Abstract
Escherichia coli and many other Gram-negative pathogenic bacteria protect themselves from the toxic effects of electrophilic compounds by using a potassium efflux system (Kef). Potassium efflux is coupled to the influx of protons, which lowers the internal pH and results in immediate protection. The activity of the Kef system is subject to complex regulation by glutathione and its S conjugates. Full activation of KefC requires a soluble ancillary protein, KefF. This protein has structural similarities to oxidoreductases, including human quinone reductases 1 and 2. Here, we show that KefF has enzymatic activity as an oxidoreductase, in addition to its role as the KefC activator. It accepts NADH and NADPH as electron donors and quinones and ferricyanide (in addition to other compounds) as acceptors. However, typical electrophilic activators of the Kef system, e.g., N-ethyl maleimide, are not substrates. If the enzymatic activity is disrupted by site-directed mutagenesis while retaining structural integrity, KefF is still able to activate the Kef system, showing that the role as an activator is independent of the enzyme activity. Potassium efflux assays show that electrophilic quinones are able to activate the Kef system by forming S conjugates with glutathione. Therefore, it appears that the enzymatic activity of KefF diminishes the redox toxicity of quinones, in parallel with the protection afforded by activation of the Kef system.
INTRODUCTION
Electrophilic compounds are often highly toxic, because they readily react with nucleophiles found both in the bases of DNA and within functionally important side chains of proteins, especially cysteine. Bacteria produce electrophiles themselves as metabolic by-products when they are unable to regulate carbon influx (39). In addition, pathogens may become exposed to electrophiles from exogenous sources (e.g., during entry into macrophages [10]). After entering the cell, electrophiles either react spontaneously with glutathione (GSH) or are conjugated to the nucleophile by glutathione S-transferase (40), which is the first step in detoxification. The glutathione adducts (GSX) can then be further detoxified (14, 27, 28). Despite active detoxification, potentially lethal direct reaction with DNA and other important macromolecules may occur, and ancillary damage may arise from depletion of the GSH pool (E. Ozyamak, C. Almeida, A. Moura, S. Miller, and I. R. Booth, unpublished data). Bacterial survival increases significantly if the cytosol is acidified during electrophile exposure (11, 13), possibly due to the reduction in nucleophile reactivity caused by protonation or partial protonation at lower pH values. Potassium efflux via KefC is accompanied by H+ and Na+ influx and hence causes acidification of the cytoplasm. Kef is activated by GSX and inactivated by GSH, binding to its cytosolic regulatory K+ transport and nucleotide binding (KTN) domain. Kef systems are found only in Gram-negative bacteria and have been most extensively studied in Escherichia coli, which has two homologues, KefB and KefC, showing differential sensitivity to electrophiles (12). Recently, we showed that GSH and GSX occupy overlapping binding pockets in KefC but that the additional steric bulk of GSX causes a significant conformational change, which results in activation of KefC (35). In addition, the KTN domain has a nucleotide binding site on a Rossman fold (36), and it has been suggested that KefC is inactivated when NADH is bound at this site (16).
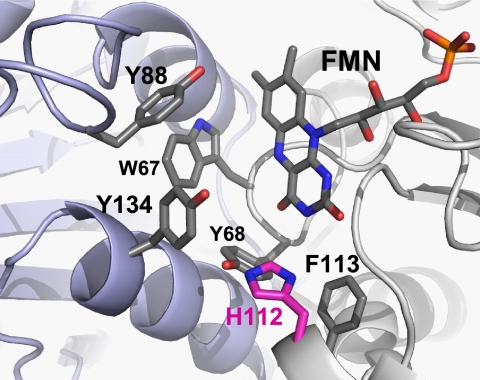
In many bacteria, including E. coli, regulation of Kef is still more complicated, as soluble KefB/KefC-binding proteins have been discovered (KefG and KefF, respectively) that are required to associate with KefB/KefC to allow full activation of these systems (29). An X-ray crystal structure of the KefC KTN domain in complex with KefF indicates that binding of the KefF dimer to KefC holds the KTN domains at a specific angle that is believed to be favorable for KefC activity (36). Surprisingly, the sequences of KefG/KefF show homology to quinone reductases, such as the human quinone reductase 1 (QR1) and QR2. Comparison of the X-ray crystal structures of KefF and human QR1 demonstrates that the sequence similarity to QR1 and QR2 is also reflected in the fold of the proteins (see Fig. S1 in the supplemental material). Furthermore, the flavin mononucleotide cofactor, FMN, required for oxidoreductase catalytic reaction, is bound to KefF in the crystal structure. Immediately adjacent to the bound FMN, there is a hydrophobic cleft suitable for substrate binding (Fig. 1). Within this cleft, electron density is present in the X-ray crystal structure, but the bound compound could not be identified (36). Thus, we sought to establish whether KefF has enzyme activity in addition to its role as an activator of KefC and whether these two functions are directly connected.
Fig. 1.
Structure of the active site in KefF. Aromatic residues of both subunits contribute to the active site of KefF; the backbone of one subunit is shown in blue and the other in gray. Histidine 112 (pink) is proposed to be important for charge stabilization on the reduced FMN during the catalytic cycle, with further delocalization to Y68 or Y134. The image was generated with PyMOL on the basis of the KefF crystal structure (36).
We demonstrate here that KefF is an effective oxidoreductase for a wide range of electron acceptors, but not for formerly known electrophiles that activate the Kef systems, like, for example, methylglyoxal (MG) and N-ethyl maleimide (NEM). The enzymatic activity of KefF is not directly required for activation of KefC. Furthermore, electrophilic quinones, which can readily form adducts with GSH, were established as potent activators of KefC and are also good substrates for the enzymatic activity of KefF.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are described in Table 1. The expression construct pTrcYabFH6, used for most experiments, carries a C-terminal His6 tag. The gene was cloned via an NcoI site at the 5′ end and an XhoI site on the 3′ end into a pTrc99A backbone, which introduced an N-terminal I2V mutation. Site-directed mutants were derived from this construct by the Stratagene QuikChange protocol. All constructs were confirmed by sequencing on both strands.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or description | Reference |
|---|---|---|
| Strains | ||
| Frag5 | F− ΔkdpABC5 thi rha lacZ | 9 |
| Frag56 | Frag5 gshA::Tn10 (kan) | 8 |
| MJF274 | F− ΔkdpABC5 thi rha lacZ trkD1 | 8 |
| MJF276 | MJF274 kefB157 kefC::Tn10 | 33 |
| MJF362 | Frag5 Δ[yabF-kefC]::kan | 29 |
| BW25113 ΔkefF | Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) lambda−rph-1 Δ(rhaD-rhaB)568 hsdR514 kefF::kan | This study |
| MJF654 | BW25113 kefF::kan kefBG::apr | This study |
| Plasmids | ||
| pTrcYabFH6 | KefF from E. coli with C-terminal His6 tag | This study |
| pYabFH6G107S | E. coli KefF with G107S mutation preventing FMN binding | This study |
| pYabFH6H112W | E. coli KefF with H112W mutation blocking substrate binding | This study |
| pYabFH6F149W | E. coli KefF with F149W mutation blocking substrate binding | This study |
| pYabFH6C147S | Mutation evaluating the regulatory role of surface-exposed cysteine | This study |
| pYabFH6C151S | Mutation evaluating the regulatory role of surface-exposed cysteine | This study |
| pYabFH6C147S/C151S | Mutation evaluating the regulatory role of surface-exposed cysteine | This study |
Strain BW25113 ΔyabF::kan was created using the method of Datsenko and Wanner (6). Briefly, the PCR primers Δyabf1 (5′-TTGTGGGAAAACAAAGGCGTAATCACGCGGGCTACCTGTGTAGGCTGGAGCTGCTTC-3′) and ΔyabF2 (5′-GAGATAAATCAGCGCCTGAACAGCGTATGGCTATCCATATGAATATCCTCCTTAG-3′) were used to amplify the kanamycin resistance gene from pKD4. The resulting DNA was purified using a Qiagen Qiaquick Kit, treated with DpnI, and transformed by electroporation into BW25113 pKD46. The transformed cells were recovered on LK medium (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter KCl) containing kanamycin (25 μg/ml) and then tested for growth on LK medium containing kanamycin or ampicillin (25 μg/ml). Colonies that retained only kanamycin resistance (thus judged to no longer carry the pKD46 plasmid) were verified for creation of kefF::kan knockout by PCR using primers designed for regions outside the kefF locus. The resultant strain, BW25113 ΔyabF::kan, was transduced to kefGB::apr using E. coli strain DY330 ΔkefGB::apr (E. Ozyamak and I. R. Booth, unpublished data) as a donor and a standard P1 phage protocol to create strain MJF654 (ΔyabF::kan kefGB::apr). MJF654 colonies were selected by resistance to 60 μg/ml apramycin (apr) and 25 μg/ml kanamycin (kan).
Measurements of KefC potassium efflux.
Potassium efflux experiments were performed as described previously (8). Briefly, freshly transformed E. coli MJF654 cells were cultured in K115 minimal medium at 37°C to an optical density at 650 nm (OD650) of ∼0.8 to 1 (9). K115 minimal medium consists of 46 mM K2HPO4, 23 mM KH2PO4, and 8 mM (NH4)2SO4 supplemented with separately autoclaved 0.4 mM MgSO4, 6 μM (NH4)2SO4·FeSO4, 1 mg/liter thiamine hydrochloride, and 2 g/liter glucose. The cell culture was harvested by filtration, and the cells were washed with K10 buffer before they were suspended in potassium-free K0 buffer. For the K0 buffer, the potassium salts of the K115 medium were replaced by sodium salts and the supplements were omitted. The K10 buffer was prepared by adding 10 mM KCl to K0 buffer. Equal volumes of cells were then poured into two insulated stirring vessels maintained at 37°C, and 0.5 mM NEM was added to one vessel. Samples were taken from each vessel at defined time intervals for measurement of the retained cellular potassium content by flame photometry. The logarithmic values of the intracellular potassium concentrations were plotted against the time after addition of NEM, and the slope was determined by linear regression. Potassium efflux triggered by quinones was measured with an ion-selective electrode as described previously (35) using the strains MJF274, MJF276, Frag56, and MJF654. Cells were grown and washed as described above. They were suspended in 5 ml potassium-free K0 buffer and added to 30 ml of K0 buffer in a single stirred vessel held at 37°C with equilibrated electrodes inserted. After 5 min, either quinones or NEM (0.5 mM final concentration) was added, and the potassium content of the medium was recorded continuously. Rate constants were obtained, using the program Origin 8.0 (Originlab) (35), by fitting exponentials to the change in potassium concentration after addition of an activator.
Protein expression and purification.
KefF constructs were transformed into the E. coli strain MJF362 for expression. The culture was grown in 0.5 liter terrific broth (TB) at 37°C. For the TB medium, 12 g tryptone, 24 g yeast extract, and 4 ml glycerol were dissolved in 900 ml deionized water and autoclaved. One hundred milliliters of separately sterilized phosphate buffer (0.17 M KH2PO4 and 0.72 M K2HPO4) was added after cooling. When the culture reached an OD650 of 0.8 to 1, expression was induced with 1.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 25°C. Cells were harvested by 15 min of centrifugation at 4,000 × g, suspended in 20 ml lysis buffer (50 mM HEPES, 150 mM NaCl, 10% glycerol), and stored at −80°C. Phenylmethylsulfonyl fluoride (PMSF) (1 mM), 10 mM benzamidine, and 10 μM FMN were added to the cell suspension before lysis in a French press (SLM Aminco) at a pressure of 18,000 lb/in2. Bacterial membranes were removed by ultracentrifugation at 100,000 × g for 1 h. The supernatant was filtered before binding to 2 ml Ni-nitrilotriacetic acid (NTA) affinity matrix (Sigma) packed in a column. After washing the column with 30 ml washing buffer (50 mM imidazole, 50 mM HEPES, 150 mM NaCl, and 10% glycerol), KefF was eluted with 5 ml elution buffer (same as washing buffer but with 300 mM imidazole). Purified protein was analyzed by SDS-PAGE using a precast 4 to 12% NuPage Novex Bis-Tris gradient gel (Invitrogen). Western blots were made on nitrocellulose and detected with anti-His6 antibody (mouse IgG2a isotype; Sigma). Images were visualized using ECL substrate (Pierce) and photographic film (Kodak). Size exclusion chromatography was carried out using a Hiload 16/60 Superdex 200 PG column (GE Healthcare) and high-performance liquid chromatography (HPLC) buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol) at a flow rate of 0.5 ml/min. The molecular mass of the KefF complex was estimated from a calibration using soluble proteins.
The protein concentration was determined using the Lowry method (25) with bovine serum albumin (BSA) as a standard, and the FMN concentration was determined using the fluorescence of the flavin. For the FMN determination, purified KefF samples and free FMN as calibration samples were diluted in buffer (50 mM MES [morpholineethanesulfonic acid]-NaOH, pH 6.0, and 50 mM NaCl) to appropriate dilutions. The samples and calibration samples were incubated for 10 min at 100°C wrapped in tin foil. After cooling, they were centrifuged at 13,000 × g for 10 min at 4°C. Fluorescence was measured with an FLS920 spectrometer (Edinburgh Instruments) at an excitation wavelength of 450 nm and an emission wavelength of 530 nm. Concentrations of KefF and FMN were confirmed by UV/visible-light (Vis) absorption. Extinction coefficients for the FMN bound to KefF were determined by comparison of absorption spectra of native KefF before and after incubation with 0.2% SDS for 10 min in the dark at room temperature (RT) to release the FMN. An extinction coefficient for free FMN of 12.5 mM−1 cm−1 at 446 nm (ε446) was assumed (26).
Enzyme kinetics.
Enzyme kinetics were measured using a Cary50 UV/Vis spectrometer (Varian) with a temperature-controlled Pelletier cell holder and stirred cells (25°C) in 50 mM HEPES, pH 7.5, with 150 mM NaCl and 3 μM FMN. A constant concentration of 0.75 mM NADH was used, and its oxidation was detected at 340 nm using an extinction coefficient (ε340) of 6.22 mM−1 cm−1, while acceptor substrates were used at a final concentration of 0.1 mM, except oxygen, which was air-saturated buffer (approximately 0.25 mM at 25°C). A stock solution of 20 mM benzoquinone was prepared in assay buffer, and activities were corrected for the spontaneous reaction of this acceptor. For other quinones, stocks were made in ethanol, keeping the final concentration of ethanol in the assay buffer 0.5%. The enzymatic reduction of the substrate thiazolylblue tetrazolium bromide (MTT) was measured at different concentrations of MTT and NADH/NADPH by the increase in the absorbance at 610 nm due to the formation of blue formazan using an extinction coefficient (ε610) of 11.3 mM−1 cm−1. Specific activities were fitted to the Michaelis-Menten equation using Origin 8.0 (OriginLab), and mean values with standard deviations for 3 independent measurements are reported.
RESULTS
Purification of KefF.
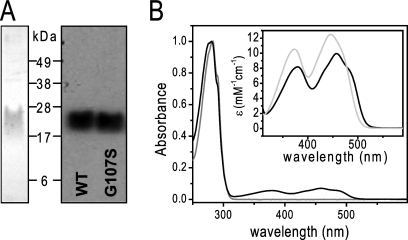
A C-terminally His-tagged construct was used to express and purify KefF in a two-step protocol. During washing of the Ni2+ immobilized-metal affinity chromatography (IMAC) column, the bed color changed to yellow due to the presumed oxidation of the FMN cofactor (this was not observed for the mutant G107S described below). After elution from the Ni2+ column, KefF was further purified and characterized by size exclusion chromatography (see Fig. S4 in the supplemental material). KefF elutes as a homogeneous peak at an elution volume corresponding to a dimeric complex, as expected from the crystal structure and homologous quinone reductases (15, 23, 35, 36). For a soluble protein, KefF shows unusual behavior on SDS gels as, even at moderate loading concentrations, it runs as a diffuse band rather than the sharp bands observed for other proteins on similar SDS-PAGE gels (Fig. 2A). Attempts to obtain sharper bands by modifying the denaturation before loading and by addition of urea to the loading buffer were unsuccessful.
Fig. 2.
Purification and characterization of KefF. (A) Coomassie-stained SDS-PAGE (left) of purified KefF wild type (WT) and Western blot (right) of WT and G107S KefF. (B) UV-Vis spectrum of purified KefF WT (black) and G107S (gray). The inset shows the visible region of the KefF WT spectrum before (black) and after (gray) addition of 0.2% SDS, representing the spectral properties of the FMN bound to KefF and free in solution, respectively.
UV/Vis spectra of purified KefF showed the typical absorption peaks of oxidized flavins, with absorption maxima at 379 and 457 nm (in comparison to 373 and 446 nm for free FMN) and extinction coefficients as follows: ε379, 8.1 mM−1 cm−1, and ε457, 10.0 mM−1 cm−1 (Fig. 2B). After purification, the flavin occupancy was never 100% and varied between 40 and 70% (Table 2). Others have made similar observations for proteins that bind FMN (34). Attempts to reconstitute FMN to full occupancy failed (data not shown); however, increases in activity were observed in assays of enzyme activity that included free FMN (see below).
Table 2.
Apparent steady-state parameters and FMN contents of KefF
| KefF | FMN/KefF ratio (mol/mol) | kcatapp(NADH) (s−1) | Kmapp (NADH) (mM) | kcatapp (MTT) (s−1) | Kmapp (MTT) (mM) |
|---|---|---|---|---|---|
| WT (NADH) | 0.4 ± 0.1 | 1.9 ± 0.6 | 0.15 ± 0.04 | 2.2 ± 0.8 | 0.4 ± 0.1 |
| WT (NADPH)a | 0.4 ± 0.1 | 1.7 ± 0.5 | 0.07 ± 0.06 | 2.7 ± 1.9 | 0.7 ± 0.6 |
| G107S | not detectable | 0.04 ± 0.03 | 1.1 ± 0.1 | 0.015 ± 0.005 | 0.13 ± 0.06 |
| H112W | 0.61 ± 0.05 | 0.28 ± 0.04 | 4.7 ± 2.5 | 0.06 ± 0.03 | 0.02 ± 0.01 |
| F149W | 0.7 ± 0.1 | 0.3 ± 0.1 | 0.05 ± 0.02 | 0.3 ± 0.1 | 0.10 ± 0.05 |
| C147S | 0.5 ± 0.1 | 2.1 ± 0.2 | 0.06 ± 0.04 | 2.7 ± 0.6 | 0.43 ± 0.06 |
| C151S | 0.53 ± 0.07 | 2.6 ± 0.7 | 0.07 ± 0.03 | 3.2 ± 0.7 | 0.33 ± 0.06 |
| C147S/C151S | 0.42 ± 0.07 | 2.0 ± 0.7 | 0.05 ± 0.03 | 2.9 ± 0.9 | 0.53 ± 0.06 |
The values in this row are for the reaction of NADPH instead of NADH.
KefF shows oxidoreductase activity.
As KefF has sequence and structural similarities to quinone reductases and retains the FMN cofactor, we evaluated its ability to act as a redox enzyme. First, we tested whether the FMN could be reduced with the physiological reductants NADH and NADPH. It was observed that both were able to reduce FMN bound to KefF, as judged from absorption spectra. The FMN reverted to its oxidized state under aerobic conditions, but not under anaerobic conditions, demonstrating that the reduced flavin cofactor in KefF showed some reactivity to oxygen (see Fig. S2A in the supplemental material).
Several electron acceptors for the oxidative half-reaction were tested in enzymatic steady-state assays. Many substrates were reduced with a high specific activity, while the turnover with oxygen was slow (Table 3). Quinones, including benzoquinone, menadione, and duroquinone, were readily reduced by KefF. For duroquinone, an apparent kcat of 99 ± 27 s−1 and Km of 14 ± 2 μM were determined (at a constant concentration of 0.75 mM NADH), which are comparable to other quinone reductases (24, 34). The 1-electron acceptors 2,6-dichloroindophenol (DCIP) and ferricyanide are good substrates for KefF (Table 3). MTT could be directly used as an electron acceptor (Table 2), while FMN, if used as a substrate, showed only low activity. On the other hand, no activity toward the nitro compound nitrofurantoin and the azo dyes Congo red and Orange II was detected. NADH and NADPH were comparable as electron donors for KefF in steady-state experiments with MTT as an acceptor (Table 2). Measurement of the enzyme rates with different substrate concentrations, for both the electron donor and acceptor, resulted in a group of parallel lines in the Lineweaver-Burk plot, indicating a Ping-Pong Bi-Bi mechanism, in which one substrate reacts with the enzyme and the product is released before a second substrate reacts with the modified enzyme (see Fig. S3 in the supplemental material). Although stable reconstitution with FMN could not be achieved, as described above, functional reconstitution was seen by a significant increase in activity if 3 μM FMN was present in the assay buffer (see Fig. S2B in the supplemental material). Similarly, the azoreductase AzoR was reported to require free FMN in the assay buffer for enzymatic activity (24). No enzymatic activity was found for typical electrophiles that activate potassium efflux through the Kef systems. NEM and MG could not directly serve as electron acceptors in a detoxification reaction catalyzed by KefF (see Fig. S2C in the supplemental material).
Table 3.
Comparison of electron acceptors for KefF
| Electron acceptor | Sp act (μmol min−1 mg−1) |
|---|---|
| Benzoquinone | 125 ± 29 |
| Methylbenzoquinone | 146 ± 22 |
| Duroquinone | 110 ± 30 |
| 2,3-Dimethoxy-5-methyl-p-benzoquinone (Q0) | 109 ± 30 |
| Menadione | 46 ± 4 |
| DCIP | 131 ± 35 |
| Ferricyanide | 25 ± 5 |
| FMN | 0.21 ± 0.02 |
| Oxygen | 0.09 ± 0.02 |
Enzymatic activity of KefF is not required for activation of KefC.
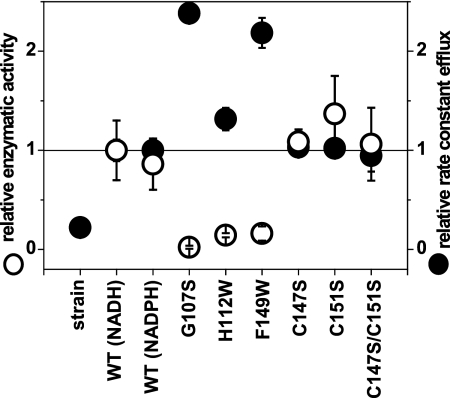
We have established that KefF shows redox enzymatic activity with a wide range of substrates. This posed the question whether there is a linkage between this enzymatic activity and the role of KefF as a modulator of potassium efflux through KefC. To address this question, we introduced mutations in the active site of KefF designed to inhibit the enzymatic redox activity. To avoid nonspecific effects on the regulation of KefC, for example, by misfolding of the KefF protein, we used the X-ray crystal structure to choose residues that when mutated were unlikely to modify the structural integrity of KefF. Three mutants, G107S, H112W, and F149W, were constructed. The first mutant was envisaged to disrupt binding of FMN to KefF, since the side chain of serine extends to a region where the ribose backbone of FMN is observed in the crystal structure. The other two mutations were intended to block the substrate-binding pocket with the more bulky tryptophan side chain. Upon purification, all of these mutated KefF constructs were obtained in yields similar to those of the wild type, and similar size exclusion profiles of the dimeric complex were observed (see Fig. S4 in the supplemental material). As expected, KefF G107S had no FMN bound to it, and accordingly, no yellow coloration was seen during purification. All three mutations caused a significant reduction of redox enzymatic activity (Table 2). These constructs were then transformed into a strain lacking KefF and tested for their abilities to activate potassium efflux via KefC. All three constructs were able to activate potassium efflux, despite their lack of enzymatic activity (Fig. 3). This observation showed clearly that the enzymatic activity was not required for activation of potassium efflux.
Fig. 3.
Comparison of enzymatic activity of KefF with potassium efflux of KefFC. Relative kcat values of purified WT KefF and mutants with the substrates NADH and MTT (open circles) are compared with the relative rate constants for potassium efflux of the plasmid-free and transformed strain MJF654 triggered by NEM (closed circles). The data were normalized with WT KefF (NADH) values. For WT KefF, the enzymatic activity with NADPH is also shown. The error bars indicate standard deviations.
On inspection of the crystal structure of KefF, we observed two cysteine residues, C147 and C151, on the protein surface close to the active site. The activity of the quinolinate synthase (NadA) from E. coli is regulated by the redox state of a surface-exposed cysteine pair (37). Similarly, surfaced-exposed cysteine residues in KefF could potentially sense oxidative stress of the cell by forming a disulfide bridge or reacting directly with electrophiles, as their surface location will likely lead to high reactivity. Therefore, we introduced the C147S and C51S mutations and measured the enzymatic activities of the resultant proteins and their abilities to activate potassium efflux. The single mutants and the double mutant displayed no significant difference from the wild type in NADH-MTT oxidoreductase activity and potassium efflux, giving no indication of a regulatory function for these cysteine residues (Fig. 3 and Table 2). Furthermore, no change in oxidoreductase activity could be seen when wild-type KefF was preincubated with NEM and MG prior to determination of the enzymatic activity (measured with NADH and MTT). Thus, modification of the cysteine residues is unlikely to modulate the behavior of KefF.
Activation of potassium efflux by electrophilic quinones.
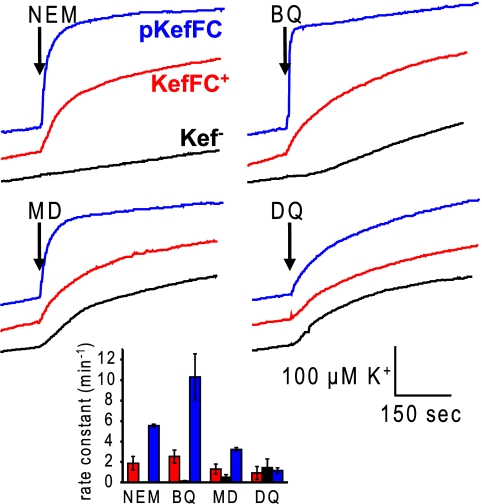
Known activators of the Kef system, such as NEM and MG, are not substrates for KefF. However, to investigate whether any cross talk between these systems exists, we considered whether typical substrates of these oxidoreductases, i.e., quinones, could activate KefC (Fig. 4) following conjugation to GSH. A subset of quinones that incorporate an enone moiety are electrophilic and can therefore undergo conjugate addition reactions with nucleophiles (4). GSH, the ligand that binds to KefC (35), spontaneously reacts with these quinones, and multiple additions at the different ortho positions can take place if the quinole is reoxidized to the quinone (22). Benzoquinone and menadione, which belong to this group of GSH-reactive quinones, elicit potassium efflux via KefC (Fig. 4B and C). When KefFC was overexpressed, the rate constant for efflux was enhanced relative to the rate observed with just the chromosomal copy of the kefFC genes (Fig. 4, inset). Benzoquinone, in particular, gave a level of activation similar to that of NEM, judged by the rate constants (Fig. 4, inset). In contrast, no increase in K+ efflux was observed for quinones that undergo conjugate addition reactions less readily, such as duroquinone. It should be noted that each of the three quinones used in our experiments triggered some loss of K+ via a mechanism that was independent of KefB and KefC (Fig. 4B to D).
Fig. 4.
Potassium efflux from E. coli is triggered by quinones. NEM (0.5 mM), benzoquinone (BQ), menadione (MD), or duroquinone (DQ) was added to the cell suspension at a time indicated by the arrows, and potassium release was recorded by an ion-selective electrode. Strains without the Kef systems (MJF276; black) and with chromosomally encoded KefFC (MJF274; red) and MJF276 complemented with a KefFC-encoding plasmid (blue) were used. The inset shows means and standard errors of rate constants obtained by fitting of the traces from three independent measurements.
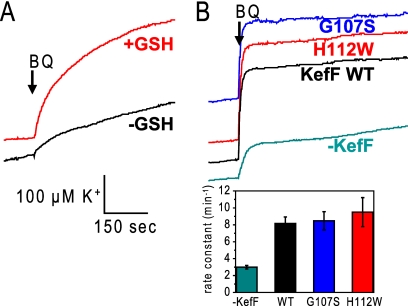
We utilized an isogenic glutathione-deficient strain (Frag56 gshA[Table 1]) to determine whether activation of KefC by electrophilic quinones was GSH dependent. When GSH was present in the growth medium, Frag56 became GSH replete, and potassium efflux was seen after the addition of benzoquinone. In contrast, in the absence of GSH, only a small effect occurred, likely due to the above-mentioned background efflux mechanism (Fig. 5 A). This difference showed that GSH was required for quinone-elicited efflux, as expected for KefC activation. Quinone-activated KefC activity was diminished in the absence of KefF, but the KefF mutants G107S and H112W could restore full activity (equivalent to wild-type KefF) in the KefF-null strain when benzoquinone was used as an electrophile (Fig. 5B).
Fig. 5.
Dependence of potassium efflux on GSH and KefF with benzoquinone as an electrophile. (A) The GSH-deficient strain Frag56 was grown in the presence (red) and absence (black) of GSH in the growth medium, and the potassium efflux triggered by benzoquinone was measured afterward. (B) The dependence of the potassium efflux on KefF was tested with the kefF mutant strain MJF654, which was used either untransformed or transformed with WT KefF-, G107S-, or H112W-bearing plasmids. Rate constants for the potassium efflux are given.
DISCUSSION
Soluble quinone reductases often have a broad substrate specificity for the oxidative half-reaction. Therefore, these enzymes have often been named according to their different acceptors, for example, azoreductase, nitroreductase, chromate reductase, ferric reductase, or flavin reductase. These enzymes have similar structures, possessing a flavin cofactor at the active site and a common flavodoxin fold, but often display low sequence similarity (7). In E. coli, several of these soluble NADH/NADPH-dependent oxidoreductases have been described: the tryptophan repressor-binding protein WrbA (34), the azoreductase AzoR (32), nitroreductases (41), and the so-called modulator of drug activity MdaB (17). With several similar oxidoreductases present with overlapping substrate selectivities, it is difficult to reveal the physiological function for each of them. In a comparison with structures in the Protein Data Bank (PDB) using the program DALI (18, 19), KefF shows much higher similarity to the human quinone reductases QR1 and QR2 than to the other oxidoreductases in E. coli. Human QR1 showed the highest similarity, with a Z score of 20.3 and a root mean square (RMS) of 2.2 Å for 167 of 272 residues (PDB code, 1QBG) (18, 38). Like QR1, KefF can accept NADH and NADPH as electron donors; it has a size similar to that of QR2 but is shorter than QR1. QR1 provides protection against oxidative stress caused by quinones, while the function of QR2 is unknown.
Comparison of the crystal structure of KefF with those of human QR1 or QR2 revealed that both the substrate-binding pocket and the flavin in KefF are much more accessible: residues 48 to 80 shield the active site in the human QRs and are missing in KefF (see Fig. S1 in the supplemental material). This difference might enable KefF to accept bigger substrates, such as MTT. The binding pocket of KefF is almost exclusively lined by aromatic residues (W67, Y68, Y88, H112, F113, and Y134) and FMN (Fig. 1). Both subunits provide residues to the active site. In QR1, the reduction of the flavin is facilitated by the provision of a proton from Y155 to O5 of the flavin adenine dinucleotide (FAD) and further charge distribution to H161 (23). The residue homologous to Y155 in KefF is F113, which is not able to provide a proton to the flavin. This function could be fulfilled by H112, which is in closer proximity to the flavin than H161 in human QRs. Mutation of H112 to tryptophan caused loss of catalytic activity in KefF, which could arise either because it simply blocks the binding site or because H112 is involved in catalysis. A charge on H112 might be stabilized by interaction with Y68 or Y134. The charge relay mechanism of human QR1 is conserved in some QRs, including E. coli MdaB (Y119) and human QR2 (Y155), but variations are seen for other QRs, for example, WrbA. Sequence alignments show that for E. coli KefG, which partners KefB, a tyrosine is conserved in this position, and the H112 of KefF is not conserved in KefG. H112, which is at the end of the binding pocket, is also well positioned to form hydrogen bonds with the substrate in a manner similar to that of H161 in human QR1 and QR2.
The binding pocket of KefF is more hydrophilic and possesses more residues capable of forming H bonds with the substrates than the human QRs. Y68 and Y134 in KefF are equivalent to F106 and F178 in human QRs (Fig. 1). Y88 of KefF is conserved in human QR1 (Y126), while a phenylalanine is found in QR2 (F126). Y134 is positioned parallel to the aromatic system of the flavin, leaving space where an aromatic substrate could fit to form a π stack, as seen in QR1 with the nicotinamide ring of NADPH (23) and in QR2 with menadione (15) and the inhibitor resveratrol (3). A phenylalanine in this position is also conserved in MdaB and AzoR from E. coli (1, 20). In E. coli WrbA, W67 fulfills the function, and a stacked structure with benzoquinone has been resolved (2, 5).
Given the structural similarities between the binding pocket of KefF and other oxidoreductases, it is perhaps no surprise that KefF is a competent enzyme, and it seems unlikely that its redox activity is an evolutionary relic. Interestingly, in a screen of environmental samples for genes with alcohol oxidoreductase activity, a gene with 90% amino acid sequence identity to E. coli KefF was identified (21). Activity measurements in extracts showed oxidation activity toward 1,2-ethanediol with NAD as a cosubstrate. This gene was not found together with the gene for KefC in the DNA fragment isolated, as is usually the case for KefF (and, similarly, for KefG with KefB). If this gene is used as a query in a BLAST search, the highest similarity (97% amino acid identity) is found for a KefF homologue from Citrobacter sp. strain 30_2. The Citrobacter KefF, however, is clustered with a kefC gene.
We wondered if the redox enzymatic activity of KefF influences the known function of KefF as the activator of KefC. Three mutants that showed no enzymatic activity, G107S, H112W, and F149W, were constructed, but all were capable of activating KefC. These mutations most likely did not disturb the structural integrity of KefF, since the mutant KefF proteins accumulated to similar levels in cells, and these proteins can activate KefC. Therefore, enzymatic activity is not required for KefC activation. Another possible link between enzymatic activity and KefC activation could be the fact that KefF reduces electrophiles and in this way detoxifies them. The Kef complex would then combine short-term rescue from the effects of electrophiles and their long-term detoxification. However, typical electrophiles that activate KefC, for example, NEM, were not reduced by KefF. In the case of this electrophile, an NEM reductase (NemA) that is homologous to the “old yellow enzyme” found in yeast has been described in E. coli (31). Previously, we established that the NEM adduct of GSH, N-ethylsuccinimido-S-glutathione, is also rapidly broken down by E. coli to release N-ethylmaleamic acid and free GSH (28). Similarly, there are multiple pathways for detoxifying MG (27, 30). Thus, E. coli has multiple mechanisms for dealing with the toxic effects of both quinone and nonquinone electrophiles.
We demonstrate here that electrophilic quinones, which form adducts with GSH, activate KefC and are also good substrates for KefF (Fig. 4 and Table 3). These compounds are ubiquitous, and it seems likely that bacteria have developed defense mechanisms against them. Through redox cycling of quinones, reactive oxygen species, including H2O2, superoxide, and hydroxyl radicals, can be formed, which causes stress for the cell. In addition, electrophilic quinones react with nucleophilic compounds, which in particular results in thiol-specific stress. Recently, it has been suggested that quinones are the native substrates for the azoreductase AzoR from E. coli (24). Our results indicate that the function of the enzymatic activity of KefF is to reduce the redox toxicity of electrophilic quinones in parallel with acting as triggers for the KefC efflux system. The two threats caused by these quinones, redox reactivity and electrophilicity, however, are interlinked, since (i) GSH not only forms adducts with them, but can also be oxidized in an nonadditive redox reaction, and (ii) the enzymatic reduction of the quinones removes their ability to undergo conjugate addition reactions.
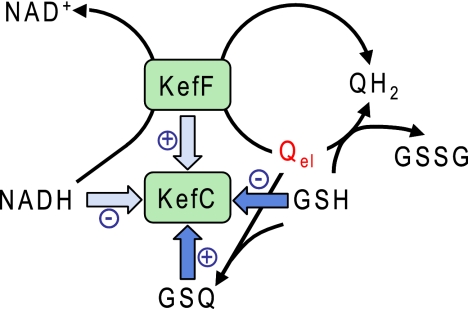
The regulation of KefC is based on three regulatory binding sites on the KTN domain: a nucleotide binding site in the Rossman fold, overlapping binding sites for GSH and GSX, and the protein-protein interface with KefF. Our new findings, presented here, indicate a further dimension to this pattern, namely, a role for KefF in activation of KefC by electrophilic quinones, summarized in Fig. 6. These quinones react with GSH, forming adducts (GSQ) that are strong activators for KefC, while GSH itself inhibits KefC. GSH can also reduce the quinones directly, resulting in the oxidation of the GSH pool. Alternatively, the quinones are reduced by NADH, a process that is catalyzed by KefF. KefF is required for full activation of KefC but does not trigger potassium efflux itself. Rather, it may increase the activity of KefC in two interdependent ways: first, by modulating the NADH pool (Fig. 6), since this nucleotide is believed to be an inhibitor of KefC (16), and second, by holding KefC KTN domains in the correct configuration to attain optimal activity (29, 36).
Fig. 6.
Regulation of KefC with electrophilic quinones as activators. Factors that regulate KefC activity are shown as blue arrows. NADH as an inhibitor and KefF as an activator are shown in light blue and have smaller effects than the other factors. Three different reactions for electrophilic quinones (Qel) are shown, and each of them has a different effect on KefC regulation.
In conclusion, we have demonstrated that the auxiliary protein KefF affects the activity of KefC and, hence, bacterial K+ efflux in two ways. KefF not only modulates the activation of KefC through physical association, but also chemically, through its redox enzyme activity. The elucidation of this complex interaction presents new avenues for manipulating the activity of KefF and, hence, bacterial K+ regulation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tarmo Roosild for helpful discussions and Gary Stuart for the initial experiments. BW25113 pKD46 and pKD4 were kind gifts from B. Wanner.
This work was supported by the Wellcome Trust (grants 040174 and 048920).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 8 July 2011.
REFERENCES
- 1. Adams M. A., Jia Z. 2006. Modulator of drug activity B from Escherichia coli: crystal structure of a prokaryotic homologue of DT-diaphorase. J. Mol. Biol. 359:455–465 [DOI] [PubMed] [Google Scholar]
- 2. Andrade S. L., Patridge E. V., Ferry J. G., Einsle O. 2007. Crystal structure of the NADH:quinone oxidoreductase WrbA from Escherichia coli. J. Bacteriol. 189:9101–9107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buryanovskyy L., et al. 2004. Crystal structure of quinone reductase 2 in complex with resveratrol. Biochemistry 43:11417–11426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Butler J., Hoey B. M. 1992. Reactions of glutathione and glutathione radicals with benzoquinones. Free Radic. Biol. Med. 12:337–345 [DOI] [PubMed] [Google Scholar]
- 5. Carey J., et al. 2007. WrbA bridges bacterial flavodoxins and eukaryotic NAD(P)H:quinone oxidoreductases. Protein Sci. 16:2301–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deller S., Macheroux P., Sollner S. 2008. Flavin-dependent quinone reductases. Cell. Mol. Life Sci. 65:141–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elmore M. J., et al. 1990. Activation of potassium efflux from Escherichia coli by glutathione metabolites. Mol. Microbiol. 4:405–412 [DOI] [PubMed] [Google Scholar]
- 9. Epstein W., Kim B. S. 1971. Potassium transport loci in Escherichia coli K-12. J. Bacteriol. 108:639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eriksson S., Lucchini S., Thompson A., Rhen M., Hinton J. C. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103–118 [DOI] [PubMed] [Google Scholar]
- 11. Ferguson G. P., McLaggan D., Booth I. R. 1995. Potassium channel activation by glutathione-S-conjugates in Escherichia coli: protection against methylglyoxal is mediated by cytoplasmic acidification. Mol. Microbiol. 17:1025–1033 [DOI] [PubMed] [Google Scholar]
- 12. Ferguson G. P., Munro A. W., Douglas R. M., McLaggan D., Booth I. R. 1993. Activation of potassium channels during metabolite detoxification in Escherichia coli. Mol. Microbiol. 9:1297–1303 [DOI] [PubMed] [Google Scholar]
- 13. Ferguson G. P., Nikolaev Y., McLaggan D., Maclean M., Booth I. R. 1997. Survival during exposure to the electrophilic reagent N-ethylmaleimide in Escherichia coli: role of KefB and KefC potassium channels. J. Bacteriol. 179:1007–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ferguson G. P., Totemeyer S., MacLean M. J., Booth I. R. 1998. Methylglyoxal production in bacteria: suicide or survival? Arch. Microbiol. 170:209–218 [DOI] [PubMed] [Google Scholar]
- 15. Foster C. E., Bianchet M. A., Talalay P., Zhao Q., Amzel L. M. 1999. Crystal structure of human quinone reductase type 2, a metalloflavoprotein. Biochemistry 38:9881–9886 [DOI] [PubMed] [Google Scholar]
- 16. Fujisawa M., Ito M., Krulwich T. A. 2007. Three two-component transporters with channel-like properties have monovalent cation/proton antiport activity. Proc. Natl. Acad. Sci. U. S. A. 104:13289–13294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hayashi M., Ohzeki H., Shimada H., Unemoto T. 1996. NADPH-specific quinone reductase is induced by 2-methylene-4-butyrolactone in Escherichia coli. Biochim. Biophys. Acta 1273:165–170 [DOI] [PubMed] [Google Scholar]
- 18. Holm L., Rosenstrom P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38:W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holm L., Sander C. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123–138 [DOI] [PubMed] [Google Scholar]
- 20. Ito K., et al. 2006. Three-dimensional structure of AzoR from Escherichia coli. An oxidereductase conserved in microorganisms. J. Biol. Chem. 281:20567–20576 [DOI] [PubMed] [Google Scholar]
- 21. Knietsch A., Waschkowitz T., Bowien S., Henne A., Daniel R. 2003. Construction and screening of metagenomic libraries derived from enrichment cultures: generation of a gene bank for genes conferring alcohol oxidoreductase activity on Escherichia coli. Appl. Environ. Microbiol. 69:1408–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lau S. S., Hill B. A., Highet R. J., Monks T. J. 1988. Sequential oxidation and glutathione addition to 1,4-benzoquinone: correlation of toxicity with increased glutathione substitution. Mol. Pharmacol. 34:829–836 [PubMed] [Google Scholar]
- 23. Li R., Bianchet M. A., Talalay P., Amzel L. M. 1995. The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: mechanism of the two-electron reduction. Proc. Natl. Acad. Sci. U. S. A. 92:8846–8850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu G., Zhou J., Fu Q. S., Wang J. 2009. The Escherichia coli azoreductase AzoR is involved in resistance to thiol-specific stress caused by electrophilic quinones. J. Bacteriol. 191:6394–6400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275 [PubMed] [Google Scholar]
- 26. Macheroux P. 1999. UV-visible spectroscopy as a tool to study flavoproteins, p. 1–7 In Chapman S. K., Reid G. A. (ed.), Flavoprotein protocols, vol. 131 Humana Press Inc., Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 27. MacLean M. J., Ness L. S., Ferguson G. P., Booth I. R. 1998. The role of glyoxalase I in the detoxification of methylglyoxal and in the activation of the KefB K+ efflux system in Escherichia coli. Mol. Microbiol. 27:563–571 [DOI] [PubMed] [Google Scholar]
- 28. McLaggan D., Rufino H., Jaspars M., Booth I. R. 2000. Glutathione-dependent conversion of N-ethylmaleimide to the maleamic acid by Escherichia coli: an intracellular detoxification process. Appl. Environ. Microbiol. 66:1393–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miller S., Ness L. S., Wood C. M., Fox B. C., Booth I. R. 2000. Identification of an ancillary protein, YabF, required for activity of the KefC glutathione-gated potassium efflux system in Escherichia coli. J. Bacteriol. 182:6536–6540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Misra K., Banerjee A. B., Ray S., Ray M. 1995. Glyoxalase III from Escherichia coli: a single novel enzyme for the conversion of methylglyoxal into D-lactate without reduced glutathione. Biochem. J. 305:999–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miura K., et al. 1997. Molecular cloning of the nemA gene encoding N-ethylmaleimide reductase from Escherichia coli. Biol. Pharm. Bull. 20:110–112 [DOI] [PubMed] [Google Scholar]
- 32. Nakanishi M., Yatome C., Ishida N., Kitade Y. 2001. Putative ACP phosphodiesterase gene (acpD) encodes an azoreductase. J. Biol. Chem. 276:46394–46399 [DOI] [PubMed] [Google Scholar]
- 33. Ness L. S., Booth I. R. 1999. Different foci for the regulation of the activity of the KefB and KefC glutathione-gated K+ efflux systems. J. Biol. Chem. 274:9524–9530 [DOI] [PubMed] [Google Scholar]
- 34. Patridge E. V., Ferry J. G. 2006. WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase. J. Bacteriol. 188:3498–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roosild T. P., et al. 2010. Mechanism of ligand-gated potassium efflux in bacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 107:19784–19789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roosild T. P., et al. 2009. KTN (RCK) domains regulate K+ channels and transporters by controlling the dimer-hinge conformation. Structure 17:893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saunders A. H., Booker S. J. 2008. Regulation of the activity of Escherichia coli quinolinate synthase by reversible disulfide-bond formation. Biochemistry 47:8467–8469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Skelly J. V., et al. 1999. Crystal structure of human DT-diaphorase: a model for interaction with the cytotoxic prodrug 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB1954). J. Med. Chem. 42:4325–4330 [DOI] [PubMed] [Google Scholar]
- 39. Tötemeyer S., Booth N. A., Nichols W. W., Dunbar B., Booth I. R. 1998. From famine to feast: the role of methylglyoxal production in Escherichia coli. Mol. Microbiol. 27:553–562 [DOI] [PubMed] [Google Scholar]
- 40. Vuilleumier S. 1997. Bacterial glutathione S-transferases: what are they good for? J. Bacteriol. 179:1431–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zenno S., Koike H., Tanokura M., Saigo K. 1996. Gene cloning, purification, and characterization of NfsB, a minor oxygen-insensitive nitroreductase from Escherichia coli, similar in biochemical properties to FRase I, the major flavin reductase in Vibrio fischeri. J. Biochem. 120:736–744 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.