Abstract
Enterovirus 71 (EV71) causes hand-foot-and-mouth disease and neurological complications in young children. Although the underlying mechanisms remain obscure, impaired or aberrant immunity is thought to play a role. In infected cells, EV71 suppresses type I interferon responses mediated by retinoid acid-inducible gene I (RIG-I). This involves the EV71 3C protein, which disrupts the formation of a functional RIG-I complex. In the present study, we report that EV71 inhibits the induction of innate immunity by Toll-like receptor 3 (TLR3) via a distinct mechanism. In HeLa cells stimulated with poly(I · C), EV71 inactivates interferon regulatory factor 3 and drastically suppresses interferon-stimulated gene expression. Notably, EV71 specifically downregulates a TRIF, TIR domain-containing adaptor inducing beta interferon (IFN-β). When expressed alone in mammalian cells, EV71 3C is capable of exhibiting these activities. EV71 3C associates with and induces TRIF cleavage in the presence of Z-VAD-FMK, a caspase inhibitor. TRIF cleavage depends on its amino acid pair Q312-S313, which resembles a proteolytic site of picornavirus 3C proteases. Further, site-specific 3C mutants with a defective protease activity bind TRIF but fail to mediate TRIF cleavage. Consequently, these 3C mutants are unable to inhibit NF-κB and IFN-β promoter activation. TRIF cleavage mediated by EV71 may be a mechanism to impair type I IFN production in response to Toll-like receptor 3 (TLR3) activation.
INTRODUCTION
Enterovirus 71 (EV71), a member of the Picornaviridae family, is a causative agent of hand-foot-and-mouth disease (HFMD) in young children and infants. Severe infection with EV71 may lead to various neurological complications, including aseptic meningitis, acute flaccid paralysis, encephalitis, and neurogenic pulmonary edema (29). EV71 outbreaks occur periodically through the world, especially in the Asia-Pacific region (1, 2, 9, 12, 19, 24, 28, 29, 51). The EV71 genome is a positive-stranded RNA molecule that encodes a single polyprotein precursor of about 2,200 amino acids. This precursor is processed into structural (VP1, VP2, VP3, and VP4) and nonstructural (2A, 2B, 2C, 3A, 3B, 3C, and 3D) proteins upon infection (29). In this process the viral proteases, 3C and 2A, work coordinately to facilitate viral replication.
The 3C protein encoded by EV71 is essential for viral replication (45, 46). In addition to its activity in viral protein processing (25), 3C participates in several other processes. The 3C protein binds to the 5′ untranslated region of viral RNA, but its effect on viral infection is unknown (45). When expressed in neuronal cells, 3C induces apoptosis through caspase activation (25). This is thought to facilitate viral spread or pathogenesis. Furthermore, 3C cleaves cellular CstF-64 protein (49). This impairs the 3′ end of host RNA processing and polyadenylation, thus providing an additional advantage for viral replication. Of note, 3C blocks type I interferon (IFN) responses that exert antiviral and immunoregulatory activities (22).
Type I IFN is induced via Toll-like receptor (TLR)-related pathways (17). It is well established that TLR3 in the endosome recognizes viral double-stranded RNA (dsRNA). Once activated, TLR3 recruits a TIR domain-containing adaptor inducing IFN-β (TRIF) (17), which together with TNF receptor-associated factor 3 (TRAF3), activates the two IKK related kinases, TANK-binding kinase 1 (TBK1) and inducible Iκ-B kinase (IKKi). These kinases phosphorylate interferon regulatory factor 3/7 (IRF3/7) (8, 11, 34, 44), leading to the expression of target genes, such as IFN-α/β (30, 35, 39, 50). Additionally, TRIF stimulates NF-κB activation via receptor-interacting protein 1 (RIP1) and TRAF6, resulting in the production of proinflammatory cytokines, such as interleukin-6 (IL-6) (14, 15). TRIF is a 712-amino-acid protein that is present in the cytoplasm. While its amino terminus interacts with TRAF6 and TRAF3, the carboxyl terminus binds to TLR3 and RIP1 (11, 14, 31, 34). Accordingly, TRIF serves as a key adaptor that transmits signals to IRF3 and NF-κB, respectively. Alternative receptors exist to detect cytosolic viral RNA (17). Examples are retinoid acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), which induce cytokine expression via the adaptor IPS-1 (17, 18, 53).
It has been reported that picornaviruses are sensed primarily by MDA5 (10, 16), but recent evidence suggests a role for RIG-I as well (37). In infected cells, several picornaviruses cleave or interact with these pattern recognition receptors (3, 4, 22, 37). TLR3 also recognizes or limits picornavirus infection (33, 41, 48). Unfortunately, little is known about how picornaviruses are involved. Here, we report that EV71 suppresses TLR3-mediated type I IFN responses by downregulation of TRIF. This requires EV71 3C, which interacts with TRIF and induces its cleavage independently of caspases. TRIF cleavage depends on its amino acid pair Q312 and S313. H40, KFRDI, and VGK motifs in 3C are not required for binding TRIF, but they are indispensable for inactivation of NF-κB and the IFN-β promoter via TRIF cleavage. These results suggest that control of TLR3 by EV71 3C may be a mechanism to disrupt type I IFN responses.
MATERIALS AND METHODS
Cell lines and viruses.
RD (rhabdomyosarcoma), 293T, and HeLa cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 10% heated-inactivated fetal bovine serum (FBS) (HyClone, Logan, UT) and penicillin-streptomycin at 37°C in a 5% CO2 humidified atmosphere. 293/TLR3 cells were a gift of Zhengfan Jiang. Enterovirus 71 (EV71) infection was carried out as follows. Briefly, cells were infected with EV71 at the indicated multiplicity of infection (MOI). Unbound virus was washed away 2 h after infection, and cells were cultured with fresh medium supplemented with 10% FBS.
Plasmids and reagents.
The plasmids pEGFP-3C and pCDNA3.1-Flag-3C and pEGFP-3C variants have been described elsewhere (22). The Flag-TRIF variants, including Δ311-336, Δ337-367, Δ368-398, Δ399-438, Δ439-485, Δ311-485, and amino acid substitution mutants, were constructed by site-directed mutagenesis using Pfu DNA polymerase (Stratagene, La Jolla, CA). All variants were confirmed by subsequent sequencing. Antibodies against Flag, Myc, green fluorescent protein (GFP), IRF3, and β-actin were purchased from Sigma (St. Louis, MO). Anti-TRIF, anti-MyD88, anti-TBK1, and anti-RIG-I antibodies were purchased from Cell Signaling Technology (Danvers, MA). Antibody against phosphorylated IRF3 (pS386) was purchased from Epitomics (Burlingame, CA) Goat anti-mouse or -rabbit secondary antibodies were purchased from Li-Cor Inc. (Lincoln, NE). The caspase inhibitor Z-VAD-FMK and poly(I · C) were purchased from Sigma (St. Louis, MO).
Reporter assays.
Reporter assays were performed as described previously (7). Briefly, 293T or 293/TLR3 cells were seeded in 24-well plates at a density of 3 × 105 cells per well. On the next day cells were transfected with a control plasmid or plasmid expressing TRIF, MyD88, and 3C variants along with pGL3-IFN β-luc, NF-κB-luc, and pRL-simian virus 40 (SV40) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The total amount of DNA was kept constant by adding empty control plasmid. At 24 h after transfection, cells were harvested and cell lysates were used to determine luciferase activities (Promega, Madison, WI).
Reverse transcription-PCR (RT-PCR).
Total RNA was extracted from cells by using TRIzol reagent (Invitrogen, Carlsbad, CA). RNA samples were treated with DNase I (Pierce, Rockford, IL), and reverse transcription was carried out using the Superscript cDNA synthesis kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. cDNA samples were subjected to PCR amplification and electrophoresis to detect ISG54, ISG56, and IL-6 expression. The primers used were as follows: human ISG54, CTGCAACCATGAGTGAGAA and CCTTTGAGGTGCTTTAGATAG; human ISG56, TACAGCAACCATGAGTACAA and TCAGGTGTTTCACATAGGC; IL-6, GCCCTGAGAAAGGAGACAT and CTGTTCTGGAGGTACTCTAGGTAT; and GAPDH (glyceraldehyde-3-phosphate dehydrogenase), CGGAGTCAACGGATTTGGTCGTA and AGCCTTCTCCATGGTGGTGAAGAC.
Immunoprecipitation.
Cells were lysed with radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris-HCl [pH 7.4] containing 150 mM NaCl, 1% NP-40, and 0.25% sodium deoxycholate) containing protease inhibitor cocktail (Roche, Indianapolis, IN). Lysates of cells were incubated with anti-Flag antibody (Sigma, St. Louis, MO) in 500 μl RIPA buffer at 4°C overnight on a rotator in the presence of protein A/G-agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA). Immunocomplexes captured on the affinity gel or protein A/G agarose beads were subjected to electrophoresis and immunoblotting analysis (47).
Western blot analysis.
Cells were pelleted by centrifugation and lysed in buffer containing 150 mM NaCl, 25 mM Tris (pH 7.4), 1% NP-40, 0.25% sodium deoxycholate, and 1 mM EDTA with protease inhibitor cocktail (Roche, Indianapolis, IN). Aliquots of cell lysates were resolved by 12% SDS-PAGE and transferred to a nitrocellulose membrane (Pall, Port Washington, NY). The membranes were blocked with 5% nonfat dry milk and then probed with the indicated primary antibodies at 4°C overnight. This was followed by incubation with the corresponding IRD Fluor 800-labeled IgG or IRD Fluor 680-labeled IgG secondary antibody (Li-Cor Inc., Lincoln, NE). After being washed, the membranes were scanned with the Odyssey infrared imaging system (Li-Cor, Lincoln, NE) at a wavelength of 700 to 800 nm, and the molecular sizes of the developed proteins were determined by comparison with prestained protein markers (Ferments, Maryland, CA).
RESULTS
EV71 suppresses the induction of innate antiviral immunity by poly(I · C) in infected cells.
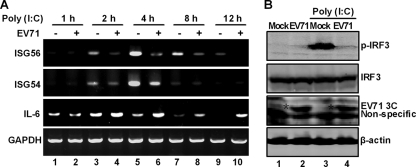
To study the effect of EV71 on the TLR3 pathway, we measured the induction of antiviral immunity by poly(I · C), a prototype TLR3 agonist. HeLa cells were initially mock infected or infected with EV71 for 4 h. At various time points after incubation with poly(I · C), the cells were examined for the expression of ISG54, ISG56, interleukin-6, and GAPDH by RT-PCR analysis. As shown in Fig. 1A, poly(I · C) induced ISG54 and ISG56 expression in mock-infected cells, which peaked at around 4 h after the treatment (lanes 1, 3, 5, 7, and 9). Such induction by poly(I · C) was sharply reduced in cells infected with EV71 at all time points examined (Fig. 1A, lanes 2, 4, 6, 8, and 10). Under these conditions, most cells were viable (96%) as measured by the trypan blue exclusion assay (data not shown). Interestingly, EV71 inhibited IRF3 phosphorylation by poly(I · C) compared to that in mock-infected cells (Fig. 1B, lanes 1 to 4). Poly(I · C) stimulated interleukin-6 expression in mock-infected cells, with an early kinetics (Fig. 1A, lanes 1, 3, 5, and 7). Intriguingly, EV71 infection enhanced this effect (Fig. 1A, lanes 4, 6, 8, and 10), suggesting that EV71 replication stimulated interleukin-6 expression. As expected, GAPDH expression remained at comparable levels in cells treated or not treated with poly(I · C) (Fig. 1A, lanes 1 to 10). Here, EV71 infection differentially modulates the expression of ISG54, ISG56, and interleukin-6.
Fig. 1.
(A) EV71 inhibits the induction of ISG54 and ISG56 by poly(I · C). HeLa cells were mock infected or infected with EV71 at an MOI of 2. At 4 h after infection, cells were incubated with or without poly(I · C). At indicated time points, total RNA extracted from cells was subjected to RT-PCR amplification and electrophoresis for ISG54, ISG56, IL-6, and GAPDH mRNAs. (B) EV71 inhibits poly(I · C)-induced phosphorylation of IRF3. HeLa cells were mock infected or infected with EV71 (MOI of 2) for 4 h. Cells were then incubated with or without poly(I · C) for additional 4 h. Cell lysates were then subjected to Western blot analysis with antibodies against IRF3 and phosphorylated IRF3, respectively.
The 3C protein of EV71 impairs the induction of antiviral molecules by poly(I · C).
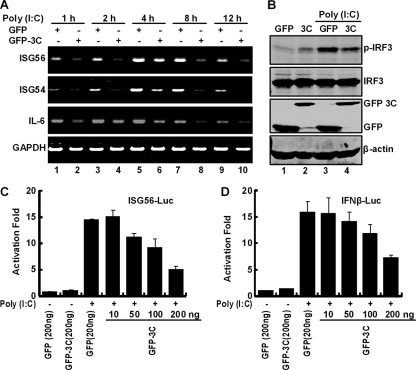
Among proteins encoded by EV71, 3C inhibits virus-induced immunity by interacting with RIG-I (22). Since EV71 blocked poly(I · C)-mediated responses, we hypothesized that 3C might contribute to this process. To test this possibility, HeLa cells were transfected with GFP or GFP-3C plasmid, with approximately 60% transfection efficiency. At 24 h after transfection, cells were incubated with poly(I · C) and the expression of ISG54, ISG56, IL-6, and GAPDH was analyzed by RT-PCR assays. As indicated in Fig. 2A, GFP-3C reduced the expression of ISG54, ISG56, and IL-6, whereas GFP displayed no inhibitory effect (lanes 1 to 10). In correlation, GFP-3C inhibited poly(I · C)-induced IRF3 phosphorylation compared to GFP (Fig. 2B, lanes 1 to 4). When expressed in 293/TLR3 cells, the 3C protein also inhibited ISG56 and IFN-β promoter activation induced by poly(I · C) compared to GFP (Fig. 2C and D). These results suggest that EV71 3C is able to inhibit dsRNA-induced innate immunity.
Fig. 2.
(A) The 3C protein inhibits the induction of ISG54, ISG56, and IL-6 by poly(I · C). HeLa cells were transfected with GFP or GFP-3C. At 24 h after transfection, cells were treated with or without poly(I · C). At the indicated time points, total RNA extracted from cells was analyzed for the expression of ISG54, ISG56, and IL-6. GAPDH was used as an internal control. (B) The 3C protein inhibits IRF3 phosphorylation induced by poly(I · C). HeLa cells transfected as described for panel A were incubated with or without poly(I · C) for 4 h. Cell lysates were subjected to Western blot analysis with antibodies against IRF3 or phosphorylated IRF3. (C) EV71 3C inhibits the ISG56 promoter activation induced by poly(I · C). 293/TLR3 cells were transfected with GFP or GFP-3C along with ISG56-Luc; pRL-SV40 was used as an internal control. At 24 h after transfection, cells were treated with or without poly(I · C) for 4 h. Cells lysates were assayed for luciferase activities. (D) EV71 3C inhibits the IFN-β promoter activation induced by poly(I · C). Assays were carried out as for panel C with the IFN-β-Luc reporter.
The 3C protein inhibits IFN-β and NF-κB activation by downregulation of TRIF.
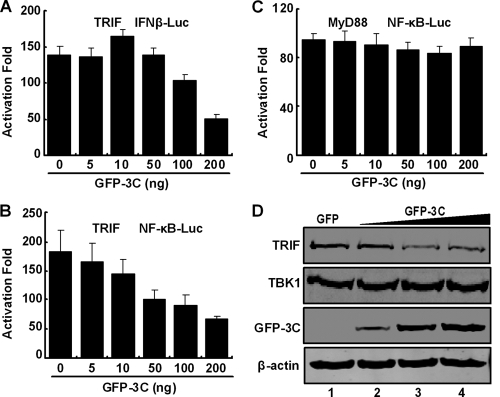
When engaged with TLR3, TRIF activates IRF3 and NF-κB, leading to type I IFN production (17). Therefore, we assessed the impact of 3C on TRIF in reporter assays. As illustrated in Fig. 3A, TRIF stimulated IFN-β promoter activation in 293T cells. Addition of 3C inhibited it in a dose-dependent manner. Likewise, 3C suppressed NF-κB activation by TRIF (Fig. 3B). Under this experimental condition, EV71 3C did not inhibit MyD88-stimulated NF-κB activation (Fig. 3C). It appears that the 3C protein inhibited TRIF but not MyD88 function, suggesting TRIF as a potential cellular target. To evaluate this, we determined whether EV71 3C affected the TRIF expression. As shown in Fig. 3D, EV71 3C reduced the level of endogenous TRIF in a dose-dependent manner when expressed in HeLa cells. This reduction was not detectable with TBK1. Thus, EV71 3C reduces TRIF expression and inhibits its activity.
Fig. 3.
Effects of 3C on NF-κB and IFN-β promoter activation. (A and B) 293T cells were transfected with TRIF and GFP-3C along with IFN-β-Luc (A) or NF-κB-Luc (B). (C) In parallel, cells were transfected with MyD88, GFP-3C, and NF-κB-Luc. A plasmid expressing GFP or pRL-SV40 was used as a control. At 24 h after transfection, cells lysates were assayed for luciferase activities. Data are representative of three independent experiments with triplicate samples. (D) Effect of 3C on TRIF expression. HeLa cells were transfected with GFP or GFP-3C. At 48 h after transfection, cells lysates were subjected to Western blot analysis with antibodies against TRIF, TBK1, GFP, and β-actin.
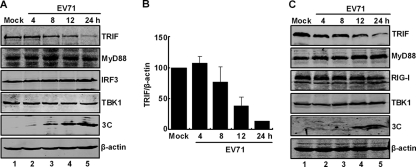
Next, we analyzed TRIF expression in EV71-infected cells. Cells were mock infected or infected with EV71. At different time points postinfection, cells were processed and examined by Western blot analysis. Figure 4A shows that HeLa cells constitutively expressed TRIF, MyD88, TBK1, IRF3, and β-actin (lane 1). EV71 infection had different effects on these proteins (lanes 2 to 5). Strikingly, the level of TRIF decreased as EV71 infection progressed. This coincided with an increase in 3C expression. In contrast, MyD88, IRF3, TBK1, and β-actin exhibited no or little reduction in EV71-infected cells. This was reproducible in several experiments. At 12 h after infection, EV71 reduced TRIF by approximately 70% (Fig. 4B). A similar pattern was also observed in RD cells (Fig. 4C, lanes 1 to 5). These results indicate that EV71 specifically downregulates TRIF in mammalian cells.
Fig. 4.
Effects of EV71 infection on protein expression. (A) HeLa cells were mock infected or infected with EV71 at an MOI of 5. At the indicated time points, cell lysates were subjected to Western blot analysis with antibodies against TRIF, MyD88, IRF3, TBK1, 3C, and β-actin. (B) Densitometry analysis. TRIF protein bands from three independent experiments from panel A were quantified and normalized to β-actin by using the Odyssey Image Software. (C) RD cells were infected and analyzed for TRIF, MyD88, RIG-I, TBK1, 3C, and β-actin as described above.
EV71 3C associates with and induces TRIF cleavage independently of caspases.
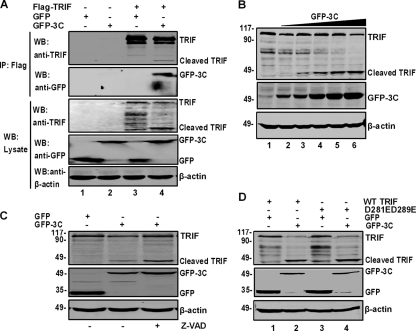
To examine the nature of 3C-TRIF interactions, we carried out immunoprecipitation analysis in 293T cells expressing Flag-TRIF, GFP, and GFP-3C. As illustrated in Fig. 5A, GFP-3C but not GFP coprecipitated with Flag-TRIF, indicating an interaction between 3C and TRIF. Notably, 3C reduced the TRIF level in the immunoprecipitates compared to GFP (lanes 3 and 4). This was accompanied by the appearance of a small species (45 kDa) (lane 4). Western blot analysis revealed comparable levels of GFP, GFP-3C, and β-actin in cell lysates (lanes 1 to 4). However, GFP-3C decreased TRIF expression (lane 4). In correlation, a small band appeared (lane 4). This is presumably a TRIF cleavage product. To further address this issue, we carried out a dose-response analysis in 293T cells cotransfected with 3C and TRIF. As shown in Fig. 5B. TRIF expression decreased as the level of GFP-3C increased. As expected, a smaller species appeared (lanes 2 to 6). Similar results were obtained with Flag-3C (data not shown). Hence, EV71 3C has the capacity to induce TRIF cleavage.
Fig. 5.
(A) The 3C protein associates with TRIF and induces TRIF cleavage. 293T cells were transfected with plasmids encoding Flag-TRIF (lanes 3 and 4), GFP (lanes 1 and 3), or GFP-3C (lanes 2 and 4). The total amount of DNA was kept constant with an empty vector. At 24 h after transfection, cell lysates were immunoprecipitated with anti-Flag antibody. Immunoprecipitates and aliquots of cell lysates were then subjected to Western blot analysis. (B) 3C induces TRIF cleavage in a dose-dependent manner. 293T cells were transfected with Flag-TRIF and increasing amounts of GFP-3C. At 24 h after transfection, cells were analyzed by Western blotting. (C) TRIF cleavage is independent of caspases. 293T cells were transfected with Flag-TRIF along with GFP or GFP-3C. After 4 h of transfection 50 μM Z-VAD was added to the medium. After 24 h of transfection, cell lysates were subjected to Western blot analysis. (D) Cleavage of TRIF bearing D281E and D289E substitutions. 293T cells were transfected with wild-type TRIF or the D281E D289E mutant along with GFP (lanes 1 and 3) or GFP-3C (lanes 2 and 4). At 24 h after transfection, cell lysates were subjected to Western blot analysis.
Previous studies showed that the 3C protein of EV71 activates caspases (25). To test whether 3C functioned via caspases, we assessed TRIF cleavage in the presence or absence of Z-VAD-FMK, a pancaspase inhibitor. Figure 5C shows that GFP-3C induced the cleavage of TRIF, resulting in a 45-kDa protein band (lane 2). Treatment with Z-VAD-FMK did not inhibit this cleavage (lane 3). The enhanced TRIF cleavage by Z-VAD-FMK is attributable to a block of apoptosis. To corroborate this result, we further analyzed a TRIF mutant carrying D281E and D289E substitutions, which is resistant to caspase cleavage (40). Similarly to wild-type TRIF, the D281E D289E mutant remained susceptible to 3C-mediated cleavage (Fig. 5D). These results indicated that 3C-mediated TRIF cleavage does not require caspases.
Glutamine 312 and serine 313 pairs within TRIF are required for 3C-induced cleavage.
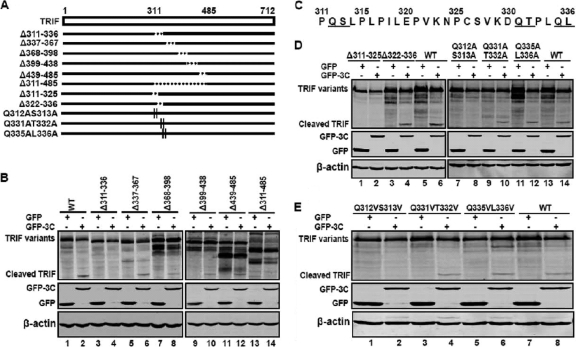
As TRIF cleavage produced a 45-kDa product, we inferred that the cleavage site(s) might fall in the central region. To test this, we analyzed a series of TRIF mutants which had deletions from amino acid 311 to 485 (Fig. 6A). These mutants were expressed along with a control GFP or GFP-3C in 293T cells. Cell lysates were subjected to Western blot analysis. As illustrated in Fig. 6B, wild-type TRIF was cleaved when ectopically expressed with GFP-3C, resulting in a 45-kDa species (lane 2). Similarly, the TRIF deletion mutants Δ337-367, Δ368-398, Δ399-438, and Δ439-485 were cleaved in the presence of GFP-3C (lanes 6, 8, 10, and 12). In contrast, Δ311-336 and Δ311-485 were resistant to the 3C cleavage (lanes 4 and 14). Thus, the TRIF cleavage site may sit between amino acids 311 and 336 (Fig. 6C). This region contains three amino acid pairs, Q312-S313, Q331-T332, and Q335-L336, which resemble a signature Q-G sequence of other picornavirus 3C proteolytic sites.
Fig. 6.
Mapping of the putative TRIF cleavage site. (A) Schematic representation of wild-type TRIF and its derivatives. The open bar denotes wild-type TRIF, with amino acid numbers shown. Solid lines underneath represent various TRIF variants. Broken lines indicate amino acid deletions. Short vertical lines indicate amino acid substitutions. (B) TRIF cleavage in mutants with amino acids 311 to 485 deleted. 293T cells were transfected with wild-type TRIF or TRIF mutants as indicated, along with GFP (lanes 1, 3, 5, 7, 9, 11, and 13) or GFP-3C (lanes 2, 4, 6, 8, 10, 12, and 14). At 24 h after transfection, cell lysates were subjected to Western blot analysis with antibodies against TRIF, GFP, or β-actin. (C) Primary sequence of amino acids 311 to 336 within TRIF. The potential cleavage sites are underlined. (D) TRIF cleavage in deletion or substitution mutants with mutations encompassing amino acids 311 to 336. (E) TRIF cleavage in substitution mutants. In both panels D and E, cells were transfected, processed, and analyzed as described for panel B.
To define the putative TRIF cleavage site, we analyzed additional TRIF mutants. As shown in Fig. 6D, like wild-type TRIF, the Δ322-336 mutant was susceptible to 3C-induced cleavage, whereas the Δ311-325 mutant was resistant (lanes 2, 4, and 6). This implies that the TRIF cleavage site is located between amino acids 311 and 325. Consistently, TRIF substitution mutants Q312A S313A (Fig. 6D, lanes 7 and 8) and Q312V S313V (Fig. 6E, lanes 1 and 2) became resistant to 3C. In contrast, Q331A T332A (Fig. 6D, lanes 9 and 10), Q335A L336A (Fig. 6D, lanes 11 and 12), Q331V T332V (Fig. 6E, lanes 3 and 4), and Q335V L336V (Fig. 6E, lanes 5 and 6) remained susceptible to the 3C cleavage. Collectively, these data indicate that the Q312-S313 junction in TRIF is critical for its cleavage mediated by EV71 3C.
3C protease activity is crucial to suppress IFN-β and NF-κB activation mediated by TRIF.
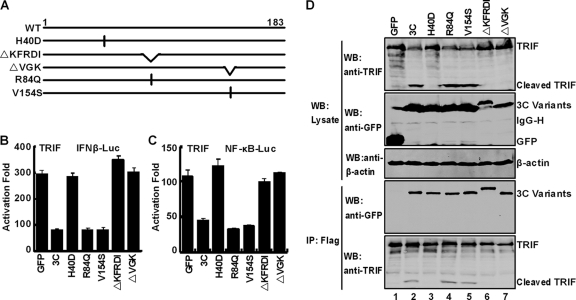
Besides being a viral protease, EV71 3C also possesses RNA binding activity (45). To further study 3C, we characterized a panel of 3C mutants (Fig. 7A). H40D has a single amino acid substitution in the catalytic center, which abrogates the 3C protease activity (45). R84Q and V154S substitution mutants are incapable of binding to RNA. ΔKFRDI has a deletion of amino acids 82 to 86, whereas ΔVGK has a deletion of amino acids 154 to 156. These last two mutants do not exhibit protease and RNA binding activities (45). As illustrated in Fig. 7B, wild-type 3C inhibited the IFN-β promoter activation mediated by TRIF. However, H40D, ΔKFRDI, and ΔVGK were unable to block the IFN-β promoter activation. Interestingly, R84Q or V154S effectively inhibited the TRIF-mediated activation. Similar phenotypes were observed for NF-κB activation (Fig. 7C). It seems that the region containing H40, KFRDI, and VGK motifs is crucial to overcome TRIF-mediated responses. Nevertheless, this activity does not involve R84 or V154, which are required for RNA binding.
Fig. 7.
Mutational analysis of 3C. (A) Schematic diagrams of 3C variants. (B and C) Effects of 3C variants on TRIF-mediated IFN-β promoter activation (B) or NF-κB promoter activation (C). 293T cells were transfected with plasmids encoding TRIF and pIFN β-Luc or NF-κB-Luc, along with GFP or GFP-3C variants. pRL-SV40 was included as an internal control. At 24 h after transfection, cells were harvested to determine luciferase activities. (D) Interaction of 3C variants with TRIF. 293T cells were transfected with Flag-TRIF (lanes 1 to 7) and GFP or GFP-3C variants as indicated. Cell lysates were immunoprecipitated with anti-Flag antibody. Immunoprecipitates and aliquots of cell lysates were subjected to Western blot analysis with antibodies against Flag, GFP, and β-actin, respectively.
We evaluated TRIF cleavage in cells expressing 3C variants by Western blot analysis. The data in Fig. 7D indicate that wild-type 3C induced a small TRIF species of 45 kDa (lane 2), whereas the control GFP did not. Similarly, R84Q and V154S were able to cause TRIF cleavage (lanes 4 and 5). In stark contrast, H40D, ΔKFRDI, and ΔVGK lost their ability to induce TRIF cleavage and a 45-kDa species (lanes 3, 6, and 7). These phenotypes correlated well with the ability of 3C variants to suppress IFN-β and NF-κB activation by TRIF. To assess protein-protein interactions, we performed an immunoprecipitation experiment. As indicated in Fig. 7D (lower panels), wild-type 3C and all mutants coprecipitated with Flag-TRIF (lanes 2 to 7). Thus, H40D, ΔKFRDI, ΔVGK, R84Q, and V154S mutations had no effect on the 3C-TRIF interaction, suggesting that association of 3C with TRIF is not sufficient to display its inhibitory effect. We conclude that the protease function associated with 3C is to block IFN-β and NF-κB activation by TRIF.
DISCUSSION
EV71 causes HFMD and neurological diseases in young children (29). Altered or impaired immunity is thought to facilitate viral pathogenesis (5, 12, 13, 26). In murine infection models, EV71 replicates in the intestine and spreads to the brain stem (6). Although inducing proinflammatory cytokines, EV71 does not elicit type I IFN expression in vivo (27). In infected cells, EV71 inhibits type I IFN induction, where 3C disrupts the RIG-I–IPS-1 complex (22). In this study, we report that EV71 inhibits TLR3-mediated immunity in infected cells through a different mechanism. In doing so, EV71 3C associates with and induces TRIF cleavage. These observations suggest that EV71 3C targets multiple components of the innate immune pathways, leading to the inhibition of type I IFN responses.
TLR3 plays a role in controlling picornaviruses, such as coxsackievirus and encephalomyocarditis virus (EMCV) (33, 41, 48). Its link to EV71 has not been reported previously. The observed inhibition of IRF3 phosphorylation and ISG induction by poly(I · C) in EV71-infected cells suggests a link between EV71 and TLR3 signaling. This activity appears to require the 3C protein. Nevertheless, the impact of this inhibition on EV71 infection remains to be established in vivo. Intriguingly, EV71 enhanced IL-6 expression in infected cells. These phenotypes mirrored those seen in mouse infection models (20, 27). The mechanism by which EV71 stimulates IL-6 induction is not known. We suspect that inhibition of type I IFN responses by EV71 may provide a growth advantage that stimulates inflammatory cytokine production. Given that 3C suppressed IL-6 induction, a separate factor or pathway might be involved to offset its inhibitory effect upon EV71 infection.
TRIF mediates IRF3 activation by associating with TBK1 via its amino terminus (30). In this process, TRAF3 and TNAK-associated protein 1 act as a bridge (11, 34, 43). On the other hand, TRIF activates NF-κB through TRAF6 and RIP, which requires both the amino terminus and the carboxyl terminus (14, 31). Additionally, TRIF binds to TLR3 via its TIR domain in the carboxyl terminus (17). A central question then arises as to how EV71 3C works. When ectopically expressed, 3C induced TRIF cleavage. This was also seen upon EV71 infection. EV71 3C formed a complex with TRIF. Accordingly, expression of 3C led to the inhibition of TRIF-induced IFN-β and NF-κB activation. Two models may explain these observations. A simple one is that TRIF cleavage by 3C may inactivate TRIF. Alternatively, 3C may associate with TRIF and impose a physical barrier that disrupts protein-protein interactions required to activate IRF3 or NF-κB. Analysis of variants of EV71 3C revealed that H40D, ΔKFRDI, and ΔVGK failed to inhibit IFN-β and NF-κB activation by TRIF. While associated with TRIF, these mutants were unable to induce TRIF cleavage. These experimental data favor the model where the 3C protease activity appears to be indispensable.
It is noteworthy that there is a putative 3C cleavage site at the Q312-S313 pair within TRIF. When expressed, wild-type 3C induced a 45-kDa species that represents the amino terminus of TRIF. This was not inhibited by the caspase inhibitor Z-VAD. Removal of amino acids 311 to 336 from TRIF inhibited the production of the 3C-induced 45-kDa species. Among the three amino acid pairs Q312-S313, Q331-T332, and Q335-L336 in this region, only the Q312-S313 pair was affected by mutations that abrogated TRIF cleavage. In this regard, it is notable that picornavirus 3C usually cleaves at amino acid pairs containing Q and G (21, 38). Alternative sites exist, such as the Q-S pair recognized by the 3C proteins of poliovirus and EMCV (21, 38). Although amino acid sequences surrounding these scissile sites vary, an alanine residue is preferred at the P4 position, the fourth amino acid residue preceding the cleavage site (36). TRIF bears three QG pairs, i.e., Q38-G39, Q190-G191, and Q480-G481. Nevertheless, mutations in these sites had no visible effect on TRIF cleavage (data not shown). We speculate that if TRIF is a cellular substrate of EV71 3C, the Q312-S313 pair may be a preferred site since an alanine residue (A309) sits at the P4 position. This specificity is probably determined by context or the accessibility of the cleavage site. Work to address this issue is in progress.
Picornaviruses possess 3C proteins that exhibit considerable amino acid sequence similarity (42). Although inhibiting innate antiviral immunity, they function differentially. It has been reported that poliovirus 3C induces MDA5 degradation through apoptosis (3). This is dependent on caspase and proteosome activity but not on the 3C protease activity (3). However, the 3C proteins encoded by poliovirus, rhinoviruses, echovirus, and encephalomyocarditis virus act to cleave RIG-I (4, 37). The 3C precursor (3ABC) of hepatitis A virus cleaves IPS-1, whereas the 3C protein of EV71 associates with RIG-I and prevents its interaction with IPS-1 (22, 52). Recent evidence suggests that the 3C protease of coxsackievirus B cleaves both TRIF and IPS-1 (32). It also is notable that the NS3/4A protease of hepatitis C virus disrupts TLR3 signaling by cleaving TRIF (23). The TRIF cleavage by EV71 3C observed in this work suggests that it may be a mechanism to impair TLR3-mediated immunity. Additional work is required to understand the mechanisms that regulate picornavirus 3C proteins.
ACKNOWLEDGMENTS
We thank Zhengfan Jiang for providing 293/TLR3 cells. We also thank Sheng Cui for reagents and helpful suggestions.
This work was supported by grants from the National Basic Research Program of China (973 Project, 2011CB504903), the Grant for Outstanding Young Talents of New Century from the Chinese Ministry of Education (NCET-07-0506), the Institute of Pathogen Biology, Chinese Academy of Medical Sciences (2009IPB112), and the U.S. National Institute of Allergy and Infectious Diseases (AI092230).
Footnotes
Published ahead of print on 22 June 2011.
REFERENCES
- 1. AbuBakar S., et al. 1999. Identification of enterovirus 71 isolates from an outbreak of hand, foot and mouth disease (HFMD) with fatal cases of encephalomyelitis in Malaysia. Virus Res. 61:1–9 [DOI] [PubMed] [Google Scholar]
- 2. AbuBakar S., et al. 2009. Enterovirus 71 outbreak, Brunei. Emerg. Infect. Dis. 15:79–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barral P. M., et al. 2007. MDA-5 is cleaved in poliovirus-infected cells. J. Virol. 81:3677–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barral P. M., Sarkar D., Fisher P. B., Racaniello V. R. 2009. RIG-I is cleaved during picornavirus infection. Virology 391:171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang L. Y., et al. 2007. Neurodevelopment and cognition in children after enterovirus 71 infection. N. Engl. J. Med. 356:1226–1234 [DOI] [PubMed] [Google Scholar]
- 6. Chen Y. C., et al. 2004. A murine oral enterovirus 71 infection model with central nervous system involvement. J. Gen. Virol. 85:69–77 [DOI] [PubMed] [Google Scholar]
- 7. Feng Z. D., Cerveny M., Yan Z. P., He B. 2007. The VP35 protein of Ebola virus inhibits the antiviral effect mediated by double-stranded RNA dependent protein kinase PKR. J. Virol. 81:182–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fitzgerald K. A., et al. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491–496 [DOI] [PubMed] [Google Scholar]
- 9. Gilbert G. L., et al. 1988. Outbreak of enterovirus 71 infection in Victoria, Australia, with a high incidence of neurologic involvement. Pediatr. Infect. Dis. J. 7:484–488 [DOI] [PubMed] [Google Scholar]
- 10. Gitlin L., et al. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U. S. A. 103:8459–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hacker H., et al. 2006. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 439:204–207 [DOI] [PubMed] [Google Scholar]
- 12. Ho M., et al. 1999. An epidemic of enterovirus 71 infection in Taiwan. Taiwan Enterovirus Epidemic Working Group. N. Engl. J. Med. 341:929–935 [DOI] [PubMed] [Google Scholar]
- 13. Huang C. C., et al. 1999. Neurologic complications in children with enterovirus 71 infection N. Engl. J. Med. 341:936–942 [DOI] [PubMed] [Google Scholar]
- 14. Jiang Z., Mak T. W., Sen G., Li X. 2004. Toll-like receptor 3-mediated activation of NF-kappaB and IRF3 diverges at Toll-IL-1 receptor domain-containing adapter inducing IFN-beta. Proc. Natl. Acad. Sci. U. S. A. 101:3533–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang Z., et al. 2003. Poly(I-C)-induced Toll-like receptor 3 (TLR3)-mediated activation of NFkappa B and MAP kinase is through an interleukin-1 receptor-associated kinase (IRAK)-independent pathway employing the signaling components TLR3-TRAF6-TAK1-TAB2-PKR. J. Biol. Chem. 278:16713–16719 [DOI] [PubMed] [Google Scholar]
- 16. Kato H., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105 [DOI] [PubMed] [Google Scholar]
- 17. Kawai T., Akira S. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131–137 [DOI] [PubMed] [Google Scholar]
- 18. Kawai T., et al. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988 [DOI] [PubMed] [Google Scholar]
- 19. Kehle J., Roth B., Metzger C., Pfitzner A., Enders G. 2003. Molecular characterization of an enterovirus 71 causing neurological disease in Germany. J. Neurovirol. 9:126–128 [DOI] [PubMed] [Google Scholar]
- 20. Khong W. X., Foo D. G., Trasti S. L., Tan E. L., Alonso S. 2011. Sustained high levels of interleukin-6 contribute to the pathogenesis of enterovirus 71 in a neonate mouse model. J. Virol. 85:3067–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kundu P., Raychaudhuri S., Tsai W., Dasgupta A. 2005. Shutoff of RNA polymerase II transcription by poliovirus involves 3C protease-mediated cleavage of the TATA-binding protein at an alternative site: incomplete shutoff of transcription interferes with efficient viral replication. J. Virol. 79:9702–9713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lei X. B., et al. 2010. The 3C protein of enterovirus 71 inhibits retinoid acid-inducible gene I-mediated interferon regulatory factor 3 activation. J. Virol. 84:8051–8061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li K., et al. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. U. S. A. 102:2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li L., et al. 2005. Genetic characteristics of human enterovirus 71 and coxsackievirus A16 circulating from 1999 to 2004 in Shenzhen, People's Republic of China. J. Clin. Microbiol. 43:3835–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li M. L., et al. 2002. The 3C protease activity of enterovirus 71 induces human neural cell apoptosis. Virology 293:386–395 [DOI] [PubMed] [Google Scholar]
- 26. Lin Y. W., et al. 2009. Lymphocyte and antibody responses reduce enterovirus 71 lethality in mice by decreasing tissue viral loads. J. Virol. 83:6477–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu M. L., et al. 2005. Type I interferons protect mice against enterovirus 71 infection. J. Gen. Virol. 86:3263–3269 [DOI] [PubMed] [Google Scholar]
- 28. McMinn P., Stratov I., Nagarajan L., Davis S. 2001. Neurological manifestations of enterovirus 71 infection in children during an outbreak of hand, foot, and mouth disease in Western Australia. Clin. Infect. Dis. 32:236–242 [DOI] [PubMed] [Google Scholar]
- 29. McMinn P. C. 2002. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol. Rev. 26:91–107 [DOI] [PubMed] [Google Scholar]
- 30. McWhirter S. M., et al. 2004. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 101:233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meylan E., et al. 2004. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat. Immunol. 5:503–507 [DOI] [PubMed] [Google Scholar]
- 32. Mukherjee A., et al. 2011. The coxsackievirus B 3Cpro protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog. 7:e1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Negishi H., et al. 2008. A critical link between Toll-like receptor 3 and type II interferon signaling pathways in antiviral innate immunity. Proc. Natl. Acad. Sci. U. S. A. 105:20446–20451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oganesyan G., et al. 2006. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 439:208–211 [DOI] [PubMed] [Google Scholar]
- 35. Oshiumi H., Matsumoto M., Funami K., Akazawa T., Seya T. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 4:161–167 [DOI] [PubMed] [Google Scholar]
- 36. Pallai P. V., et al. 1989. Cleavage of synthetic peptides by purified poliovirus 3C proteinase. J. Biol. Chem. 264:9738–9741 [PubMed] [Google Scholar]
- 37. Papon L., et al. 2009. The viral RNA recognition sensor RIG-I is degraded during encephalomyocarditis virus (EMCV) infection. Virology 393:311–318 [DOI] [PubMed] [Google Scholar]
- 38. Parks G. D., Baker J. C., Palmenberg A. C. 1989. Proteolytic cleavage of encephalomyocarditis virus capsid region substrates by precursors to the 3C enzyme. J. Virol. 63:1054–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peters K. L., Smith H. L., Stark G. R., Sen G. C. 2002. IRF-3-dependent, NFkappa B- and JNK-independent activation of the 561 and IFN-beta genes in response to double-stranded RNA. Proc. Natl. Acad. Sci. U. S. A. 99:6322–6327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rebsamen M., Meylan E., Curran J., Tschopp J. 2008. The antiviral adaptor proteins Cardif and Trif are processed and inactivated by caspases. Cell Death Differ. 15:1804–1811 [DOI] [PubMed] [Google Scholar]
- 41. Richer M. J., Lavallée D. J., Shanina I., Horwitz M. S. 2009. Toll-like receptor 3 signaling on macrophages is required for survival following coxsackievirus B4 infection. PLoS One 4:e4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ryan M. D., Flint M. 1997. Virus-encoded proteinases of the picornavirus super-group. J. Gen. Virol. 78:699–723 [DOI] [PubMed] [Google Scholar]
- 43. Sasai M., et al. 2005. NF-kB-activated kinase-associated protein 1 participates in TLR3/Toll-IL-1 homology domain-containing adapter molecule-1 mediated IFN regulatory factor 3 activation. J. Immunol. 174:27–30 [DOI] [PubMed] [Google Scholar]
- 44. Sharma S., tenOever B. R., Grandvaux N., Zhou G. P., Lin R., Hiscott J. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148–1151 [DOI] [PubMed] [Google Scholar]
- 45. Shih S. R., et al. 2004. Mutations at KFRDI and VGK domains of enterovirus 71 3C protease affect its RNA binding and proteolytic activities. J. Biomed. Sci. 11:239–248 [DOI] [PubMed] [Google Scholar]
- 46. Sim A. C., Luhur A., Tan T. M., Chow V. T., Poh C. L. 2005. RNA interference against enterovirus 71 infection. Virology 341:72–79 [DOI] [PubMed] [Google Scholar]
- 47. Verpooten D., Ma Y., Hou S., Yan Z., He B. 2009. Control of TANK-binding kinase 1-mediated signaling by the gamma(1)34.5 protein of herpes simplex virus 1. J. Biol. Chem. 284:1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Q., et al. 2009. Role of double-stranded RNA pattern recognition receptors in rhinovirus-induced airway epithelial cell responses. J. Immunol. 183:6989–6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weng K. F., Li M. L., Huang C. T., Shih S. R. 2009. Enterovirus 71 3C protease cleaves a novel target CstF-64 and inhibits cellular polyadenylation. PLoS Pathog. 5:e1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yamamoto M., et al. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301:640–643 [DOI] [PubMed] [Google Scholar]
- 51. Yang F., et al. 2009. Enterovirus 71 outbreak in the People's Republic of China in 2008. J. Clin. Microbiol. 47:2351–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang Y., et al. 2007. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc. Natl. Acad. Sci. U. S. A. 104:7253–7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yoneyama M., et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737 [DOI] [PubMed] [Google Scholar]