Abstract
The chromosomes of eukaryotes are organized into structurally and functionally discrete domains that provide a mechanism to compact the DNA as well as delineate independent units of gene activity. It is believed that insulator/boundary elements separate these domains. Here we report the identification and characterization of boundary elements that flank the transcriptionally repressed HMR locus in the yeast Saccharomyces cerevisiae. Deletion of these boundary elements led to the spread of silenced chromatin, whereas the ectopic insertion of these elements between a silencer and a promoter blocked the repressive effects of the silencer on that promoter at HMR and at telomeres. Sequence analysis indicated that the boundary element contained a TY1 LTR, and a tRNA gene and mutational analysis has implicated the Smc proteins, which encode structural components of chromosomes, in boundary element function.
Keywords: Heterochromatin, silencing, HMR, boundary elements, S. cerevisiae
In eukaryotic cells the nuclear DNA is packaged into chromatin, which compacts the length of the chromosomal DNA molecule into a substantially more condensed form. This compaction involves a hierarchical level of organization in which the first and second levels of packaging involve the formation of nucleosomes and then the 30-nm fiber, respectively (Widom 1998). The third and higher levels of compaction possibly involve the formation of topologically constrained loop domains (Saitoh and Laemmli 1993).
Cytological and molecular studies of chromosomes suggest that the domains may be due to the formation of chromosome loops attached to a proteinaceous chromosome scaffold (Gasser and Laemmli 1987). Genetic studies on position-effect variegation (PEV) in Drosophila indicate that the chromosomes have both transcriptionally active and inactive domains (Elgin 1996). The identification of both structural and functional domains has led to models in which specific DNA elements act as insulators or boundaries, functionally separating one domain from another. Studies in Drosophila and chickens have identified DNA elements that possess insulator function and impart position-independent expression of a transgene when the gene is inserted into either euchromatin or heterochromatin (Kellum and Elgin 1998). These insulator elements function to constrain regulatory elements such as silencers and enhancers. Silencers and enhancers modulate promoter activity in an orientation-, distance-, and gene-promoter-independent manner (Kamakaka 1997). Functionally discrete domains could serve to constrain silencers and enhancers in one domain from adventitious interactions with genes in neighboring domains. Indeed, Drosophila insulator elements block enhancer-promoter interactions when interposed between two such elements (Geyer and Corces 1992; Kellum and Schedl 1992).
The HMR locus is a well-characterized transcriptionally silenced locus in the yeast Saccharomyces cerevisiae. Silencing at this locus is achieved by the concerted action of proteins (Rap1p, Abf1p, Sir1p, and ORC) that bind inactivation centers called silencers as well as proteins (Sir2p, Sir3p, and Sir4p) that interact with nucleosomes in the silent domain (Loo and Rine 1995). DNA sequence elements called silencers flank the silent loci and are necessary for silencing. Two silencers called E and I flank HMR, and each silencer contains binding sites for various proteins. The HMR-E silencer is sufficient on its own for silencing the HMR locus on a chromosome (Brand et al. 1985). One of the roles of the silencers is to recruit the Sir proteins to the silent loci. The recruitment and consequent binding of the Sir proteins to nucleosomes generates a chromatin domain that is inaccessible to various enzymatic probes and is transcriptionally repressed. Differential restriction enzyme accessibility studies demonstrate that the heterochromatic domain at HMR extends beyond the silencers but for a limited distance (Singh and Klar 1992; Loo and Rine 1994). The mechanism that prevents the further spread of heterochromatin into neighboring euchromatin is not known.
We have, therefore, undertaken a study to determine both whether boundary elements exist in S. cerevisiae at the silenced HMR locus and to understand the molecular mechanism of how such elements function.
Results
The silenced domain emanates bidirectionally from the silencers
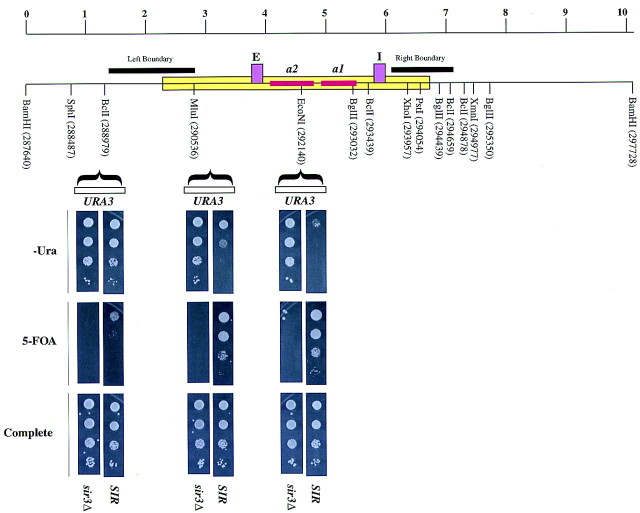
The silent HMR domain (Fig. 1) is refractory to digestion by various restriction endonucleases in wild-type cells but is accessible to these enzymes in sir3 mutants (Loo and Rine 1994) (see Fig. 2B). This inaccessible domain (pink box in Fig. 1) is not limited to the region between the two silencers but extends several hundred base pairs beyond the silencers.
Figure 1.
The silenced domain emanates bi-directionally from the silencers. (A) A schematic representation of the 10-kb BamHI fragment encompassing the HMR locus with the sites of insertion of the URA3 gene shown. All coordinates used in this study are based on the Saccharomyces Genome Database (SGD) coordinates. Strain numbers are shown in parentheses. (B) Three sir3Δ strains, ROY656, ROY834, and ROY836, in which the URA3 gene was inserted at SGD coordinates 292140 (∼640 bp to the right of HMR-E), 290921 (∼475 bp to the left of HMR-E), and 288489 (∼2840 bp to the left of HMR-E), respectively, were generated. Strains ROY648, ROY508, and ROY513 are Sir+ derivatives of ROY656, ROY834, and ROY836, respectively. Cells were grown in liquid media, and 3 μl of 10-fold serial dilutions were spotted on YPD plates (complete), on supplemented YMD plates lacking uracil (−Ura), or on supplemented YMD plates containing 1 mg/ml 5-FOA to assay for URA3 expression.
Figure 2.
(A) The 3.5-kb silenced HMR domain can be expanded to 7 kb. sirΔ strains carrying insertions (at SGD coordinate 293032) of either (1) 1-kb of the TRP1 gene (ROY49 and ROY1075) or (2) 1 kb of the TRP1 gene plus 2.5 kb of the HMG2 coding sequence (ROY55 and ROY1080) were generated. Strains ROY803, ROY84, ROY1076, and ROY 1079 are Sir+ derivatives of strains ROY49, ROY55, ROY1075, and ROY1080, respectively. All cells were grown in liquid media and 3 μl of 10-fold serial dilutions were spotted on either YPD plates (+Trp) or on YMD plates lacking tryptophan (−Trp) to assay for TRP1 expression. Strain numbers are shown in parentheses. (B) Differential restriction endonuclease digestion analysis of the expanded silenced HMR domain. Nuclei isolated from wild-type and sirΔ strains were digested with various restriction endonucleases. The DNA following purification was digested with a second restriction endonuclease and analyzed by DNA blot hybridization. For each site tested the band corresponding to wild-type cells is present on the left and the sirΔ strain is on the right.
We employed an in vivo assay to refine the boundary of the silenced domain and to test how these boundary elements might function. A set of isogenic strains was created in which the URA3 gene was inserted at three specific sites on chromosome III, at varying distances from the silencer (Fig. 1). Transcriptional repression was measured by the degree of silencing of the URA3 gene.
Placing the URA3 gene between the two silencers (∼640 bp to the right of HMR-E) led to near complete repression of the gene. When placed 475 bp to the left of HMR-E, URA3 was also repressed, whereas when placed 2840 bp to the left of HMR-E, URA3 was derepressed completely. Furthermore, transcriptional repression was SIR3 dependent (cf. SIR with sir3Δ in Fig. 1). These results support the previous conclusion about the silenced domain extending beyond the silencer and provide a convenient genetic assay for factors or mutants affecting the limits of the silenced domain.
The silenced HMR domain could be doubled in size
The heterochromatic domain at HMR spans ∼3.5 kb of DNA. One hypothesis for the size of the silenced domain is that the pools of Sir proteins in the cell limit its size (Renauld et al. 1993). We tested this model by determining whether an increase in the distance between HMR-E and HMR-I would lead to concomitant derepression of HMR and loss of the silenced domain.
Strains were constructed wherein the HMR domain was increased from 3.5 to 4.5 kb or 7 kb, by inserting either the TRP1 gene alone, or TRP1 with one copy of an unrelated fragment of yeast DNA from the coding region of the HMG2 gene. Examining the expression of the TRP1 gene monitored silencing in these strains. TRP1 remained repressed even when the domain was expanded from 3.5 to 7.0 kb (Fig. 2A). This repression was dependent on the Sir proteins, suggesting that the size of the silent domain was not due to limiting pools of the Sir proteins, as it can be increased.
To confirm that the repression observed in vivo was due to the expansion of the silenced domain we utilized a differential restriction endonuclease accessibility assay (Loo and Rine 1994). In this assay, nuclei isolated from wild-type and sirΔ strains were partially digested with various restriction enzymes to assay the accessibility of the DNA to these enzymatic probes. Following digestion, the DNA was analyzed by DNA blot hybridization as described (Loo and Rine 1994). Mapping the expanded domain using the differential restriction endonuclease accessibility assay demonstrated that the entire expanded domain was inaccessible to the enzymes in wild-type cells but not sirΔ cells (Fig. 2B), confirming the observations made in vivo with expression of the TRP1 gene. Interestingly, results from the restriction endonuclease assay also demonstrated that the expanded domain still terminated at the same DNA sequence as at the wild-type HMR locus (cf. Fig. 2B, top and bottom). These results suggest that the silent domain was restricted in space by DNA elements that flank the HMR locus.
Deletion of the right boundary leads to a spread of silencing
Because the boundaries of the silent domain were unchanged even when its size was doubled we speculated that the presence of a boundary element would restrict the spread of the silenced domain, and conversely, deletion of this element would lead to the spread of the domain.
Previous studies implicated a 500-bp region to the right of HMR-I as a boundary of the silenced chromatin (Loo and Rine 1994). To test whether deleting the boundary element led to a spread of silenced chromatin, we generated strains containing an insertion of the URA3 gene ∼1420 bp to the right of HMR-I. The putative right boundary (∼1 kb) was either left intact or was replaced with a 1-kb fragment of unrelated DNA. Additionally, we overexpressed SIR3 in these strains to see whether this overexpression led to the expansion of the silenced domain, as has been observed at telomeres (Renauld et al. 1993). The effect of deleting this putative boundary element was analyzed by monitoring the expression of the URA3 gene (Fig. 3). In the presence of an intact boundary element, the URA3 gene was insulated from the repressing chromatin and was active (Fig. 3). However, when the putative element was deleted, the URA3 gene was significantly repressed, indicating that the silenced chromatin had expanded to repress the URA3 gene. The silencing of the URA3 gene was dependent on the normal mechanisms that function at HMR, as it required the presence of the Sir proteins.
Figure 3.
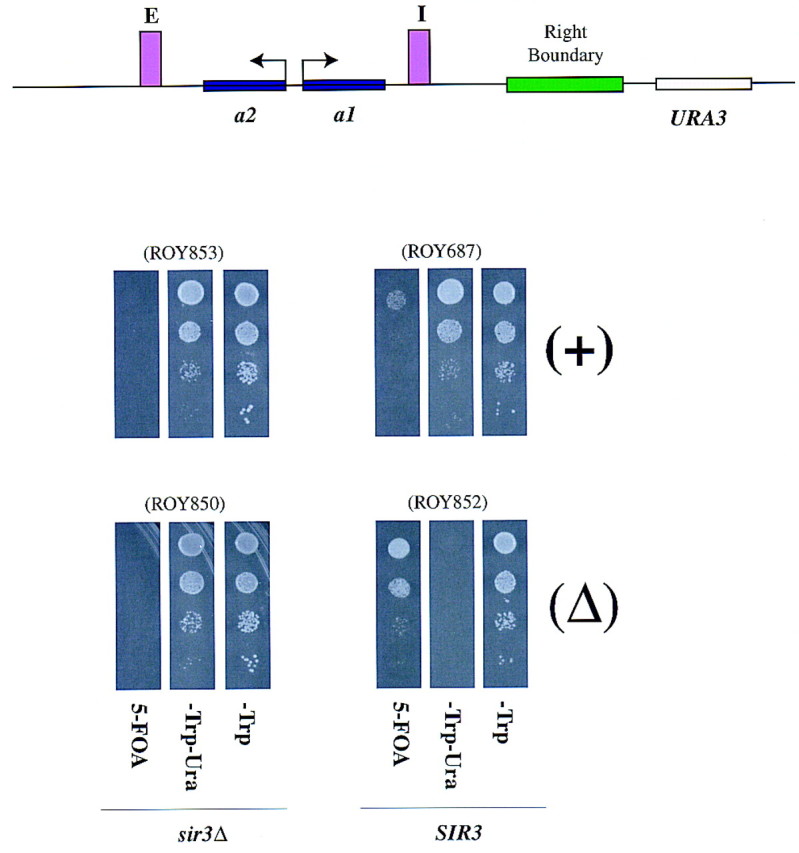
Deletion of the right boundary leads to a spread of silencing. (A) A schematic depiction of the URA3 insertion at HMR in strains with (ROY687 and ROY853) or without (ROY852 and ROY850) the right boundary element. (B) ROY853 is a sir3Δ strain carrying a URA3 insertion between SGD coordinates 295027 and 295277 (∼1420 bp to the right of HMR-I) with an intact right boundary element. ROY687 is isogenic to ROY853 except that it is Sir+. In strain ROY850 (sir3Δ), the putative right boundary element (between nucleotides 293957 and 294977) was deleted and the URA3 gene with a 1-kb fragment of pUC18 stuffer DNA inserted between nucleotide 295027 and 295277. ROY852 is a Sir+ version of ROY850. All the strains were also transformed with SIR3 on a 2μ-based plasmid (pRO146). The cells were grown in liquid medium and 3 μl of 10-fold serial dilutions was spotted on either supplemented YMD plates lacking tryptophan (−Trp) to select for the plasmid, or supplemented YMD plates lacking tryptophan and uracil (−Trp−Ura), or on supplemented YMD plates lacking tryptophan but containing 1 mg/ml 5-FOA to assay for URA3 expression. Strain numbers are shown in parentheses.
Boundary elements block the spread of silenced chromatin
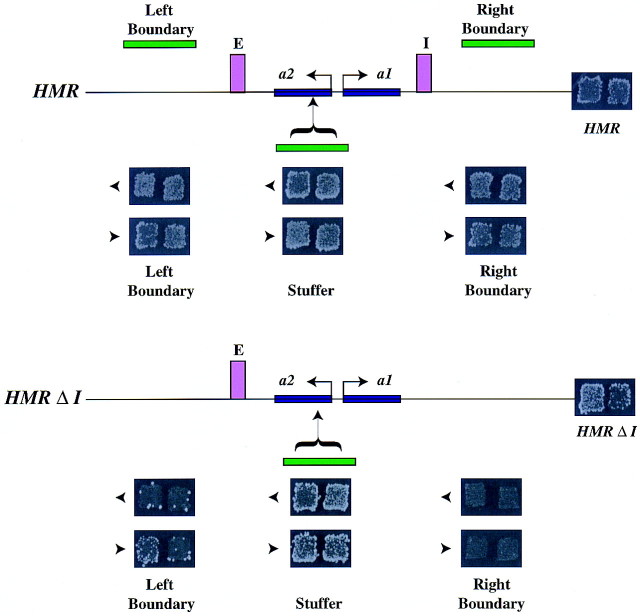
A key characteristic of a boundary element is the ability of these elements to confer position-independent expression on a gene by blocking the spread of silenced chromatin. We therefore asked whether the boundary element could block the spread of silencing when inserted between a silencer and a gene promoter.
In the first series of experiments (Fig. 4A), we inserted DNA fragments containing either the left or right boundary elements or a similarly sized stuffer fragment in both orientations between the HMR-E silencer and the MATa1 gene at the wild-type HMR locus on a plasmid. To determine whether the putative boundary elements were capable of blocking the spread of silencing from the HMR-E silencer, we monitored expression of the MATa1 gene by a mating assay. In a wild-type MATα cell, the MATa1 gene at HMR is repressed and the MATα cell is capable of mating with a MATa cell, giving rise to diploids. However, if the MATa1 gene at HMR is derepressed, the MATα cell is unable to mate and fails to form diploid colonies. Monitoring the expression of the MATa1 gene indicated that the gene remained repressed in all of the cases analyzed. This result indicated either that HMR-I was sufficient to silence MATa1, or neither boundary was capable of blocking repression from HMR-E.
Figure 4.
Boundary elements block the spread of silenced chromatin. (A) (Top) A schematic representation of boundary element insertions at the EcoNI site of the MATa2 gene at nucleotide 292140 of wild-type HMR. (B) ROY113 (MATα HMRΔ, nucleotide 288980–295350) was transformed with a 10-kb fragment of wild-type HMR carrying either (1) a 1 kb stuffer fragment of the TRP1 gene, or (2) a 1.6-kb fragment of the putative left boundary element (from nucleotide 289256 to 290846), or (3) a 1-kb fragment of the putative right boundary element (from nucleotide 293695 to 294686) at the EcoNI site of the MATa2 gene (at nucleotide 292140). Expression of the MATa1 gene was monitored by patch-mating assays using JRY19 as the mating tester lawn. All matings were performed under conditions that constantly selected for the plasmid. (B) (Top) A schematic representation of boundary element insertions at the EcoNI site of the MATa2 gene at nucleotide 292140 of HMRΔI. (Bottom) ROY113 (MATα HMRΔ) transformants carrying an 8-kb fragment of HMRΔI with the same set of putative boundary insertions as described in A (bottom) were used to monitor expression of MATa1.
Previous experiments established that silencing of HMR does not require HMR-I in strains with a fully functional HMR-E (Brand et al. 1985). Therefore, in the second set of experiments, we deleted HMR-I such that silencing could initiate only from the HMR-E silencer. Inserting the left or right boundary elements between the HMR-E silencer and the MATa1 gene resulted in the expression of the MATa1 gene, suggesting that both elements blocked the spread of silencing (Fig. 4; bottom). In contrast, the stuffer fragment was unable to block the spread of silencing. Further analysis indicated that the right boundary fragment was more proficient at mediating boundary function than was the left fragment (Fig. 4, bottom). Potentially, this difference in effectiveness could result from either inefficient insulator function of the left boundary, or perhaps the fragment used did not encompass the entire boundary element.
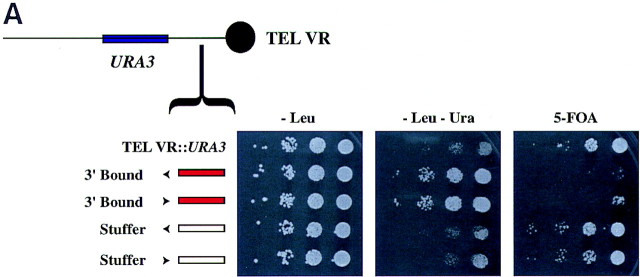
The HMR boundary element blocked the spread of telomere position effect
We tested whether the right boundary element could function to block the spread of telomere position effect. S. cerevisiae telomeres possess silenced chromatin, which emanates from the TG1–3 repeats and spreads for ∼4.5 kb. The URA3 gene was silenced when inserted close to the telomere on chromosome 5 as reported previously (see Fig. 5A; Gottschling et al. 1990). We inserted the right boundary element between the telomere and the URA3 gene and analyzed URA3 expression. The presence of the boundary element in either orientation blocked the spread of silenced chromatin, leading to the expression of the URA3 gene, whereas insertion of a stuffer fragment of the same size did not (Fig. 5A).
Figure 5.
(A) The HMR boundary element blocked the spread of telomere position effect. ROY783 contained a synthetic telomere on chromosome VR with an URA3 insertion in close proximity to the telomere. Strains ROY791 and ROY787 carried 1-kb insertions of the right boundary element, whereas ROY838 and ROY844 carried 1-kb insertions of the TRP1 gene in both orientations. All strains were transformed with SIR3 on a 2μ-based plasmid (pRO329). The strains were grown in liquid media lacking leucine, and 3 μl of 10-fold serial dilutions were spotted on supplemented YMD plates lacking leucine (−Leu) to select for the plasmid, on supplemented YMD plates lacking leucine and uracil (−Leu−Ura), or on supplemented YMD plates lacking leucine but containing 1 mg/ml 5-FOA to assay for URA3 expression. (B) The right boundary element does not function in trans. Diploid strains bearing different versions of the HMR-URA3 locus were analyzed for expression of the URA3 gene as described in Fig. 3.
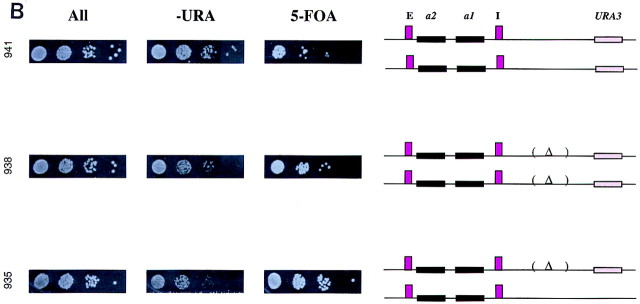
The boundary element did not function in trans
In Drosophila it has been reported that insulator activity can function in trans such that the Su(Hw) protein bound to the gypsy insulator on one chromosome can act in trans to inactivate enhancers located in the other homolog. We therefore determined whether the boundary element identified in S. cerevisiae was also capable of functioning in trans in a diploid cell.
To perform this analysis, expression of the URA3 gene located adjacent to the HMR locus was monitored in three diploid strains. In one of these strains (ROY938) the right boundary element was deleted in both HMR alleles, whereas the second strain (ROY935) was heterozygous for the right boundary element, carrying one wild-type allele of HMR and one allele with the boundary deleted. In the third strain (ROY941), both alleles of HMR–URA3 had the wild-type boundary element. If the boundary element was capable of functioning in trans, the URA3 gene in strain ROY935 would be insulated from HMR and thus be active. However, as is evident in Figure 5B, the URA3 gene in ROY935 is significantly repressed in a manner similar to strain ROY938. These results indicate that in S. cerevisiae the boundary elements are unable to function in trans to insulate the reporter gene.
A deletion analysis of the right boundary element
Because the right boundary element identified above resides in a 1-kb fragment of DNA, we decided to perform a deletion analysis of this fragment to identify the minimum fragment that is necessary for boundary function. The results of this analysis are presented in Figure 6. The analysis was performed by inserting DNA fragments, derived from the 1-kb right boundary element, between the HMR-E silencer and the MATa1 gene and monitoring expression by the mating assay (described in Fig. 4). The 1-kb right boundary fragment contains a Ty1 LTR and a tRNA gene. A systematic deletion analysis of the element from one end indicated that a DNA fragment containing just the Ty1 LTR (pRO367) had some boundary function but was not sufficient for mediating robust boundary function. In addition, the 5′ end of the LTR alone (pRO273) did not have any boundary activity on its own.
Figure 6.
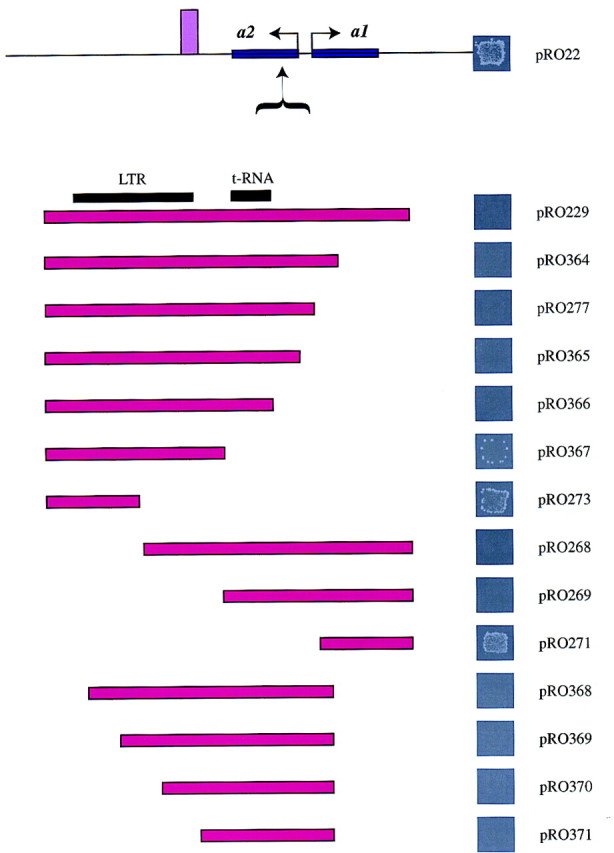
A deletion analysis of the boundary element. ROY113 (MATα hmrΔ) transformants carrying an 8-kb fragment of HMRΔI with a series of right boundary element deletions inserted at the EcoNI site of the MATa2 gene at nucleotide 292140 were used to monitor expression of the MATa1 gene as described in Fig. 4.
A systematic deletion from the other end of the 1-kb DNA fragment indicated that deletion of the LTR did not result in complete loss of boundary function, whereas deletion of a fragment of DNA encompassing the tRNA gene caused a significant loss of boundary element function (cf. pRO269 to pRO271). Further deletion analysis indicated that a small DNA fragment (<400 bp) lacking the LTR but encompassing the tRNA gene (pRO370) possessed significant boundary element function. These data suggest that robust boundary element function was mediated by a combination of both the LTR-containing fragment and the tRNA-containing fragment.
Extragenic mutations that affect boundary function
In Drosophila a multiprotein complex mediates insulator function. We therefore also began to look for mutations in genes that would lead to loss of boundary element function. Because the 1-kb right boundary element encompassed an LTR element, we tested mutations identified previously as being necessary for LTR function for their effects on boundary function (Fig. 7). spt3, spt4, and spt8 mutants are defective in LTR function, but these mutations had no major effect on boundary function. We also analyzed boundary function in cells carrying mutations in proteins that have putative binding sites in the 1-kb boundary fragment—specifically tec1, mig1, pho4, and gcr1 (data not shown). Because Gcr1p has been shown to interact with Rap1p we also tested mutations in RAP1. However, individual mutations in these proteins did not result in loss of boundary element function. Then, we tested mutations in chromatin assembly proteins—cac1, hat1, and rlf6 and found that single mutations in these proteins also had no effect on boundary function. Insulator elements have been suggested to function by anchoring DNA loops to the chromosomal scaffold. We therefore tested several mutants defective in higher order chromosome structure—smc1, smc3 (data not shown), mcd1, pds1, and top2. Interestingly, of these mutants, only loss of SMC1 and SMC3 disrupted boundary function significantly . It is not clear whether this effect is allele specific or strain specific as only smc1-2 but not smc1-259 affected boundary function (data not shown). These data implied a role for a structural protein of chromosomes in boundary function.
Figure 7.
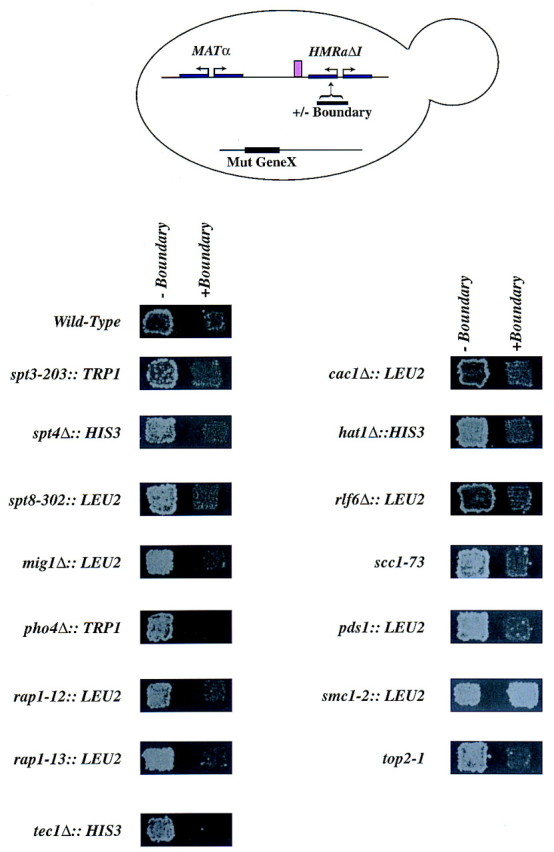
Extragenic mutations that affect boundary function. MATα strains, with either HMRΔI or HMRΔI with the right boundary element inserted between HMR-E and the MATa1 promoter at nucleotide 292140 and carrying mutations in various genes as indicated, were generated. The strains were monitored for expression of the MATa1 gene using a patch-mating assay as described in Fig. 4.
Discussion
Studies on the silenced HMR domain have shown that silencing is mediated by a repressive chromatin structure that covers the entire locus and extends beyond the silencers (Singh and Klar 1992; Loo and Rine 1994). Incidentally, the limits of the silent domain appear to coincide with previously mapped DNase I hypersensitive sites (Nasmyth 1982). Our study identified DNA segments in S. cerevisiae that restricted the spread of silenced chromatin and functioned as boundary elements of chromatin domains. We identified boundary elements by virtue of the spread of silenced chromatin in their absence and by their ability when inserted between a silencer and a reporter gene to block expression of the reporter.
The native HMR domain spans ∼3.5 kb of DNA. The relatively small size of this domain was not due to limiting amounts of the Sir proteins, as the domain could be expanded to two times its normal size and still remained repressed. These results were consistent with Ty insertions into HML that expanded that locus to 14 kb yet did not interfere with silencing (Mastrangelo et al. 1992). Biochemical analysis using a differential restriction endonuclease accessibility assay confirmed the observation that the entire expanded domain was inaccessibile to digestion by various restriction enzymes. Interestingly, in the expanded domain the boundaries of the domain remained constant with respect to the underlying DNA sequence. Taken together, these observations suggest the existence of boundary or insulator elements that restrict the spread of silenced chromatin.
We hypothesized that sequence near the border between silenced and nonsilenced domains would include a boundary element. We deleted a presumptive boundary element and discovered that neighboring reporter genes became inactive, presumably because of the spread of the silent chromatin from HMR. Previous studies of chromosomal translocations and PEV in Drosophila led to the suggestion that boundary elements limited the spread of inactive chromatin (Kellum and Elgin 1998). The results of this deletion analysis provided direct support for this model.
The boundary element interfered with silencer–promoter interactions in a position-dependent manner. The boundary appeared to function only when inserted between a silencer and a promoter. When two silencers flank a reporter, inserting the boundary element between one of these silencers and the promoter was not sufficient to insulate the promoter. The boundary element function in S. cerevisiae was position dependent with respect to the silencers and promoters. This behavior was similar to insulator elements from larger eukaryotes that are able to confer position-independent expression of the reporter gene only when they bracket the gene (Kellum and Schedl 1991; Roseman et al. 1993) and also disrupted the action of an enhancer only when positioned between the enhancer and the promoter. Our results also suggest that the primary function of the HMR boundary element was to limit the activity of silencers to specific domains of the chromosome.
Studies in Drosophila have also shown that boundary elements are not enhancer- or promoter-specific (Gdula et al. 1996). Similarly, we have found that the S. cerevisiae boundary element could insulate a reporter gene (URA3) from silenced chromatin at both the HMR locus and the chromosome VR telomeric locus. It will be of interest to determine whether any of the native telomeres are flanked by insulator elements and whether insulator elements are present at other loci in the cell.
An analysis of the sequences of the left and right boundary elements indicates that both elements contain a LTR. However, the results also indicate that the left element is not very efficient at boundary function, whereas a deletion analysis of the right element suggests that the LTR within this fragment on its own is also a poor boundary element. These results suggest that although the LTR contributes to boundary function, it is not sufficient. Further deletion analysis of the right boundary element demonstrated that deletion of the tRNA gene within this fragment resulted in a significant loss of boundary function. These data together suggest that the robust nature of the right boundary was probably due to a combined action of the Ty1 LTR and the tRNA gene, as both of these elements can function individually to varying degrees as boundary elements.
The correlation between Ty elements and boundary function, although surprising, was not unexpected, as in Drosophila the gypsy retrotransposon contains an insulator function in vivo (Corces and Geyer 1991). At this point we do not know whether all classes of Ty elements affect boundary function in S. cerevisiae.
The demonstration that a tRNA gene fragment acts as a boundary element is intriguing because it has been been demonstrated previously that a tRNA gene located at the HMR locus becomes silenced (Schnell and Rine 1986). In a second study, the insertion of a tRNA gene upstream of a RNA polymerase II (Pol II) promoter caused the repression of the Pol II promoter (Hull et al. 1994). However, we find that the insertion of a DNA fragment containing a tRNA gene results in the disruption of silenced chromatin and the consequent activation of the Pol II reporter gene (MATa1 or URA3). Further studies should help resolve this paradox. It has been shown previously that transposable elements preferentially integrate adjacent to tRNA genes and the integration is mediated by interactions between the proteins bound to the LTR and the tRNA locus (Voytas and Boeke 1993). The boundary function observed in our study could therefore be due to a cooperation between the proteins at the LTR and the tRNA, which could form a large multiprotein complex that would act as a barrier to the spread of heterochromatin.
The demonstration that DNA sequences flanking the HMR locus contained boundary element function led us to test mutations in proteins that affect boundary function. The 1-kb fragment of DNA that contains boundary element function contains putative binding sites for Mig1p, Gcn4p, Pho4p, and Gcr1p. The fragment of DNA also contained a Ty1 LTR that is regulated by the Spt gene products. There was, however, no detectable defect on boundary element function in spt3, spt4, spt8, mig1, pho4, rap1, tec1, and gcr1 mutants. Because some of these genes have been implicated in LTR-mediated transcription (Eisenmann et al. 1992, 1994; Swanson and Winston 1992; Laloux et al. 1994; Turkel et al. 1997), the results suggest that the boundary element function was not due to transcription from the LTR. Of the chromosome structure mutants tested, only SMC1 and SMC3 (data not shown) mutants had a significant effect on boundary function. It is not clear whether this effect is allele specific or strain specific, as the defect seen in the smc3-1 strain varied from strain to strain. The Smc proteins (Koshland and Strunnikov 1996) are a family of proteins required for chromosome condensation and cohesion. Relatives of this family are involved in dosage compensation in Caenorhabditis elegans (Chuang et al. 1994), are part of the nuclear scaffold (Hirano and Mitchison 1994), and are thought to be involved in chromosome loop organization. Our demonstration that insulator function is disrupted in smc1 and smc3 mutants is consistent with models in which insulator element function is dependent on chromosome architecture. Our results suggest that proteins involved in higher order chromosome structure might be involved in the functional delineation of the chromosome. Further studies should help clarify the connection between these proteins and boundary function.
In conclusion, we propose that the formation of large multiprotein complexes would prevent the spread of heterochromatin and thus act as a boundary element. It is possible that enzymatic activities such as those mediated by the Swi/Snf complex or acetyltransferases could be recruited by these complexes to the boundary to actively remodel the repressed chromatin, thereby preventing its repressive effects from spreading. Alternatively boundary elements could actively sequester specific domains into subregions of the nucleus where the effective concentration of activators and repressors would determine the transcription state of the gene.
Materials and methods
Yeast transformations and integrations
PCR-based integrations were performed with oligonucleotides containing sequences corresponding to the marker gene flanked by sequences corresponding to the 5′ or 3′ sequence of the site of integration. All fragments of DNA amplified by PCR utilized Taq DNA polymerase. Plasmid-based integrations were performed with plasmid fragments isolated by gel purification. For integrations into yeast either 5 μg of a specific gel-eluted DNA fragment or 250 μl of PCR product was concentrated to 5 μl and used for a single transformation as described (Kamakaka and Rine 1998).
Serial dilutions
A single colony of yeast cells was used to innoculate 5 ml of liquid YPD or YM medium with the appropriate supplements to allow maintenance of a plasmid. The cells were grown overnight at 30°C to an A600 of ∼1.0. All cells were diluted to an initial concentration of 1.0 A600/ml in YMD medium and serially diluted 10-fold. Approximately 3 μl of each serial dilution was spotted onto appropriately supplemented plates using a cell spotter, and the cells were allowed to grow at 30°C for 48 hr prior to photography.
Patch mating
Patches of the appropriate strains were grown on YMD plates with selection for the plasmid for 1–2 days at either 23°C or 30°C. The mating potential of the cells was monitored by replica plating the patches onto selective YMD plates spread with a mating lawn (JRY19), maintaining the selection for the plasmids following mating.
Strains
Yeast strains and plasmids are given in Tables 1 and 2, respectively.
Table 1.
Yeast strains
| Strain
|
Genotype
|
Strain
|
Genotype
|
|---|---|---|---|
| JRY19 | MATahis4-519 ura3Δ52 leu2-3,112 trp1 can1 | ROY1041* | MATα ADE2 lys2Δ HMRΔI mig1Δ::LEU2 |
| JRY3009 ROY113 | MATα ade2-1 MATα ade2-1 HMRΔ (nt. 288980 to 295350) | ROY1042* | MATα ADE2 lys2Δ HMRΔI–(right bound insert)– mig1Δ::LEU2 |
| ROY49 | MATα lys2Δ sir4Δ::LEU2 HMR::TRP1 | ROY1044* | MATα ADE2 LYS2 HMRΔI spt8-302::LEU2 l |
| ROY508 | MATα ade2 LYS URA3–HMR ppr1Δ::HIS3 | ROY1046* | MATα ade2-1 LYS HMRΔI rlf6Δ::LEU2 |
| ROY513 ROY55 | MATα ade2 lys2Δ URA3–HMR ppr1Δ::HIS3 MATα lys2Δ sir4Δ::LEU2 HMR::TRP1::HMG2 | ROY1049* ROY1051* | MATαade2-1 LYS HMRΔI tec1Δ::HIS3 MATα ade2-1 LYS HMRΔI–(right bound insert)– |
| ROY648 | MATα ade2 LYS HMR::URA3 ppr1Δ::HIS3 | tec1Δ::HIS3 | |
| ROY652 | MATα ade2 lys2Δ HMR::URA3 | ROY1052* | MATα ADE2 lys2Δ HMRΔI spt4Δ::HIS3 ppr1Δ::HIS3 |
| ROY656 | MATa ADE2 lys2Δ sir3Δ::TRP1 HMR::URA3ppr1Δ::HIS3 | ROY1054* | MATαade2-1 LYS HMRΔI top2-1 |
| ROY687 ROY783 | MATα ADE LYS ppr1Δ::HIS3 HMR–URA3 MATa ADE ppr1Δ::HIS3 URA3–TEL–VR | ROY1059* ROY1060* | MATαade2-1 LYS HMRΔI pds1::LEU2 MATα ade2-1 LYS HMRΔI smc3-1::LEU2 |
| ROY787 | MATa ade2 ppr1Δ::HIS3 URA3–HMR–right bound–TEL–VR | ROY1061* | MATα ade2-1 LYS HMRΔI–(right bound insert)– smc3-1::LEU2 |
| ROY 791 | MATa ADE ppr1Δ::HIS3 URA3–HMR–right | ROY1063* | MATα ADE2 lys2Δ HMRΔI scc1-73 |
| bound–TEL–VR | ROY1065 | MATαade2-1 LYS HMRΔI pho4Δ::TRP1 | |
| ROY803 ROY834 | MATα ade2 LYS HMR::TRP1 MATα ADE lys2Δ URA3–HMR ppr1Δ::HIS3 sir3Δ::TRP1 | ROY1067 | MATα ade2-1 LYS2 HMRΔI–(right bound insert) pho4Δ::TRP1 |
| ROY836 | MATaade2 lys2Δ URA3–HMR ppr1Δ::HIS3 sir3Δ::TRP1 | ROY1075 | MATa ade2-1 LYS2 HMR::TRP1 sir3Δ::HIS3 |
| ROY838 | MATα ADE URA3–TRP1–TEL–VR–ppr1Δ::HIS3 | ROY1076 | MATa ade2-1 lys2Δ HMR::TRP1 |
| ROY84 ROY844 | MATα lys2Δ ade2 HMR::TRP1::HMG2 MATα ADE URA3–TRP1–TEL–VR–ppr1Δ::HIS3 | ROY1079 ROY1080 | MATaADE2 LYS2 HMR::TRP1::HMG2 MATa ADE2 LYS2 HMR::TRP1::HMG2 sir3Δ::HIS3 |
| ROY850 | MATalys2Δ ADE HMR–(right bound delete)–URA3 | ROY1088 | MATα ADE2 lys2Δ HMRΔI(right bound insert)– |
| sir3Δ::TRP1ppr1Δ::HIS3 | hat1Δ::HIS3 | ||
| ROY852 | MATα lys2D ADE2 HMR–(right bound delete)–URA3 | ROY1089 | MATα ADE2 lys2Δ HMRΔI–(right bound insert)– |
| ppr1Δ::HIS3 | cac1Δ::LEU2 | ||
| ROY853 ROY935 | MATα lys2Δ ADE HMR–URA3 sir3Δ::TRP1 ppr1Δ::HIS3 MATa/MATα ppr1Δ::HIS3/ppr1Δ::HIS3 ADE/ADE | ROY1090 | MATα ADE2 lys2Δ HMRΔI–(right bound insert)– rap1-12::LEU2 |
| LYS/LYS HMR/HMR–(right bound delete)–URA3 | ROY1094* | MATα ADE2 lys2Δ HMRΔI–(right bound insert)– | |
| ROY938 | MATa/MATα ppr1Δ::HIS3/ppr1Δ::HIS3 ADE/ADE | rlf6Δ::LEU2 | |
| LYS/LYS HMR–(right bound delete)–URA3/HMR–(right bound delete)–URA3 | ROY1096* ROY1099* | MATαADE2 lys2Δ HMRΔI–(right bound insert)–top2-1 MATα ADE2 lys2Δ HMRΔI–(right bound insert)– | |
| ROY941 | MATa/MATα ppr1Δ::HIS3/ppr1Δ::HIS3 ADE/ADE LYS/LYS HMR–URA3/HMR–URA3 | ROY1101* | spt3-203::TRP1 MATα ADE2 lys2Δ HMRΔI–(right bound insert)– |
| ROY951 | MATα ade2-1 LYS2 HMRΔI | spt8-302::LEU2 | |
| ROY961 ROY1025 | MATα ADE2 lys2Δ HMRΔI–(right bound insert) MATα ade2-1 LYS HMRΔI cac1Δ::LEU2 | ROY1102* | MATα ADE2 lys2Δ HMRΔI–(right bound insert)– pds1::LEU2 |
| ROY1027 ROY1029 | MATα ade2-1 LYS HMRΔI rap1-12::LEU2 MATα ADE2 LYS2 HMRδI hat1Δ::HIS3 | ROY1104* | MATα ADE2 lys2Δ HMRΔI–(right bound insert)– scc1-73::TRP1 |
| ROY1032* ROY1034 | MATα ade2-1 LYS HMRΔI smc1-2::LEU2 MATα ADE2 lys2Δ HMRΔI rap1-13::LEU2 | ROY1108* | MATα ADE2 lys2Δ HMRΔI–(right bound insert)– smc1-2::LEU2 |
| ROY1036 | MATα ADE2 lys2Δ HMRΔI–(right bound insert) –rap1-13::LEU2 | ROY1112* | MATαADE2 lys2Δ HMRΔI–(right bound insert)– spt4Δ::HIS3 |
| ROY1039* | MATα ADE2 lys2Δ HMRΔI spt3-203::TRP1 |
All strains used in this study are either isogenic to W-303 or have been backcrossed six times with W-303 (*) and are therefore leu2-3,112 his3-11,15 ura3-1 trp1-1.
Table 2.
Plasmids
| pJR987 | pJJ244 |
| pJR988 | pJJ246 |
| pJR1380 | pv-UCA (V–R–URA3–TEL) |
| pJR1571 | EcoRI–HindIII fragment of HMRa cloned into pUC18 |
| pRO4 | 10-kb BamHI fragment of HMRa cloned into pRS406 |
| pRO10 | BamHI–BglII fragment of TRP1 from pJR989 cloned into the BglII site of pJR1571 in the Mata1 gene |
| pRO11 | 2.5-kb BamHI fragment containing the coding region of HMG2 cloned into the BglII site of pRO10 |
| pRO13 | A 7-kb BclI–BglII fragment (nt. 288980–295350) was deleted from pRO4 |
| pRO22 | pRO4 with a deletion of the BclI fragment from nt 293439–294878 |
| pRO146 | pHR67-23 |
| pRO224 | left boundary insert PCR amplified using Roog 28–29 and cloned into the EcoNI site of pRO22 |
| pRO225 | left boundary insert PCR amplified using Roog 28–29 and cloned into the EcoNI site of pRO22 but in the opposite orientation to pRO224 |
| pRO229 | right boundary insert PCR amplified using Roog 75–86 and cloned into the EcoNI site of pRO22 |
| pRO230 | right boundary insert PCR amplified using Roog 75–86 and cloned into the EcoNI site of pRO22 but in the opposite orientation to pRO229 |
| pRO233 | right boundary insert PCR amplified using Roog 75–86 and cloned into the EcoNI site of pRO4 |
| pRO234 | right boundary insert PCRd using Roog 75–86 and cloned into the EcoNI site of pRO4 but in the opposite orientation to pRO233 |
| pRO237 | left boundary insert PCRd using Roog 28–29 and cloned into the EcoNI site of pRO4 |
| pRO239 | left boundary insert PCR amplified using Roog 28–29 and cloned into the EcoNI site of pRO4 but in the opposite orientation to pRO237 |
| pRO241 | (pΔppr1::HIS3) |
| pRO248 | XhoI–XmnI deletion of pRO247 (which is identical to pRO4 except that the XhoI site in the polylinker is deleted) |
| pRO252 | A PvuII fragment of the URA3 gene from pJR987 was cloned into the EcoNI site of pRO4 |
| pRO256 | A PvuII fragment of TRP1 from pJR988 cloned into the EcoNI site of pRO4 |
| pRO258 | A PvuII fragment of TRP1 from pJR988 cloned into the EcoNI site of pRO4 but in the opposite orientation to pRO256 |
| pRO260 | A PvuII fragment of TRP1 from pJR988 cloned into the EcoNI site of pRO22 |
| pRO262 | A PvuII fragment of TRP1 fom pJR988 cloned into the EcoNI site of pRO22 but in the opposite orientation to pRO260 |
| pRO268 | right boundary insert PCR amplified using Roog 75–210 and cloned into the EcoNI site of pRO22 |
| pRO269 | right boundary insert PCR amplified using Roog 75–211 and cloned into the EcoNI site of pRO22 |
| pRO271 | right boundary insert PCR amplified using Roog 75–212 and cloned into the EcoNI site of pRO22 |
| pRO273 | right boundary insert PCR amplified using Roog 86–192 and cloned into the EcoNI site of pRO22 |
| pRO277 | right boundary insert PCR amplified using Roog 86–194 and cloned into the EcoNI site of pRO22 |
| pRO303 | right boundary PCR amplified using Roog 213–214 and cloned into BamHI site of pJR1380 |
| pRO304 | right boundary PCR amplified using Roog 213–214 and cloned into BamHI site of pJR1380 but in the opposite orientation to pRO303 |
| pRO305 | TRP1 gene was PCR amplified using Roog 240–241 and cloned into pJR1380 |
| pRO306 | TRP1 gene was PCR amplified using Roog 240–241 and cloned into pJR1380 but in the opposite orientation to pRO305 |
| pRO329 | HpaI fragment of SIR3 cloned into pRS425 |
| pRO363 | HMRΔI SacI–SalI fragment with a BamHI site engineered in the Mata2 gene cloned into pRS406 |
| pRO364 | right boundary insert PCR amplified using Roog 277–291 and cloned into the BamHI site of pRO363 |
| pRO365 | right boundary insert PCR amplified using Roog 277–284 and cloned into the BamHI site of pRO363 |
| pRO366 | right boundary insert PCR amplified using Roog 277–278 and cloned into the BamHI site of pRO363 |
| pRO367 | right boundary insert PCR amplified using Roog 277–279 and cloned into the BamHI site of pRO363 |
| pRO368 | right boundary insert PCR amplified using Roog 291–286 and cloned into the BamHI site of pRO363 |
| pRO369 | right boundary insert PCR amplified using Roog 291–287 and cloned into the BamHI site of pRO363 |
| pRO370 | right boundary insert PCR amplified using Roog 291–288 and cloned into the BamHI site of pRO363 |
| pRO371 | right boundary insert PCR amplified using Roog 291–285 and cloned into the BamHI site of pRO363 |
Oligonucleotides
The sequences of the various oligonucleotides used in this study will be provided upon request.
Acknowledgments
We thank D. Gottschling, R. Sternglanz, K. Nasmyth, M. Johnston, D. Shore, J. Berman, P. Kaufman, E. O’Shea, F. Winston, D. Koshland, M. Carlson, and G. Fink for generously providing various strains and plasmids. We thank N. Dhillon for her help and encouragement and A. Ehrehenhofer-Murray, A. Dillin, C. Fox, P. Wade, B. Blackwood, J. Kadonaga, and N. Dhillon for comments on the manuscript. A National Institutes of Health (NIH) grant to J.R. and a NIH grant (ZO1HD01904-01) to R.T.K. supported this research.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Rohinton@helix.nih.gov; FAX (301) 402-1323.
References
- Brand AH, Breeden L, Abraham J, Sternglanz R, Nasmyth K. Characterization of a ‘silencer’ in yeast: A DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985;41:41–48. doi: 10.1016/0092-8674(85)90059-5. [DOI] [PubMed] [Google Scholar]
- Chuang PT, Albertson DG, Meyer BJ. DPY-27: A chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell. 1994;79:459–474. doi: 10.1016/0092-8674(94)90255-0. [DOI] [PubMed] [Google Scholar]
- Corces VG, Geyer PK. Interactions of retrotransposons with the host genome: The case of the gypsy element of Drosophila. Trends Genet. 1991;7:86–90. doi: 10.1016/0168-9525(91)90277-W. [DOI] [PubMed] [Google Scholar]
- Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes & Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- Eisenmann DM, Chapon C, Roberts SM, Dollard C, Winston F. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics. 1994;137:647–657. doi: 10.1093/genetics/137.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin SCR. Heterochromatin and gene regulation in Drosophila. Curr Opin Genet Dev. 1996;6:193–202. doi: 10.1016/s0959-437x(96)80050-5. [DOI] [PubMed] [Google Scholar]
- Gasser SM, Laemmli UK. A glimpse at chromosomal order. Trends Genet. 1987;3:16–22. [Google Scholar]
- Gdula DA, Gerasimova TI, Corces VG. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc Natl Acad Sci. 1996;93:9378–9383. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophilazinc finger protein. Genes & Dev. 1992;6:1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Hull MW, Erickson J, Johnston M, Engelke DR. tRNA genes as transcriptional repressor elements. Mol Cell Biol. 1994;14:1266–1277. doi: 10.1128/mcb.14.2.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka RT. Silencers and locus control regions: Opposite sides of the same coin. Trends Biochem Sci. 1997;22:124–128. doi: 10.1016/s0968-0004(96)10074-8. [DOI] [PubMed] [Google Scholar]
- Kamakaka RT, Rine J. Sir- and silencer-independent disruption of silencing in Saccharomyces by sas10p. Genetics. 1998;149:903–914. doi: 10.1093/genetics/149.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R, Elgin S. Chromatin boundaries: Punctuating the genome. Curr Biol. 1998;8:521–524. doi: 10.1016/s0960-9822(07)00337-5. [DOI] [PubMed] [Google Scholar]
- Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- ————— A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D, Strunnikov A. Mitotic chromosome condensation. Annu Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Laloux I, Jacobs E, Dubois E. Involvement of SRE element of Ty1 transposon in TEC1-dependent transcriptional activation. Nucleic Acids Res. 1994;22:999–1005. doi: 10.1093/nar/22.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo S, Rine J. Silencers and domains of generalized repression. Science. 1994;264:1768–1771. doi: 10.1126/science.8209257. [DOI] [PubMed] [Google Scholar]
- ————— Silencing and heritable domains of gene expression. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- Mastrangelo MF, Weinstock KG, Shafer BK, Hedge A-M, Garfinkel DJ, Strathern JN. Disruption of a silencer domain by a retrotransposon. Genetics. 1992;131:519–529. doi: 10.1093/genetics/131.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth KA. The regulation of yeast mating-type chromatin structure by SIR: An action at a distance affecting both transcription and transposition. Cell. 1982;30:567–578. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- Renauld H, Aparicio OM, Zierath PD, Billington BL, Chhablani SK, Gottschling DE. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes & Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- Roseman RR, Pirrotta V, Geyer PK. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 1993;12:435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh Y, Laemmli UK. From the chromosomal loops and the scaffold to the classic bands of metaphase chromosomes. Cold Spring Harbor Symp Quant Biol. 1993;58:755–765. doi: 10.1101/sqb.1993.058.01.083. [DOI] [PubMed] [Google Scholar]
- Schnell R, Rine J. A position effect on the expresion of a tRNA gene mediated by the SIR genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:494–501. doi: 10.1128/mcb.6.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Klar AJS. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: A novel in vivo probe for chromatin structure of yeast. Genes & Dev. 1992;6:186–196. doi: 10.1101/gad.6.2.186. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Winston F. SPT4, SPT5 and SPT6 interactions: Effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992;132:325–336. doi: 10.1093/genetics/132.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkel S, Liao X-B, Farabaugh PJ. GCR1-dependent transcriptional activation of yeast retrotransposon Ty2-917. Yeast. 1997;13:917–930. doi: 10.1002/(SICI)1097-0061(199708)13:10<917::AID-YEA148>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Voytas DF, Boeke JD. Yeast retrotransposons and tRNAs. Trends Genet. 1993;9:421–427. doi: 10.1016/0168-9525(93)90105-q. [DOI] [PubMed] [Google Scholar]
- Widom J. Structure, dynamics, and function of chromatin in vitro. Annu Rev Biophys Biomol Struct. 1998;27:285–327. doi: 10.1146/annurev.biophys.27.1.285. [DOI] [PubMed] [Google Scholar]