Abstract
The two hemolysin gene clusters previously identified in Vibrio anguillarum, the vah1 cluster and the rtxACHBDE cluster, are responsible for the hemolytic and cytotoxic activities of V. anguillarum in fish. In this study, we used degenerate PCR to identify a positive hemolysin regulatory gene, hlyU, from the unsequenced V. anguillarum genome. The hlyU gene of V. anguillarum encodes a 92-amino-acid protein and is highly homologous to other bacterial HlyU proteins. An hlyU mutant was constructed, which exhibited an ∼5-fold decrease in hemolytic activity on sheep blood agar with no statistically significant decrease in cytotoxicity of the wild-type strain. Complementation of the hlyU mutation restored both hemolytic activity and cytotoxic activity. Both semiquantitative reverse transcription-PCR (RT-PCR) and quantitative real-time RT-PCR (qRT-PCR) were used to examine expression of the hemolysin genes under exponential and stationary-phase conditions in wild-type, hlyU mutant, and hlyU complemented strains. Compared to the wild-type strain, expression of rtx genes decreased in the hlyU mutant, while expression of vah1 and plp was not affected in the hlyU mutant. Complementation of the hlyU mutation restored expression of the rtx genes and increased vah1 and plp expression to levels higher than those in the wild type. The transcriptional start sites in both the vah1-plp and rtxH-rtxB genes' intergenic regions were determined using 5′ random amplification of cDNA ends (5′-RACE), and the binding sites for purified HlyU were discovered using DNA gel mobility shift experiments and DNase protection assays.
INTRODUCTION
Vibrio anguillarum is a marine member of the class Gammaproteobacteria. This highly motile Gram-negative bacterium is the causative agent of warm-water vibriosis, a fatal hemorrhagic septicemic disease in fish, crustaceans, and bivalves (1). The mortality rate from V. anguillarum infections ranges from 30% to 100% (1). Infections by these bacteria have resulted in severe economic losses to aquaculture worldwide (1, 21) and affect many farm-raised fish, including Pacific salmon, Atlantic salmon, sea bass, cod, and eel (1, 4, 5, 21).
Hemolytic activity has been considered to be a virulence factor for V. anguillarum and is thought to contribute to the hemorrhagic septicemia characteristic of vibriosis (7, 16). Rock and Nelson (16) reported that the vah1 hemolysin gene cluster contains at least two genes, vah1 and plp, that affect hemolytic activity. Vah1 is a putative pore-forming hemolysin that causes vacuolization of target cells (10). It was suggested that pore-forming hemolysins, like HlyA in Escherichia coli, cause direct lysis of blood cells by disrupting the membrane integrity (13). Mutations in the divergently transcribed plp gene result in both increased expression of vah1 and increased hemolysis, suggesting that Plp is a putative repressor of vah1 transcription (16). Additionally, restoration of plp by complementation restores the wild-type level of vah1 transcription and hemolysis (16). Plp is a phosphatidylcholine (PC)-specific phospholipase A2 (PLA2), which causes lysis of PC-rich fish erythrocytes (9).
Besides the vah1 cluster, a second hemolysin gene cluster, rtxACHBDE, was identified in the V. anguillarum (10). This gene cluster contains rtxA, which encodes a potent MARTX toxin and the specialized type I secretion system (T1SS) genes (rtxDBE) responsible for the secretion of the RtxA hemolysin/cytotoxin. A mutant containing mutations in both vah1 and rtxA completely lost hemolytic activity on sheep blood agar (10). Additionally, RtxA exhibits cytotoxic activity and causes Atlantic salmon kidney (ASK) cells to round and die (10).
HlyU, a member of the SmtB/ArsR family, is a metal-regulated transcriptional regulatory protein (17). It has been reported that HlyU is a positive regulator of hemolysin and toxin genes in Vibrio species. In Vibrio cholerae, the HlyU protein positively regulates expression of hemolysin gene hlyA, as well as the HlyA-coregulated gene hcp (22, 23). Williams et al. (22) reported that a mutation in hlyU attenuates V. cholerae O17 in the infant mouse cholera infection model. Recently, HlyU was also identified in Vibrio vulnificus, and it appears to be a positive regulator of virulence genes (8, 11). Kim et al. (8) reported that HlyU of V. vulnificus may be one of the master regulators of in vivo virulence gene expression. Specifically, in a V. vulnificus hlyU mutant, cytotoxic activity against HeLa cells was nearly abolished, and the 50% lethal dose (LD50) of V. vulnificus in mice by intraperitoneal infection was increased by 10- to 50-fold (8). Liu et al. (11) also demonstrated that HlyU was required for virulence of RtxA1, a homologue of RtxA of V. anguillarum in V. vulnificus CMCP6. In V. vulnificus, HlyU acted as a competitor that antagonized the binding of H-NS, a repressor of rtxA1, in the upstream region of the rtxA1 operon so that the presence of HlyU resulted in derepression of rtxA1 (12).
In this report, we identified the hlyU homologue in V. anguillarum by degenerate PCR and constructed an hlyU mutant strain and its complement. The hemolytic activity and cytotoxicity of the mutant were determined and compared to those of the wild-type and complemented strains. We also identified the transcriptional start site of genes in both the vah1 cluster and rtxA operon and localized the HlyU binding sites to the upstream region of the two hemolysins by gel mobility shift and DNase protection assays. Additionally, the amounts of transcription from various hemolysin genes, including vah1, plp, rtxA, rtxH, and rtxB, were determined in the hlyU mutant and its isogenic wild-type parent and complement by real-time reverse transcription-PCR (RT-PCR).
MATERIALS AND METHODS
Fish cell line, bacterial strains, plasmids, and growth conditions.
Atlantic salmon kidney (ASK) cells (ATCC CRL-2747) were cultured at 20°C in Leibovitz-15 medium containing 100 μg/ml ampicillin, 100 μg/ml streptomycin, and 20% fetal bovine serum (FBS) (Invitrogen). All bacterial strains and plasmids used in this report are listed in Table 1. V. anguillarum strains were routinely grown in Luria-Bertani broth plus 2% NaCl (LB20) (6), supplemented with the appropriate antibiotic, in a shaking water bath at 27°C. Overnight cultures of V. anguillarum, grown in LB20, were harvested by centrifugation (8,000 × g, 10 min), and the pelleted cells were washed twice with nine-salt solution (NSS) (6). Washed cells were resuspended to appropriate cell densities in experimental media. Specific conditions for each experiment are described in the text. E. coli strains were routinely grown in Luria-Bertani broth plus 1% NaCl (LB10) (18). Antibiotics were used at the following concentrations: streptomycin, 200 μg/ml (Sm200); ampicillin, 100 μg/ml (Ap100); chloramphenicol, 20 μg/ml (Cm20) for E. coli and 5 μg/ml (Cm5) for V. anguillarum; kanamycin, 50 μg/ml (Km50) for E. coli and 80 μg/ml (Km80) for V. anguillarum; and tetracycline, 15 μg/ml (Tc15) for E. coli and 2 μg/ml (Tc2) for V. anguillarum.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and features | Source or reference |
|---|---|---|
| Strains | ||
| V. anguillarum | ||
| M93Sm | Spontaneous Smr mutant of M93 (serotype J-O-1) | 3, 4 |
| S305 | Smr Cmr; M93Sm hlyU mutant | This study |
| S307 | Smr Cmr Tetr; M93Sm hlyU complement | This study |
| S183 | Smr Cmr Kanr; M93Sm rtxA vah1 double mutant | 10 |
| E. coli | 14 | |
| Sm10 | thi thr leu tonA lacY supE recA RP4-2-Tc::Mu::Km (λ pir) | |
| M15 | Nals Strs Rifsthilacara+gal+mtl F−recA+uvr+lon+ (pREP4; Kmr) | Qiagen |
| Plasmids | ||
| pNQ705-1 | Cmr; suicide vector with R6K origin | 14 |
| pSUP202 | E. coli-V. anguillarum shuttle vector | 14 |
| PCR2.1 | Cloning vector | Invitrogen |
| pQE30UA | Expression vector with N-terminal His6 tag | Qiagen |
Degenerate PCR.
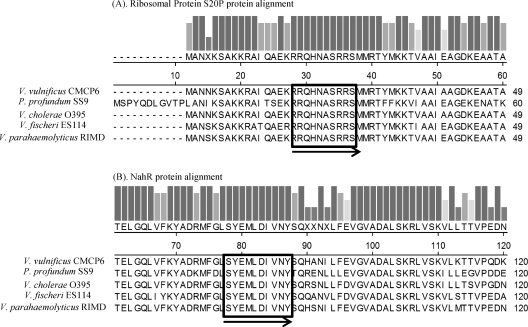
Degenerate PCR was used to identify the hlyU gene in V. anguillarum. Previously sequenced hlyU genes, including their flanking genes from various Vibrio species, were obtained from the NCBI website (http://www.ncbi.nlm.nih.gov) and aligned using the ClustalW program (20). Degenerate primers (Table 2) were designed from the conserved regions (Fig. 1) and used to amplify the possible hlyU gene from V. anguillarum M93Sm genomic DNA. The PCR products were separated and purified from a 1% agarose gel and then subcloned into pCR2.1 vector (Invitrogen). Colonies containing the cloned hlyU gene in pCR2.1 were selected on LB10 plates plus Ap100 and Km50, and the presence of hlyU was confirmed by plasmid purification and DNA sequencing.
Table 2.
Primers used in this study
| Primer | Sequence (5′ to 3′)a | Description |
|---|---|---|
| Pm301 | AGYTAYGARATGYTNGATACGTNAAYTA | Degenerate hlyU F |
| Pm302 | CGTCGYCAGCAYAAYGCTAGCCGTCGYTC | Degenerate hlyU R |
| Pm297 | ACTGAGAGCTCGGTGTTGTTAAAGGCTATGGC | hlyU insertional mutation F |
| Pm298 | ATCGATCTAGAGTATCCACTAACCCATCTCTT | hlyU insertional mutation R |
| R vah1 RT (BF) | GGCTCAACCTCTCCTTGTAACCAA | 5′-RACE vah1 |
| plpF RT | CAGACGACCACCAGTAACCACTAA | 5′-RACE plp |
| Pm112 | TGGTTGTAAGCCGCAGCAC | 5′-RACE rtxH |
| Pm163 | GGGGTATCTGAGTCACATGGATGAATT | 5′-RACE rtxB |
| Primer AP | GACCACGCGTATCGATGTCGACTTTTTTTTTTTTTTTTV | 5′-RACE anchor primer |
| Pm303 | ATGGAAAAAAATTCCGCTAAAGCA | Entire HlyU protein F |
| Pm304 | CTAGCGGCAGTATAAACCGTGTAA | Entire HlyU protein R |
| Pm305 | CCCGGATCCGCAACTTATCGGTCAGATTGATG | hlyU complementation F |
| Pm306 | CCCGGATCCGATGCGCACTTACATGAAGAAAAC | hlyU complementation R |
Restriction sites for SacI (GAGCTC), XbaI (TCTAGA), and EcoRI (GGATCC) are underlined. V=A, C, or G; N=A, C, T, or G; R=A or G; and Y=C or T.
Fig. 1.
Protein alignments of the hlyU flanking genes, encoding ribosomal protein S20P (A) and the transcriptional regulator NahR (B), in five Vibrio-related bacterial species. The bars above the alignment indicate the relative amount of conservation of amino acid residues. The regions enclosed in black boxes were used to design the degenerate primers according to their original DNA sequences. The black arrows show the orientation of primers.
Insertional mutagenesis of hlyU.
Insertional mutagenesis by homologous recombination was used to create a gene interruption within the hlyU gene by using a modification of the procedure described by Milton and Wolf-Watz (14). Briefly, primers (Table 2) were designed based on the hlyU gene sequence of M93Sm (GenBank accession no. HQ149334). Then a 161-bp hlyU DNA fragment was PCR amplified by using primer pair Pm297 and Pm298 (Table 2) and cloned into the suicide vector pNQ705 by using SacI and XbaI restriction sites to yield the pNQ705 derivative plasmid, which was confirmed by both PCR amplification and restriction analysis. The mobilizable suicide vector was transferred from E. coli Sm10 (λ pir) into V. anguillarum M93Sm by conjugation (14). Transconjugants were selected by utilizing the chloramphenicol resistance gene located on the suicide plasmid. The incorporation of the suicide vector into the hlyU gene was confirmed by PCR analysis, as described previously (14). The resulting V. anguillarum hlyU mutant was designated S305 (Table 1) for future use.
Complementation of the hlyU mutant.
The mutant was complemented by cloning the appropriate hlyU gene fragment into the shuttle vector pSUP202 (GenBank accession no. AY428809), as described previously by Rock and Nelson (16). Briefly, primers (Table 2) were designed and EcoRI sites were introduced at the 5′ end of the primers. The primer pair was then used to amplify the entire hlyU gene plus ∼500 bp of the 5′ and 3′ flanking regions from genomic DNA of V. anguillarum M93Sm. The PCR product was cloned into the pCR2.1 vector (Invitrogen) and digested with EcoRI restriction enzyme, and the DNA fragments were separated on a 1% agarose gel. Subsequently, the gel-purified PCR fragment was ligated into pSUP202 after digestion with EcoRI and the ligation mixture was introduced into E. coli Sm10(λ pir) by electroporation with Bio-Rad Gene Pulser II. Transformants were selected on LB10-Ap100 agar plates. The complementing plasmid, pSUP202-hlyU, was transferred from E. coli Sm10 into the V. anguillarum hlyU mutant (S305) by conjugation using the procedures described previously (16). The transconjugants were confirmed by PCR amplification and restriction digestion.
Hemolytic activity assay.
Hemolytic activities of various V. anguillarum strains were determined by measuring the diameter of beta-hemolysis on plates containing Trypticase soy agar (TSA) plus sheep blood agar after 24 h at 27°C, as previously described (16).
Cytotoxicity assay.
Cytotoxic activity of V. anguillarum strains was determined by changes to cell morphology or by measurement of released lactate dehydrogenase (LDH). ASK cells were seeded into a six-well microtiter plate (Costar) in Leibovitz's L-15 medium supplemented with 20% fetal bovine serum and grown at 20°C to a cell density of ∼2 × 105 cells ml−1. V. anguillarum cultures grown overnight were harvested, washed twice in NSS, and resuspended in NSS (at a cell density of ∼2 × 109 cells ml−1). Washed bacterial cells were added to ASK cells at multiplicities of infection (MOI) of 100 and incubated at 20°C for 4 h. Changes in cell morphology were assessed and photographed by viewing live cells with an inverted microscope (Nikon TE2000 model). To determine the released LDH, a CytoTox-ONE homogeneous membrane integrity assay kit (Promega) was used. Briefly, ASK cells were seeded into a 96-well white-wall microtiter plate (Costar), as described above, at 20,000 cells/well. NSS-washed bacterial cells were added to each well at MOI of 20, 50, 100, and 200 and then incubated at 20°C for 4 h. The assay measures the generation of the fluorescent resorufin product, which is proportional to the amount of LDH at excitation/emission of 560 nm//590 nm.
RNA isolation.
Exponential-phase cells (∼0.5 × 108 CFU ml−1) and stationary-phase cells (2 × 109 CFU ml−1) of various V. anguillarum strains were harvested by centrifugation. Total RNA was isolated using the RNeasy kit (Qiagen) according to the manufacturer's instructions. All purified RNA samples were quantified spectrophotometrically by measuring absorption at 260 and 280 nm using a NanoDrop spectrophotometer and stored at −75°C for future use.
Semiquantitative RT-PCR and real-time qRT-PCR.
Total RNA was isolated from exponential- and stationary-growth-phase V. anguillarum cells as described above. All RNA samples were treated with DNase, and 100 μg of RNA was used as the template for reverse transcription-PCR (RT-PCR). RT-PCR was performed using Brilliant II SYBR green single-step quantitative RT-PCR (qRT-PCR) master mix (Stratagene). Briefly, gene-specific primers (Table 2) were used to reverse transcribe the specific cDNA from RNA templates, and the resulting cDNA was used as the template with which to amplify the specific DNA product, using 25-cycle regular PCR to give a semiquantitative determination of the original RNA amount. Genomic DNA (100 μg) extracted from wild-type strain M93Sm was used as the positive control. The thermal profile was 50°C for 30 min and 95°C for 15 min and then 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. The PCR product was visualized in a 1% agarose gel using a 100-bp DNA molecular weight ladder (Promega) as a standard. All real-time quantitative RT-PCRs (qRT-PCRs) were performed using an Mx3005 or Mx4000 multiplex quantitative PCR system (Stratagene). The primers used were the same as the semiquantitative RT-PCR (Table 2). Quantitation of various mRNAs was performed using Brilliant II SYBR green single-step qRT-PCR master mix (Stratagene) with 10 ng of total RNA in 25-μl reaction mixtures. The thermal profile was 50°C for 30 min, 95°C for 15 min, and then 40 cycles of 95°C for 30 s and 55°C for 30 s. Fluorescence was measured at the end of the 55°C step during every cycle. Samples were run in triplicate plus a no-RT control and no-template control.
5′-RACE assay.
Total RNA was isolated from exponential-phase V. anguillarum cells grown in LB20 using the RNeasy kit (Qiagen). To identify the transcriptional start site, RNA was subjected to 5′ rapid amplification of cDNA ends (5′-RACE) using the 2nd generation 5′-RACE kit (2). Primers used in RT-PCR are listed in Table 2. Briefly, 5 μg of RNA was used to generate specific first-strand cDNA from target mRNA (vah1, plp, rtxH, or rtxB) in a reverse transcriptase reaction with a gene-specific primer. Poly(A) tails were added to the 3′ cDNA end using dATP and terminal deoxynucleotidyl transferase [or in some cases, poly(G) tails were added with dGTP and terminal deoxynucleotidyl transferase]. A PCR product was amplified from the tailed cDNA by using a 5′-RACE anchor primer (AP) (Table 2) and the primer specific for that sequence. The PCR product was cloned into PCR2.1 cloning vector (Invitrogen), and plasmids from appropriate transformants were purified and sequenced.
DNA sequence and analysis.
All DNA sequencing was done at the RI Genomics and Sequencing Center (University of Rhode Island, Kingston), using an ABI 3170xl genetic analyzer unit (Applied Biosystems). Multiple alignments and phylogenic trees were analyzed using the ClustalW method in the DNASTAR Lasergene 7 program.
Overexpression and purification of the V. anguillarum HlyU protein.
The DNA fragment encoding HlyU was PCR amplified using primers Pm303 and Pm304 (Table 2) and cloned into the 6×His (His6) tag expression plasmid pQE30-UA (Qiagen, Inc.), generating the plasmid pQE30-UA/HlyU, which encodes HlyU with an N-terminal fusion tag. The correct recombinant clone confirmed by sequencing was used for expression of His-tagged HlyU protein in E. coli M15 (S301). Ten milliliters of overnight bacterial culture growing at 37°C in Luria broth supplemented with 50 μg/ml kanamycin and 100 μg/ml ampicillin was inoculated into 250 ml of the same fresh medium. When the optical density at 600 nm (OD600) reached 0.6, 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to induce the expression of HlyU protein. After bacteria were grown for an additional 5 h at 37°C, the cells were collected by centrifugation (8,000 × g, 10 min) and the cell pellets were resuspended in 5 ml lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0]). The cell suspension was incubated with lysozyme (0.5 mg/ml) on ice for 30 min and then sonicated (six bursts at 20 s per burst with 30-s intervals on ice). The resulting cell lysate was centrifuged (10,000 × g, 20 min), and the soluble supernatant containing HlyU-His6 was collected. The recombinant protein was then purified from this fraction by affinity chromatography using Ni-nitrilotriacetic acid resin (Qiagen, Inc.) according to the manufacturer's instructions. The concentration of the purified HlyU protein was determined by measuring the absorbance at 280 nm using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific).
Gel mobility shift assay.
The gel mobility shift assay was performed using a 2nd generation digoxigenin (DIG) gel shift kit (Roche, Indianapolis, IN). Three fragments (a, b, and c) from the rtxH-rtxB intergenic region and five fragments (d, e, f, g, and h) from the plp-vah1 intergenic region were amplified by PCR and then 3′ end labeled with digoxigenin-11-ddUTP using terminal deoxynucleotidyl transferase. After the labeling efficiency was determined, each of the labeled probes (0.4 ng for fragments a, b, and c and 0.2 ng for fragments d, e, f, g, and h) was incubated with 350 ng purified HlyU protein in 20 μl binding buffer [100 mM HEPES (pH 7.6), 5 mM EDTA, 50 mM (NH4)2SO4, 5 mM dithiothreitol, 1% (wt/vol) Tween 20, 150 mM KCl]. For competition analysis, labeled probe (0.4 ng for fragments a, b, and c and 0.2 ng for fragments d, e, f, g, and h) and 350 ng HlyU protein were incubated with 100 ng/μl unlabeled specific probe. The binding reactions were carried out at room temperature for 15 min, and then samples were separated by 6% polyacrylamide DNA retardation gel (Invitrogen, Carlsbad, CA). The DNA-protein complex was transferred to positively charged nylon membrane by electroblotting, and then immunological detection and chemiluminescent signal detection were carried out according to the instructions of the manufacturer (Roche, Indianapolis, IN).
DNase I protection assay.
The DNA probes for the DNase I protection assay were amplified from V. anguillarum genomic DNA by PCR using primers (Integrated DNA Technologies, Inc.) shown in Table 3. Thus, two rtxH-rtxB intergenic region probes (4 and 5) were labeled with 6-carboxyfluorescein (6-FAM) at the 5′ end on the upper strand and the lower strand, respectively. The two plp-vah1 intergenic probes (2 and 3) were also labeled with 6-FAM at the 5′ end on the upper strand and lower strand, respectively. The assay was carried out using a method modified from Zianni et al. (25). Briefly, 40 ng of a DNA probe and various amounts of recombinant HlyU (rHlyU) (up to 1.88 μg) were incubated at 37°C in a total volume of 20 μl, containing binding buffer (4 μl, 5× concentration) from the 2nd generation DIG gel shift kit (Roche Applied Science), for 1 h. The DNA-protein complex was then digested by adding 0.001 U RQ1 RNase-free DNase (Promega Corporation) in a total volume of 23 μl containing reaction buffer (2.3 μl, 10× concentration) at 37°C for 1 min. The reaction was stopped by adding 2.6 μl stop solution (10× concentration) followed by heating (95°C, 10 min). The DNA was purified with a QIAquick PCR purification kit (Qiagen, Inc.), using a QIAcube and its standard protocol, except that the elution volume was adjusted to 30 μl. The DNA in the elutant (5 μl) was added to 10 μl Hi-Di formamide containing 0.1 μl GeneScan 600 LIZ size standard (Applied Biosystems), and the mixture was submitted to capillary electrophoresis fragment analysis (Rhode Island Genomics and Sequencing Center).
Table 3.
Primers used to amplify DNA probes in the DNase I protection assay
| Intergenic region | Probe no. (length [bp]) | Primer name | Primer sequence | Primer 5′ label | Primer strand |
|---|---|---|---|---|---|
| rtxH-rtxB | 4 (336) | Pm414 | CAGTGGCTCATAAAAGCAGTTGC | 6-FAM | rtxH sense |
| Pm318 | CAGCGGTAAGTAGACTGATA | None | rtxB sense | ||
| 5 (394) | Pm315 | CTCAGACATAAATAAATCACC | None | rtxH sense | |
| Pm415 | CAGCGGTAAGTAGACTGATAAGCAATG | 6-FAM | rtxB sense | ||
| plp-vah1 | 2 (496) | Pm412 | CCGTATTTTCTGCAATCGCCATGG | 6-FAM | plp sense |
| Pm322 | AAAATAAAAGGACATTGGTTTTTTGG | None | vah1 sense | ||
| 3 (468) | Pm327 | GTATTTTCTGCAATCGCCATG | None | plp sense | |
| Pm413 | CACCTTTGTGGCGAATTATTAATAGATCTT | 6-FAM | vah1 sense |
RESULTS
Identification of the hlyU gene in V. anguillarum.
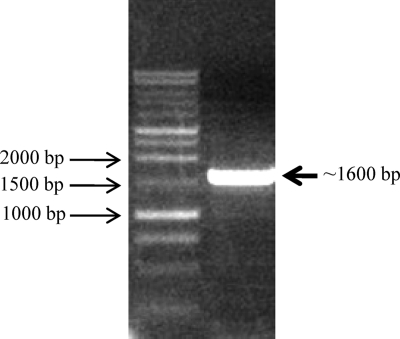
Previous studies indicated that the hlyU gene is a conserved transcriptional regulator in many Vibrio species (11, 12, 22, 23). We hypothesized that HlyU could be a putative regulator of the two hemolysin gene clusters in V. anguillarum. In order to identify the unknown hlyU gene in V. anguillarum, several hlyU genes from Vibrio species, including V. cholerae, V. vulnificus, V. fischeri, and V. parahaemolyticus, were compared using freely available software and database from the Integrated Microbial Genomes (IMG) website (http://img.jgi.doe.gov). The comparison revealed that the flanking genes of hlyU were identical among these Vibrio species and encoded a transcriptional activator protein, NhaR, and a ribosomal protein, S20P (Fig. 1). Conserved regions (sequence data obtained from http://www.ncbi.nlm.nih.gov) from both flanking regions were aligned (Fig. 1), and degenerate PCR primers (Table 2) were designed and used to perform degenerate PCR to amplify the putative hlyU gene and flanking DNA from V. anguillarum. A single PCR product was obtained by degenerate PCR (Fig. 2) and was purified, cleaned, cloned, and sequenced. As expected, DNA sequence data revealed that the PCR product included the intact 294-bp hlyU gene homologue (GenBank accession no. HQ149334), which encodes a predicted protein with 97 amino acids, a molecular mass of 11,095 Da, and strong homology to HlyU proteins found in a variety of Vibrio species, including Vibrio furnissii (97% similarity and 92% identity), Vibrio coralliilyticus (95% similarity and 91% identity), V. cholerae (93% similarity and 86% identity), V. parahaemolyticus (92% similarity and 88% identity), and V. vulnificus (94% similarity and 87% identity).
Fig. 2.
The degenerate PCR product was amplified from V. anguillarum M93Sm genomic DNA using primer pair Pm301/Pm302. The PCR product was separated and visualized in a 1% agarose gel using a Promega 1-kb DNA ladder as the size standard and was 1.6 kbp long. The PCR product was purified, cleaned, and cloned into pCR2.1 vector and then transformed into the E. coli DH5α strain. The plasmid, purified from the appropriate colony, was sequenced.
Mutation in hlyU decreases hemolytic activity.
An insertional mutation by single-crossover homologous recombination in the hlyU gene was obtained. The hemolytic activity of the hlyU mutant was determined and found to decrease about 5-fold compared with wild-type strain M93Sm on sheep blood agar (Fig. 3). Complementation of the hlyU mutant restored the hemolytic activity, which was even higher than wild type (Fig. 3), indicating that HlyU is a positive regulator of hemolysis in V. anguillarum.
Fig. 3.
Hemolytic activity of V. anguillarum strains M93Sm (wild type), S305 (hlyU mutant) colonies, and S307 (hlyU complement) transferred onto 5% TSA-sheep blood agar and incubated at 27°C for 24 h. Relative hemolytic activity was determined by measuring the beta-hemolysis zone surrounding each colony.
Mutation in hlyU has no significant effect on cytotoxicity.
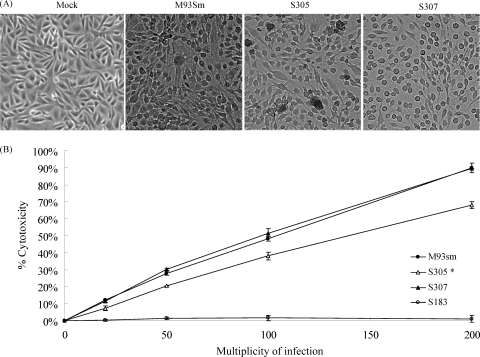
One hemolysin gene, rtxA, has been shown to be a major virulence factor for V. anguillarum (10). Previous studies revealed that RtxA has strong cytotoxic activity against Atlantic salmon kidney (ASK) cells and causes cells to round-up, detach, and die (10). However, experiments showed that ASK cells still rounded up and died when incubated with S305, M93Sm, or S307 cells (Fig. 4A) at an MOI of 100 for 4 h, indicating the mutation in hlyU did not completely knock out the cytotoxicity of V. anguillarum. Indeed, the LDH release assay revealed that S305 retained ∼75 to 80% (P > 0.1) of cytotoxicity at all MOI values compared to the wild-type strain M93Sm (Fig. 4B), confirming that the mutation of hlyU had only a small, statistically insignificant, effect on cytotoxicity. As a negative control, the rtxA vah1 double mutant strain S183 exhibited no cytotoxicity compared to the wild-type strain M93Sm (Fig. 4B), confirming that rtxA and vah1 are the major cytotoxins in V. anguillarum (10). When strain S307 was assayed for cytotoxic activity by the LDH release assay, the activity was restored to the same levels seen in M93Sm (Fig. 4B).
Fig. 4.
The hlyU mutation and its complement did not significantly affect the cytotoxicity of V. anguillarum against ASK cells. (A) ASK cells were incubated with various strains of V. anguillarum at an MOI of 100 for 4 h at 20°C. The data showed that strain S305 (hlyU mutant) still caused ASK cells to round-up, as did wild-type strain M93Sm and strain S307 (hlyU complement). ASK cells treated with NSS buffer (mock) exhibited no rounding or detachment during the course of the experiment. The photograph magnification is 100×. (B) LDH release from ASK cells treated with wild-type M93Sm (•), hlyU strain S305 (▵), hlyU complement S307 (▴), and rtxA vah1 double mutant S183 (○) at various MOI (20, 50, 100, and 200) for 4 h. LDH release was measured in relative fluorescence units (RFU) and then calculated to yield the percentage of cytotoxicity according to the instructions of the manufacturer. The incubation time at 0 was the base level of LDH in ASK cells treated with NSS buffer. Data are the representative of three separate experiments done with three replicates. Error bars show the standard deviation of the average. *, statistical difference of strain S305 from the wild-type treated data (P > 0.1).
HlyU positively regulates hemolysin genes at the transcriptional level.
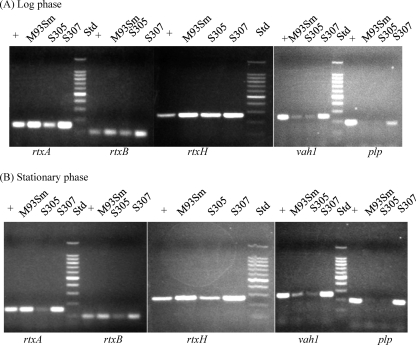
Semiquantitative RT-PCR and real-time qRT-PCR were performed to determine the expression levels of hemolysin genes, including vah1, plp, rtxA, rtxH, and rtxB, in the wild-type strain (M93Sm), hlyU mutant (S305), and the hlyU complement (S307) during both the exponential and stationary growth phases. Previously, we demonstrated that the rtx gene cluster contains two divergently cotranscribed sets of genes: rtxHCA, with the rtxH promoter proximal, and rtxBDE, with the rtxB promoter proximal (10). As shown in Fig. 5, RNA expression of rtxH, rtxA, and rtxB decreased in the hlyU mutant compared to wild-type expression levels during both exponential and stationary phases, indicating that the transcriptional levels of rtx genes were downregulated in the absence of HlyU. Indeed, complementation of the hlyU mutation upregulated the expression of rtx genes back to wild-type levels (or higher), indicating that HlyU positively regulates the expression of rtx genes. Real-time qRT-PCR data also revealed that in the hlyU mutant during the exponential and stationary phases, respectively, expression of rtxA decreased by 7.94- and 20-fold; expression of rtxB decreased by 3.56- and 8.07-fold; and expression of rtxH decreased by 5.9- and 15.1-fold (Table 4). The data strongly suggest that HlyU is a positive regulator of rtx gene expression, playing an important role in the expression of rtx genes during both exponential and stationary phases. In fact, the data show that the mutation in hlyU has a larger effect on stationary-phase expression of rtx genes than on exponential-phase expression. Additionally, expression of the same rtx genes increased to levels higher than wild type in the hlyU complement (Table 4), indicating the overexpression of hlyU positively regulates the expression of rtx genes.
Fig. 5.
HlyU regulates both hemolysin clusters positively at the transcriptional level. Semiquantitative RT-PCR was used to determine the expression levels of rtxA, rtxB, rtxH, vah1, and plp with 100 μg of RNA extracted from V. anguillarum cells grown to the exponential phase (A) and stationary phase (B). Data show that the expression of rtxA, rtxB, and rtxH decreased in the hlyU mutant (S305). However, expression of vah1 and plp did not decrease in S305, but did increase in the hlyU complement (S307). The M93Sm genomic DNA was used as a positive control (+). RT-PCRs with no added reverse transcriptase used as negative controls showed no amplified bands (data not shown). The DNA molecular size standards (Std) are a 100-bp Promega DNA ladder.
Table 4.
Real time qRT-PCR analysisa
| Parameter and gene expressed | V. anguillarum strainb | Expression at: |
|
|---|---|---|---|
| Log phase | Stationary phase | ||
| rtxA | |||
| RNA copy no. | M93Sm | 2.74 × 104 ± 1,300 | 8.88 × 103 ± 252 |
| S305 | 3.45 × 103 ± 800 | 4.47 × 102 ± 15 | |
| S307 | 1.18 × 105 ± 424 | 1.48 × 104 ± 1,237 | |
| Relative change (fold) in expressionc | M93Sm | 1.00 | 1.00 |
| S305 | −7.94 ± 0.4 | −20.00 ± 0.6 | |
| S307 | 4.30 ± 0.01 | 1.68 ± 0.14 | |
| rtxB | |||
| RNA copy no. | M93Sm | 3.40 × 104 ± 2,298 | 2.92 × 104 ± 736 |
| S305 | 9.54 × 103 ± 596 | 3.62 × 103 ± 141 | |
| S307 | 9.83 × 104 ± 6,045 | 3.60 × 104 ± 7,771 | |
| Relative change (fold) in expressionc | M93Sm | 1.00 | 1.00 |
| S305 | −3.56 ± 0.2 | −8.07 ± 0.3 | |
| S307 | 2.89 ± 0.18 | 1.23 ± 0.3 | |
| rtxH | |||
| RNA copy no. | M93Sm | 4.74 × 104 ± 0 | 1.93 × 104 ± 9,333 |
| S305 | 7.43 × 103 ± 643 | 1.18 × 103 ± 410 | |
| S307 | 1.46 × 105 ± 10,606 | 2.96 × 105 ± 707 | |
| Relative change (fold) in expressionc | M93Sm | 1.00 | 1.00 |
| S305 | −5.90 ± 0.5 | −15.1 ± 6 | |
| S307 | 3.08 ± 0.2 | 14.6 ± 0.03 | |
| vah1 | |||
| RNA copy no. | M93Sm | 3.17 × 102 ± 56 | 5.57 × 102 ± 302 |
| S305 | 3.36 × 102 ± 17 | 6.92 × 102 ± 58 | |
| S307 | 3.66 × 103 ± 556 | 1.47 × 104 ± 1,572 | |
| Relative change (fold) in expressionc | M93Sm | 1.00 | 1.00 |
| S305 | 1.06 ± 0.05 | 1.24 ± 0.1 | |
| S307 | 11.6 ± 1.7 | 26.3 ± 2.7 | |
| plp | |||
| RNA copy no. | M93Sm | 2.70 × 102 ± 34 | 6.83 × 102 ± 46 |
| S305 | 4.22 × 102 ± 62 | 3.27 × 102 ± 53 | |
| S307 | 2.25 × 103 ± 445 | 5.92 × 104 ± 3,467 | |
| Relative change (fold) in expressionc | M93Sm | 1.00 | 1.00 |
| S305 | 1.56 ± 0.05 | −2.08 ± 0.01 | |
| S307 | 8.32 ± 1.1 | 86.2 ± 7.8 | |
The data presented are from a representative experiment of two independent experiments. Each sample is the average of three replicates.
M93Sm is the wild type, S305 is the hlyU mutant, and S307 is the hlyU complement.
Gene expression is shown as either upregulated (positive number) or downregulated (negative number) compared to expression in M93Sm.
In contrast to rtx genes, expression of genes in the vah1 cluster, including vah1 and plp, exhibited little or no decrease in the hlyU mutant by the semiquantitative RT-PCR experiments (Fig. 5). Measurements of expression of vah1 and plp by real-time qRT-PCR were consistent with data from the semiquantitative experiments showing no significant changes in expression in the hlyU mutant (Table 4), indicating that the absence of HlyU does not affect either vah1 or plp expression. However, when the expression of vah1 and plp was examined by both semiquantitative RT-PCR (Fig. 6) and qRT-PCR (Table 4) in the hlyU complement (S307), we observed that expression of both genes increased. Specifically, expression of vah1 in S307 increased over wild-type (M93Sm) levels by 11.6- and 26.3-fold during the exponential and stationary phases, respectively, and expression of plp increased over levels in M93Sm by 8.32- and 86.2-fold during the exponential and stationary phases, respectively. These data indicate that overexpression of HlyU can positively regulate expression of vah1 and plp.
Fig. 6.
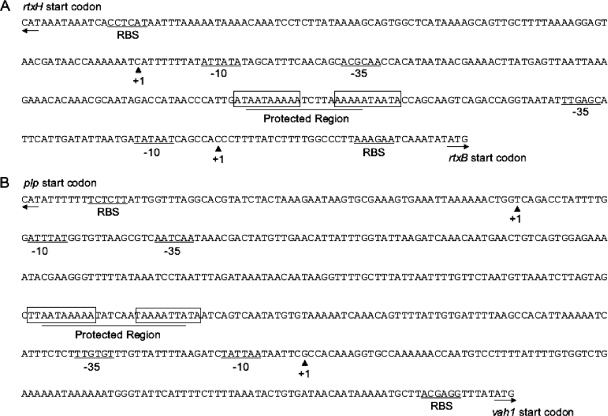
Intergenic regions of the rtxACHBDE gene cluster (A) and vah1-plp gene cluster (B). The transcriptional start sites of hemolysin genes are indicated by solid triangles marked with +1. The −10 and −35 promoter sequences of hemolysin genes are indicted by underlined nucleotides labeled as −10 and −35 and are predicted to be σ70 promoters. Black arrows indicate the start codons of genes. Lines indicating ribosomal binding sites of hemolysin genes are labeled “RBS.” The boxed sequences represent inverted repeats located with the HlyU-DNase I protected sequences, which are indicated as a protected region.
Mapping transcriptional start sites of hemolysin genes.
Since it had been reported that HlyU is a DNA binding protein (17) that positively regulates hlyA (homologue of vah1) in V. cholerae (23) and rtxA1 (homologue of rtxA) in V. vulnificus (11), we wanted to determine possible HlyU binding sites in the vah1 gene cluster and rtxACHBDE cluster in V. anguillarum. The transcriptional start sites of both hemolysin clusters were identified using 5′-RACE. In the vah1 gene cluster, there is a 508-bp intergenic region between the divergent plp and vah1 genes. The 5′-RACE results demonstrated that the region between the +1 sites of plp and vah1 was 318 bases long (Fig. 6B). The +1 transcriptional start site (A) of plp is 73 bases prior to its start codon, with a predicted −35 and −10 promoter sequence of TTGATT-N13-ATAAAT (Fig. 6B). The divergent hemolysin gene, vah1, had a transcriptional start site (G) 119 bases before the vah1 start codon, with a predicted −35 and −10 promoter sequence of TTGTGT-N16-TATTAA (Fig. 6B).
For the rtx gene cluster, the intergenic space between the divergent rtxH and rtxB genes is 325 bp. 5′-RACE results show that the region between the transcriptional start sites of rtxH and rtxB is 187 bp (Fig. 6A). The +1 transcriptional start site (G) of rtxH is 103 bases prior to its start codon, with a predicted −35 and −10 promoter sequence of TTGCGT-N15-TATAAT (Fig. 6A). The divergent rtxA transporter gene, rtxB, was found to have a transcriptional start site (C) 34 bases before the rtxB start codon, with a predicted −35 and −10 promoter sequence of TTGAGC-N18-TATAAT (Fig. 6A). Analysis of the predicted promoter regions of these two hemolysin clusters revealed strong similarities to a σ70 consensus promoter, TTGACA-N17-TATAAT. Additionally, the putative ribosomal binding site (RBS) for all genes was also located upstream of the ATG start codons (Fig. 6A and B).
HlyU binds to the intergenic promoter regions of the hemolysin gene clusters.
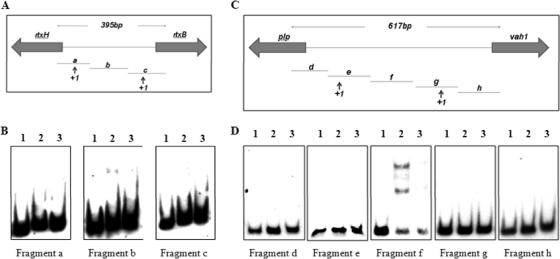
Previously, Liu et al. (11) demonstrated that HlyU binds to the promoter region of the rtxA1 operon of V. vulnificus. In an effort to determine whether HlyU acted in a similar fashion to help regulate expression of the hemolysin gene clusters in V. anguillarum, we carried out gel mobility shift experiments using purified HlyU-His6 protein. Briefly, the purified protein (350 ng) was reacted with each of the three DIG-labeled DNA subfragments amplified from the intergenic region between rtxH and rtxB (Fig. 7A) and with each of the five DIG-labeled DNA subfragments amplified from the intergenic region between plp and vah1 (Fig. 7C). DNA mobility shift experiments were performed on the mixtures containing HlyU plus DIG-labeled DNA. The results revealed that HlyU bound to fragment b of the rtxH-rtxB intergenic region (Fig. 7B) and to fragment f of the plp-vah1 intergenic region (Fig. 7D). When unlabeled competitor DNA was added to each of these reactions, binding was decreased or abolished.
Fig. 7.
DNA gel mobility shift demonstrating binding of purified HlyU to intergenic regions of the rtxACHBDE operon (A and B) and the vah1-plp gene cluster (C and D). DIG-labeled DNA fragments of the intergenic region between rtxH and rtxB (A) and between plp and vah1 (C) were obtained by PCR amplification. Individual DIG-labeled fragments (0.4 ng) were reacted with no additions (lane 1), 350 ng HlyU (lane 2), and 350 ng HlyU plus 100 ng unlabeled DNA fragment (lane 3). (B) Fragments a, b, and c from the intergenic regions of the rtxACHBDE operon.(D) Fragments d, e, f, g, and h from the intergenic regions of the vah1-plp gene cluster.
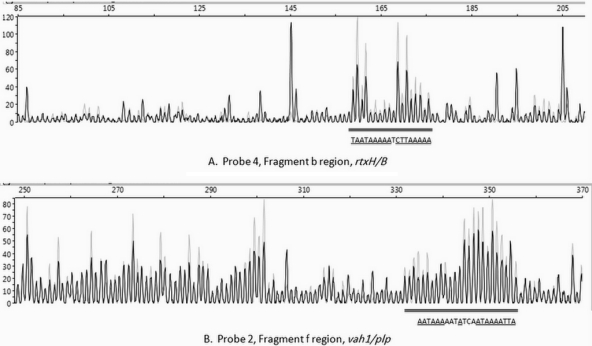
In an effort to more closely characterize the binding sites of HlyU for each hemolysin gene cluster, each of the two DNA subfragments that bound HlyU was examined by a DNase I protection assay, as described in Materials and Methods. The results of these experiments revealed that HlyU protected an 18-bp region (5′-TAATAAAAATCTTAAAAA-3′) in fragment b (Fig. 8A) with two 5-bp direct repeats of TAAAA. This region starts 103 bp upstream of the +1 site of rtxH and 67 bp upstream from rtxB. Similarly, HlyU protected a 22-bp region (5′-AATAAAAATATCAATAAAATTA 3′) in fragment f (Fig. 8B) with the same two 5-bp direct repeats of TAAAA. The binding region in subfragment f starts 192 bp upstream of plp and 106 bp upstream of the +1 site for vah1.
Fig. 8.
Capillary electrophoresis of 6-FAM-labeled DNA fragments b (A) and f (B) from DNase protection assays in the presence (gray traces) and absence (black traces) of HlyU, demonstrating that HlyU binds to specific sequences in fragments b and f of the rtxACHBDE and plp-vah1 intergenic regions, respectively, and protects against DNase I digestion. DNA fragments b and f were prepared and labeled with 6-FAM, reacted with HlyU (0 or 1.88 μg) followed by DNase I, and then analyzed by DNA fragment analysis as described in Materials and Methods. The double black lines show the binding regions. The binding region sequences are shown below the double black line. Underlined bases indicate those that are higher in the presence of HlyU (gray trace) than in its absence (black trace).
DISCUSSION
Hemolytic activity of V. anguillarum has been considered the virulence factor responsible for hemorrhagic septicemia during infection (1, 5). We previously reported that there are two major hemolysin gene clusters in V. anguillarum M93Sm (10, 16). The vah1 cluster consists of four genes, plp, vah1, llpA, and llpB. Vah1 is a putative pore-forming hemolysin, which shows strong homology to HlyA of V. cholerae. HlyA integrates into the erythrocyte membrane to cause lysis (13). The plp gene, divergent from the vah1 gene, encodes a hemolysin with phospholipase A2 activity specific for phosphatidylcholine and is highly conserved among members of the Vibrionaceae as a lecithinase/thermolabile hemolysin. Plp has the ability to lyse fish erythrocytes because of the abundance of phosphatidylcholine in their membranes (9). Additionally, mutations in plp result in increased expression of vah1 (16). The second hemolysin gene cluster in V. anguillarum is the rtxACHBDE cluster (10), in which rtxHAC is divergently transcribed from rtxBDE. The V. anguillarum RtxA is a major virulence factor for V. anguillarum with both hemolytic and cytotoxic activities (10). While mutations of both vah1 and rtxA are required for the complete loss of hemolytic activity on sheep blood agar (10), mutations in plp, vah1, and rtxA are required for a 90% loss of hemolytic activity against fish erythrocytes. Thus, all three genes encode proteins that are major hemolysins in the fish host (9). However, prior to this study, little was known about the regulation of these hemolysins in V. anguillarum.
It has been reported that HlyU regulates the expression of hemolysins in Vibrio species. In V. cholerae, HlyU positively regulates the expression of hemolysin HlyA (23) and the HlyA-coregulated gene hcp (24). It was also suggested that mutation of hlyU attenuates the virulence of V. cholerae O17 in the infant mouse cholera infection model (22). Recent evidence suggests that HlyU is a master regulator of virulence in V. vulnificus, as several virulence factors, including vvh1 and rtxA1, a homologue of rtxA of V. anguillarum, appear to be regulated by HlyU (8, 11). Therefore, we hypothesized that the hlyU gene in V. anguillarum might encode a regulator for both hemolysin clusters in V. anguillarum. In this study, we used degenerate PCR to discover the unknown hlyU gene from the V. anguillarum genome. The experiment successfully identified an hlyU gene (Fig. 2) from V. anguillarum with strong homology to other hlyU genes in Vibrio species.
HlyU is a member of SmtB-ArsR protein family. Some members of this transcriptional regulator family, such as NolR of Rhizobium meliloti, SmtB of Synechococcus sp. strain PCC 7942, and ArsR of Staphylococcus aureus, act to repress target gene expression by binding metal ions to a metal binding site located on the repressor protein to enhance binding to the DNA binding site (19). However, recent studies suggested that HlyU in V. cholerae acts as a positive regulator because of the absence of the metal binding site on HlyU (17). Furthermore, the crystal structure of HlyU from V. vulnificus strain CMCP6, recently solved by Nishi et al. (15), confirmed that HlyU has no metal binding site. Analysis of the HlyU amino acid sequence using the ClustalW program reveals that the V. anguillarum HlyU, as well as other Vibrio species (V. cholerae, V. vulnificus, V. fischeri, and V. parahemolyticus), does not contain a metal binding site and probably has similar binding characteristics to the homologues found in V. cholerae and V. vulnificus.
Evidence suggests that mutation of hlyU has a strong effect on virulence. For example, a mutation in hlyU attenuates V. cholerae O17 virulence in the infant mouse cholera infection model (22). In V. vulnificus, the LD50 increased about 104-fold in an hlyU mutant using the iron-overloaded mouse infection model (11) or the iron-normal mouse infection model (8). Additionally, cytotoxic activity was lost in an hlyU mutant of V. vulnificus (11). However, we found that in V. anguillarum, cytotoxicity of the hlyU mutant remained relatively high according to both the LDH release assay and observations of morphological changes in ASK cells exposed to the hlyU mutant (Fig. 4). These observations indicate that rtxA was still expressed in the hlyU mutant, even though rtxA expression was significantly decreased in the mutant (Fig. 5 and Table 4). While our data indicate that HlyU is a positive regulator of rtxA, rtxH, and rtxB, these genes are still expressed in the absence of HlyU in V. anguillarum. It is interesting to note that levels of transcription of rtxA, rtxH, and rtxB in the wild-type strain and hlyU mutant all decrease during stationary phase (Table 4). This may suggest that either greater amounts of HlyU are required during stationary phase or hlyU expression may be repressed during stationary phase.
Additionally, cytotoxicity data were consistent with the hemolytic activity assay, in which the hlyU mutant did not completely eliminate the hemolysis on the sheep blood agar (Fig. 3), indicating that the hemolysins were expressed in the mutant. Interestingly, real-time RT-PCR data showed that the hlyU mutant did not affect the expression of vah1 and plp compared to the wild-type strain (Table 4). However, the observation that overexpression of HlyU in the hlyU complement dramatically increased expression of both vah1 and plp suggests that vah1 and plp are regulated by HlyU in a different manner than the rtx gene cluster. This is supported by our previous observation that plp null mutations increase hemolysin activity and vah1 transcription (16). Thus, the results presented here suggest that in addition to HlyU, one or more other factors may regulate the hemolysins/cytotoxins of V. anguillarum.
While HlyU is a positive regulator of V. cholerae hlyA (22, 23), there is no experimental evidence to demonstrate that HlyU binds to the hlyA promoter region. Therefore, it is still unclear if HlyU is a direct transcriptional activator binding to the hlyA promoter region or if it interferes with an unknown repressor of hlyA to cause derepression of hlyA by HlyU (17). The latter assumption was recently supported by the study of V. vulnifucus, where HlyU was found to bind to the upstream region of rtxH, which competes with the binding site of rtxA1 repressor H-NS (12). Similar to V. anguillarum, expression of rtxH and rtxA1 in V. vulnificus is regulated by the same rtxH proximal promoter. Therefore, it was suggested that the absence of HlyU would increase the H-NS binding, which repressed the expression of rtxA1 in V. vulnificus (12). It is reasonable to think that a similar situation might exist in the both hemolysin clusters of V. anguillarum.
In this study, transcriptional start sites of both hemolysin clusters were identified, and promoter regions for the potential HlyU binding were targeted (Fig. 6). We found that the central regions of the intergenic sequence for each hemolysin gene cluster contain a conserved binding site for HlyU, as determined by both DNA mobility shift experiments (Fig. 7) and DNase I protection assays (Fig. 8). The two binding sites are quite similar (Fig. 8): the intergenic rtxH-rtxB protected binding region is 18 bp long, while the intergenic plp-vah1 region is 22 bp long, and both have identical 5-bp direct repeats of TAAAA, strongly suggesting that HlyU binds as a dimer, as suggested by Saha and Chakrabarti (17). In fact, the direct repeat may be a bit longer than 5 bp. If one uses an imperfect match, the direct repeat is 7 bp: (A/T)TAAAA(A/T). Additionally, examination of the sequences immediately adjacent to the protected regions reveals that both the rtxH-rtxB and the plp-vah1 intergenic regions contain 25- to 26-bp regions that are nearly identical with 10-bp inverted repeats at each end (Fig. 6). The rtxH-rtxB intergenic region contains identical 10-bp inverted repeats at each end of its 25-bp region with a nucleotide sequence of ATAATAAAAA. Similarly, the plp-vah1 intergenic region has nearly identical 10-bp inverted repeats at each end of the 26-bp region. The nucleotide sequence of the plp proximal repeat is 5′-TTAATAAAAA-3′. The nucleotide sequence of the vah1 proximal repeat is 3′-ATATTAAAAT-5′. Furthermore, comparison between the HlyU binding sites identified here and the site identified by Liu et al. (12) reveals that in both cases HlyU binds to AT-rich regions upstream of the transcriptional start sites of the regulated hemolysin genes. However, Liu et al. (10, 12) found that HlyU bound far upstream (bp −376 to −417) of the transcriptional start site of the rtxA1 operon. In contrast, we have located HlyU binding somewhat closer to the start transcription sites of rtxH-rtxB and plp-vah1. The binding sites of HlyU are 104 bp and 68 bp upstream of the rtxH and rtxB +1 sites, respectively, and 150 bp and 145 bp upstream of the plp and vah1 +1 sites, respectively. While we have not yet demonstrated H-NS repressor binding to these regulatory regions in V. anguillarum, the shorter distance between HlyU binding sites and the transcriptional start sites in V. anguillarum compared to V. vulnificus may indicate that V. anguillarum has fewer H-NS binding sites than the five sites found for the V. vulnificus rtxA1 regulatory region (12).
ACKNOWLEDGMENT
This work was supported by the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service, grant no. 2008-35204-04605, awarded to D.R.N.
This research was based in part upon work conducted using the Rhode Island Genomics Sequencing Center, which is supported in part by the National Science Foundation under EPSCoR grant 0554548. We thank Maureen Varina Driscoll for her help with this study.
Footnotes
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Austin B., Austin D. A. 1999. Bacterial fish pathogens: disease of farmed and wild fish, 3rd ed Praxis Publishing Co., London, United Kingdom [Google Scholar]
- 2. Bowen D. J., et al. 2003. Genetic and biochemical characterization of PrtA, an RTX-like metalloprotease from Photorhabdus. Microbiology 149:1581–1591 [DOI] [PubMed] [Google Scholar]
- 3. Denkin S. M., Nelson D. R. 1999. Induction of protease activity in Vibrio anguillarum by gastrointestinal mucus. Appl. Environ. Microbiol. 65:3555–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Denkin S. M., Nelson D. R. 2004. Regulation of Vibrio anguillarum empA metalloprotease expression and its role in virulence. Appl. Environ. Microbiol. 70:4193–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Egidius E. 1987. Vibriosis: pathogenicity and pathology. Aquaculture 7:15–28 [Google Scholar]
- 6. Garcia T., Otto K., Kjelleberg S., Nelson D. R. 1997. Growth of Vibrio anguillarum in Salmon intestinal mucus. Appl. Environ. Microbiol. 63:1034–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hirono I., Masuda T., Aoki T. 1996. Cloning and detection of the hemolysin gene of Vibrio anguillarum. Microb. Pathog. 21:173–182 [DOI] [PubMed] [Google Scholar]
- 8. Kim Y. R., et al. 2003. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect. Immun. 71:5461–5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li L. 2009. Identification and regulation of hemolysin genes of Vibrio anguillarum. Dissertation. University of Rhode Island, Kingston [Google Scholar]
- 10. Li L., Rock J. L., Nelson D. R. 2008. Identification and characterization of a repeat-in-toxin gene cluster in Vibrio anguillarum. Infect. Immun. 76:2620–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu M., Alice A. F., Naka H., Crosa J. H. 2007. The HlyU protein is a positive regulator of rtxA1, a gene responsible for cytotoxicity and virulence in the human pathogen Vibrio vulnificus. Infect. Immun. 75:3282–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu M., Naka H., Crosa J. H. 2009. HlyU acts as an H-NS antirepressor in the regulation of the RTX toxin gene essential for the virulence of the human pathogen Vibrio vulnificus CMCP6. Mol. Microbiol. 72:491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Menestrina G., Moser C., Pellet S., Welch R. 1994. Pore-formation by Escherichia coli hemolysin (HlyA) and other members of the RTX toxins family. Toxicology 87:249–267 [DOI] [PubMed] [Google Scholar]
- 14. Milton D. L., O'Toole R., Horstedt P., Wolf-Watz H. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nishi K., et al. 2010. Crystal structure of the transcriptional activator HlyU from Vibrio vulnificus CMCP6. FEBS Lett. 584:1097–1102 [DOI] [PubMed] [Google Scholar]
- 16. Rock J. L., Nelson D. R. 2006. Identification and characterization of a hemolysin gene cluster in Vibrio anguillarum. Infect. Immun. 74:2777–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saha R. P., Chakrabarti P. 2006. Molecular modeling and characterization of Vibrio cholerae transcription regulator HlyU. BMC Struct. Biol. 6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sezonov G., Joseleau-Petit D., D'Ari R. 2007. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189:8746–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shi W., Wu J., Rosen B. P. 1994. Identification of a putative metal binding site in a new family of metalloregulatory proteins. J. Biol. Chem. 269:19826–19829 [PubMed] [Google Scholar]
- 20. Thompson J. D., Gibson T. J., Higgins D. G. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics Chapter 2:Unit 2.3 [DOI] [PubMed] [Google Scholar]
- 21. Toranzo A. E., Barha J. L. 1993. Virulence factors of bacteria pathogenic for coldwater fish. Annu. Rev. Fish Dis. 3:5–36 [Google Scholar]
- 22. Williams S. G., Attridge S. R., Manning P. A. 1993. The transcriptional activator HlyU of Vibrio cholerae: nucleotide sequence and role in virulence gene expression. Mol. Microbiol. 9:751–760 [DOI] [PubMed] [Google Scholar]
- 23. Williams S. G., Manning P. A. 1991. Transcription of the Vibrio cholerae haemolysin gene, hlyA, and cloning of a positive regulatory locus, hlyU. Mol. Microbiol. 5:2031–2038 [DOI] [PubMed] [Google Scholar]
- 24. Williams S. G., Varcoe L. T., Attridge S. R., Manning P. A. 1996. Vibrio cholerae Hcp, a secreted protein coregulated with HlyA. Infect. Immun. 64:283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zianni M., Tessanne K., Merighi M., Laguna R., Tabita F. R. 2006. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J. Biomol. Tech. 17:103–113 [PMC free article] [PubMed] [Google Scholar]