Abstract
We have examined the expression of the rpsO-pnpoperon in an RNase III (rnc) mutant of Streptomyces coelicolor. Western blotting demonstrated that polynucleotide phosphorylase (PNPase) levels increased in the rncmutant, JSE1880, compared with the parental strain, M145, and this observation was confirmed by polymerization assays. It was observed that rpsO-pnpmRNA levels increased in the rncmutant by 1.6- to 4-fold compared with M145. This increase was observed in exponential, transition, and stationary phases, and the levels of the readthrough transcript, initiated upstream of rpsOin the rpsO-pnpoperon; the pnptranscript, initiated in the rpsO-pnpintergenic region; and the rpsOtranscript all increased. The increased levels of these transcripts in JSE1880 reflected increased chemical half-lives for each of the three. We demonstrated further that overexpression of the rpsO-pnpoperon led to significantly higher levels of PNPase activity in JSE1880 compared to M145, reflecting the likelihood that PNPase expression is autoregulated in an RNase III-dependent manner in S. coelicolor. To explore further the increase in the level of the pnptranscript initiated in the intergenic region in JSE1880, we utilized that transcript as a substrate in assays employing purified S. coelicolorRNase III. These assays revealed the presence of hitherto-undiscovered sites of RNase III cleavage of the pnptranscript. The position of those sites was determined by primer extension, and they were shown to be situated in the loops of a stem-loop structure.
INTRODUCTION
Polynucleotide phosphorylase (PNPase) is a 3′-5′-exoribonuclease that functions in the phosphorolytic degradation of RNA molecules in bacteria and in eukaryotic organelles (14, 21). In Escherichia coliand other bacteria, PNPase plays an important role in the degradation of mRNAs. Thus, endonucleolytic cleavage of RNA molecules generates 3′ ends that are substrates for the action of PNPase and RNase II, an exonuclease that functions hydrolytically (8, 12, 28). PNPase plays another role in E. coli, at least under some circumstances. As is the case in eukaryotes, the 3′ ends of at least some RNA molecules in bacteria are polyadenylated (31, 32). Polyadenylation facilitates the degradation of RNAs in bacteria (22, 24, 27). While the major enzyme responsible for RNA polyadenylation in E. coliis poly(A) polymerase I (PAP I [7]), mutants of E. colilacking PAP I still retain the ability to polyadenylate RNAs (26), indicating that there is at least one other polyadenylating enzyme in those cells. Mohanty and Kushner have presented evidence indicating that the second PAP in E. coliis none other than PNPase (25). They argue that under appropriate conditions in vivo, PNPase can serve to degrade RNAs or synthesize poly(A) tails and that this enzyme is responsible for the G, C, and U residues that are found at low frequency in the poly(A) tails of RNAs from wild-type E. coli(25).
PNPase structure, function, and expression have been studied extensively in species of the soil-dwelling, antibiotic-producing genus Streptomyces. Available data indicate that Streptomycesspecies do not contain PAP I and that the enzyme responsible for the synthesis of RNA 3′ tails in members of that genus is PNPase (1, 33). In Streptomyces coelicolorand Streptomyces antibioticus, the phosphorolytic activity of PNPase is modulated by nucleoside diphosphates (11) and both phosphorolysis and polymerization are modulated by the alarmone, (p)ppGpp (13). As is the case in E. coli, the PNPase gene, pnp, is a part of an operon that includes the rpsOgene, which encodes ribosomal protein S15 (2). In E. coli, there is an intergenic hairpin, situated between rpsOand pnp, that is a site for processing by the double-strand-specific endonuclease, RNase III (17, 29, 30). That processing is responsible for autoregulation of PNPase expression in E. coli, and we have shown that a similar hairpin exists in the intergenic region of the rpsO-pnpoperon of Streptomyces(10). That hairpin is cleaved by StreptomycesRNase III (10), but little is known about the impact of this cleavage on the stability and function of the cleaved transcripts or about the larger role that RNase III may play in the expression of rpsO-pnpin Streptomyces.
Given that RNase III plays an important role in RNA processing and in the regulation of gene expression in bacteria and that PNPase is an important participant in RNA decay in Streptomyces, it was of interest to examine the effects of RNase III processing of pnptranscripts on pnpexpression. To this end, we have examined expression of the rpsO-pnpoperon in an rncnull mutant of S. coelicolorin comparison with the parental strain from which the mutant was derived. We describe here, among other findings, the discovery of heretofore-unreported RNase III cleavage sites, located within the coding region of the pnptranscripts.
MATERIALS AND METHODS
Growth of organisms.
S. coelicolorM145 and the rncnull mutant, JSE1880, were grown on SFM (soy-flour-mannitol) agar to obtain spores or, for liquid cultures, on Streptomycesminimal medium containing carboxymethyl cellulose as a dispersant (13, 19). Spores were pregerminated as described previously (4), and growth of cultures was monitored by measuring absorbance at 450 nm.
Western blotting and PNPase enzyme assays.
Liquid cultures of strains M145 and JSE1880 (100 ml) were grown as described above, and 25-ml portions were removed at times corresponding to exponential phase (A450of ca. 0.40), transition phase (A450of ca. 0.7, just prior to the onset of antibiotic production in M145), and stationary phase (A450of ca. 1.0, after antibiotic production by M145 had begun). Mycelium was collected by brief centrifugation, washed with buffer I (50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 1 mM dithiothreitol, 5% glycerol), and sonicated twice for 30 s in 2 ml of buffer I with the sample tubes immersed in an ice-salt bath. Sonicates were centrifuged for 5 min at 13,000 rpm, and the supernatants were used for Western blotting and for polymerization assays.
For Western blots, samples of 1 and 2 μg of extract protein were fractionated by electrophoresis on 10% SDS-polyacrylamide gels. Proteins were transferred to nitrocellulose by electroblotting, and the blots were developed using the Bio-Rad Opti-4CN detection kit, according to the manufacturer's instructions. Affinity-purified, polyclonal, rabbit anti-PNPase antibody was prepared by Invitrogen Corporation, and a 1/25,000 dilution of the antibody was used for blotting. Goat anti-rabbit antibody–horseradish peroxidase conjugate was used as the second antibody and was supplied by Bio-Rad. To quantify PNPase levels, blots were scanned by computer and the intensities of the protein bands were determined by densitometry using the Scion Imaging software for Windows.
Mycelial extracts were also used for PNPase polymerization assays with [3H]ADP as substrate. Assays were performed as described previously (11, 18), and duplicate 60-μl reaction mixtures contained up to 150 μg of extract protein.
RNA preparation and Northern blotting.
Cultures (100 ml) of S. coelicolorstrains M145 and JSE1880 were grown as described above, and 40-ml portions were removed at A450values of ca. 0.4 (exponential phase) and 0.7 (transition phase) for the preparation of total RNA. Mycelium was collected by suction filtration and was washed with 0.9% NaCl. Pellets were resuspended in 2 ml of buffer composed of 10 mM Tris-HCl, pH 8.0, 1 mM Na2-EDTA, and 10.3% sucrose. To this suspension were added 1 ml of a 1.5-mg/ml solution of lysozyme and several 2- to 3-mm-diameter glass beads. This suspension was mixed briefly on a Vortex mixer and incubated for 10 min at 37°C when 3 ml of 2× Kirby mix (20) was added. This suspension was mixed vigorously for 2 min and then centrifuged for 6 min at 7,000 rpm in a Sorvall refrigerated centrifuge. The supernatants from this centrifugation were extracted with equal volumes of phenol-chloroform–isoamyl alcohol until the interface regions were free of visible protein. Nucleic acids were collected by isopropanol precipitation, and DNA was removed by DNase treatment as described previously (16).
Northern blotting was performed after fractionation of 5- or 10-μg portions of each RNA preparation on 5% polyacrylamide gels containing 7 M urea. RNAs were transferred to Brightstar-Plus membranes (Ambion) by electroblotting, and membranes were hybridized to DNA probes. One probe was obtained by digestion of the plasmid pJSE600 (10), containing the rpsO-pnpoperon of S. coelicolor, with BamHI and RsrII. This digest produced a ca.-500-bp fragment extending from 87 bp upstream of the rpsOstart codon to 126 bp past the rpsOstop codon. This probe was specific for transcripts containing rpsO. A second probe was prepared by digesting plasmid pJSE3512 (11), containing the S. coelicolorPNPase gene, with NdeI and SalI, producing a ca.-1.8-kb fragment which hybridized to transcripts containing pnp. Radioactive probes were prepared by random priming and hybridization and washing were carried out at 42°C according to the instructions provided in the Ambion NorthernMax-Gly kit. Size standards for these experiments were prepared by runoff transcription of two plasmid templates. A ca.-2,900-base, nonradioactive transcript was prepared by transcription of pJSE602 that was linearized with BplI. pJSE602 is a pBluescript SK+ derivative containing the entire S. coelicolorrpsO-pnpregion, flanked by 224 additional bp at the 5′ end and 81 bp at the 3′ end. A ca.-2,500-base transcript was synthesized using pJSE3512 (11) which had been linearized with BamHI as the template for T7 RNA polymerase. A ca.-716-base transcript was prepared by linearizing pJSE602 with RsrII, and a ca.-594-base transcript was prepared by linearizing pJSE602 with NheI.
For half-life measurements, 100-ml liquid cultures were grown to A450values of ca. 0.4, when 20 ml was removed and the mycelium was collected and washed as described above. Actinomycin D was then added to the remaining cultures to a final concentration of 75 μg/ml, 20-ml samples were removed, and the mycelium was collected at 4, 8, and 12 min after actinomycin addition. RNA was prepared from the mycelium as described above except that the lysozyme digestion was omitted and the mycelium was extracted immediately by vigorous vortex mixing in 2× Kirby mix with glass beads. Half-lives were determined via Northern blotting, performed as described above. Blots were probed with the DNA fragments described above to identify the readthrough, pnp, and rpsOtranscripts. To control for RNA loading, blots were subsequently stripped according to the manufacturer's instructions and reprobed with a ca.-767-bp SmaI fragment that is completely internal to the 16S rRNA gene from Streptomyces nodosus(4, 36). Blots were scanned, and the densities of the mRNA bands were normalized to those of the 16S rRNA band from the same time point. Half-lives were calculated from least squares regression plots of the log of normalized band densities versus time after actinomycin addition.
Overexpression of PNPase.
The pnpopen reading frame (ORF) was excised from the overexpression construct, pJSE3512 (11), as an NdeI/BamHI fragment and cloned into the streptomycete expression vector pIJ8600 (34) to produce pJSE353. That plasmid was transferred to S. coelicolorM145 and JSE1880 by conjugation from E. coli. Integration of the plasmid into the S. coelicolorchromosome was verified by PCR. For overexpression of PNPase, the resulting strains, M145/pJSE353 and JSE1880/pJSE353, were grown to an A450of ca. 0.4, when thiostrepton was added to 25 μg/ml to induce the tipApromoter. Samples (15 ml) were removed 0, 1, 2, 3, 4, and 6 h after thiostrepton addition; mycelium was collected, washed, and sonicated as described above; and PNPase was measured in polymerization assays. Control strains contained pIJ8600 alone.
RNase III digestions and primer extension analysis.
To obtain a transcript lacking part of the rpsO-pnpintergenic hairpin, a PCR product was prepared using 5′-TGGCCCCCGGGGGCAGATCCC-3′ as the 5′ primer and 5′-CTCGTCGCGGGATCCGACGTG-3′ as the 3′ primer. The resulting fragment extends from the 5′ RNase III cleavage site in the rpsO-pnpintergenic region (see Fig. 1) to 206 bases beyond the pnpstop codon. This fragment was cloned in pCR2.1-TOPO (Novagen) to produce pJSE5675. The plasmid was linearized with BamHI and was used for transcription with T7 RNA polymerase as described previously (10), producing the 5675 transcript. The 5675 transcript, radiolabeled using [α-32P]dCTP, was used as a substrate for RNase III in reactions performed as described previously (15) and contained 2 μg of labeled transcript (ca. 1.1 × 106cpm) and 0 to 50 ng of His-tagged, affinity-purified RNase III. Digestion products were separated on 5% polyacrylamide-7 M urea gels as described previously (15).
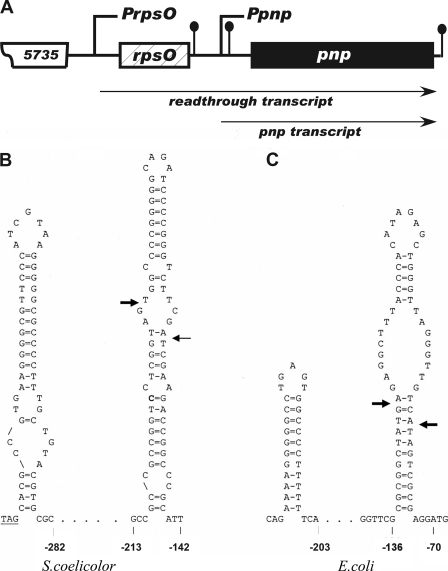
Fig. 1.
(A) Schematic representation of the rpsO-pnpoperon of S. coelicolor. PrpsOand Ppnprepresent the upstream and intergenic promoters of the operon, respectively. The ball-and-stick structures immediately following rpsOand pnpare rho independent terminators. The ball-and-stick structure just upstream of pnpis the intergenic hairpin. SCO5735 is a probable integral membrane protein. The diagram is not drawn to scale. (B) Sequence and structure of the S. coelicolorrpsOterminator and intergenic hairpin. (C) Sequence and structure of the E. colirpsOterminator and intergenic hairpin. The bases are numbered reckoning from the first base of the pnpstart codon designated +1. The arrows indicate the sites of RNase III cleavage. The rpsOstop codon is underlined in panel B.
For primer extension, unlabeled 5675 RNA was prepared and 3-μg samples were digested with 37.5, 75, and 150 ng of RNase III. Samples were prepared for primer extension as described previously (10), and extensions were performed using the primer 5′-GCC-GAG-GGC-CTC-GGA-CAC-CTG-3′. This primer extends from nucleotides 1371 to 1351, reckoned from the first base in the pnpstart codon. The primer was also used for sequencing the pJSE5675 plasmid to generate a ladder for the primer extension analysis.
RESULTS
Organization of the rpsO-pnpregion in S. coelicolor.
The pertinent features of the rpsO-pnpregion of S. coelicolorare shown in Fig. 1A. As is the case in E. coliand other bacteria, pnpis a part of an operon that includes rpsO, the gene that encodes ribosomal protein S15. As is also the case in E. coli, pnpis transcribed from two promoters in S. coelicolor, PrpsO, situated upstream of rpsOand producing a readthrough transcript, and Ppnp, situated between rpsOand pnpand producing a transcript that contains pnpbut lacks rpsO(3). The latter transcript is referred to here as the pnptranscript. The precise endpoints of the rpsO-pnptranscripts have not been determined, but we estimate that the readthrough transcript is ca. 2,950 bases in length while the pnptranscript is ca. 2,400 bases in length. PrpsOalso produces a transcript of the rpsOgene only; thus, the stem-loop structures at the left of each diagram in Fig. 1B and C are the rpsOterminators. We estimate the rpsOtranscript to be ca. 480 bases in length in S. coelicolor. The rightmost stem-loop in each case is an intergenic hairpin, shown to contain cleavage sites for RNase III. The structure of the S. coelicolorintergenic hairpin (Fig. 1B) was verified by RNase probing (11). Arrows in Fig. 1indicate the RNase III cleavage sites, and it is noteworthy that the cleavage pattern in S. coelicoloris more akin to that observed in the processing of bacteriophage T7 mRNAs (6) than to the cleavage pattern for the E. colirpsO-pnpoperon (arrows in Fig. 1B and C).
Western blotting and enzyme assays to assess PNPase levels.
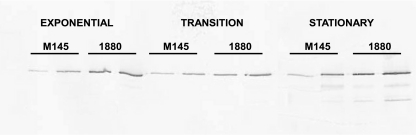
In an earlier study, Takata et al. reported an increase in PNPase levels in an rncmutant of E. coliand this increase was attributed to the stabilization of pnpmRNA in the absence of RNase III (35). It was therefore of interest to determine whether PNPase levels increased in the rncmutant of S. coelicolor. To this end, Western blotting was performed using mycelial extracts of M145 and JSE1880 as described in Materials and Methods and with the results shown in Fig. 2. For this analysis, mycelium was harvested from cultures during exponential, transition, and stationary phases at A450values of ca. 0.4, 0.7, and 1.0. PNPase protein was easily detected using polyclonal antibody prepared against S. coelicolorPNPase (Fig. 2). It is also apparent from Fig. 2that higher levels of PNPase protein were present in extracts from JSE1880 than in extracts prepared from M145. This was the case for all three stages of growth analyzed in this experiment. A quantitative estimate of the differences in PNPase levels was obtained by densitometry of the bands shown in Fig. 2. Based on this analysis, mycelial extracts from JSE1880 contained 1.5- to 2-fold more PNPase than did extracts of M145 (see Fig. 5). Figure 2also shows bands migrating more rapidly than the PNPase band in both the M145 and JSE1880 extracts obtained from stationary-phase cultures. These bands presumably represent PNPase degradation products which have been observed before in stationary-phase Streptomycescultures (unpublished results).
Fig. 2.
Western blotting of mycelial extracts. Blotting was performed as described in Materials and Methods using extracts from exponential-, transition-, and stationary-phase cells. Extracts were fractionated by SDS-PAGE, and proteins were transferred to nitrocellulose filters by electroblotting. Each pair of lanes represents 1 and 2 μg of extract protein fractionated on the gel.
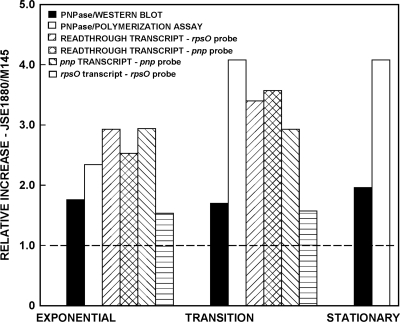
Fig. 5.
Quantification of the Western blotting, polymerization assay, and Northern blotting results. Each bar in the figure shows the ratio of the level of each parameter measured in experiments using JSE1880 to the corresponding level of that parameter in experiments using M145. Thus, the filled bar for exponential phase indicates that the average ratio of the densities of the protein bands in the Western blot (the average of the values obtained with 1 and 2 μg of extract protein), i.e., JSE1880/M145, was ca. 1.8. Since the readthrough transcript was detected using both the rpsOand pnpprobes, data for that transcript were analyzed from both panels of Fig. 4and included in this figure.
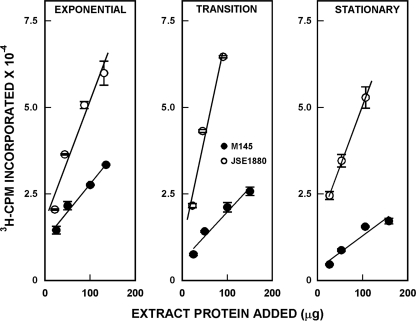
To confirm the results of Western blotting, we determined the levels of PNPase in the mycelial extracts via the polymerization assay using [3H]ADP as substrate. Results of this analysis are shown in Fig. 3. It is apparent again that there is significantly increased PNPase activity in extracts of JSE1880 compared with M145 and that this increase is manifested during exponential, transition, and stationary phases of growth. PNPase activity levels that were ca. 2.5- to 4-fold higher in JSE1880 than in M145 were observed (see Fig. 5). It is not surprising that the PNPase activity levels were higher than the protein levels measured via Western blotting, as it is known that proteolytic fragments of PNPase, like those observed in the experiments of Fig. 2, can retain PNPase activity (14, 21).
Fig. 3.
PNPase polymerization assays. The mycelial extracts used for Western blotting were assayed for PNPase as described in Materials and Methods. Data in the figure show 3H cpm incorporated into trichloroacetic acid-insoluble material in the polymerization assays and are averages of duplicate assays ± standard errors of the means. Lines were fitted to the data points by linear regression.
Taken together, these experiments show clearly that the absence of RNase III leads to a significant increase in PNPase levels in JSE1880.
Northern blotting and quantification of readthrough, pnp, and rpsOtranscripts.
It was next of interest to determine whether the increase in PNPase levels just described reflected an increase in the levels of pnpmRNA in JSE1880 compared with M145. To examine this question, Northern blotting was performed using RNAs isolated from both strains.
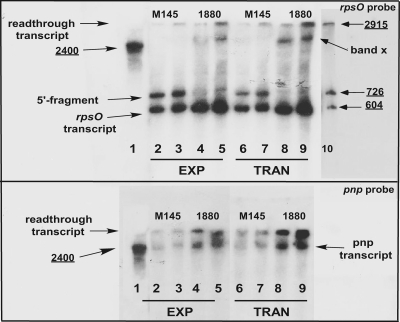
Because of the possibility that degradation of pnpmRNAs might occur during stationary phase, we analyzed only exponential- and transition-phase transcripts in these experiments. Results of typical Northern blot analyses are shown in Fig. 4. The top panel shows a blot probed with a DNA fragment specific for rpsO. Thus, this probe would identify only the readthrough and rpsOtranscripts derived from the operon, not the pnptranscript. Lane 1 of Fig. 4shows the pnpRNA standard derived from pJSE3512 (see Materials and Methods). Each pair of succeeding lanes represents 5 or 10 μg of total RNA isolated from M145 or JSE1880 during exponential and transition phases. Lane 10 shows the other size standards described in Materials and Methods. The band near the very top of the gel has the same mobility as the ca.-2,900-base standard and hybridizes to both the rpsOand pnpprobes. Thus, we identified this band as the readthrough transcript. This transcript is present in larger amounts in the RNA pool from JSE1880 than in RNA from M145 (compare, e.g., lanes 3 and 5 and lanes 7 and 9).
Fig. 4.
Northern blotting of S. coelicolorRNAs. RNA preparation and blotting procedures were as described in Materials and Methods. The blot in the top panel of the figure was probed with a DNA fragment specific for rpsOonly. Lane 1 of the top panel contains a pnpstandard. As that standard does not hybridize to the rpsOprobe, the region of the blot containing that lane was cut away and hybridized separately with the pnpprobe. Marks on the two pieces of the blot were used to align them for autoradiography. Lanes 2 to 5 contained RNAs isolated from exponential-phase cultures, and lanes 6 to 9 contained RNAs from transition-phase cultures. Each pair of lanes beginning at lane 2 contained 5 and 10 μg of total RNA, respectively. Lane 10 contains a set of standard transcripts prepared as described in Materials and Methods, and this lane was pasted onto the figure from a film of the gel that had been exposed for a longer period of time. The bottom panel of the figure is identical to the top panel except that the blot was probed with a DNA fragment that hybridizes to pnpbut not to rpsOand the gel did not contain the standards shown in lane 10 of the top panel.
The rpsOprobe identified two other transcripts in the RNA preparations. The band nearest the bottom of the gel hybridizes to the rpsOprobe but not to the pnpprobe and is ca. 500 bases in length. We thus identified this band as the rpsOtranscript. The level of the rpsOtranscript also increased in JSE1880 compared with M145. Just above the putative rpsOtranscript is a band which we believe represents the 5′ fragment produced by RNase III cleavage of the intergenic hairpin shown in Fig. 1B. That fragment would hybridize to the rpsOprobe, while the 3′ fragment produced by RNase III cleavage would not. Moreover, it is apparent from Fig. 4that this fragment is present in M145 but is absent from JSE1880. Since JSE1880 does not contain RNase III, the 5′ fragment would be absent from the transcript pool in that strain. The predicted size of the 5′ fragment is 575 bases. The bottom panel of Fig. 4is identical to the top panel except that a fragment of the pnpgene was used as a probe. This probe thus identified both the readthrough transcript and the transcript from Ppnp, the band migrating with the 2,500-base standard. This blot confirms the increased levels of the readthrough transcript in JSE1880 compared with M145. Moreover, this analysis shows that there are also increased levels of the pnptranscript in JSE1880 compared with its parent.
We also observed a band, designated band x in Fig. 4, that was present in the RNA pool obtained from JSE1880 but not from M145. This band cannot be the pnptranscript since (a) the rpsOprobe cannot hybridize to that transcript and (b) the band consistently migrated more slowly than the pnpstandard. We speculate that band x is a degradation product of the readthrough transcript produced by an alternative decay pathway that becomes operative in JSE1880 when RNase III is absent.
We determined the relative amounts of the various transcripts in the Northern blots by densitometry. Results of the densitometric analysis, of a similar analysis of the Western blots, and of the polymerization assays are summarized in Fig. 5. This figure depicts the ratio of the level of enzyme or RNA in JSE1880 compared with M145. Thus, the filled bar for exponential phase indicates that the average ratio of the densities of the protein bands in the Western blot, JSE1880/M145, was ca. 1.8, i.e., there was 1.8-fold more PNPase in JSE1880 than in M145, as measured by Western blotting at that time point. As has been mentioned, higher PNPase levels were measured via the polymerization assays than via the Western blot assays. Thus, the ratios, JSE1880/M145, of PNPase activities measured for 100 μg of extract protein in reaction mixtures (Fig. 3) were 2.3, 4.1, and 4.1 for exponential, transition, and stationary phases, respectively. As shown in Fig. 5, densitometry gave similar results from Northern blot analyses performed with each of the two probes that were employed. Thus, the ratio for readthrough transcripts obtained using exponential-phase cultures was 2.9 as determined with the rpsOprobe and 2.5 as determined with the pnpprobe. Similarly, for transition-phase cultures the respective ratios were 3.4 and 3.6. The rpsOtranscript increased by ca. 1.6-fold in JSE1880 compared with M145.
mRNA half-life measurements.
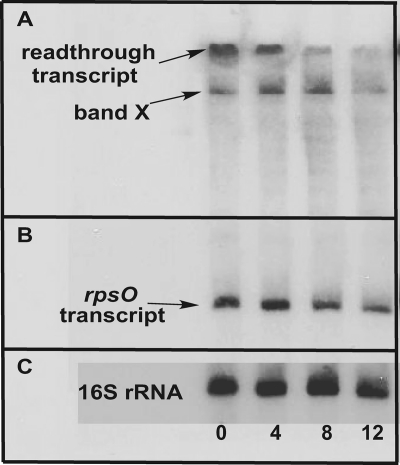
The data of Fig. 5indicated similar increases in rpsO-pnptranscript levels and in the levels of PNPase measured in S. coelicolorduring various stages of growth (with the caveat stated above regarding the differences between the results of the Western blotting and polymerization assays). It seemed likely that these increased transcript levels reflected an increase in the stability of those transcripts in the absence of RNase III. To examine this possibility, the half-lives of the rpsO-pnptranscripts were determined by Northern blotting as described in Materials and Methods, using 16S rRNA as an internal control. A typical Northern blot is shown in Fig. 6, and results of the half-life determinations are summarized in Table 1. In the parental strain, M145, the readthrough and pnptranscripts decayed rapidly, with half-lives that were much less than 4 min. At the 4-min time point, these transcripts were virtually undetectable. In contrast, the readthrough and pnptranscripts decayed with half-lives of ca. 7 and 3 min, respectively, in S. coelicolorJSE1880. Both the 5′ fragment produced by RNase III cleavage and the rpsOtranscript were more stable than the other rpsO-pnptranscripts in M145. Interestingly, the half-life of the rpsOtranscript increased in JSE1880, from ca. 4 min to 7 min. The possible significance of this increase will be discussed in detail below. The half-life measurements confirm the hypothesis that the increased levels of the rpsO-pnptranscripts observed in the S. coelicolorrncmutant are due, at least in part, to an increase in the stability of those transcripts in the absence of RNase III.
Fig. 6.
Measurement of transcript half-lives. RNA samples were prepared from cultures of S. coelicolorM145 and JSE1880 0, 4, 8, and 12 min after adding actinomycin D as described in Materials and Methods. Electrophoresis and blotting were performed as described in Materials and Methods. This figure shows results for a blot of JSE1880 RNAs hybridized to the rpsOprobe (A and B) and to a probe specific for 16S rRNA (C). Panel A represents a 3-h exposure of the blot, while panels B and C represent a 45-min exposure of the blot.
Table 1.
Chemical half-lives of rpsO-pnptranscriptsa
| Transcript | Half-life (min) for strain: |
|
|---|---|---|
| M145 | JSE1880 | |
| Readthrough | ≪4 | 6.9 ± 1.1 |
| pnp | ≪4 | 3.3 ± 0.4 |
| 5′ fragment | 6.6 ± 0.6 | |
| rpsO | 4.1 ± 1.3 | 7.1 ± 1.2 |
Half-lives were calculated from Northern blotting results as described in Materials and Methods.
Evidence for autoregulation of PNPase expression in S. coelicolor.
In E. coli, PNPase expression is autoregulated in an RNase III-dependent fashion. RNase III cleavages lead to the rapid decay of both the readthrough and pnptranscripts, and this decay is in turn dependent on PNPase (17, 29, 30). To determine whether RNase III-dependent autoregulation of PNPase expression occurs in S. coelicolor, the pnpopen reading frame, lacking both the rpsOand pnppromoters, was cloned and overexpressed in the streptomycete expression vector pIJ8600 (34). Cultures were grown in the presence of thiostrepton to induce the tipApromoter in pIJ8600. Thus, it is likely that pnpexpression was driven only by the tipApromoter in strains containing pJSE353, grown with thiostrepton. Thiostrepton was added to relevant cultures grown to an A450value of ca. 0.4 (zero time), and samples were removed from thiostrepton-treated and control cultures 1, 2, 3, 4, and 6 h later and assayed for PNPase activity.
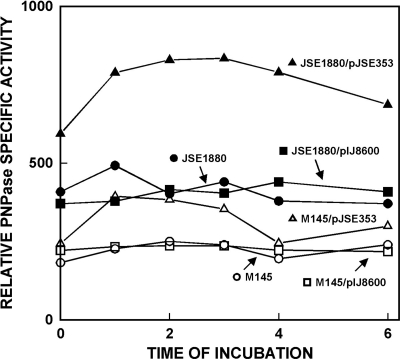
Results of these overexpression experiments are shown in Fig. 7. In S. coelicolorM145 and in M145 containing the cloning vector alone, the levels of PNPase activity did not change significantly over the 6-h course of the experiment. As expected, thiostrepton had no effect on PNPase levels in M145/pIJ8600. In M145/pJSE353, the strain containing the cloned pnpORF, the addition of thiostrepton led to a ca.-60% increase in PNPase activity, compared with the parental strain and the strain containing pIJ8600 only, within an hour following that addition. Over the next 4 h, the PNPase level gradually returned to near preinduction levels. Similarly, the levels of PNPase activity were essentially the same in JSE1880 and JSE1880 containing the cloning vector alone, although these levels were approximately twice the levels observed in the corresponding M145 derivatives, reflecting the absence of RNase III from JSE1880. Overexpression of pnpin JSE1880/pJSE353 led to an additional 2-fold increase in PNPase activity levels compared with the control strains (Fig. 7). The fact that only a 60% increase in PNPase activity was observed in M145/JSE353 compared with controls while a 2-fold increase was observed in JSE1880/pJSE353 strongly suggests that an RNase III-dependent autoregulation mechanism is operative in S. coelicolor. Compared with M145, overexpression of the rspO-pnpoperon led to a 4-fold increase in the levels of PNPase activity in JSE1880 (Fig. 7). The levels of PNPase decreased slightly in JSE1880/pJSE353 at later times after thiostrepton addition (Fig. 7). We are unable to explain this decrease at this point, but it might involve either an RNase III-independent decay of the pnptranscript or an increased level of degradation of the PNPase protein.
Fig. 7.
Overexpression of the rpsO-pnpoperon in S. coelicolorM145 and JSE1880. The overexpression construct in pIJ8600 was prepared as described in Materials and Methods. Samples were assayed for PNPase via the polymerization assay. The zero time point represents the time of addition of thiostrepton to cultures containing pIJ8600 derivatives. Specific activities were determined for 50 μg of extract protein and are expressed as cpm incorporated per μg of extract protein.
Identification of an RNase III cleavage site in the coding region of the pnptranscripts.
The results shown in Fig. 4and 5indicated that both the readthrough transcript and the transcript derived from Ppnpincreased in JSE1880 compared with M145. A straightforward explanation for the observation that the levels of the pnptranscript are RNase III dependent in S. coelicolorwould be the existence of a hitherto-undiscovered RNase III site in that transcript.
To examine this possibility, we cloned a PCR fragment that began at the 5′ site of RNase III cleavage in the rpsO-pnpintergenic region (Fig. 1) and extended 200 bp downstream of the pnpopen reading frame. That fragment was cloned in pCR2.1-TOPO to produce pJSE5675, and that plasmid was used as a template for in vitrotranscription. The 5675 transcript was treated with purified S. coelicolorRNase III, and cleavage of that transcript was demonstrated by gel electrophoresis of reaction mixtures (data not shown). Using the uncut 5675 transcript and the 5601 transcript (a ca.-500-base transcript containing the rpsO-pnpintergenic region) and its cleavage products (15) as size standards, we estimated that the RNase III cleavage site in the 5675 transcript occurred somewhere between 1,000 and 1,500 bases downstream of the pnpstart codon.
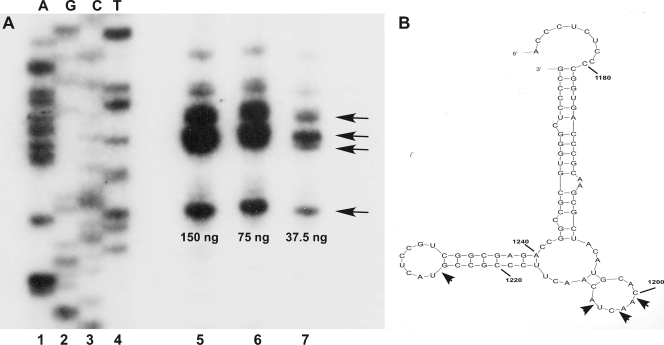
To determine the precise location of the cleavage site, primer extension analysis was performed using the primer described in Materials and Methods and nonradioactive 5675 transcript that had been cleaved with various amounts of RNase III. The extension products were fractionated on a urea-polyacrylamide gel, with the results shown in Fig. 8A. Several cleavage sites were observed as a result of this analysis and were mapped to specific bases in the 5675 transcript using the sequence ladder. Those bases are indicated in the model shown in Fig. 8B, which was obtained using the RNA folding program Mfold (37). Our analysis suggests four major cleavage sites indicated by the arrows in the model shown in the figure. It is noteworthy that, while this model has not at this point been verified by RNase probing, all of the cleavage sites occur in loops of a stem-loop structure of the model, as is the case for cleavage of the intergenic hairpin (10).
Fig. 8.
(A) Primer extension analysis of the products of RNase III digestion of the pnptranscript. Digestions and primer extension were performed as described in Materials and Methods, and products were separated on a 7 M urea-5% polyacrylamide gel. The amounts of RNase III used in the digests are shown below the lanes containing the extension products (lanes 5 to 7). The primer used for extension was also employed to generate the sequencing ladder shown in the figure, in the order A, G, C, T. The ladder is somewhat underexposed in the figure to show the extension products more clearly. Arrows indicate the bands that correspond to the four major cleavage sites. (B) Mfold model of the region containing the internal cleavage sites in the pnptranscript. Arrows show the cleavage sites revealed by the primer extension analysis in panel A. The bases are numbered reckoning from the first base of the pnpstart codon designated +1.
To search for additional RNase III cleavage sites, we tested the 716-base transcript of pJSE602 described in Materials and Methods (Fig. 4) as a substrate for RNase III. That transcript contains the rpsOopen reading frame, the rpsOterminator, and a portion of the rpsO-pnpintergenic region. No RNase III cleavage sites were observed in that transcript (data not shown).
DISCUSSION
This report marks the first detailed study of the expression of the rpsO-pnpoperon in Streptomyces coelicolor, and our results indicate differences and similarities between the Streptomycesand E. colisystems. In both systems, expression of the operon involves RNase III cleavages, and an intergenic hairpin is a target for RNase III in both. We have shown that an additional RNase III cleavage site exists within the pnpopen reading frame in S. coelicolor. To our knowledge, no such site has been demonstrated in E. coli. Although the start point for the pnptranscript has not been determined in S. coelicolor, studies from Streptomyces antibioticussuggest that the pnptranscript may not be cleaved by RNase III upstream of the coding region (2). If this is the case in S. coelicolor, the internal cleavage site explains the observation that the level of the pnptranscript, like that of the readthrough transcript, increases in the absence of RNase III (Fig. 4and 5).
In E. coli, RNase III processing of the transcripts from the rpsO-pnpoperon is responsible for autogenous regulation of pnpexpression (17, 29, 30). In the most recent model proposed for this process, PNPase digests the 5′ fragment produced by RNase III cleavage at the intergenic hairpin and the residual pnpmRNAs are degraded in an RNase E-dependent pathway (9). Data presented here indicate that autoregulation of pnpexpression also occurs in S. coelicolor. We observed only a 60% increase in PNPase activity levels upon overexpression of the pnpopen reading frame in S. coelicolorM145, whereas a 2-fold increase was observed under similar conditions in JSE1880, the rncmutant strain. Thus, RNase III-dependent modulation of PNPase levels does obtain in S. coelicoloras in E. coli. Since the S. coelicolorpnptranscripts contain an internal RNase III cleavage site, additional to the site in the intergenic region, it is possible that the mechanism of autoregulation, including the possible role of PNPase in that mechanism, is different in S. coelicolorcompared with E. coli. An additional observation suggesting a role for PNPase in autoregulation is the absence from Northern blots of bands corresponding to fragments produced by internal cleavage of the pnptranscript. Based on the position of the internal cleavage sites, bands of ca. 1,200 bases would be predicted. The absence of such bands (Fig. 4) suggests that the cleavage fragments are rapidly degraded following that cleavage in M145.
It is noteworthy that evidence for autoregulation of pnpexpression was observed in a previous study in S. antibioticus. Overexpression of the rpsO-pnpoperon in S. antibioticusled initially to a 2- to 3-fold increase in PNPase levels compared with uninduced cultures (3). This increase was observed using an overexpression plasmid containing either the entire rpsO-pnpoperon, including the intergenic region, or with a plasmid containing only the pnpcoding region. Interestingly, with either construct, PNPase levels gradually returned to preinduction values over time (3), suggesting again the existence of mechanisms to regulate intracellular concentrations of PNPase. It should be noted that the S. antibioticusstrain used in these studies did contain RNase III.
As shown in Fig. 4and 5, the rpsOtranscript level increased in JSE1880 compared with M145, and the half-life of that transcript also increased in the rncmutant strain (Table 1). This observation is in contrast to the situation in E. coli, where no increase in the half-life of the rpsOtranscript was observed in the rncmutant (35). This difference could be rationalized if an additional RNase III site existed upstream of the intergenic hairpin in S. coelicolor; however, we found no such site in transcripts representing this region. We suggest two possible explanations for the increase in the level of the rspOtranscript in JSE1880. One possibility is that the rpsOtranscript is degraded by one or more additional nucleases whose expression depends on RNase III. In the absence of RNase III that pathway would be inoperative, or would operate at a reduced level, and the level of the rpsOtranscript would increase. A second possibility relates to the observation that PNPase itself has been shown to stabilize transcripts in E. coli(5, 23), including the rpsOtranscript (5). While the exact mechanism of this stabilization is unknown, it is conceivable that the increased levels of PNPase in the absence of RNase III result in the stabilization of the rpsOtranscript, and thus its increase, in S. coelicolor.
It is also noteworthy that both the 5′ fragment produced by RNase III cleavage and the rpsOtranscript itself are significantly more stable in S. coelicolorM145 than are the readthrough and pnptranscripts (Fig. 4and Table 1). One possible explanation for this observation relates to the function of these transcripts. Even though the 5′ fragment is produced by RNase III cleavage, that RNA fragment might still be translated to produce ribosomal protein S15. It is likely that significant quantities of that protein and other ribosomal proteins are required to support vegetative growth of S. coelicolormycelium. Having several sources of the mRNAs that can be translated to produce that protein would help to ensure that sufficient quantities of it are available to participate in ribosome biogenesis.
As indicated above, the rpsO-pnpoperon is transcribed from at least two promoters in S. coelicolor. We are currently performing a detailed analysis of rpsO-pnptranscription and its regulation.
ACKNOWLEDGMENTS
This work was supported by grant number MCB 0817177from the National Science Foundation.
Footnotes
Published ahead of print on 8 July 2011.
REFERENCES
- 1. Bralley P., Gust B., Chang S. A., Chater K. F., Jones G. H. 2006. RNA 3′-tail synthesis in Streptomyces: in vitroand in vivoactivities of RNase PH, the SCO3896gene product and PNPase. Microbiology 152:627–636 [DOI] [PubMed] [Google Scholar]
- 2. Bralley P., Jones G. H. 2004. Organization and expression of the polynucleotide phosphorylase gene (pnp) of Streptomyces: processing of pnptranscripts in Streptomyces antibioticus. J. Bacteriol. 186:3160–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bralley P., Jones G. H. 2003. Overexpression of the polynucleotide phosphorylase gene (pnp) of Streptomyces antibioticusaffects mRNA stability and poly(A) tail length but not ppGpp levels. Microbiology 149:2173–2182 [DOI] [PubMed] [Google Scholar]
- 4. Bralley P., Jones G. H. 2001. Poly(A) polymerase activity and RNA polyadenylation in Streptomyces coelicolorA3(2). Mol. Microbiol. 40:1155–1164 [DOI] [PubMed] [Google Scholar]
- 5. Briani F., et al. 2008. Polynucleotide phosphorylase hinders mRNA degradation upon ribosomal protein S1 overexpression in Escherichia coli. RNA 14:2417–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Calin-Jageman I., Nicholson A. W. 2003. Mutational analysis of an RNA internal loop as a reactivity epitope for Escherichia coliRNase III substrates. Biochemistry 42:5025–5034 [DOI] [PubMed] [Google Scholar]
- 7. Cao G. J., Sarkar N. 1992. Identification of the gene for an Escherichia colipoly(A) polymerase. Proc. Natl. Acad. Sci. U. S. A. 89:10380–10384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carpousis A. J., Vanzo N. F., Raynal L. C. 1999. mRNA degradation: a tale of poly (A) and multiprotein machines. Trends Genet. 15:24–28 [DOI] [PubMed] [Google Scholar]
- 9. Carzaniga T., et al. 2009. Autogenous regulation of Escherichia colipolynucleotide phosphorylase expression revisited. J. Bacteriol. 191:1738–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang S. A., Bralley P., Jones G. H. 2005. The absBgene encodes a double strand-specific endoribonuclease that cleaves the read-through transcript of the rpsO-pnpoperon in Streptomyces coelicolor. J. Biol. Chem. 280:33213–33219 [DOI] [PubMed] [Google Scholar]
- 11. Chang S. A., Cozad M., Mackie G. A., Jones G. H. 2008. Kinetics of polynucleotide phosphorylase: comparison of enzymes from Streptomyces and Escherichia coli and effects of nucleoside diphosphates. J. Bacteriol. 190:98–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coburn G. A., Mackie G. A. 1999. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol. 62:55–105 [DOI] [PubMed] [Google Scholar]
- 13. Gatewood M. L., Jones G. H. 2010. (p)ppGpp inhibits polynucleotide phosphorylase from Streptomycesbut not from Escherichia coliand increases the stability of bulk mRNA in Streptomyces coelicolor. J. Bacteriol. 192:4275–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Godefroy-Colburn T., Grunberg-Manago M. 1972. Polynucleotide phosphorylase. Enzymes 7:533–574 [Google Scholar]
- 15. Gravenbeek M. L., Jones G. H. 2008. The endonuclease activity of RNase III is required for the regulation of antibiotic production by Streptomyces coelicolor. Microbiology 154:3547–3555 [DOI] [PubMed] [Google Scholar]
- 16. Hsieh C.-J., Jones G. H. 1995. Nucleotide sequence, transcriptional analysis, and glucose regulation of the phenoxazinone synthase gene from Streptomyces antibioticus. J. Bacteriol. 177:5740–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jarrige A. C., Mathy N., Portier C. 2001. PNPase autocontrols its expression by degrading a double-stranded structure in the pnp mRNA leader. EMBO J. 20:6845–6855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jones G. H., Symmons M. F., Hankins J. S., Mackie G. A. 2003. Overexpression and purification of untagged polynucleotide phosphorylases. Protein Expr. Purif. 32:202–209 [DOI] [PubMed] [Google Scholar]
- 19. Kieser Y., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. 2000. Practical Streptomycesgenetics. The John Innes Foundation, Norwich, England [Google Scholar]
- 20. Kirby K. S., Fox-Carter E., Guest M. 1967. Isolation of DNA and rRNA from bacteria. Biochem. J. 104:258–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Littauer U. Z., Soreq H. 1982. Polynucleotide phosphorylase. Enzymes 15:517–553 [Google Scholar]
- 22. Mohanty B. K., Kushner S. R. 1999. Analysis of the function of Escherichia colipoly(A) polymerase in RNA metabolism. Mol. Microbiol. 34:1094–1108 [DOI] [PubMed] [Google Scholar]
- 23. Mohanty B. K., Kushner S. R. 2003. Genomic analysis in Escherichia colidemonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol. Microbiol. 50:645–658 [DOI] [PubMed] [Google Scholar]
- 24. Mohanty B. K., Kushner S. R. 2002. Polyadenylation of Escherichia colitranscripts plays an integral role in regulating intracellular levels of polynucleotide phosphorylase and RNase E. Mol. Microbiol. 45:1315–1324 [DOI] [PubMed] [Google Scholar]
- 25. Mohanty B. K., Kushner S. R. 2000. Polynucleotide phosphorylase functions both as a 3′-5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:11966–11971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mohanty B. K., Kushner S. R. 1999. Residual polyadenylation in poly(A) polymerase I (pcnB) mutants of Escherichia colidoes not result from the activity encoded by the f310gene. Mol. Microbiol. 34:1109–1119 [DOI] [PubMed] [Google Scholar]
- 27. O'Hara E. B., et al. 1995. Polyadenylation helps regulate mRNA decay in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 92:1807–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rauhut R., Klug G. 1999. mRNA degradation in bacteria. FEMS Microbiol. Rev. 23:353–370 [DOI] [PubMed] [Google Scholar]
- 29. Robert-Le Meur M., Portier C. 1992. E.colipolynucleotide phosphorylase expression is autoregulated through an RNase III-dependent mechanism. EMBO J. 11:2633–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Robert-Le Meur M., Portier C. 1994. Polynucleotide phosphorylase of Escherichia coli induces the degradation of its RNase III processed messenger by preventing its translation. Nucleic Acids Res. 22:397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sarkar N. 1996. Polyadenylation of mRNA in bacteria. Microbiology 142:3125–3133 [DOI] [PubMed] [Google Scholar]
- 32. Sarkar N. 1997. Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem. 66:173–197 [DOI] [PubMed] [Google Scholar]
- 33. Sohlberg B., Huang J., Cohen S. N. 2003. The Streptomyces coelicolorpolynucleotide phosphorylase homologue, and not the putative poly(A) polymerase, can polyadenylate RNA. J. Bacteriol. 185:7273–7278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun J., Kelemen G. H., Fernandez-Abalos J. M., Bibb M. J. 1999. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolorA3(2). Microbiology 145:2221–2227 [DOI] [PubMed] [Google Scholar]
- 35. Takata R., Mukai T., Hori K. 1987. RNA processing by RNase III is involved in the synthesis of Escherichia colipolynucleotide phosphorylase. Mol. Gen. Genet. 209:28–32 [DOI] [PubMed] [Google Scholar]
- 36. Yap W. H., Wang Y. 1999. Molecular cloning and comparative sequence analyses of rRNA operons in Streptomyces nodosusATCC 14899. Gene 232:77–85 [DOI] [PubMed] [Google Scholar]
- 37. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]