Abstract
The Kruppel-like factor Klf4 is implicated in tumorigenesis and maintaining stem cell pluripotency, and Klf4 can both activate and repress gene expression. We show that the Pbx1 and Meis2 homeodomain proteins interact with Klf4 and can be recruited to DNA elements comprising a Klf4 site or GC box, with adjacent Meis and Pbx sites. Meis2d and Pbx1a activate expression of p15Ink4a and E-cadherin, dependent on the Meis2d transcriptional activation domain. In HepG2 cells, reducing expression of endogenous Meis2 or Pbx1 decreases p15 gene expression and increases the number of cells entering S phase. Although DNA binding by all three proteins contributes to full cooperative activation, the sequence requirements for binding by Meis2 and Pbx1 are variable. In the E-cadherin promoter, a Pbx-like site is required for full activation, whereas in the p15 promoter, the Klf4 site appears to play the major role. Through a bioinformatics search we identified additional genes with conserved binding sites for Klf4, Meis2, and Pbx1 and show that at least some of these genes can be activated cooperatively by Klf4 and Meis2/Pbx1. We suggest a model in which genes with Klf4 sites can be cooperatively activated by Meis2/Pbx1 and Klf4, dependent primarily on recruitment by Klf4. This provides a mechanism to modulate transcriptional regulation by the multifunctional Klf4 transcription factor.
INTRODUCTION
Homeodomain proteins comprise a large evolutionarily conserved family of DNA binding proteins, with diverse functions in organisms from yeast to mammals (17, 39, 40). The homeodomain is an approximately 60 amino-acid domain consisting of three alpha helices. The third helix is primarily responsible for DNA binding, whereas helices 1 and 2 play a structural role and are responsible for protein-protein interactions (10, 18, 49, 52). Many homeodomain proteins bind to DNA in complex with other proteins (9, 29, 30, 37). Meis2 and Pbx1 are members of the TALE superfamily of homeodomains, in which alpha helices 1 and 2 are separated by an extra three-amino-acid loop extension (hence TALE) (5, 7, 44). The TALE superfamily includes proteins which are transcriptional activators, such as Meis1 and Meis2, and repressors, such as Tgif1 and Tgif2 (5, 20, 41, 64, 65). In mice and humans, there are three Meis paralogs (Meis1, 2, and 3), as well as the related Prep1 and Prep2 (4, 15). Meis/Prep proteins share a highly conserved TALE homeodomain and a second conserved domain, termed the homothorax homology domain (Hth), which is present in the fly homothorax protein (42, 53). The Hth domain of Meis family proteins mediates interaction with Pbx proteins via the conserved PBC-A and PBC-B domains (22, 54, 57). Interaction of Meis and Pbx partners with each other facilitates their cooperative binding to a composite DNA element. Additionally, Meis proteins can be recruited to DNA by interaction with a Pbx partner and other homeodomain proteins, with out the need for Meis to bind DNA (34, 57). Meis and Pbx family proteins can contribute to the formation of DNA-bound transcription factor complexes with non-homeodomain proteins. For example, Pbx1 can recruit MyoD to promoter elements at which Pbx1 is prebound, prior to MyoD expression (2, 28). Some members of the Meis family, including Meis1 and Meis2, have transcriptional activation domains (20, 21), whereas Prep1 and 2 appear to be much weaker activators and may contribute primarily to the formation of a DNA-bound complex (3). Pbx proteins are unable to activate transcription alone but contribute to the binding of Meis proteins to DNA and may facilitate transcriptional activation by Meis partner proteins (35). Multiple splice variants of Meis2 have been identified (65). Most of the alternate splicing of Meis2 affects the transcriptional activation domain. However, one major splice variant (Meis2e) inserts a translational stop close to the amino terminus of the homeodomain, generating a variant lacking the activation domain and most of the homeodomain (65).
Mammalian Meis1 (myeloid ecotropic insertion site 1) was identified as a common site of viral integration in mouse myeloid leukemia cells (43), and the related Meis2 and Meis3 genes were identified based on sequence similarity (46, 48). Meis1 plays a key role in the progression of acute myeloid leukemia (AML) and mixed lineage leukemia (MLL), and fusion proteins generated by chromosomal rearrangements in MLL can induce increased expression of Meis1 (55, 61, 63). The Meis relative Prep1 plays a role in hematopoietic stem cell function and in early T cell development (12, 50, 51). Pbx proteins, which are common partners of Meis family members, have also been implicated in tumorigenesis. The Pbx1 gene is fused to the transcription factor E2A as a result of the t(1;19) translocation in pre-B cell leukemia (25, 26). This fusion prevents interaction with Meis proteins and converts Pbx1 to a transcriptional activator. Thus, there is significant evidence for deregulation of Meis and Pbx family proteins promoting tumorigenesis in lymphoid and hematopoietic cells.
Sp1 is a zinc finger-containing transcription factor which regulates a large number of genes via binding to a GC-rich consensus site (6, 13, 24). The related Sp3 protein binds to the same sequence and is highly homologous over the carboxyl-terminal zinc finger domain but is less well conserved outside this region (27). Sp1 has been shown to regulate the p15 promoter and to activate p15 cooperatively with transforming growth factor β (TGF-β)-regulated Smads (32, 33). Sp1 and Sp3 belong to a larger family of Kruppel-related zinc finger transcription factors that includes more than 15 KLF (for Kruppel-like factor) proteins (23, 60). The KLF family includes GKLF (gut-enriched KLF), also known as Klf4, which although highly expressed in gut epithelium is also expressed in a wide variety of other tissues (14) and plays a role in the maintenance of pluripotent stem cells (36, 47, 62). Klf4 has the characteristic three zinc fingers of the KLF family located at its carboxyl terminus and contains both a transcriptional repression domain and an activation domain that can interact with p300/CBP (19, 66). Thus, Klf4 can activate or repress transcription, possibly depending on the context. Klf4 also plays apparently contradictory roles in tumorigenesis, as there is evidence for Klf4 being both a tumor suppressor and an oncogene (14, 38). For example, Klf4 levels are decreased in colorectal cancer and in medulloblastoma, both by hypermethylation and by mutation (45, 68). In contrast, Klf4 expression in keratinocytes can induce squamous epithelial dysplasia (16). Consistent with a role as an oncogene, Klf4 expression is increased in a number of cancers, including mammary carcinomas and some squamous cell cancers (38).
Here we show that Klf4 can recruit Meis2 and Pbx1 to a suboptimal Meis/Pbx site adjacent to a GC box in the p15 promoter. The GC box is essential for Meis2 and Pbx1 recruitment and for the transcriptional activation by Meis/Pbx, which requires the Meis2 transcriptional activation domain. Using a combination of bioinformatics and functional analysis we identified a number of other genes that are candidates for cooperative regulation by Klf4-Meis/Pbx complexes. This work suggests that Meis/Pbx dimers may promote transcriptional activation over repression by DNA-bound Klf4, dependent on adjacent Meis or Pbx binding sites.
MATERIALS AND METHODS
Plasmids and oligonucleotides.
TGIF expression and short hairpin RNA (shRNA) plasmids have been described previously (1). p15 reporter constructs were created in pGL2 or pGL3 (Promega) or in pGL2 basic into which a minimal TATA element from the adenovirus major late promoter (MLP) had been inserted. The E-cad-luc reporter contains sequences from 178 bp upstream of the transcriptional start to +92 from the mouse gene. The four-copy SBR2 and two-copy Meis/Pbx reporters are as described previously (21, 56). Meis2, Pbx1, KLF, and Sp1 expression constructs were created in a modified pCMV5 with either a Flag or T7 epitope tag. KLF4 was also expressed from within pCDNA3. Meis2 and Pbx1 mutants and deletion constructs are as described previously (21). Pax3 and Etv1 luciferase reporters are as described previously (8, 11).
Cell culture and siRNA knockdown.
HepG2 (ATCC HB-8065), Mcf7, NMuli, and HeLa cells were maintained in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS), and COS1 cells were grown in DMEM with 10% bovine growth serum (BGS). For knockdown, cells were plated in 12-well plates and transfected with Dharmacon SMARTpool oligonucleotides by using DharmaFECT reagent 1, according to the manufacturer's instructions. Small interfering RNA (siRNA) sequences are available on request. RNA was isolated 60 h after transfection. The control pool (mouse siGENOME nontargeting siRNA pool 3) was used for the nontargeting control. For EdU labeling, cells were labeled with 10 μM EdU for 1 h at 37°C, and after fixation in 4% paraformaldehyde, were permeabilized with Triton X-100 for 30 min at room temperature. EdU was detected with an Alexa Fluor 488 EdU detection kit (Click-iT EdU [Molecular Probes]), according to the manufacturer's protocol, and DNA was stained with Hoechst 33342. Images were captured on a Zeiss AxioObserver with Volocity.
RNA analysis.
RNA was isolated and purified using an Absolutely RNA kit (Stratagene). For quantitative real-time PCR (qRT-PCR), cDNA was generated using Superscript III (Invitrogen) and analyzed in triplicate by real-time PCR using a Bio-Rad MyIQ cycler and Sensimix Plus SYBR green plus fluorescein isothiocyanate (FITC) mix (Quantace). Intron-spanning primer pairs were selected using Primer3 (http://frodo.wi.mit.edu/). Primer sequences for qRT-PCR are available on request. Expression was normalized to cyclophilin using the cycle threshold (ΔΔCT) method and is shown as the mean plus standard deviation (SD) from triplicate experiments.
Immunoprecipitation and Western blotting.
COS1 cells were transfected using Lipofectamine (Invitrogen). Forty hours after transfection, cells were lysed by sonication in phosphate-buffered saline (PBS) with 1% NP-40, 1 mM dithiothreitol (DTT), and protease inhibitors. Immunocomplexes were precipitated with Flag M2-agarose (Sigma). Following SDS-polyacrylamide gel electrophoresis, proteins were electroblotted to Immobilon-P (Millipore) and incubated with antisera specific for Flag (Sigma) or T7 (Novagen). Proteins were visualized with horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit Ig (Pierce) and ECL (Amersham Pharmacia Biotech). For testing dependence on DNA binding, ethidium bromide was added to a concentration of 1 μM to cell lysates prior to precipitation.
DNA affinity precipitation.
For isolation of protein complexes on double-stranded DNA oligonucleotides, lysates were prepared from 75% of a confluent 15-cm dish of COS1 cells for each condition, in MSLD (100 mM NaCl, 20 mM HEPES, pH 7.8, 10% glycerol, 0.1% Tween 20, with 1 mM DTT) and protease inhibitors. Lysates were precleared with protein A-agarose (Pierce) and incubated in 1 ml with 100 ng of biotinylated double-stranded oligonucleotide and 1 μg poly(dI-dC)·poly(dI-dC). Complexes were isolated on streptavidin-agarose, washed 4 times in binding buffer, and then subjected to analysis by SDS-PAGE and Western blotting. Sp1 antibody is from Upstate (07-645), Meis2 is from Abnova (H00004212-M01), and Pbx1 is from Abnova (H00005087-M01).
Luciferase assays.
HepG2 cells were transfected using Exgen 500 (MBI Fermentas) according to the manufacturer's instructions. Cells were transfected with firefly luciferase reporters, a Renilla transfection control (phCMVRLuc [Promega]), and the indicated expression constructs. After 40 h, firefly luciferase activity was assayed using firefly substrate (Biotium) and Renilla luciferase was assayed with 0.09 μM coelenterazine (Biosynth), by using a Berthold LB953 luminometer. Mithramycin was added to a final concentration of 200 nM, 24 h prior to analysis, where indicated.
ChIP.
For transfected chromatin immunoprecipitation (ChIP) HeLa cells were transfected with Exgen 500 (Fermentas) in 60-mm dishes and 5 μg of DNA. Two days after transfection, cells were washed with PBS, fixed in PBS with 1% formaldehyde for 15 min, and then quenched with 0.125 M glycine for 5 min. Plates were washed twice with cold PBS, scraped into 1 ml of cold radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris pH 8, 5 mM EDTA) with protease inhibitors, and sonicated 15 times for 10 s. Lysates were centrifuged at 21,000 relative centrifugal force (RCF) for 15 min to remove cell debris and then precleared with protein G-agarose (Pierce) for 2 h. Immunoprecipitation was carried out overnight with 15 μl Flag-agarose (in PBS, 1 mg/ml bovine serum albumin [BSA], 0.3 mg/ml salmon sperm DNA). Precipitates were washed twice with RIPA, 4 times with Szak's IP wash buffer (100 mM Tris-HCl, pH 8.5, 500 mM LiCl, 1% NP-40, 1% deoxycholate), twice more with RIPA, and twice with 1× Tris-EDTA (TE). One hundred microliters of 1.5× Talianidis elution buffer (70 mM Tris-HCl, pH 8, 1 mM EDTA, 1.5% SDS, 300 mM NaCl) was added to precipitates (and inputs) in 50 μl TE and samples were incubated at 65°C for 5 h. Samples were treated with 10 μg of proteinase K for 30 min at 45°C, and DNA was isolated using QIAquick columns (Qiagen) in 100 μl of water. Amounts of immunoprecipitated DNA were analyzed by qPCR on a Bio-Rad MyIQ cycler with Sensimix Plus SYBR green plus FITC mix (Quantace). Primer sequences for ChIP are available on request. Signal was expressed as bound versus input by the ΔΔCT method. For endogenous ChIP, Mcf7 cells were fixed and harvested as described above. One 15-cm dish was used per sample. Immunoprecipitation was carried out overnight using 3 μg of antibody and 15 μl of protein G-agarose (Pierce) with BSA and salmon sperm DNA, as described above. Antibody for Klf4 is from Santa Cruz (sc-20691) and Pbx1 was from Abnova (H00005087-M01). Immunoprecipitated fractions were washed and analyzed as described above.
In silico site search.
Mouse and human genomic databases were searched using the Site Search program (59): http://www.sitesearch.mshri.on.ca/Genome/index.html. We searched 2 kb upstream of the predicted start site of each gene, for the combination of a Klf4 site (RRGGYSY [58]) with both Meis and Pbx consensus sites (TGACA and CAATC) within 40 bp either side of the Klf4 site. This combination had to be present in both mouse and human, and we then accepted only those in which the orientations were the same in both mouse and human. We then ranked the hits by the total difference in spacing between the sites between mouse and human.
RESULTS
Activation of the p15 promoter by Meis2d and Pbx1a.
As shown in Fig. 1 A, the proximal 1 kb of the human p15 promoter contains four close matches to the Meis/Tgif consensus binding site. One of these (a 6/7 match) is present within the SBR2 that contributes to p15 expression in response to TGF-β signaling (56). We were therefore interested to know whether the p15 gene is a target for direct activation by Meis2 and repression by Tgif1, as has been proposed for the dopamine 1A receptor gene (65). In the context of the 1-kb p15 reporter construct, we observed little effect of Tgif1 overexpression or knockdown in these cells (data not shown). TALE homeodomains, including members of the Meis and Prep family, often bind to DNA together with other homeodomain proteins, including Hox and Pbx proteins. We therefore tested whether any response of the p15 promoter to Meis2d might be affected by Pbx1a. Coexpression of Meis2d with a reporter in which luciferase activity is driven by a 1-kb fragment of the p15 promoter resulted in around 3.5-fold activation, and coexpression of Pbx1a increased p15 activity to around 16-fold (Fig. 1A and B). We next analyzed a series of p15 promoter deletions to identify the region that responds to Meis2 and Pbx1 (Fig. 1A). As shown in Fig. 1A and B (also data not shown), a region containing only the 113 bases of the proximal promoter responded to Meis2 and Pbx1, suggesting that the four consensus sites are dispensable for this activity. This analysis suggests that the p15 promoter can be activated cooperatively by Meis2d and Pbx1a and that this response depends on sequences between −113 and −15 relative to the transcriptional start site. This region lacks a good match to a Meis or Pbx binding site but contains two previously characterized Sp1 binding sites.
Fig. 1.
The p15 promoter responds to Meis2d and Pbx1a. (A) The human p15 promoter is shown schematically, with the positions of potential Meis/TGIF binding sites, and consensus Sp1 and Initiator elements indicated. The reporters used are shown schematically, together with the base numbers relative to the transcriptional start site. The fold induction with Meis2d and Pbx1a together is shown for each reporter. For the four-copy SBR2 reporter this number is bracketed, since addition of Pbx1a had no further effect over what was seen with Meis2d alone. (B) HepG2 cells were transfected with two p15 reporters and assayed for luciferase activity in the presence of Meis2d and Pbx1a as indicated. Luciferase activity was measured after 40 hours and is shown normalized to a Renilla transfection control, as mean + SD from duplicate transfections. (C) HepG2 cells were transfected with the shorter (−113/+78) p15 promoter reporter, a p21 promoter reporter, a thymidine kinase (TK) promoter reporter, and an E-cadherin promoter reporter. Meis2d and Pbx1a were coexpressed as indicated. (D and E) HepG2 cells were transfected with siRNA duplexes targeting either Meis2 or Pbx1 alone, in combination, or with a control pool of nontargeting siRNAs. mRNA was isolated after 60 hours and analyzed by qRT-PCR for expression of the indicated genes. Relative expression is shown as the average + SD of triplicates. (F) HepG2 cells were transfected with the indicated siRNA pools and after 60 hours were incubated with EdU for 1 hour. EdU incorporation was determined by fluorescence microscopy and is presented as the percentage of cells incorporating EdU (average + SD of triplicates). Significance levels, as determined by Student's t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We next compared the effects of expression of Meis2d and Pbx1a on transcriptional reporters containing promoter regions from the p15 gene (two Sp1 sites), p21 (four Sp1 sites), thymidine kinase (TK) (one Sp1 site), and E-cadherin (one Sp1 site). As shown in Fig. 1C, the TK-luc and p21-luc reporters did not respond to Meis2d and Pbx1a, whereas the E-cadherin reporter was activated around 7-fold with both Meis2d and Pbx1a. We repeated these assays in the human SK-HEP-1 cell line, which was derived from a patient with adenocarcinoma, and again observed activation of the p15 and E-cadherin reporters by Meis2d and Pbx1a (data not shown). For the E-cadherin reporter, expression of either Meis2 or Pbx alone activates less than 2-fold, but expression of both results in around 15-fold activation. These data suggest that Meis2 and Pbx1 may activate expression of a subset of Sp1 site-containing promoters.
Regulation of p15 expression and cell cycle progression and Meis/Pbx.
To test whether the regulation of p15 and E-cadherin genes by Meis2d and Pbx1a occurred at the endogenous level, we knocked down Meis2 and Pbx1 and analyzed gene expression by qRT-PCR. HepG2 cells were transfected with siRNA oligonucleotides: a nonspecific pool as control or siRNAs to Meis2, Pbx1, or both. Targeting Meis2 resulted in around 60% reduction in Meis2 mRNA levels, and Pbx1 knockdown decreased its expression by up to 75% (Fig. 1D). Analysis of p15 expression revealed a significant decrease when Meis2 or Pbx1 was knocked down and a similar effect of the double Meis2/Pbx1 knockdown. We also observed a significant decrease in E-cadherin expression with knockdown of either Meis2 or Pbx1 or with knockdown of both together (Fig. 1D). We next analyzed expression of p21 and several other cell cycle regulators in cells with Meis2 and Pbx1 knockdown. p21 expression was significantly reduced in the knockdown, but only by about 40%. In contrast, we observed little effect on expression of other cell cycle regulators, although expression of cyclin A2 was somewhat increased (Fig. 1E). Since we observed decreased expression of p15, as well as some reduction in p21 expression, we next tested whether this affected cell cycle progression. Following knockdown of Meis2, Pbx1, or both together, cells were incubated with EdU to identify those that were undergoing DNA synthesis. As shown in Fig. 1F, there was a significant increase in the proportion of EdU-positive cells when either Meis2 or Pbx1 was knocked down, consistent with the idea that these cells were more rapidly transiting G1 and entering S phase. Together, these results suggest that Meis2 and Pbx1 can contribute to the activation of the p15 and E-cadherin genes in HepG2 cells and that reducing Meis2 and Pbx1 levels promotes proliferation.
The Meis2 activation domain is required for activation of p15.
To test the requirements for activation of p15 and E-cadherin expression by Meis2d and Pbx1a, we tested a panel of Meis2d and Pbx1a mutants in HepG2 cells (Fig. 2 A). We tested two truncation mutants of Meis2d, which affect either Pbx1 interaction or remove the transcriptional activation domain, and a mutant in which one of the DNA contact residues is altered from arginine to methionine (R332M) (21). We also tested a DNA binding point mutant form of Pbx1a (N286S) and a deletion mutant that has reduced interaction with Meis2d. We first verified the effects of these mutations on the activity of the Pbx/Meis consensus site reporter. Interfering with the interaction of Meis2d and Pbx1a with each other, or with DNA binding, severely reduced transcriptional activity, as did deletion of the Meis2d activation domain (Fig. 2B). Disruption of the interaction of Pbx1a with either DNA or Meis2d reduced the activation of the p15 reporter by about half, and preventing DNA binding by Meis2d effectively abolished activation by Meis2d and Pbx1a (Fig. 2C). A similar pattern was seen with the E-cadherin reporter, although the effect of deleting the Meis2 interaction domain of Pbx1a was more dramatic with this reporter (Fig. 2D). Importantly, deletion of the Meis2 activation domain abolished activation of either reporter by coexpressed Meis2d and Pbx1a, suggesting that the Meis2d activation domain is essential for the observed effects of Meis2d and Pbx1a on p15 and E-cadherin expression. Additionally, these data suggest that DNA binding by both Meis2d and Pbx1a is required for activation of p15 and E-cadherin expression.
Fig. 2.
Requirements for activation by Meis2/Pbx1. (A) The proteins encoded by the Pbx1a and Meis2d expression constructs used in panels B to D are shown schematically. The effects of the mutations or deletions on interaction with Meis2 or Pbx1 or on DNA binding are summarized, and the presence of the activation domain (Tx AD) in the Meis2d constructs is indicated. (B to D) HepG2 cells were transfected with the Pbx/Meis reporter (B), the p15 reporter (C), or the E-cadherin luciferase reporter (D), together with the indicated Pbx1a and Meis2d expression constructs. Luciferase activity was measured after 40 h and is presented in arbitrary units, as the mean plus SD of duplicate transfections.
GC boxes contribute to Meis/Pbx activation of the p15 and E-cadherin promoters.
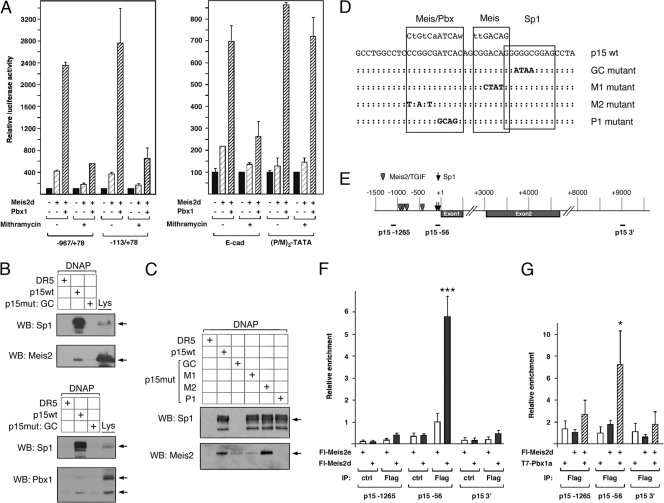
To test whether activation by Meis2d and Pbx1a required the Sp1 sites in the p15 promoter, we analyzed the activity of the −967/+78 and −113/+78 reporters in HepG2 cells treated with mithramycin, which is used as a relatively broad specificity inhibitor of transcription factor binding to GC-rich elements, such as Sp1 sites. As shown in Fig. 3 A, the activation of both p15 reporters by Meis2d and Pbx1a was significantly reduced in cells treated with mithramycin. As with the p15 reporter constructs, we observed a reduction in the activation of the E-cadherin reporter by Meis2d/Pbx1a in the presence of mithramycin (Fig. 3A). In contrast, the consensus Pbx/Meis site reporter, which is not GC rich and is bound directly by Pbx1a and Meis2d, was unaffected by mithramycin treatment. To test whether the Sp1 sites in the p15 promoter could contribute to recruitment of Meis2, we used a series of biotinylated DNA duplexes to isolate the endogenous Sp1 and Meis2 proteins from COS1 cells. We focused on the region of the p15 promoter which contains the 5′-most Sp1 site and the sequences with partial matches to Meis and Pbx sites (Fig. 3D, oligonucleotide sequences). As shown in Fig. 3B, both Sp1 and Meis2 bound specifically to the p15 wild-type oligonucleotide but not to the unrelated DR5 retinoic acid response element. Similar results were seen with the binding of endogenous Pbx1 to this element. Importantly, mutation of the Sp1 site (GC mutant) abolished binding of Sp1 as well as Meis2 and Pbx1 to the p15 sequence (Fig. 3B). To further delineate the requirements for recruitment of Meis2 to this site, we tested three other mutant forms of this element, in which the closest matches to Pbx and Meis consensus binding sites had been mutated. Sp1 clearly bound to all of the p15 elements tested except for the GC mutant (Fig. 3C). In contrast, we observed greatly reduced binding of Meis2 when either the Sp1 site or the Sp1-proximal Meis-like site was mutated (Fig. 3C, GC and M1 mutants). Mutation of a second Meis-like site had no effect on Meis2 binding, whereas mutating the closest match to a Pbx consensus abolished recruitment of Meis2 (p15mut P1). Together, these data suggest that endogenously expressed Meis2 and Pbx1 can form a complex on DNA at a suboptimal site, dependent on the adjacent GC-rich sequence.
Fig. 3.
GC boxes contribute to Meis/Pbx activation of the p15 promoter. (A) HepG2 cells were transfected with the indicated p15 (−967/+78 and −113/+78), E-cadherin, or (P/M)2-TATA luciferase reporters with Meis2d or Meis2d and Pbx1a expression constructs as indicated. Twenty-four hours after transfection, mithramycin was added and cells were assayed 24 h later. Results are shown normalized to a Renilla transfection control as mean + SD from duplicate transfections. (B and C) COS1 cell lysates were used to test binding of endogenous Sp1 and Meis2 or Pbx1 to biotinylated double-stranded oligonucleotides. Lysates were incubated with a control sequence (a DR5 retinoic acid response element), the wild-type sequence from the p15 promoter (p15wt), or a mutant form which disrupts the GC box (p15mut: GC). Bound proteins were analyzed for the presence of Sp1 and Meis2 (B, upper panel) or Sp1 and Pbx1 (C, lower panel). Expression in the lysate is also shown (Lys). Specific bands are indicated with arrows. In panel C, a series of mutant oligonucleotides was used to test for binding of Sp1 and Meis2. (D) The sequences of the oligonucleotides used in panels B and C are shown, together with the similarity to consensus Sp1, Meis, or Meis/Pbx sites. Colons indicate unchanged sequence in the mutants; altered bases are shown. (E) The p15 locus is shown schematically, with positions (in base pairs) relative to the start site shown above. The positions of the amplicons tested by ChIP are shown below. (F) Cells were transfected with the indicated Flag-Meis2 constructs and subjected to ChIP with anti-Flag-agarose or with anti-GluGlu-agarose as a control. (G) ChIP was performed as for panel F, with Flag-Meis2d and T7-Pbx1a as indicated. Relative enrichment is shown (in arbitrary units, as the mean + SD from triplicates. ***, P < 0.001; *, P < 0.05, as determined by Student's t test.
We next wanted to test whether Meis2 and Pbx1 were present at the endogenous p15 locus. We expressed Flag-epitope-tagged versions of each protein and performed ChIP with anti-Flag agarose. We analyzed three regions of the p15 locus (Fig. 3E), including the proximal promoter region, centered around 56 bp upstream of the transcriptional start site, a second region further upstream (at bp −1265 from the start) and a region 9 kb 3′ of the start. As shown in Fig. 3F, Meis2d was found at the p15 proximal promoter but was not significantly enriched either at sequences further upstream or at a far downstream region. In contrast, Meis2e, which lacks the homeodomain was not enriched at the p15 promoter. We next performed similar experiments with low levels of transfected Flag-Meis2d, with or without coexpression of T7-tagged Pbx1a. As shown in Fig. 3G, coexpression of Pbx1a increased recruitment of Meis2d to the p15 promoter, consistent with the formation of a cocomplex on DNA. Together, these data suggest that Meis2d and Pbx1a can be recruited to the proximal p15 promoter, dependent on the presence of an Sp1 binding site.
Pbx1a interacts with Klf4.
Since the Sp1 sites in the p15 promoter appeared to be critical for the transcriptional response to Meis2d and Pbx1a and for their binding to DNA, we wondered whether Meis2d or Pbx1a might interact with Sp1. COS1 cells were transfected with expression constructs encoding T7 epitope-tagged Sp1 or Sp3, together with Flag-tagged Meis2d, Meis2e, or Pbx1a. Protein complexes were collected on anti-Flag agarose and analyzed for the presence of coprecipitating T7-Sp1 or T7-Sp3. Meis2d coprecipitated with both Sp1 and Sp3, whereas the Meis2e splice variant did not (Fig. 4 A). Additionally, we observed some interaction with Pbx1a. To identify the region of Meis2 responsible for interaction with Sp1, we tested a deletion mutant (containing amino acids 2 to 345) lacking the Meis2d transcriptional activation domain, and Meis2d(R332M) that is unable to bind DNA. Amino acids 2 to 345 of Meis2 were still able to interact with Sp1 in this assay, whereas the R332M point mutation completely abolished interaction with Sp1, raising the possibility that Meis2d-Sp1 interaction is dependent on DNA binding (data not shown). To test whether the interaction of Meis2 with Sp1 was dependent on DNA binding, we performed coimmunoprecipitation experiments and incubated lysates with ethidium bromide prior to precipitation with anti-Flag agarose. The inclusion of ethidium bromide effectively reduced the Meis2d-Sp1 interaction (Fig. 4B). Thus, the apparent interaction of Meis2d with Sp1 is mediated by binding to DNA.
Fig. 4.
Meis2d and Pbx1 interact with Klf4. (A) COS1 cells were transfected with T7-tagged Sp1 or Sp3 and Flag- Meis2 and Pbx1a expression constructs as indicated. Cells were lysed, and protein complexes collected on anti-Flag-agarose and analyzed by Western blotting for the presence of coprecipitating T7-tagged Sp1 or Sp3. For panels A to E, specific coprecipitating bands are indicated with arrows and expression in the lysates is shown below. (B) COS1 cells were transfected with T7-Sp1 and Flag-Meis2d as indicated. Lysates were subjected to Flag immunoprecipitation in the presence or absence of ethidium bromide (as indicated), and coprecipitating Sp1 was identified by T7 Western blotting. (C and D) COS1 cells were transfected with T7 epitope-tagged Klf4 or Klf3 and Flag-tagged Meis2d and Pbx1a expression constructs as indicated. Cells were lysed, and protein complexes were collected on anti-Flag-agarose and analyzed by Western blotting for the presence of coprecipitating T7-tagged proteins. In panel C, ethidium bromide was added prior to immunoprecipitation, as indicated. (E) COS1 cells were transfected with T7 epitope-tagged Klf4 and Flag epitope-tagged mutations of Pbx1 as indicated. Lysates were then analyzed as for panel A. (F) HepG2 cells were transfected with the indicated p15 promoter reporter and Meis2d and Pbx1a as indicated, together with increasing amounts (as shown in ng) of a Klf4 expression plasmid. Luciferase activity assayed after 40 hours is shown normalized to a Renilla transfection control, as mean + SD from triplicates. (G) Binding to the p15 locus of Flag-tagged Klf4 and Sp1 as indicated was analyzed by ChIP, as in Figure 3. (H) HepG2 cells were transfected with siRNA duplexes targeting Klf4 or a control pool of nontargeting siRNAs. mRNA was isolated after 60 hours and analyzed by qRT-PCR for expression of the indicated genes. Relative expression is shown as the average + SD of triplicates. Significance levels, as determined by Student's t test are as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Since the GC-rich Sp1 sites in the p15 promoter contribute to the Meis2d/Pbx1a response and to binding to DNA, we tested whether other members of the SP/KLF family could interact with Meis2d or Pbx1a. We first tested for effects of Klf4 on p15 expression and interaction with Meis2d and Pbx1a. As shown in Fig. 4C, we observed interaction of both Meis2d and Pbx1a with Klf4, although the interaction was more readily detectable with Pbx1a. Importantly, this interaction was not affected by inclusion of ethidium bromide. The interaction was not affected by the introduction of a DNA binding point mutation (N286S) into Pbx1a, further suggesting that it was not entirely dependent on DNA binding (Fig. 4D). Additionally, comparison of the interaction between Meis2d and Sp1 or Klf4 revealed that the Klf4 interaction was much more readily detectable (data not shown). We also tested interaction with Klf3 and Klf5 but did not detect a clear interaction with either Meis2d or Pbx1a (Fig. 4D and data not shown). As shown in Fig. 4E, deletion of the Pbx1a homeodomain (construct containing amino acids 2 to 233) had little effect on interaction with Klf4, whereas removal of the amino-terminal 90 amino acids (Δ90) clearly reduced the interaction. Taken together, these results suggest that Pbx1a and Klf4 interact and that this interaction does not depend on DNA binding.
To test whether Klf4 could activate the p15 promoter, we expressed increasing amounts of Klf4 with or without Meis2d and Pbx1a. As shown in Fig. 4F, we observed a small but significant increase in reporter activity with increasing Klf4 in the presence of Meis2d and Pbx1a, whereas there was no effect of Klf4 on basal activity. In contrast, coexpression of Sp1 or Sp3 did not increase activity (data not shown). To test whether Klf4 could bind to the endogenous p15 locus, we performed ChIP experiments using Flag-tagged Klf4 or Sp1 and analyzed recruitment to the p15 locus as before. We observed significant enrichment of both Sp1 and Klf4 to the p15 proximal promoter but not to other regions of the p15 locus, consistent with the location of the known Sp1 sites (Fig. 4G). To test whether endogenously expressed Klf4 could play a role in the activation of p15, we transiently knocked down Klf4 in HepG2 cells and tested expression of Klf4, p15 and E-cadherin by qRT-PCR. Klf4 expression was reduced by about 80%, and we observed a significant reduction in expression of the endogenous p15 and E-cadherin genes on Klf4 knockdown (Fig. 4H). Taken together, these results suggest that Klf4 and a Meis2/Pbx1 complex are recruited to the p15 proximal promoter to activate p15 expression.
Pbx1 and Klf4 are recruited to the p15 and E-cadherin promoters.
The E-cadherin gene has recently been shown to bind endogenous Klf4 in breast cancer cell lines (67). We therefore tested whether modulation of Klf4 and Meis2 or Pbx1 levels affected p15 and E-cadherin expression in MCF7 cells. As shown in Fig. 5 A, siRNA-mediated knockdown of Pbx1 and Klf4 in these cells resulted in a clear reduction in both mRNA and protein levels, although Meis2 knockdown in MCF7 had little effect (data not shown). As with our previous analysis in HepG2 cells, we also observed a significant reduction in p15 and E-cadherin mRNA levels as with knockdown of either Pbx1 or Klf4 (Fig. 5B). We next tested whether Pbx1 and Klf4 could be detected at the p15 and E-cadherin promoters in MCF7 cells, but this time we analyzed binding of endogenous proteins. Chromatin was precipitated with antibodies against Pbx1 or Klf4, or with an IgG as a control, and the presence of the E-cadherin promoter and the −56 region of the p15 promoter analyzed by PCR. As shown in Fig. 5C, Klf4 clearly bound to both the p15 and E-cadherin promoters, whereas it was not detected at the p15 3′ region. We also observed binding of Pbx1 to both promoters, although this was somewhat harder to detect. Together, these data suggest that the endogenous Klf4-Pbx1 complex can bind to both p15 and E-cadherin promoters and that the cooperative regulation of gene expression by Klf4-Pbx1 complexes can occur in multiple cell types.
Fig. 5.
Pbx1 and Klf4 regulate p15 and E-cadherin in MCF7 cells. (A and B) MCF7 cells were transfected with the indicated siRNA duplexes, or a nontargeting control, and assayed by qRT-PCR and Western blotting for Klf4 and Pbx1 (A) and by qRT-PCR for p15 and E-cadherin (B). mRNA was isolated after 60 h and analyzed by qRT-PCR for expression of the indicated genes. Relative expression is shown as the average + SD of triplicates. Significance levels, as determined by Student's t test, were as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) ChIP was performed on chromatin from MCF7 cells using control IgG or antibodies to Pbx1 or Klf4. ChIP fractions were analyzed by PCR using primer sets to the E-cadherin promoter, the p15 proximal promoter (p15 −56), or as a negative control, a 3′ region of p15 (p15 3′). PCR on input chromatin is shown in the right lane.
Identification of novel potential targets for Klf4-Meis/Pbx activation.
We next created two mutant forms of the p15 −967/+78 luciferase reporter: the first contained mutations in both of the GC boxes, and the second contained mutations in the M1 Meis site and the Pbx-like site (Fig. 3D). Mutation of the GC boxes resulted in an increase in basal activity and a decrease in the response to coexpressed Meis2d and Pbx1a (Fig. 6 A and C). In contrast, the mutations which disrupt the Meis- and Pbx-like sites (M/P mutant) had relatively little effect on activity either with or without coexpressed Meis2d and Pbx1a (Fig. 6A). To examine whether the E-cadherin promoter responded similarly, we made mutations in the GC box and an adjacent Pbx-like site. As shown in Fig. 6B and C, the E-cadherin reporter with a mutated GC box had lower basal activity but a similar level of induction with Meis2 and Pbx1. However, unlike the p15 promoter, mutation of the Pbx site resulted in both lower activity and reduced induction by Meis2 and Pbx1 (Fig. 6B and C). These data suggest that there is a variable requirement for Meis and Pbx sites in the p15 and E-cadherin promoters and that the sequence requirements for cooperative activation by Meis/Pbx and Klf4 are quite variable, at least in terms of the Meis and Pbx binding sequences.
Fig. 6.
Analysis of sequence requirements for activation by Meis2 and Pbx1. (A) HepG2 cells were transfected with the indicated p15 luciferase reporters: p15 wild type (−967/+78) or reporters that had mutated GC boxes or Meis- and Pbx-like sites. Klf4, Pbx1a, and Meis2d were expressed as indicated. Activity is shown in arbitrary units as mean + SD from duplicate transfections. (B) HepG2 cells were transfected as in panel A with the indicated E-cadherin luciferase reporters: E-cadherin wild type (E-cad) or reporters that had a mutated GC box or Pbx-like site. (C) Fold induction from panels A and B is shown for p15 and E-cadherin. (D) The outline of the site search protocol for Klf4-Meis/Pbx elements is shown schematically, with the number of hits shown to the right. (E and F) HepG2 cells were transfected with luciferase reporters for the Pax3 (E) or Etv1 (F) promoters. Meis2d, Pbx1a, and Klf4 were added as indicated. Activity is shown in arbitrary units as mean + SD from triplicate transfections. Maximum fold activation by Meis/Pbx for each set of four conditions is shown above. Significance (by Student's t test; P < 0.05) is shown: #, comparison of Meis/Pbx to control; *, comparison of equal amounts of Meis/Pbx with or without Klf4. Amounts of expression plasmids transfected in E and F: triangle symbols for Meis2d and Pbx1a represent 25, 50, and 100 ng per well, and for Klf4 plus and double-plus symbols represent 2.5 and 5 ng per well, respectively.
To further test the possibility that Meis/Pbx and Klf4 cooperatively activate gene expression, we performed an in silico search for genes with conserved binding sites for all three proteins (http://www.sitesearch.mshri.on.ca/Genome/index.html) (59). We were unable to search for degenerate Meis and Pbx sites since this would provide no selectivity, so we used the minimal 5-base consensus for each, combined with the sequence RRGGYSY based on the Klf4 consensus. As outlined in Fig. 6D, we restricted the search to the proximal 2 kb upstream of the predicted transcriptional start site and searched for a Klf4 site with both Meis and Pbx sites present within 40 bp. This search yielded 484 hits with the combination of all three sites in the upstream region of the same gene in both mouse and human. To reduce this number, we first took only those hits in which the sites were in the same order and same relative orientation to each other in both species and ranked them by how similar the spacing between the sites was in mouse and human. By this ranking, the top 17 had exactly the same spacing between the three sites in both species, and the top 35 had a total difference of only 6 bp or less. Of these top 35, published reporters were available for two of them. Pax3 has been shown to be regulated by Meis and Pbx, and a luciferase construct containing the potential Klf4-Meis/Pbx element was shown to respond to Meis2 and Pbx1 expression (11). Although a luciferase reporter for Etv1 has been characterized, it had not been tested with either Meis/Pbx or Klf4 (8). We obtained the reporters for Pax3 and Etv1 and tested them for cooperative activation by Klf4 and Meis/Pbx. The Pax3-luc reporter was clearly activated by coexpression of Meis2d and Pbx1a, whereas expression did not increase with Klf4 expression alone (Fig. 6E). Coexpression of all three proteins together resulted in a significant increase in activity over that seen with Meis2d and Pbx1a. With Etv1 we observed similar results, although Meis2d and Pbx1a had less effect in the absence of Klf4 and the cooperative activation by all three proteins was more pronounced (Fig. 6F). Taken together, these data suggest that Meis2, Pbx1, and Klf4 can activate gene expression cooperatively and that Meis and Pbx proteins may modulate the transcriptional activity of Klf4.
DISCUSSION
We demonstrate that Meis2d and Pbx1a functionally cooperate with Klf4 to activate gene expression. The recruitment of Meis2d and Pbx1a allows transcriptional activation by the complex, dependent on the Meis2d activation domain.
This work uncovers a mechanism by which Meis and Pbx proteins can be preferentially recruited to a subset of target genes via interaction with Klf4, resulting in the fine-tuning of the Klf4 response. Klf4 is a multifunctional protein with both transcriptional activation and repression domains. It therefore is not surprising that interactions with other transcription factors may contribute to whether Klf4 activates or represses transcription. We show that the Meis2d activation domain is required for activation of the p15 gene in the context of a Klf4-Meis/Pbx complex, suggesting that one way in which Klf4 can switch from repression to activation is via the recruitment of a Meis2-containing homeodomain protein complex. For other reporters tested, Klf4 reduced activity when expressed alone but allowed greater activation by Meis/Pbx coexpression. Thus, Klf4 bound to a promoter element could maintain the gene in a relatively inactive state but allow greater activation on expression of Meis and Pbx cofactors. We have identified a number of candidate genes for this mode of regulation and have begun to validate two of them functionally in cultured cells. However, further work will be required to determine which are genuine in vivo targets of a Klf4-Meis/Pbx complex. Given the complex roles of Klf4 in tumorigenesis, it is possible that differential interactions with additional factors including Meis and Pbx proteins help determine whether Klf4 acts as a tumor suppressor or an oncogene. In addition to a potential role in tumorigenesis based on the regulation of p15 expression, a recent report supports our notion that E-cadherin is targeted for activation by Klf4 in breast cancer cells and suggests that Klf4 modulates epithelial-to-mesenchymal transition (EMT) by this mechanism. Additionally, we show that Klf4 cooperates with Pbx to activate E-cadherin in MCF7 cells and that both Klf4 and Pbx1 bind the E-cadherin promoter. We suggest that Pbx, and possibly Meis, proteins may play a role in modulating the ability of Klf4 to control EMT via the regulation of E-cadherin. In addition to being expressed in breast cancer cell lines, analysis of the Oncomine database (www.oncomine.org) suggests that Meis2, Pbx1, and Klf4 are expressed in prostate cancers and Pbx1 expression has been shown to be regulated by androgen signaling (67).
Both Pbx1a and Meis2d can interact with Klf4, although the interaction of Pbx1a appears to be more readily detectable. Thus, it is possible that Meis2 may interact with Klf4 primarily via Pbx1, since these two homeodomain proteins dimerize. Based on the conservation among Meis and Pbx family members, we expect that other members of these families will function similarly. Pbx1 has been shown to bind DNA together with MyoD, although in this case it appears that Pbx binds first and recruits MyoD (2), and Pbx proteins are cofactors for more than just other homeodomain proteins (31). We have not been able to show that other Sp/KLF family members are able to recruit Meis2 and Pbx1, although this is a relatively large family of proteins and we have not tested all of them. However, outside the conserved zinc finger domain, members of the Sp/KLF family are quite divergent, consistent with the possibility that Meis/Pbx interaction may be specific for Klf4. We therefore suggest a model in which the Klf component of this complex is limited to Klf4 but that Klf4 can likely recruit multiple members of the Meis/Pbx family. Our data suggest that the interaction of Pbx1 or Meis2 with Klf4 is not absolutely dependent on DNA binding, but that cooperative activation of gene expression requires DNA binding by all three proteins. For p15, it appears that the GC box provides the major component of complex binding, since mutations in the GC box reduce Meis2 and Pbx1 binding. However, DNA binding requirements at other promoters may be different. For example, the E-cadherin has a Klf4 site and a degenerate Pbx site but does not have an obvious Meis2 binding site. Based on the very loose match to the Meis and Pbx sites found in the p15 promoter, it is possible that binding of Meis2d to the E-cadherin promoter may be to a very degenerate site. It is also possible that at some promoters recruitment of Meis or Pbx proteins may not require DNA binding at all. This is not unprecedented, as composite Hox-Pbx binding elements can be activated by a Meis2d mutant that is unable to bind to DNA (21).
Since the sequence requirements for binding of Meis2 and Pbx1 to a composite Klf4 element appear to be relatively relaxed, the identification of potential response elements is difficult. Our bioinformatics approach relied on the presence of a perfect match to the minimal 5-bp consensus for both Meis2 and Pbx, since any further relaxation of the sequence provides too little selectivity. Most of the genes identified are not known to be Meis/Pbx responsive, with the exception of Pax3 (11). However, we show that a Pax3 reporter responded cooperatively to Meis2d/Pbx1a and Klf4, suggesting that the search for composite elements was valid. Among the other top hits from this search, we were able to show cooperative activation by Meis2d/Pbx1a and Klf4 of a reporter containing the Etv1 promoter, which was not previously known to respond to any of the three proteins. This clearly supports the idea that there are additional targets of activation by Klf4-Meis/Pbx complexes.
In summary, we propose a model in which Klf4 recruits a complex of Meis and Pbx proteins to DNA, resulting in Meis2 transcriptional activation domain-dependent activation of a subset of Klf4 target genes. This may represent a mechanism by which expression of Meis/Pbx proteins can alter the Klf4 transcriptional program.
ACKNOWLEDGMENTS
We thank J. Massagué, G. K. Owens, M. Mouradian, P. Kamps, M. Cleary, A. Cano, L. Shemshedini, and J. Steinke for providing plasmids.
This work was supported by NIH grant HD39926 to D.W.
Footnotes
Published ahead of print on 11 July 2011.
REFERENCES
- 1. Bartholin L., et al. 2006. TGIF inhibits retinoid signaling. Mol. Cell. Biol. 26:990–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berkes C. A., et al. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14:465–477 [DOI] [PubMed] [Google Scholar]
- 3. Berthelsen J., Zappavigna V., Ferretti E., Mavilio F., Blasi F. 1998. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 17:1434–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berthelsen J., Zappavigna V., Mavilio F., Blasi F. 1998. Prep1, a novel functional partner of Pbx proteins. EMBO J. 17:1423–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bertolino E., Reimund B., Wildt-Perinic D., Clerc R. 1995. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J. Biol. Chem. 270:31178–31188 [DOI] [PubMed] [Google Scholar]
- 6. Briggs M. R., Kadonaga J. T., Bell S. P., Tjian R. 1986. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science 234:47–52 [DOI] [PubMed] [Google Scholar]
- 7. Burglin T. R. 1997. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25:4173–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cai C., et al. 2007. ETV1 is a novel androgen receptor-regulated gene that mediates prostate cancer cell invasion. Mol. Endocrinol. 21:1835–1846 [DOI] [PubMed] [Google Scholar]
- 9. Chang C. P., Brocchieri L., Shen W. F., Largman C., Cleary M. L. 1996. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol. Cell. Biol. 16:1734–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang C. P., et al. 1995. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 9:663–674 [DOI] [PubMed] [Google Scholar]
- 11. Chang C. P., et al. 2008. Pbx1 functions in distinct regulatory networks to pattern the great arteries and cardiac outflow tract. Development 135:3577–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Di Rosa P., et al. 2007. The homeodomain transcription factor Prep1 (pKnox1) is required for hematopoietic stem and progenitor cell activity. Dev. Biol. 311:324–334 [DOI] [PubMed] [Google Scholar]
- 13. Dynan W. S., Tjian R. 1983. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell 35:79–87 [DOI] [PubMed] [Google Scholar]
- 14. Evans P. M., Liu C. 2008. Roles of Krupel-like factor 4 in normal homeostasis, cancer and stem cells. Acta Biochim. Biophys. Sin. (Shanghai) 40:554–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fognani C., et al. 2002. Characterization of PREP2, a paralog of PREP1, which defines a novel sub-family of the MEINOX TALE homeodomain transcription factors. Nucleic Acids Res. 30:2043–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Foster K. W., et al. 2005. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene 24:1491–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gehring W. J., Affolter M., Burglin T. 1994. Homeodomain proteins. Annu. Rev. Biochem. 63:487–526 [DOI] [PubMed] [Google Scholar]
- 18. Gehring W. J., et al. 1994. Homeodomain-DNA recognition. Cell 78:211–223 [DOI] [PubMed] [Google Scholar]
- 19. Geiman D. E., Ton-That H., Johnson J. M., Yang V. W. 2000. Transactivation and growth suppression by the gut-enriched Kruppel-like factor (Kruppel-like factor 4) are dependent on acidic amino acid residues and protein-protein interaction. Nucleic Acids Res. 28:1106–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang H., et al. 2005. MEIS C termini harbor transcriptional activation domains that respond to cell signaling. J. Biol. Chem. 280:10119–10127 [DOI] [PubMed] [Google Scholar]
- 21. Hyman-Walsh C., Bjerke G. A., Wotton D. 2010. An autoinhibitory effect of the homothorax domain of Meis2. FEBS J. 277:2584–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jacobs Y., Schnabel C. A., Cleary M. L. 1999. Trimeric association of hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol. Cell. Biol. 19:5134–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaczynski J., Cook T., Urrutia R. 2003. Sp1- and Kruppel-like transcription factors. Genome Biol. 4:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. 1987. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 51:1079–1090 [DOI] [PubMed] [Google Scholar]
- 25. Kamps M. P., Look A. T., Baltimore D. 1991. The human t(1;19) translocation in pre-B ALL produces multiple nuclear E2A-Pbx1 fusion proteins with differing transforming potentials. Genes Dev. 5:358–368 [DOI] [PubMed] [Google Scholar]
- 26. Kamps M. P., Murre C., Sun X. H., Baltimore D. 1990. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell 60:547–555 [DOI] [PubMed] [Google Scholar]
- 27. Kingsley C., Winoto A. 1992. Cloning of GT box-binding proteins: a novel Sp1 multigene family regulating T-cell receptor gene expression. Mol. Cell. Biol. 12:4251–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Knoepfler P. S., et al. 1999. A conserved motif N-terminal to the DNA-binding domains of myogenic bHLH transcription factors mediates cooperative DNA binding with pbx- Meis1/Prep1. Nucleic Acids Res. 27:3752–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Knoepfler P. S., Calvo K. R., Chen H., Antonarakis S. E., Kamps M. P. 1997. Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc. Natl. Acad. Sci. U. S. A. 94:14553–14558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Knoepfler P. S., Kamps M. P. 1995. The pentapeptide motif of Hox proteins is required for cooperative DNA binding with Pbx1, physically contacts Pbx1, and enhances DNA binding by Pbx1. Mol. Cell. Biol. 15:5811–5819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laurent A., Bihan R., Omilli F., Deschamps S., Pellerin I. 2008. PBX proteins: much more than Hox cofactors. Int. J. Dev. Biol. 52:9–20 [DOI] [PubMed] [Google Scholar]
- 32. Li J.-M., Nichols M. A., Chandrasekharan S., Xiong Y., Wang X.-F. 1995. Transforming growth factor β activates the promoter of cyclin-dependent kinase inhibitor p15Ink4B through an Sp1 consensus site. J. Biol. Chem. 270:26750–26753 [DOI] [PubMed] [Google Scholar]
- 33. Li J. M., et al. 1998. Sp1, but not Sp3, functions to mediate promoter activation by TGF-beta through canonical Sp1 binding sites. Nucleic Acids Res. 26:2449–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu Y., MacDonald R. J., Swift G. H. 2001. DNA binding and transcriptional activation by a PDX1.PBX1b.MEIS2b trimer and cooperation with a pancreas-specific basic helix-loop-helix complex. J. Biol. Chem. 276:17985–17993 [DOI] [PubMed] [Google Scholar]
- 35. Lu Q., Kamps M. P. 1996. Selective repression of transcriptional activators by Pbx1 does not require the homeodomain. Proc. Natl. Acad. Sci. U. S. A. 93:470–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maherali N., et al. 2007. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1:55–70 [DOI] [PubMed] [Google Scholar]
- 37. Mann R. S., Affolter M. 1998. Hox proteins meet more partners. Curr. Opin. Genet. Dev. 8:423–429 [DOI] [PubMed] [Google Scholar]
- 38. McConnell B. B., Ghaleb A. M., Nandan M. O., Yang V. W. 2007. The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays 29:549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McGinnis W., Garber R. L., Wirz J., Kuroiwa A., Gehring W. J. 1984. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell 37:403–408 [DOI] [PubMed] [Google Scholar]
- 40. McGinnis W., Levine M. S., Hafen E., Kuroiwa A., Gehring W. J. 1984. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature 308:428–433 [DOI] [PubMed] [Google Scholar]
- 41. Melhuish T. A., Gallo C. M., Wotton D. 2001. TGIF2 interacts with histone deacetylase 1 and represses transcription. J. Biol. Chem. 276:32109–32114 [DOI] [PubMed] [Google Scholar]
- 42. Moens C. B., Selleri L. 2006. Hox cofactors in vertebrate development. Dev. Biol. 291:193–206 [DOI] [PubMed] [Google Scholar]
- 43. Moskow J. J., Bullrich F., Huebner K., Daar I. O., Buchberg A. M. 1995. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH- 2 mice. Mol. Cell. Biol. 15:5434–5443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mukherjee K., Burglin T. R. 2007. Comprehensive analysis of animal TALE homeobox genes: new conserved motifs and cases of accelerated evolution. J. Mol. Evol. 65:137–153 [DOI] [PubMed] [Google Scholar]
- 45. Nakahara Y., et al. 2010. Genetic and epigenetic inactivation of Kruppel-like factor 4 in medulloblastoma. Neoplasia 12:20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakamura T., Jenkins N. A., Copeland N. G. 1996. Identification of a new family of Pbx-related homeobox genes. Oncogene 13:2235–2242 [PubMed] [Google Scholar]
- 47. Okita K., Ichisaka T., Yamanaka S. 2007. Generation of germline-competent induced pluripotent stem cells. Nature 448:313–317 [DOI] [PubMed] [Google Scholar]
- 48. Oulad-Abdelghani M., et al. 1997. Meis2, a novel mouse Pbx-related homeobox gene induced by retinoic acid during differentiation of P19 embryonal carcinoma cells. Dev. Dyn. 210:173–183 [DOI] [PubMed] [Google Scholar]
- 49. Passner J. M., Ryoo H. D., Shen L., Mann R. S., Aggarwal A. K. 1999. Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex [see comments]. Nature 397:714–719 [DOI] [PubMed] [Google Scholar]
- 50. Penkov D., et al. 2005. Involvement of Prep1 in the alphabeta T-cell receptor T-lymphocytic potential of hematopoietic precursors. Mol. Cell. Biol. 25:10768–10781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Penkov D., Palazzolo M., Mondino A., Blasi F. 2008. Cytosolic sequestration of Prep1 influences early stages of T cell development. PLoS One 3:e2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Piper D. E., Batchelor A. H., Chang C.-P., Cleary M. L., Wolberger C. 1999. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96:587–597 [DOI] [PubMed] [Google Scholar]
- 53. Rieckhof G. E., Casares F., Ryoo H. D., Abu-Shaar M., Mann R. S. 1997. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell 91:171–183 [DOI] [PubMed] [Google Scholar]
- 54. Ryoo H. D., Marty T., Casares F., Affolter M., Mann R. S. 1999. Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle complex. Development 126:5137–5148 [DOI] [PubMed] [Google Scholar]
- 55. Schnabel C. A., Jacobs Y., Cleary M. L. 2000. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene 19:608–616 [DOI] [PubMed] [Google Scholar]
- 56. Seoane J., et al. 2001. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat. Cell Biol. 3:400–408 [DOI] [PubMed] [Google Scholar]
- 57. Shanmugam K., Green N. C., Rambaldi I., Saragovi H. U., Featherstone M. S. 1999. PBX and MEIS as non-DNA-binding partners in trimeric complexes with HOX proteins. Mol. Cell. Biol. 19:7577–7588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shields J. M., Yang V. W. 1998. Identification of the DNA sequence that interacts with the gut-enriched Kruppel-like factor. Nucleic Acids Res. 26:796–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Silvestri C., et al. 2008. Genome-wide identification of Smad/Foxh1 targets reveals a role for Foxh1 in retinoic acid regulation and forebrain development. Dev. Cell 14:411–423 [DOI] [PubMed] [Google Scholar]
- 60. Suske G., Bruford E., Philipsen S. 2005. Mammalian SP/KLF transcription factors: bring in the family. Genomics 85:551–556 [DOI] [PubMed] [Google Scholar]
- 61. Thorsteinsdottir U., Kroon E., Jerome L., Blasi F., Sauvageau G. 2001. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol. Cell. Biol. 21:224–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wernig M., et al. 2007. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448:318–324 [DOI] [PubMed] [Google Scholar]
- 63. Wong P., Iwasaki M., Somervaille T. C., So C. W., Cleary M. L. 2007. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 21:2762–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wotton D., Lo R. S., Swaby L. A., Massague J. 1999. Multiple modes of repression by the smad transcriptional corepressor TGIF. J. Biol. Chem. 274:37105–37110 [DOI] [PubMed] [Google Scholar]
- 65. Yang Y., et al. 2000. Tale homeodomain proteins Meis2 and TGIF differentially regulate transcription. J. Biol. Chem. 275:20734–20741 [DOI] [PubMed] [Google Scholar]
- 66. Yet S. F., et al. 1998. Human EZF, a Kruppel-like zinc finger protein, is expressed in vascular endothelial cells and contains transcriptional activation and repression domains. J. Biol. Chem. 273:1026–1031 [DOI] [PubMed] [Google Scholar]
- 67. Yori J. L., Johnson E., Zhou G., Jain M. K., Keri R. A. 2010. Kruppel-like factor 4 inhibits epithelial-to-mesenchymal transition through regulation of E-cadherin gene expression. J. Biol. Chem. 285:16854–16863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhao W., et al. 2004. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene 23:395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]