Abstract
PagL and LpxO are enzymes that modify lipid A. PagL is a 3-O deacylase that removes the primary acyl chain from the 3 position, and LpxO is an oxygenase that 2-hydroxylates specific acyl chains in the lipid A. pagL and lpxO homologues have been identified in the genome of Bordetella bronchiseptica, but in the current structure for B. bronchiseptica lipid A the 3 position is acylated and 2-OH acylation is not reported. We have investigated the role of B. bronchiseptica pagL and lpxO in lipid A biosynthesis. We report a different structure for wild-type (WT) B. bronchiseptica lipid A, including the presence of 2-OH-myristate, the presence of which is dependent on lpxO. We also demonstrate that the 3 position is not acylated in the major WT lipid A structures but that mutation of pagL results in the presence of 3-OH-decanoic acid at this position, suggesting that lipid A containing this acylation is synthesized but that PagL removes most of it from the mature lipid A. These data refine the structure of B. bronchiseptica lipid A and demonstrate that pagL and lpxO are involved in its biosynthesis.
INTRODUCTION
Lipopolysaccharide (LPS) is the major component of the outer leaflet of the outer membrane of Gram-negative bacteria. The lipid A region contains the endotoxin activity of LPS (30), which arises when lipid A binds to a host membrane complex that includes Toll-like receptor 4 (TLR4) and activates signaling by TLR4 (1), resulting in the expression of many proinflammatory cytokines and chemokines in cells of the innate immune system. TLR4 signaling is also important for activation of adaptive immunity, particularly through the activation of dendritic cells (DC), which act as antigen-presenting cells (APC) for T cells. Thus, lipid A-TLR4 interactions are central to the host immune response to Gram-negative bacteria. In addition, given the importance of LPS as a structural component of the outer membrane, lipid A has an important role in maintaining the function of this membrane.
The basic structure of most lipid As can be described as a bisphosphorylated 1-6-linked diglucosamine backbone in which the 2 and 3 positions of each glucosamine are variably acylated. The number, nature, and arrangement of acyl chains vary enormously between different bacteria, and individual strains synthesize a heterogeneous mixture of variably acylated molecules (39). The biological activity of lipid A, including its potency for stimulation of TLR4, is heavily influenced by this acylation pattern, as well as by the presence/absence and substitution (see below) of the phosphates, such that the overall activity of a bacterium's lipid A is the sum of the activities of the different lipid A molecules that it expresses.
Some bacteria regulate their lipid A structure through a number of covalent modifications (40) (Fig. 1). They include addition of palmitate by PagP (3), deacylation at the 3 position by PagL (48), 2-hydroxylation of myristate by LpxO (22), dephosphorylation at the 1 position by LpxE (49), and addition of aminohexose to the lipid A phosphates by ArnT (49). Some of these modifications may alter host-pathogen interactions (17, 24, 27). Although lipid A modifications occur in a number of important pathogens, there is surprisingly little direct evidence for the roles of lipid A modifications in vivo.
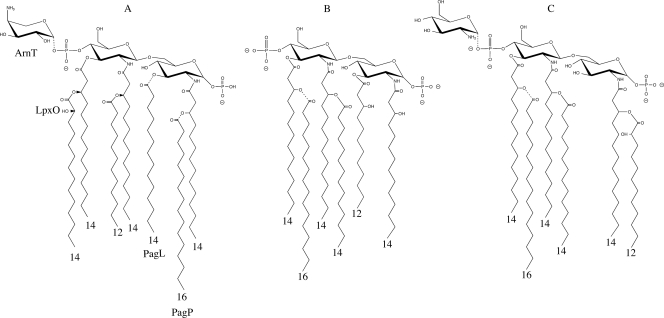
Fig. 1.
(A) Structure of Salmonella enterica serovar Typhimurium lipid A. The variable modifications effected by ArnT, PagP, LpxO, and PagL are indicated by dashed bond lines. (B) Previously reported structure of B. bronchiseptica RB50 lipid A (35). (C) Structure of the predominant lipid A species determined in this study, with the variable addition of palmitate, glucosamine, and 2-hydroxylation and the variable presence of the 1-phosphate indicated by dashed bond lines.
Bordetella bronchiseptica infects many different mammals. It causes disease in some hosts, for example, infectious tracheobronchitis/kennel cough in dogs and atrophic rhinitis in pigs (23, 28); however, even though B. bronchiseptica infections can be lifelong in some hosts, the great majority of them are asymptomatic (23). We have demonstrated multiple different roles for B. bronchiseptica LPS, including the lipid A region, in the infection biology of B. bronchiseptica using a mouse model, for example (25, 32, 33, 36).
In addition, we and others have studied the genetic basis for B. bronchiseptica lipid A biosynthesis. The work of Caroff and colleagues has defined B. bronchiseptica lipid A to comprise a bisphosphorylated diglucosamine backbone acylated with 3-OH-C14 at the 2, 2′, and 3′ positions, acylated with C12 or C12-OH at the 3 position, and secondarily acylated with C14 at the 2′ position and C16 at the 3′ position (5) (Fig. 1B). Interestingly, homologues of lipid A modification genes, including pagP, arnT, lpxO, pagL, and lpxE, have been identified in the genomes of Bordetella, including B. bronchiseptica. ArnT-dependent addition of glucosamine to the Bordetella lipid A phosphates and its effect on lipid A stimulation of TLR4-mediated responses have been characterized (20, 34, 35). Previously we described the role of PagP in palmitoylation of B. bronchiseptica RB50 lipid A to produce hexa-acylated molecules and in protecting the bacteria from antibody-dependent, complement-mediated killing (36, 38). Expression of pagP is regulated by the two-component system Bvg (38). The Bvg system is regarded as the global regulator of Bordetella gene expression, with the Bvg regulon appearing to comprise several hundred genes, including many of those whose products are involved in infecting a host during infection, such as adhesins and toxins (12). The precise environmental stimuli to which the system responds in vivo are unknown, but in vitro the system is fully activated (the Bvg+ phase) by growth of the bacterium at 37°C in the absence of inhibitors (see below), is fully inactivated (the Bvg− phase) by growth below 25°C or by the presence of the inhibitor nicotinate or sulfate ions at concentrations above 8 mM or 50 mM, respectively, or has variable intermediate activity (Bvgi phase) at reduced concentrations of these ions or at temperatures between 25°C and 30°C (9). Bvg-phase-locked strains have helped greatly in the study of the Bvg system. pagP is fully expressed in the Bvg+ phase (38).
The presence of an lpxO homologue suggests that Bordetella synthesizes lipid A containing 2-OH acylations. 2-OH-C12 was observed in Bordetella pertussis lipid A, but only in a mutant overexpressing the lipid A secondary acyltransferase, LpxL1 (19). The role of pagL in Bordetella lipid A biosynthesis has not been addressed directly, but B. bronchiseptica PagL was demonstrated to deacylate B. pertussis lipid A when the B. bronchiseptica pagL gene was heterogeneously expressed in this species (21). Here, we sought to define the role of lpxO and pagL in B. bronchiseptica biosynthesis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
B. bronchiseptica wild-type (WT) strain RB50 and the B. bronchiseptica Bvg-phase-locked strains RB53, RB53i, and RB54 have been described previously (10, 11). Escherichia coli XL1-Blue (Stratagene) and Top10 (Invitrogen) were used for cloning and maintenance of plasmids. E. coli SM10 (44) was used as a donor strain in conjugations. The pCR8 gateway entry vector (Invitrogen) was used to clone PCR products. pEX100T (42) converted to a Gateway destination vector was used as a suicide vector for allelic exchange mutagenesis in B. bronchiseptica.
Bacterial growth media and conditions.
B. bronchiseptica strains were grown on charcoal agar (Difco) supplemented with 15% defibrinated horse blood (Oxoid) at 37°C. Under these conditions, WT B. bronchiseptica and the B. bronchiseptica lpxO and pagL mutants adopt the Bvg+ phase. E. coli strains were grown on LB agar at 37°C. For liquid culture, B. bronchiseptica was grown in Stainer-Scholte broth (46) modified by the addition of Casamino Acids (50 g liter−1) at 37°C with shaking and E. coli was grown in LB broth at 37°C with aeration. Streptomycin (200 μg ml−1), ampicillin (100 μg ml−1), and kanamycin (50 μg ml−1) were used where appropriate.
Chemicals and reagents.
Chemicals and reagents were from Difco, Fisher Scientific, or Sigma.
DNA purification.
Plasmid DNA was purified using an Invitrogen plasmid DNA purification kit according to the manufacturer's instructions. Genomic DNA was purified using the agarose plug method (41).
DNA manipulation.
DNA manipulations were performed according to standard methods. DNA restriction and -modifying enzymes were from New England BioLabs, Roche, or Gibco Life Technologies.
PCR and primers.
The genomic DNA template was made by resuspending plate-grown bacteria to an optical density at 600 nm (OD600) of ∼1.0 in 0.5 ml of water, boiling in a water bath for 5 min, spinning at top speed in a benchtop microcentrifuge for 2 min, and taking 0.2 ml of supernatant. One microliter of supernatant was used per PCR. Each 50-μl PCR mixture comprised genomic DNA template, buffer as directed by the manufacturer, deoxynucleoside triphosphates (dNTPs) (25 mM each), 20 ng of each primer, 5% (vol/vol) dimethyl sulfoxide (DMSO), 5 mM MgCl2, and 2.5 units of Taq DNA polymerase (Promega). Primers used to amplify the B. bronchiseptica lpxO and pagL loci were LpxOF (5′-ATTTATCCCACGGATGCCGC-3′), LpxOR (5′-CGCGGTAGGGGGGCCGTCC-3′), PagLF (5′-GGCTGCCCGCCGGCAGTGCC-3′), and PagLR (5′-ATCGGCGGCCTTGCTCAGAAC-3′). PCR mixtures were incubated at 94°C for 5 min, followed by 30 cycles of 94°C for 75 s, 60°C for 75 s, and 72°C for 90 s, followed by a final step of 72°C for 7 min.
Conjugations.
Conjugations were performed as described previously (37).
Lipid A purification.
LPS was purified from 1-liter overnight B. bronchiseptica cultures (late log phase, OD600 ∼ 2.5) as described previously (50). Further treatment of LPS with RNase A, DNase I, and proteinase K ensured removal of contaminating nucleic acids and proteins (18). Hydrolysis of LPS to isolate lipid A was accomplished with 1% sodium dodecyl sulfate (SDS) at pH 4.5 as described previously (6).
GC.
Gas chromatography (GC) analyses of lipid A fatty acids were performed as described previously (45).
ESI LTQ-FT MS.
Lipid A was analyzed by electrospray ionization (ESI) in the negative-ion mode on an LTQ-FT linear ion trap Fourier transform ion cyclotron resonance (ICR) mass spectrometer (Thermo Scientific, San Jose, CA). Samples were diluted to 1.0 mg/ml in chloroform-methanol (1:1, vol/vol) and infused at a rate of 1.0 ml/min via a fused silica capillary (75-μm inner diameter [i.d.], 360-μm outer diameter [o.d.]) with a 30-μm spray tip (New Objective, Woburn, MA). Instrument calibration and tuning parameters were optimized using a solution of Ultramark 1621 (Lancaster Pharmaceuticals, PA) in both positive- and negative-ion modes. For experiments acquired in the ICR cell, the mass resolving power was set to 100,000 and ion populations were held constant by automatic gain control at 1.0 × 106 for mass spectrum (MS1) acquisition and at 5.0 × 105 for tandem mass spectra (MSn). For tandem mass spectra, the precursor ion selection window was set to 4 Da and the collision energy was set to 30% on the instrument scale. The collision-induced dissociation (CID) MSn analyses in the linear ion trap were done with an ion population of 1.0 × 104 and maximum fill time of 200 ms. The subsequent MS2, MS3, and MS4 events had an isolation window of 2 Da with a collision energy of 25%. All spectra were acquired over a time period of 1 to 2 min and averaged. Data were acquired and processed using Xcalibur, version 1.4 (Thermo Scientific), utilizing 7-point Gaussian smoothing.
RESULTS
Mutation of B. bronchiseptica pagL and lpxO.
To investigate the role of B. bronchiseptica pagL and lpxO in lipid A biosynthesis, mutants of each were constructed in which the relevant open reading frame BB3371 (pagL) or BB3402 (lpxO) was disrupted by insertion of a nonpolar kanamycin resistance cassette, using allelic exchange mutagenesis as described previously (37). Briefly, each gene was amplified by PCR, and the amplicons were cloned. The kanamycin resistance cassette was ligated into a unique SacI (lpxO) or BamHI (pagL) restriction endonuclease site within the coding region of the gene, and this mutated region was cloned into an allelic exchange vector and introduced into B. bronchiseptica RB50 by conjugation. The expected genomic rearrangements were confirmed by PCR and Southern hybridization analyses (data not shown).
GC analyses.
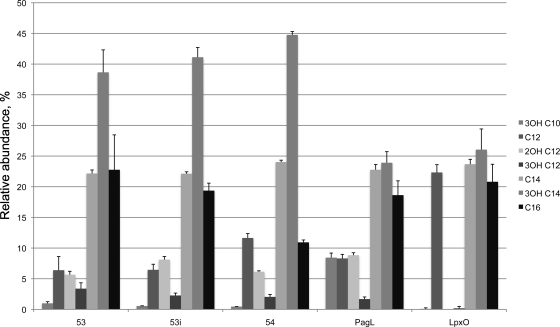
Previously, the fatty acid content of WT strain RB50 grown under Bvg+-phase conditions was analyzed and C12, C12-OH, C14, C14-OH, and C16 were detected in the ratio 0.3:0.5:1:2.8:1 (38). That same study identified that the addition of C16 to the lipid A was regulated by the Bvg system. However, the effect of Bvg activity on other lipid A acylations has not been investigated. Thus, the nature and relative abundance of the acyl chains present in bacteria grown in three different Bvg phases were analyzed by GC (Fig. 2). To do this, Bvg-phase-locked mutants were utilized. RB53 and RB53i contain point mutations in bvgS and are locked in the Bvg+ and Bvgi phases, respectively. RB54 contains a deletion of the bvgAS locus and is thus locked in the Bvg− phase. This allowed stable Bvg activity to be maintained for each culture, permitting the influence of Bvg on lipid A structure to be observed. For the purposes of this analysis, these strains are regarded as “WT” for lipid A and display the spectrum of WT lipid A structures synthesized by B. bronchiseptica.
Fig. 2.
Gas chromatography analysis of the fatty acid compositions of lipid As from B. bronchiseptica Rb53 (Bvg+ phase), Rb53i (Bvg-intermediate phase), Rb54 (Bvg− phase), and pagL and lpxO mutants. The pagL lipid A contains higher levels of 3OH-C10 than the WT, and lpxO lipid A is devoid of 2OH-C12.
In agreement with our previous study (38), from these analyses it was observed that the level of palmitate present in the lipid A was Bvg phase dependent. However, the presence or absence of the other fatty acids detected was not. Thus, further studies were conducted on Bvg+-phase bacteria, as this is the phase adopted by bacteria during infection of a host and the results could be compared to those from other analyses of B. bronchiseptica lipid A structure in which the bacteria were grown under conditions in which they would adopt this phase (5, 47).
In all three samples representative of WT lipid A, low levels of 3-OH-C10 were observed, regardless of Bvg activity. 3-OH-C10 is present in B. pertussis lipid A as a primary acylation at the 3 position but has not been previously reported for B. bronchiseptica. In addition, both 3-OH-C12 and 2-OH-C12 were identified in the WT lipid A samples, also independent of Bvg activity.
The fatty acid profiles of lpxO and pagL mutants grown under Bvg+ conditions were also analyzed. In the pagL mutant, the main difference from the WT was the increase in the relative abundance of 3OH-C10. In the lpxO mutant, 2-OH-C12 was completely absent and 3-OH-C12 was nearly absent (a very low level was detected in just one of three samples). In addition, 3-OH-C10 was also nearly absent (again, a very low level was detected in just one of three samples, but a different sample from the one in which 3-OH-C12 was detected).
The low levels of 3-OH-C10 in the WT and the increase in its level in the pagL mutant suggests that WT lipid A containing this acylation is synthesized but that PagL removes most of it from the mature lipid A. The absence of 2-OH-C12 from the lpxO mutant, with the concomitant increase in the relative level of C12, suggests that LpxO hydroxylates a portion of the C12 present in the lipid A.
Determination of lipid A structures.
To confirm these hypotheses and to elucidate the positions of these modifications, the structures of the lipid A molecules from the WT and the pagL and lpxO mutants (all grown under Bvg+-phase conditions) were elucidated using ESI LTQ-FT MS and MSn.
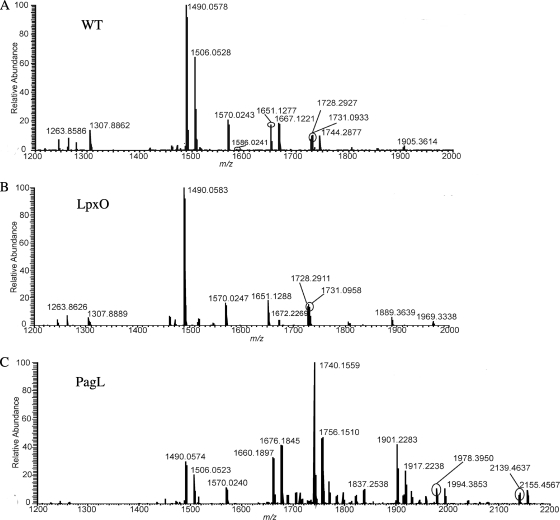
The negative-ion mode mass spectra of lipid As isolated from WT, lpxO mutant, and pagL mutant LPSs are shown in Fig. 3 A to C. The most abundant ion at m/z 1490 from the WT mass spectrum (Fig. 3A) corresponded to a singly deprotonated lipid A structure that contained one phosphate group and five acyl chains. Structure determination based on accurate mass measurement indicated that this lipid A structure had three primary acyl chains (all 3-hydroxymyristic acid [3-OH-C14] acyl chains) and two secondary acyl chains (one myristic acid [C14] acyl chain and one lauric acid [C12] acyl chain). Initial characterization based on elemental composition from accurate mass measurement of the 16-mass-unit modification between ions at m/z 1490 and 1506 revealed the composition of the 16-mass-unit modification to be a hydroxyl modification to an acyl chain from the ion at m/z 1490. The elemental composition difference between a nonhydroxylated fatty acid and a hydroxylated fatty acid (e.g., C12 and 2-OH-C12) is one oxygen atom. The accurate mass of an oxygen atom is 15.994915 Da, and the mass difference between the ions at m/z 1490.0578 and 1506.0528 was 15.9950 Da, resulting in a mass unit difference of 0.001. Accurate mass measurement indicated that the lipid A structure at m/z 1506 had three primary acyl chains (all 3-OH-C14) and two secondary acyl chains (one C14 and one 2-hydroxylauric acid [2-OH-C12]). Of note, the designation of 2-OH-C12 having a hydroxyl group at its carbon 2 position was not based on tandem mass spectrometric experiments. Rather, 2-OH-C12 was determined by identification via GC (see above). Several other prominent singly charged ions were recorded at m/z values greater than 1490 and corresponded to lipid A structures containing the addition of a hydroxyl group, hexosamine, palmitic acid, phosphate, and/or a combination of these modifications to the structure at m/z 1490. Figure 4 shows the correlation of lipid A structure to m/z value.
Fig. 3.
Negative-ion mode ESI FT-ICR mass spectra of lipid As isolated from WT B. bronchiseptica WT (A), B. bronchiseptica lpxO (B), and B. bronchiseptica pagL (C) LPSs. All ions are singly charged.
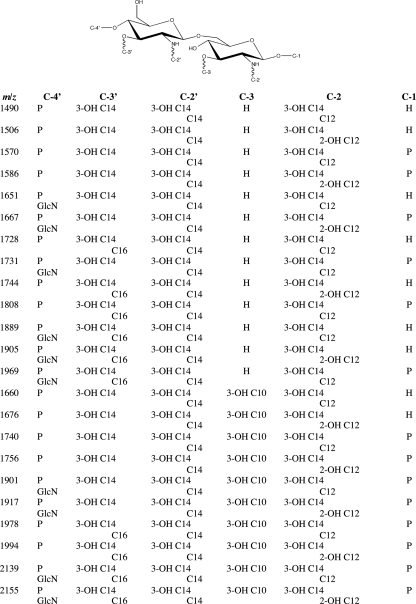
Fig. 4.
Proposed structures for precursor ions from Fig. 1.
Analogous to the WT mass spectrum, the mass spectrum for lipid A from the lpxO mutant (Fig. 3B) displayed the most abundant ion at m/z 1490, corresponding to a singly deprotonated lipid A structure that contained one phosphate group and five acyl chains. Initial structure determination revealed the ion at m/z 1490 had three primary acyl chains (all 3-OH-C14) and two secondary acyl chains (one C14 and one C12), identical to the ion at m/z 1490 from the WT. In contrast to that of the WT, the lpxO mutant mass spectrum was devoid of an ion peak at m/z 1506, which convincingly demonstrated that the lpxO mutant did not have a lipid A structure associated with a hydroxyl addition to that of the most abundant ion peak at m/z 1490. In the WT spectrum ion peaks at m/z 1667 and 1744 also arise from addition of hydroxyl to the ions at m/z 1651 and 1728, respectively, and these are also absent from the lpxO mutant spectrum. Except for this absence of hydroxyl addition, the lpxO mutant mass spectrum displayed ion peaks similar to those of the WT mass spectrum which were associated with the addition of hexosamine, palmitic acid, phosphate, and/or a combination of the these modifications to the structure at m/z 1490. Figure 4 shows the correlation of lipid A structure to m/z value.
In contrast to both the WT and lpxO mutant mass spectra, the pagL mutant mass spectrum (Fig. 3C) exhibited the most abundant ion at m/z 1740, which corresponded to a singly deprotonated lipid A structure that had six acyl chains and two phosphate groups. Initial characterization based on accurate mass measurement indicated that this lipid A structure had four primary acyl chains (three 3-OH-C14 and one 3-hydroxycapric acid [3-OH-C10]) and two secondary acyl chains (one C14 and one C12). Thus, the most abundant ion detected from B. bronchiseptica pagL lipid A contained the addition of a 3-OH-C10 acyl chain and a second phosphate group compared to the most abundant ion observed from WT lipid A. In comparison to the WT mass spectrum, the pagL mutant mass spectrum displayed ion peaks associated with the addition of a hydroxyl group, hexosamine, palmitic acid, and/or a combination of these modifications to the structure at the base peak ion at m/z 1740. Of note, the ion at m/z 1490 was present in the pagL mutant mass spectrum but at moderate abundance compared to in both the WT and lpxO mutant mass spectra. Figure 4 shows the correlation of lipid A structure to m/z value.
Tandem mass spectrometry.
Tandem mass spectrometry experiments were performed on the lipid A precursor ion at m/z 1490 isolated from WT, lpxO mutant, and pagL mutant LPSs to confirm the locations of the five fatty acids and the one phosphate group. As mentioned above, elemental composition based on accurate mass measurements of the ion at m/z 1490 revealed the composition of the five fatty acids, comprised of three primary fatty acids (all 3-OH-C14 acyl chains) and two secondary fatty acids (one C14 acyl chain and C12 acyl chain). Of particular note, the ion at m/z 1490 displayed identical tandem mass spectra for all three LPSs (data not shown), thus identifying that the lipid A structure corresponding to this particular ion was the same for the WT and the lpxO and pagL mutants.
Figure 5 a displays the dissociation mass spectrum (MS2) of the ion at m/z 1490 from WT LPS. The major dissociations of the precursor ion at m/z 1490 involved competitive neutral loss of primary and secondary acyl chains and diagnostic cross-ring product ions. For example, abundant product ions at m/z 1290, 1264, 1262, and 1246 corresponded to neutral loss of [C12], [3-OH-C14ketene], [C14], and [3-OH-C14], respectively. Although useful, these abundant product ions did not allow unambiguous assignment of acyl chains to specific locations.
Fig. 5.
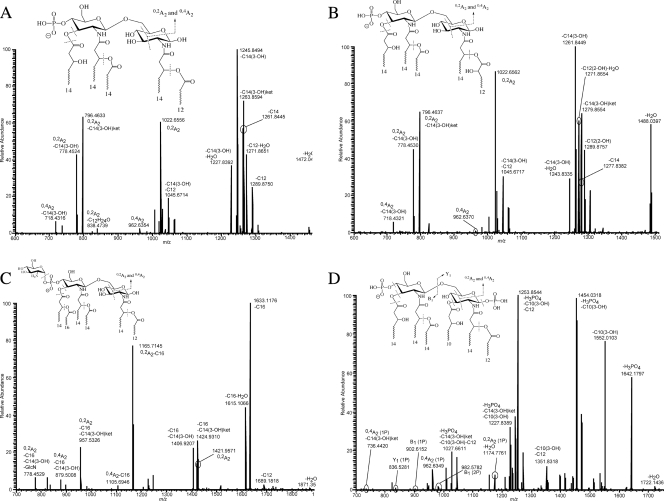
Negative-ion mode ESI FT-ICR CID MS2 of the precursor ions at m/z 1490 from the WT (A), at m/z 1506 from the WT (B), at m/z 1889 from the WT (C), and at m/z 1740 from the pagL mutant (D). The insets display the proposed structures for each ion, with dashed lines across likely bond cleavage sites. The nomenclature associated with each m/z value indicates neutral loss or cross-ring designation as described in the text.
Consequently, acyl chain positioning was determined by identifying cross-ring product ions. Figure 5A highlights a series of A-type product ions in the m/z region below 1030 that allowed the assignment of the fatty acids to specific locations. The product ions at m/z values 1022.6, 962.6, 796.5, 778.4, and 718.4 represent a series of A-type product ions and were assigned the designations 0,2A2, 0,4A2, 0,2A2-3OH-C14ket, 0,2A2-3-OH-C14, and 0,4A2-3-OH-C14, respectively. Cross-ring product ions were named following the nomenclature outlined previously (8, 13, 14), with the slight modification of designating the loss of neutral acyl chains if applicable.
The A-type product ions at m/z 1022.6 and 962.6 allowed the assignment of three acyl chains to the nonreducing end of the disaccharide backbone based on accurate mass measurements. These three fatty acids were determined to be two primary fatty acids (both 3-OH-C14) and one secondary fatty acid (C14). The following logic was applied in order to assign specific locations to the nonreducing-end acyl chains. Starting with the ion at m/z 1022.6 (0,2A2) and following its consecutive dissociations, neutral losses of 3)H-C14ket and of 3-OH-C14 were observed. These indicate that the fatty acid lost is a 3-OH-C14 and, as documented, lipid A primary fatty acids are almost exclusively hydroxylated (39). This confirms the loss of the 3-OH-C14 acyl chain from a primary position (either C-2′ or C-3′). In addition, ester-linked fatty acids are more labile than amide-linked fatty acids upon gas-phase dissociation of lipid A anions (4, 7, 29). Consequently, the loss of the 3-OH-C14 fatty acid must be at the C-3′ ester-linked position. In turn, this localized the secondary C14 to the C-2′ 3-OH-C14 primary fatty acid.
With regard to the acylation configuration at the reducing end, based on the observation and subsequent analysis of the A-type product ions, it was concluded that two acyl chains are present on the reducing end of the lipid A disaccharide backbone. Accurate mass measurements revealed that the two fatty acids are 3-OH-C14 and C12. The 3-OH-C14 acyl chain is a primary fatty acid based on the explanation above, which leaves the C12 as a secondary fatty acid. Reducing-end product ions were not observed, which would have provided direct evidence for acyl chain positioning. However, the presence of the abundant 0,2A2 ions emphasized the absence of a fatty acid at the C-3 position. Abundant 0,2A2 product ions are only observed when the C-3 position of the lipid A backbone is not occupied by a fatty acid. Conversely, when the C-3 position is occupied, 0,4A2 product ions are substantially more abundant than 0,2A2 product ions. The reasoning behind why 0,2A2 product ions are preferentially observed in gas-phase dissociation of lipid A anions with a vacated C-3 position is beyond the scope of this paper, but it does provide critical insight into the structural dependence of the gas-phase fragmentation process. In addition, corroborating evidence for a vacated C-3 position is seen in the absence of neutral loss of more than one 3-OH-C14 fatty acid. If a primary 3-OH-C14 acyl chain was present at the C-3 position, it would be an ester-linked fatty acid and therefore considered a likely candidate for neutral loss upon gas-phase fragmentation. As detailed above, the C-3′ position has a primary 3-OH-C14 fatty acid, and if the C-3 position contained a primary fatty acid, the observation of product ions resulting from the neutral loss of two primary 3-OH-C14 acyl chains would be expected; this was not the case. In conclusion, the reducing end has a primary 3-OH-C14 at the C-2 position with a corresponding secondary C12 acyl chain.
The location of the monophosphate in the precursor ion at m/z 1490 was predominately at the C-4′ position. Evidence for this assertion can be seen by lack of any glycosidic or cross product ions with a C-1 position phosphate (e.g., Y1 ions), observation of very abundant A-type ions which squarely put a phosphate at the C-4′ position, and the relatively high ratio of neutral loss of the primary C-3′ acyl chain as a ketene and free fatty acid (15, 29, 31, 43). Based on all of the experimental data, it was concluded that the lone phosphate group was positioned primarily at the C-4′ position (i.e., C-4′ monophosphate). However, the possibility of a small percentage of this lipid A structure having a C-1 position monophosphate cannot be conclusively eliminated. Figure 4 and the inset structure in Fig. 5A show the proposed structure of the precursor ion at m/z 1490. Further evidence supporting structure assignment for the precursor ion at m/z 1490 was achieved via additional MSn experiments (data not shown).
Determination of the structure of lipid A modified by hydroxylation.
The hydroxylated modification describes the presence of a 16-mass-unit addition to an ion of similar structure. The modification was determined to be the difference between a hydroxylated fatty acid and a nonhydroxylated fatty acid. In all cases, the lipid A structures devoid of the hydroxylated modification had a secondary C12 fatty acid at the C-2 position. Conversely, all ions containing the hydroxylated modification had a secondary 2-OH-C12 fatty acid at the C-2 position. The hydroxylated modification was present in similar abundance and identical positioning in both WT and pagL mutant lipid A. In contrast, the hydroxylated modification was absent from lpxO mutant lipid A.
The presence and location of the hydroxylated fatty acid were confirmed by tandem mass spectrometric analysis of the precursor ion at m/z 1506 from the WT. Figure 5B displays the dissociation mass spectrum (MS2) of the ion at m/z 1506. Similar to the case for the precursor ion at m/z 1490, the major dissociations of the precursor ion at m/z 1506 involved competitive neutral loss of primary and secondary acyl chains and diagnostic cross-ring product ions. Systematic identification of these product ions demonstrated the absence of a C12 acyl chain. Conversely, several product ions corresponding to the loss of a 2-OH-C12 fatty acid were recorded in moderate abundance. Localization of the 2-OH-C12 acyl chain was performed by identifying the presence and absence of diagnostic glycosidic, cross-ring, and neutral loss of acyl chains, similar to as outlined for the precursor ion at m/z 1490. In addition, the location of the monophosphate in the precursor ion at m/z 1506 was predominately at the C-4′ position, identical to the phosphate location in the structure corresponding to the ion at m/z 1490. Figure 4 and the inset structure in Fig. 5B show the proposed structure of the precursor ion at m/z 1506. Further evidence supporting the structure assignment for the precursor ion at m/z 1506 was achieved via additional MSn experiments (data not shown).
Determination of the structure of lipid A modified by addition of a secondary C16 acyl chain and/or hexosamine residue.
The secondary C16 acyl chain addition was present in similar abundances among WT, lpxO mutant, and pagL mutant LPSs. In all circumstances, the secondary C16 fatty acid was identified and localized exclusively as a secondary fatty acid ester linked to the primary C-3′ 3-OH-C14 acyl chain. Figure 5C shows the dissociation mass spectrum (MS2) of a precursor ion that contained a secondary C16 acyl chain. Figure 4 and the inset structure in Fig. 5C display the proposed lipid A structures for precursor ions having the C16 modification. Further evidence supporting the structure assignment for precursor ions having the C16 addition was achieved via additional MSn experiments (data not shown).
The identification and location of the hexosamine modification were consistent among all the lipid A structures isolated from WT and lpxO mutant LPSs. The hexosamine modification was found to be solely linked to the lipid A disaccharide backbone via a phosphodiester linkage at either the C-1 or C-4′ phosphate position. Figure 5c shows the dissociation mass spectrum (MS2) of a precursor ion that contained the hexosamine modification. For those lipid A structures containing a monophosphate, the hexosamine residue was linked to either the C-1 or C-4′ monophosphate. As detailed above, the most abundant ion from the WT and the lpxO mutant contained a C-4′ monophosphate, and thus the principal location for the hexosamine modification in the WT and the lpxO mutant was at the C-4′ monophosphate position. Pinpointing the exact location of the hexosamine residue for diphosphoryl lipid A structures was not as straightforward. The diphosphoryl lipid A structures were a heterogeneous mixture of bisphosphate and pyrophosphate configurations, which convoluted the gas-phase dissociation of diphosphoryl lipid A precursor ions. In turn, the dissociation mass spectra of diphosphoryl lipid A precursor ions were not easily interpreted and tended to yield ambiguity in structure assignment of phosphate configurations and modifications to the phosphate groups. All mass spectrometric evidence suggested that only one hexosamine modification was present in all lipid A structures from WT and lpxO mutant LPSs. At this level of MS sophistication, it was not possible to determine if the hexosamine modification was biased toward the C-1 or C-4′ monophosphate position in diphosphoryl lipid A, and it was assumed that the hexosamine modification could either be at the C-1 or C-4′ monophosphate in equal proportions. For lipid A structures with the pyrophosphate configuration, the hexosamine modification was assumed to be attached to the lipid A backbone via the pyrophosphate substituent at either the C-1 or C-4′ position. Conclusive evidence for localizing the pyrophosphate moiety to either the C-1 or C-4′ position and/or determining the amount of pyrophosphate was not achieved. It is worth mentioning the amount of pyrophosphate was considered to be small (roughly 5% of the total diphosphoryl lipid A based on estimates from diphosphoryl lipid A isolated from Yersinia pestis LPS), and therefore this was not considered a dominant structure (26).
Determination of the structure of lipid A from the pagL mutant.
Tandem mass spectrometric experiments on the lipid A precursor ion at m/z 1740 isolated from pagL mutant LPS were carried out to confirm the locations of the six fatty acids and the two phosphate groups. Elemental composition based on accurate mass measurements of the ion at m/z 1740 revealed the composition of the six fatty acids, comprised of four primary fatty acids (three 3-OH-C14 acyl chains and one 3-OH-C10 acyl chain) and two secondary fatty acids (one C14 acyl chain and one C12 acyl chain). Of specific note, the ion at m/z 1740 was present only in the pagL mutant mass spectrum and was not present in either the WT or lpxO mutant mass spectrum. Two distinct differences between the most abundant lipid A structure from the pagL mutant and those from the other two LPSs were the presence of a primary 3-OH-C10 acyl chain and a second phosphate group. Figure 5D displays the dissociation mass spectrum (MS2) of the ion at m/z 1740 from pagL mutant LPS. The primary 3-OH-C10 acyl chain was determined to be situated at the C-3 position. This was in direct contrast to the lipid A structures from the WT and the lpxO mutant, where their C-3 positions lacked a primary fatty acid. Similar to the diphosphoryl lipid A structures from the WT and the lpxO mutant, the diphosphoryl lipid A structures from the pagL mutant were a heterogeneous mixture of bisphosphate and pyrophosphate. The major difference between the phosphorylation pattern from the pagL mutant and the other two variants was that the pagL mutant lipid A structures were predominantly diphosphorylated as opposed to monophosphorylated. Figure 4 and the inset structure in Fig. 5d show the proposed structure of the precursor ion at m/z 1740. Further evidence supporting structure assignment for the precursor ion at m/z 1740 was achieved via additional MSn experiments (data not shown).
DISCUSSION
Here, the structure of B. bronchiseptica lipid A is demonstrated to be different from those reported previously (5), including for the same strain background, RB50, that was analyzed in this report. The use of detailed tandem mass spectrometry analyses allowed unambiguous determination of the acylation pattern and revealed that in the most abundant lipid A species detected, C12 or 2-OH-C12 was present as a secondary acylation at the 2 position and the 3 position was empty.
We have also shown for the first time that the lipid A modification genes pagL and lpxO are involved in Bordetella lipid A biosynthesis. We have demonstrated that lpxO is required for the presence of 2-OH-C12 in B. bronchiseptica lipid A, and we suggest that LpxO hydroxylates a portion of the C12 that is a secondary acylation at the 2 position.
The secondary acyl chains of E. coli lipid A are transferred to the molecule by two acyl transferases, LpxL and LpxM (39). Bordetella homologues of lpxL and lpxM (named lpxL1 and lpxL2) have been identified (19). In B. pertussis, LpxL2 is responsible for addition of the secondary myristate at the 2′ position. In B. pertussis, LpxL1 appeared to mediate addition of 2-OH-C12 at the 2 position, but only when the lpxL1 gene was overexpressed (19), as this acylation was not observed in WT lipid A. That study could not distinguish between LpxL1 transferring 2-OH-C12 or it transferring C12 that was subsequently hydroxylated by LpxO. Here we show that in B. bronchiseptica the latter occurs, and this is also very likely the case in B. pertussis, as these genes are paralogous between the two species.
Interestingly, GC analyses identified that WT lipid As also contained low levels of 3-OH-C12, which was also absent from the lpxO mutant lipid A, but the position of this acylation was not determined. Low levels of 3-OH-C12 were also detected in the fatty acid profile of lipid As from B. pertussis WT and an lpx1 mutant (19). The authors suggested that 3-OH-C12 was a minor component, either as a secondary acylation of one of the primary 3-OH-C14s or possibly in place of one. That study did not construct a B. pertussis lpxO mutant, and thus it is not known whether the presence of 3-OH-C12 is lpxO dependent in B. pertussis.
We have demonstrated that mutation of pagL results in higher levels of 3-OH-C10 in the lipid A and that in pagL mutant lipid A it is present at the 3 position. Analysis of WT lipid A revealed that most molecules of WT B. bronchiseptica are not acylated at the 3 position, but in GC analyses of the fatty acid content of WT lipid A, low levels of 3-OH-C10 were detected, suggesting that some WT molecules, albeit a small minority, likely do contain some acylation at the 3 position. We propose that B. bronchiseptica lipid A is synthesized with 3-OH-C10 at the 3 position but that PagL removes most of it from the mature lipid A.
B. parapertussis also contains pagL, and its lipid A is also not acylated at the 3 position (16), suggesting that B. parapertussis PagL has the same function as the B. bronchiseptica enzyme. B. pertussis pagL has suffered a frameshift mutation and is thus predicted to be nonfunctional. In support of this, most WT B. pertussis lipid A molecules contain 3-OH-C10 at the 3 position.
We have also demonstrated that B. bronchiseptica RB50 expresses a heterogeneous mixture of lipid A molecules. In addition to the different modifications discussed above, we also observed the variable presence of a hexosamine as a substitution of the lipid A phosphates. Modification of lipid A phosphates by glucosamine in B. pertussis has been reported and is effected by ArnT (34, 35). It has been also observed in B. parapertussis (20). Hexosamine modification was observed in B. bronchiseptica but not confirmed as glucosamine (47). In B. pertussis both phosphates can be modified (35). We have demonstrated that the modification in B. bronchiseptica RB50 is also glucosamine and is also arnT dependent (A. Preston and R. Ernst, unpublished data). Interestingly, here we observed that in B. bronchiseptica a single phosphate is modified with glucosamine, partly due to the fact that abundant lipid A molecules were monophosphorylated and thus provided only one acceptor site for this modification. We also observed that the variable presence of palmitate that we have shown previously is added by PagP (38).
During preparation of this paper, Basheer et al. published the lipid A structures of 3 other B. bronchiseptica strains (2). They too observed the 3 position to be empty and 2-OH-C12 as a secondary acylation at the 2 position. However, they inferred that both phosphates were substituted with GlcN and that secondary palmitates were present, one at the 3 position as reported here and as an alternative secondary acylation at the 2 position. We did not find any evidence for these modifications in our data. While this does not rule out their existence, it means that they would be present at very low abundance in our analyses (i.e., precursor ions corresponding to lipid A structures with the addition of a second GlcN or palmitate were not distinguishable at the existing signal-to-noise ratio). While the conclusions of Basheer et al. (2) are broadly in agreement with the findings reported here, detailed data supporting the identification of the second GlcN and palmitate modifications were not presented in the paper. We suggest the use of high-resolution mass spectrometry, including tandem mass spectrometry, as a requirement for accurate lipid A structure determination from mass spectrometry-based experiments.
Lipid A is an important component of the outer membrane, and changes to it can alter the function of the membrane in terms of its permeability and ability to withstand certain insults. The endotoxicity of lipid A is also important to host-pathogen interactions during an infection. These biological activities of lipid A are determined by its structure, and it is likely that the lipid A repertoire of B. bronchiseptica is shaped by the functional constraints for both maintaining a viable outer membrane and balancing the bacterium's interactions with the host during colonization or infection. The abilities to precisely define lipid A structures and to construct mutants with characterized alterations to their lipid As allow investigation of the role of specific lipid A structures in an organism's biology.
ACKNOWLEDGEMENT
This work was supported in part by National Institutes of Health grant 1U54 AI57141 (R.K.E.).
Footnotes
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Akira S., Takeda K. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4:499–511 [DOI] [PubMed] [Google Scholar]
- 2. Basheer S. M., Guiso N., Tirsoaga A., Caroff M., Novikov A. 2011. Structural modifications occurring in lipid A of Bordetella bronchiseptica clinical isolates as demonstrated by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid. Commun. Mass. Spectrom. 25:1075–1081 [DOI] [PubMed] [Google Scholar]
- 3. Bishop R. E., et al. 2000. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 19:5071–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boue S. M., Cole R. B. 2000. Confirmation of the structure of lipid A from Enterobacter agglomerans by electrospray ionization tandem mass spectrometry. J. Mass. Spectrom. 35:361–368 [DOI] [PubMed] [Google Scholar]
- 5. Caroff M., et al. 2001. Structural variability and originality of the Bordetella endotoxins. J. Endotoxin Res. 7:63–68 [PubMed] [Google Scholar]
- 6. Caroff M., Tacken A., Szabo L. 1988. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr. Res. 175:273–282 [DOI] [PubMed] [Google Scholar]
- 7. Chan S., Reinhold V. N. 1994. Detailed structural characterization of lipid A: electrospray ionization coupled with tandem mass spectrometry. Anal. Biochem. 218:63–73 [DOI] [PubMed] [Google Scholar]
- 8. Costello C. E., Vath J. E. 1990. Tandem mass spectrometry of glycolipids. Methods Enzymol. 193:738–768 [DOI] [PubMed] [Google Scholar]
- 9. Cotter P. A., Jones A. M. 2003. Phosphorelay control of virulence gene expression in Bordetella. Trends Microbiol. 11:367–373 [DOI] [PubMed] [Google Scholar]
- 10. Cotter P. A., Miller J. F. 1994. BvgAS-mediated signal-transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 62:3381–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cotter P. A., Miller J. F. 1997. A mutation in the Bordetella bronchiseptica bvgS gene results in reduced virulence and increased resistance to starvation, and identifies a new class of Bvg-regulated antigens. Mol. Microbiol. 24:671–685 [DOI] [PubMed] [Google Scholar]
- 12. Cummings C. A., Bootsma H. J., Relman D. A., Miller J. F. 2006. Species- and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. J. Bacteriol. 188:1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Domon B., Costello C. E. 1988. Systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate 5:397–409 [Google Scholar]
- 14. Domon B., Vath J. E., Costello C. E. 1990. Analysis of derivatized ceramides and neutral glycosphingolipids by high-performance tandem mass spectrometry. Anal. Biochem. 184:151–164 [DOI] [PubMed] [Google Scholar]
- 15. El-Aneed A., Banoub J. 2005. Elucidation of the molecular structure of lipid A isolated from both a rough mutant and a wild strain of Aeromonas salmonicida lipopolysaccharides using electrospray ionization quadrupole time-of-flight tandem mass spectrometry. Rapid. Commun. Mass. Spectrom. 19:1683–1695 [DOI] [PubMed] [Google Scholar]
- 16. El Hamidi A., Novikov A., Karibian D., Perry M. B., Caroff M. 2009. Structural characterization of Bordetella parapertussis lipid A. J. Lipid Res. 50:854–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ernst R. K., et al. 2003. Pseudomonas aeruginosa lipid A diversity and its recognition by Toll-like receptor 4. J. Endotoxin Res. 9:395–400 [DOI] [PubMed] [Google Scholar]
- 18. Fischer W., Koch H. U., Haas R. 1983. Improved preparation of lipoteichoic acids. Eur. J. Biochem. 133:523–530 [DOI] [PubMed] [Google Scholar]
- 19. Geurtsen J., et al. 2007. A novel secondary acyl chain in the lipopolysaccharide of Bordetella pertussis required for efficient infection of human macrophages. J. Biol. Chem. 282:37875–37884 [DOI] [PubMed] [Google Scholar]
- 20. Geurtsen J., et al. 2009. Identification of a novel lipopolysaccharide core biosynthesis gene cluster in Bordetella pertussis: influence of core structure and lipid A glucosamine substitution on endotoxic activity. Infect. Immun. 77:2602–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Geurtsen J., et al. 2006. Expression of the lipopolysaccharide-modifying enzymes PagP and PagL modulates the endotoxic activity of Bordetella pertussis. Infect. Immun. 74:5574–5585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gibbons H. S., Lin S., Cotter R. J., Raetz C. R. 2000. Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, A new Fe 2+/alpha-ketoglutarate-dependent dioxygenase homologue. J. Biol. Chem. 275:32940–32949 [DOI] [PubMed] [Google Scholar]
- 23. Goodnow R. A. 1980. Biology of Bordetella bronchiseptica. Microbiol. Rev. 44:722–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hajjar A. M., Ernst R. K., Tsai J. H., Wilson C. B., Miller S. I. 2002. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat. Immunol. 3:354–359 [DOI] [PubMed] [Google Scholar]
- 25. Harvill E. T., et al. 2000. Multiple roles for Bordetella lipopolysaccharide molecules during respiratory tract infection. Infect. Immun. 68:6720–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jones J. W., Shaffer S. A., Ernst R. K., Goodlett D. R., Turecek F. 2008. Determination of pyrophosphorylated forms of lipid A in Gram-negative bacteria using a multivaried mass spectrometric approach. Proc. Natl. Acad. Sci. U. S. A. 105:12742–12747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kawasaki K., Ernst R. K., Miller S. I. 2004. 3-O-deacylation of lipid A by PagL, a PhoP/PhoQ-regulated deacylase of Salmonella typhimurium, modulates signaling through Toll-like receptor 4. J. Biol. Chem. 279:20044–20048 [DOI] [PubMed] [Google Scholar]
- 28. Keil D. J., Fenwick B. 1998. Role of Bordetella bronchiseptica in infectious tracheobronchitis in dogs. J. Am. Vet. Med. Assoc. 212:200–207 [PubMed] [Google Scholar]
- 29. Kussak A., Weintraub A. 2002. Quadrupole ion-trap mass spectrometry to locate fatty acids on lipid A from Gram-negative bacteria. Anal. Biochem. 307:131–137 [DOI] [PubMed] [Google Scholar]
- 30. Luderitz O., et al. 1973. Lipid A: chemical structure and biological activity. J. Infect. Dis. 128:S17–S29 [DOI] [PubMed] [Google Scholar]
- 31. Madalinski G., Fournier F., Wind F.-L., Alfonso C., Tabet J. C. 2006. Gram-negative bacterial lipid A analysis by negative electrospray ion trap mass spectrometry: step-wise dissociations of deprotonated species under low energy CID conditions. Int. J. Mass. Spectrom. 249-250:77–92 [Google Scholar]
- 32. Mann P. B., Elder K. D., Kennett M. J., Harvill E. T. 2004. Toll-like receptor 4-dependent early elicited tumor necrosis factor alpha expression is critical for innate host defense against Bordetella bronchiseptica. Infect. Immun. 72:6650–6658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mann P. B., Kennett M. J., Harvill E. T. 2004. Toll-like receptor 4 is critical to innate host defense in a murine model of bordetellosis. J. Infect. Dis. 189:833–836 [DOI] [PubMed] [Google Scholar]
- 34. Marr N., et al. 2010. Substitution of the Bordetella pertussis lipid A phosphate groups with glucosamine is required for robust NF-kappaB activation and release of proinflammatory cytokines in cells expressing human but not murine Toll-like receptor 4-MD-2-CD14. Infect. Immun. 78:2060–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marr N., Tirsoaga A., Blanot D., Fernandez R., Caroff M. 2008. Glucosamine found as a substituent of both phosphate groups in Bordetella lipid A backbones: role of a BvgAS-activated ArnT ortholog. J. Bacteriol. 190:4281–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pilione M. R., Pishko E. J., Preston A., Maskell D. J., Harvill E. T. 2004. pagP is required for resistance to antibody-mediated complement lysis during Bordetella bronchiseptica respiratory infection. Infect. Immun. 72:2837–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Preston A., et al. 1999. Genetic basis for lipopolysaccharide O-antigen biosynthesis in bordetellae. Infect. Immun. 67:3763–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Preston A., et al. 2003. Bordetella bronchiseptica PagP is a Bvg-regulated lipid A palmitoyl transferase that is required for persistent colonization of the mouse respiratory tract. Mol. Microbiol. 48:725–736 [DOI] [PubMed] [Google Scholar]
- 39. Raetz C. R., Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Raetz C. R. H. 2001. Regulated covalent modifications of lipid A. J. Endotoxin Res. 7:73–78 [PubMed] [Google Scholar]
- 41. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 42. Schweizer H. P., Hoang T. T. 1995. An improved system for gene replacement and XylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15–22 [DOI] [PubMed] [Google Scholar]
- 43. Shaffer S. A., Harvey M. D., Goodlett D. R., Ernst R. K. 2007. Structural heterogeneity and environmentally regulated remodeling of Francisella tularensis subspecies novicida lipid A characterized by tandem mass spectrometry. J. Am. Soc. Mass. Spectrom. 18:1080–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Simon R., Priefer U., Pühler A. 1982. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology 1:784–791 [Google Scholar]
- 45. Somerville J. E., Jr., Cassiano L., Bainbridge B., Cunningham M. D., Darveau R. P. 1996. A novel Escherichia coli lipid A mutant that produces an antiinflammatory lipopolysaccharide. J. Clin. Invest. 97:359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stainer D. W., Scholte M. J. 1970. A simple chemically defined medium for the production of phase I Bordetella pertussis. J. Gen. Microbiol. 63:211–220 [DOI] [PubMed] [Google Scholar]
- 47. Tirsoaga A., El Hamidi A., Perry M. B., Caroff M., Novikov A. 2007. A rapid, small-scale procedure for the structural characterization of lipid A applied to Citrobacter and Bordetella strains: discovery of a new structural element. J. Lipid Res. 48:2419–2427 [DOI] [PubMed] [Google Scholar]
- 48. Trent M. S., Pabich W., Raetz C. R., Miller S. I. 2001. A PhoP/PhoQ-induced lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J. Biol. Chem. 276:9083–9092 [DOI] [PubMed] [Google Scholar]
- 49. Trent M. S., Stead C. M., Tran A. X., Hankins J. V. 2006. Diversity of endotoxin and its impact on pathogenesis. J. Endotoxin Res. 12:205–223 [DOI] [PubMed] [Google Scholar]
- 50. Westphal O., Jann K. 1965. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 5:83–91 [Google Scholar]