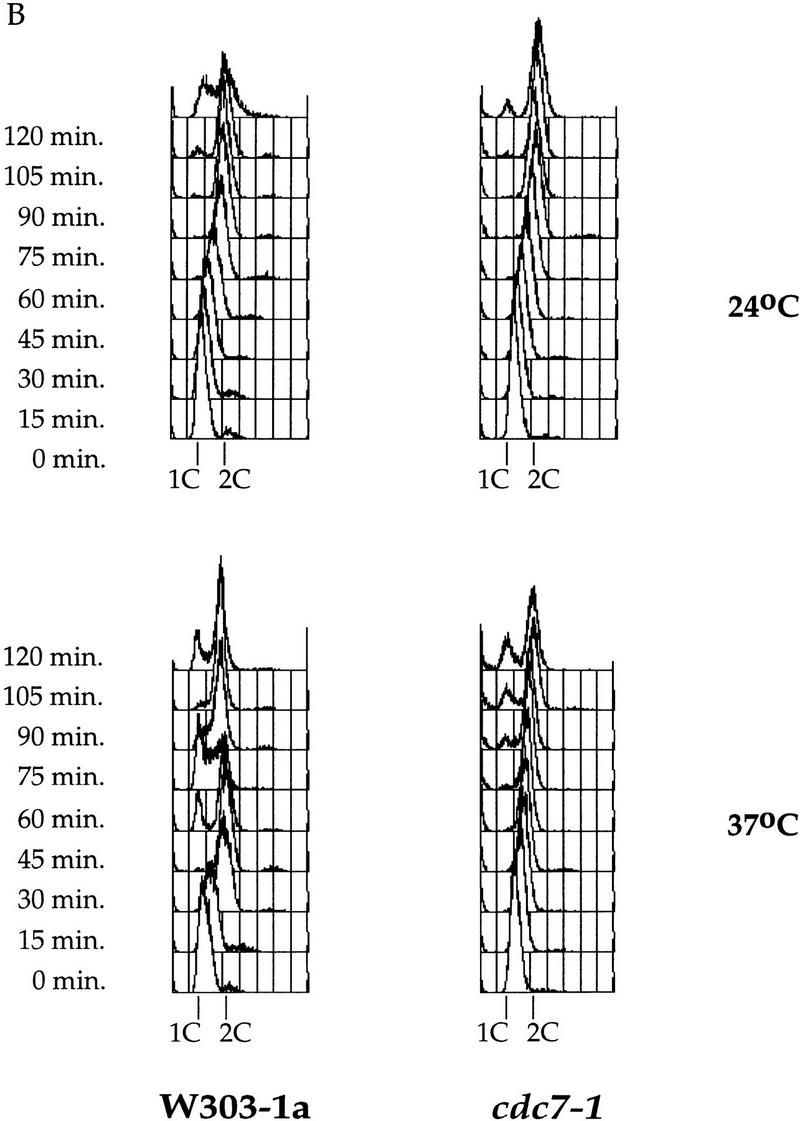
We identified 28°C and 33°C as semipermissive temperatures for two cdc7 mutant alleles, cdc7-1 and cdc7-4, respectively. After arrest at 24°C in G1 with α factor-mating pheromone, cultures were incubated at either the permissive, semipermissive, or restrictive temperature for 30 min and then released from the α factor block at the same temperature. DNA content was monitored by flow cytometry at different times after release. At 24°C, cdc7-4 mutant cells entered and proceeded through S phase like the parental wild-type strain (Fig. 1A). By 60 min after release from the G1 arrest, both wild-type and mutant cultures had reached a 2C DNA content. At 38°C, the wild-type strain proceeded even faster through S phase, reaching a 2C DNA content in 40–60 min, whereas the cdc7-4 mutant arrested with a 1C DNA content. At the semipermissive temperature, the wild-type culture reached a 2C DNA content in ∼40–60 min after release. The cdc7-4 mutant culture, however, required much longer, 140 min, to reach the 2C DNA content at the semipermissive temperature. Figure 1A shows that this delay is not caused by a delay in S-phase entry, but rather, is the result of slow progression through S phase.
Figure 1.
cdc7 mutants are defective in S-phase progression, not G1–S transition. (A,B) Flow cytometric analysis of either cdc7-4 (A) or the cdc7-1 (B) temperature-sensitive cells (top) and the parental wild-type strain (bottom). Cells blocked in G1 with α factor at 24°C were shifted to the indicated temperatures for 30 min and released from the block. Samples were collected every 20 min. Positions of 1C and 2C DNA contents are indicated below each histogram.
At 24°C, cdc7-1 mutant cells already showed defects in cell growth (not shown) and S-phase progression, requiring 80–100 min to accumulate a 2C DNA content compared with 60 min for the parental wild-type strain (Fig. 1B). At 28°C, this effect was enhanced and the cdc7-1 mutant cells required 120 min to accumulate a 2C DNA content. Again, as with the cdc7-4 mutant, the delay was in S-phase progression rather than S-phase entry.
Thus, for both cdc7 mutant alleles, semipermissive conditions did not result in transient accumulation of cells with 1C DNA content, but instead, resulted in slow progression through S phase. These results are not consistent with the hypothesis that Cdc7 is only required for the G1–S transition. Instead, these results indicate that Cdc7 is required for some aspect of S-phase progression.
Cdc7 is important but not essential after a HU block
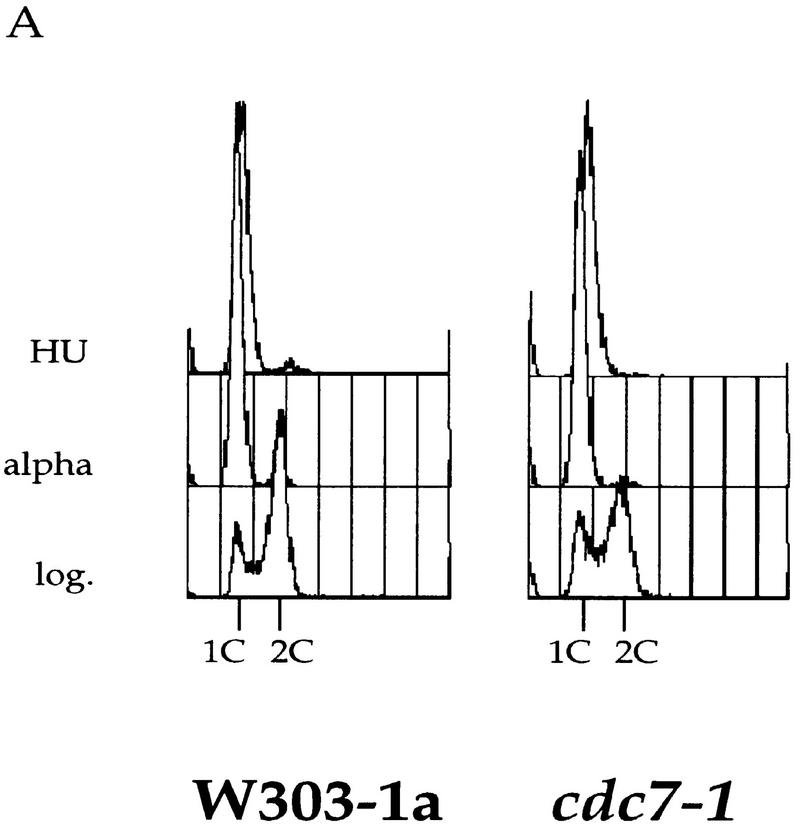
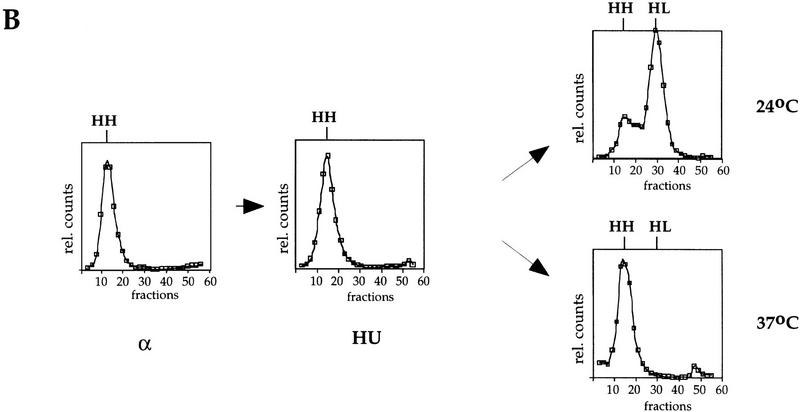
The results described in the previous section are consistent with two possibilities: Either Cdc7 acts throughout S phase to promote S-phase progression, or Cdc7 acts at the G1–S transition, and this action is subsequently manifested during S-phase progression. To begin to distinguish between these two possibilities, we set out to determine whether Cdc7 executes any function in S-phase progression after the G1–S transition. HU is an inhibitor of ribonucleotide reductase that causes replication forks to stall, presumably because of depletion of deoxynucleoside triphosphates. If cells are first synchronized in G1 with α factor and then released into HU, they arrest with a near 1C DNA content (Fig. 2A). By use of genomic footprinting, we have found that under these conditions, early origins are converted to the postreplicative state, suggesting that they have fired, whereas late origins remain for long periods of time in the prereplicative state suggesting that they do not fire (C. Santocanale and J. Diffley, unpubl.). Therefore, we tested whether Cdc7 was required after this point for completion of S phase. Wild-type or cdc7-1 mutant cells were first synchronized in G1 with α factor at 24°C and released into HU at 24°C. Cells were then released from the HU block at 24°C or 37°C and the rate of S-phase progression was measured by FACS analysis. As shown in Figure 2B, both the wild type and mutant completed S phase with similar kinetics at 24°C. At 37°C, the parental wild-type strain accumulated a 2C DNA content within ∼45 min after release from HU. By 60 min after release, a significant proportion of cells completed the cell cycle and entered the next G1 phase. In contrast, the cdc7 mutant cells were much slower, requiring ∼75 min to complete S phase. By 90 min after release, a fraction of the mutant cells also complete the cell cycle and enter the subsequent G1 phase. Very similar results were obtained for cells containing the cdc7-4 allele (data not shown). These experiments show that although Cdc7 is not essential for completion of the cell cycle after an HU block, the completion of S phase in the cdc7 mutant is substantially slowed relative to a wild-type control. Because some, but not all, replication origins have already fired in the HU block, a simple explanation for these results is that Cdc7 is essential for firing late replication origins during S phase, but replication forks established from early firing replication origins are sufficient to passively replicate the entire genome, albeit more slowly, after the HU block.
Figure 2.
Cdc7 is important, but not essential, after a HU block. (A) Flow cytometric analysis of cdc7-1 cells (right) and the parental wild-type cells (left) is shown. Cells were grown in logarithmic phase (log.), arrested in G1 with α factor (alpha), and then released into HU for 90 min at 24°C. (B) The cells arrested with HU (A) were held in HU at either 24°C (top) or 37°C (bottom) for 30 min to inactivate Cdc7 in the cdc7-1 strain. They were then washed (0 min sample) and released from the HU block at either 24°C or 37°C, respectively. Every 15 min, aliquots were withdrawn to follow the cell-cycle progression.
Cdc7 is required for replication of a late, but not an early, origin-containing plasmid after HU
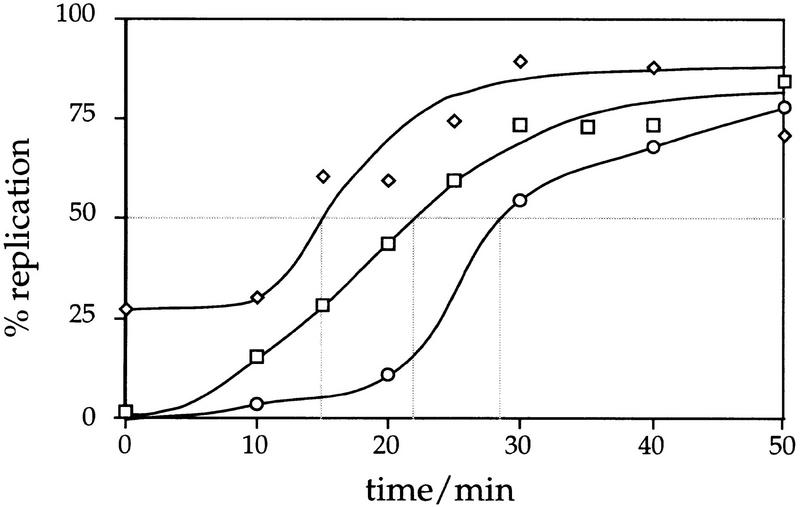
To test the possibility that Cdc7 is required for replication from origins that have not fired previously in the HU block, we investigated whether the replication of plasmids containing either an early or a late origin of replication depends on Cdc7 activity after release from the HU block. By use of plasmids with a single origin of replication, we could avoid the complication of passive replication from origins that had fired before the HU block. Previous analysis has shown that whereas early firing origins like ARS305 have returned to the postreplicative state in the HU block, ARS301 in a plasmid context remains prereplicative for long periods of time (C. Santocanale and J. Diffley, unpubl.). The basis of this observation might be that early origins fire before the HU block is established, and thereby change to a postreplicative state, whereas later origins stay prereplicative because they are prevented from firing by an unknown mechanism. To test whether ARS301 on a plasmid is a late-firing origin of replication in an undisturbed S phase, we compared the replication time of an ARS301 plasmid (pCS1) with the early replicating ARS305 on chromosome III and the late-replicating sequence R11 on chromosome V (McCarroll and Fangman 1988; Ferguson et al. 1991b; Friedman et al. 1996) after release from a cdc7 block as described in Materials and Methods. In this experiment, a small portion of ARS305 is already replicated in the cdc7 block as reported previously (Fig. 3; Reynolds et al. 1989). We calculated replication times for ARS305 and R11 as 14.7 and 28.5 min, respectively, similar to those reported previously by Friedman and coworkers of 14.7 and 33.0 min for ARS305 and R11, respectively (Friedman et al. 1996).
Figure 3.
A plasmid containing ARS301 as sole origin of replication replicates late in S phase. Replication of ARS301 on pCS1 was followed through S phase in comparison with the early-firing origin ARS305 on chromosome III and the late-replicating sequence R11 on chromosome V by density transfer as described in Materials and Methods. Percentage of replication of pCS1 (□), ARS305 (⋄), and R11 (○) is plotted against the time after release from a cdc7 block. Dotted (shaded) lines indicate the replication times trep for the three sequences of interest. See Table 1 for the times determined. trep is defined as the time when half of a specific sequence has replicated (i.e., shifted to the HL peak).
The plasmid containing ARS301 (pCS1) replicates 7.2 min later than ARS305 with a replicating time of 21.9 min (Fig. 3; Table 1). This is 6.6 min earlier than R11. Also, the late origin ARS501, at which replication of R11 is initiated, replicates several minutes before R11 (Ferguson et al. 1991b; Ferguson and Fangman 1992). The replication time of ARS301 (plasmid) in relation to that of the early and late markers ARS305 and R11, respectively, is reflected in its replication index (Friedman et al. 1996) of 0.52 (Table 1). In comparison, the relative early firing origin ARS1 has a replication index of 0.27 and the late-replicating origins ARS1413 and ARS1412 in a plasmid context have indices of 0.57 and 0.72, respectively (Friedman et al. 1996). ARS301 on pCS1 fires after early to mid–early origins like ARS305 and ARS1 and can, therefore, be considered as a later firing origin of replication. Although ARS301 has been shown to be inactive in its chromosomal location (Dubey et al. 1991) it fires efficiently on a plasmid (Fig. 3; Santocanale and Diffley 1996).
Table 1.
Plasmid-borne ARS301 replicates after ARS305 during S phase
| Sequence
|
Location
|
trep/mina
|
Replication indexb
|
|---|---|---|---|
| ARS305 | chromosome III | 14.7 | 0 |
| R11 | chromosome V | 28.5 | 1 |
| ARS301 | plasmid pCS1 | 21.9 | 0.52 |
Replication times (trep) and replicative indices of the early chromosomal origin ARS305, the late replicating sequence R11 on chromosome V, and ARS301 on a plasmid (pCS1) as determined in Fig. 3.
trep is defined as the time when 50% of a specific sequence is replicated.
The replicative index for ARS305 and R11 is defined as 0 and 1, respectively (Friedman et al. 1996). The replication index of ARS301 (plasmid) = trep(ARS301) − trep(ARS305)/trep(R11) − trep(ARS305).
Therefore, we compared the ARS301 containing plasmid pCS1 with a plasmid containing the early replicating ARS305 (p305.2) for their dependence on Cdc7 activity to replicate after release from a HU block.
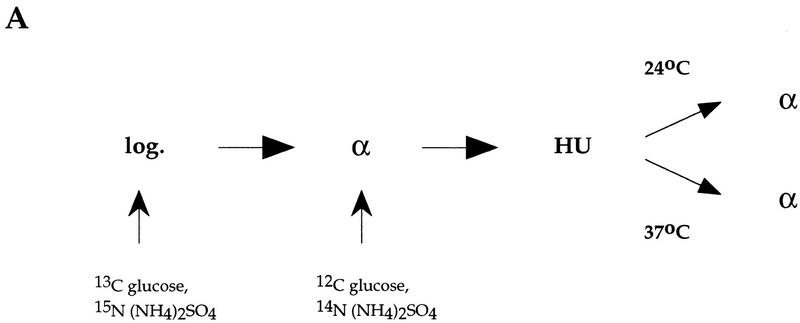
The experimental procedure is outlined in Figure 4A. Cells containing the cdc7-1 mutation and a plasmid harboring either ARS305 (Fig. 4) or ARS301 (Fig. 5) were grown for more than seven generations in heavy medium [containing 13C glucose and 15N (NH4)2SO4] to fully substitute the DNA with dense isotopes. After G1 arrest at 24°C with α factor, cells were transferred to light medium [containing [12C]glucose and 14N (NH4)2SO4] and held in α factor for 30 min to allow accumulation of pools of light nucleotides. Cells were released into HU at 24°C as in Figure 2 and then released from the HU block at either 24°C or 37°C. To prevent entry into the next S phase, α factor was added to the medium. DNA was digested with restriction enzymes and separated on CsCl gradients. The positions of individual sequences within the gradient were determined by DNA blot hybridization with specific DNA probes. Replication is seen as transfer of specific sequences from the heavy–heavy (HH) peak in which both DNA strands are substituted with dense isotope to the heavy–light (HL) peak in which only the parental strand is substituted with dense isotope.
Figure 4.
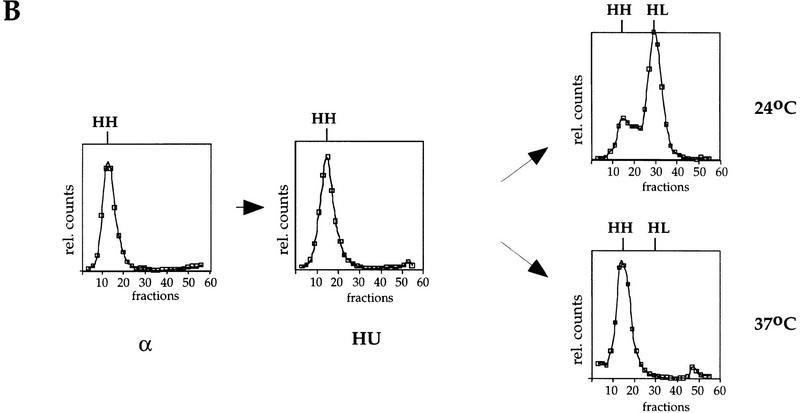
Cdc7 is not required for replication of a plasmid containing the early replication origin ARS305 after HU. (A) Outline of the experimental procedure (see text for details). (B) RM14-3a cells (cdc7-1) containing an ARS305 plasmid (p305.2) were treated as illustrated in A. Cells were grown in heavy medium and arrested with α factor (α). They were subsequently released into HU in light medium (HU), and then released from HU at either 24°C or 37°C for 3 hr as described in Materials and Methods. DNA was isolated and analyzed for replication. Replication of ARS305 on the plasmid was analyzed by DNA blot hybridization with a plasmid-specific probe (see Materials and Methods). The relative amounts of radioactivity are plotted against the fraction numbers. Location of HH and HL DNA are indicated. Additionally, the HH peak as it appears in the G1 block is superimposed onto the other plots as a shaded line.
Figure 5.
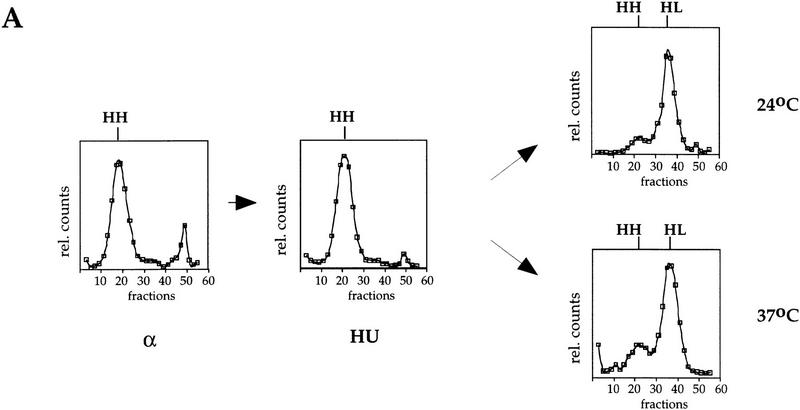
Cdc7 is required for replication of a plasmid containing ARS301 after HU. RM14-3a cells (cdc7-1) containing the ARS301 plasmid (pCS1) were treated and their DNA was analyzed as described for Fig. 4. (A) Replication of a late-replicating sequence on chromosome V was analyzed by DNA blot hybridization with the R11 probe. (B) Replication of a plasmid containing ARS301 as the sole origin was analyzed by probing the same fractions with a probe specific for the vector (see Materials and Methods). The relative amounts of radioactivity is plotted against the fraction number. Location of HH and HL DNAs are indicated.
We first examined a plasmid containing the early firing ARS305 (p305.2). As shown in Figure 4B, p305.2 DNA is in the HH peak in α factor. After release into HU, the DNA shifts slightly in the direction of the HL peak consistent with the interpretation that ARS305 has already fired in HU, and that replication of the plasmid is incomplete because of the elongation block. After release from the HU block at either 24°C or 37°C, p305.2 DNA is fully transferred to the HL peak indicating that replication of this plasmid containing an early firing origin is independent of Cdc7 activity.
We then investigated the late-replicating plasmid containing ARS301 (pCS1) under the same conditions. As a control for this experiment, we first examined the late replicating region of chromosome V (R11), which normally replicates after pCS1 (Fig. 3). As shown in Figure 5A, R11 DNA is in the HH peak in α factor and remains HH after release into HU. After release from the HU block at either 24°C or 37°C, R11 is efficiently replicated as indicated by the shift from HH to HL. This is consistent with our flow cytometric analysis (Fig. 2) showing that the entire genome replicates after release from HU at the cdc7 restrictive temperature.
The ARS301 plasmid (Fig. 5B) is also HH in both the α factor block and after release into HU. Like R11, the ARS301 plasmid replicates efficiently after release from the HU block at the cdc7 permissive temperature. In contrast to R11, however, the ARS301 plasmid does not replicate after release from the HU block at the restrictive temperature. Taken together, these results strongly suggest that whereas Cdc7 is neither required to complete replication of the genome nor for replicating a plasmid with an early origin of replication after the HU block, late origins cannot initiate replication after release from the HU block in the absence of Cdc7 function.
Cdc7 is required to convert late origins to the postreplicative state after HU
The fact that Cdc7 is required to replicate the ARS301-containing plasmid after the HU block but is not essential for the completion of genomic replication strongly suggests that Cdc7 is essential for the firing of late origins after the HU block and that, in the absence of late origin firing, the genome can be replicated passively from origins that have fired before HU. Assays such as density substitution and two-dimensional gels provide a somewhat indirect measurement of origin firing because they require the accumulation of replication products and intermediates, respectively. Genomic footprinting provides a potentially more direct assay for origin-specific events. Therefore, we sought to examine the role of Cdc7 in converting origins from the prereplicative to the postreplicative state. cdc7-1 mutant cells harboring the ARS301-containing plasmid were arrested in α factor and HU at 24°C as described above and then released into nocodazole at either 24°C or 37°C. The state of ARS301 was examined by genomic footprinting. ORC binding to ARS301 induces two DNase I hypersensitive sites (asterisks in Fig. 6) adjacent to the ARS consensus sequence (ACS) that can be seen in vitro with purified ORC (Bell et al. 1993) as well as in vivo in G2 blocked cells (Fig. 6, lanes 3,4; Santocanale and Diffley 1996). The pre-RC at ARS301 is primarily seen as the Cdc6-dependent suppression of these two hypersensitive sites (Fig. 6, lanes 5,6; Santocanale and Diffley 1996). After release from the G1 block into HU, ARS301 remains prereplicative for long periods of time (Fig. 6, lanes 7,8; C. Santocanale and J. Diffley, unpubl.). Under these conditions, an early firing origin, ARS1, was converted to the postreplicative state (data not shown). After release from the HU block at the permissive temperature 24°C, ARS301 is converted to the postreplicative state within 1 hr (Fig. 6, lanes 9,10), consistent with the fact that the origin is activated under these conditions (Fig. 5B). Figure 6, lanes 13–16, shows that after release from the HU block at the restrictive temperature, 37°C, ARS301 remains in the prereplicative state consistent with the fact that it doesn’t replicate. Results from this experiment have been quantified by densitometric scanning of the autoradiogram and are presented in Figure 6B. This experiment shows that Cdc7 is not only required for the replication of the ARS301-containing plasmid after the HU block, but is also required for conversion of this origin to the postreplicative state.
Figure 6.
Cdc7 is required to convert plasmid-borne ARS301 to the postreplicative state after HU. (A) cdc7-1 cells (2032) containing the ARS301 plasmid (pCS1) were arrested in G2/M by nocodazole (lanes 3,4) or in G1 by α factor (lanes 5,6). G1-arrested cells were released into HU (lanes 7,8). The culture was split in two. Each half was held in HU and incubated at either 24°C or 37°C for 30 min before release into nocodazole at these temperatures. Samples were taken for genomic footprinting analysis 1 hr and 3 hr after release from HU into nocodazole (lanes 9–16). The primer (JD63b) is specific for the plasmid borne ARS301. Lanes 1 and 2 show primer extensions performed on naked DNA. Exposure levels were adjusted because plasmid recovery was lower from G1 and S-phase cells and on release from HU at 37°C. (B) Densitometric quantification of Fig. 4A, lanes 4, 6, 8, 10, 12, 14, and 16. Levels were adjusted to a background band marked by an arrowhead. The ORC-induced hypersensitive sites of the post-RC are indicated by asterisks. The functional ARS element is marked by an open box. The position of the ACS is indicated.
Discussion
Cdc7 is required during S phase for origin firing
The results presented in this paper provide four lines of evidence indicating that Cdc7 is required for the firing of replication origins during S phase. (1) cdc7 mutants do not show a defect in the G1–S transition at semipermissive temperatures, but, instead, show a substantial reduction in the rate of S-phase progression consistent with replication from a reduced number of origins. (2) cdc7 mutants complete S phase at a reduced rate after release from an HU block at the restrictive temperature indicating that inactivation of Cdc7 during S phase prevents the timely replication of the genome. (3) After release from a HU block, cdc7 mutants complete S phase at the restrictive temperature and a plasmid with an early firing origin of replication (ARS305), which had already fired in HU, completes replication indicating that Cdc7 is not essential for the elongation phase of DNA replication. In contrast, a plasmid containing a later firing origin (ARS301) shows no evidence of replication under these conditions, consistent with the idea that Cdc7 is required for firing this origin after release from HU. (4) Finally, Cdc7 is required to convert late origins from the prereplicative to the postreplicative state after the HU block arguing that it acts directly on replication origins.
In Donaldson et al. (1998) very similar conclusions are drawn, using a different approach. They found that a short pulse of Cdc7 kinase at the beginning of S phase was sufficient to activate early- but not late-firing replication origins. This is unlikely to be caused by a greater requirement for Cdc7 by late-firing origins because the loss rates of plasmids containing both early and late origins were affected similarly at semipermissive temperatures. Taken together, our results argue strongly that Cdc7 is required during S phase for origin firing.
Previous experiments have indicated that Cdc7 is not required for completion of the cell cycle after an HU block (Hereford and Hartwell 1974; Hartwell 1976). The results presented in this paper are essentially in agreement with this result. We find a significant fraction of cells complete the cell cycle and enter G1 after release of the cdc7-1 mutant at the restrictive temperature. Thus, an important corollary that emerges from our experiments is that origins that fire after the HU block must not be essential to complete replication of the genome. Previous analysis by Newlon and co-workers has shown that most origins from a derivative of chromosome III are dispensable for efficient chromosome maintenance (Newlon et al. 1993) arguing that budding yeast chromosomes contain an excess of replication origins. Our results are consistent with this interpretation. A second important corollary from our work is that firing of new origins is not required to resume DNA synthesis after the HU block. Presumably then, replication forks from origins that had already fired in the HU block are competent to resume replication after release from the block.
The Cdc7p-associated protein kinase activity is relatively high in cells blocked early in S1 with HU (Jackson et al. 1993). Our results show that Cdc7 is still required after release from such a block. Therefore, the active Cdc7 kinase in HU blocked cells is, in some way, unable to execute its function. We have found that the inability of late origins to fire in HU-arrested cells is an active process that requires the Rad53p checkpoint protein (C. Santocanale and J. Diffley, data not shown). Therefore, either directly or indirectly, the Rad53 checkpoint pathway is responsible for preventing the active Cdc7 protein kinase from firing these late replication origins.
We cannot rule out the possibility that the requirement for Cdc7 after the HU block is a nonessential checkpoint-induced adaptation to aid recovery from the drug. The defect in S-phase progression at cdc7 semipermissive temperatures, however, together with the results of Donaldson et al. (1998) and the fact that Dbf4 interacts with origins in vivo suggest that it is more likely that Cdc7 acts throughout S phase to activate individual origins.
What is the nature of the G1–S transition?
Because Cdc7 is required during S phase, something upstream of Cdc7 must either activate Cdc7 in the first place or make origins competent to be activated by Cdc7. Such an activity might be more properly thought of as catalyzing the G1–S transition. Downstream of Start, two protein kinases must be activated for entry into S phase: Cdc28 and Cdc7. Both proteins are activated by subunits (Clb5,6 and Dbf4, respectively) that are unstable proteins and are synthesized late in G1. Although it is unclear at the moment how these two kinases act relative to each other (discussed in Diffley 1995), preliminary experiments indicate that, like Cdc7, Cdc28–Clb kinase is also required to complete S phase after a HU block (C. Santocanale, K. Bousset, and J. Diffley, unpubl.). Thus, activities involved in global regulation of the G1–S transition must lie upstream of both Cdc7 and Cdc28. We suggest that the G1–S transition may not be a simple switch, but, instead, occurs with the convergence of several pathways that are activated at Start. These include (1) targeting the Clb–Cdc28 inhibitor Sic1 and the Cdc6 protein for ubiquitin-mediated degradation via the Cdc4–Cdc34–Cdc53 pathway, (2) inactivation of the anaphase-promoting complex to allow subsequent accumulation of Clbs, and (3) activation of the late G1 transcription pathway leading to synthesis of Clb5,6 and Dbf4.
Materials and methods
Strains and plasmids
Yeast strains used in this study were RM14-3a (MATa his6 trp1-289 leu2-3,112 ura3-52 bar1 cdc7-1), W303-1a (MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100), and their derivatives. The strain 2032 is a W303-1a derivative containing the cdc7-1 mutation and was provided by K. Nasmyth (Amon et al. 1992). YKB2 (W303-1a cdc7-4) was constructed as follows: The entire cdc7-4-coding region was amplified from a cdc7-4 mutant congenic to A364A (MATa ura1 ade1 ade2 tyr1 his7 lys2 gal1-1) by PCR with the primers 5′-TAGAGAATTCATAATGACAAGCAAAAC and 5′-GAAATTTTTATTGAATTCTTTCACTATGC (EcoRI sites introduced for subsequent cloning are underlined) and was cloned into pRS306 (Sikorski and Hieter 1989). Integration was directed to the CDC7 locus by digestion of the plasmid with BsrGI before transformation into W303-1a. This results in a strain with a duplication of the CDC7 gene, one wild type and one cdc7-4, separated by the URA3-containing vector. Recombination between the two CDC7 genes could then be selected for by isolating uracil auxotrophs. Colonies were grown on YEPD for several generations and uracil auxotrophs were selected on 5-fluoroorotic acid. KB2 is a temperature-sensitive strain that arose from this selection. The temperature-sensitive phenotype of KB2 was fully complemented by a centromeric plasmid containing a single copy of the CDC7 gene.
The ARS301-containing plasmid pCS1 was described before (Santocanale and Diffley 1996). The plasmid p305.2 was provided by Anne Donaldson. It consists of a 17,216-bp fragment of chromosome III containing ARS305 as sole origin of replication in a YIp5-5 backbone (Ferguson et al. 1991b).
Flow cytometric analysis
For flow cytometric analysis, cells were grown in YPD. Samples were collected and processed as described before (Santocanale et al. 1995) and analyzed with a Becton-Dickinson FACScan.
Dense isotope substitution experiments
Dense isotope substitution experiments were performed essentially as described earlier (McCarroll and Fangman 1988) with the strain RM14-3a. For the HU release experiments, cells were grown at 24°C for more than seven generations in heavy minimal medium containing 0.1% 13C glucose and 0.01% 15N (NH4)2SO4 (CK Gas Products Ltd, UK) as carbon and nitrogen source, respectively. They were arrested in G1 by α factor (200 nm). After the block was complete (∼3 hr, 15 min) they were transferred to minimal medium [2% 12C glucose, 0.01% 15N (NH4)2SO4] and kept in α factor for 30 min. They were then washed and released into minimal medium containing 0.2 m HU and Pronase (0.01–0.05 mg/ml). After >90% of the cells had medium sized buds (100–110 min), the culture was split into two and incubated in minimal medium containing 0.2 m HU for 30 min at either 37°C or 24°C. Cells were then washed and released from the block for 3 hr at either 37°C or 24°C, respectively. α factor was added to prevent entry into the next S phase.
For the replication timing experiment, cells were grown in heavy medium and blocked with α factor as described above. They were transferred to 37°C prewarmed light medium, and kept in α factor for 40 min. Pronase was then added to release the cells into the cdc7 block. The 0 min time point was taken before the flask was chilled to 24°C in ice water, and further incubated at 24°C. Samples were withdrawn for analysis after the indicated time points.
For density shift analysis, 3.2 × 107 cells were collected on frozen EDTA (0.2 m) sodium azide (0.1%), vortexed until the ice broke, centrifuged, washed with cold water, and the pellet was frozen on dry ice/ethanol. Cells were lysed by bead beating and DNA was precipitated after phenol extraction. The DNA was treated with RNase A, digested with EcoRI, and separated on CsCl gradients (McCarroll and Fangman 1988). The gradients were fractionated and every second fraction of a refractive index between 1.4065 and 1.403 was blotted onto Hybond N+ membranes (Amersham) and hybridized with specific probes (Church and Gilbert 1984). Probes were labeled with a [α32P]dCTP with a Boehringer DNA-labeling kit. For quantification, a Molecular Dynamics PhosphorImager was used.
The specific probes recognizing a late-replicating sequence of chromosome V (R11) and the early origin ARS305 on Chromosome III (305), respectively, were described before (Ferguson et al. 1991b). The 1913 base pair PvuI fragment of pRS306 was used as a probe for the plasmid pCS1. As a specific probe for the plasmid p305.2, EcoRI-cleaved pBR322 was used. To determine replication times, the areas under the HH and HL peak were quantified in NIH image and percent replication was calculated as 0.5× area(HL)/area(HH) + 0.5× area (HL). The replication time of a specific sequence is defined as time after the cdc7 block when half of the specific DNA in a culture is replicated (McCarroll and Fangman 1988).
Genomic footprinting
For genomic footprinting, cells were grown for at least one generation in YPD before arresting cells in either G2/M or G1 by incubating in nocodazole (5 μg/ml) or α factor (2 μg/ml) for ∼2 hr and 30 min. The release from G1 into 0.2 m HU was done for 90–100 min until >90% of the cells had medium-sized buds. Cells were then treated as for the density transfer experiment, except that YPD was used and nocodazole (5 μg/ml) was added to release into G2/M. Cell lysis for genomic footprinting and primer extension was performed as described before (Santocanale and Diffley 1996). JD63b (Rowley et al. 1995) is a primer specific for the plasmid.
Acknowledgments
We thank Anne Donaldson, Walt Fangman, and Bonny Brewer for communicating results before publication; we thank M.K. Raghuraman and Corrado Santocanale for many helpful discussions and Miguel Godinho Ferreira for help with quantifying the timing experiment. K.B. was supported by a fellowship of the Deutsche Forschungsgemeinschaft.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL J.Diffley@icrf.icnet.uk; FAX +44-171-269-3801.
References
- Amon A, Surana U, Muroff I, Nasmyth K. Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae. Nature. 1992;355:368–371. doi: 10.1038/355368a0. [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM complexes and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Bahman M, Buck V, White A, Rosamond J. Characterisation of the CDC7 gene product of Saccharomyces cerevisiae as a protein kinase needed for the initiation of mitotic DNA synthesis. Biochim Biophys Acta. 1988;951:335–343. doi: 10.1016/0167-4781(88)90104-2. [DOI] [PubMed] [Google Scholar]
- Bell SP, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Diller JD, Friedman KL, Kolor KM, Raghuraman MK, Fangman WL. The topography of chromosome replication in yeast. Cold Spring Harbor Symp Quant Biol. 1993;58:425–442. doi: 10.1101/sqb.1993.058.01.049. [DOI] [PubMed] [Google Scholar]
- Buck V, White A, Rosamond J. CDC7 protein kinase activity is required for mitosis and meiosis in Saccharomyces cerevisiae. Mol & Gen Genet. 1991;227:452–457. doi: 10.1007/BF00273937. [DOI] [PubMed] [Google Scholar]
- Carpenter PB, Mueller PR, Dunphy WG. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature. 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- Chong JPJ, Mahbubani HM, Khoo C-Y, Blow JJ. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature. 1995;375:418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JFX. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Diffley JFX, Nasmyth KA. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- Diffley JFX. The initiation of DNA replication in the budding yeast cell division cycle. Yeast. 1995;11:1651–1670. doi: 10.1002/yea.320111608. [DOI] [PubMed] [Google Scholar]
- ————— Once and only once upon a time: Specifying and regulating origins of DNA replication in eukaryotic cells. Genes & Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- Diffley JFX, Cocker JH. Protein-DNA interactions at a yeast replication origin. Nature. 1992;357:169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- Diffley JFX, Cocker JH, Dowell SJ, Rowley A. two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Dixon WJ, Campbell JL. Preparation of active cdc7/dbf4 kinase from yeast-cells. Methods Enzymol. 1997;283:390–397. doi: 10.1016/s0076-6879(97)83032-7. [DOI] [PubMed] [Google Scholar]
- Donaldson, A.D., W.L. Fangman, and B.J. Brewer. 1998. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Donovan S, Harwood J, Drury LS, Diffley JFX. Cdc6-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell SJ, Romanowski P, Diffley JFX. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science. 1994;265:1243–1246. doi: 10.1126/science.8066465. [DOI] [PubMed] [Google Scholar]
- Dubey DD, Davis LR, Greenfeder SA, Ong LY, Zhu JG, Broach JR, Newlon CS, Huberman JA. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol Cell Biol. 1991;11:5346–5355. doi: 10.1128/mcb.11.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman WL, Brewer BJ. A question of time—replication origins of eukaryotic chromosomes. Cell. 1992;71:363–366. doi: 10.1016/0092-8674(92)90505-7. [DOI] [PubMed] [Google Scholar]
- Ferguson BM, Brewer BJ, Fangman WL. Temporal control of DNA replication in yeast. Cold Spring Harbor Symp Quant Biol. 1991a;56:293–302. doi: 10.1101/sqb.1991.056.01.036. [DOI] [PubMed] [Google Scholar]
- Ferguson BM, Brewer BJ, Reynolds AE, Fangman WL. A yeast origin of replication is activated late in S phase. Cell. 1991b;65:507–515. doi: 10.1016/0092-8674(91)90468-e. [DOI] [PubMed] [Google Scholar]
- Ferguson BM, Fangman WL. A position effect on the time of replication origin activation in yeast. Cell. 1992;68:333–339. doi: 10.1016/0092-8674(92)90474-q. [DOI] [PubMed] [Google Scholar]
- Fox CA, Loo S, Dillin A, Rine J. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes & Dev. 1995;9:911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]
- Friedman KL, Diller JD, Ferguson BM, Nyland SV, Brewer BJ, Fangman WL. Multiple determinants controlling activation of yeast replication origins late in S phase. Genes & Dev. 1996;10:1595–1607. doi: 10.1101/gad.10.13.1595. [DOI] [PubMed] [Google Scholar]
- Hardy C. Characterization of an essential Orc2p-associated factor that plays a role in DNA replication. Mol Cell Biol. 1996;16:1832–1841. doi: 10.1128/mcb.16.4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CF, Dryga O, Seematter S, Pahl PM, Sclafani RA. Mcm5/Cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH. Saccharomyces cerevisiae cell cycle. Bacteriol Rev. 1974;38:164–198. doi: 10.1128/br.38.2.164-198.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol. 1976;104:803–817. doi: 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- Hereford LM, Hartwell LH. Sequential gene function in the initiation of Saccharomyces cerevisiae DNA synthesis. J Mol Biol. 1974;84:445–461. doi: 10.1016/0022-2836(74)90451-3. [DOI] [PubMed] [Google Scholar]
- Hollingsworth RE, Sclafani R. DNA metabolism gene CDC7 from yeast encodes a serine (threonine) protein kinase. Proc Natl Acad Sci. 1990;87:6272–6276. doi: 10.1073/pnas.87.16.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Pahl PM, Harrison K, Rosamond J, Sclafani RA. Cell cycle regulation of the yeast CDC7 protein kinase by association with the DBF4 protein. Mol Cell Biol. 1993;13:2899–2908. doi: 10.1128/mcb.13.5.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LH, Thomas AP. A further two mutants defective in initiation of the S phase in the yeast Saccharomyces cerevisiae. Mol & Gen Genet. 1982;186:445–448. doi: 10.1007/BF00729467. [DOI] [PubMed] [Google Scholar]
- Kitada K, Johnson AL, Johnston LH, Sugino A. A multicopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is identified as CDC5. Mol Cell Biol. 1993;13:4445–4457. doi: 10.1128/mcb.13.7.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S-I, Takisawa H, Nojima H. Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell. 1995;81:601–609. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- Lei M, Kawasaki Y, Young MR, Kihara M, Sugino A, Tye BK. Mcm2 is target of regulation by Cdc7-Dbf4 during the initiation DNA synthesis. Genes & Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo S, Fox CA, Rine J, Kobayashi R, Stillman B, Bell S. The origin recognition complex in silencing, cell-cycle progression, and DNA-replication. Mol Biol Cell. 1995;6:741–756. doi: 10.1091/mbc.6.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madine MA, Khoo C-Y, Mills AD, Laskey RA. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature. 1995a;375:421–424. doi: 10.1038/375421a0. [DOI] [PubMed] [Google Scholar]
- Madine MA, Khoo C-Y, Mills AD, Mushal C, Laskey RA. The nuclear envelope prevents reinitiation of replication by regulating the binding of MCM3 to chromatin in Xenopus egg extracts. Curr Biol. 1995b;5:1270–1279. doi: 10.1016/s0960-9822(95)00253-3. [DOI] [PubMed] [Google Scholar]
- Masai H, Miyake T, Arai K-I. hsk1+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J. 1995;14:3094–3104. doi: 10.1002/j.1460-2075.1995.tb07312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll RM, Fangman WL. Time of replication of yeast centromeres and telomeres. Cell. 1988;54:505–513. doi: 10.1016/0092-8674(88)90072-4. [DOI] [PubMed] [Google Scholar]
- Muzi-Falconi M, Brown GW, Kelly TJ. Controlling initiation during the cell cycle. DNA replication. Curr Biol. 1996;6:229–233. doi: 10.1016/s0960-9822(02)00464-5. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Viewpoint: Putting the cell cycle in order. Science. 1996;274:1643–1645. doi: 10.1126/science.274.5293.1643. [DOI] [PubMed] [Google Scholar]
- Newlon CS, Collins I, Dershowitz A, Deshpande AM, Greenfeder SA, Ong LY, Theis JF. Analysis of replication origin function on chromosome III of Saccharomyces cerevisiae. Cold Spring Harbor Symp Quant Biol. 1993;58:415–423. doi: 10.1101/sqb.1993.058.01.048. [DOI] [PubMed] [Google Scholar]
- Patterson M, Sclafani RA, Fangman WL, Rosamond J. Molecular characterisation of the cell cycle gene CDC7 from Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:1590–1598. doi: 10.1128/mcb.6.5.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Bohm T, Cocker JH, Diffley JFX, Nasmyth K. Activation of S-phase promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes & Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- Raghuraman MK, Brewer BJ, Fangman WL. Cell cycle-dependent establishment of a late replication program. Science. 1997;276:806–809. doi: 10.1126/science.276.5313.806. [DOI] [PubMed] [Google Scholar]
- Reynolds AE, McCarroll RM, Newlon CS, Fangman WL. Time of replication of ARS elements along yeast chromosome III. Mol Cell Biol. 1989;9:4488–4494. doi: 10.1128/mcb.9.10.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA-replication and mcm binding to chromatin. Curr Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- Rowles A, Chong JPJ, Brown L, Howell M, Evan GI, Blow JJ. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- Rowley A, Cocker JH, Harwood J, Diffley JFX. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J. 1995;14:2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JFX. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J. 1996;15:6671–6679. [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Neecke H, Longhese MP, Lucchini G, Plevani P. Mutations in the gene encoding the 34 kDa subunit of yeast replication protein A cause defective S phase progression. J Mol Biol. 1995;254:595–607. doi: 10.1006/jmbi.1995.0641. [DOI] [PubMed] [Google Scholar]
- Sato N, Arai K, Masai H. Human and Xenopus cDNAs encoding budding yeast cdc7-related kinases–in-vitro phosphorylation of mcm subunits by a putative human homolog of cdc7. EMBO J. 1997;16:4340–4351. doi: 10.1093/emboj/16.14.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E, Bohm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon NA, Wright MB, Chang S, Buckley AM, Dumas LB, Gaber RF. Genetic and molecular analysis of DNA43 and DNA52: Two new cell-cycle genes in Saccharomyces cerevisiae. Yeast. 1992;8:273–289. doi: 10.1002/yea.320080405. [DOI] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA-replication origins is regulated by Cdc6p and Cdk. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Wuarin J, Nurse P. Regulating S phase: CDKs, licensing and proteolysis. Cell. 1996;85:785–787. doi: 10.1016/s0092-8674(00)81261-1. [DOI] [PubMed] [Google Scholar]