Abstract
Summary: Members of the large superclass of P-loop GTPases share a core domain with a conserved three-dimensional structure. In eukaryotes, these proteins are implicated in various crucial cellular processes, including translation, membrane trafficking, cell cycle progression, and membrane signaling. As targets of mutation and toxins, GTPases are involved in the pathogenesis of cancer and infectious diseases. In prokaryotes also, it is hard to overestimate the importance of GTPases in cell physiology. Numerous papers have shed new light on the role of bacterial GTPases in cell cycle regulation, ribosome assembly, the stress response, and other cellular processes. Moreover, bacterial GTPases have been identified as high-potential drug targets. A key paper published over 2 decades ago stated that, “It may never again be possible to capture [GTPases] in a family portrait” (H. R. Bourne, D. A. Sanders, and F. McCormick, Nature 348:125-132, 1990) and indeed, the last 20 years have seen a tremendous increase in publications on the subject. Sequence analysis identified 13 bacterial GTPases that are conserved in at least 75% of all bacterial species. We here provide an overview of these 13 protein subfamilies, covering their cellular functions as well as cellular localization and expression levels, three-dimensional structures, biochemical properties, and gene organization. Conserved roles in eukaryotic homologs will be discussed as well. A comprehensive overview summarizing current knowledge on prokaryotic GTPases will aid in further elucidating the function of these important proteins.
INTRODUCTION
GTPases are widely distributed molecular switches that generally cycle between a GDP-bound “off” state and a GTP-bound “on” state. The conformational changes associated with these different molecular states are involved in the regulation of multiple cellular processes. In general, GTPases interact with downstream effectors when bound to GTP. GTP hydrolysis proceeds by a nucleophilic water molecule attacking the GTP γ-phosphate, leaving the protein in a GDP-bound state. The accompanying conformational changes reduce the affinity for effector molecules. Both GDP and GTP can dissociate from the GTPase, leaving the protein in the empty or apo state (23, 267).
Classification and Distribution of GTPases
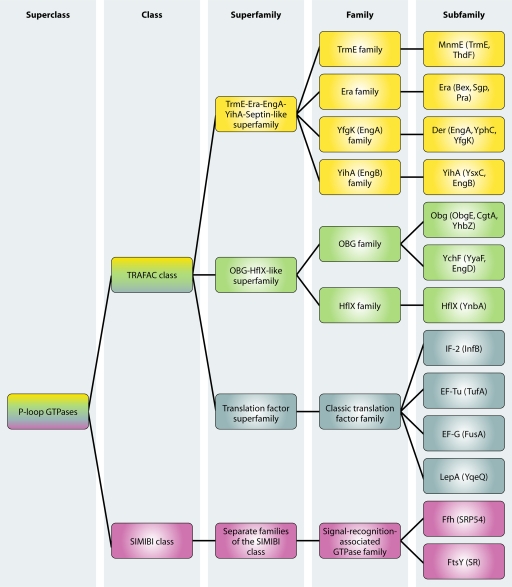
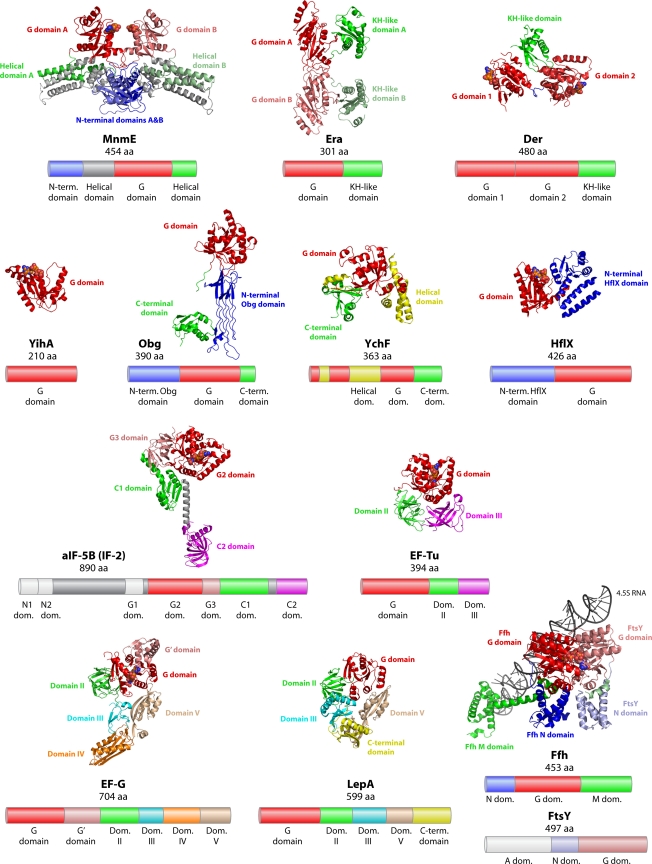
Based on sequence and structure similarities, the GTPase superfamily can be divided into two large classes (Fig. 1). The TRAFAC class (translation factors) includes proteins involved in translation, signal transduction, cell motility, and intracellular transport. The SIMIBI class (signal recognition particle, MinD, and BioD) contains the signal-recognition-associated GTPases, the MinD-like ATPases, and a group of proteins with kinase or phosphate transferase activity. Both classes comprise seven large superfamilies that are further subdivided into families and subfamilies, based on sequence, structure, and domain architecture (153). Within these families, a core group of eight universally conserved GTPases are found in all domains of life, including YihA, YchF, HflX, IF-2, EF-Tu, EF-G, Ffh, and FtsY. Three additional GTPases (Era, Der, and Obg) are conserved in prokaryotes and eukaryotes but not in archaea. Finally, MnmE and LepA are found in all bacteria and in eukaryotic mitochondria and chloroplasts, indicating a bacterial origin (42, 43, 234). These 13 conserved GTPases are the subject of this review, and their main characteristics are listed in Table 1. In addition to these proteins, most bacteria encode several other GTPases. In Bacillus subtilis, for example, 21 GTPases have been identified, 12 of which (Era, Der, YihA, Obg, IF-2, EF-Tu, EF-G, Ffh, FtsY, FtsZ, YlqF, and YqeH) are essential for cell viability (157, 173).
Fig. 1.
Classification of GTPases described in this review. The superclass of P-loop GTPases is subdivided into two classes. The TRAFAC class is comprised of universally conserved protein families belonging to the TrmE-Era-EngA-YihA-septin-like superfamily (yellow), the OBG-HflX-like superfamily (green), and the translation factor superfamily (blue). From the SIMIBI class, only GTPases belonging to the signal-recognition-associated GTPase family (violet) are universally conserved. (Data adapted from reference 153.)
Table 1.
Characteristics of GTPases described in this review
| GTPase | Crystal structure reference(s) | Molecular mass of E. coli homolog (kDa) | Major process(es) involved | Phylogenetic distribution(s)a | Essentialb | Cellular localization |
|---|---|---|---|---|---|---|
| MnmE | 178, 228 | 49.3 | tRNA modification | B, E* | +/− | Cytoplasmic, partially membrane associated |
| Era | 47 | 33.9 | Cell cycle regulation, assembly and maturation of 30S ribosomal subunit, energy metabolism | B, E | +/− | Cytoplasmic, partially membrane associated |
| Der | 214 | 55.1 | Cell cycle regulation, assembly and maturation of 50S ribosomal subunit | B, E | Yes | Cytoplasmic, partially membrane associated |
| YihA | 64, 216 | 23.7 | Cell cycle regulation, assembly and maturation of 50S ribosomal subunit | B, E, A | +/− | Cytoplasmic |
| Obg | 34, 144 | 43.4 | Cell cycle regulation, assembly and maturation of 50S ribosomal subunit, stringent response, stress response, morphological development, sporulation | B, E | Yes | Cytoplasmic, possibly partially membrane associated |
| YchF | 251 | 39.8 | Translation, stress response in plants, antioxidant response in humans | B, E, A | No | Cytoplasmic |
| HflX | 276 | 48.4 | Assembly and maturation of 50S ribosomal subunit | B, E, A | No | Cytoplasmic, partially membrane associated |
| IF-2 | 215 | 97.5 | Assembly of the translation initiation complex | B, E, A | Yes | Cytoplasmic |
| EF-Tu | 1 | 43.4 | Delivery of aminoacyl-tRNAs to the ribosome | B, E, A | Yes | Cytoplasmic |
| EF-G | 1 | 77.7 | Ribosomal translocation | B, E, A | Yes | Cytoplasmic |
| LepA | 79 | 66.7 | Ribosomal back-translocation | B, E* | No | Cytoplasmic |
| Ffh | 84, 134 | 49.9 | Cotranslational targeting of membrane-bound proteins | B, E, A | Yes | Cytoplasmic |
| FtsY | 185, 186 | 54.6 | Membrane-bound signal recognition particle receptor | B, E, A | Yes | Cytoplasmic, partially membrane associated |
B, bacteria; E, eukaryotes; E*, eukaryotic organelles of prokaryotic origin; A, archaea.
+/−, essential in certain genetic backgrounds or strains.
Conserved Sequence Motifs
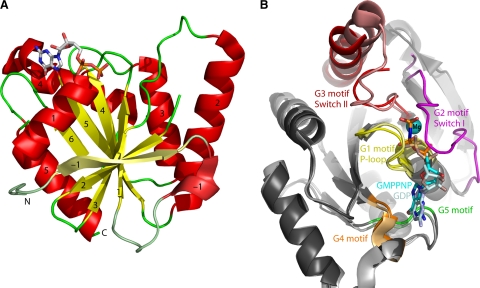
GTPases comprise a superclass of P-loop (phosphate-binding loop) NTPases that share a domain often called the G domain. The G domain typically adopts an α/β fold with a central β-sheet of at least 6 (mostly parallel) β-strands surrounded on both sides by α-helices (Fig. 2 A). The G domain is further characterized by the occurrence of conserved amino acids sequences called the G motifs (G1 through G5) (Fig. 2B and 3). G1 [GX4GK(S/T)] is also known as the Walker A motif or P-loop. This motif is shared with other proteins that bind purine nucleotide triphosphates, including ATPases and some kinases, and is involved in the binding of the phosphates of GTP and GDP. The G2 region (also known as the effector region) is highly conserved within each GTPase family but not among different families. Members of the TRAFAC class are characterized by a conserved threonine in this region. G2 interacts with effector molecules, and it is responsible for coordinating a Mg2+ ion that binds to the β- and γ-phosphates. This region often shows large structural differences between the GTP- and GDP-bound states and is therefore also referred to as the switch I region. The G3 region is also known as the Walker B motif. It comprises a typical DX2G motif that is involved in Mg2+ coordination and binding to the γ-phosphate. Like G2, this domain shows large conformational changes between the GDP- and GTP-bound forms and is therefore also called the switch II region. The G4 region consists of four hydrophobic or apolar amino acids followed by (N/T)(K/Q)XD. It determines nucleotide specificity by forming hydrogen bonds exclusively with guanine rings. Last, the G5 region interacts with guanine via water-mediated hydrogen bonds. The G5 motif is not strictly conserved across GTPases, as, in general, the contacts between the protein and the nucleotide involve only main-chain atoms (24, 115, 132, 182, 216). (For a detailed description of the structure and regulators of GTPases, see references 200 and 267.)
Fig. 2.
Overall structure of the G domain of P-loop GTPases. (A) Ribbon plot of a G domain. The structure of B. subtilis YihA in complex with GDP is shown (Protein Data Bank [PDB] accession number 1SVI). β-Strands are shown in yellow, α-helices are in red, and connecting loops are in green. GDP is shown in a stick representation. YihA contains an extra N-terminal β-strand and α-helix compared to the minimal 6-stranded mixed β-sheet of Ras. Conforming to the secondary structure numbering of Ras, these extra elements have been numbered β-strand and α-helix −1. A peptide region connecting α-helix 1 and β-strand 2 (corresponding to switch I) is disordered in this structure and is not shown. (B) Conserved GTPase motifs. The figure shows a superposition of B. subtilis YihA in the GDP-bound “off” state (PDB accession number 1SVI) and B. subtilis YihA bound to the GTP analog GMPPNP, mimicking the “on” state (PDB accession number 1SVW). The conserved sequence elements and the switch regions are shown in different colors, as indicated. YihA-GMPPNP is shown in dark-shaded colors, while YihA-GDP is shown in the corresponding lighter-shaded colors. In YihA-GDP, switch I and switch II are disordered and ordered, respectively, while in YihA-GMPPNP, switch I and switch II are ordered and disordered, respectively (216).
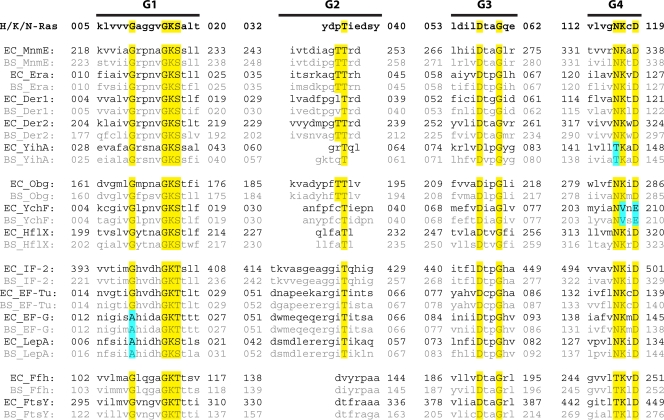
Fig. 3.
Sequence alignment of conserved G motifs. The amino acid sequence of human Ras G motifs is shown in boldface type. Other proteins are grouped according to their classifications in superfamilies (153). Sequences for E. coli (EC) and B. subtilis (BS) homologs are shown in black and gray, respectively. Residues conserved in all GTPases are highlighted in yellow. Residues deviating from the consensus sequence are marked in blue. The length of the G motifs was chosen as described previously (24). Sequences of the respective G2 motifs were obtained from previous work (24, 34, 43, 97, 115, 188, 207, 293).
The GTPase Cycle
GTPases are known to bind and hydrolyze GTP, leaving the protein in the GDP-bound state. GDP can subsequently be released, resetting the protein for another round of GTP binding and hydrolysis. Intrinsic properties of a GTPase determine the duration of the GTP, GDP, and apo states during the GTPases cycle. However, other proteins can provide additional levels of regulation. GTPase-activating proteins (GAPs) promote GTP hydrolysis, and guanine nucleotide exchange factors (GEFs) catalyze the exchange of GDP for GTP, whereas guanine nucleotide dissociation inhibitors (GDIs) inhibit the release of GDP (17, 22, 49, 124, 243). The deformation of the phosphate- and Mg2+-binding site may be key to GEF-catalyzed nucleotide exchange, and as a general mechanism, GTP hydrolysis is stimulated by GAPs via direct interactions with the conformationally labile switch regions of the GTPase and/or by providing catalytic residues in trans. GAPs associated with Ras family members have been studied most thoroughly. They typically stabilize the “closed” conformation of amino acids located in switches I and II and provide a catalytic arginine residue in trans (“arginine finger”) that counters negative-charge development at the phosphate groups of GTP during the hydrolysis reaction (224, 243).
Although ubiquitous in eukaryotes, regulation by GAPs and GEFs seems to be rare in prokaryotes. With the notable exception of EF-Tu, bacterial GTPases often have a low nucleotide affinity, foregoing the need for GEFs. Only very recently, YihI was identified as the first prokaryotic GAP, stimulating the GTPase activity of Der (114). However, interactions with ribosomal proteins, rRNA (e.g., Era), or specific ions (e.g., MnmE) as well as the dimerization of the G domains (e.g., MnmE) seem to enhance the GTPase activity of certain prokaryotic GTPases (164, 176, 229). GTPases activated by nucleotide-dependent dimerization (GADs) are not restricted to prokaryotes, and dimer formation can proceed between two distinct GTPases of the same family with identical active-site residues (e.g., Ffh-FtsY) or between monomers of the same GTPase (e.g., MnmE). The interaction with effector molecules is coupled to and regulated by the GTPase reaction (87).
Cellular Functions
Eukaryotic GTPases have long been known to play roles in protein synthesis, transmembrane signaling via receptor-mediated communication, the translocation of proteins, vesicular traffic, cytoskeleton organization, differentiation, and cell proliferation. As targets of mutation and toxins, GTPases have previously described roles in the pathogenesis of cancer and infectious diseases (23, 243). All of the universally conserved bacterial GTPases have been implicated in ribosome assembly or protein synthesis. Moreover, bacterial GTPases represent the largest class of essential ribosome assembly factors (132). However, their exact function in ribosome assembly remains elusive. Probable roles are (i) the recruitment or displacement of ribosomal proteins onto the nascent ribosome, (ii) the recruitment or regulation of an assembly factor, (iii) the prevention of premature ribosomal protein binding onto the nascent ribosome by acting as a reversible placeholder, (iv) the coupling of the assembly of ribosomal subunits in the cell with intracellular GTP concentrations, and (v) the induction of conformational rearrangements within the nascent ribosome (RNA chaperone activity) (28, 132). Besides a role in ribosome assembly, most GTPases have been additionally implicated in other cellular processes, including DNA replication, cell division, the stress response, sporulation, and pathogenesis.
THE TrmE-Era-EngA-YihA-SEPTIN-LIKE SUPERFAMILY
Within the TRAFAC class of P-loop GTPases, the TrmE-Era-EngA-YihA-Septin-like superfamily contains four universally conserved families, namely, MnmE (TrmE), Era, Der (EngA), and YihA (Fig. 1). Proteins in this superfamily generally show sequence conservation in the region between the Walker A and B motifs and as such can be distinguished from other GTPases (42, 153). For representative proteins of each family, the genetic organization, cellular localization, protein structure, biochemical characteristics, role in cell cycle regulation, role in translation, and other potential functions are described below. When appropriate, functions conserved in eukaryotic homologs are discussed as well.
MnmE (TrmE or ThdF)
The universally conserved GTPase MnmE (methylaminomethyl E) has been implicated in tRNA modifications in both prokaryotes and eukaryotes. Originally, the mnmE locus was described to encode a protein with a postulated role in thiophene and furan oxidation by Escherichia coli (5). The GTPase encoded on this locus was hence called ThdF, for thiophene degrading F (37), although later research could not confirm the relationship between this locus and the oxidation of thiophenes (43). The MnmE protein is also known as TrmE (tRNA modification E). It is widely conserved in eukaryotes and in eubacteria but missing in Chlamydia and Mycobacterium (188). Homologs have not been found in archaea (40, 283), suggesting that this gene has been acquired by eukaryotes from a promitochondrial endosymbiont (153). The encoding gene is not essential for growth in B. subtilis (188), but it appears to be indispensable for viability in certain genetic backgrounds of E. coli (40).
Genetic organization.
In E. coli, mnmE is in a genetic locus with rpmH (ribosomal protein L34), rnpA (RNase P) (which is responsible for the removal of the 5′-leader sequence from pre-tRNA), yidD, and oxaA (an inner membrane protein required for the insertion of integral membrane proteins into the membrane) (Fig. 4). This gene organization is well conserved in the Betaproteobacteria, Gammaproteobacteria, Deltaproteobacteria, and Epsilonproteobacteria. mnmE has its own promoter region and is preceded by a terminator located between oxaA and mnmE (37, 40).
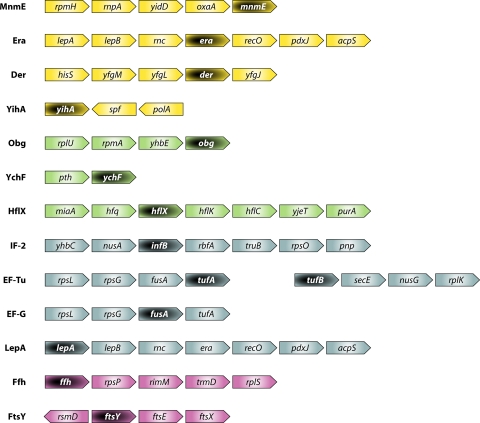
Fig. 4.
Gene organization of E. coli GTPases described in this review. Color coding corresponds to the GTPase classification shown in Fig. 1. Genes encoding GTPases are indicated in light lettering on a dark background. The figure was constructed by using the Search Tool for the Retrieval of Interacting Genes (STRING) database (247). For YihA, no neighboring genes were identified by using STRING, and the gene organization is shown as described in reference 129.
Cellular localization.
E. coli MnmE is a cytoplasmic protein that was also reported to be partially associated with the inner membrane (40). MnmE has a conserved C(I/L/V)GK motif at its extreme C terminus, which shows resemblance to the CAAX motif of Ras proteins. Although the CAAX motif is responsible for anchoring RAS to the cell membranes of eukaryotic cells, the C-terminal cysteine residue of MnmE is not involved in subcellular distribution or membrane association. Rather, the cysteine residue plays a direct catalytic role in the MnmE-catalyzed tRNA modification reaction (see below) (282).
Protein structure.
Crystal structures of MnmE proteins from Thermotoga maritima, Chlorobium tepidum, and Nostoc species have been solved. The protein is a homodimer, with each monomer consisting of an N-terminal domain, a central all-α-helical domain, and the G domain, which is in the primary structure inserted into the central α-helical domain (Fig. 5) (178, 228).
Fig. 5.
Structures of GTPases described in this review. Structural domains are indicated and colored as follows: red, G domains; blue and gray, domains N terminal to the G domain; yellow, domains integrated into the G domain; green, magenta, violet, cyan, orange, brown, and yellow, domains C terminal to the G domain. Nucleotides bound to the active site of the GTPase are shown in a sphere representation. One representative of each subfamily is shown: MnmE, structure of MnmE from Nostoc sp. in complex with GDP (PDB accession number 3GEH); Era, structure of Era from E. coli in the apo form (PDB accession number 1EGA); Der, structure of Der from B. subtilis in complex with GDP (PDB accession number 2HJG); YihA, structure of YihA from B. subtilis in complex with GDP (PDB accession number 1SVI); Obg, Obg from T. thermophilus in the apo form (PDB accession number 1UDX); YchF, YchF from H. influenzae in the apo form (PDB accession number 1JAL); HflX, homolog of HflX from the archaeon S. solfataricus in complex with GDP (PDB accession number 2QTH); IF-2, homolog of IF-2 from the archaeon M. thermautotrophicus (aIF5b) in complex with GMPPNP (PDB accession number 1G7T); EF-Tu, EF-Tu from T. thermophilus in complex with GMPPNP (PDB accession number 1EXM); EF-G, slow mutant of EF-G from T. thermophilus in complex with GTP (PDB accession number 2BV3); LepA, LepA from E. coli in the apo form (PDB accession number 3CB4); Ffh-FtsY, E. coli Ffh in complex with E. coli FtsY and the 4.5S RNA from D. radiodurans, with both proteins bound to the nonhydrolyzable GTP analog β, γ-methylene-GTP (GMPPCP) (PDB accession number 2XXA). Below each structure, the domain structure of the corresponding E. coli homolog is shown with the corresponding color coding. Additional E. coli domains are marked in light gray. Pfam accession numbers are P25522 (MnmE), P06616 (Era), P0A6P5 (Der), P0A6P7 (YihA), P42641 (Obg), P0ABU2 (YchF), P25519 (HflX), P0A6N1 (EF-Tu), P0A6M8 (EF-G), P0A705 (IF-2), P60785 (LepA), P0AGD7 (Ffh), and P10121 (FtsY). aa, amino acids.
The N-terminal domain shows structural similarity to the tetrahydrofolate-binding domain of N,N-dimethylglycine oxidase (228). This domain induces the permanent, GTP/GDP-independent homodimerization of MnmE. The interface of the N-terminal domains also harbors two binding pockets for a tetrahydrofolate derivative, which might be used as the donor of the first methyl group in the formation of the carboxymethylaminomethyl-uridine tRNA modification (see below) (189, 228). In spite of significant sequence similarity shared by the G domains of MnmE and Era, a detailed comparison of the secondary structure of the MnmE G domain shows more similarities to Ras than to Era (184). The amino acid sequence of the α-helical domain is less well conserved, apart from some small patches at the tip of the central 4-helix bundle and the C-terminal C(I/L/V)GK motif.
In addition to the structures of the full-length protein, crystal structures of the individual G domain of E. coli MnmE in complex with GDP-AlFx, a transition-state analog of GTP hydrolysis, and various ions have been solved. These structures show that the G domains of MnmE homodimerize (independently of the N-terminal domain) in the GTP-bound state in a potassium-dependent manner (229). This G domain dimerization was also confirmed later via pulse electron paramagnetic resonance spectroscopy measurements (178).
GTPase cycle.
The nucleotide-binding specificity and the kinetic properties of the GTPase reaction have been investigated for the MnmE proteins from E. coli and T. maritima. Both proteins show specificity toward guanine nucleotides. MnmE has a relatively low affinity for GTP and GDP combined with a very high intrinsic GTP hydrolysis rate, which is stimulated by a factor of 20 in the presence of potassium ions (Table 2) (40, 180, 183, 278). Considering this combination of parameters, it is predicted that the GTPase cycle can proceed without any external GAPs or GEFs. GTP hydrolysis by MnmE, but not GTP binding or the formation of a transition-state complex using GDP and AlFx, is impaired at an acidic pH, suggesting that the chemistry of the transition-state mimic is different from that of the true transition state and that some residues critical for GTP hydrolysis are severely affected by a low pH (183).
Table 2.
Biochemical parameters of GTPases described in this review
| GTPase | KdGTP (μM) | KdGDP (μM) | kcat (min−1) | Km (μM) | Reference(s) |
|---|---|---|---|---|---|
| MnmE (with K+) | 1.5–280 | 0.62–4.1 | 7.8–26 | 12–833 | 40, 171, 180, 183, 228, 229, 278, 282 |
| MnmE (without K+) | 5.82 | 0.57 | 0.33 | 53 | 180, 229 |
| Era | 2.8–5.5 | 0.49–1 | 0.0029–0.2 | 9–430 | 46, 244, 291 |
| Der | 4.7–8.3 | 1.6–1.8 | 0.11–1.17 | 110–143 | 20, 115, 148, 214 |
| YihA | 27 | 3 | Extremely low | ND | 152 |
| Obg | 1.2–9.4 | 0.5 | 0.0061–0.312 | 5.4–18 | 159, 236, 250, 272 |
| YchF | ND | ND | 0.213–0.329 | 25,100–57,100 | 97 |
| HflX | 180–194 | 2.8–3.6 | 0.061–0.065 | 12.1–16.1 | 207, 234, 276 |
Ranges of values (strongly dependent on experimental conditions) found in the literature are shown. Kd, dissociation constant; ND, not determined.
Like most bacterial GTPases, MnmE is a so-called HAS (hydrophobic amino acid substituted for catalytic glutamine)-GTPase, which means that it has a hydrophobic amino acid in lieu of the catalytic glutamine of classical GTPases (Q61 in Ras) (Fig. 3). The hydrolysis of GTP proceeds by a nucleophilic attack of a water molecule. In canonical GTPases, the catalytic glutamine stabilizes the transition state and orients the attacking water molecule (182). Mutations of this highly conserved glutamine residue have been reported to be oncogenic. A Q61E mutant of H-Ras p21 has a 20-fold-higher rate of GTP hydrolysis than the wild-type protein, whereas the substitution of Q61 by other amino acids reduces the GTPase rate (251). Although in HAS-GTPases, the substituted hydrophobic residue is positioned away from GTP (182), hydrolysis proceeds efficiently (8). In general, in HAS-GTPases, the potentially catalytic residue may be presented (i) from a different region of the G domain (in cis), (ii) from a domain adjacent to the G domain (in cis), or (iii) from an interacting protein (in trans) (182). In the case of MnmE, a catalytic glutamic acid at position 282 in the G domain that activates or orients the nucleophilic water via a bridging water molecule was identified (229).
In most Ras family GTPases, the transition state is further stabilized by a so-called arginine finger that in most members is supplied by a GAP. The positively charged arginine reduces the flexibility of amino acids located in switches I and II and counters the development of a negative charge at the phosphate groups of GTP during the hydrolysis reaction. In α subunits of trimeric G proteins, the catalytic arginine is provided in cis from a helical domain of the GTPase polypeptide (229). MnmE does not use an arginine finger to drive catalysis, although R252 as well as other residues in the G2 motif (249GTTRD253) may play a role in stabilizing the transition state (171, 183). However, the GTP hydrolysis reaction by the G domains of the E. coli and T. maritima MnmE proteins is stimulated in an analogous way by potassium ions and homodimerization (180, 278). Potassium provides a positive charge into the catalytic site in a position analogous to the arginine finger in the Ras-RasGAP system, thereby stabilizing the transition state. Residues 245TDIA248 in the switch I region are termed the K-loop and are responsible for coordinating the potassium ion and shielding it from the solvent (229). Potassium can be replaced by monovalent cations with an ionic radius in the range of 138 to 152 pm. Cations that are smaller (sodium) or larger (cesium) either do not bind or do not have the effects described for potassium. In the GDP-bound state, MnmE forms a homodimer (via its N-terminal domain) in which the highly mobile G domains face each other in various orientations but are not in close contact (178). The dimerization of the G domains occurs in the GTP-bound state, in the presence of potassium ions, and in turn leads to the activation of the GTPase reaction (180). Dimerization stabilizes the switch regions and orients the catalytic E282 residue, which in turn positions and stabilizes the attacking water (229). These GTPase-driven conformational changes are also necessary for in vivo functioning. Hence, MnmE belongs to the expanding class of GTPases activated by nucleotide-dependent dimerization (87, 178).
The dimerization of the G domains with the concomitant activation of the GTPase reaction is further enhanced by complex formation with MnmG (also known as GidA, for glucose-inhibited division A). MnmG interacts with MnmE to form an α2β2 heterotetramer that is stabilized when MnmE is bound to a GTP analog or to GDP-AlFx, mimicking the transition state (179, 283). The other way around, MnmG binding induces large conformational changes in MnmE, thereby stabilizing the GTP-bound form, inducing G domain dimerization, and stimulating GTP hydrolysis (180). In the α2β2 complex, an extensive patch of positive charges is apparent, which is absent on the surface of MnmE, suggesting that in the complex, MnmG is mainly responsible for tRNA binding (179, 198, 199). MnmG can thus be considered a new type of regulatory protein that acts to coordinate the GTPase cycle of MnmE while MnmE tunes the enzymatic modification of tRNA by the MnmG/MnmE complex (21).
The effects of potassium and MnmG on the GTPase activity are additive. However, potassium stimulates GTP hydrolysis much more potently than MnmG (21, 180). B. subtilis YqeH, another member of the HAS-GTPases, also uses potassium to achieve GTP hydrolysis, but it does not require dimerization for activity (8). Since Era and Der also belong to the HAS-GTPases, it was suggested that they too show potassium-dependent GTPase activation (229).
Growth rate.
A Tn10 transposon insertion mutant of mnmE in E. coli has a reduced growth rate in LB or LB containing glucose. In minimal medium (conditions of slower growth), there was no significant difference between the parental and the mnmE::Tn10 strains (26).
Role in tRNA modification.
tRNA contains a high proportion and a large variety of modified nucleosides. Some positions are more prone to modification than others, and especially position 37 (immediately 3′ of the anticodon) and position 34 (wobble base) show a wide variety of often complex modifications. In E. coli, uridine at position 34 is always modified, and the modification is of either the xo5U type (derivatives of 5-hydroxyuridine) or the xm5(s2)U(m) type (derivatives of 5-methyluridine, 5-methyl-2-thiouridine, or 5-methyl-2′-O-methyluridine). These wobble base modifications likely function in the codon recognition process. Modified nucleosides of the xm5(s2)U(m) type are found in tRNAs reading A or G in the third position of the codon in mixed-codon family boxes that code for two amino acids (101).
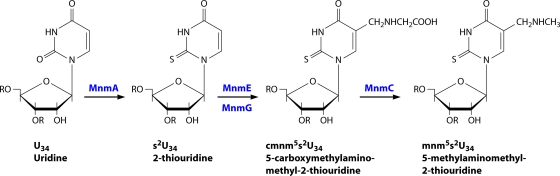
In E. coli, MnmE and MnmG form a functional α2β2 heterotetrameric complex that controls the formation of 5-carboxymethylaminomethyluridine (cmnm5U) in the wobble position of , , , , , and . In certain tRNAs, the cmnm5 group is demodified to 5-aminomethyl (nm5) and subsequently methylated in an S-adenosyl-l-methionine-dependent step to produce methylaminomethyl (mnm5). Both reactions are carried out by the same enzyme, called MnmC (35). Thiolation in the 2-position of the wobble uridine is carried out by the mnmA gene product (Fig. 6) (189).
Fig. 6.
Uridine modification by MnmE. MnmA (in collaboration with other proteins) carries out thiolation at the 2-position of the wobble uridine, whereas an α2β2 heterotetrameric complex formed by MnmE and MnmG independently catalyzes the first step of the cmnm5 modification at the 5-position. MnmC has two enzymatic activities that transform the cmnm5 intermediate into the final mnm5 modification in certain bacteria. (Adapted from reference 283 by permission of Oxford University Press.)
An initial model for the tRNA modification reaction catalyzed by MnmE/MnmG proposed an MnmE-bound 5-formyl-tetrahydrofolate as the donor of the first carbon of the cmnm5 group, where the carboxymethylamino group is due to the subsequent incorporation of glycine via a Schiff base intermediate (228). More recently, another model was proposed, where glycine reacts first on a 5,10-methylene-tetrahydrofolate derivative, after which the total cmnm is transferred to the C5 position of the wobble uridine (189). The FAD/NADH bound to MnmG catalyzes subsequent oxidoreduction steps. Both models proposed a cysteine, coming from either MnmE [C(I/L/V)GK motif] or MnmG, as a catalytic residue that activates the C5 of the uridine via a nucleophilic attack on the C6 position (199). Active GTP hydrolysis by the MnmE G domains, rather then just the binding of GTP, is required for the tRNA modification reaction (171, 180). The binding of nonhydrolyzable GTP analogs or a mutation that impairs the GTPase activity abolishes tRNA modifications in vivo and in vitro. However, it is assumed that the G domains do not participate directly in tRNA binding or in the chemical steps of the tRNA modification reaction. Rather, a model has been proposed where the conformational changes triggered by GTP hydrolysis (i.e., G domain opening and closing) are relayed to the C-terminal C(I/L/V)GK motif of MnmE and/or throughout the MnmE-MnmG heterotetramer to tune certain events in the tRNA modification reaction.
tRNA from E. coli mnmE mutants was shown to carry a 2-thiouridine (s2U) instead of mnm5s2U at the wobble position. The general view is that these modifications of nucleotides in the anticodon loop are important for tRNA recognition by cognate aminoacyl-tRNA (aa-tRNA) synthetases and for accurate mRNA decoding (2). This is extremely important in mixed-codon box families (glutamic acid, glutamine, lysine, leucine, and arginine), for which the base pairing of U with C or U would lead to a misincorporation of amino acids. According to the “4-way wobbling” theory, an unmodified U34 can recognize all four codons. The wobble modifications restrict and facilitate the codon recognition to NNA/NNG in the ribosomal A site, where the xm5U modification especially contributes to increasing the codon interaction for NNG, while 2-thiolation favors the interaction with NNA (77, 142). However, the lack of the mnm5U modification in an E. coli mnmE mutant does not reduce the in vivo aminoacylation levels of tRNAGlu, tRNALys, and tRNAGln. The lack of the s2U34 modification causes a 40% reduction in the charging level of tRNAGlu, while the charging of tRNALys and tRNAGln is less affected. The lack of either modification does not affect mischarging or mistranslation (101, 143). Curiously, the misreading of asparagine codons (AAU/C) by tRNALys(AAA/G) was greatly reduced in E. coli mnmE mutants containing the hypomodified s2U34 instead of the fully modified mnm5s2U34 (101). It has been suggested that tRNAs containing hypomodified U34 are slow in certain steps of the translation cycle prior to peptidyl transfer, allowing a longer time for proofreading. The modifications would then increase the efficiency (“speed”) of the tRNA in these steps.
tRNA modifications also improve reading frame maintenance and prevent errors due to translational +1 frameshifting. tRNA hypomodification in mnmE mutants enhances peptidyl-tRNA slippage by decreasing the rate at which the complex is recruited to the A site (26, 27, 263). In contrast to their impact on +1 frameshifting, tRNA modifications have no or only a minimal stimulatory effect on −1 frameshifting (264).
Other functions.
MnmE has been implicated in other cellular functions, possibly as a secondary effect of altered translation efficiencies.
MnmE is involved in cold adaptation, as a transposon insertion mutant in mnmE of Pseudomonas syringae causes a cold-sensitive growth phenotype, and mnmE expression is induced at low temperatures (240, 241).
Furthermore, MnmE regulates glutamate-dependent acid resistance in LB with glucose (96, 222). Glutamate decarboxylase (GadA/GadB) is used to consume intracellular protons, and a glutamate:γ-amino butyric acid (GABA) antiporter (GadC) expels GABA in exchange for extracellular glutamate. GadA/B transcription and translation are regulated by MnmE, and both types of regulation require GTPase activity. MnmE regulates transcription by affecting the expression of GadE (an essential activator of the gadA and gadBC genes) from its P2 promoter. Furthermore, MnmE affects gadA and gadB expression posttranscriptionally. The MnmE requirement for GadA/B production is dependent on the presence of glucose, suggesting that glucose metabolism represses a separate, MnmE-independent, induction pathway (96).
Finally, MnmG and MnmE are necessary for Streptococcus pyogenes virulence. mnmG and mnmE mutants have no obvious in vitro growth defect and a nearly normal global transcription profile, but their expression levels of multiple secreted virulence factors are reduced due to the impaired translation efficiencies of at least one key transcription regulator (RopB) (53).
Role of MnmE in tRNA modification is conserved in eukaryotes.
In Saccharomyces cerevisiae, the homologs of MnmE and MnmG are Mss1 and Mto1, which also form a complex (26). Mss1 was first identified as a nuclear-encoded mitochondrial GTPase involved in the splicing of COX1 introns (subunit of cytochrome c oxidase). In an mss1 mutant, maturases encoded in the intronic regions of the COX1 pre-mRNA are not translated, leading to an accumulation of incompletely spliced transcripts (69). Mss1 and Mto1 are responsible for modifications of the wobble uridine of mitochondrial tRNALeu and tRNATrp to the corresponding cmnm5Um derivative. This modification enhances the capacity of these tRNAs to translate codons terminating in either A or G, and hypomodified tRNAs show a reduced cognate codon-decoding efficiency and are less efficient at decoding codons ending in G. The disruption of yeast mss1 or mto1 results in reduced oxygen consumption, but growth is not altered (262). A PR454 mutation (paromomycin resistance) in the 15S mitochondrial rRNA (mt-rRNA) gene combined with a mutation in either mss1 or mto1 renders yeast cells respiratory deficient (69). This is probably due to a less accurate translation in the paromomycin-resistant background compounded by the effects on the translation of the mss1 and mto1 mutations (26, 56, 262). Alternatively, the Mss1/Mto1 complex was also reported to interact with 15S mt-rRNA and might play a role in optimizing mitochondrial protein synthesis in yeast, possibly by a proofreading mechanism (56). The combination of mss1 or mto1 null mutations with mutations in other genes involved in the decoding process also has a synergistic effect (269).
The human homologs of MnmE and MnmG are called GTPBP3 and MTO1, respectively. The respective human genes can complement the respiratory-deficient phenotype of yeast mss1 and mto1 mutant cells carrying the PR454 mutation, implying that human GTPBP3 and MTO1 are structural and functional homologs of yeast Mss1 and Mto1. GTPBP3 and MTO1 localize in the mitochondria and are ubiquitously expressed in various human tissues, with a markedly elevated expression level in tissues with high metabolic rates (158). Intriguingly, however, the human homologs incorporate a taurine molecule rather then a glycine molecule into human mitochondrial tRNAs, leading to a taurinomethyluridine (τmU) derivative (246).
The GTPBP3 N-terminal domain mediates potassium-independent dimerization. Like its bacterial homolog, GTPBP3 exhibits a moderate affinity for guanine nucleotides, although it hydrolyzes GTP at a 100-fold-lower rate. GTP and potassium induce the dimerization of the GTPBP3 G domain, but the dimerization of the G domain does not stimulate GTPase activity. The partial inactivation of GTPBP3 by small interfering RNA (siRNA) reduces oxygen consumption, ATP production, and mitochondrial protein synthesis, while the degradation of these proteins increases slightly. It also results in mitochondria with defective membrane potential and increased superoxide levels (269).
C5 taurine modifications at the wobble U position do not occur in mitochondrial tRNALeu(UUR) with either an A3243G or U3271C mutation or in mitochondrial tRNALys with an A8344G mutation. The resulting interference with the translation process leads to mitochondrial encephalomyopathic diseases, namely, MELAS (mitochondrial encephalomyopathy and lactic acidosis with stroke-like episodes) and MERRF (myoclonic epilepsy and ragged red fiber) (280, 281). Hypomodification does not lead to the mistranslation of noncognate codons by the tRNA (e.g., phenylalanine for lysine), but it does cause an almost complete loss of translational activity for cognate codons. The anticodon base modification defect disturbs codon-anticodon pairing in the mutant tRNALys, leading to a severe reduction in mitochondrial translation that eventually results in the onset of MERRF (137, 246, 279). Furthermore, the hypomodification of mitochondrial tRNA caused by a GTPBP3 or MTO1 mutation leads to nonsyndromic deafness in patients with an A1555G mutation in 12S mt-rRNA (corresponding to the 15S mt-rRNA PR454 mutation in yeast), providing further evidence that the modification of mitochondrial tRNA plays a role in human diseases (39).
Era (Bex, Sgp, or Pra)
Era is one of the best-studied bacterial GTPases with described roles in cell cycle regulation, carbon and nitrogen metabolism, and ribosome assembly. The protein has been suggested to cycle between the bacterial membrane and the ribosomes in response to certain trigger factors, thereby providing a checkpoint for ribosome maturation. Era stands for E. coli Ras-like protein (3), although studies have shown that it is not more closely related to eukaryotic Ras proteins than are other GTP-binding proteins, such as EF-Tu (16, 46). The B. subtilis homolog is called Bex, a homonym of BecS (Bacillus era–complementing segment), and Era is also known as Sgp (Streptococcus GTP-binding protein) and Pra (Pseudomonas Ras-like protein). Era is found in eubacteria (except Chlamydia and Mycobacterium), archaea, and higher eukaryotes but not in fungi (32, 153, 188). Although it was reported to be an essential protein in E. coli (170, 249), it seems to be dispensable for growth in Streptococcus pneumoniae and Staphylococcus aureus (285) and in certain strains of B. subtilis (181). Era is highly conserved, illustrated by the fact that an E. coli mutant can be complemented by era from Francisella tularensis, Streptococcus mutans, or Coxiella burnetii (203, 293).
Genetic organization.
In the Betaproteobacteria and Gammaproteobacteria, including E. coli, era is transcribed from the rnc operon, encoding RNase III, Era, and RecO (Fig. 4). RNase III is a double-stranded-RNA-specific endoribonuclease involved in rRNA and mRNA processing and degradation, while RecO is involved in the recombination of circular plasmids (RecF recombination pathway) and the repair of UV damage to DNA (16, 249). The era gene is essential, while rnc and recO are dispensable (249). Furthermore, the structure, sequence, function, and regulation of the Salmonella enterica serovar Typhimurium and E. coli rnc-era-recO operons are conserved (9).
RNase III and Era are coupled not only transcriptionally but also translationally, as the era Shine-Dalgarno (SD) sequence is located inside rnc. The rnc operon is autoregulated by RNase III, which cleaves a site in a 5′-noncoding stem-loop region, thereby reducing the half-life of the transcript. Other RNAs compete with rnc for processing by RNase III, suggesting that Era synthesis is correlated to both the growth rate and the level of macromolecular synthesis (16). However, the synthesis of RNase III and Era was later shown to increase with the growth rate, and the regulation of synthesis is posttranscriptional and independent of RNase III (32).
Cellular localization and concentration.
The mean number of Era molecules per B. subtilis cell is estimated to be 3,000 (188). In E. coli, Era comprises 0.01% of the total protein synthesis (46), leading to fewer than 500 molecules per cell (16, 172), amounts that are typical for regulatory proteins (32).
E. coli Era is located primarily in the cytosol, with an estimated 20 to 30% of all proteins associated with the membrane fraction (3). Era is localized at the cytoplasmic surface of the inner membrane, a location characteristic for membrane signaling proteins (94, 163, 170). In addition, Era occurs in patches that correspond to potential sites of septation (poles, midpoints, and halfway between poles and midpoints) (95). Era from S. mutans shows an increased association with the membrane fraction under conditions of elevated temperatures, acidic pH, or stationary-phase growth (13). Studies of S. pneumoniae showed that the C-terminal part but not GTPase activity is required for binding to the cytoplasmic membrane (103, 291).
Protein structure.
E. coli Era is a 34-kDa protein (3) comprising two functional domains: an N-terminal GTP-binding domain that resembles p21 Ras and a unique C-terminal domain containing a KH domain with an αββααβ topology (Fig. 5) (47). KH stands for K homology, referring to the presence of this domain in heterogeneous nuclear ribonucleoprotein K (98). E. coli Era occurs as a dimer in the crystal structure, and dimerization was hypothesized to be required for functional signaling through interactions with rRNA (47). More recently, structures of Era in complex with GDP (from E. coli) and a ternary complex with a GTP analog and a short RNA fragment (from Aquifex aeolicus) have been solved (258). These structures show that the release of GDP does not cause significant conformational changes. In contrast, GTP binding or hydrolysis causes dramatic changes in the conformation of the switch loops of the G domain as well as in the RNA-binding KH domain.
Era is autophosphorylated in a GTP-dependent manner at either T36 or S37 (242) and possibly at S34 (203). Autophosphorylation was also shown for the Pseudomonas aeruginosa homolog (54).
The QTTR sequence in the G2 motif of the G domain (Fig. 3) is characteristic of the Era GTPase subfamily (181). An E. coli mutant lacking the G2 motif due to a deletion from A40 to G49 displays a dominant negative phenotype. The deletion decreases the affinity for GTP and increases the Km 5-fold (235). T42A and T43A substitutions do not impede GTP binding but increase the Km 12-fold (235). A P17R substitution in the G1 motif causes a 4- to 5-fold decrease in GTPase activity compared to that of wild-type Era while not altering GDP and GTP binding (30, 31). Interestingly, proline is the only amino acid that can replace glycine in the corresponding position 12 of Ras without causing the protein to become oncogenic. An E. coli strain expressing only the P17R-mutated allele has a reduced growth rate at 25°C, 37°C, and 42°C, and these cells are defective in the cell cycle (reflected by the presence of four nucleoids), possibly at the initiation of cell division. An Era P17V mutant was shown to be cold sensitive for GTPase activity and growth (156). Furthermore, Pillutla and colleagues described K21R, S34P, I35F, P211T, and S213P mutations and the N18D-K282R double mutation as being inhibitory to Era functioning (203).
The Era C terminus contains a KH domain with a type II fold that has been implicated in 16S rRNA binding (102, 258). Mutations in the Era KH motif decrease rRNA binding and abolish Era function in vivo (128). Also, the structure of A. aeolicus Era showed that a small fragment of the 16S rRNA is bound to the C-terminal KH domain in a GTP-dependent way (258). Moreover, studies using a mutant Era protein lacking the C terminus showed that this domain is needed for binding to the cytoplasmic membrane (103). rRNA binding is inhibited by liposomes, implying that the membrane- and rRNA-binding sites of Era overlap (103), and it was suggested previously that the C terminus is responsible for sequestering Era in the cell membrane when it is not bound to rRNA (103, 175).
The presence of the C terminus is required for the complementation of an E. coli era mutant (102, 291), and both GTPase activity and the RNA- and membrane-binding activities of Era are essential for proper functioning (103). There seems to be a complicated interplay between Era's G domain and the C terminus. rRNA binding by Era was reported not to require GTPase activity (103), but it seems to depend on GTP binding (258). Mutations in L66 (located in switch II of the G domain) decrease rRNA binding but not GTPase activity in E. coli, suggesting that the G domain regulates RNA binding in response to cellular cues (128). On the other hand, the C terminus is involved in the regulation of the GTPase activity, as the removal of the KH domain reduces GTP hydrolysis activity but not GTP binding (103). Curiously, an Era E200K mutant has an impaired function in ribosome biogenesis without losing its ribosome binding, autophosphorylation, or GTPase activity (122).
GTPase cycle.
Purified Era from E. coli shows specificity for GDP, dGTP, and GTP but not for UTP, CTP, ATP, GMP, cGMP, or dATP (46). Guanine nucleotide binding and exchange by E. coli Era are fast (exemplified by the 10- to 50-times-higher dissociation constants for GDP and GTP and the 20-times-higher Km than those of Ras), while hydrolysis proceeds much more slowly (Table 2) (46). Moreover, guanine nucleotide association and exchange are independent of Mg2+ (244). This suggests that the exchange of guanine nucleotides plays a significant role in Era function (46, 244). The intrinsic GTP-hydrolyzing activity of Era is stimulated 3- to 12-fold in the presence of 16S rRNA (176, 258) and by the addition of 30S ribosomal subunits (164). Era binding to 16S rRNA was reported not to increase GTP binding, seemingly conflicting with the finding that GTP binding increases the affinity for rRNA (175, 258).
Growth rate and cell cycle regulation.
Although Era overexpression reduces the expression levels of other cellular proteins (46), overproduction up until it comprised 5% of the total cellular proteins was reported not to alter growth (170). On the other hand, the disruption of the Listeria monocytogenes era-like gene lmo1462 reduced the growth rate (12), and a B. subtilis era deletion mutant showed a severe growth defect (181). Furthermore, era antisense RNA expression in S. mutans results in decreased growth under environmental stress conditions (44°C, an acidic pH, or high osmolarity) (219).
Two chromosomal mutations, rnc15 and era1, were shown to suppress the defect in chromosome partitioning observed for E. coli carrying the mutations dnaG2903 and parB in the dnaG gene, encoding DNA primase (30, 31). rnc15 carries an insertion of an IS1 element in the leader region of the rnc operon. This insertion reduces cellular levels of Era, RNase III, and RecO. The era1 mutant has a P17R substitution that causes a decrease in the GTPase activity compared to that of wild-type Era while not altering GDP and GTP binding. The suppression of the dnaG mutant phenotype is not fully understood. It has been suggested that the era mutations result in the overexpression of dnaG or play a more direct role in DNA replication. Alternatively, the mutations could cause a general slowdown of cell growth and delay progression through the cell cycle, thereby providing time for the defects caused by dnaG mutations to be corrected (30, 31). Interestingly, although the era1 mutation suppresses cell cycle mutations affecting chromosome replication and partitioning, the P17R substitution cannot suppress cell division mutations or mutations resulting in a defect in DNA replication initiation (32).
Several studies pointed to a role for Era in cell cycle regulation. B. subtilis cells depleted of Era are elongated 1.5 to 2 times compared to the corresponding wild-type cells and are filled with diffuse nucleoid material. Depletion mutants showed an excess initiation of DNA replication, suggesting that Era negatively controls the initiation of chromosome replication (181, 188). In E. coli, the depletion of Era resulted in a loss of viability, growth inhibition, elongation, and a defect in the formation of septa and constrictions. DNA segregation seems to proceed normally. This phenotype resembles that of an ftsZ mutant, suggesting a role in cell division (94).
An E. coli rnc40 mutant carries an insertion in the leader region of the rnc operon that renders the expression of the operon dependent on the presence of tetracycline (248). Phenotypic characterization of the rnc40 mutant and an era1 mutant carrying a P17R substitution showed that the limiting of Era functioning causes a stop in cell division at the predivisional two-cell stage. Cells exhibit temporary growth arrest at this stage, corresponding to an increased generation time. This suggests that Era is required for progression through the cell cycle and that it functions as a checkpoint for cell division at a point after nucleoid segregation but before cell division (32). It was suggested that Era determines the time for cell division to be completed under different growth conditions and that a threshold level of Era GTPase activity is required to initiate septation (32). Strains defective for both RNase III and Era have additional defects in chromosome partitioning, leading to DNA being condensed as one large nucleoid in the cell center. Furthermore, FtsZ rings are rarely present in double mutants (32). In E. coli rnc40 and era1 mutants, the cell division defect is different from the phenotype associated with fts mutations that typically result in elongated, multinucleate filaments (32).
Role in ribosome assembly.
In Era-depleted cells, a 16S rRNA precursor accumulates, and the intracellular levels of unassociated 30S and 50S subunits increase relative to levels of 70S ribosomes (121). The addition of Era to a cell-free protein synthesis system obtained from Era-depleted E. coli cells did not restore the translation defects associated with the depletion of Era, implying that Era is not directly involved in protein synthesis (221). Era was shown to bind 16S rRNA and the 30S ribosomal subunit in vitro, and both GTP and GDP were first reported to inhibit this interaction (221). However, GTP binding was later shown to be a prerequisite for rRNA recognition by Era (258). 16S rRNA binding of Era proceeds mainly via its C-terminal KH domain (176), and recently, a detailed description of the interactions between the 30S ribosomal subunit and both the N- and C-terminal domains was reported (233). The observed interaction with several assembly elements suggests that Era is involved in the assembly and maturation of the 30S subunit (233). Era does not seem to facilitate the binding of specific proteins but rather seems to moderate a global maturation event (36). In this respect, Era was shown to serve as a chaperone for the processing and maturation of 16S rRNA (258).
Era and S1 (a ribosomal protein known to directly influence SD/anti-SD interactions) cannot coexist on the 30S subunit, and the binding of Era prevents the recruitment of mRNA. Therefore, the binding of Era locks the subunit in a conformation that is not favorable for an association with the 50S subunit (233). Interactions with the 30S subunit increase the GTPase activity of Era (176, 258), and it was suggested that GTP hydrolysis is required for the release of Era from the fully assembled 30S subunit, allowing the incorporation of S1, which ultimately leads to the formation of the 70S complex (233). In conclusion, Era is involved in the processing and maturation of the 30S subunit to a conformational state that is suitable for an association with the 50S subunit, and it serves as a checkpoint for ribosome assembly (233, 258). Era was suggested to play a central role in signaling between cell division and the initiation of protein synthesis (221).
Other clues for the role of Era in ribosome maturation come from suppressor studies. A mutation in era suppresses the phenotype of a temperature-sensitive mutant of rpsL, encoding the 30S ribosomal subunit protein S12 (192). A cold-sensitive era mutant showing a defect in cell division at low temperatures, while DNA replication and nucleoid segregation stay normal (154), was rescued by the overexpression of the 16S rRNA methyltransferase gene ksgA (165). KsgA does not influence cellular concentrations of Era, and the exact cause of the suppressor phenotype remains unknown. Possibly, high levels of KsgA enhance the cell division frequency or rescue the phenotype by protein-protein interactions (165). The overproduction of Era (the wild type or a mutant lacking the G2 effector domain) partly suppresses the cold-sensitive growth phenotype of an rbfA mutant (121). Like that of Era, the KH domain of RbfA associates with the 30S subunit, and an rbfA deletion mutant accumulates a precursor of 16S rRNA, with a corresponding decrease in the levels of polysomes and an increase in levels of 30S and 50S subunits relative to those of 70S monosomes (121). RbfA binds in the immediate vicinity of Era's binding position on the 30S subunit, suggesting that Era can partially compensate for RbfA function through an interaction with common structural elements of the ribosome (68). However, Era and RbfA were shown to interact with 16S rRNA at different sites, and RbfA cannot replace Era, suggesting that the latter is involved not only in 16S rRNA maturation but also in other essential cellular functions. Furthermore, rbfA but not era can complement a rimM (ribosome maturation factor M) deletion mutant. RimM has also been described to bind the free 30S subunit, and a deletion mutant accumulates 16S rRNA precursors. These findings suggest a hierarchy among these three proteins (Era, RfbA, and RimM) in the 16S rRNA maturation process (121). The observation that rbfA is essential for cell growth only at low temperatures can be explained by the fact that Era can complement its function only at high temperatures (122).
Role in energy metabolism.
Measurements of intracellular guanine nucleotide pools indicated that the presence of a functional era gene enables S. mutans to maintain high-energy GTP as the major guanine nucleotide (13). Era associates with Ndk (nucleoside diphosphate kinase) and Scs (succinyl coenzyme A [CoA] synthetase), two enzymes involved in high-energy phosphate metabolism (128). During the exponential growth phase, Ndk is cytoplasmic and produces all of the nucleoside triphosphates (NTPs). In the stationary phase, Ndk is membrane associated and predominantly performs GTP synthesis (54). At moderate concentrations of Era, complex formation between Era and Ndk increases the formation of GTP and dGTP and diminishes the synthesis of CTP, UTP, dCTP, and dTTP (54). Era proteins from both E. coli and P. aeruginosa interact with Pk (pyruvate kinase), and Era-Pk complex formation results in increased GTP synthesis (54). These findings suggest that Era is involved in GTP generation by the membrane-associated Ndk-Pk complex. Furthermore, it is likely that the association of Era with Ndk and/or Pk restricts its intrinsic GTPase activity (54).
Other findings linking Era to cell metabolism have been described. The phenotype of a temperature-sensitive era mutant with reduced GDP and GTP binding (119) was partly rescued in a strain with a truncated rpoN or a ptsN null mutation (208). RpoN is required for the transcription of genes needed for nitrogen assimilation and fixation, while ptsN encodes IIANtr, homologous to IIAFru of the phosphoenolpyruvate:sugar phosphotransferase system. The suppression of the phenotype implies a role for Era in both carbon and nitrogen metabolism (208). However, ptsN is located in the rpoN operon, and complementation analyses implicated ptsN as playing the major role in suppression (208). Era was demonstrated to reduce the capacity for the utilization of carbohydrate intermediates such as pyruvate (155), while IIANtr enhances this capacity (208). Furthermore, 3-phosphoglycerate and glyceraldehyde-3-phosphate, key intermediates in glycolysis, modulate the GTPase activity of Era (176).
Other functions.
Era has been implicated in other cellular functions. The L. monocytogenes era-like lmo1462 gene is required for cells to adhere to stainless steel surfaces (12). In B. subtilis, stationary-phase era expression is Spo0A dependent, and deletion mutants exhibit severely impaired spore formation. It is not clear whether this is due to a specific role in spore formation or whether it is a nonspecific consequence of the severely impaired growth of the deletion mutant (181). E. coli YggG, a membrane-associated heat shock protein and a putative zinc metalloprotease, was shown to interact with wild-type Era and with the P17R substitution mutant Era1. Furthermore, the transcription of yggG was upregulated in response to stress caused by the Era1 mutant protein, thereby promoting the growth of E. coli. The disruption of yggG enhances the stress caused by Era1 (110, 111). Era was also shown to interact with the nucleoside triphosphate pyrophosphohydrolase MazG, and the interaction was stronger in the presence of GDP. The interaction between Era and MazG does not modulate their individual GTP hydrolysis activities (286). Finally, the depletion of Era resulted in the depressed synthesis of the heat shock proteins DnaK, GroEL, GroES, D33.4, and C62.5 and a lack of thermal induction of ppGpp levels (155).
Eukaryotic homologs.
Many properties of bacterial Era seem to be conserved in eukaryotes. The human Era homolog is called H-ERA (human Era) (4) or ERAL1 (Era G protein-like 1) (29). The gene encoding ERAL1 is located in a chromosomal region where the loss of heterozygosity is often associated with various types of cancer, making it an attractive candidate for a tumor suppressor gene (29). ERAL1 comprises three domains: an N-terminal MSD (mammalian ERA-specific domain) that is possibly cleaved by posttranslational modifications, a GTP-binding domain, and a C-terminal KH domain that binds RNA (4). ERAL1 is located in the mitochondrial matrix, where it was shown to act as a mitochondrial RNA chaperone for 12S mt-rRNA and to be involved in the assembly of the 28S small mitochondrial ribosomal subunit (70, 260). The siRNA knockdown of ERAL1 causes a myriad of defects, including decreased mitochondrial translation, redistributed ribosomal small subunits, reduced levels of 12S mt-rRNA, elevated levels of mitochondrial superoxide production, decreased mitochondrial membrane potential, and inhibited growth of HeLa cells with an accumulation of apoptotic cells (260). Moreover, the overexpression of ERAL1 carrying substitutions in the G1 motif induces the apoptosis of HeLa cells, suggesting that ERAL1 is an apoptosis regulator. Apoptosis is dependent on the presence of the ERAL1 C terminus and is suppressed by the expression of the antiapoptotic proteins BCL-XL and BCL-2 (4). ERAL1 depletion leads to apoptosis, but cell death was shown not to be the result of any appreciable loss of mitochondrial protein synthesis or a reduction in the stability of mitochondrial mRNA (70).
Chicken ERA is located in the cytosol and bound to RNA. It regulates G1-phase progression via an as-yet-unknown molecular mechanism that involves RNA recognition. The depletion of chicken ERA diminishes the growth rate and increases apoptosis (93).
In plants, the Antirrhinum majus (snapdragon) Era homolog ERG (ERA-related GTPase) is expressed in dividing or metabolically active cells and is required for embryonic viability. It has a crucial role in plant growth and development, possibly by influencing mitochondrial division. ERG is also found in Arabidopsis thaliana (120). Erl1 (Era-like 1) from the rice fungus Magnaporthe oryzae localizes to the nucleus and is required for root virulence (106).
Der (EngA, YphC, or YfgK)
Members of the Der subfamily of GTPases contain a unique structure in which two G domains are tandemly repeated. These proteins are required for large-ribosomal-subunit biogenesis, and the ribosome association is fine-tuned by the nucleotide occupancy of the G domains. Der (double-Era-like domains) is also known as YphC, YfgK, and EngA (essential neisserial GTP-binding protein A). Der was shown to be essential in E. coli (115), Neisseria gonorrhoeae (174), B. subtilis (188), S. pneumoniae, S. aureus, and Haemophilus influenzae (285). It is conserved in all eubacteria (188) and some eukaryotes, but it is absent in archaea (115, 277).
Genetic organization.
The genomic context of der is not conserved. In most members of the Gammaproteobacteria, including E. coli, der is in a locus with hisS (histidyl-tRNA synthetase), yfgM (uncharacterized), yfgL (lipoprotein), and yfgJ (uncharacterized) (Fig. 4) (7). In B. subtilis, der is predicted to be the first gene of a two-gene operon with gpsA, encoding glycerol-3-phosphate dehydrogenase. gpsA has no effect on cell growth when depleted (223). N. gonorrhoeae der is in an operon with the rdgC gene, which is involved in pilus-dependent colony phase variation and in pilin antigenic variation (174).
Cellular localization and concentration.
A Der-green fluorescent protein (GFP) fusion protein localizes to the cytoplasm in E. coli (270). However, a recent study showed E. coli Der to localize to the membrane (150). The mean number of Der molecules per B. subtilis cell is estimated to be 7,000 (188).
Protein structure.
Der contains two homologous GTPase domains (GD1 and GD2) connected by a highly acidic linker peptide of 22 to 46 amino acids. In addition to the tandemly repeated G domains, the protein contains a unique C terminus of 90 to 122 amino acid residues (Fig. 5) (115).
In T. maritima, the two Der G domains are 31% identical in primary structure. Furthermore, the G2 motif is well conserved between Der proteins and between both G domains inside a single Der protein (Fig. 3), suggesting that both G domains bind the same effector molecule (115). Both GD1 and GD2 are required for cell growth at low temperatures (117), while at high temperatures (42°C), either one of the two domains is dispensable (118). However, neither an S16A nor an S217A (S216A in E. coli MG1655) substitution in GD1 or GD2 supports viability in a conditional der null mutant, and mutations targeting either G domain have a significant and cooperative impact on the GTPase activity of the protein as a whole (20). Curiously, the T. maritima NKAE sequence of the G4 motif in GD1 deviates from the nearly universal (N/T)KXD sequence. This is true for approximately half of the members of the Der GTPase family (214). A similar mutation in Ras and EF-Tu changes the specificity of the proteins from GTP to XTP (98).
The C-terminal domain of Der is highly basic in amino acid composition and has a high pI value, suggesting that this region is involved in an interaction with nucleic acids (118). Furthermore, it was suggested previously that members of this class of domains facilitate protein-protein interactions (214).
The crystal structures of Der from T. maritima and B. subtilis show a three-domain architecture with two G domains that do not interact directly with each other but pack at either side of the C-terminal KH-like domain (Fig. 5) (214). The N-terminal G domain is composed of six β-strands and five α-helices, while the second G domain consists of seven β-strands and six α- helices, in a structural arrangement typical of TRAFAC GTPases. The C-terminal central domain is composed of a 3-stranded β-sheet with two α-helices stacked to one side of the sheet in an αββαβ topology. Interactions between GD1 and the KH domain are influenced directly by the GTP/GDP state of the protein (190). These conformational changes expose a large patch of positive charge in the presumed “on” state, which is absent in the presumed “off” state (it should be noted, however, that no structures in the presence of a GTP analog have been reported thus far). These conformational changes might thus regulate the binding of RNA. In contrast, the GD2-KH domain interface is distal to the GTP/GDP-binding site of GD2, suggesting that the two G domains make different contributions to the regulation of RNA binding by Der (190, 214).
GTPase cycle.
T. maritima Der shows specificity for the binding of GTP, GDP, and dGTP but not GMP, ATP, CTP, and UTP (115). Optimal conditions for the GTPase assay were determined to be pH 7.5 in 400 mM KCl and 5 mM MgCl2 at 70°C (Table 2) (115). Both G domains show GTPase activity, but the hydrolysis activity of isolated GD1 approximates that of the full-length protein and is twice that of isolated GD2. An asparagine-to-aspartate substitution in the G4 motif of GD1 significantly decreases the GTPase activity of the protein, whereas a similar substitution in GD2 slightly increases the overall GTPase activity (214). S. Typhimurium Der binds GDP with a 20-fold-higher affinity than that for GTP, and the two GTPase domains in Der show a 5.3-fold difference in the affinity for GDP (148), with GD2 having a higher affinity for nucleotides (214, 253). As opposed to results obtained with T. thermophilus Der, steady-state kinetic studies using S. Typhimurium Der mutants with serine-to-alanine substitutions in the G1 motif of GD1 and/or GD2 suggest a strong positive cooperativity between both GTPase domains, whereby binding or hydrolysis in one domain considerably stimulates the other (20).
Although pathogenic bacteria such as Pseudomonas, Salmonella, and Yersinia encode cytotoxins (ExoS, ExoT, SptP, and YopE) functioning as GAPs for their host Rho GTPase, bacterial GTPases generally do not require GAPs. YihI was recently reported to be the first prokaryotic GAP, although it stimulates Der's GTPase activity only weakly (about 2-fold). YihI seems to recognize the GTP-bound state of both G domains. It is specific for Der, as it does not stimulate GTP hydrolysis by Era or Obg. The deletion of yihI accelerates growth, and the yihI gene product negatively regulates cell growth during the lag phase. Overexpression causes an accumulation of rRNA precursors and an aberrant ribosome profile that is similar to that of Der-depleted cells. A model has been presented in which YihI associates with GTP-bound Der at the beginning of exponential growth. This interaction activates GTP hydrolysis, which causes Der to dissociate from the ribosomes. yihI expression is suppressed later in the cell cycle, allowing Der to associate with 50S ribosomal subunits, where it performs a role in ribosome maturation (114). In contrast to Der, YihI is not conserved in all eubacteria, and so far, it has been identified only in members of the Gammaproteobacteria (247).
Growth rate and cell cycle regulation.
The E. coli growth rate correlates with the amount of Der in the cell (118), and the depletion of Der leads to elongation, filamentation, and defective chromosome segregation (115). Furthermore, B. subtilis der knockout mutants showed an increase in cell length, nucleoid condensation, and an abnormally curved cell shape (188).
Role in ribosome assembly.
A first clue linking Der to ribosome assembly came from a complementation study of an rrmJ mutant. The E. coli RrmJ (FtsJ) heat shock protein functions as an rRNA methyltransferase that modifies position U2552 of 23S rRNA in intact 50S ribosomal subunits (41). An in-frame deletion of the rrmJ gene leads to severe growth defects and causes a significant accumulation of ribosomal subunits at the expense of functional 70S ribosomes (33). The overexpression of Der or Obg (but not Era) complements the phenotype of an E. coli rrmJ mutant, causing a wild-type ribosome profile and normal growth (250). ΔrrmJ suppression by Der requires two intact G domains (116).
Later, Der was shown to interact with ribosomes in vitro, and this interaction was stabilized by the nonhydrolyzable GTP analog guanosine-5′-(β,γ)-imidotriphosphate (GMPPNP) but not GDP or GTP, which is probably rapidly hydrolyzed by Der (223). E. coli Der interacts with the 50S ribosomal subunit and copurifies with five ribosomal proteins (20, 38). The 50S interaction proceeds via the KH domain of Der (116). The depletion of Der leads to the accumulation of precursors of both 23S and 16S rRNAs and of 50S and 30S ribosomal subunits, with a concomitant reduction of polysomes and 70S ribosomes (20, 118). Cells depleted of Der have 50S subunits in which L16, L27, and L36 are missing (223). Furthermore, the depletion of Der affects late assembly proteins (L9, L2, L6, and L18) in the 50S subunit, suggesting that it is involved in the biogenesis and stability of this subunit (118). Upon the maturation of the 50S subunit and binding to the 30S subunit, Der dissociates from the ribosome (118). Both GTPase domains have a cooperative function in ribosome stability and/or biogenesis (20). S. Typhimurium Der interacts with the ribosomal structural proteins S7 and S9 and was suggested to ensure the proper delivery of rRNA-modifying enzymes to the appropriate regions of the ribosome (148).
Apart from its interaction with 50S subunits, Der was shown to interact with the 30S subunit, the 70S ribosome, 23S rRNA, and 16S rRNA. The fine-tuning of the ribosome interactions seems to proceed through the GTP binding of the different G domains of Der. The key requirement for any Der-ribosome association is GTP binding to GD2. In this state, Der displays a weak 50S association, which is stabilized when, additionally, GD1 binds GTP. The exchange of bound GTP with GDP at GD1 results in interactions with the 50S, 30S, and 70S subunits. Therefore, it appears that GD1 employs GTP hydrolysis as a means to regulate the differential specificity of Der for either the 50S subunit alone or the 50S, 30S, and 70S subunits (253). Der was suggested to interact with the membrane when bound to GDP in both domains or bound to GDP in one G domain with the other domain in the empty state. A model was proposed in which Der cycles between a binding site on the ribosome that is recognized by Der·GTP and a binding site on the membrane that is recognized by Der·GDP (150).
Der and the stringent response.
Upon nutrient limitation, bacteria initiate a complex set of cellular responses called the stringent response. This results in increased protein degradation, amino acid synthesis, and carbohydrate metabolism and reduced protein and nucleic acid synthesis. The stringent response is modulated by ppGpp (GDP 3′-diphosphate) and pppGpp (GTP 3′-diphosphate), often collectively denoted (p)ppGpp. These nucleotides are synthesized by the phosphorylation of GDP and GTP, respectively, using ATP as a phosphate donor. During nutrient limitation, the cellular level of (p)ppGpp increases significantly. In E. coli, two closely related enzymes, RelA and SpoT, are responsible for the synthesis and degradation of (p)ppGpp. RelA is a synthetase, whereas SpoT is a bifunctional enzyme with both hydrolase and synthetase activities (25, 130).
E. coli Der N321D or N118D mutants show polysome defects and do not support growth at low temperatures (118). This growth defect is restored by the overexpression of relA. The overproduction of RelA does not affect the expression of Der N321D. Moreover, (p)ppGpp synthesis but not the interaction of RelA with ribosomes is essential for the suppressor phenotype. Possibly, the downregulation of rRNA synthesis by excess (p)ppGpp concentrations allows for the partly defective Der N321D protein to process rRNA properly. In addition, an excessive amount of (p)ppGpp effectively inhibits the GTPase activity of E. coli Der, suggesting that (p)ppGpp is able to bind to Der at the GTP-binding site and regulate its activity under stress conditions. RelA cannot suppress a null mutation of der (117).
Other functions.
A recent study pointed toward a possible role for Der in cell wall assembly (150). Furthermore, Der seems to be involved in pathogenesis. A polar mutation in yfgL from S. enterica serovar Enteritidis showed decreased colonization of chicken spleen and cecum, reduced invasiveness of macrophages and enterocytes, lower virulence, reduced secretion of the SPI-1 and flagellar proteins, and reduced motility (7). Also, intracellular Chlamydophila pneumoniae and Chlamydia trachomatis serovar D persisters showed an increased transcription of der. Chlamydial persisters are morphologically distinct, nonreplicative cells that show a loss of infectivity with the capacity to reactivate to productive infection once the persistence-inducing conditions are removed. In vitro, chlamydial persistence has been induced by the addition of antibiotics; the depletion of iron, glucose, and essential amino acid levels; and treatment with cytokines such as gamma interferon (205).
Eukaryotic homologs.
Der homologs are present in some but not all eukaryotes. Examples include slime molds, diatoms, oomycetes, green and brown algae, mosses, and flowering plants. No functional studies have been reported so far.
YihA (YsxC or EngB)
Like Era, YihA was suggested to participate in a checkpoint mechanism that ensures a correct coordination of cell cycle events. It also participates in the assembly of the 50S ribosomal subunit. YihA is widely distributed among the three domains of life but appears to be absent from high-GC Gram-positive eubacteria and Mycobacterium tuberculosis (65, 152, 188). yihA is essential for growth in B. subtilis (188, 209), E. coli (11, 65), S. pneumoniae, S. aureus, and H. influenzae (58, 285) but not in Mycoplasma genitalium or Mycoplasma pneumoniae (113).
Genetic organization.
In E. coli, yihA is found on the chromosome in proximity of polA (encoding DNA polymerase I) and spf (encoding an unstable RNA that modulates the level of DNA polymerase I activity), with yihA being transcribed in opposite direction (Fig. 4) (129). In all Gram-positive bacteria, yihA is located downstream of lonA or clpX, both of which encode class III ATP-dependent heat shock proteases (209). Bacillus brevis (123) and B. subtilis (213, 225) yihA genes are located at the end of a predicted operon with the lonA gene, encoding an ATP-dependent serine endopeptidase. B. subtilis yihA is likely to be transcribed together with lonA, since its start codon overlaps with the lonA coding sequence, and no yihA-specific promoter or transcriptional initiation site has been detected. Furthermore, lonA and yihA show similar transcription patterns, including induction by heat and other stresses (213). Given the connection to LonA, it was suggested that YihA is involved in an intracellular signaling process that controls protein turnover in response to changing environmental conditions (216).
Cellular localization and concentration.
An E. coli YihA-GFP fusion protein localizes diffusely throughout the cytoplasm (270). The mean number of YihA molecules per B. subtilis cell is estimated to be 1,000 (188).
Protein structure.
The primary structure of E. coli YihA reveals that it contains a single GTPase domain flanked on both sides by extra stretches of 30 and 60 amino acids at the N and C termini, respectively (Fig. 5) (152). YihA appears to be a monomer in solution. All YihA proteins share a DXXG (F/Y)G sequence in their G3 motif (Fig. 3) (153). The crystal structure of B. subtilis YihA has been solved in the apo form, in complex with GDP, and in complex with the nonhydrolyzable GTP analog GMPPNP. These structures show that the protein folds into a single globular domain with a fold that resembles those of other TRAFAC class GTPases. However, the central β-sheet contains seven β-strands, with an additional strand provided by the N-terminal residues of the protein. The switch I and switch II regions of B. subtilis YihA become ordered and disordered, respectively, in the “closed” or “on” GTP-bound state and disordered and ordered, respectively, in the “open” or “off” GDP-bound conformation. There is a unique, conserved cluster of basic residues that lies adjacent to the nucleotide-binding site. Similar clusters are known to interact with hydroxyl groups or with ion species such as phosphate or sulfate (216). The C-terminal 23 residues of B. subtilis YihA comprise a highly charged region and are essential for protein function (209).
GTPase cycle.
The kinetic and mechanistic properties of the GTPase reaction of YihA homologs have not been studied in detail. E. coli YihA binds guanine nucleotides specifically and does not bind adenine nucleotides (Table 2) (152).
Growth rate and cell cycle regulation.
Knockouts of B. subtilis yihA show an increase in cell length, nucleoid condensation, and an abnormally curved cell shape (188). However, no distinguishable alterations were observed for YihA-depleted S. aureus cells by light or transmission electron microscopy (58). In E. coli, yihA is necessary for normal cell division, and YihA depletion leads to a severe reduction in the growth rate and extensive filamentation. The filaments display no defect in chromosome partitioning, indicating that the defect is primarily in septation. Filamentation is due mainly to the insufficient synthesis or stability of FtsZ and can be suppressed by the overexpression of ftsQ, ftsA, and ftsZ and, to some extent, by ftsZ alone. FtsQ, FtsA, and FtsZ do not suppress the slow growth of cells depleted of YihA. It was suggested that YihA participates in a checkpoint mechanism that ensures a correct coordination of cell cycle events (65).
Role in ribosome assembly.
S. aureus YihA copurifies with ribosomes (especially the 50S subunit) and interacts with S2, S10, and L17 and also with the β′ subunit of the RNA polymerase. The depletion of YihA leads to a decrease in mature ribosomes, suggesting that YihA is essential for ribosome assembly or stability (58). YihA is also required for large ribosomal subunit biogenesis in B. subtilis, and depleted cells have 50S ribosomal subunits that lack the L16, L27, and L36 proteins. It was suggested previously that the association of YihA with the 50S presubunit allows the incorporation of the missing proteins (273). Purified B. subtilis YihA binds 70S ribosomes and, preferably, 50S subunits (more specifically, L1, L6, and L7/L12 proteins) but not 30S subunits. The strength of these interactions is increased in the presence of either GTP or GDP (223), with the strongest interaction established in the presence of a nonhydrolyzable GTP analog (273). The charged amino acids in the C terminus of B. subtilis YihA were suggested to mediate interactions with RNA (216). YihA most probably participates in the assembly and/or processing steps of the 50S subunit (273).
Eukaryotic homologs.
The plant A. thaliana has at least two homologs of YihA, one 219 and the other 318 amino acid residues in length. The length of the smaller homolog is in the range of prokaryotic YihA proteins (190 to 219 amino acids). The length of the longer YihA homolog is similar to that of the human and S. cerevisiae YihA orthologs. The N terminus of the longer YihA homolog contains a putative transmembrane helix between amino acid residues 11 and 29, and the protein is currently the only putative membrane-bound member of the YihA family. The human YihA ortholog harbors a specific deletion within the G1-G3 effector region (209).
THE OBG-HflX SUPERFAMILY
The OBG-HflX superfamily comprises a second large group of proteins of the TRAFAC class of P-loop GTPases (Fig. 1). The OBG family has four subfamilies: Obg, Nog1, DRG, and YchF. Except for Nog1 homologs, all members share a glycine-rich sequence (GAX2GXGXGX3l, where l is one of the aliphatic residues I, L, or V) immediately after the G3 motif DX2G in the switch II region. Obg, DRG, YchF, and HflX also share a YXFXTX5G sequence in the G2 motif switch I region between the Walker A and B motifs (153). Bacteria encode two OBG proteins (Obg and YchF), whereas archaea possess YchF and two OBG proteins of unknown function that are related to DRG and Nog1. In addition to YchF, eukaryotes generally encode four OBG proteins. The bacterial and the eukaryotic mitochondrial Obg proteins are likely to be homologs, as sequences flanking the GTP-binding domain are conserved (162). Eukaryotes also possess a nucleolar OBG protein, Nog1, that is critical for the biogenesis of the 60S ribosomal subunit. Two DRG proteins that are associated with translating ribosomes were identified in S. cerevisiae (275). Plants encode an additional protein from the Obg subfamily that localizes to the chloroplast (14), while mammals encode an additional nucleolus-localized Obg (108).
Obg (ObgE, CgtA, or YhbZ)
Obg is a versatile GTPase that has been implicated in the stress response, ribosome assembly, DNA replication, sporulation, and morphological development. Obg stands for spo0B-associated GTP-binding protein (254). The protein is also known as ObgE (Obg of E. coli) (138) and CgtA (Caulobacter GTP-binding protein A [166] or common GTP-binding protein [61]). Obg is conserved in eubacteria and widely present across eukaryotes (188). The gene has been shown to be essential for viability in B. subtilis (81, 254), Vibrio cholerae (231), Vibrio harveyi (237), Streptomyces coelicolor (197), Caulobacter crescentus (166), E. coli (138), S. pneumoniae, S. aureus, and H. influenzae (285).
Genetic organization.
In virtually all bacteria, the obg gene is physically linked to rplU and rpmA, encoding the ribosomal proteins L21 and L27, respectively. In the Gammaproteobacteria, the order is rplU–rpmA–yhbE (predicted inner membrane permease)-obg (Fig. 4). obg is not preceded by yhbE in other Proteobacteria. In B. subtilis, the order is rplU-open reading frame (ORF)-rpmA–spo0B–obg (61, 81, 166, 196, 237). The significance of this chromosomal arrangement is unknown, and the downstream genes are not conserved.
Cellular localization and concentration.
Although C. crescentus Obg shows no transmembrane segments (166) and B. subtilis Obg is probably not membrane associated (227), S. coelicolor and M. tuberculosis Obg proteins are membrane-bound proteins (197, 217). E. coli Obg is localized in the cytosol (218) and is partly associated with the membrane (138).
The B. subtilis spo0B locus is maximally expressed during the mid-log phase of growth, and protein levels decline slightly as cultures reach the stationary phase (81). The cellular concentrations of Obg molecules in E. coli and V. harveyi also decrease upon entry into the stationary phase (218, 237), but E. coli Obg levels are not largely altered with the initiation of the stringent response (202). In S. coelicolor, there is a sharp decrease in cellular Obg concentrations just after the onset of aerial mycelium development and at the end of vegetative growth (197). C. crescentus Obg levels remain at a low, constant level throughout the cell cycle, indicating that in this species, the regulation of Obg function is not at the level of total proteins in the cell but is likely due to differences in the activation state of Obg (166). Curiously, M. tuberculosis Obg expression levels increase markedly from the early log phase to the stationary phase, with a drop in expression levels at the late stationary phase (217).
It was estimated that 50 to 100 copies of B. subtilis Spo0B are present in the stationary phase (81). On the other hand, there are approximately 200 and 6,000 Obg molecules per cell in stationary-phase cells of C. crescentus and exponential-phase cells of B. subtilis, respectively (161, 188). Obg is abundant in E. coli, with 34,000 molecules in log-phase cells and 5,600 molecules in stationary-phase cells, numbers as high as those of ribosomes and nucleoid proteins (138).
Protein structure.
Within the Obg subfamily, there are two distinct classes of proteins: (i) an N-terminally extended form that possesses a 150-amino-acid glycine-rich N terminus preceding the Ras-like domain and (ii) a C-terminally extended form that lacks the N-terminal extension but has additional residues at the C terminus. All bacterial Obg proteins known to date possess the N-terminal extension, while both classes of Obg-like proteins have been identified in eukaryotes (159). Bacterial Obg proteins usually also contain an additional C-terminal domain (Fig. 5). Crystal structures of the C-terminally truncated Obg protein from B. subtilis in the apo form and bound to GDP and ppGpp and of full-length Obg from Thermus thermophilus in the apo form have been solved. The latter structure shows a three-domain arrangement with an N-terminal glycine-rich domain, a central G domain, and a C-terminal domain that is highly variable among Obg proteins from different species (Fig. 5) (34, 144).
The bacterial N-terminal glycine-rich domain does not share structural, functional, or sequence similarity to any other known protein, and therefore, it is also referred to as the Obg fold. It consists of six left-handed type II helices that pack together in both parallel and antiparallel fashions and an eight-stranded β-barrel that forms the contacts between the N-terminal domain and the G domain. Few surface residues are conserved within the Obg fold, indicating that the overall structure and shape of this domain are more important than the surface-exposed residues (34). The Obg fold contains a region with high similarity to the α1 chain of mammalian collagen that possibly serves to anchor Obg at a particular location in the cell (254). Using molecular dynamic simulations, the N terminus was recently shown to be the most favored part for potential protein-protein interactions (151). The overexpression of B. subtilis Obg carrying a G92D mutation (G93D in E. coli) impairs cell growth and the ability of Obg to associate with ribosomes (145). An E. coli Obg mutant with an N-terminal deletion of 5 amino acids, including the conserved D5 residue, does not support growth (138). Moreover, a temperature-sensitive B. subtilis obg allele carrying two missense mutations (G79E and D84N) in the Obg fold has been described. The D84N mutation is not sufficient for the temperature-sensitive phenotype and might even be dispensable (139). The observed effects of Obg(Ts) are not caused by impaired GTPase activity, and the mutation most probably impairs the ability of Obg to interact with other proteins of a downstream signal transduction pathway (272). More recently, it was demonstrated that C. crescentus Obg lacking the 160 N-terminal amino acid residues does not support growth, although this terminus plays no significant role in guanine nucleotide binding or GTPase activity. Here too, the N terminus was suggested to be involved in the anchoring of Obg to its cellular target via protein-protein interactions (160).
The central G domain of Obg is highly conserved (Fig. 3) (34), and the binding of guanine nucleotides is critical for viability in C. crescentus (67). The G domain consists of the classical six-stranded β-sheet and five α-helices. The switch elements mediate contacts between the G domain and the N- and C-terminal domains, suggesting mechanisms that transduce the nucleotide-bound state of the G domain to the other domains (34). The structural changes in B. subtilis Obg due to GTP binding were studied by using molecular dynamic simulations, which suggested that the angle between the N-terminal domain and the G domain is significantly different in the GTP-bound form compared to other nucleotide-bound forms (apo, GDP, and GDP plus Pi) (151). However, the interaction between putative targets of Obg and the N-terminal domain of C. crescentus Obg is not affected by structural differences between the GTP- and GDP-bound states (160).
Several mutations within the G domain have been characterized. In C. crescentus Obg, T192A and/or T193A substitutions within the G2 motif modestly reduce binding to GDP and significantly reduce the binding affinity for GTP, especially in the T193A mutant. GTP hydrolysis is impaired in a T193A mutant but not in a T192A mutant, and a T193A mutant does not support growth in vivo, suggesting that hydrolysis plays a critical role in Obg functioning (161). An S. coelicolor P168V mutant was assumed to have a defect in hydrolysis (197), but this was later contradicted (67). Unlike the analogous mutation in Ras that significantly impairs both intrinsic and GAP-stimulated hydrolysis (91, 141), the P168V mutation in C. crescentus Obg does not affect GTP hydrolysis and is therefore not activating. The P168V protein shows a modest reduction in the binding affinity for GDP as well as a modest increase in GDP exchange. This reduced affinity does not influence Obg's essential function, as both P168V and P168G mutants support growth. Both mutants, however, show a modest cold-sensitive phenotype. The expression of Obg with a P168R, G171A, K172N, S173N, N280Y (N283Y in E. coli), or N280K substitution does not support the growth of a C. crescentus strain carrying a repressed chromosomal copy of obg. Similarly, a G171A-K172A-S173A triple mutant and a D213A-G216A double mutant do not support growth. An S173N mutant is significantly impaired for GDP and GTP binding in vitro, consistent with a critical role for this residue in guanine nucleotide binding (67). In general, conserved amino acids in the GTP-binding pocket are important for viability, but analogous mutations in Obg have distinctly different effects on biochemical properties compared to those of their Ras counterparts (67).
In vitro, less than 1% of B. subtilis Obg autophosphorylates with GTP (not ATP), most probably at a histidine residue in switch II (272). However, most Obg homologs do not have a histidine residue in switch II (166). Correspondingly, C. crescentus Obg phosphorylation is base labile, indicating that Obg is phosphorylated on either a serine or a threonine residue. Neither T192 nor T193 is the site of autophosphorylation in C. crescentus Obg (161). Recently, M. tuberculosis Obg was demonstrated to be autophosphorylated as well (217).
The Obg C-terminal domain is not widely conserved between Obg family members. In B. subtilis, the C terminus contains a TGS motif, named after its occurrence in members of three protein families (threonyl-tRNA synthetase ThrRS, GTPase, and SpoT) (34). The T. thermophilus HB8 C-terminal domain of Obg has a unique fold (the Obg C–terminal, or OCT, fold) that does not show sequence similarity to TGS domains. The C-terminal domain was suggested to function as a GEF under some biological conditions (144). E. coli Obg with a C-terminal deletion of 34 amino acids supports growth (138). In contrast, C. crescentus Obg lacking the C terminus does not support growth, and the C-terminal addition of a hemagglutinin tag results in retarded growth (162). The C-terminal end of Obg was demonstrated to be important for cell elongation upon the overexpression of obg in V. cholerae (231).
Noteworthy, the reported structure of T. thermophilus Obg is considerably different from the structure of B. subtilis Obg (34), with significant conformational changes in the switch regions and drastic differences in the orientation of the Obg fold vis-à-vis the G domain. In the crystal packing, the T. thermophilus G domain interacts with the C-terminal domain of the adjacent molecule in the crystal by a head-to-tail contact. This interaction might indicate multimerization in the crystal, but analytical ultracentrifugation revealed that Obg is a monomer in solution (144). Less than 10% of purified E. coli Obg migrates as a dimer, and GTP, rather than GDP, facilitates Obg dimerization (218, 274). Multimers are not detected in a crude extract, suggesting that in vivo, the Obg protein is monomeric (138). V. cholerae Obg was also demonstrated to form dimers (211).
GTPase cycle.
Obg has a relatively low GTP hydrolysis rate, and the rate of turnover is much lower than the rates obtained for the α subunits of heterotrimeric G proteins, which are in the range of 3 to 5 min−1. Obg binds both GTP and GDP with a moderate affinity, it shows a high guanine nucleotide exchange rate, and the spontaneous dissociation of both GTP and GDP is extremely rapid (103- to 105-fold faster than that of eukaryotic Ras-like GTPases) (Table 2). These features suggest that the nucleotide occupancy of Obg is controlled mainly by the intracellular levels of guanine nucleotides (159, 250, 274). Rapid exchanges of guanine nucleotides and a modest affinity for nucleotides are universal properties of the Obg family. Similar biochemical properties have also been found for other bacterial GTPases predicted to play roles in ribosome function, such as Era, Der, YihA, and YjeQ (236). Interestingly, V. harveyi Obg displays a 10-fold more rapid GTP hydrolysis rate than is typical for other family members, perhaps reflecting the diversity and specificity of bacterial ecological niches (236). In S. Typhimurium, Obg binds GDP with a 3.8-fold-higher affinity than that for GTP (148).
The GTPase activity of Obg is inhibited by GDP, while KCl enhances the hydrolysis rate, and Mg2+ is essential for the binding of GTP (but not GDP) and hydrolysis. Obg is a specific GTP- and GDP-binding protein, and ATP cannot substitute as a substrate (236, 272). Remarkably, ppGpp was found in the active site of the G domain in the crystal structure of B. subtilis Obg. As ppGpp copurified with the protein, this could be explained either by specificity for ppGpp or by an artifact of the starvation conditions encountered upon the induction of Obg expression (34). However, E. coli Obg was later found to bind ppGpp with a biologically relevant affinity in vitro, implicating ppGpp as an in vivo ligand of Obg (202). Strangely, in one study, ppGpp appeared to increase the rate of GTP hydrolysis at low concentrations and to inhibit hydrolysis at higher ppGpp concentrations (34). In a later study, ppGpp was found to have a purely inhibitory effect on GTPase activity, which can be explained most simply through competitive binding inhibition (202).
Growth rate and cell cycle regulation.
In E. coli, the growth rate correlates with the intracellular Obg concentration (218). Moreover, the depletion of Obg affects the growth of V. cholerae. The overexpression of V. cholerae Obg also affects the growth and cell morphology of E. coli but only the cell morphology (not the growth) of V. cholerae (231).
A first observation hinting at a possible role for Obg in DNA replication came from studies with a heat-sensitive B. subtilis obg mutant. At the nonpermissive temperature, this mutant continues to divide for two generations and stops growing only after three doublings. Therefore, the mutation was suggested not to impair septation and the basic processes of RNA and protein synthesis, implicating a possible role in chromosome replication (139). As discussed below, later findings showed that Obg is involved in the first two steps of the cell cycle, namely, the initiation of DNA replication and nucleoid segregation, but not in cell division per se. Moreover, Obg is required for progression through the cell cycle.
(i) Role in DNA replication.
The synthesis of DNA, RNA, and proteins is not significantly affected in E. coli cells overexpressing obg. However, overexpression leads to elongated cells with an impaired regulation of synchronization of DNA replication initiation (73). Later, high expression levels of obg in E. coli were reported to produce mild overreplication and the accumulation of an odd number of chromosomes. Obg overexpression also causes SeqA foci, normally localized to replication forks (220), to spread extensively within the cell (83). Moreover, E. coli obg mutant cells have a higher DNA content than do wild-type cells, and Obg was suggested previously to regulate the total DNA content within E. coli cells (202).
Derivatives of plasmids λ, F, R1, R6K, and RK2 show diminished DNA replication in an E. coli strain expressing a dysfunctional Obg. The replication initiator protein DnaA is involved in the replication of these plasmids, and levels of DnaA proteins were demonstrated to decrease in Obg-depleted cells (261). The expression of dnaA was also shown to be impaired in a temperature-sensitive E. coli obg mutant, and the resulting deficiency in DnaA activity causes an inhibition of chromosomal DNA replication initiation (238). The ectopic expression of dnaA partially restores plasmid replication as well as growth in liquid medium and DNA synthesis defects but not the elongated morphology of a temperature-sensitive obg mutant (238, 261).
Obg most probably does not affect DNA synthesis per se. It is more likely to influence replication initiation indirectly by regulating the synthesis or activity of one or more replication factors, including DnaA (261). (p)ppGpp couples DNA replication with growth rate control through the expression of the replication initiator protein DnaA, and therefore, the role of Obg may be indirect (67, 127). However, the temperature-sensitive obg mutant is capable of evoking the stringent response, suggesting a more direct regulation of DnaA by Obg (238). Obg senses the physiological state of the cell by responding to the GTP/GDP ratio, and it may control the efficiency of ribosome biogenesis and the translation of crucial regulatory genes, including dnaA. This control mechanism, enhanced by the regulation of dnaA transcription, might couple DNA replication to ribosome biogenesis and translation (238).
(ii) Role in chromosome segregation.
The depletion of B. subtilis Obg results in an increased cell length, an abnormally curved cell shape, and nucleoid condensation (188). At the restrictive temperature, E. coli cells with a temperature-sensitive obg allele show a deficient partition of nucleoids. Mutant cells are elongated, with the nucleoid located in the middle of the cells. The overexpression of obg also leads to aberrant chromosome partitioning, which results in elongated and anucleate cells (73, 138). These findings suggest that Obg is involved directly in chromosome partitioning (138). However, the phenotype might also result from a decreased expression of the dnaA operon (67, 238) or from impaired ribosome function, resulting in the inhibition of protein synthesis in Obg depletion strains (218), indicating an indirect role for Obg in chromosome partitioning.
Both (p)ppGpp+ and (p)ppGpp0 cells of E. coli show elongation phenotypes upon Obg overexpression, suggesting that the effect on cell morphology may not be due to the alteration of cellular (p)ppGpp levels (231). Moreover, the E. coli Obg depletion phenotype is not equivalent to that produced by a sustained stringent response. The induction of the stringent response blocks DNA replication in E. coli, whereas Obg deletion strains continue to replicate, producing polyploid filamentous cells. Although this does not rule out a role for E. coli Obg in the regulation of the stringent response, it does suggest an additional and essential function for the E. coli protein (82).
In V. harveyi and V. cholerae, the depletion of Obg does not result in either cell elongation or a DNA-partitioning phenotype. Therefore, it was suggested previously that V. cholerae Obg is functionally similar to V. harveyi Obg and that a role in cell division and/or DNA replication is not a core function for all Obg proteins (211, 237).
(iii) Progression through the cell cycle.
E. coli cells depleted of Obg have intact chromosomes, as evidenced by the absence of SOS response induction. However, some signal for cell cycle progression is evidently lacking. DNA replication and cell growth continue throughout the depletion to generate elongated and extensively polyploid cells. It was concluded that Obg is required to allow chromosome segregation and subsequent cell cycle events (82). Furthermore, C. crescentus cells carrying a temperature-sensitive obg allele show a G1-to-S-phase arrest at the nonpermissive temperature, suggesting that Obg is necessary not only for DNA replication but also for progression through the cell cycle (67). The temperature-sensitive mutant does not exhibit additional ribosome defects at the nonpermissive temperature. Therefore, the essential function of C. crescentus Obg appears to lie in ensuring progression through the cell cycle rather than correct ribosome assembly (67). This was also concluded for E. coli Obg (82).
(iv) Model for the role of Obg in cell cycle regulation.
In high-nutrient environments, bacteria shorten their cell cycle by the initiation of new rounds of replication before the completion of previous rounds (220). The addition of a replication inhibitor (for example, hydroxyurea) causes the rapid depletion of deoxynucleotide pools, resulting in replication fork arrest (252). The arrest of a replication fork risks the convergence of later forks to the stall site, which results in double-strand chromosomal breaks (83).
A model for the role of Obg in the cell cycle was proposed based on studies with replication inhibitors. The depletion of Obg renders V. cholerae cells highly sensitive to hydroxyurea (231). In E. coli, Obg carrying a Tn5 insertion in its C-terminal domain causes increased sensitivity to replication inhibitors. A high proportion of hydroxyurea-treated mutant cells are extended cell filaments with uneven DNA masses in segregated chains. Chromosome segregation occurs aberrantly, and cell division is blocked. Sensitivity to general DNA-damaging agents such as UV radiation is not altered. The phenotype is not caused by the deletion of the last 9 amino acids of the protein but rather by the 68-amino-acid peptide fused to Obg. B. subtilis Obg(Δ22) (22 amino acids of the chromosomal Obg C terminus replaced with 26 amino acids) does not display an enhanced sensitivity to hydroxyurea (145). Wild-type Obg was shown not to merely increase deoxynucleotide pools. It has a direct role in promoting replication fork stability, and it acts in a way complementary to the RecA-dependent SOS response to promote bacterial survival of replication fork arrest. The Obg::Tn5 protein is less stable and present in the cell at lower concentrations than wild-type Obg. Although it has no effect on growth, a P168V mutation also enhances sensitivity to replication inhibitors. Both obg mutants show an accumulation of chromosome breaks and regressed forks and exhibit asynchronous overreplication during normal growth (83).
A model has been proposed in which Obg·GDP mediates interactions that bring about an S-phase checkpoint-like response. Mutants failing to sense this signal cause aberrant replication and cell cycle progression. The likely function of Obg·GTP is to permit chromosome segregation and cell division (83). Obg controls replication initiation, potentially in response to the status of existing replication forks. However, a role for (p)ppGpp levels in controlling replication fork progression was reported previously, and therefore, the role of Obg in replication fork stabilization may be indirect (127).
Role in ribosome assembly.
Several findings link Obg to ribosomes. First, the overexpression of Obg suppresses the phenotype of an E. coli rrmJ deletion mutant (250). In addition, Obg proteins are relatively abundant in log phase and gradually decrease in number toward stationary phase. This expression pattern is reminiscent of the change in ribosomal proteins whose synthesis rate is well correlated with the cell growth rate (218). Furthermore, Obg was shown to physically interact with ribosomes in the following different bacteria.
V. harveyi Obg is associated with the 50S ribosomal subunit (237).
E. coli Obg cosediments with free 50S subunits, with 70S monosomes, and with polysomes. It was shown to interact specifically with L13 (274). In a different study, E. coli Obg was shown to bind to 30S and, preferably, 50S ribosomal subunits but not to 70S ribosomes (218). Purified Obg associates with ribosomal particles only in the GTP-bound form (126), and likewise, rRNA binding by Obg proceeds only in the GTP-bound form (218). A GTP-dependent interaction (either direct or indirect) was established with 16S and 23S rRNAs and with the ribosomal proteins S3, S4, S5, S13, S16, L2, L4, L16, and L17. Other reported interaction partners include the RNA helicase CsdA, ClpA, hypothetical protein 274#5, and the RNA polymerase β and β′ subunits (218).
C. crescentus Obg cosediments with 50S subunits but not with 70S monosomes or translating ribosomes. The interaction is moderately weak and sensitive to the salt concentration and medium composition but is not dependent on the presence of either GDP or GTP. The Obg C terminus is critical for ribosome interaction (162).
S. Typhimurium Obg interacts with the 30S ribosomal proteins S3, S5, and S9 and with the rRNA-modifying pseudouridine synthase enzyme RluD. The interaction of RluD with Obg does not stimulate the GTPase activity of Obg, and the presence of GTP has no effect on the binding of these two proteins. Obg was suggested previously to aid in the specific targeting of rRNA-modifying enzymes to their site of action (148).
B. subtilis Obg coelutes with ribosomal subunits (presumably the 50S subunit) and binds specifically to the ribosomal protein L13 (227). The loss of L11 does not influence the Obg-ribosome interaction (289). The addition of GTP, GDP, or ATP prolongs the Obg-ribosome association, while the inclusion of a nonhydrolyzable GTP analog preserves it. This suggests that in B. subtilis the ribosome association is stabilized by GTP and that Obg is released from the ribosome as a consequence of GTP hydrolysis, possibly induced by the ribosome (289).
M. tuberculosis Obg is present in all three (30S, 50S, and 70S) ribosomal fractions, in more or less equal amounts (217).
Obg is probably not a structural component of the ribosome. The protein is more likely to play a role in ribosomal assembly/stability, or it is involved in monitoring the assembly state of the ribosomes (126, 161, 218). A C. crescentus Obg depletion strain shows a perturbed polysome profile with reduced levels of 70S monosomes and polyribosomes (162). Similarly, a temperature-sensitive mutation in E. coli Obg causes a significant reduction in 70S ribosome levels with a concomitant increase in the levels of the 30S and 50S subunits (126, 218). Obg affects rRNA processing, and depletion strains accumulate a 50S intermediate that lacks L33, L34, and L16 (and perhaps L23) (126, 218). Therefore, Obg seems to be required for the optimal incorporation of certain late-assembly ribosomal proteins into the large ribosomal subunit. Obg either plays a direct role in the assembly pathway or may be required for the efficient recruitment of ribosomal proteins (126). The increasing sedimentation rate observed with 50S intermediates accumulating when YihA, RbgA, Der, and Obg are depleted suggests a preliminary order to the action of these GTPases, with YihA acting before RbgA and Der and Obg being the last GTPase to act on assembling 50S subunits (132). The essential nature of the obg gene does not appear to result from its ribosome function. This was shown by using a temperature-sensitive Obg G80E C. crescentus mutant. At the permissive temperature, the mutant grows slowly and shows a reduction in 50S ribosomal subunits. Surprisingly, at the nonpermissive temperature, G80E cells rapidly lose viability and yet do not display an additional ribosome defect (66).
In eukaryotes, there is a large protein network (involving at least one Obg homolog) that connects ribosome biogenesis, DNA replication, and chromosome segregation (72). Similarly, bacterial Obg was presented as a good candidate to link DNA replication with protein synthesis via its association with the ribosome, the chromosome, and/or the replication forks (19).
Role in the stringent response.
E. coli Obg interacts with full-length SpoT, a protein involved in the regulation of the stringent response through a dual activity of (p)ppGpp synthesis and hydrolysis. More specifically, it was shown that Obg interacts with the N-terminal (p)ppGpp synthetase/hydrolase domain and with the putatively regulatory C terminus of SpoT (274). Obg's ribosome association makes it well positioned to be involved in the control of SpoT function (126, 274), and since Obg was shown previously to bind ppGpp (34, 202), SpoT could affect the activity of Obg by presenting ppGpp (274). These findings raise the possibility that Obg is involved in the stringent response to nutrient limitation.
More recently, a postulated regulatory role for Obg in the stringent response was established (127, 211, 231). In V. cholerae, the depletion of Obg causes increased (p)ppGpp levels and global changes in gene expression that are consistent with the induction of the stringent response (211, 231). Similarly, (p)ppGpp levels increase in an E. coli obg mutant, but the accumulation of (p)ppGpp during amino acid starvation is not affected, providing evidence that Obg regulates (p)ppGpp levels during exponential growth but not during the stringent response. Moreover, Obg is not associated with the 50S subunit or SpoT under conditions in which (p)ppGpp accumulates, and the loss of Obg from the ribosome is necessary for maximal (p)ppGpp accumulation. Obg dissociation under stress conditions might be due to a lower GTP/GDP ratio in the cells, as it was shown previously that E. coli Obg associates with ribosomes and rRNA in a GTP-bound form (126, 218). Alternatively, bound (p)ppGpp might induce GTPase activity (34), or it could be that (p)ppGpp-bound Obg is not ribosome associated (127). A model was proposed in which Obg promotes SpoT (p)ppGpp degradation activity on the ribosome when bacteria are growing in nutrient-rich environments, thereby repressing the stringent response (127).
However, a direct role for Obg in regulating SpoT activity has not been demonstrated, and high (p)ppGpp levels in Obg-depleted strains could be an indirect effect of growth problems in these mutants (202). Interestingly, an E. coli obg mutant shows an increased ratio of pppGpp to ppGpp within the cell during the stringent response, although the total level of ppGpp plus pppGpp is not detectably altered. Like the translation factors IF-2, EF-Tu, and EF-G (see below), Obg may hydrolyze pppGpp directly. An altered pppGpp-to-ppGpp ratio results in a delayed inhibition of DNA replication initiation, a delayed resumption of DNA replication after the release of serine hydroxamate (an inducer of the stringent response), and decreased survival after amino acid deprivation. Therefore, Obg was suggested to be an in vivo effector of the response to amino acid starvation, regulating the timing of the onset and termination of the stringent response (202). The correlation of the pppGpp-to-ppGpp ratio with a delayed change in DNA replication further supports a connection between two functions of Obg often considered to be disparate, namely, its involvement in DNA replication control and its connection to pppGpp (202).
Obg was reported previously to no longer be essential in a V. cholerae relA deletion mutant (211), although this finding was subsequently contradicted (231). Moreover, in the absence of spoT and relA, obg is still an essential gene in B. subtilis (145) and E. coli (82, 127). In conclusion, the essential function of the Obg protein remains unknown.
Role in the response to UV irradiation.
In Deinococcus radiodurans, the deletion of the nos gene (encoding a homolog of mammalian nitric oxide synthase) compromises recovery from UV irradiation, and this defect is substantially alleviated by the overexpression of Obg. Furthermore, NO generated by Nos after UV exposure induces the obg gene (201). The expression of the obg gene is also enhanced after the UV irradiation of E. coli and V. harveyi cells, and moderate overexpression in E. coli enhances survival in both wild-type and dnaQ strains but not in uvrA, uvrB, umuC, and recA mutant hosts. Moreover, levels of the RecA protein are lower in an E. coli obg mutant, and recA gene expression is not increased after the UV irradiation of this mutant. It was suggested that Obg is involved in DNA repair processes by the stimulation of recA gene expression and the resultant activation of RecA-dependent DNA repair pathways (292). Obg is unlikely to be part of the SOS regulon because the increased transcription of the obg gene in UV-irradiated E. coli cells was found to be independent of lexA gene function (59).
Role in sporulation in B. subtilis.
The initiation of sporulation in B. subtilis is controlled in part by the phosphorylation of the transcription factor Spo0A. The depletion of Obg causes a defect in sporulation and in the expression of sporulation genes that are activated by phosphorylated Spo0A, defects that are all relieved by mutations in spo0A that bypass the need for the phosphorelay (268). Obg seems to be necessary for the transition from vegetative growth to stage 0 or stage II of sporulation, but subsequent sporulation stages are unaffected by Obg. The protein was suggested to communicate the intracellular levels of GTP to the sporulation initiation machinery (139). Spores of a temperature-sensitive Obg mutant germinated normally at the nonpermissive temperature but failed to outgrow (139). Interestingly, the N-terminal domain of B. subtilis Obg interacts with CotN, a protein that is secreted into the medium early in sporulation and also incorporated into the endospore (34).
Role in the general stress response in B. subtilis.
In B. subtilis, the stress response σ factor σB is released from its anti-σ factor after a drop in intracellular ATP levels or under environmental stress conditions. σB binds RNA polymerase and directs the resulting holoenzyme to promoters of the general stress regulon. This activation proceeds through a collection of regulatory kinases and phosphatases, the Rsb proteins (regulator of sigma B). Obg interacts with several Rsb proteins, and the depletion of Obg suppresses the activation of σB in response to environmental stress factors but still allows activation by a drop in ATP levels. Obg (or a process under its control) is thought to play a role in the activation of RsbT, the most upstream effector of the stress-induced pathway of σB. The overexpression of Obg has no effect on σB activation, and the requirement for Obg in σB activation is not due to an effect on the transcription factor Spo0A (226). Interestingly, M. tuberculosis Obg was recently demonstrated to interact with the σB anti-sigma factor RsbW homolog UsfX (217).
The loss of ribosomal protein L11 was previously demonstrated to reduce the growth rate significantly and to eliminate the stringent response. Interestingly, a B. subtilis mutant lacking ribosomal protein L11 is blocked in the stress-induced but not in the energy-dependent activation of σB. The Rsb proteins are present in the mutant but fail to be activated by stress (290). Components of the σB stress activation pathway (RsbR, RsbS, and RsbT) were reported previously to coelute with Obg and the 50S ribosomal subunit (227). However, Obg, but not RsbR, RsbS, or RsbT, was later shown to be ribosome associated (146). The interaction of Obg with ribosomes is a possible mediator of the activity of Obg in the stress-dependent induction of σB (227). According to one theory, Obg communicates cellular energy levels to the ribosome. Alternatively, the ribosome might sense regulatory inputs in the cell and communicate these to other components via its interaction with Obg (227). In support of the second theory, the ribosome has been described to function as a sensor for stress factors. The blocking of E. coli translation by the addition of ribosome-specific antibiotics increases the expression levels of heat and cold shock proteins in a manner similar to that resulting from stress conditions (265). Furthermore, amino acid starvation induces the ribosome-mediated activation of relA, leading to the stringent response. These signals could be communicated, perhaps through ribosome-associated Obg, to other components of the cell, including the Rsb proteins (227).
The growth-promoting and stress response activities of B. subtilis Obg can be uncoupled by mutation. Obg G92D overexpression impairs cell growth and the ability of Obg to associate with ribosomes (giving rise to an altered ribosome profile) but fails to block sporulation or the induction of the general stress response. Obg(Δ22) (22 amino acids of the chromosomal Obg C terminus replaced by 26 unrelated amino acids) cofractionates with ribosomes and allows normal growth but blocks sporulation by preventing the activation of Spo0A. This protein also restricts the RsbT-dependent process that activates σB, thereby impairing the induction of the general stress response. Obg likely plays distinct roles in growth promotion and the stress response, and the failure of B. subtilis to properly initiate stress responses in the absence of Obg is not merely the consequence of a growth defect (145).
Role in morphological development in Streptomyces.
The overexpression of Streptomyces griseus obg suppresses the development of the aerial mycelium on solid medium and spore formation in liquid cultures of S. griseus, S. coelicolor, and Streptomyces lividans. Decreasing cellular GTP levels by the addition of decoyinine restores morphological development. Furthermore, the suppression of aerial mycelium formation is enhanced by use of an Obg P168V mutant and diminished by use of an Obg G171A mutant. Although Obg P168V was assumed to have a defect in hydrolysis, this is probably not the case. Obg P168V instead has a decreased affinity for GDP (67). A Ras G15A mutation (corresponding to Obg G171A) confers a dominant negative effect by depleting a guanine nucleotide dissociation stimulator from wild-type Ras, leaving the protein in the GDP-bound state. Neither of these Obg mutants can support growth in an obg deletion strain (197). Physiological differentiation [a direct function of (p)ppGpp] as measured by the production of streptomycin is not influenced by Obg (196), and the overexpression of obg reduces the production of the antibiotic actinorhodin but not undecylprodigiosin (197). It was suggested that by monitoring the intracellular GTP pool size, the Obg protein is involved in sensing changes in the nutritional environment, ultimately leading to morphological differentiation (196, 197).
Other functions.
C. pneumoniae and C. trachomatis serovar D persisters show increased transcription levels of obg. The activation of the stringent response in bacteria leads to the inhibition of DNA replication and cell division and the upregulation of amino acid biosynthesis genes, outcomes that are similarly described for chlamydial persistence. It was hypothesized previously that Obg coordinates the development of persistence after sensing the GTP energy levels of the host cell (18, 204, 205). Chlamydia abortus Obg lacks the Obg C-terminal domain but still cofractionates with the E. coli 50S subunit. The overexpression of chlamydial Obg in E. coli causes growth defects and elongation, although the allele is not capable of complementing an E. coli temperature-sensitive mutant. Obg is thought to have a role in linking the chlamydial stress response to ribosome function and cellular growth (206).
Obg is a broadly conserved and indispensable protein with a putative role in ribosome assembly, and as such, it was suggested to be a promising therapeutic target (57). M. tuberculosis Obg has been considered a potential drug target based on the observation that its C terminus (not present in, for example, E. coli) appears to be disordered. Intrinsically disordered proteins or domains have a significant or near-complete lack of folded structure and an extended conformation with high intramolecular flexibility and little secondary structure. Protein interactions can cause disorder-to-order transitions (48, 75). The M. tuberculosis proteome has a large proportion of disordered regions, probably resulting from the pathogen trying to mimic the host machinery and successfully evading the defense mechanism. Intrinsically disordered proteins are emerging as interesting drug targets, as they have a disproportionately large binding surface and multiple contact points (10).
Eukaryotic homologs.
The yeast Obg homolog is called Mtg2 (mitochondrial GTPase 2). Mtg2 is peripherally localized to the mitochondrial inner membrane facing the matrix compartment and associates with the mitochondrial 54S large ribosomal subunit in a salt-dependent manner. The protein possesses 88 N-terminal amino acids not found in bacterial Obg proteins that are predicted to contain a mitochondrial targeting sequence, two short insertions in the Obg fold, and no additional C-terminal domain. Mtg2 is essential for mitochondrial ribosome function, as a loss of the encoding gene leads to a decrease in mitochondrial translation and the subsequent loss of mitochondrial DNA and lowered levels of mitochondrial ribosomal subunits. Moreover, elevated levels of Mtg2 partially suppress the thermosensitive loss of mitochondrial DNA caused by a mutation in mrm2, encoding an RrmJ ortholog involved in 21S mt-rRNA methyltransferase. Mtg2 does not function in early ribosome assembly. It either is involved directly in the late steps of the biogenesis of the mitochondrial large ribosomal subunit or (less likely) plays an as-yet-undefined role in translation (66). A Schizosaccharomyces pombe homolog (gtp1) was found downstream of the stf1 locus (112).
In humans, two Obg homologs (OBGH1 and OBGH2) are present. Both are capable of complementing the Obg function in E. coli ribosome maturation. The knockdown of the OBGH1 gene induces the elongation of mitochondria. OBGH1 contains a long N-terminal region with putative mitochondrion localization signal, and the protein was shown to localize to mitochondria. On the other hand, OBGH2 localizes to the dense fibrillar compartment region of the nucleolus, and knockdown results in the disorganization of the nucleolar architecture (108). Both OBGH1 and OBGH2 show similar rates of GTP hydrolysis (0.014 ± 0.005 min−1 and 0.010 ± 0.002 min−1, respectively) compared to that of the B. subtilis homolog (Table 2) (34, 108).
There are two Obg homologs in A. thaliana (OBGC and OBGM, localizing to the chloroplasts and the mitochondria, respectively) and three in rice (OBGC1, OBGC2, and OBGM). Chloroplast OBG is much more closely related to bacterial Obg than to eukaryotic homologs, which suggests that among all eukaryotic organisms, their existence is unique to photosynthesizing eukaryotes (14). No Obg subfamily protein in A. thaliana functions in the nucleolus, suggesting that the nucleolus-localizing Obg protein is specific to animals. In plants and yeast, the function of the nucleolar Obg protein is most likely performed by other proteins belonging to the Nog1, DRG, YchF, or HflX subfamily (108).
OBGC (At5g18570, CPSAR1, or AtOBGL [Obg-like GTPase]) is nuclear encoded and expressed in embryos and green tissues throughout development (86). It contains a long N terminus (comprising 207 amino acids) for chloroplast targeting that is absent from bacterial Obg (14, 52). OBGC has a dual localization in the stroma and the inner envelope chloroplast membrane but not in thylakoid membranes. Although it does not contain any transmembrane domain, OBGC could be recruited to the inner envelope membrane by interactions with a membrane protein. Alternatively, it could form an oligomeric complex capable of attaching to the membrane (52, 86). The N terminus (but not the GTP-binding domain) is important for the observed oligomerization by mediating protein-protein interactions (14). The protein has intrinsic GTPase activity that is comparable to that of other Obg subfamily proteins (14, 86). It was suggested that OBGC has a function in chloroplast ribosome biogenesis or protein synthesis (14). However, there are no indications for the colocalization of OBGC with chloroplast ribosomes, since these are found mainly in the stroma and associated with the thylakoid membrane, and there is no clear evidence for their association with the envelope membrane where OBGC is found (52, 86). An obgc null mutant exhibits an embryonic lethal phenotype, suggesting that OBGC is essential for early embryogenesis (14, 52, 86). OBGC is required for the normal organization of mature thylakoid stacks and, ultimately, for embryo development. Mutant embryos are unable to develop thylakoid membranes, resulting in the absence of thylakoid stacks in plastids (52). Also, OBGC is a good candidate for a role in the vesicular traffic between the inner envelope membrane and thylakoids (52).
YchF (YyaF or EngD)
YchF has developed an altered NTP specificity within the family of GTPases and shows a preference for ATP over GTP hydrolysis. The protein is involved in translation, and it functions as a negative regulator of the stress response in plants and the antioxidant response in humans. YchF is widely conserved in all eubacteria, eukaryotes, and archaea (50, 188). The encoding gene is not essential for growth in B. subtilis (188) and S. pneumoniae (80). Previously, the Obg family had been divided into five subfamilies (Obg, Nog1, DRG, YchF, and Ygr210), with the Ygr210-like proteins defining an independent subfamily. Phylogenetic comparisons revealed that the Ygr210 branch and the YchF branch are closely related and group together into one subfamily (140). The YchF and Ygr210 subfamilies form one branch of the family with the YchF proteins present in bacteria and eukaryotes and the Ygr210 proteins present in archaea and fungi (251).
Genetic organization.
E. coli ychF forms a single operon with pth (peptidyl-tRNA hydrolase) (which cleaves peptidyl-tRNAs released abortively from ribosomes during protein synthesis) (Fig. 4). Two transcriptional promoters were identified by primer extension experiments: one located upstream of pth, which presumably gives rise to both the mono- and bicistronic pth transcripts, and the other, preceding ychF, which generates its monocistronic message. RNase E regulates the expression of pth and ychF (60). The gene sequence pth-ychF observed for E. coli is typical of the Proteobacteria, whereas the rest of the sequenced bacterial genomes have pth- and ychF-homologous genes dispersed in different regions (60).
Protein structure.
The crystal structures of H. influenzae YchF (251) and of the human homolog hOLA1 (human Obg-like ATPase 1) (140) show that the protein folds into three distinct structural domains (Fig. 5). The N-terminal domain consists of a classical TRAFAC class G domain with a six-stranded, mostly parallel β-sheet flanked by α-helices on both sides. A second α-helical domain, consisting of two long coiled-coil α-helices and a short α-helix, is inserted between β5 and β6 of the G domain. The third C-terminal α/β domain consists of a mixed β-sheet curved as a half-barrel around an α-helix. This C-terminal domain is completed by an α-helix and a short β-strand, coming from an extension of a loop following β2 of the G domain. The C-terminal domain has a topology typical of the ubiquitin-like superfamily and is structurally related to the TGS domain also found in SpoT and Obg. The homology of the C-terminal and α-helical domains to proteins involved in nucleic acid binding and the presence of clusters of positive charges suggest a binding site for double-stranded nucleic acid in the cleft between these domains (251). Teplyakov et al. suggested previously that GTP/GDP induces conformational changes that could be transmitted to the nucleic acid-binding site to regulate nucleic acid binding (251).
GTPase cycle.
A peculiar feature of YchF is the replacement of the conserved aspartate with glutamate and the lack of lysine in the (N/T)KXD G4 motif (Fig. 3), which normally determines the guanosine specificity of G proteins. This raised questions regarding the nucleotide specificity of YchF (251). In agreement, ATPase activity is a general feature of the YchF subfamily of Obg-like GTPases, and homologs from human, H. influenzae, and S. cerevisiae (called hOLA1, YchF, and Ola1p [Ybr025c], respectively) preferentially hydrolyze ATP compared with GTP (Table 2) (140). Similarly, Trypanosoma cruzi YchF also hydrolyzes ATP to a greater extent than GTP (97).
The G4 motif in the YchF subfamily is quite variable, e.g., NVNE in E. coli, NMSE in yeast, and NLSE in human. Curiously, when the G4 motif of hOLA1 is reverted to the G4 consensus NKXD by site-directed mutagenesis, the hOLA1-NKXD mutant retains specificity for ATP binding (140).
Role in translation.
The YchF α-helical and C-terminal domains show resemblance to RNA-binding proteins, and the crab-like three-domain architecture of YchF suggests a binding site for a double-stranded nucleic acid in the cleft between these two domains. Moreover, several findings point to a role for YchF in translation. First, the yeast homolog interacts with translation elongation factor eEF1 (88). In addition, E. coli ychF expression is downregulated in response to DNA damage, which is also the case for genes related to protein biosynthesis (135). Finally, the ychF gene is located next to the pth gene, and the phylogenetic patterns of both genes are similar, suggesting that the proteins may be functionally related (60). It was suggested previously that YchF is involved in translation as part of a nucleoprotein complex and may function as a GTP-dependent translation factor (251).
Other functions.
YchF has been implicated in pathogenesis. C. pneumoniae and C. trachomatis serovar D persisters show increased transcription levels of ychF (205). Furthermore, an S. pneumoniae ychF mutant has a reduced growth rate in vitro and a proportionally reduced invasiveness in an intranasal murine challenge model, while it is still capable of colonizing the upper airways. However, the observed phenotype might be due to polar effects (80).
In Brucella melitensis 16M, the YchF homolog DugA (DHBA utilization GTPase homolog A) is involved in iron utilization. However, a dugA Tn5 transposon insertion mutant is not attenuated compared to the wild-type strain (63).
Eukaryotic homologs.
The NTPase activity of rice YCHF1 is enhanced by its regulatory protein GAP1, which is specific for higher plants and which contains protein kinase C conserved region 2 (51). YCHF1 binds 26S rRNA via its TGS domain, and 26S rRNA binding is negatively regulated by GAP1 (50). YCHF1 (and its A. thaliana homolog) and GAP1 are involved in the plant defense response: at low YCHF1 or high GAP1 levels, a higher level of resistance against the pathogen P. syringae pv. tomato DC3000 can be observed. At high levels of YCHF1, the opposite results were obtained. It was suggested previously that YCHF1 acts as a repressor to prevent the unnecessary provoking of the detrimental defense response (50, 51). A model has been proposed for the regulation of YCHF1 by GAP1. Under normal conditions, YCHF1 and GAP1 localize to the cytosol, and GAP1 levels are low; therefore, most YCHF1 molecules are in their active form, bound to 26S rRNA. Upon wounding or pathogen challenge, GAP1 levels increase. GAP1 activates the NTPase activity of YCHF1, thereby converting it to the inactive form. Furthermore, GAP1 blocks the binding of YCHF1 to 26S rRNA and sequesters YCHF1 in the plasma membrane by binding to phospholipids via its C2 region (50).
Human GBP45 (GTP-binding protein with a molecular weight of 45 kDa) shows resemblance to the human YchF homolog hOLA1. The encoding gene is strongly expressed in neuronal tissues and the pancreas, and the protein was suggested previously to play important roles in cell proliferation and cell death related to mitochondrial function (136). hOLA1 does not contact the ribosome (140), but the yeast homolog (Ybr025c or yOla1p) interacts with components of the translation elongation factor eEF1 (88). Moreover, T. cruzi YchF is associated with ribosomal subunits and polysomes and with the proteasome. The inactivation of YchF inhibits growth, suggesting that the protein plays an important role in the translation machinery of trypanosomes (97). The proteasome interaction was also established for Ybr025c in yeast (100). The upregulation of the gene encoding Ybr025c was observed during the adaptive stress response to H2O2 in yeast (92), suggesting that Ybr025c, through an interaction with the proteasome, is involved in the degradation of damaged proteins in cells subjected to oxidative stress (97). In contrast, the overexpression of hOLA1 in human cells increases cellular sensitivity to oxidative stress. Moreover, the knockdown of hOLA1 elicits increased resistance to peroxide oxidants and thiol-depleting agents without affecting cell proliferation, baseline apoptosis, or sensitivity to other cytotoxic agents that target the mitochondria, cytoskeleton, or DNA. Several anticancer treatments induce high levels of ROS (reactive oxygen species), suppressing tumor metastasis by destroying cancer cells directly or through the activation of cell death pathways. Conversely, a number of studies suggested that moderate levels of ROS stimulate cancer cell proliferation, migration, and invasion. Therefore, hOLA1 seems to function as a negative regulator of the cellular antioxidant response, and it was reported to have a specific therapeutic effect on the antioxidant defense system, with a low probability of adverse side effects (287). The knockdown of hOLA1 inhibits the motility and invasion of breast cancer cells through a mechanism that involves the modulation of intracellular ROS levels (possibly affecting actin cytoskeleton polymerization) or the redox status. hOLA1 knockdown reduces ROS production but does not alter cell growth (288).
HflX (YnbA)
Surprisingly little is known about the HflX (high frequency of lysogenization protein X) GTPase, which shows a nucleotide-dependent association with ribosomes. HflX is widely distributed and conserved among nearly all bacterial species. It is also found in eukaryotes and archaea (153). It is not essential in B. subtilis (188) and E. coli (89), but it might play a vital role under stress conditions (234).
Genetic organization.
The E. coli amiB-mutL-miaA-hfq-hflX-hflK-hflC superoperon contains important genes involved in several fundamental cellular processes, including cell wall hydrolysis (N-acetylmuramoyl-l-alanine amidase II [amiB]), DNA repair (mutL), tRNA modification (tRNA dimethylallyl diphosphate transferase [miaA]), small-RNA-mediated pleiotropic regulation (hfq), and proteolysis (hflX-hflK-hflC). In the Gammaproteobacteria, yjeT (inner membrane protein) and purA (adenylosuccinate synthetase) are also found in the vicinity of hflX (Fig. 4). hflX expression levels vary in response to stress situations, with an upregulation under heat shock and osmotic stress and a downregulation in the presence of ofloxacin (55, 131, 212, 271). The transcription regulation of the operon in response to heat shock has been well documented (55, 255–257). Mutations in the hflA or the hflB locus result in a very high frequency of bacteriophage λ lysogeny, which has been correlated with an increased stability of cII (15, 195). The hflA locus contains the genes hflX, hflK, and hflC. Both hflK and hflC encode membrane proteins, and the C-terminal region of HflC contains a domain resembling the catalytic domain of ClpP (the proteolytic subunit of the ATP-dependent protease ClpCP) (195). HflC and HflK interact to form a multimeric complex, and free subunits are unstable (15). In Corynebacterium glutamicum, ClgR controls the transcription of hflX. ClgR also enhances the transcription of clpCP, while at the same time, it is a substrate for the ClpCP1 and/or ClpCP2 protease (78).
Cellular localization and concentration.
The C. glutamicum HflX pool is distributed equally between the cytoplasm and the plasma membrane, although HflX is not predicted to contain transmembrane segments (78). C. pneumoniae HflX partly localizes to the membrane (207), as does a portion of overexpressed E. coli HflX (74). The in vivo expression level of hflX is very low, as is the intracellular concentration of the protein (255).
Protein structure.
E. coli and Sulfolobus solfataricus HflX are monomeric proteins in solution (74, 276). The crystal structure of S. solfataricus HflX has been solved in the apo and GDP-bound forms, showing a two-domain arrangement (Fig. 5). The N-terminal domain does not show any similarity to known proteins and is known as the HflX domain. This domain consists of a four-stranded parallel β-sheet flanked by two helices on either side and an antiparallel coiled coil of two α-helices. The latter two helices contain many positively charged residues, making up a positive patch on the surface of HflX. A long flexible linker, rich in glycines, connects the HflX domain to a canonical G domain (with six β-strands and five α-helices). The C-terminal GTPase domain is highly conserved, with some GTPase family-specific variations. The G2 motif is strongly conserved within HflX subfamily members and includes a distinct phenylalanine residue that is shared by members of the OBG family (207). In E. coli, an extra, poorly conserved C-terminal domain of about 50 amino acids is present, which is absent in B. subtilis and S. solfataricus HflX proteins (125, 207, 276).
GTPase cycle.
C. pneumoniae HflX shows specificity for guanine nucleotides and exhibits low intrinsic GTPase activity (Table 2) (207). S. solfataricus HflX also has low GTPase activity and a relatively low affinity for GTP. An HflX mutant lacking the N-terminal domain exhibits a 24-fold-enhanced turnover rate, suggesting a reduction of the activity of the G domain by the N-terminal domain (276). Despite the presence of the G4 specificity motif (N/T)KXD, E. coli HflX binds and hydrolyzes both ATP and GTP (74). E. coli HflX has a preference for nucleotide diphosphates, with a slightly higher affinity for guanine nucleotides than for adenine nucleotides. Given the cellular concentrations of the two purine nucleotides, an estimated 80% of HflX is bound to guanine nucleotides, indicating that HflX may function as a guanine nucleotide-dependent enzyme in vivo (234). The intrinsic GTPase activity of HflX is very low and can be stimulated ∼1,000-fold by 50S and 70S (but not 30S) ribosomal particles. ATPase activity is also stimulated in the presence of these particles (234). Ribosome-dependent GTPase stimulation is inhibited by chloramphenicol, which binds to the large ribosomal subunit, but not by kanamycin, an aminoglycoside targeting the small ribosomal subunit. This may hint at a previously unknown mechanism of antibiotic action through the inhibition of the ribosome-associated activity of HflX (234). The relatively fast dissociation of nucleotides from the complex ensures the rapid exchange of the bound nucleotides, and thus, no GEF is required for HflX function (234).
HflX is a HAS-GTPase in which the classical catalytic glutamine residue following the DXXG G3 motif is replaced by phenylalanine. In S. solfataricus, the conserved F236 residue in the DXXGF sequence is involved in the regulation of the interaction between the N-terminal and G domains and of GTPase activity. The role in aligning a nucleophilic water molecule played by the Ras Q61 residue is replaced by the backbone amide group of G235 (109).
Cell cycle regulation.
A C. glutamicum ΔhflX strain has the same cell morphology and growth behavior as those of the wild type (78).
Role in ribosome assembly.
The ectopic expression of full-length C. pneumoniae hflX in E. coli revealed a cosedimentation of HflX with the E. coli 50S large ribosomal subunit (207). E. coli HflX binds 16S and 23S rRNA in a nucleotide-independent manner. In contrast to most other GTPases that interact with ribosomes only in a GTP-bound state, HflX was reported to interact with 50S subunits in the presence of GTP, GDP, ATP, or ADP but not in the absence of nucleotides. Full-length HflX is required for the interaction (125). The conserved, positively charged surface patches of the S. solfataricus N-terminal domain may mediate interactions with the large ribosomal subunit (276).
Other functions.
C. pneumoniae and C. trachomatis serovar D persisters show a decreased transcription of hflX (18, 205).
Based on its presence in an operon with the proteolysis-regulatory genes hflK and hflC (200) and its coregulation with these genes (78), HflX was suggested to be involved in the regulation of proteolysis. Moreover, a Tn5 insertion in E. coli hflX causes an hfl mutant phenotype (15). However, this is probably due to the polar nature of the mutation. By using a deletion strain as well as a strain overexpressing hflX, it was shown that E. coli HflX has no role in λ lysogeny and that HflX does not interact with HflC or HflK (74). Also, E. coli hflX was identified in a large-scale screening for genes influencing transposition (259), but this too was later attributed to the effect on one or both of the downstream genes hflK and hflC rather than hflX (74).
Eukaryotic homologs.
The human HflX homolog PGPL (pseudoautosomal GTP-binding protein-like) or GTPBP6 (GTP–binding protein 6) is a pseudoautosomal gene on the short arms of the sex chromosomes that is highly conserved and expressed in all tissues. The N-terminal sequence was suggested previously to function as a mitochondrial matrix-targeting sequence (90). The exact function of PGPL remains unknown (104), but in Klinefelter's syndrome, the expression of the human HflX homolog is inversely correlated with verbal intelligence quotient (IQ) and four other measures of verbal ability (266).
THE TRANSLATION FACTOR SUPERFAMILY
The remaining universally conserved GTPases of the TRAFAC class group together in the translation factor superfamily (Fig. 1). To understand the importance of GTPases belonging to this superfamily, a short summary describing the different steps in bacterial protein synthesis is in order. The translation cycle is divided into three parts: initiation, elongation, and termination. In the initiation phase, the ribosomal subunits, the initiator tRNA (fMet-tRNAfMet), and the mRNA that is to be translated are assembled in a process that is coordinated by the initiation factors (IFs) IF-1, IF-2, and IF-3. At this stage, fMet-tRNAfMet is bound to the P (peptidyl) site of the large ribosomal subunit. Elongation refers to the polymerization of amino acids into a nascent polypeptide. It starts with the incorporation of the first aminoacyl tRNA (aa-tRNA) into the A (aminoacyl) site of the ribosome. aa-tRNA is transferred to the ribosome in a ternary complex together with GTP-bound EF-Tu (eEF1A in eukaryotes). Upon cognate codon-anticodon interactions in the small ribosomal subunit, GTP hydrolysis by EF-Tu is triggered, which causes the GTPase to dissociate from the ribosome. After peptide bond formation in the PTC (peptidyl transferase center) of the large ribosomal subunit, the ribosome is in a hybrid configuration containing a peptidyl-tRNA in the A site and a deacylated tRNA in the P site. The binding of the GTPase EF-G (eEF2 in eukaryotes) catalyzes the translocation of the mRNA-tRNA complex, preparing the ribosome for another round of elongation. All three steps of the elongation cycle (decoding, peptide bond transfer, and translocation) are repeated until the complete mRNA sequence has been read and a stop codon marking termination is sensed by the small subunit's decoding center. Termination includes the sequence of events following the recognition of the stop codon up to the disassembly of the ribosome into subunits and the subsequent dissociation of translation factors, tRNA, and mRNA (1, 6).
Several GTPases are involved in the protein synthesis pathway in bacteria, namely, IF-2, EF-Tu, EF-G, LepA, SelB, and RF3. The role of GTP hydrolysis in translation is not completely understood, but GTPases likely provide energy for the translation process. Alternatively, GTP hydrolysis could be regarded as a means to recycle factors involved in protein synthesis (147). The universally conserved GTPases belonging to the translation factor superfamily have been thoroughly reviewed elsewhere during the last few years (1, 6, 147, 149, 193, 194, 239, 284) and will therefore be discussed only briefly here.
IF-2 (InfB)
In eukaryotes and archaea, 10 IFs act in concert to enhance the rate of formation of the translation initiation complex (6). In contrast, just three IFs with distinct but coordinated functions are required in prokaryotes, namely, IF-1, IF-2, and IF-3 (239). IF-2 is the largest of the bacterial IFs. It is encoded by the infB gene, which in the Gammaproteobacteria is part of the polycistronic nusA operon containing metY (minor form of the initiator tRNA), yhbC (protein of unknown function), nusA (a transcriptional termination factor), infB, rbfA (ribosome-binding factor A), truB (tRNA pseudouridine 5S synthase), rpsO (ribosomal protein S15), and pnp (polynucleotide phosphorylase) (Fig. 4) (149). In the Enterobacteriaceae, three IF-2 isoforms exist, which are translated from three independent but in-frame translational start sites of the infB mRNA (149). infB is essential in bacteria (45) and is conserved among all three domains of life (6). In archaea and eukaryotes, IF-2 is referred to as aIF5B and eIF5B, respectively (149).
IF-2 is a large protein that has a central GTP-binding domain consisting of three separate subdomains (G1, G2, and G3). The G2 domain contains the complete GTP/GDP-binding motif characteristic of many GTPases, while G3 is structurally homologous to domain II of the elongation factor proteins. The C-terminal region of IF-2 consists of two subdomains (C1 and C2), while the less conserved N-terminal region encompasses domains N1 and N2 (Fig. 5). The N terminus functions to enhance the interaction of IF-2 with the 30S and 50S ribosomal subunits (147, 239). G2 and G3 also play a role in ribosome binding, whereas G1 and the C-terminal region do not show an affinity for the ribosome (45). While structural (mainly nuclear magnetic resonance [NMR]) data are available for individual domains of bacterial IF-2 (e.g., N-terminal, C1, C2, and G2 domains), no structure of a full-length bacterial IF-2 has been reported so far. There is, however, a solved structure of the homologous archaeal aIF5B from Methanothermobacter thermautotrophicus (Fig. 5) (215).
The three IF (IF-1, IF-2, and IF-3) are involved in the assembly of the prokaryotic initiation complex, consisting of the two ribosomal subunits, mRNA, and the initiator fMet-tRNAfMet. IF-2·GTP recognizes the formyl group on fMet-tRNAfMet and stimulates the binding of the initiator tRNA to the P site of the small subunit to form the 30S initiation complex. IF-1 binds to the A site and interacts specifically with IF-2, while IF-3 acts as an antiassociation factor, keeping the large and small subunits apart prior to the correct association of the fMet-tRNAfMet anticodon to the P site. The hydrolysis of GTP on IF-2 and the subsequent dissociation of the IFs from the ribosome and the binding of the 50S subunit comprise the final initiation steps: the ribosome is now primed for elongation (6, 147, 187, 239). The presence of IF-2·GTP in the 30S initiation complex and GTP hydrolysis during 70S complex formation are essential for the initiation of protein biosynthesis. IF-2 has no intrinsic GTPase activity, and the hydrolysis of GTP depends on the presence of the ribosomes. The protein associates only 10-fold more strongly with GDP than with GTP, and a GEF is not required (149). Besides its function as a translation factor, IF-2 has the properties of a chaperone, promoting the functional folding of proteins and forming stable complexes with unfolded proteins. Furthermore, the expression of E. coli IF-2 was demonstrated to be upregulated during the cold shock response, and the factor is important for the translation of leaderless transcripts (149).
More comprehensive descriptions of prokaryotic IFs have been reported previously (6, 149, 239).
EF-Tu (TufA)
Elongation factor Tu (EF-Tu) is one of the most abundant cellular proteins, with a stoichiometry of 7/1 relative to ribosomes. The intracellular concentrations of EF-Tu and tRNAs are strongly correlated under a variety of growth conditions. In the cell, most of the tRNAs are charged and bound to EF-Tu·GTP, forming a reactive ternary complex (147). EF-Tu is encoded by tufA and tufB (Fig. 4), whose gene products differ by only 1 amino acid at their C termini and exhibit identical physical, chemical, and catalytic properties (147). The protein consists of three domains, with N-terminal GTP-binding domain I being highly conserved among all GTPases (Fig. 5). The acceptor arm of the tRNA is known to interact primarily with domain III, while the CCA sequence present at the 3′ terminus of all mature tRNAs resides in the crevice between domains I and II (1).
EF-Tu has a relatively low intrinsic GTPase activity, which is enhanced 105-fold in the presence of ribosomes (1, 147). It displays a high affinity for GDP (in the nM range) and a 100-fold-lower affinity for GTP. Once EF-Tu·GDP has dissociated from the translating ribosome, GDP is exchanged for GTP by the GEF EF-Ts (EF-1Bα in eukaryotes) (194).
It has been estimated that proteins are synthesized in vivo at a rate of 15 to 20 amino acids per second, with an error rate below 10−4. The high fidelity of translation is achieved in part by EF-Tu. A ternary complex containing EF-Tu·GTP and a near-cognate tRNA dissociates from the ribosomes more rapidly than a complex containing a cognate tRNA. Therefore, the stimulation of the GTPase activity of EF-Tu, initiated by the codon-anticodon interaction, proceeds faster for a cognate ternary complex. This ensures a high probability for the acceptance of a cognate ternary complex. Upon GTP hydrolysis, EF-Tu departs from the ribosome, and the 3′-CCA end of the tRNA is accommodated in the A site, leading to rapid peptide transfer (1).
EF-Tu is the target of at least four classes of antibiotics and antibacterials that work by preventing either the binding of EF-Tu to aa-tRNA (pulvomycin and GE2270A) or the release of EF-Tu·GDP from the ribosome (kirromycin and enacyloxin IIa) (194).
EF-G (FusA)
EF-G is encoded by the fusA gene (Fig. 4), and one molecule of EF-G is present in the cell for every ribosome (147). The protein consists of six domains: the G domain, an insertion in the G domain (G′ or domain I), and domains II to V (Fig. 5) (1). The elongation factors EF-G and EF-Tu are structural homologs that both bind to ribosomes at the GTPase-associated center (GAC) and the sarcin-ricin loop (SRL). The orientation of the mobile GAC relative to the fixed SRL determines whether EF-G or EF-Tu will bind to the ribosome (230).
EF-G has a weak affinity for both GDP and GTP in solution, and therefore, it does not need any GEF to switch from the GDP to the GTP state (147). GTP binding is stabilized by 4 orders of magnitude upon binding to the ribosome, indicating that EF-G binds to the ribosome in the GTP-bound form (194).
The movement of tRNAs from A and P to P and E (exit) sites during translocation is an intrinsic capacity of ribosomes under certain experimental conditions. However, EF-G plays a catalytic role, increasing the translocation rate approximately 500-fold (147). EF-G·GTP binding to the ribosome promotes the release of the deacylated tRNA from the P site and the translocation of the peptidyl-tRNA to this location. GTP hydrolysis by EF-G precedes translocation, and therefore, it has been postulated that energy from GTP hydrolysis directly drives translocation. Moreover, GTP hydrolysis seems to confer conformational changes in the ribosome that stimulate translocation. GTP hydrolysis is also required for the recycling of EF-G as the protein is ejected from the ribosome in its GDP-bound form (1, 147, 194). Apart from a role in translocation, EF-G is involved in the recycling of ribosomal components after a successful round of protein synthesis. Upon peptide release, the ribosome-recycling factor (RRF) binds to the ribosomal A site. Next, EF-G translocates RRF to the P site, which results in the ejection of deacylated tRNA from the ribosome, ultimately leading to dissociation into mRNA, tRNA, and the ribosomal subunits ready for recycling (130, 187). EF-G is the target of the antibacterial compound fusidic acid, which binds to EF-G and prevents the release of EF-G·GDP from the ribosome. Similarly, sordarin blocks the release of eEF2·GDP from the eukaryotic ribosome (194).
For a more elaborate overview of EF-Tu and EF-G, the reader is referred to some excellent reviews on the subject (1, 107, 147, 193, 194).
LepA (YqeQ)
LepA is involved in ribosomal back-translocation. Its orthologs are highly conserved, and they are found exclusively in bacteria and eukaryotic cell organelles of prokaryotic origin (i.e., mitochondria and chloroplasts) (284). There are numerous rare codons in the LepA open reading frame, suggesting that LepA is normally expressed at a low level (168). The overexpression of LepA is toxic in E. coli, but the deletion of LepA in bacteria does not cause any discernible phenotype (284).
LepA has five distinct domains (Fig. 5). It is highly related to EF-G (169) and hydrolyzes GTP in a ribosome-dependent manner. LepA catalyzes unexpected one-codon backward movement on the ribosome: it causes ribosomes to “back up,” placing the codon that was just translated (and the peptidyl-tRNA with which it is still associated) back into the A site (210). Youngman and Green summarized a number of theories explaining the significance of this action (284). LepA can increase the fidelity of translocation by back-translocating ribosomes that have been in some way imperfectly translocated by EF-G. If this is indeed the role of LepA, it is not clear why such a central and conserved function was discarded by eukaryotes. Alternatively, LepA could be recruited to promote ribosome stalling on particular messages or under particular cellular conditions. Also, LepA could block the activation of the stringent response upon the detection of deacylated tRNAs in the ribosomal A site by RelA. This is advantageous when a ribosome encounters a rare codon resulting in an empty A site. Finally, lepA is promoter proximal in an operon with lepB, encoding the leader peptidase or signal peptidase I (Fig. 4) (168), and since LepA is preferentially localized in the cytoplasmic membrane and periplasm (168), it was suggested that LepA could be involved (for example, by pausing translation) in coupling events on the ribosome to signal peptide processing and secretion (167, 284).
SIGNAL-RECOGNITION-ASSOCIATED GTPase FAMILY
Of the SIMIBI class of P-loop GTPases, only the signal-recognition-associated GTPases are universally conserved (Fig. 1). These proteins are involved in the cotranslational targeting of membrane-bound or secreted proteins. Homologs of the signal recognition particle (SRP) and the SRP receptor (SR) have been identified in all living cells analyzed to date (133). Mammalian SRP contains six polypeptides (SRP9, SRP14, SRP19, SRP54, SRP68, and SRP72) and one RNA molecule (7SL RNA or SRP-RNA). Signal sequence recognition is mediated by the SRP54 subunit, which also binds directly to SRP-RNA. The membrane-bound SR is comprised of a peripheral subunit (SRα) and a transmembrane subunit (SRβ) (62, 133). In bacteria, the SRP contains the GTPase Ffh (SRP54 or fifty-four homolog) and SRP-RNA (termed 4.5S RNA in E. coli). Bacterial SR consists of a single polypeptide, FtsY (133). FtsY is either located cytoplasmically or loosely associated with the bacterial inner membrane (133), but the mechanism by which FtsY is targeted, assembled, and released from the membrane is not fully understood (105). Both bacterial signal-recognition-associated GTPases are essential for cell growth (105).
Mode of action.
SRP directs proteins destined for either secretion or membrane integration to the SR in the endoplasmic reticulum (eukaryotes) or the plasma membrane (prokaryotes). The depletion of bacterial Ffh, SRP-RNA, or FtsY most strongly affects the insertion of integral membrane proteins, whereas the secretion of soluble periplasmic proteins is only weakly impaired (133). Integral membrane proteins are specifically recognized by a direct interaction between their noncleaved hydrophobic signal anchor sequences and the bacterial SRP consisting of Ffh and SRP-RNA. The primary sequence of the signal peptide recognized by SRP is not well conserved, although it is always highly hydrophobic. Exported proteins may not interact with Ffh because their signal peptide is less hydrophobic than the membrane-spanning region of membrane proteins. Furthermore, a ribosomal chaperone named trigger factor prevents the recognition of exported proteins by SRP by binding earlier to the nascent chains of exported proteins (62). Recognition occurs during peptide synthesis at the ribosome and leads to cotranslational targeting to the SecYEG translocon that is mediated by FtsY·GTP and the hydrolysis of GTP by the G domains of FtsY and Ffh. The mechanism by which the membrane protein is transferred from FtsY to the SecYEG machinery is still unknown (62, 191). In eukaryotes, the interaction of SRP with the nascent polypeptide chain causes elongation arrest, and it was suggested that a similar elongation arrest exists in prokaryotes (105). Interaction with its receptor releases SRP from the ribosome-nascent-chain complex, enabling it to participate in another round of protein targeting while simultaneously allowing the ribosome to resume translation (133, 232).
An SRP-like component has been identified in chloroplasts, where, together with an organellar FtsY homolog, it targets proteins for translocation across the thylakoid membrane. Chloroplast SRP contains no known RNA subunit. Some proteins targeted to the thylakoid membrane are first imported into chloroplasts from the cytosol. Therefore, SRP in chloroplasts can also act posttranslationally (99, 133).
Protein structure.
Ffh contains two domains: an NG domain (consisting of a helical N domain and a GTPase G domain) and a C-terminal M domain (Fig. 5). The NG domain contains the GTP-binding site and is involved in the interaction with a similar NG domain of FtsY. The M domain (containing a high percentage of methionine residues) is implicated in the interaction with the nascent polypeptide chain and the SRP-RNA. Unlike its mammalian homologs, bacterial FtsY usually contains two distinct domains: an NG domain and an N-terminal acidic (A) domain that has been suggested to anchor FtsY peripherally to the membrane via an interaction with phospholipids (Fig. 5). Both FtsY and Ffh contain homologous GTPase domains, and these domains also have similar tertiary structures (105). SRP and SR interact primarily through their respective NG domains, and one function of the N domain might be to sense or regulate the GTP-binding state of the G domain (133, 232). In the Ffh-FtsY complex, the NG domains face each other, with their nucleotides in an antiparallel orientation. The G proteins mutually activate each other via the positioning of catalytic residues in each other's active site and through an interaction of the ribose 3′-OH with the phosphate of the neighboring GTP molecule (76). The orchestrated structural rearrangements of Ffh and FtsY in response to their interaction partners (the signal sequence, the ribosome, the translocation channel, membrane lipids, and guanine nucleotides) have been reviewed elsewhere (99, 232).
SRP-RNA was suggested to stabilize the structure of the Ffh M domain and to enhance both the association and dissociation of the Ffh-FtsY complex (133). However, recent studies showed that the postulated role of SRP-RNA in Ffh-FtsY complex formation is strongly dependent on the experimental conditions used (99). SRP-RNA contains only one domain (termed domain IV) that is universally conserved. This part of the SRP-RNA participates in the interaction with both the Ffh/SRP54 protein and the hydrophobic nascent chain and therefore plays a pivotal role in the bacterial SRP pathway. Mammalian SRP-RNA contains an Alu sequence implicated in translation arrest. This Alu sequence is absent in E. coli but present in B. subtilis SRP-RNA, although it is not known whether it is involved in translation arrest in prokaryotes (105).
GTPase cycle.
SRP and FtsY interact in their GTP-bound forms, and GTP hydrolysis by both GTPases is dramatically increased upon the formation of the complex with the concomitant heterodimerization of their G domains. Other components, including the ribosomes and the signal sequence, are known to regulate GTPase activity (133, 232). Ffh and FtsY have a relatively weak affinity for GDP and therefore do not require an external GEF. Whereas most GTPases are unstable in the nucleotide-free state, in Ffh several charged residues that normally interact with a bound nucleotide serve as hydrogen-bonding partners for each other when the binding site is empty. Hence, the apo state was shown previously to be a functionally relevant intermediate (133, 232, 243). GTP hydrolysis can function to provide unidirectionality to the interactions in the targeting reaction, but it remains unclear why a mechanism utilizing the hydrolysis of two (and in eukaryotes, even three) GTP molecules has evolved (133). Also, the question of exactly how GTP hydrolysis is finally triggered by the insertion of the signal sequence into the translocation channel remains unanswered (99).
The reader is referred to some excellent reviews on this subject (62, 71, 99, 105, 133, 191, 232).
CONCLUDING REMARKS
The studies described here illustrate that bacterial GTPases comprise a vast superclass of proteins with putative roles in diverse cellular processes, such as cell cycle regulation (Era, Der, YihA, and Obg), the assembly and maturation of ribosomes (Era, Der, YihA, Obg, and HflX), translation (IF-2, EF-Tu, EF-G, LepA, Ffh, and FtsY), the stringent response (Era, Der, and Obg), the stress response (Obg and YchF), pathogenesis (MnmE, Der, and YchF), tRNA modifications (MnmE), energy metabolism (Era), and morphological differentiation (Obg) (Table 1). Although the available biochemical data on protein activities may be influenced by variations in experimental setups, in general, bacterial GTPases seem to have a low affinity for both GDP and GTP, causing rapid nucleotide binding and exchange. Moreover, with the notable exception of MnmE, bacterial GTPases have low intrinsic GTP hydrolysis rates (Table 2). These findings suggest that nucleotide occupancy is controlled mainly by the intracellular levels of guanine nucleotides, perhaps allowing bacterial GTPases to detect intracellular energy levels and respond to changes in the nutritional environment.
Since all P-loop GTPases descend from a common ancestor with a generic regulatory role in translation (153), it is not surprising that several GTPases are implicated in similar cellular processes. Similarly, certain GTPases have been described to suppress phenotypes caused by the depletion of other superclass members. For example, both Era and IF-2 are multicopy suppressors of the slow-growth phenotype, and they altered the polysome profile of an E. coli strain lacking the GTPase YjeQ (44). However, diversification has also resulted in the acquisition of diverse cellular functions that in some cases appear to be unique for certain GTPase families (e.g., tRNA modification by the MnmE-MnmG complex).
Future Areas of Research
In view of their role in the development of cancer, eukaryotic GTPases have been studied thoroughly for several decades. However, with the exception of translation factors and signal-recognition-associated GTPases, the exact cellular role of the universally conserved GTPases in bacterial physiology remains puzzling. While recent studies have contributed to our understanding of the apparent plethora of roles carried out by MnmE, Era, Der, and Obg, surprisingly little is known about bacterial homologs from the YihA, YchF, and HflX families. Furthermore, a clear view on how bacterial GTPases mediate their cellular functions is lacking, and additional cellular targets and effectors undoubtedly await discovery.
The regulation of GTPase activity is another point of interest that requires further attention. Unlike eukaryotic GTPases, whose regulation by GAPs, GEFs, and GDIs has been well documented, the regulation of the GTPase cycle in prokaryotes remains largely elusive. YihI was only recently identified as the first prokaryotic GAP, stimulating the GTPase activity of Der. Furthermore, interactions with ribosomal proteins or rRNA (Era and HflX) as well as cis-multimerization (MnmE) or pseudo-trans-dimerization (Ffh or FtsY) are known to enhance GTPase activity in prokaryotic GTPases. Moreover, several in vitro studies pointed toward the importance of potassium ions in GTP hydrolysis by certain HAS-GTPase, raising the possibility that these ions mediate GTPase activity in an in vivo setting. However, because those studies were conducted mostly under in vitro conditions, in vivo assays to assess nucleotide binding and GTPase activity inside bacterial cells are warranted. Furthermore, sequestration in the membrane has been postulated to confer a level of regulation to S. pneumoniae Era (103, 175) and A. thaliana YCHF1 (50), emphasizing the need for proper localization studies. Other signals modulating GTPase activity, perhaps originating from the extracellular environment, remain to be discovered.
GTPases as Drug Targets
The translation factors EF-Tu and EF-G are known to be the targets of several antibiotics (194). Furthermore, other bacterial GTPases are emerging as promising novel drug targets to combat infections by pathogenic bacteria for several reasons. First, many GTPases are broadly conserved and indispensable proteins that are crucial for fundamental cellular processes, including ribosome assembly (57). Second, M. tuberculosis Obg has been named a potential target because of its disordered C terminus (10). Intrinsically disordered proteins exist as dynamic structural ensembles without fixed secondary structures that were suggested to undergo disorder-to-order transitions upon binding to ligands (75). Such interactions can be exploited in drug design, as they have unusually low binding free energy per unit area of interaction and are therefore relatively easy to block with small molecules (48, 177). Furthermore, GTPases were previously shown to be amenable to inhibition by small-molecule compounds (85, 245), illustrating their potential as therapeutic targets. Finally, it was suggested that selective toxicity toward bacterial GTPase proteins is achievable, just as has been accomplished for bacterial protein translation (57).
Target-based drug discovery requires a full characterization in terms of the structure and function of the potential drug target. With regard to bacterial GTPases, additional studies are needed to elucidate their central role in cell physiology. Downstream factors in the regulatory circuit need to be identified, and three-dimensional structures need to be determined. These studies will pave the way for rational drug design, retaining specificity for the pathogen's target molecule without affecting host proteins. In the future, all this can be expected to lead to the development of new classes of drugs to combat pathogenic infections.
ACKNOWLEDGMENTS
N.V. received a fellowship from the Fund for Scientific Research-Flanders (FWO). W.V. received a postdoctoral fellowship from the Fund for Scientific Research-Flanders (FWO). The work was supported by grants from the K. U. Leuven Research Council (IDO/07/010 and IDO/09/010) and the FWO (G.0299.07, G.0413.10, and G.0259.09).
Biographies

Natalie Verstraeten received her master's degree in Applied Biosciences and Engineering (specialization in Cell and Gene Biotechnology) in 2006 from the Katholieke Universiteit Leuven, Leuven, Belgium. Since October 2006, she has been working as a research associate in the Symbiotic and Pathogenic Interactions group of the Centre of Microbial and Plant Genetics, K. U. Leuven. Since October 2007, her work has been funded by the Fund for Scientific Research (FWO-Vlaanderen). Currently, she is finishing her Ph.D. research on a conserved GTPase involved in bacterial antibiotic tolerance.

Maarten Fauvart graduated as a master in Applied Biosciences and Engineering, with specialization in Cell and Gene Biotechnology, in 2003 and as a Ph.D. in Bioscience Engineering in 2008. Currently, he is a postdoctoral fellow at the Centre of Microbial and Plant Genetics, K. U. Leuven, where he supervises research carried out in the Symbiotic and Pathogenic Interactions group.

Wim Versées received his Ph.D. in Applied Biological Sciences in 2002 at the Free University of Brussels, Brussels, Belgium. From 2003 onward, he worked as a postdoctoral fellow at the Structural Biology Brussels Laboratory (Free University Brussels) and the Department of Structural Biology at the Max Planck Institute of Molecular Physiology (Dortmund, Germany). Since 2007 he has been an assistant professor at the Free University Brussels and a staff scientist within the Flanders Institute for Biotechnology (VIB). His research focuses on the elucidation of the mechanistic and structural principles that underlie the tremendous catalytic power and regulation of enzymes and enzyme complexes.

Jan Michiels is full professor at the K. U. Leuven Faculty of Bioscience Engineering. He is head of the Symbiotic and Pathogenic Interactions group of the Centre of Microbial and Plant Genetics. He teaches the courses molecular biology, genetics, and bacterial physiology. His research focuses on molecular aspects of microbe-host interactions and stress resistance in bacteria, in particular the antibiotic tolerance of pathogenic bacteria. He has published over 60 articles in international journals with peer review.
REFERENCES
- 1. Agirrezabala X., Frank J. 2009. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q. Rev. Biophys. 42:159–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agris P. F. 2004. Decoding the genome: a modified view. Nucleic Acids Res. 32:223–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahnn J., March P. E., Takiff H. E., Inouye M. 1986. A GTP-binding protein of Escherichia coli has homology to yeast RAS proteins. Proc. Natl. Acad. Sci. U. S. A. 83:8849–8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Akiyama T., et al. 2001. Mammalian homologue of E. coli Ras-like GTPase (ERA) is a possible apoptosis regulator with RNA binding activity. Genes Cells 6:987–1001 [DOI] [PubMed] [Google Scholar]
- 5. Alam K. Y., Clark D. P. 1991. Molecular cloning and sequence of the thdF gene, which is involved in thiophene and furan oxidation by Escherichia coli. J. Bacteriol. 173:6018–6024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Allen G. S., Frank J. 2007. Structural insights on the translation initiation complex: ghosts of a universal initiation complex. Mol. Microbiol. 63:941–950 [DOI] [PubMed] [Google Scholar]
- 7. Amy M., et al. 2004. Identification of a new Salmonella enterica serovar Enteritidis locus involved in cell invasion and in the colonisation of chicks. Res. Microbiol. 155:543–552 [DOI] [PubMed] [Google Scholar]
- 8. Anand B., Surana P., Prakash B. 2010. Deciphering the catalytic machinery in 30S ribosome assembly GTPase YqeH. PLoS One 5:e9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anderson P. E., Matsunaga J., Simons E. L., Simons R. W. 1996. Structure and regulation of the Salmonella typhimurium rnc-era-recO operon. Biochimie 78:1025–1034 [DOI] [PubMed] [Google Scholar]
- 10. Anurag M., Dash D. 2009. Unraveling the potential of intrinsically disordered proteins as drug targets: application to Mycobacterium tuberculosis. Mol. Biosyst. 5:1752–1757 [DOI] [PubMed] [Google Scholar]
- 11. Arigoni F., et al. 1998. A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 16:851–856 [DOI] [PubMed] [Google Scholar]
- 12. Auvray F., Chassaing D., Duprat C., Carpentier B. 2007. The Listeria monocytogenes homolog of the Escherichia coli era gene is involved in adhesion to inert surfaces. Appl. Environ. Microbiol. 73:7789–7792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baev D., England R., Kuramitsu H. K. 1999. Stress-induced membrane association of the Streptococcus mutans GTP-binding protein, an essential G protein, and investigation of its physiological role by utilizing an antisense RNA strategy. Infect. Immun. 67:4510–4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bang W. Y., et al. 2009. AtObgC, a plant ortholog of bacterial Obg, is a chloroplast-targeting GTPase essential for early embryogenesis. Plant Mol. Biol. 71:379–390 [DOI] [PubMed] [Google Scholar]
- 15. Banuett F., Herskowitz I. 1987. Identification of polypeptides encoded by an Escherichia coli locus (hflA) that governs the lysis-lysogeny decision of bacteriophage lambda. J. Bacteriol. 169:4076–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bardwell J. C., et al. 1989. Autoregulation of RNase III operon by mRNA processing. EMBO J. 8:3401–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barr F., Lambright D. G. 2010. Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22:461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Belland R. J., et al. 2003. Transcriptome analysis of chlamydial growth during IFN-γ-mediated persistence and reactivation. Proc. Natl. Acad. Sci. U. S. A. 100:15971–15976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Berthon J., Fujikane R., Forterre P. 2009. When DNA replication and protein synthesis come together. Trends Biochem. Sci. 34:429–434 [DOI] [PubMed] [Google Scholar]
- 20. Bharat A., Jiang M., Sullivan S. M., Maddock J. R., Brown E. D. 2006. Cooperative and critical roles for both G domains in the GTPase activity and cellular function of ribosome-associated Escherichia coli EngA. J. Bacteriol. 188:7992–7996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Böhme S., et al. 2010. Stabilization of G domain conformations in the tRNA-modifying MnmE-GidA complex observed with double electron electron resonance spectroscopy. J. Biol. Chem. 285:16991–17000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bos J. L., Rehmann H., Wittinghofer A. 2007. GEFs and GAPs: critical elements in the control of small G proteins. Cell 129:865–877 [DOI] [PubMed] [Google Scholar]
- 23. Bourne H. R., Sanders D. A., McCormick F. 1990. The GTPase superfamily: a conserved switch for diverse cell functions. Nature 348:125–132 [DOI] [PubMed] [Google Scholar]
- 24. Bourne H. R., Sanders D. A., McCormick F. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117–127 [DOI] [PubMed] [Google Scholar]
- 25. Braeken K., Moris M., Daniels R., Vanderleyden J., Michiels J. 2006. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol. 14:45–54 [DOI] [PubMed] [Google Scholar]
- 26. Brégeon D., Colot V., Radman M., Taddei F. 2001. Translational misreading: a tRNA modification counteracts a +2 ribosomal frameshift. Genes Dev. 15:2295–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brierley I., Meredith M. R., Bloys A. J., Hagervall T. G. 1997. Expression of a coronavirus ribosomal frameshift signal in Escherichia coli: influence of tRNA anticodon modification on frameshifting. J. Mol. Biol. 270:360–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Britton R. A. 2009. Role of GTPases in bacterial ribosome assembly. Annu. Rev. Microbiol. 63:155–176 [DOI] [PubMed] [Google Scholar]
- 29. Britton R. A., et al. 2000. Isolation and preliminary characterization of the human and mouse homologues of the bacterial cell cycle gene era. Genomics 67:78–82 [DOI] [PubMed] [Google Scholar]
- 30. Britton R. A., Lupski J. R. 1997. Isolation and characterization of suppressors of two Escherichia coli dnaG mutations, dnaG2903 and parB. Genetics 145:867–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Britton R. A., Powell B. S., Court D. L., Lupski J. R. 1997. Characterization of mutations affecting the Escherichia coli essential GTPase era that suppress two temperature-sensitive dnaG alleles. J. Bacteriol. 179:4575–4582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Britton R. A., et al. 1998. Cell cycle arrest in Era GTPase mutants: a potential growth rate-regulated checkpoint in Escherichia coli. Mol. Microbiol. 27:739–750 [DOI] [PubMed] [Google Scholar]
- 33. Bügl H., et al. 2000. RNA methylation under heat shock control. Mol. Cell 6:349–360 [DOI] [PubMed] [Google Scholar]
- 34. Buglino J., Shen V., Hakimian P., Lima C. D. 2002. Structural and biochemical analysis of the Obg GTP binding protein. Structure 10:1581–1592 [DOI] [PubMed] [Google Scholar]
- 35. Bujnicki J. M., et al. 2004. Identification of a bifunctional enzyme MnmC involved in the biosynthesis of a hypermodified uridine in the wobble position of tRNA. RNA 10:1236–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bunner A. E., Nord S., Wikstrom P. M., Williamson J. R. 2010. The effect of ribosome assembly cofactors on in vitro 30S subunit reconstitution. J. Mol. Biol. 398:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burland V., Plunkett G., III, Daniels D. L., Blattner F. R. 1993. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics 16:551–561 [DOI] [PubMed] [Google Scholar]
- 38. Butland G., et al. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531–537 [DOI] [PubMed] [Google Scholar]
- 39. Bykhovskaya Y., et al. 2004. Phenotype of non-syndromic deafness associated with the mitochondrial A1555G mutation is modulated by mitochondrial RNA modifying enzymes MTO1 and GTPBP3. Mol. Genet. Metab. 83:199–206 [DOI] [PubMed] [Google Scholar]
- 40. Cabedo H., et al. 1999. The Escherichia coli trmE (mnmE) gene, involved in tRNA modification, codes for an evolutionarily conserved GTPase with unusual biochemical properties. EMBO J. 18:7063–7076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Caldas T., et al. 2000. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S rRNA methyltransferase. J. Biol. Chem. 275:16414–16419 [DOI] [PubMed] [Google Scholar]
- 42. Caldon C. E., March P. E. 2003. Function of the universally conserved bacterial GTPases. Curr. Opin. Microbiol. 6:135–139 [DOI] [PubMed] [Google Scholar]
- 43. Caldon C. E., Yoong P., March P. E. 2001. Evolution of a molecular switch: universal bacterial GTPases regulate ribosome function. Mol. Microbiol. 41:289–297 [DOI] [PubMed] [Google Scholar]
- 44. Campbell T. L., Brown E. D. 2008. Genetic interaction screens with ordered overexpression and deletion clone sets implicate the Escherichia coli GTPase YjeQ in late ribosome biogenesis. J. Bacteriol. 190:2537–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Caserta E., et al. 2010. Ribosomal interaction of Bacillus stearothermophilus translation initiation factor IF2: characterization of the active sites. J. Mol. Biol. 396:118–129 [DOI] [PubMed] [Google Scholar]
- 46. Chen S. M., et al. 1990. Expression and characterization of RNase III and Era proteins. Products of the rnc operon of Escherichia coli. J. Biol. Chem. 265:2888–2895 [PubMed] [Google Scholar]
- 47. Chen X., Court D. L., Ji X. 1999. Crystal structure of ERA: a GTPase-dependent cell cycle regulator containing an RNA binding motif. Proc. Natl. Acad. Sci. U. S. A. 96:8396–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cheng Y., et al. 2006. Rational drug design via intrinsically disordered protein. Trends Biotechnol. 24:435–442 [DOI] [PubMed] [Google Scholar]
- 49. Cherfils J., Chardin P. 1999. GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem. Sci. 24:306–311 [DOI] [PubMed] [Google Scholar]
- 50. Cheung M. Y., et al. 2010. An ancient P-loop GTPase in rice is regulated by a higher plant-specific regulatory protein. J. Biol. Chem. 285:37359–37369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cheung M. Y., et al. 2008. Constitutive expression of a rice GTPase-activating protein induces defense responses. New Phytol. 179:530–545 [DOI] [PubMed] [Google Scholar]
- 52. Chigri F., Sippel C., Kolb M., Vothknecht U. C. 2009. Arabidopsis OBG-like GTPase (AtOBGL) is localized in chloroplasts and has an essential function in embryo development. Mol. Plant 2:1373–1383 [DOI] [PubMed] [Google Scholar]
- 53. Cho K. H., Caparon M. G. 2008. tRNA modification by GidA/MnmE is necessary for Streptococcus pyogenes virulence: a new strategy to make live attenuated strains. Infect. Immun. 76:3176–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chopade B. A., Shankar S., Sundin G. W., Mukhopadhyay S., Chakrabarty A. M. 1997. Characterization of membrane-associated Pseudomonas aeruginosa Ras-like protein Pra, a GTP-binding protein that forms complexes with truncated nucleoside diphosphate kinase and pyruvate kinase to modulate GTP synthesis. J. Bacteriol. 179:2181–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chuang S. E., Blattner F. R. 1993. Characterization of twenty-six new heat shock genes of Escherichia coli. J. Bacteriol. 175:5242–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Colby G., Wu M., Tzagoloff A. 1998. MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J. Biol. Chem. 273:27945–27952 [DOI] [PubMed] [Google Scholar]
- 57. Comartin D. J., Brown E. D. 2006. Non-ribosomal factors in ribosome subunit assembly are emerging targets for new antibacterial drugs. Curr. Opin. Pharmacol. 6:453–458 [DOI] [PubMed] [Google Scholar]
- 58. Cooper E. L., Garcia-Lara J., Foster S. J. 2009. YsxC, an essential protein in Staphylococcus aureus crucial for ribosome assembly/stability. BMC Microbiol. 9:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Courcelle J., Khodursky A., Peter B., Brown P. O., Hanawalt P. C. 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cruz-Vera L. R., Galindo J. M., Guarneros G. 2002. Transcriptional analysis of the gene encoding peptidyl-tRNA hydrolase in Escherichia coli. Microbiology 148:3457–3466 [DOI] [PubMed] [Google Scholar]
- 61. Czyż A., Zielke R., Konopa G., We¸grzyn G. 2001. A Vibrio harveyi insertional mutant in the cgtA (obg, yhbZ) gene, whose homologues are present in diverse organisms ranging from bacteria to humans and are essential genes in many bacterial species. Microbiology 147:183–191 [DOI] [PubMed] [Google Scholar]
- 62. Dalbey R. E., Chen M. 2004. Sec-translocase mediated membrane protein biogenesis. Biochim. Biophys. Acta 1694:37–53 [DOI] [PubMed] [Google Scholar]
- 63. Danese I., et al. 2004. The Ton system, an ABC transporter, and a universally conserved GTPase are involved in iron utilization by Brucella melitensis 16M. Infect. Immun. 72:5783–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Das S. K., et al. 2004. Expression, purification, crystallization and preliminary crystallographic analysis of a putative GTP-binding protein, YsxC, from Bacillus subtilis. Acta Crystallogr. D Biol. Crystallogr. 60:166–168 [DOI] [PubMed] [Google Scholar]
- 65. Dassain M., Leroy A., Colosetti L., Carole S., Bouche J. P. 1999. A new essential gene of the ‘minimal genome’ affecting cell division. Biochimie 81:889–895 [DOI] [PubMed] [Google Scholar]
- 66. Datta K., Fuentes J. L., Maddock J. R. 2005. The yeast GTPase Mtg2p is required for mitochondrial translation and partially suppresses an rRNA methyltransferase mutant, mrm2. Mol. Biol. Cell 16:954–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Datta K., Skidmore J. M., Pu K., Maddock J. R. 2004. The Caulobacter crescentus GTPase CgtAC is required for progression through the cell cycle and for maintaining 50S ribosomal subunit levels. Mol. Microbiol. 54:1379–1392 [DOI] [PubMed] [Google Scholar]
- 68. Datta P. P., et al. 2007. Structural aspects of RbfA action during small ribosomal subunit assembly. Mol. Cell 28:434–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Decoster E., Vassal A., Faye G. 1993. MSS1, a nuclear-encoded mitochondrial GTPase involved in the expression of COX1 subunit of cytochrome c oxidase. J. Mol. Biol. 232:79–88 [DOI] [PubMed] [Google Scholar]
- 70. Dennerlein S., Rozanska A., Wydro M., Chrzanowska-Lightowlers Z. M., Lightowlers R. N. 2010. Human ERAL1 is a mitochondrial RNA chaperone involved in the assembly of the 28S small mitochondrial ribosomal subunit. Biochem. J. 430:551–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Doudna J. A., Batey R. T. 2004. Structural insights into the signal recognition particle. Annu. Rev. Biochem. 73:539–557 [DOI] [PubMed] [Google Scholar]
- 72. Du Y. C., Stillman B. 2002. Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell 109:835–848 [DOI] [PubMed] [Google Scholar]
- 73. Dutkiewicz R., We¸grzyn M. Słomińska G., Czyż A. 2002. Overexpression of the cgtA (yhbZ, obgE) gene, coding for an essential GTP-binding protein, impairs the regulation of chromosomal functions in Escherichia coli. Curr. Microbiol. 45:440–445 [DOI] [PubMed] [Google Scholar]
- 74. Dutta D., Bandyopadhyay K., Datta A. B., Sardesai A. A., Parrack P. 2009. Properties of HflX, an enigmatic protein from Escherichia coli. J. Bacteriol. 191:2307–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dyson H. J., Wright P. E. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6:197–208 [DOI] [PubMed] [Google Scholar]
- 76. Egea P. F., et al. 2004. Substrate twinning activates the signal recognition particle and its receptor. Nature 427:215–221 [DOI] [PubMed] [Google Scholar]
- 77. Elseviers D., Petrullo L. A., Gallagher P. J. 1984. Novel E. coli mutants deficient in biosynthesis of 5-methylaminomethyl-2-thiouridine. Nucleic Acids Res. 12:3521–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Engels S., et al. 2005. The transcriptional activator ClgR controls transcription of genes involved in proteolysis and DNA repair in Corynebacterium glutamicum. Mol. Microbiol. 57:576–591 [DOI] [PubMed] [Google Scholar]
- 79. Evans R. N., Blaha G., Bailey S., Steitz T. A. 2008. The structure of LepA, the ribosomal back translocase. Proc. Natl. Acad. Sci. U. S. A. 105:4673–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Fernebro J., et al. 2008. The influence of in vitro fitness defects on pneumococcal ability to colonize and to cause invasive disease. BMC Microbiol. 8:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ferrari F. A., Trach K., Hoch J. A. 1985. Sequence analysis of the spo0B locus reveals a polycistronic transcription unit. J. Bacteriol. 161:556–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Foti J. J., Persky N. S., Ferullo D. J., Lovett S. T. 2007. Chromosome segregation control by Escherichia coli ObgE GTPase. Mol. Microbiol. 65:569–581 [DOI] [PubMed] [Google Scholar]
- 83. Foti J. J., Schienda J., Sutera V. A., Jr., Lovett S. T. 2005. A bacterial G protein-mediated response to replication arrest. Mol. Cell 17:549–560 [DOI] [PubMed] [Google Scholar]
- 84. Freymann D. M., Keenan R. J., Stroud R. M., Walter P. 1997. Structure of the conserved GTPase domain of the signal recognition particle. Nature 385:361–364 [DOI] [PubMed] [Google Scholar]
- 85. Gao Y., Dickerson J. B., Guo F., Zheng J., Zheng Y. 2004. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. U. S. A. 101:7618–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Garcia C., Khan N. Z., Nannmark U., Aronsson H. 2010. The chloroplast protein CPSAR1, dually localized in the stroma and the inner envelope membrane, is involved in thylakoid biogenesis. Plant J. 63:73–85 [DOI] [PubMed] [Google Scholar]
- 87. Gasper R., Meyer S., Gotthardt K., Sirajuddin M., Wittinghofer A. 2009. It takes two to tango: regulation of G proteins by dimerization. Nat. Rev. Mol. Cell Biol. 10:423–429 [DOI] [PubMed] [Google Scholar]
- 88. Gavin A. C., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141–147 [DOI] [PubMed] [Google Scholar]
- 89. Gerdes S. Y., et al. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185:5673–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Gianfrancesco F., et al. 1998. A novel pseudoautosomal gene encoding a putative GTP-binding protein resides in the vicinity of the Xp/Yp telomere. Hum. Mol. Genet. 7:407–414 [DOI] [PubMed] [Google Scholar]
- 91. Gibbs J. B., Sigal I. S., Poe M., Scolnick E. M. 1984. Intrinsic GTPase activity distinguishes normal and oncogenic ras p21 molecules. Proc. Natl. Acad. Sci. U. S. A. 81:5704–5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Godon C., et al. 1998. The H2O2 stimulon in Saccharomyces cerevisiae. J. Biol. Chem. 273:22480–22489 [DOI] [PubMed] [Google Scholar]
- 93. Gohda J., et al. 2003. Elimination of the vertebrate Escherichia coli Ras-like protein homologue leads to cell cycle arrest at G1 phase and apoptosis. Oncogene 22:1340–1348 [DOI] [PubMed] [Google Scholar]
- 94. Gollop N., March P. E. 1991. A GTP-binding protein (Era) has an essential role in growth rate and cell cycle control in Escherichia coli. J. Bacteriol. 173:2265–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Gollop N., March P. E. 1991. Localization of the membrane binding sites of Era in Escherichia coli. Res. Microbiol. 142:301–307 [DOI] [PubMed] [Google Scholar]
- 96. Gong S., Ma Z., Foster J. W. 2004. The Era-like GTPase TrmE conditionally activates gadE and glutamate-dependent acid resistance in Escherichia coli. Mol. Microbiol. 54:948–961 [DOI] [PubMed] [Google Scholar]
- 97. Gradia D. F., et al. 2009. Characterization of a novel Obg-like ATPase in the protozoan Trypanosoma cruzi. Int. J. Parasitol. 39:49–58 [DOI] [PubMed] [Google Scholar]
- 98. Grishin N. V. 2001. KH domain: one motif, two folds. Nucleic Acids Res. 29:638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Grudnik P., Bange G., Sinning I. 2009. Protein targeting by the signal recognition particle. Biol. Chem. 390:775–782 [DOI] [PubMed] [Google Scholar]
- 100. Guerrero C., Tagwerker C., Kaiser P., Huang L. 2006. An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol. Cell. Proteomics 5:366–378 [DOI] [PubMed] [Google Scholar]
- 101. Hagervall T. G., Pomerantz S. C., McCloskey J. A. 1998. Reduced misreading of asparagine codons by Escherichia coli tRNALys with hypomodified derivatives of 5-methylaminomethyl-2-thiouridine in the wobble position. J. Mol. Biol. 284:33–42 [DOI] [PubMed] [Google Scholar]
- 102. Hang J. Q., Meier T. I., Zhao G. 2001. Analysis of the interaction of 16S rRNA and cytoplasmic membrane with the C-terminal part of the Streptococcus pneumoniae Era GTPase. Eur. J. Biochem. 268:5570–5577 [DOI] [PubMed] [Google Scholar]
- 103. Hang J. Q., Zhao G. 2003. Characterization of the 16S rRNA- and membrane-binding domains of Streptococcus pneumoniae Era GTPase: structural and functional implications. Eur. J. Biochem. 270:4164–4172 [DOI] [PubMed] [Google Scholar]
- 104. Helena Mangs A., Morris B. J. 2007. The human pseudoautosomal region (PAR): origin, function and future. Curr. Genomics 8:129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Herskovits A. A., Bochkareva E. S., Bibi E. 2000. New prospects in studying the bacterial signal recognition particle pathway. Mol. Microbiol. 38:927–939 [DOI] [PubMed] [Google Scholar]
- 106. Heupel S., et al. 2010. Erl1, a novel Era-like GTPase from Magnaporthe oryzae, is required for full root virulence and is conserved in the mutualistic symbiont Glomus intraradices. Mol. Plant Microbe Interact. 23:67–81 [DOI] [PubMed] [Google Scholar]
- 107. Hilgenfeld R. 1995. Regulatory GTPases. Curr. Opin. Struct. Biol. 5:810–817 [DOI] [PubMed] [Google Scholar]
- 108. Hirano Y., Ohniwa R. L., Wada C., Yoshimura S. H., Takeyasu K. 2006. Human small G proteins, ObgH1, and ObgH2, participate in the maintenance of mitochondria and nucleolar architectures. Genes Cells 11:1295–1304 [DOI] [PubMed] [Google Scholar]
- 109. Huang B., et al. 2010. Functional study on GTP hydrolysis by the GTP-binding protein from Sulfolobus solfataricus, a member of the HflX family. J. Biochem. 148:103–113 [DOI] [PubMed] [Google Scholar]
- 110. Huang Y., et al. 2008. Expression and regulation of the yggG gene of Escherichia coli. Curr. Microbiol. 56:14–20 [DOI] [PubMed] [Google Scholar]
- 111. Huang Y., et al. 2007. Up-regulation of yggG promotes the survival of Escherichia coli cells containing Era-1 mutant protein. FEMS Microbiol. Lett. 275:8–15 [DOI] [PubMed] [Google Scholar]
- 112. Hudson J. D., Young P. G. 1993. Sequence of the Schizosaccharomyces pombe gtp1 gene and identification of a novel family of putative GTP-binding proteins. Gene 125:191–193 [DOI] [PubMed] [Google Scholar]
- 113. Hutchison C. A., et al. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165–2169 [DOI] [PubMed] [Google Scholar]
- 114. Hwang J., Inouye M. 2010. A bacterial GAP-like protein, YihI, regulating the GTPase of Der, an essential GTP-binding protein in Escherichia coli. J. Mol. Biol. 399:759–772 [DOI] [PubMed] [Google Scholar]
- 115. Hwang J., Inouye M. 2001. An essential GTPase, Der, containing double GTP-binding domains from Escherichia coli and Thermotoga maritima. J. Biol. Chem. 276:31415–31421 [DOI] [PubMed] [Google Scholar]
- 116. Hwang J., Inouye M. 2010. Interaction of an essential Escherichia coli GTPase, Der, with the 50S ribosome via the KH-like domain. J. Bacteriol. 192:2277–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hwang J., Inouye M. 2008. RelA functionally suppresses the growth defect caused by a mutation in the G domain of the essential Der protein. J. Bacteriol. 190:3236–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Hwang J., Inouye M. 2006. The tandem GTPase, Der, is essential for the biogenesis of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 61:1660–1672 [DOI] [PubMed] [Google Scholar]
- 119. Inada T., et al. 1989. Temperature-sensitive lethal mutant of era, a G protein in Escherichia coli. J. Bacteriol. 171:5017–5024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ingram G. C., Simon R., Carpenter R., Coen E. S. 1998. The Antirrhinum ERG gene encodes a protein related to bacterial small GTPases and is required for embryonic viability. Curr. Biol. 8:1079–1082 [DOI] [PubMed] [Google Scholar]
- 121. Inoue K., Alsina J., Chen J., Inouye M. 2003. Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli. Mol. Microbiol. 48:1005–1016 [DOI] [PubMed] [Google Scholar]
- 122. Inoue K., Chen J., Tan Q., Inouye M. 2006. Era and RbfA have overlapping function in ribosome biogenesis in Escherichia coli. J. Mol. Microbiol. Biotechnol. 11:41–52 [DOI] [PubMed] [Google Scholar]
- 123. Ito K., Udaka S., Yamagata H. 1992. Cloning, characterization, and inactivation of the Bacillus brevis lon gene. J. Bacteriol. 174:2281–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Iwashita S., Song S. Y. 2008. RasGAPs: a crucial regulator of extracellular stimuli for homeostasis of cellular functions. Mol. Biosyst. 4:213–222 [DOI] [PubMed] [Google Scholar]
- 125. Jain N., et al. 2009. E. coli HflX interacts with 50S ribosomal subunits in presence of nucleotides. Biochem. Biophys. Res. Commun. 379:201–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Jiang M., et al. 2006. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 188:6757–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Jiang M., Sullivan S. M., Wout P. K., Maddock J. R. 2007. G-protein control of the ribosome-associated stress response protein SpoT. J. Bacteriol. 189:6140–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Johnstone B. H., et al. 1999. The widely conserved Era G-protein contains an RNA-binding domain required for Era function in vivo. Mol. Microbiol. 33:1118–1131 [DOI] [PubMed] [Google Scholar]
- 129. Joyce C. M., Grindley N. D. 1982. Identification of two genes immediately downstream from the polA gene of Escherichia coli. J. Bacteriol. 152:1211–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kaczanowska M., Rydén-Aulin M. 2007. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 71:477–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kaldalu N., Mei R., Lewis K. 2004. Killing by ampicillin and ofloxacin induces overlapping changes in Escherichia coli transcription profile. Antimicrob. Agents Chemother. 48:890–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Karbstein K. 2007. Role of GTPases in ribosome assembly. Biopolymers 87:1–11 [DOI] [PubMed] [Google Scholar]
- 133. Keenan R. J., Freymann D. M., Stroud R. M., Walter P. 2001. The signal recognition particle. Annu. Rev. Biochem. 70:755–775 [DOI] [PubMed] [Google Scholar]
- 134. Keenan R. J., Freymann D. M., Walter P., Stroud R. M. 1998. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell 94:181–191 [DOI] [PubMed] [Google Scholar]
- 135. Khil P. P., Camerini-Otero R. D. 2002. Over 1000 genes are involved in the DNA damage response of Escherichia coli. Mol. Microbiol. 44:89–105 [DOI] [PubMed] [Google Scholar]
- 136. Kira Y., Nishikawa M. 2008. The identification and characterization of a new GTP-binding protein (Gbp45) involved in cell proliferation and death related to mitochondrial function. Cell. Mol. Biol. Lett. 13:570–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Kirino Y., Suzuki T. 2005. Human mitochondrial diseases associated with tRNA wobble modification deficiency. RNA Biol. 2:41–44 [DOI] [PubMed] [Google Scholar]
- 138. Kobayashi G., Moriya S., Wada C. 2001. Deficiency of essential GTP-binding protein ObgE in Escherichia coli inhibits chromosome partition. Mol. Microbiol. 41:1037–1051 [DOI] [PubMed] [Google Scholar]
- 139. Kok J., Trach K. A., Hoch J. A. 1994. Effects on Bacillus subtilis of a conditional lethal mutation in the essential GTP-binding protein Obg. J. Bacteriol. 176:7155–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Koller-Eichhorn R., et al. 2007. Human OLA1 defines an ATPase subfamily in the Obg family of GTP-binding proteins. J. Biol. Chem. 282:19928–19937 [DOI] [PubMed] [Google Scholar]
- 141. Krengel U., et al. 1990. Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell 62:539–548 [DOI] [PubMed] [Google Scholar]
- 142. Krüger M. K., Pedersen S., Hagervall T. G., Sørensen M. A. 1998. The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J. Mol. Biol. 284:621–631 [DOI] [PubMed] [Google Scholar]
- 143. Krüger M. K., Sørensen M. A. 1998. Aminoacylation of hypomodified tRNAGlu in vivo. J. Mol. Biol. 284:609–620 [DOI] [PubMed] [Google Scholar]
- 144. Kukimoto-Niino M., et al. 2004. Crystal structure of the GTP-binding protein Obg from Thermus thermophilus HB8. J. Mol. Biol. 337:761–770 [DOI] [PubMed] [Google Scholar]
- 145. Kuo S., Demeler B., Haldenwang W. G. 2008. The growth-promoting and stress response activities of the Bacillus subtilis GTP binding protein Obg are separable by mutation. J. Bacteriol. 190:6625–6635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Kuo S., Zhang S., Woodbury R. L., Haldenwang W. G. 2004. Associations between Bacillus subtilis σB regulators in cell extracts. Microbiology 150:4125–4136 [DOI] [PubMed] [Google Scholar]
- 147. Laalami S., Grentzmann G., Bremaud L., Cenatiempo Y. 1996. mRNA translation in prokaryotes: GTPase centers associated with translational factors. Biochimie 78:577–589 [DOI] [PubMed] [Google Scholar]
- 148. Lamb H. K., et al. 2007. Functional analysis of the GTPases EngA and YhbZ encoded by Salmonella typhimurium. Protein Sci. 16:2391–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Laursen B. S., Sorensen H. P., Mortensen K. K., Sperling-Petersen H. U. 2005. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 69:101–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Lee R., Aung-Htut M. T., Kwik C., March P. E. 2011. Expression phenotypes suggest that Der participates in a specific, high affinity interaction with membranes. Protein Expr. Purif. 78:102–112 [DOI] [PubMed] [Google Scholar]
- 151. Lee Y., et al. 2010. Molecular modeling study for interaction between Bacillus subtilis Obg and nucleotides. PLoS One 5:e12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Lehoux I. E., Mazzulla M. J., Baker A., Petit C. M. 2003. Purification and characterization of YihA, an essential GTP-binding protein from Escherichia coli. Protein Expr. Purif. 30:203–209 [DOI] [PubMed] [Google Scholar]
- 153. Leipe D. D., Wolf Y. I., Koonin E. V., Aravind L. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317:41–72 [DOI] [PubMed] [Google Scholar]
- 154. Lerner C. G., Gulati P. S., Inouye M. 1995. Cold-sensitive conditional mutations in Era, an essential Escherichia coli GTPase, isolated by localized random PCR mutagenesis. FEMS Microbiol. Lett. 126:291–298 [DOI] [PubMed] [Google Scholar]
- 155. Lerner C. G., Inouye M. 1991. Pleiotropic changes resulting from depletion of Era, an essential GTP-binding protein in Escherichia coli. Mol. Microbiol. 5:951–957 [DOI] [PubMed] [Google Scholar]
- 156. Lerner C. G., Sood P., Ahnn J., Inouye M. 1992. Cold-sensitive growth and decreased GTP-hydrolytic activity from substitution of Pro17 for Val in Era, an essential Escherichia coli GTPase. FEMS Microbiol. Lett. 74:137–142 [DOI] [PubMed] [Google Scholar]
- 157. Levdikov V. M., et al. 2004. The crystal structure of YloQ, a circularly permuted GTPase essential for Bacillus subtilis viability. J. Mol. Biol. 340:767–782 [DOI] [PubMed] [Google Scholar]
- 158. Li X., Guan M. X. 2002. A human mitochondrial GTP binding protein related to tRNA modification may modulate phenotypic expression of the deafness-associated mitochondrial 12S rRNA mutation. Mol. Cell. Biol. 22:7701–7711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Lin B., Covalle K. L., Maddock J. R. 1999. The Caulobacter crescentus CgtA protein displays unusual guanine nucleotide binding and exchange properties. J. Bacteriol. 181:5825–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Lin B., Maddock J. R. 2001. The N-terminal domain of the Caulobacter crescentus CgtA protein does not function as a guanine nucleotide exchange factor. FEBS Lett. 489:108–111 [DOI] [PubMed] [Google Scholar]
- 161. Lin B., et al. 2001. Alanine scan mutagenesis of the switch I domain of the Caulobacter crescentus CgtA protein reveals critical amino acids required for in vivo function. Mol. Microbiol. 39:924–934 [DOI] [PubMed] [Google Scholar]
- 162. Lin B., Thayer D. A., Maddock J. R. 2004. The Caulobacter crescentus CgtAC protein cosediments with the free 50S ribosomal subunit. J. Bacteriol. 186:481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Lin Y. P., Sharer J. D., March P. E. 1994. GTPase-dependent signaling in bacteria: characterization of a membrane-binding site for Era in Escherichia coli. J. Bacteriol. 176:44–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Loh P. C., Morimoto T., Matsuo Y., Oshima T., Ogasawara N. 2007. The GTP-binding protein YqeH participates in biogenesis of the 30S ribosome subunit in Bacillus subtilis. Genes Genet. Syst. 82:281–289 [DOI] [PubMed] [Google Scholar]
- 165. Lu Q., Inouye M. 1998. The gene for 16S rRNA methyltransferase (ksgA) functions as a multicopy suppressor for a cold-sensitive mutant of Era, an essential RAS-like GTP-binding protein in Escherichia coli. J. Bacteriol. 180:5243–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Maddock J., Bhatt A., Koch M., Skidmore J. 1997. Identification of an essential Caulobacter crescentus gene encoding a member of the Obg family of GTP-binding proteins. J. Bacteriol. 179:6426–6431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. March P. E. 1992. Membrane-associated GTPases in bacteria. Mol. Microbiol. 6:1253–1257 [DOI] [PubMed] [Google Scholar]
- 168. March P. E., Inouye M. 1985. Characterization of the lep operon of Escherichia coli. Identification of the promoter and the gene upstream of the signal peptidase I gene. J. Biol. Chem. 260:7206–7213 [PubMed] [Google Scholar]
- 169. March P. E., Inouye M. 1985. GTP-binding membrane protein of Escherichia coli with sequence homology to initiation factor 2 and elongation factors Tu and G. Proc. Natl. Acad. Sci. U. S. A. 82:7500–7504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. March P. E., Lerner C. G., Ahnn J., Cui X., Inouye M. 1988. The Escherichia coli Ras-like protein (Era) has GTPase activity and is essential for cell growth. Oncogene 2:539–544 [PubMed] [Google Scholar]
- 171. Martínez-Vicente M., et al. 2005. Effects of mutagenesis in the switch I region and conserved arginines of Escherichia coli MnmE protein, a GTPase involved in tRNA modification. J. Biol. Chem. 280:30660–30670 [DOI] [PubMed] [Google Scholar]
- 172. Matsunaga J., Dyer M., Simons E. L., Simons R. W. 1996. Expression and regulation of the rnc and pdxJ operons of Escherichia coli. Mol. Microbiol. 22:977–989 [DOI] [PubMed] [Google Scholar]
- 173. Matsuo Y., Oshima T., Loh P. C., Morimoto T., Ogasawara N. 2007. Isolation and characterization of a dominant negative mutant of Bacillus subtilis GTP-binding protein, YlqF, essential for biogenesis and maintenance of the 50 S ribosomal subunit. J. Biol. Chem. 282:25270–25277 [DOI] [PubMed] [Google Scholar]
- 174. Mehr I. J., Long C. D., Serkin C. D., Seifert H. S. 2000. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154:523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Meier T. I., Peery R. B., Jaskunas S. R., Zhao G. 1999. 16S rRNA is bound to era of Streptococcus pneumoniae. J. Bacteriol. 181:5242–5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Meier T. I., Peery R. B., McAllister K. A., Zhao G. 2000. Era GTPase of Escherichia coli: binding to 16S rRNA and modulation of GTPase activity by RNA and carbohydrates. Microbiology 146:1071–1083 [DOI] [PubMed] [Google Scholar]
- 177. Metallo S. J. 2010. Intrinsically disordered proteins are potential drug targets. Curr. Opin. Chem. Biol. 14:481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Meyer S., et al. 2009. Kissing G domains of MnmE monitored by X-ray crystallography and pulse electron paramagnetic resonance spectroscopy. PLoS Biol. 7:e1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Meyer S., Scrima A., Versées W., Wittinghofer A. 2008. Crystal structures of the conserved tRNA-modifying enzyme GidA: implications for its interaction with MnmE and substrate. J. Mol. Biol. 380:532–547 [DOI] [PubMed] [Google Scholar]
- 180. Meyer S., Wittinghofer A., Versées W. 2009. G-domain dimerization orchestrates the tRNA wobble modification reaction in the MnmE/GidA complex. J. Mol. Biol. 392:910–922 [DOI] [PubMed] [Google Scholar]
- 181. Minkovsky N., et al. 2002. Bex, the Bacillus subtilis homolog of the essential Escherichia coli GTPase Era, is required for normal cell division and spore formation. J. Bacteriol. 184:6389–6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Mishra R., Gara S. K., Mishra S., Prakash B. 2005. Analysis of GTPases carrying hydrophobic amino acid substitutions in lieu of the catalytic glutamine: implications for GTP hydrolysis. Proteins 59:332–338 [DOI] [PubMed] [Google Scholar]
- 183. Monleón D., et al. 2007. Structural insights into the GTPase domain of Escherichia coli MnmE protein. Proteins 66:726–739 [DOI] [PubMed] [Google Scholar]
- 184. Monleón D., Yim L., Martínez-Vicente M., Armengod M. E., Celda B. 2004. Backbone 1H, 13C and 15N resonance assignments for the 18.7 kDa GTPase domain of Escherichia coli MnmE protein. J. Biomol. NMR 28:307–308 [DOI] [PubMed] [Google Scholar]
- 185. Montoya G., Svensson C., Luirink J., Sinning I. 1997. Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature 385:365–368 [DOI] [PubMed] [Google Scholar]
- 186. Montoya G., Svensson C., Luirink J., Sinning I. 1997. Expression, crystallization and preliminary X-ray diffraction study of FtsY, the docking protein of the signal recognition particle of E. coli. Proteins 28:285–288 [DOI] [PubMed] [Google Scholar]
- 187. Moreno J. M., Sorensen H. P., Mortensen K. K., Sperling-Petersen H. U. 2000. Macromolecular mimicry in translation initiation: a model for the initiation factor IF2 on the ribosome. IUBMB Life 50:347–354 [DOI] [PubMed] [Google Scholar]
- 188. Morimoto T., et al. 2002. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology 148:3539–3552 [DOI] [PubMed] [Google Scholar]
- 189. Moukadiri I., et al. 2009. Evolutionarily conserved proteins MnmE and GidA catalyze the formation of two methyluridine derivatives at tRNA wobble positions. Nucleic Acids Res. 37:7177–7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Muench S. P., Xu L., Sedelnikova S. E., Rice D. W. 2006. The essential GTPase YphC displays a major domain rearrangement associated with nucleotide binding. Proc. Natl. Acad. Sci. U. S. A. 103:12359–12364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Müller M., Koch H. G., Beck K., Schäfer U. 2001. Protein traffic in bacteria: multiple routes from the ribosome to and across the membrane. Prog. Nucleic Acid Res. Mol. Biol. 66:107–157 [DOI] [PubMed] [Google Scholar]
- 192. Nashimoto H., Miura A., Saito H., Uchida H. 1985. Suppressors of temperature-sensitive mutations in a ribosomal protein gene, rpsL (S12), of Escherichia coli K12. Mol. Gen. Genet. 199:381–387 [DOI] [PubMed] [Google Scholar]
- 193. Nilsson J., Nissen P. 2005. Elongation factors on the ribosome. Curr. Opin. Struct. Biol. 15:349–354 [DOI] [PubMed] [Google Scholar]
- 194. Noble C. G., Song H. 2008. Structural studies of elongation and release factors. Cell. Mol. Life Sci. 65:1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Noble J. A., et al. 1993. The Escherichia coli hflA locus encodes a putative GTP-binding protein and two membrane proteins, one of which contains a protease-like domain. Proc. Natl. Acad. Sci. U. S. A. 90:10866–10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Okamoto S., Itoh M., Ochi K. 1997. Molecular cloning and characterization of the obg gene of Streptomyces griseus in relation to the onset of morphological differentiation. J. Bacteriol. 179:170–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Okamoto S., Ochi K. 1998. An essential GTP-binding protein functions as a regulator for differentiation in Streptomyces coelicolor. Mol. Microbiol. 30:107–119 [DOI] [PubMed] [Google Scholar]
- 198. Osawa T., Inanaga H., Numata T. 2009. Crystallization and preliminary X-ray diffraction analysis of the tRNA-modification enzyme GidA from Aquifex aeolicus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65:508–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Osawa T., et al. 2009. Conserved cysteine residues of GidA are essential for biogenesis of 5-carboxymethylaminomethyluridine at tRNA anticodon. Structure 17:713–724 [DOI] [PubMed] [Google Scholar]
- 200. Paduch M., Jeleń F., Otlewski J. 2001. Structure of small G proteins and their regulators. Acta Biochim. Pol. 48:829–850 [PubMed] [Google Scholar]
- 201. Patel B. A., et al. 2009. Endogenous nitric oxide regulates the recovery of the radiation-resistant bacterium Deinococcus radiodurans from exposure to UV light. Proc. Natl. Acad. Sci. U. S. A. 106:18183–18188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Persky N. S., Ferullo D. J., Cooper D. L., Moore H. R., Lovett S. T. 2009. The ObgE/CgtA GTPase influences the stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 73:253–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Pillutla R. C., et al. 1995. Cross-species complementation of the indispensable Escherichia coli era gene highlights amino acid regions essential for activity. J. Bacteriol. 177:2194–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Polkinghorne A. 2005. Ph.D. thesis Queensland University of Technology, Queensland, Australia [Google Scholar]
- 205. Polkinghorne A., Hogan R. J., Vaughan L., Summersgill J. T., Timms P. 2006. Differential expression of chlamydial signal transduction genes in normal and interferon gamma-induced persistent Chlamydophila pneumoniae infections. Microbes Infect. 8:61–72 [DOI] [PubMed] [Google Scholar]
- 206. Polkinghorne A., Vaughan L. 2011. Chlamydia abortus YhbZ, a truncated Obg family GTPase, associates with the Escherichia coli large ribosomal subunit. Microb. Pathog. 50(3–4):200–206 [DOI] [PubMed] [Google Scholar]
- 207. Polkinghorne A., et al. 2008. Chlamydophila pneumoniae HflX belongs to an uncharacterized family of conserved GTPases and associates with the Escherichia coli 50S large ribosomal subunit. Microbiology 154:3537–3546 [DOI] [PubMed] [Google Scholar]
- 208. Powell B. S., et al. 1995. Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli. Enzyme IIA Ntr affects growth on organic nitrogen and the conditional lethality of an erats mutant. J. Biol. Chem. 270:4822–4839 [DOI] [PubMed] [Google Scholar]
- 209. Prágai Z., Harwood C. R. 2000. YsxC, a putative GTP-binding protein essential for growth of Bacillus subtilis 168. J. Bacteriol. 182:6819–6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Qin Y., et al. 2006. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell 127:721–733 [DOI] [PubMed] [Google Scholar]
- 211. Raskin D. M., Judson N., Mekalanos J. J. 2007. Regulation of the stringent response is the essential function of the conserved bacterial G protein CgtA in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 104:4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Richmond C. S., Glasner J. D., Mau R., Jin H., Blattner F. R. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Riethdorf S., et al. 1994. Cloning, nucleotide sequence, and expression of the Bacillus subtilis lon gene. J. Bacteriol. 176:6518–6527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Robinson V. L., Hwang J., Fox E., Inouye M., Stock A. M. 2002. Domain arrangement of Der, a switch protein containing two GTPase domains. Structure 10:1649–1658 [DOI] [PubMed] [Google Scholar]
- 215. Roll-Mecak A., Cao C., Dever T. E., Burley S. K. 2000. X-Ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell 103:781–792 [DOI] [PubMed] [Google Scholar]
- 216. Ruzheinikov S. N., et al. 2004. Analysis of the open and closed conformations of the GTP-binding protein YsxC from Bacillus subtilis. J. Mol. Biol. 339:265–278 [DOI] [PubMed] [Google Scholar]
- 217. Sasindran S. J., Saikolappan S., Scofield V. L., Dhandayuthapani S. 2011. Biochemical and physiological characterization of the GTP-binding protein Obg of Mycobacterium tuberculosis. BMC Microbiol. 11:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Sato A., et al. 2005. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells 10:393–408 [DOI] [PubMed] [Google Scholar]
- 219. Sato T., Wu J., Kuramitsu H. 1998. The sgp gene modulates stress responses of Streptococcus mutans: utilization of an antisense RNA strategy to investigate essential gene functions. FEMS Microbiol. Lett. 159:241–245 [DOI] [PubMed] [Google Scholar]
- 220. Sawitzke J., Austin S. 2001. An analysis of the factory model for chromosome replication and segregation in bacteria. Mol. Microbiol. 40:786–794 [DOI] [PubMed] [Google Scholar]
- 221. Sayed A., Matsuyama S., Inouye M. 1999. Era, an essential Escherichia coli small G-protein, binds to the 30S ribosomal subunit. Biochem. Biophys. Res. Commun. 264:51–54 [DOI] [PubMed] [Google Scholar]
- 222. Sayed A. K., Foster J. W. 2009. A 750 bp sensory integration region directs global control of the Escherichia coli GadE acid resistance regulator. Mol. Microbiol. 71:1435–1450 [DOI] [PubMed] [Google Scholar]
- 223. Schaefer L., et al. 2006. Multiple GTPases participate in the assembly of the large ribosomal subunit in Bacillus subtilis. J. Bacteriol. 188:8252–8258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Scheffzek K., et al. 1997. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277:333–338 [DOI] [PubMed] [Google Scholar]
- 225. Schmidt R., Decatur A. L., Rather P. N., Moran C. P., Jr., Losick R. 1994. Bacillus subtilis lon protease prevents inappropriate transcription of genes under the control of the sporulation transcription factor σG. J. Bacteriol. 176:6528–6537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Scott J. M., Haldenwang W. G. 1999. Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor σB. J. Bacteriol. 181:4653–4660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Scott J. M., Ju J., Mitchell T., Haldenwang W. G. 2000. The Bacillus subtilis GTP binding protein Obg and regulators of the σB stress response transcription factor cofractionate with ribosomes. J. Bacteriol. 182:2771–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Scrima A., Vetter I. R., Armengod M. E., Wittinghofer A. 2005. The structure of the TrmE GTP-binding protein and its implications for tRNA modification. EMBO J. 24:23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Scrima A., Wittinghofer A. 2006. Dimerisation-dependent GTPase reaction of MnmE: how potassium acts as GTPase-activating element. EMBO J. 25:2940–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Sergiev P. V., Bogdanov A. A., Dontsova O. A. 2005. How can elongation factors EF-G and EF-Tu discriminate the functional state of the ribosome using the same binding site? FEBS Lett. 579:5439–5442 [DOI] [PubMed] [Google Scholar]
- 231. Shah S., Das B., Bhadra R. K. 2008. Functional analysis of the essential GTP-binding-protein-coding gene cgtA of Vibrio cholerae. J. Bacteriol. 190:4764–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Shan S. O., Walter P. 2005. Co-translational protein targeting by the signal recognition particle. FEBS Lett. 579:921–926 [DOI] [PubMed] [Google Scholar]
- 233. Sharma M. R., et al. 2005. Interaction of Era with the 30S ribosomal subunit: implications for 30S subunit assembly. Mol. Cell 18:319–329 [DOI] [PubMed] [Google Scholar]
- 234. Shields M. J., Fischer J. J., Wieden H. J. 2009. Toward understanding the function of the universally conserved GTPase HflX from Escherichia coli: a kinetic approach. Biochemistry 48:10793–10802 [DOI] [PubMed] [Google Scholar]
- 235. Shimamoto T., Inouye M. 1996. Mutational analysis of Era, an essential GTP-binding protein of Escherichia coli. FEMS Microbiol. Lett. 136:57–62 [DOI] [PubMed] [Google Scholar]
- 236. Sikora A. E., Datta K., Maddock J. R. 2006. Biochemical properties of the Vibrio harveyi CgtAV GTPase. Biochem. Biophys. Res. Commun. 339:1165–1170 [DOI] [PubMed] [Google Scholar]
- 237. Sikora A. E., Zielke R., Datta K., Maddock J. R. 2006. The Vibrio harveyi GTPase CgtAV is essential and is associated with the 50S ribosomal subunit. J. Bacteriol. 188:1205–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Sikora A. E., Zielke R., Wegrzyn A., Wegrzyn G. 2006. DNA replication defect in the Escherichia coli cgtA(ts) mutant arising from reduced DnaA levels. Arch. Microbiol. 185:340–347 [DOI] [PubMed] [Google Scholar]
- 239. Simonetti A., et al. 2009. A structural view of translation initiation in bacteria. Cell. Mol. Life Sci. 66:423–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Singh A. K., Pindi P. K., Dube S., Sundareswaran V. R., Shivaji S. 2009. Importance of trmE for growth of the psychrophile Pseudomonas syringae at low temperatures. Appl. Environ. Microbiol. 75:4419–4426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Singh A. K., Shivaji S. 2010. A cold-active heat-labile t-RNA modification GTPase from a psychrophilic bacterium Pseudomonas syringae (Lz4W). Res. Microbiol. 161:46–50 [DOI] [PubMed] [Google Scholar]
- 242. Sood P., Lerner C. G., Shimamoto T., Lu Q., Inouye M. 1994. Characterization of the autophosphorylation of Era, an essential Escherichia coli GTPase. Mol. Microbiol. 12:201–208 [DOI] [PubMed] [Google Scholar]
- 243. Sprang S. R. 1997. G proteins, effectors and GAPs: structure and mechanism. Curr. Opin. Struct. Biol. 7:849–856 [DOI] [PubMed] [Google Scholar]
- 244. Sullivan S. M., Mishra R., Neubig R. R., Maddock J. R. 2000. Analysis of guanine nucleotide binding and exchange kinetics of the Escherichia coli GTPase Era. J. Bacteriol. 182:3460–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Surviladze Z., et al. 2010. Identification of a small GTPase inhibitor using a high-throughput flow cytometry bead-based multiplex assay. J. Biomol. Screen. 15:10–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Suzuki T., Suzuki T., Wada T., Saigo K., Watanabe K. 2002. Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J. 21:6581–6589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Szklarczyk D., et al. 2011. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39:D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Takiff H. E., Baker T., Copeland T., Chen S. M., Court D. L. 1992. Locating essential Escherichia coli genes by using mini-Tn10 transposons: the pdxJ operon. J. Bacteriol. 174:1544–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Takiff H. E., Chen S. M., Court D. L. 1989. Genetic analysis of the rnc operon of Escherichia coli. J. Bacteriol. 171:2581–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Tan J., Jakob U., Bardwell J. C. 2002. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 184:2692–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Teplyakov A., et al. 2003. Crystal structure of the YchF protein reveals binding sites for GTP and nucleic acid. J. Bacteriol. 185:4031–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Timson J. 1975. Hydroxyurea. Mutat. Res. 32:115–132 [DOI] [PubMed] [Google Scholar]
- 253. Tomar S. K., Dhimole N., Chatterjee M., Prakash B. 2009. Distinct GDP/GTP bound states of the tandem G-domains of EngA regulate ribosome binding. Nucleic Acids Res. 37:2359–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Trach K., Hoch J. A. 1989. The Bacillus subtilis spo0B stage 0 sporulation operon encodes an essential GTP-binding protein. J. Bacteriol. 171:1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Tsui H. C., Feng G., Winkler M. E. 1996. Transcription of the mutL repair, miaA tRNA modification, hfq pleiotropic regulator, and hflA region protease genes of Escherichia coli K-12 from clustered Esigma32-specific promoters during heat shock. J. Bacteriol. 178:5719–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Tsui H. C., Leung H. C., Winkler M. E. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 13:35–49 [DOI] [PubMed] [Google Scholar]
- 257. Tsui H. C., Winkler M. E. 1994. Transcriptional patterns of the mutL-miaA superoperon of Escherichia coli K-12 suggest a model for posttranscriptional regulation. Biochimie 76:1168–1177 [DOI] [PubMed] [Google Scholar]
- 258. Tu C., et al. 2009. Structure of ERA in complex with the 3′ end of 16S rRNA: implications for ribosome biogenesis. Proc. Natl. Acad. Sci. U. S. A. 106:14843–14848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Twiss E., Coros A. M., Tavakoli N. P., Derbyshire K. M. 2005. Transposition is modulated by a diverse set of host factors in Escherichia coli and is stimulated by nutritional stress. Mol. Microbiol. 57:1593–1607 [DOI] [PubMed] [Google Scholar]
- 260. Uchiumi T., et al. 2010. ERAL1 is associated with mitochondrial ribosome and elimination of ERAL1 leads to mitochondrial dysfunction and growth retardation. Nucleic Acids Res. 38:5554–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Ulanowska K., Sikora A., We¸grzyn G., Czyż A. 2003. Role of the cgtA gene function in DNA replication of extrachromosomal elements in Escherichia coli. Plasmid 50:45–52 [DOI] [PubMed] [Google Scholar]
- 262. Umeda N., et al. 2005. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 280:1613–1624 [DOI] [PubMed] [Google Scholar]
- 263. Urbonavičius J., Qian Q., Durand J. M., Hagervall T. G., Björk G. R. 2001. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 20:4863–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Urbonavičius J., et al. 2003. tRNA modifications that alter +1 frameshifting in general fail to affect −1 frameshifting. RNA 9:760–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. VanBogelen R. A., Neidhardt F. C. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 87:5589–5593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Vawter M. P., Harvey P. D., DeLisi L. E. 2007. Dysregulation of X-linked gene expression in Klinefelter's syndrome and association with verbal cognition. Am. J. Med. Genet. B Neuropsychiatr. Genet. 144B:728–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Vetter I. R., Wittinghofer A. 2001. The guanine nucleotide-binding switch in three dimensions. Science 294:1299–1304 [DOI] [PubMed] [Google Scholar]
- 268. Vidwans S. J., Ireton K., Grossman A. D. 1995. Possible role for the essential GTP-binding protein Obg in regulating the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 177:3308–3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Villarroya M., et al. 2008. Characterization of human GTPBP3, a GTP-binding protein involved in mitochondrial tRNA modification. Mol. Cell. Biol. 28:7514–7531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Watt R. M., et al. 2007. Visualizing the proteome of Escherichia coli: an efficient and versatile method for labeling chromosomal coding DNA sequences (CDSs) with fluorescent protein genes. Nucleic Acids Res. 35:e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Weber A., Kogl S. A., Jung K. 2006. Time-dependent proteome alterations under osmotic stress during aerobic and anaerobic growth in Escherichia coli. J. Bacteriol. 188:7165–7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Welsh K. M., Trach K. A., Folger C., Hoch J. A. 1994. Biochemical characterization of the essential GTP-binding protein Obg of Bacillus subtilis. J. Bacteriol. 176:7161–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Wicker-Planquart C., Foucher A. E., Louwagie M., Britton R. A., Jault J. M. 2008. Interactions of an essential Bacillus subtilis GTPase, YsxC, with ribosomes. J. Bacteriol. 190:681–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Wout P., et al. 2004. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J. Bacteriol. 186:5249–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Wout P. K., Sattlegger E., Sullivan S. M., Maddock J. R. 2009. Saccharomyces cerevisiae Rbg1 protein and its binding partner Gir2 interact on polyribosomes with Gcn1. Eukaryot. Cell 8:1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Wu H., et al. 2010. Structure of the ribosome associating GTPase HflX. Proteins 78:705–713 [DOI] [PubMed] [Google Scholar]
- 277. Xu L., Muench S. P., Roujeinikova A., Sedelnikova S. E., Rice D. W. 2006. Cloning, purification and preliminary crystallographic analysis of the Bacillus subtilis GTPase YphC-GDP complex. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62:435–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Yamanaka K., Hwang J., Inouye M. 2000. Characterization of GTPase activity of TrmE, a member of a novel GTPase superfamily, from Thermotoga maritima. J. Bacteriol. 182:7078–7082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Yasukawa T., Suzuki T., Ishii N., Ohta S., Watanabe K. 2001. Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. EMBO J. 20:4794–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Yasukawa T., et al. 2000. Defect in modification at the anticodon wobble nucleotide of mitochondrial tRNA(Lys) with the MERRF encephalomyopathy pathogenic mutation. FEBS Lett. 467:175–178 [DOI] [PubMed] [Google Scholar]
- 281. Yasukawa T., Suzuki T., Ueda T., Ohta S., Watanabe K. 2000. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAs(Leu)(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J. Biol. Chem. 275:4251–4257 [DOI] [PubMed] [Google Scholar]
- 282. Yim L., et al. 2003. The GTPase activity and C-terminal cysteine of the Escherichia coli MnmE protein are essential for its tRNA modifying function. J. Biol. Chem. 278:28378–28387 [DOI] [PubMed] [Google Scholar]
- 283. Yim L., Moukadiri I., Bjork G. R., Armengod M. E. 2006. Further insights into the tRNA modification process controlled by proteins MnmE and GidA of Escherichia coli. Nucleic Acids Res. 34:5892–5905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Youngman E. M., Green R. 2007. Ribosomal translocation: LepA does it backwards. Curr. Biol. 17:R136–R139 [DOI] [PubMed] [Google Scholar]
- 285. Zalacain M., et al. 2003. A global approach to identify novel broad-spectrum antibacterial targets among proteins of unknown function. J. Mol. Microbiol. Biotechnol. 6:109–126 [DOI] [PubMed] [Google Scholar]
- 286. Zhang J., Inouye M. 2002. MazG, a nucleoside triphosphate pyrophosphohydrolase, interacts with Era, an essential GTPase in Escherichia coli. J. Bacteriol. 184:5323–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Zhang J., Rubio V., Lieberman M. W., Shi Z. Z. 2009. OLA1, an Obg-like ATPase, suppresses antioxidant response via nontranscriptional mechanisms. Proc. Natl. Acad. Sci. U. S. A. 106:15356–15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Zhang J. W., Rubio V., Zheng S., Shi Z. Z. 2009. Knockdown of OLA1, a regulator of oxidative stress response, inhibits motility and invasion of breast cancer cells. J. Zhejiang Univ. Sci. B 10:796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Zhang S., Haldenwang W. G. 2004. Guanine nucleotides stabilize the binding of Bacillus subtilis Obg to ribosomes. Biochem. Biophys. Res. Commun. 322:565–569 [DOI] [PubMed] [Google Scholar]
- 290. Zhang S., Scott J. M., Haldenwang W. G. 2001. Loss of ribosomal protein L11 blocks stress activation of the Bacillus subtilis transcription factor sigma(B). J. Bacteriol. 183:2316–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Zhao G., Meier T. I., Peery R. B., Matsushima P., Skatrud P. L. 1999. Biochemical and molecular analyses of the C-terminal domain of Era GTPase from Streptococcus pneumoniae. Microbiology 145(Pt. 4):791–800 [DOI] [PubMed] [Google Scholar]
- 292. Zielke R., Sikora A., Dutkiewicz R., We¸grzyn G., Czyż A. 2003. Involvement of the cgtA gene function in stimulation of DNA repair in Escherichia coli and Vibrio harveyi. Microbiology 149:1763–1770 [DOI] [PubMed] [Google Scholar]
- 293. Zuber M., Hoover T. A., Dertzbaugh M. T., Court D. L. 1997. A Francisella tularensis DNA clone complements Escherichia coli defective for the production of Era, an essential Ras-like GTP-binding protein. Gene 189:31–34 [DOI] [PubMed] [Google Scholar]