Abstract
Us3 is a serine-threonine protein kinase encoded by herpes simplex virus 1 (HSV-1). We have identified UL47, a major virion protein, as a novel physiological substrate of Us3. In vitro kinase assays and systematic analysis of mutations at putative Us3 phosphorylation sites near the nuclear localization signal of UL47 showed that serine at residue 77 (Ser-77) was required for Us3 phosphorylation of UL47. Replacement of UL47 Ser-77 by alanine produced aberrant accumulation of UL47 at the nuclear rim and impaired the nuclear localization of UL47 in a significant fraction of infected cells. The same defect in UL47 localization was produced by an amino acid substitution in Us3 that inactivated its protein kinase activity. In contrast, a phosphomimetic mutation at UL47 Ser-77 restored wild-type nuclear localization. The UL47 S77A mutation also reduced viral replication in the mouse cornea and the development of herpes stromal keratitis in mice. In addition, UL47 formed a stable complex with Us3 in infected cells, and nuclear localization of Us3 was significantly impaired in the absence of UL47. These results suggested that Us3 phosphorylation of UL47 Ser-77 promoted the nuclear localization of UL47 in cell cultures and played a critical role in viral replication and pathogenesis in vivo. Furthermore, UL47 appeared to be required for efficient nuclear localization of Us3 in infected cells. Therefore, Us3 protein kinase and its substrate UL47 demonstrated a unique regulatory feature in that they reciprocally regulated their subcellular localization in infected cells.
INTRODUCTION
Us3 is a serine/threonine protein kinase encoded by herpes simplex virus 1 (HSV-1) with an amino acid sequence that is conserved in the subfamily Alphaherpesvirinae (11, 40, 56). RnX(S/T)YY is the consensus target sequence of an HSV-1 Us3 homologue encoded by pseudorabies virus (PRV), where n is ≥2; X can be Arg, Ala, Val, Pro, or Ser; and Y can be any amino acid except an acidic residue (33, 34, 55). This sequence also appears to be the target sequence of HSV-1 Us3, based on reports that the phosphorylation sites of HSV-1 Us3 identified to date match the consensus target sequence (24–26, 47, 62). The phosphorylation target site specificity of HSV-1 Us3 has also been reported to be similar to that of protein kinase A (PKA), a cellular cyclic AMP-dependent protein kinase (2), and Akt (4). Some antibodies to the phosphorylated substrate sequences of PKA can also react with Us3 phosphorylation sites (2, 24, 25). The Us3 protein and its catalytic activity have been suggested to play a critical role in HSV-1 replication and pathogenicity, based on studies showing that recombinant Us3-null mutant viruses and recombinant viruses encoding catalytically inactive Us3 have impaired growth properties in cell cultures and reduced virulence, pathogenicity, and replication in mouse models (41, 44, 60, 62, 63).
In general, the activity of a protein kinase is tightly regulated; e.g., by autophosphorylation, transphosphorylation by other protein kinases, or interaction with other proteins (61). The activated protein kinase phosphorylates a substrate(s), and as a result, the phosphorylated substrate(s) expresses its functions. Although it has been reported recently that the Us3 kinase activity in infected cells is, in part, regulated by autophosphorylation (25, 63), it remains largely unknown how HSV-1 Us3 kinase activity is regulated in infected cells. In contrast, numerous studies have elucidated the potential downstream effects of HSV-1 Us3, including blocking apoptosis (35, 48, 49, 50), promoting nuclear egress of progeny nucleocapsids through the nuclear membrane (NM) (47, 60, 62, 72), redistributing and phosphorylating NM-associated viral nuclear egress factors UL31 and UL34 and cellular factors lamin A/C and emerin (26, 32, 45–47, 58, 59), mediating the phosphorylation of histone deacetylases (HDACs) and promoting gene expression by blocking histone deacetylation (53, 54), controlling infected-cell morphology (25, 49, 70), downregulating the expression of viral envelope glycoprotein B (gB) on the cell surface by promoting gB endocytosis (18, 24), and stimulating mRNA translation by mimicking Akt and activating mTORC1 (4). These data suggest that Us3 is a multifunctional protein that plays various roles in viral replication by phosphorylating a number of viral and cellular substrates. In agreement with this hypothesis, it has been reported that HSV-1 Us3 is a promiscuous protein kinase and may phosphorylate more substrates than originally predicted (46). Therefore, there may be Us3 substrates other than those reported to date, and their identification and characterization are required in order to determine the functions of Us3 and understand their mechanisms.
UL47, another protein encoded by HSV-1, is a major structural protein in the virion tegument (37). HSV-1 UL47 is posttranslationally modified by phosphorylation in infected cells (42), and its amino acid sequence is conserved in the subfamily Alphaherpesvirinae (12, 16). Deletion of the UL47 gene from HSV-1, PRV, Marek's disease virus 1, avian infectious laryngotracheitis virus (ILTV), or bovine herpesvirus 1 (BHV-1) usually impairs viral replication in cell cultures (9, 16, 30, 36, 74), and UL47-null mutants of PRV, ILTV, and BHV-1 have attenuated virulence in mouse models and their natural hosts (16, 29, 36). From these observations, UL47 proteins have been considered to be positive regulators of alphaherpesvirus replication and pathogenicity. Although the precise function(s) of UL47 in viral replication and virulence remains largely unknown at present, the mechanisms by which UL47 proteins act in infected cells have been gradually elucidated. Early studies revealed that HSV-1 UL47 is involved in modulating the activity of tegument protein VP16, which upon HSV-1 infection is released into the cytoplasm, is transported into the nucleus, and transactivates the expression of α genes, the class of HSV-1 genes expressed first in infected cells (74). Later studies have shown that the UL47 proteins of HSV-1 and BHV-1 are nucleocytoplasmic shuttling proteins (6, 7, 69), functionally similar to human immunodeficiency virus type 1 (HIV-1) Rev, HSV-1 ICP27, and influenza virus NS2 (43, 51, 52). These viral proteins also have RNA binding activity (8), suggesting a role in RNA biogenesis in infected cells similar to that of other virally encoded RNA binding proteins.
In the present study, we identify the UL47 protein as a novel physiological substrate of Us3 in HSV-1-infected cells. Studies investigating the effect(s) of Us3 phosphorylation of UL47 suggest that this phosphorylation promotes the nuclear localization of UL47 in cell cultures and plays a critical role in viral replication and pathogenesis in vivo. In addition, we show that UL47 formed a stable complex with Us3 in infected cells, and we present data suggesting that UL47 is required for the efficient nuclear localization of Us3 in infected cells. Thus, it appears that Us3 and UL47 reciprocally regulate their subcellular localization in infected cells.
MATERIALS AND METHODS
Cells and viruses.
Vero and rabbit skin cells have been described previously, as has the wild-type HSV-1 strain HSV-1(F) (68). The recombinant virus YK501, encoding Us3 fused to the fluorescent protein Venus with an A206K mutation (VenusA206K), and the recombinant virus YK603, encoding UL47 fused to monomeric red fluorescent protein 1 (mRFP1), have been described previously (19, 67).
Plasmids.
To generate a fusion protein consisting of maltose binding protein (MBP) and part of UL47, plasmid pMAL-UL47-P1 was constructed by amplifying the domains encoding UL47 codons 15 to 120 from pBC1007 by PCR (28) and cloning the DNA fragments into pMAL-c (New England BioLabs) in frame with the MBP. To generate a fusion protein consisting of MBP fused to UL47 carrying an S77A or S88A mutation, pMAL-UL47-P1-S77A or pMAL-UL47-P1-S88A, respectively, was constructed as described previously (24). pRSETB-mRFP1-Kan was generated by amplifying a fragment encoding the I-SceI site and the kanamycin resistance gene from pEPkan-S (25), cloning it into the PstI site of pRSETB-mRFP1 (67), and using it as a template for the two-step Red-mediated mutagenesis procedure described below. The pBS-mCherry-UL37 transfer plasmid, used for generating recombinant HSV-1 expressing UL37 fused to mCherry (mCherry-UL37), was constructed as described previously (67).
Mutagenesis of viral genomes in Escherichia coli and generation of recombinant HSV-1.
To generate YK524, encoding UL47 fused to the fluorescent protein mRFP1 (mRFP1-UL47) (Fig. 1), the two-step Red-mediated mutagenesis procedure was carried out as described previously (25), except that the primers listed in Table 1 and the pRSETB-mRFP1-Kan template were used. Recombinant virus YK532, encoding mRFP1-UL47 with an alanine substituted for serine at residue Ser-77 (mRFP1-UL47S77A), and YK534, encoding mRFP1-UL47 with an aspartic acid substituted for serine at residue Ser-77 (mRFP1-UL47S77D) (Fig. 1), were constructed by the two-step Red-mediated mutagenesis procedure described previously (25), except that the primers listed in Table 1 and E. coli containing a YK524 genome were used. Recombinant virus YK533, in which the mRFP1-UL47S77A mutation in YK532 was repaired (mRFP1-UL47S77A-repair) (Fig. 1), was generated with the primers listed in Table 1 as described previously (25). Recombinant virus YK527, a Us3 kinase-dead mutant virus in which UL47 was tagged with mRFP1 and Us3 Lys-220 was replaced with methionine (mRFP1-UL47/Us3K220M), and YK528, in which Us3K220M in YK527 was repaired (Fig. 1), were generated as described above using the primers listed in Table 1. Recombinant virus YK521, in which Us3 was tagged with Venus fluorescent protein carrying the mutation A206K (67) and the UL47 gene was disrupted by the insertion, just downstream of the UL47 start codon, of a foreign gene cassette carrying a stop codon (TGA), an I-SceI site, a kanamycin resistance gene, and 60 bp of a sequence upstream of the second codon of the UL47 gene (VenusA206K-Us3/ΔUL47) (Fig. 1), was generated by the Red-mediated mutagenesis procedure as described previously (25), except that the primers listed in Table 1 and E. coli containing the YK501 genome were used (19). Recombinant viruses into which the same foreign gene cassette was inserted just downstream of the UL41 or UL49 start codon were not able to express UL41 or UL49 protein in infected cells (M. Tanaka and Y. Kawaguchi, unpublished observations).
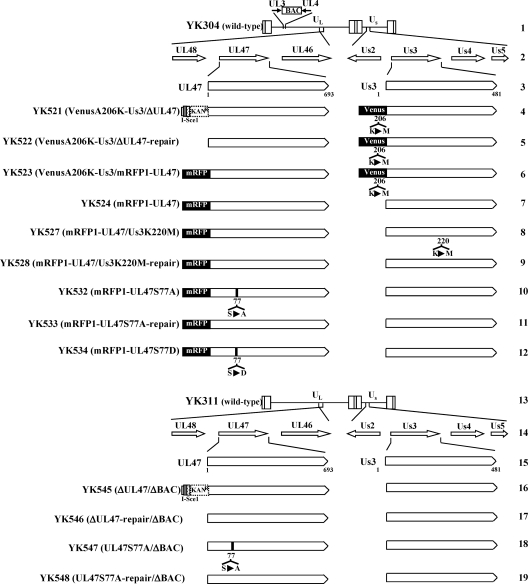
Fig. 1.
Schematic diagram of the genomes of wild-type YK304 and YK311 viruses and the relevant domains of the recombinant viruses used in this study. Line 1, linear representation of the YK304 genome carrying a bacmid (BAC) in the intergenic region between UL3 and UL4. Lines 2 and 14, domains encoding the UL46 to UL48 and Us2 to Us5 open reading frames. Lines 3 and 15, domains of the UL47 and Us3 genes. Lines 4 to 12, schematic diagrams of the recombinant viruses carrying the BAC that were used in this study. Line 13, linear representation of the genome of YK311, in which the BAC sequence was excised from the YK304 genome. Lines 16 to 19, schematic diagrams of the recombinant viruses from which the BAC sequence was excised that were used in this study.
Table 1.
Primer sequences used for the construction of recombinant viruses
| Mutation | Sequence |
|---|---|
| mRFP-UL47 | 5′-TTCTTTTTTGGGGGGTAGCGGACATCCGATAACCCGCGTCTATCGCCACCATGGCCTCCTCCGAGGACGT-3′ |
| 5′-CGGGGGCGGGTGGATGCGCGCCTCCTGCGCCCCGCGGGTTCGCGAGCCGACAAGGCGCCGGTGGAGTGGC-3′ | |
| UL47-S77A | 5′-TCGACGGGAAGGCCCGCGAGCCCGACGACGGCGCGCGGCGGAAGCCCCGCCCACATCCCAAGGATGACGACGATAAGTAG-3′ |
| 5′-GCCGGGACGCTCGGCGATGGGATGTGGGCGGGGCTTCCGCCGCGCGCCGTCGTCGGGCTCCAACCAATTAACCAATTCTG-3′ | |
| UL47-S77A-repair | 5′-TCGACGGGAAGGCCCGCGAGCCCGACGACGGCGCGCGTCGGAAGCCCCGCCCACATCCCAAGGATGACGACGATAAGTAG-3′ |
| 5′-GCCGGGACGCTCGGCGATGGGATGTGGGCGGGGCTTCCGACGCGCGCCGTCGTCGGGCTCCAACCAATTAACCAATTCTG-3′ | |
| UL47-S77D | 5′-TCGACGGGAAGGCCCGCGAGCCCGACGACGGCGCGCGGACGAAGCCCCGCCCACATCCCAAGGATGACGACGATAAGTAG-3′ |
| 5′-GCCGGGACGCTCGGCGATGGGATGTGGGCGGGGCTTCGTCCGCGCGCCGTCGTCGGGCTCCAACCAATTAACCAATTCTG-3′ | |
| ΔUL47 | 5′-GGTCTTTTTCTTTTTTGGGGGGTAGCGGACATCCGATAACCCGCGTCTATCGCCACCATGTAAAGGATGACGACGATAAGTAGGG-3′ |
| 5′-ATGCGCGCCTCCTGCGCCCCGCGGGTTCGCGAGCCGACATGGTGGCGATAGACGCGGGTTCAACCAATTAACCAATTCTGATTAG-3′ | |
| Us3-K220 M | 5′-CAGCAGCCATCCAGATTACCCCCAACGGGTAATCGTGATGGCGGGGTGGTACACGAGCACAGGATGACGACGATAAGTAGGG-3′ |
| 5′-GCAGTCGCGCCTCGTGGCTCGTGCTCGTGTACCACCCCGCCATCACGATTACCCGTTGGGCAACCAATTAACCAATTCTGATTAG-3′ | |
| Us3-K220 M-repair | 5′-CAGCAGCCATCCAGATTACCCCCAACGGGTAATCGTGAAGGCGGGGTGGTACACGAGCACAGGATGACGACGATAAGTAGGG-3′ |
| 5′-GCAGTCGCGCCTCGTGGCTCGTGCTCGTGTACCACCCCGCCTTCACGATTACCCGTTGGGCAACCAATTAACCAATTCTGATTAG-3′ |
Recombinant virus YK522, in which the foreign gene cassette inserted into the UL47 locus of YK521 was excised (VenusA206K-Us3/ΔUL47-repair) (Fig. 1), was generated by the Red-mediated mutagenesis procedure as described above using the primers listed in Table 1. Recombinant virus YK523, encoding VenusA206K-Us3 and mRFP1-UL47 (VenusA206K-Us3/mRFP1-UL47) (Fig. 1), was generated by coinfection with YK501 (VenusA206K-Us3) (19) and YK603 (mRFP1-UL47) (67) as described previously (67). Recombinant viruses YK541, in which nontagged UL47 was disrupted by insertion of the foreign gene cassette described above just downstream of the UL47 start codon (ΔUL47), YK542, in which the foreign gene cassette inserted into the UL47 gene of YK541 was excised (ΔUL47-repair), YK543, in which the Ser-77 in nontagged UL47 was replaced with alanine (UL47S77A), and YK544, in which the S77A mutation in nontagged UL47 of YK543 was repaired (UL47S77A-repair), were constructed as described above using the primers listed in Table 1. Recombinant viruses YK545 (ΔUL47/ΔBAC), YK546 (ΔUL47-repair/ΔBAC), YK547 (UL47S77A/ΔBAC), and YK548 (UL47S77A-repair/ΔBAC) (Fig. 1), in which the bacmids were excised from YK541 (ΔUL47), YK542 (ΔUL47-repair), YK543 (UL47S77A), and YK544 (UL47S77A-repair), respectively, were constructed by coinfection of Vero cells with YK541, YK542, YK543, or YK544 and recombinant adenovirus AxCANCre, expressing Cre recombinase, as described previously (68). Recombinant virus YK615, encoding mCherry-UL37, was generated as described previously (67) except that the transfer plasmid pBS-mCherry-UL37 was used.
Production and purification of MBP fusion proteins in E. coli.
MBP fusion proteins MBP-UL47-P1, MBP-UL47-P1-S77A, MBP-UL47-P1-S88A, and MBP-LacZ were expressed in E. coli that had been transformed with pMAL-UL47-P1, pMAL-UL47-P1-S77A, pMAL-UL47-P1-S88A, or pMAL-c, respectively, and were purified as described previously (26).
In vitro kinase assays.
MBP fusion proteins were captured on amylose beads (New England BioLabs) and were used as substrates in in vitro kinase assays with purified glutathione S-transferase-tagged Us3 (GST-Us3) and GST-Us3K220M as described previously (26).
Antibodies.
Rabbit polyclonal antibodies to Us3, UL46, and UL48 have been described previously (25, 27, 44). A rabbit monoclonal antibody against the phospho-PKA substrate (clone 100G7) was purchased from Cell Signaling Technology. A rabbit polyclonal antibody against RFP and a mouse monoclonal antibody against RFP (3G5) coupled to agarose beads were purchased from MBL. A rabbit polyclonal antibody against DsRed was purchased from Clontech; this antibody recognizes mRFP1 and mCherry, since these fluorescent proteins were generated by modification of DsRed (3, 64).
Immunoblotting, immunoprecipitation, and live-cell imaging.
Immunoblotting, immunoprecipitation, and live-cell imaging were performed as described previously (28, 67). The proteins in immunoblot bands were quantitated using the Dolphin-Doc image capture system with Dolphin-1D software (Wealtec). For quantitation of the fluorescence intensity of VenusA206K-Us3 in live Vero cells infected with YK501 (VenusA206K-Us3), YK521 (VenusA206K-Us3/ΔUL47), or YK522 (VenusA206K-Us3/ΔUL47-repair), images of 10 cells were randomly selected for each viral infection; three circular areas of each infected cell were randomly selected; and the fluorescence intensity of each area was determined using the software supplied with the LSM5 confocal microscope (Zeiss). The sizes of all 30 areas examined were equal.
Phosphatase treatment.
MBP fusion proteins after in vitro kinase assays and immunoprecipitates were treated with λ protein phosphatase (λ-PPase) (New England BioLabs) as described previously (26).
Animal studies.
Female ICR mice were purchased from Charles River. For ocular infection, 5-week-old female mice were infected with each HSV-1 strain as described previously (63). The scoring scale for the severity of herpes stromal keratitis (HSK) has been described previously (63). Viral titers in tear films were determined as described previously (63). All animal studies were carried out with the approval of the Ethics Committee for Animal Experimentation of the University of Tokyo.
RESULTS
Us3-dependent phosphorylation of UL47 in infected cells.
We previously identified physiological substrates of Us3 by a combination of bioinformatics analysis, in vitro kinase assays using Us3 and specific substrates, and detection of phosphorylation in infected cells with an anti-phospho-PKA substrate antibody (100G7), which detects proteins containing a phosphorylated serine or threonine residue with an arginine at residues −2 and −3 (RRXS or RRXT) (24, 25). Based on the consensus sequence of the Us3 phosphorylation site, we identified four putative Us3 phosphorylation sites in UL47, at codons 10 to 14 (RRRAS), 73 to 77 (RRRAS), 85 to 88 (RRAS), and 682 to 685 (RRAT), all of which would be expected to be detected by the anti-phospho-PKA substrate antibody (100G7) if they were phosphorylated. In order to monitor UL47 in infected cells easily, we generated the recombinant virus YK524, in which UL47 was tagged with the fluorescent protein mRFP1, by the two-step Red-mediated mutagenesis procedure. The growth curve of YK524 (mRFP1-UL47) was similar to that of wild-type HSV-1(F) in Vero cells infected at a multiplicity of infection (MOI) of 5, but YK524 had a slightly lower (3.8-fold) progeny virus yield than wild-type HSV-1(F) at 24 h postinfection (Fig. 2A), suggesting that the tagging of UL47 with mRFP1 had a slight effect on viral growth. This was consistent with our previous observation with recombinant virus YK603, encoding mRFP1-UL47, which was constructed by the homologous recombination procedure (67). To investigate the effects of Us3 protein kinase activity on UL47 in infected cells, we also constructed YK527, a Us3 kinase-dead mutant virus that had a K220M mutation in Us3 (25) and expressed mRFP1-UL47 (mRFP1-UL47/Us3K220M), and YK528, the corresponding virus in which the Us3 K220M mutation in YK527 was repaired (mRFP1-UL47/Us3K220M-repair). The growth curve of YK527 was almost identical to those of YK524 and YK528 in Vero cells infected at an MOI of 5 (Fig. 2B), indicating that the kinase activity had no effect on the growth of the virus in which UL47 was tagged with mRFP1.
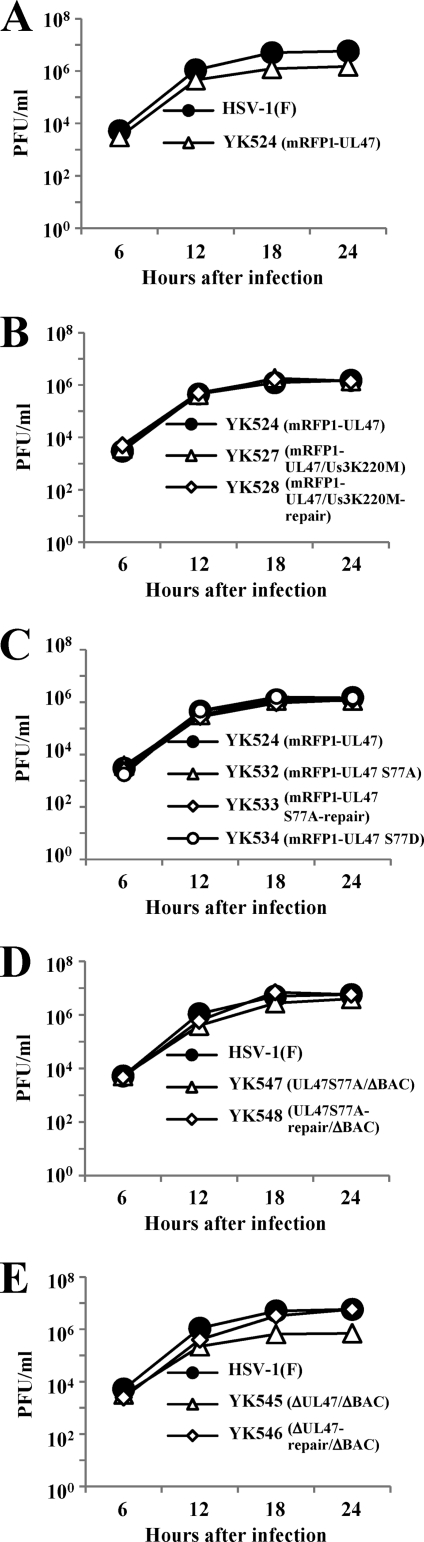
Fig. 2.
Growth curves of recombinant viruses. Vero cells were infected at an MOI of 5 with wild-type HSV-1(F) (A, D, and E), YK524 (mRFP1-UL47) (A, B, and C), YK527 (mRFP1-UL47/Us3K220M) (B), YK528 (mRFP1-UL47/Us3K220M-repair) (B), YK532 (mRFP1-UL47S77A) (C), YK533 (mRFP1-UL47S77A-repair) (C), YK534 (mRFP1-UL47S77D) (C), YK547 (UL47S77A/ΔBAC) (D), YK548 (UL47S77A-repair/ΔBAC) (D), YK545 (ΔUL47/ΔBAC) (E), or YK546 (ΔUL47-repair/ΔBAC) (E). Total virus from the cell culture supernatants and the infected cells was harvested at the indicated times and was assayed on Vero cells.
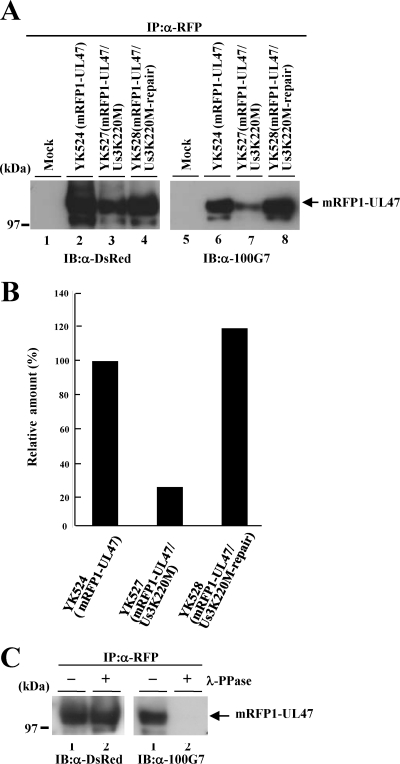
To examine whether Us3 mediated the phosphorylation of UL47 in infected cells, Vero cells were infected with YK524 (mRFP1-UL47), YK527 (mRFP1-UL47/Us3K220M), or YK528 (mRFP1-UL47/Us3K220M-repair) at an MOI of 3, harvested at 24 h postinfection, solubilized, immunoprecipitated with the anti-RFP antibody coupled to agarose beads, and analyzed by immunoblotting with the anti-DsRed antibody and the anti-phospho-PKA substrate antibody (100G7). As shown in Fig. 3A, the anti-phospho-PKA substrate antibody reacted with mRFP1-UL47 purified from YK524-infected Vero cells by immunoprecipitation (Fig. 3A, lane 6). To confirm that the detection of mRFP1-UL47 by the anti-phospho-PKA substrate antibody was due to phosphorylation of UL47, mRFP1-UL47 from YK524-infected cells was purified by immunoprecipitation with an anti-RFP antibody and was treated with λ-PPase. As shown in Fig. 3C, the reactivity of mRFP1-UL47 with the anti-phospho-PKA substrate antibody was eliminated by phosphatase treatment, indicating that the anti-phospho-PKA substrate antibody had reacted with phosphorylated mRFP1-UL47. Furthermore, the level of phosphorylation of mRFP1-UL47 detected by the anti-phospho-PKA substrate antibody in YK527 (mRFP1-UL47/Us3K220M)-infected cells was significantly lower (Fig. 3A, lane 7, and B) than that of mRFP1-UL47 in YK524 (mRFP1-UL47)- or YK528 (mRFP1-UL47/Us3K220M-repair)-infected cells (Fig. 3A, lanes 6 and 8, respectively, and B). These results indicate that UL47 is phosphorylated in infected cells at the epitope(s) of the anti-phospho-PKA substrate antibody (100G7) and that Us3 protein kinase mediates most of this phosphorylation. We noted that phosphorylation of mRFP1-UL47 was detected at low levels but consistently by the anti-phospho-PKA substrate antibody in the absence of Us3 kinase activity in infected cells (Fig. 3B), indicating that a protein kinase(s) other than Us3 mediated this phosphorylation in infected cells. In agreement with this hypothesis, the phosphorylation target site specificity of HSV-1 Us3 has been reported to be similar to those of cellular protein kinases PKA and Akt (2, 4).
Fig. 3.
(A) Immunoblots (IB) of electrophoretically separated mRFP1-UL47 immunoprecipitates (IP) from Vero cells that were either mock infected (lanes 1 and 5) or infected with YK524 (mRFP1-UL47) (lanes 2 and 6), YK527 (mRFP1-UL47/Us3K220M) (lanes 3 and 7), or YK528 (mRFP1-UL47/Us3K220M-repair) (lanes 4 and 8) at an MOI of 3, harvested at 24 h postinfection, immunoprecipitated with an anti-RFP (α-RFP) antibody, and analyzed by immunoblotting with an anti-DsRed antibody (left) or an anti-phospho-PKA substrate antibody (100G7) (right). The position of a molecular mass marker is given on the left. (B) Quantitation of phosphorylated mRFP1-UL47, determined from the anti-phospho-PKA substrate antibody (100G7) immunoblot in panel A, relative to the amount of total mRFP1-UL47, determined from the anti-DsRed antibody immunoblot in panel A. The data were normalized to the relative amount in YK524-infected cells. (C) RFP1 immunoprecipitates of YK524-infected cells prepared as for panel A, lane 2, were either mock treated (lanes 1) or treated with λ-PPase (lanes 2) and were then analyzed by immunoblotting with an anti-DsRed antibody (left) or an anti-phospho-PKA substrate antibody (right). A molecular mass marker is indicated on the left.
Effect of Us3 protein kinase activity on the localization of UL47 in infected cells.
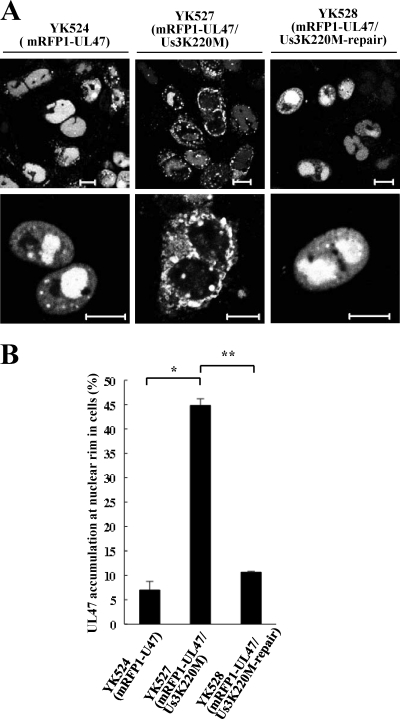
Three of the four putative Us3 phosphorylation sites (Ser-77, Ser-88, and Thr-685) in UL47 described above are located close to the protein's nuclear localization signal (NLS) (codons 50 to 68) and nuclear export signal (NES) (codons 658 to 667) (8, 71). This led us to examine whether Us3 is involved in regulation of the subcellular localization of UL47 in infected cells. For this purpose, Vero cells were infected with either YK524 (mRFP1-UL47), YK527 (mRFP1-UL47/Us3K220M), or YK528 (mRFP1-UL47/Us3K220M-repair) at an MOI of 1 for 18 h and were then examined by confocal microscopy. In agreement with previous reports (6), mRFP1-UL47 was localized predominantly in the nuclei of cells infected with YK524 (Fig. 4A). In contrast, the distribution of mRFP1-UL47 was significantly different in approximately 45% of 200 cells infected with the Us3 kinase-dead mutant YK527, with aberrant accumulation of mRFP1-UL47 at the nuclear rim and impaired nuclear localization (Fig. 4A). The percentage of YK527-infected cells with aberrant accumulation of mRFP1-UL47 at the nuclear rim was approximately 6-fold higher than the corresponding percentage of YK524-infected cells (Fig. 4B). However, nuclear localization of mRFP1-UL47 was restored in cells infected with YK528, in which the Us3K220M mutation was repaired (Fig. 4A and B). These results indicate that Us3 protein kinase activity is required for proper nuclear localization of UL47 in a fraction (almost half) of infected cells.
Fig. 4.
(A) Digital confocal microscope images showing the localization of mRFP1-UL47 proteins in infected Vero cells. Vero cells were infected with either YK524 (mRFP1-UL47), YK527 (mRFP1-UL47/Us3K220M), or YK528 (mRFP1-UL47/Us3K220M-repair) at an MOI of 1 and were examined by confocal microscopy at 18 h postinfection. Bars, 10 μm. (B) Quantitation of infected cells exhibiting aberrant accumulation of mRFP1-UL47 at the nuclear rim. Infected Vero cells were examined by confocal microscopy as described for panel A, and the percentages of cells showing aberrant accumulation of mRFP1-UL47 at the nuclear rim were determined for 200-cell samples, each infected with one of the recombinant viruses. Data are means and standard errors from three independent experiments. Asterisks indicate significant differences (∗, P = 3.5 × 10−5; ∗∗, P = 7.8 × 10−6) by a two-tailed Student t test.
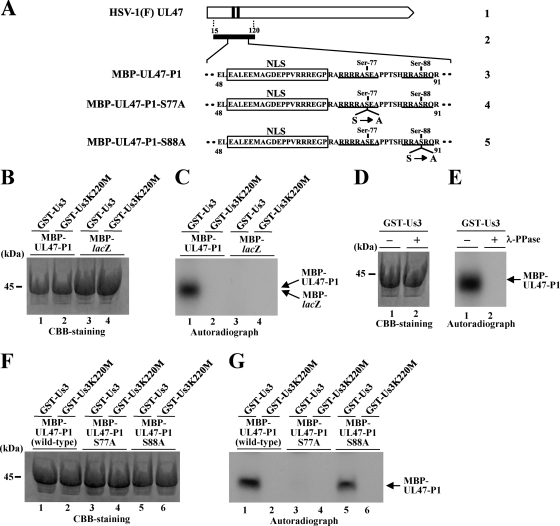
Us3 phosphorylation of the amino-terminal domain of UL47 in vitro.
It has been reported that the nuclear localization of viral and cellular proteins is controlled by phosphorylation in or close to the NLS of the protein (21, 22). Therefore, in view of our results described above, we hypothesized that Us3 phosphorylation at a site(s) close to the UL47 NLS may regulate the nuclear localization of UL47. To test this hypothesis, we examined whether Us3 can phosphorylate the amino-terminal domain of UL47 containing its NLS in vitro. For this study, we generated and purified a chimeric protein consisting of MBP fused to a peptide encoded by UL47 codons 15 to 120 (MBP-UL47-P1) (Fig. 5A) and tested it as a substrate in in vitro kinase assays with purified wild-type GST-Us3 and the kinase-negative mutant GST-Us3K220M. As shown in Fig. 5C, MBP-UL47-P1 was labeled with [γ-32P]ATP in kinase assays using GST-Us3 (Fig. 5C, lane 1), while MBP-LacZ was not (Fig. 5C, lane 3). When the kinase-negative mutant GST-Us3K220M was used, neither MBP fusion protein was labeled (Fig. 5C, lanes 2 and 4). To confirm that the labeling of MBP-UL47-P1 by GST-Us3 was due to phosphorylation, the labeled MBP-UL47-P1 was treated with λ-PPase. As shown in Fig. 5E, MBP-UL47-P1 labeling by GST-Us3 was eliminated by phosphatase treatment, indicating that MBP-47-P1 was labeled by phosphorylation. The presence of each MBP fusion protein was verified by staining with Coomassie brilliant blue (CBB) (Fig. 5B and D). These results indicate that Us3 specifically and directly phosphorylates the UL47 amino-terminal peptide encoded by codons 15 to 120 in vitro.
Fig. 5.
(A) Schematic diagram of UL47. Line 1, structure of the UL47 open reading frame. The shaded areas represent arginine-rich motifs in the amino-terminal domain of the protein. Line 2, amino-terminal domain of UL47 containing residues 15 to 120, which was used in these studies to generate the MBP-UL47-P1 fusion protein. Lines 3 to 5, amino acid sequences of UL47 residues 48 to 91 in MBP-UL47-P1 and its S77A and S88A mutants. Two sites with the consensus sequence for phosphorylation by Us3 are underlined, and the UL47 NLS is boxed. (B) Purified MBP-UL47-P1 (lanes 1 and 2) and MBP-LacZ (lanes 3 and 4) were incubated in a kinase buffer containing [γ-32P]ATP and either purified GST-Us3 (lanes 1 and 3) or GST-Us3K220M (lanes 2 and 4) for 30 min, separated on a denaturing gel, and stained with CBB. A molecular mass marker is indicated on the left. (C) Autoradiograph of the gel shown in panel B. (D) Purified MBP-UL47-P1 was incubated in a kinase buffer containing [γ-32P]ATP and purified GST-Us3 for 30 min and was then either mock treated (lane 1) or treated with λ-PPase (lane 2), separated on a denaturing gel, and stained with CBB. A molecular mass marker is indicated on the left. (E) Autoradiograph of the gel shown in panel D. (F) Purified MBP-UL47-P1 (lanes 1 and 2), MBP-UL47-P1 S77A (lanes 3 and 4), and MBP-UL47-P1 S88A (lanes 5 and 6) were incubated in a kinase buffer containing [γ-32P]ATP and either purified GST-Us3 (lanes 1, 3, and 5) or GST-Us3K220M (lanes 2, 4, and 6) for 30 min, separated on a denaturing gel, and stained with CBB. A molecular mass marker is indicated on the left. (G) Autoradiograph of the gel shown in panel F.
Identification of the amino acids in the amino-terminal domain of UL47 required for Us3 phosphorylation in vitro.
As described above, the UL47 amino-terminal peptide encoded by codons 15 to 120 contains two putative Us3 phosphorylation sites, at codons 74 to 77 (RRAS) and 85 to 88 (RRAS), that may be epitopes recognized by the anti-phospho-PKA substrate antibody (100G7). Therefore, we substituted alanine for the serine in each MBP-UL47-P1 phosphorylation site and tested these mutants as substrates in in vitro Us3 kinase assays. As shown in Fig. 5G, MBP-UL47-P1 carrying an S77A mutation was not phosphorylated by Us3, whereas MBP-UL47-P1 carrying an S88A mutation was efficiently phosphorylated by Us3. These results indicate that Ser-77 is required for Us3 phosphorylation of the amino-terminal domain of UL47 in vitro.
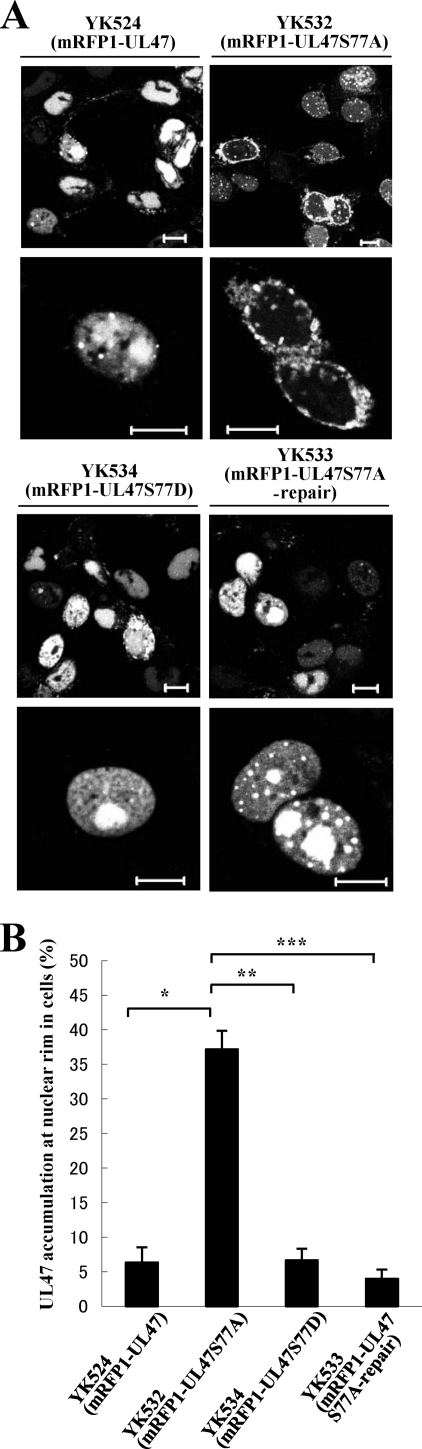
Effect of amino acid substitution of UL47 Ser-77 on the localization of UL47 in infected cells.
To investigate the linkage between Us3 phosphorylation in the amino-terminal domain of UL47 in vitro and the regulation of UL47 nuclear localization in infected cells, we constructed recombinant virus YK532 (mRFP1-UL47S77A), encoding mRFP1-UL47 in which UL47 Ser-77 was replaced with alanine (Fig. 1). We also generated recombinant virus YK533 (mRFP1-UL47S77A- repair), in which the UL47 S77A mutation in YK532 was repaired (Fig. 1). We then examined the localization of mRFP1-UL47 in Vero cells infected with either YK524 (mRFP1-UL47), YK532, or YK533 at an MOI of 1 for 18 h. As was found with the Us3 kinase-dead virus YK527 (mRFP1-UL47/Us3K220M) (Fig. 4), in approximately 37% of 200 cells infected with YK532, the intracellular distribution of mRFP1-UL47S77A was different from that of mRFP1-UL47 in cells infected with YK524, with mRFP1-UL47S77A showing aberrant accumulation at the nuclear rim and impaired nuclear localization (Fig. 6A). The percentage of YK532-infected cells with aberrant accumulation of mRFP1-UL47S77A at the nuclear rim was approximately 6-fold higher than the percentage of YK524-infected cells with aberrant accumulation of mRFP1-UL47 (Fig. 6B). However, nuclear localization of mRFP1-UL47 was restored in cells infected with YK533, in which the S77A mutation in UL47 was repaired. These results indicate that UL47 Ser-77 is required for proper nuclear localization of UL47 in a significant fraction of infected cells and that the effect of the UL47 S77A mutation in impairing proper UL47 nuclear localization is similar to that observed in cells infected with a Us3 kinase-inactive virus.
Fig. 6.
Digital confocal microscope images showing the localization of mRFP1-UL47 proteins in infected Vero cells. Vero cells were infected with either YK524 (mRFP1-UL47), YK532 (mRFP1-UL47S77A), YK533 (mRFP1-UL47S77A-repair), or YK534 (mRFP1-UL47S77D) at an MOI of 1, and the cells were examined by confocal microscopy at 18 h postinfection. Bars, 10 μm. (B) Quantitation of infected cells exhibiting aberrant accumulation of mRFP1-UL47 and its mutants at the nuclear rim. Infected Vero cells were examined by confocal microscopy as described for panel A, and the percentages of cells showing aberrant accumulation of mRFP1-UL47 at the nuclear rim were determined for 200-cell samples, each infected with one of the recombinant viruses. Data are means and standard errors from three independent experiments. Asterisks indicate significant differences (∗, P = 1.8 × 10−4; ∗∗, P = 3.7 × 10−4; ∗∗∗, P = 4.4 × 10−5) by a two-tailed Student t test.
To further investigate the hypothesis that phosphorylation of UL47 at Ser-77, which is close to the UL47 NLS, is critical for proper nuclear localization of UL47, we generated recombinant virus YK534, encoding mRFP1-UL47 in which Ser-77 was replaced with aspartic acid (mRFP1-UL47S77D), and examined the localization of mRFP1-UL47S77D in Vero cells infected with YK534 at an MOI of 1 for 18 h. It is known that an acidic amino acid, such as glutamic or aspartic acid, mimics the negative charge produced by phosphorylation (18, 24, 47, 73). As shown in Fig. 6A and B, mRFP1-UL47S77D was localized predominantly in the nuclei of YK534-infected cells, and its localization was indistinguishable from that of mRFP1-UL47 in YK524-infected cells. These results indicate that a negatively charged amino acid at UL47 residue 77, due to either phosphorylation of Ser-77 or an S77D substitution, is required for proper nuclear localization of UL47 in infected cells, further supporting the hypothesis that phosphorylation of UL47 at Ser-77 is critical for proper nuclear localization of UL47 in infected cells.
Effect of the UL47 S77A or UL47-null mutation on viral replication and pathogenesis in mice.
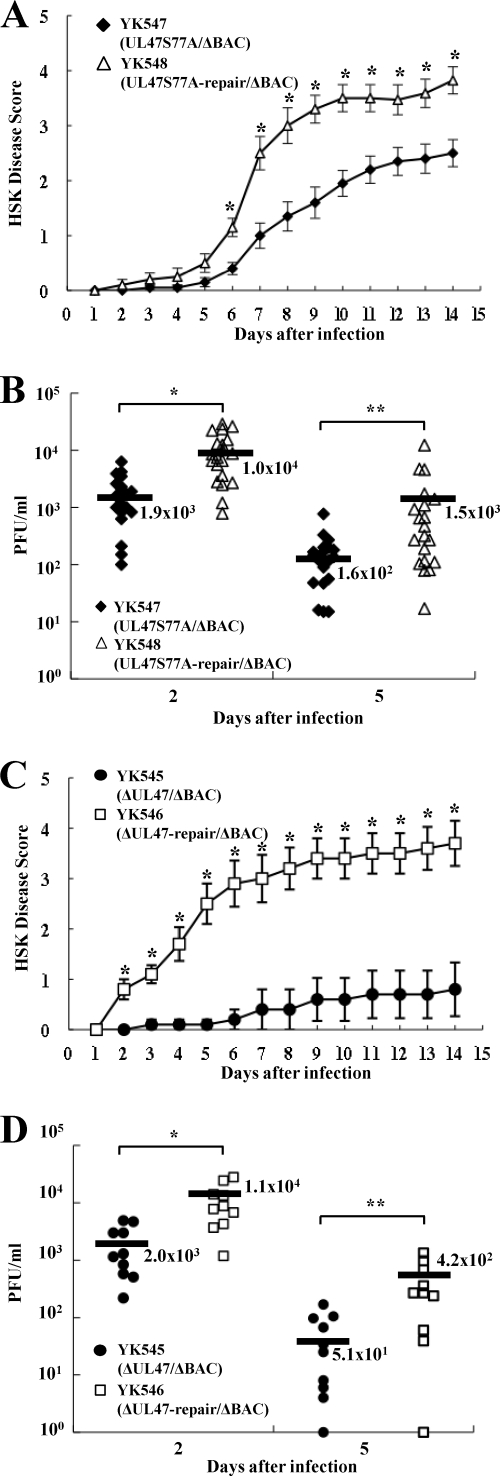
The growth curve of YK532 (mRFP1-UL47S77A) was almost identical to those of YK524 (mRFP1-UL47), YK533 (mRFP1-UL47S77A-repair), and YK534 (mRFP1-UL47S77D) in Vero cells infected at an MOI of 5 (Fig. 2C), indicating that phosphorylation of UL47 at Ser-77 does not have a measurable effect on viral growth in Vero cells. To investigate the clinical relevance of the phosphorylation of UL47 Ser-77 in HSV-1 infection, we examined the effect of the UL47 S77A mutation in a mouse herpes stromal keratitis (HSK) model for HSV-1 infection. In addition, we examined the effect of a UL47-null mutation in the mouse model, since HSV-1 UL47 has not been reported to play a role in viral replication and pathogenesis in vivo. Although the mRFP1 tag on UL47 had a small effect on viral growth in cell cultures, as described above, our earlier mouse experiments (19, 63) indicated that the mRFP1 tag might have a more significant effect in the mouse model. Therefore, we generated recombinant virus YK547, in which Ser-77 in nontagged UL47 was replaced with alanine and the bacmid was excised from the viral genome (UL47S77A/ΔBAC), YK548, in which the UL47 S77A mutation in YK547 was repaired (UL47S77A-repair/ΔBAC), YK545, in which the UL47 gene was inactivated by insertion of a foreign gene cassette and the bacmid was excised from the viral genome (ΔUL47/ΔBAC), and YK546, in which the inactivated UL47 gene in YK545 was repaired (ΔUL47-repair/ΔBAC) (Fig. 1). The growth curve of YK547 in Vero cells infected at an MOI of 5 was almost identical to that of YK548 (Fig. 2D), in agreement with the results for the mRFP1-tagged viruses described above, including YK532 (mRFP1-UL47S77A), YK524 (mRFP1-UL47), and YK533 (mRFP1-UL47S77A-repair). In contrast, YK545 (ΔUL47/ΔBAC) had an 8.2-fold lower progeny yield than YK546 (UL47S77A-repair/ΔBAC) in Vero cells at 24 h after infection at an MOI of 5 (Fig. 2E). This result is in agreement with an earlier report that a UL47 deletion impaired viral growth in cell cultures (74).
When these recombinant viruses were tested in the mouse HSK model, mice infected with YK547 (UL47S77A/ΔBAC) or YK545 (ΔUL47/ΔBAC) exhibited significantly less severe HSK than mice infected with YK548 (UL47S77A-repair/ΔBAC) or YK546 (ΔUL47-repair/ΔBAC), respectively (Fig. 7 A and C). In addition, YK547 and YK545 replicated significantly less efficiently in the tear films of these infected mice than did YK548 and YK546, respectively (Fig. 7B and D). These results indicate that UL47 is required for efficient viral replication and development of HSK in mice and that one of the sites in UL47 responsible for its function(s) in mice is the Us3 phosphorylation site at UL47 Ser-77.
Fig. 7.
Effects of the UL47 S77A substitution and the UL47-null mutation on HSK and virus growth in the tear films in mice. Twenty (A and B) or 10 (C and D) 5-week-old female ICR mice were infected with 1 × 106 PFU YK547 (UL47S77A/ΔBAC) or YK548 (UL47S77A-repair/ΔBAC) (A and B) or with 1 × 106 PFU YK545 (ΔUL47/ΔBAC) or YK546 (ΔUL47-repair/ΔBAC) (C and D) per eye by corneal scarification. (A and C) Mice were scored for HSK every other day for 14 days. Each data point is the mean and standard error for the observations. Asterisks indicate P values of 6.2 × 10−4 (A) and 3.1 × 10−3 (C) by a two-tailed Student t test. (B and D) Viral titers in the tear films of infected mice at 2 and 5 days postinfection were determined by standard plaque assays. Each data point represents the titer in the tear film of one mouse. The average titer for each group is marked by a horizontal bar and is given below the bar. Asterisks indicate significant differences by a two-tailed Student t test. (B) ∗, P = 2.8 × 10−4; ∗∗, P = 2.5 × 10−2. (D) ∗, P = 4.7 × 10−3; ∗∗, P = 1.7 × 10−2.
Interaction of Us3 with UL47 in infected cells.
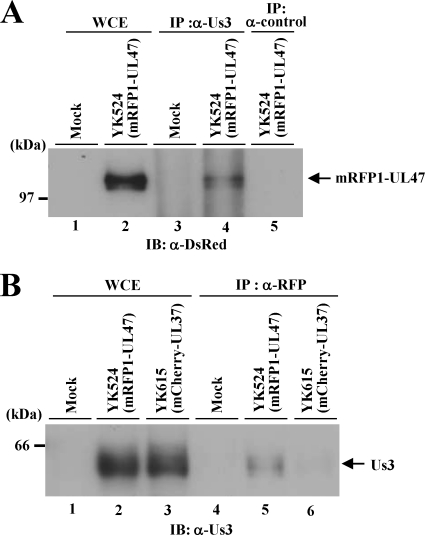
Herpesvirus protein kinases sometimes form a stable complex with their substrates (1). Therefore, to investigate whether this occurs with Us3 and UL47, Vero cells were either mock infected or infected with YK524 (mRFP1-UL47) at an MOI of 3 for 24 h. Then the whole-cell lysates of mock-infected or YK524-infected Vero cells were immunoprecipitated with anti-rabbit Us3 or a control rabbit antibody, and the immunoprecipitates were analyzed by immunoblotting with an anti-DsRed antibody. As shown in Fig. 8A, the anti-Us3 antibody coprecipitated mRFP1-UL47 from the lysates of YK524-infected cells (Fig. 8A, lane 4), whereas the control antibody did not (Fig. 8A, lane 5). In a reciprocal experiment, an anti-mRFP1 antibody coprecipitated Us3 from the lysates of YK524-infected cells (Fig. 8B, lane 5) but not from the lysates of cells infected with YK615, expressing UL37 tagged with mCherry (Fig. 8B, lane 6). These results indicate that Us3 forms a stable complex with UL47 in infected cells.
Fig. 8.
Interaction of Us3 with UL47. (A) Vero cells that were either mock infected (lanes 1 and 3) or infected with YK524 (mRFP1-UL47) (lanes 2, 4, and 5) at an MOI of 3 for 24 h were harvested, immunoprecipitated (IP) with anti-Us3 (α-Us3) (lanes 3 and 4) or with normal rabbit serum (lane 5), and analyzed by immunoblotting (IB) with an anti-DsRed antibody. One percent of the amount of Vero whole-cell extract (WCE) used in the reaction mixture for lane 3 was loaded in lane 1, and 1% of the WCE used for lanes 4 and 5 was loaded in lane 2. A molecular mass marker is indicated on the left. (B) Vero cells that were either mock infected (lanes 1 and 4) or infected with YK524 (lanes 2 and 5) or YK615 (mCherry-UL37) (lanes 3 and 6) at an MOI of 3 for 24 h were harvested, immunoprecipitated with anti-RFP, and analyzed by immunoblotting with an anti-Us3 antibody. One percent of the amount of Vero WCE used in the reaction mixtures for lanes 4, 5, and 6 was loaded in lanes 1, 2 and 3, respectively.
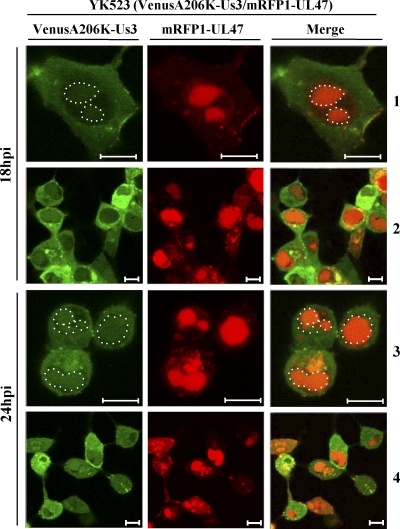
Next, to examine the subcellular localization of Us3 and UL47 in infected cells, we generated recombinant virus YK523, expressing mRFP1-UL47 and Us3 tagged with the fluorescent protein VenusA206K (VenusA206K-Us3/mRFP1-UL47), and examined the localization of mRFP1-UL47 and VenusA206K-Us3 in Vero cells infected with YK523 at an MOI of 1 at 18 and 24 h postinfection. As shown in Fig. 9, at 18 h postinfection, VenusA206K-Us3 was localized predominantly in the cytoplasm, as reported previously (25), but was also faintly detectable in the nucleus. At 24 h postinfection, nuclear localization of VenusA206K-Us3 was more evident than at 18 h postinfection. At both time points, nuclear VenusA206K-Us3 colocalized with mRFP1-UL47, and VenusA206K-Us3 in some cytoplasmic areas colocalized with mRFP1-UL47, although most of the Us3 and UL47 did not colocalize in the cytoplasm. These results support our conclusion that Us3 forms a stable complex with UL47 in infected cells.
Fig. 9.
Digital confocal microscope images showing the localization of the mRFP1-UL47 and VenusA206K-Us3 proteins in infected Vero cells. Vero cells were infected with YK523 (VenusA206K-Us3/mRFP1-UL47) at an MOI of 1, and cells were examined by confocal microscopy at 18 and 24 h postinfection (hpi). Nuclear Us3 in the VenusA206K-Us3 and merge columns is circled. Bars, 10 μm.
Effect of UL47 on the subcellular localization of Us3 in infected cells.
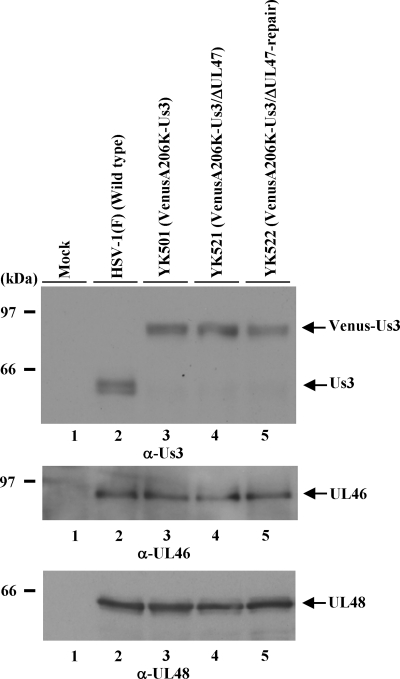
The results discussed above, showing that Us3 formed a stable complex with UL47 in infected cells and that Us3 colocalized with UL47, led us to examine whether UL47 regulated the localization of Us3 in infected cells. For this purpose, we constructed recombinant virus YK521, in which the UL47 gene was inactivated by insertion of a foreign gene cassette and Us3 was tagged with VenusA206K (VenusA206K-Us3/ΔUL47), and YK522, in which the UL47-null mutation in YK521 was repaired (VenusA206K-Us3/ΔUL47-repair) (Fig. 1). Insertion of the foreign gene cassette into UL47 in YK521 had no effect on the expression of the UL47-neighboring genes UL46 and UL48, based on the observation that wild-type HSV-1(F), YK501 (VenusA206K-Us3), YK521, and YK522 produced indistinguishable levels of UL46 and UL48 in infected Vero cells, as determined by immunoblotting (Fig. 10). However, at 24 h postinfection, in Vero cells infected at an MOI of 5, YK521 had a progeny virus yield 15.2-fold lower than that of YK501 and 8.1-fold lower than that of YK522 (data not shown). These results are consistent with those for YK545 (ΔUL47/ΔBAC) and YK546 (ΔUL47-repair/ΔBAC) described above.
Fig. 10.
Effect of the UL47-null mutation on the expression of the neighboring UL46 and UL48 genes. Vero cells infected with either wild-type HSV-1(F), YK501 (VenusA206K-Us3), YK521 (VenusA206K-Us3/ΔUL47), or YK522 (VenusA206K-Us3/ΔUL47-repair) at an MOI of 1 for 24 h were harvested and were analyzed by immunoblotting with anti-Us3 (α-Us3), anti-UL46, and anti-UL48 antibodies. The positions of molecular mass markers are shown on the left.
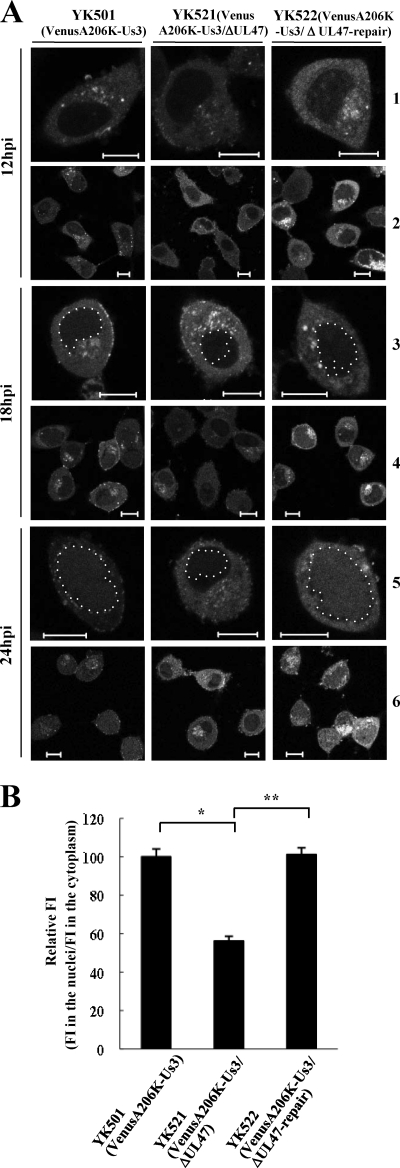
Vero cells were infected at an MOI of 1 with YK501 (VenusA206K-Us3), YK521 (VenusA206K-Us3/ΔUL47), or YK522 (VenusA206K-Us3/ΔUL47-repair), and the localization of VenusA206K-Us3 was examined at 12, 18, and 24 h postinfection. In agreement with the results shown in Fig. 9, VenusA206K-Us3 localized predominantly in the cytoplasm of cells infected with YK501 and YK522 at 12 and 18 h postinfection, while nuclear localization of VenusA206K-Us3 was evident at 24 h postinfection (Fig. 11A). In contrast, nuclear localization of VenusA206K-Us3 was significantly impaired in YK521-infected cells at 24 h postinfection (Fig. 11A). Quantitation of VenusA206K-Us3 fluorescence in cells infected with YK501, YK521, or YK522 at 24 h postinfection confirmed the significant decrease in the nuclear accumulation of VenusA206K-Us3 in cells infected with YK521 compared to that in cells infected with YK501 or YK522. These results indicate that UL47 is required for the efficient nuclear localization of Us3 in infected cells.
Fig. 11.
(A) Digital confocal microscope images showing the localization of VenusA206K-Us3 proteins in infected Vero cells. Vero cells were infected with either YK501 (VenusA206K-Us3), YK521 (VenusA206K-Us3/ΔUL47), or YK522 (VenusA206K-Us3/ΔUL47-repair) at an MOI of 1, and the infected cells were examined by confocal microscopy at 12, 18, and 24 h postinfection (hpi). Nuclear Us3 is circled in rows 3 and 5. Bars, 10 μm. (B) Quantitation of the fluorescence intensity (FI) of VenusA206K-Us3 in the nuclei relative to that in the cytoplasm of infected Vero cells at 24 h postinfection. Infected Vero cells were examined by confocal microscopy as described for panel A. Images of 10 infected cells were randomly selected for each virus infection. Three circular areas of the same size were randomly selected in the cytoplasm and in the nucleus of each of the 10 infected cells, and the FI of each circular area was analyzed. For each virus infection, the relative FI was calculated as the mean FI of the 30 circular areas in the nuclei divided by the mean FI of the 30 circular areas in the cytoplasm. The data are means and standard errors from three independent experiments. Asterisks indicate significant differences (*, P = 1.6 × 10−8; **, P = 3.5 × 10−9) by a two-tailed Student t test. The mean relative FI for YK501-infected cells was normalized to 100%.
DISCUSSION
More than 15 putative HSV-1 Us3 substrates have been reported to date (4, 24–26, 32, 45–47, 49, 54, 57, 58). However, only a few of these substrates have been shown both to be physiological substrates of Us3 in infected cells and to be directly linked with Us3 functions in infected cells. These include gB, UL31, Us3 itself, and tuberous sclerosis complex 2 (TSC2) (4, 18, 24, 47, 63). It has been reported that Us3 phosphorylation of these substrates appears to regulate the expression of gB on the cell surface (18, 24), the nuclear egress of nucleocapsids (47, 72), the catalytic activity of Us3 (63), and mRNA translation (4) in infected cells. Furthermore, phosphorylation by Us3 of both gB and Us3 itself appears to play critical roles in viral replication in vivo and HSV-1 pathogenesis, based on data showing that viral replication and pathogenicity in mice were attenuated in infections with recombinant HSV-1 viruses in which the Us3 phosphorylation site in gB or Us3 was mutated, or in infections with an HSV-1 Us3 kinase-dead mutant virus (19, 63).
An important finding of this study is that UL47 is a physiological substrate of Us3 in infected cells and that Us3 phosphorylation of UL47 appears to be critical for viral replication and pathogenesis in vivo. This conclusion is based on the following results: (i) purified UL47 was phosphorylated by purified Us3 in vitro; (ii) detection of UL47 in infected cells with an anti-phospho-PKA substrate antibody (100G7) was dependent on Us3 kinase activity; and (iii) substitution of alanine for UL47 Ser-77, which was identified as a Us3 phosphorylation site in in vitro kinase assays, significantly reduced viral replication and pathogenesis in a mouse HSK model. Similar reductions in viral replication and pathogenesis were reported in studies of mice infected with a Us3 kinase-dead virus (63). Our conclusion is also supported by the recent report that the BHV-1 Us3 homologue phosphorylated the BHV-1 UL47 homologue in vitro, although the biological significance of the phosphorylation remains to be addressed (31). Thus, our present study adds UL47 to the known physiological substrates of Us3 that are biologically relevant in HSV-1 infection. However, as is usual with such mutational analyses, we cannot completely exclude the possibility that the UL47 S77A mutation caused a conformational change in UL47 that directly impaired UL47 function(s) in vivo rather than that the impairment resulted from blocking a site of Us3-mediated phosphorylation. Therefore, further analyses of the functions of UL47 and their relation to Us3 phosphorylation of UL47 Ser-77 will be necessary in order to define the precise role(s) of UL47 phosphorylation in viral infection and disease in vivo. However, taking the results described above together with the data showing that the UL47-null mutant virus significantly attenuated viral replication and pathogenicity in mice, this study is the first report demonstrating that HSV-1 UL47 plays a critical role in HSV-1 replication and pathogenesis in vivo. As described above, other alphaherpesvirus homologues of UL47, including PRV UL47, BHV-1 UL47, and ILTV UL47, have been reported to be important for viral pathogenesis in vivo (16, 29, 36). Therefore, UL47 homologues appear to be common virulence factors in alphaherpesviruses.
The UL47 NLS has been mapped to UL47 residues 50 to 68, which contain a cluster of arginine residues (8). The arginine-rich nature of the UL47 NLS suggests that it is not related to the classical monopartite and bipartite NLS sequences that contain a lysine-rich domain but that it is a member of the class of NLSs that contain an arginine-rich domain, as reported for HIV-1 Rev and Tat and for human T-lymphotropic virus 1 (HTLV-1) Rex (14, 15, 38, 65, 66). In the nuclear transport of proteins carrying a lysine- or arginine-rich NLS, proteins are recognized in the cytoplasm by an importin α/importin β heterodimer or importin β alone, respectively (21, 22). In either case, importin β mediates passage through the nuclear pore and release into the nucleus, dependent on the GTP-binding protein Ran (21, 22). Phosphorylation of a protein near its NLS, mediated by many different protein kinases, is a key and common mechanism by which the transport of NLS-containing proteins into the nucleus can be regulated (20–22). As with other proteins carrying a phosphorylation-regulated NLS, Us3 phosphorylation of UL47 Ser-77, which is adjacent to the UL47 NLS, appears to promote nuclear localization of UL47 in a significant fraction of infected cells. This conclusion is based on the following results: (i) alanine substitution of the Us3 physiological phosphorylation site Ser-77 in UL47 induced aberrant accumulation of UL47 at the nuclear rim and impaired nuclear localization of UL47 in a significant fraction of infected cells; (ii) aberrant accumulation of UL47 at the nuclear rim and impaired nuclear localization of UL47 were also observed in a significant fraction of cells infected with a Us3 kinase-dead mutant virus; and (iii) a phosphomimetic substitution at UL47 Ser-77 (S77D), which mimicked constitutive phosphorylation (18, 46, 47, 73), restored wild-type localization of UL47 in infected cells. The aberrant accumulation of UL47 at the nuclear rim is assumed to reflect inhibition of UL47 passage through the nuclear pore, probably mediated by importin(s), although it remains to be shown whether importin(s) mediates the nuclear transport of UL47. It has been reported that phosphorylation of sites adjacent to the NLS of simian virus 40 T antigen enhanced the binding of importins to the NLS, leading to efficient nuclear localization of the protein (17, 21). Similarly, Us3 phosphorylation of UL47 near its NLS might enhance the affinity of an import receptor(s), such as an importin, resulting in the promotion of nuclear transport of UL47 in infected cells. At present, we cannot exclude the possibility that a protein kinase(s) other than Us3 also phosphorylates UL47 Ser-77 in infected cells and is involved in the nuclear transport of UL47, based on the observations that the phosphorylation target site specificity of HSV-1 Us3 appears to be similar to those of cellular protein kinases PKA and Akt (2, 4) and that in the absence of Us3 kinase activity in infected cells, low-level but consistent phosphorylation of UL47 was detected. It is noteworthy that in this study, the S77A mutation in UL47 not only altered the subcellular localization of UL47 in infected cells but also reduced viral replication and pathogenesis in a mouse HSK model, suggesting that proper nuclear localization of UL47 in infected cells is critical for HSV-1 replication and pathogenesis in vivo. Furthermore, it has been reported that the UL47 NLS also functions as an RNA binding domain (8). Therefore, it is of interest to investigate whether Us3 phosphorylation of UL47 near its NLS (Ser-77) affects the ability of UL47 to bind RNAs in infected cells.
It was surprising that aberrant accumulation of UL47 at the nuclear rim was observed in only a fraction (∼40%) of cells infected with the recombinant virus with a UL47 S77A mutation. Interestingly, this seems to be consistent with the observation that the HSK produced by YK547 (UL47S77A) in mice was twice as severe as that produced by YK545 (ΔUL47/ΔBAC). At present, it is not known why the S77A mutation in UL47 affected UL47 localization only in some infected cells. However, a similar observation has recently been reported for histone deacetylase 5 (HDAC5): alanine substitutions in the physiological phosphorylation sites within its NLS impaired nuclear localization only in some cells (13). One possible explanation of these observations may be that subcellular localization of these proteins is regulated by multiple factors. In agreement with this hypothesis, it has been reported that, when expressed in transiently transfected cells, UL47 shuttled rapidly between the nucleus and cytoplasm, with an overall localization in the nucleus (7, 71), suggesting that at least two cellular functions, including those involved in nuclear import and export, may regulate UL47 localization in transfected cells. It has also been reported that UL47 was localized in both the nucleus and the cytoplasm in HSV-1-infected cells, suggesting that a viral factor(s) and/or a cellular factor(s) induced by viral infection may play a role in keeping UL47 in the cytoplasm of infected cells (6). In addition, in this study we have shown that phosphorylation of UL47 Ser-77, near the UL47 NLS, by Us3 optimally regulated its localization in infected cells. It is also possible that both Us3 and cellular protein kinases with a phosphorylation target site specificity similar to that of Us3, such as PKA and Akt, may phosphorylate the same multiple sites, including Ser-77 in UL47, and the multiple phosphorylation events mediated by the multiple protein kinases may regulate the subcellular localization of UL47 in infected cells. In agreement with this hypothesis, we identified a putative Us3 phosphorylation site in UL47 at codons 682 to 686, which is close to the UL47 NES (codons 658 to 667), as described above. The subcellular localization of UL47 in infected cells may be determined by the balance among these and/or a yet to be recognized cellular and/or viral function(s). Blocking the phosphorylation of UL47 Ser-77 may cause a subtle imbalance among the factors that determine the localization of UL47 in infected cells, resulting in aberrant localization of UL47 in only a fraction of infected cells.
In the present study, we showed that the HSV-1 protein kinase Us3 formed a stable complex with its UL47 substrate in infected cells. It appears that the stable interaction between Us3 and UL47 is conserved in other alphaherpesviruses, based on the observation that the BHV-1 Us3 homologue was coprecipitated with the transiently expressed BHV-1 UL47 homologue from BHV-1-infected cells (31). These observations raise the possibility that UL47 affects Us3 function(s) in infected cells. In agreement with this possibility, we also showed that the absence of UL47 in infected cells significantly impaired the nuclear localization of Us3. However, we should note that most of the Us3 remained in the cytoplasm throughout infection regardless of the presence of UL47, indicating that UL47 was in part required for proper nuclear localization of Us3 in infected cells. Therefore, Us3 and UL47 exhibited a unique feature in that they regulated each other's subcellular localization in infected cells. Since some functions of HSV-1 Us3, including regulation of the nuclear egress of nucleocapsids by phosphorylation of NM-associated viral and cellular proteins, are carried out in the nuclei of infected cells, Us3 needs to localize in the nuclei of infected cells. It has been reported recently that the kinase activity of HSV-2 Us3 is required for efficient nuclear localization of the protein when expressed in transiently transfected cells (10). This observation is consistent with earlier reports showing that the kinase activity of HSV-1 Us3 is required for proper localization of Us3 in infected cells (25, 62). However, it remains to be elucidated whether HSV-1 Us3 kinase activity is in fact involved in the regulation of its nuclear localization in infected cells, since it has been reported that the roles of HSV-2 Us3 kinase activity in infected cells are remarkably different from those of HSV-1 Us3 (44). Therefore, there is a lack of information on how the nuclear localization of HSV-1 Us3 is regulated in infected cells. In the present study, we have identified UL47 as a factor in the regulation of the proper nuclear localization of Us3 in infected cells. It has been reported that HSV-1 ICP27 associated with, and translocated to the nuclei of HSV-1-infected cells, the major form of polyadenylate-binding protein (PABP), PABPC1, which localizes predominantly in the cytoplasm of uninfected cells and plays central roles in posttranscriptional gene regulation via modulation of translation initiation and mRNA metabolism (5, 23, 39). Similarly, UL47 might associate with Us3 in the cytoplasm of infected cells and might take the viral protein kinase to the nucleus with it. As reported previously (41, 44, 63), HSV-1 Us3 plays critical roles in viral replication and pathogenesis in mice. Therefore, the involvement of UL47 in the proper nuclear localization of Us3 in cultured cells may in part account for reduced viral replication and pathogenesis in mice infected with the UL47-null mutant virus.
ACKNOWLEDGMENTS
We thank Shihoko Koyama for excellent technical assistance.
This study was supported by the Funding Program for Next Generation World-Leading Researchers and Grants for Scientific Research from the Japan Society for the Promotion of Science (JSPS), a Grant for Scientific Research in Priority Areas, a contract research fund for the Program of Japan Initiative for Global Research Network on Infectious Diseases and the Global COE Program “Center of Education and Research for the Advanced Genome-Based Medicine–For personalized medicine and the control of worldwide infectious diseases” from the Ministry of Education, Culture, Science, Sports and Technology (MEXT) of Japan, and grants from the Takeda Science Foundation, the Daiichi-Sankyo Foundation of Life Science, the Uehara Memorial Foundation, and the Asahi Glass Foundation. K.S., J.A., and T.I. were supported by JSPS Research Fellowships for Young Scientists.
Footnotes
Published ahead of print on 6 July 2011.
REFERENCES
- 1. Asai R., Kato A., Kawaguchi Y. 2009. Epstein-Barr virus protein kinase BGLF4 interacts with viral transactivator BZLF1 and regulates its transactivation activity. J. Gen. Virol. 90:1575–1581 [DOI] [PubMed] [Google Scholar]
- 2. Benetti L., Roizman B. 2004. Herpes simplex virus protein kinase US3 activates and functionally overlaps protein kinase A to block apoptosis. Proc. Natl. Acad. Sci. U. S. A. 101:9411–9416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Campbell R. E., et al. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 99:7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chuluunbaatar U., et al. 2010. Constitutive mTORC1 activation by a herpesvirus Akt surrogate stimulates mRNA translation and viral replication. Genes Dev. 24:2627–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dobrikova E., Shveygert M., Walters R., Gromeier M. 2010. Herpes simplex virus proteins ICP27 and UL47 associate with polyadenylate-binding protein and control its subcellular distribution. J. Virol. 84:270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Donnelly M., Elliott G. 2001. Fluorescent tagging of herpes simplex virus tegument protein VP13/14 in virus infection. J. Virol. 75:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Donnelly M., Elliott G. 2001. Nuclear localization and shuttling of herpes simplex virus tegument protein VP13/14. J. Virol. 75:2566–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Donnelly M., Verhagen J., Elliott G. 2007. RNA binding by the herpes simplex virus type 1 nucleocytoplasmic shuttling protein UL47 is mediated by an N-terminal arginine-rich domain that also functions as its nuclear localization signal. J. Virol. 81:2283–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dorange F., Tischer B. K., Vautherot J. F., Osterrieder N. 2002. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76:1959–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Finnen R. L., Roy B. B., Zhang H., Banfield B. W. 2010. Analysis of filamentous process induction and nuclear localization properties of the HSV-2 serine/threonine kinase Us3. Virology 397:23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frame M. C., Purves F. C., McGeoch D. J., Marsden H. S., Leader D. P. 1987. Identification of the herpes simplex virus protein kinase as the product of viral gene US3. J. Gen. Virol. 68(Pt 10): 2699–2704 [DOI] [PubMed] [Google Scholar]
- 12. Fuchs W., Granzow H., Mettenleiter T. C. 2003. A pseudorabies virus recombinant simultaneously lacking the major tegument proteins encoded by the UL46, UL47, UL48, and UL49 genes is viable in cultured cells. J. Virol. 77:12891–12900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greco T. M., Yu F., Guise A. J., Cristea I. M. 2011. Nuclear import of histone deacetylase 5 by requisite nuclear localization signal phosphorylation. Mol. Cell. Proteomics 10:M110.004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hammerschmid M., et al. 1994. Scanning mutagenesis of the arginine-rich region of the human immunodeficiency virus type 1 Rev trans activator. J. Virol. 68:7329–7335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hauber J., Malim M. H., Cullen B. R. 1989. Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. J. Virol. 63:1181–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Helferich D., Veits J., Teifke J. P., Mettenleiter T. C., Fuchs W. 2007. The UL47 gene of avian infectious laryngotracheitis virus is not essential for in vitro replication but is relevant for virulence in chickens. J. Gen. Virol. 88:732–742 [DOI] [PubMed] [Google Scholar]
- 17. Hübner S., Xiao C. Y., Jans D. A. 1997. The protein kinase CK2 site (Ser111/112) enhances recognition of the simian virus 40 large T-antigen nuclear localization sequence by importin. J. Biol. Chem. 272:17191–17195 [DOI] [PubMed] [Google Scholar]
- 18. Imai T., et al. 2011. Role of the herpes simplex virus 1 Us3 kinase phosphorylation site and endocytosis motifs in the intracellular transport and neurovirulence of envelope glycoprotein B. J. Virol. 85:5003–5015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Imai T., Sagou K., Arii J., Kawaguchi Y. 2010. Effects of phosphorylation of herpes simplex virus 1 envelope glycoprotein B by Us3 kinase in vivo and in vitro. J. Virol. 84:153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jans D. A. 1995. The regulation of protein transport to the nucleus by phosphorylation. Biochem. J. 311(Pt 3):705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jans D. A., Hübner S. 1996. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol. Rev. 76:651–685 [DOI] [PubMed] [Google Scholar]
- 22. Jans D. A., Xiao C. Y., Lam M. H. 2000. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays 22:532–544 [DOI] [PubMed] [Google Scholar]
- 23. Kahvejian A., Svitkin Y. V., Sukarieh R., M'Boutchou M. N., Sonenberg N. 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 19:104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kato A., et al. 2009. Herpes simplex virus 1 protein kinase Us3 phosphorylates viral envelope glycoprotein B and regulates its expression on the cell surface. J. Virol. 83:250–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kato A., et al. 2008. Identification of a physiological phosphorylation site of the herpes simplex virus 1-encoded protein kinase Us3 which regulates its optimal catalytic activity in vitro and influences its function in infected cells. J. Virol. 82:6172–6189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kato A., et al. 2005. Identification of proteins phosphorylated directly by the Us3 protein kinase encoded by herpes simplex virus 1. J. Virol. 79:9325–9331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kato K., et al. 2000. Synthesis, subcellular localization and VP16 interaction of the herpes simplex virus type 2 UL46 gene product. Arch. Virol. 145:2149–2162 [DOI] [PubMed] [Google Scholar]
- 28. Kawaguchi Y., Van Sant C., Roizman B. 1997. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328–7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Klopfleisch R., et al. 2004. Influence of tegument proteins of pseudorabies virus on neuroinvasion and transneuronal spread in the nervous system of adult mice after intranasal inoculation. J. Virol. 78:2956–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kopp M., Klupp B. G., Granzow H., Fuchs W., Mettenleiter T. C. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 76:8820–8833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Labiuk S. L., Babiuk L. A., van Drunen Littel-van den Hurk S. 2009. Major tegument protein VP8 of bovine herpesvirus 1 is phosphorylated by viral US3 and cellular CK2 protein kinases. J. Gen. Virol. 90:2829–2839 [DOI] [PubMed] [Google Scholar]
- 32. Leach N., et al. 2007. Emerin is hyperphosphorylated and redistributed in herpes simplex virus type 1-infected cells in a manner dependent on both UL34 and US3. J. Virol. 81:10792–10803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leader D. P. 1993. Viral protein kinases and protein phosphatases. Pharmacol. Ther. 59:343–389 [DOI] [PubMed] [Google Scholar]
- 34. Leader D. P., Deana A. D., Marchiori F., Purves F. C., Pinna L. A. 1991. Further definition of the substrate specificity of the alpha-herpesvirus protein kinase and comparison with protein kinases A and C. Biochim. Biophys. Acta 1091:426–431 [DOI] [PubMed] [Google Scholar]
- 35. Leopardi R., Van Sant C., Roizman B. 1997. The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc. Natl. Acad. Sci. U. S. A. 94:7891–7896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lobanov V. A., et al. 2010. A UL47 gene deletion mutant of bovine herpesvirus type 1 exhibits impaired growth in cell culture and lack of virulence in cattle. J. Virol. 84:445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Loret S., Guay G., Lippe R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605–8618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Malim M. H., Bohnlein S., Hauber J., Cullen B. R. 1989. Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell 58:205–214 [DOI] [PubMed] [Google Scholar]
- 39. Mangus D. A., van Hoof A. 2003. Making and breaking the message. Genome Biol. 4:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McGeoch D. J., Davison A. J. 1986. Alphaherpesviruses possess a gene homologous to the protein kinase gene family of eukaryotes and retroviruses. Nucleic Acids Res. 14:1765–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Meignier B., Longnecker R., Mavromara-Nazos P., Sears A. E., Roizman B. 1988. Virulence of and establishment of latency by genetically engineered deletion mutants of herpes simplex virus 1. Virology 162:251–254 [DOI] [PubMed] [Google Scholar]
- 42. Meredith D. M., Lindsay J. A., Halliburton I. W., Whittaker G. R. 1991. Post-translational modification of the tegument proteins (VP13 and VP14) of herpes simplex virus type 1 by glycosylation and phosphorylation. J. Gen. Virol. 72(Pt 11):2771–2775 [DOI] [PubMed] [Google Scholar]
- 43. Meyer B. E., Malim M. H. 1994. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 8:1538–1547 [DOI] [PubMed] [Google Scholar]
- 44. Morimoto T., et al. 2009. Differences in the regulatory and functional effects of the Us3 protein kinase activities of herpes simplex virus 1 and 2. J. Virol. 83:11624–11634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Morris J. B., Hofemeister H., O'Hare P. 2007. Herpes simplex virus infection induces phosphorylation and delocalization of emerin, a key inner nuclear membrane protein. J. Virol. 81:4429–4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mou F., Forest T., Baines J. D. 2007. US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 81:6459–6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mou F., Wills E., Baines J. D. 2009. Phosphorylation of the UL31 protein of herpes simplex virus 1 by the US3-encoded kinase regulates localization of the nuclear envelopment complex and egress of nucleocapsids. J. Virol. 83:5181–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Munger J., Chee A. V., Roizman B. 2001. The US3 protein kinase blocks apoptosis induced by the d120 mutant of herpes simplex virus 1 at a premitochondrial stage. J. Virol. 75:5491–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Munger J., Roizman B. 2001. The US3 protein kinase of herpes simplex virus 1 mediates the posttranslational modification of BAD and prevents BAD-induced programmed cell death in the absence of other viral proteins. Proc. Natl. Acad. Sci. U. S. A. 98:10410–10415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ogg P. D., McDonell P. J., Ryckman B. J., Knudson C. M., Roller R. J. 2004. The HSV-1 Us3 protein kinase is sufficient to block apoptosis induced by overexpression of a variety of Bcl-2 family members. Virology 319:212–224 [DOI] [PubMed] [Google Scholar]
- 51. O'Neill R. E., Talon J., Palese P. 1998. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 17:288–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Phelan A., Clements J. B. 1997. Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J. Gen. Virol. 78(Pt 12):3327–3331 [DOI] [PubMed] [Google Scholar]
- 53. Poon A. P., Gu H., Roizman B. 2006. ICP0 and the US3 protein kinase of herpes simplex virus 1 independently block histone deacetylation to enable gene expression. Proc. Natl. Acad. Sci. U. S. A. 103:9993–9998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Poon A. P., Liang Y., Roizman B. 2003. Herpes simplex virus 1 gene expression is accelerated by inhibitors of histone deacetylases in rabbit skin cells infected with a mutant carrying a cDNA copy of the infected-cell protein no. 0. J. Virol. 77:12671–12678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Purves F. C., Deana A. D., Marchiori F., Leader D. P., Pinna L. A. 1986. The substrate specificity of the protein kinase induced in cells infected with herpesviruses: studies with synthetic substrates [corrected] indicate structural requirements distinct from other protein kinases. Biochim. Biophys. Acta 889:208–215 [DOI] [PubMed] [Google Scholar]
- 56. Purves F. C., Longnecker R. M., Leader D. P., Roizman B. 1987. Herpes simplex virus 1 protein kinase is encoded by open reading frame US3 which is not essential for virus growth in cell culture. J. Virol. 61:2896–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Purves F. C., Ogle W. O., Roizman B. 1993. Processing of the herpes simplex virus regulatory protein α22 mediated by the UL13 protein kinase determines the accumulation of a subset of α and γ mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. U. S. A. 90:6701–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Purves F. C., Spector D., Roizman B. 1991. The herpes simplex virus 1 protein kinase encoded by the US3 gene mediates posttranslational modification of the phosphoprotein encoded by the UL34 gene. J. Virol. 65:5757–5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Reynolds A. E., et al. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reynolds A. E., Wills E. G., Roller R. J., Ryckman B. J., Baines J. D. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Roux P. P., Blenis J. 2004. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 68:320–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ryckman B. J., Roller R. J. 2004. Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 78:399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sagou K., Imai T., Sagara H., Uema M., Kawaguchi Y. 2009. Regulation of the catalytic activity of herpes simplex virus 1 protein kinase Us3 by autophosphorylation and its role in pathogenesis. J. Virol. 83:5773–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shaner N. C., et al. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 [DOI] [PubMed] [Google Scholar]
- 65. Siomi H., Shida H., Maki M., Hatanaka M. 1990. Effects of a highly basic region of human immunodeficiency virus Tat protein on nucleolar localization. J. Virol. 64:1803–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Siomi H., et al. 1988. Sequence requirements for nucleolar localization of human T cell leukemia virus type I pX protein, which regulates viral RNA processing. Cell 55:197–209 [DOI] [PubMed] [Google Scholar]
- 67. Sugimoto K., et al. 2008. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J. Virol. 82:5198–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tanaka M., Kagawa H., Yamanashi Y., Sata T., Kawaguchi Y. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 77:1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Verhagen J., Donnelly M., Elliott G. 2006. Characterization of a novel transferable CRM-1-independent nuclear export signal in a herpesvirus tegument protein that shuttles between the nucleus and cytoplasm. J. Virol. 80:10021–10035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Walters M. S., Kinchington P. R., Banfield B. W., Silverstein S. 2010. Hyperphosphorylation of histone deacetylase 2 by alphaherpesvirus US3 kinases. J. Virol. 84:9666–9676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Williams P., Verhagen J., Elliott G. 2008. Characterization of a CRM1-dependent nuclear export signal in the C terminus of herpes simplex virus type 1 tegument protein UL47. J. Virol. 82:10946–10952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wisner T. W., et al. 2009. Herpesvirus gB-induced fusion between the virion envelope and outer nuclear membrane during virus egress is regulated by the viral US3 kinase. J. Virol. 83:3115–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yokoyama A., et al. 2001. Identification of major phosphorylation sites of Epstein-Barr virus nuclear antigen leader protein (EBNA-LP): ability of EBNA-LP to induce latent membrane protein 1 cooperatively with EBNA-2 is regulated by phosphorylation. J. Virol. 75:5119–5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang Y., Sirko D. A., McKnight J. L. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in αTIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65:829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]