Abstract
The life cycle of hepatitis C virus (HCV) is highly dependent on cellular factors. Using small interfering RNA (siRNA) library screening, we identified peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (Pin1) as a host factor involved in HCV propagation. Here we demonstrated that silencing of Pin1 expression resulted in decreases in HCV replication in both HCV replicon cells and cell culture-grown HCV (HCVcc)-infected cells, whereas overexpression of Pin1 increased HCV replication. Pin1 interacted with both the NS5A and NS5B proteins. However, Pin1 expression was increased only by the NS5B protein. Both the protein binding and isomerase activities of Pin1 were required for HCV replication. Juglone, a natural inhibitor of Pin1, inhibited HCV propagation by inhibiting the interplay between the Pin1 and HCV NS5A/NS5B proteins. These data indicate that Pin1 modulates HCV propagation and may contribute to HCV-induced liver pathogenesis.
INTRODUCTION
Hepatitis C virus (HCV) is a major etiologic agent of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (HCC) worldwide. This virus is the sole member of the genus Hepacivirus within the family Flaviviridae (6, 9, 15, 28). HCV is an enveloped virus with a positive-sense, single-stranded RNA genome of ∼9.6 kb. The HCV genome encodes a single precursor polyprotein, which is cleaved by both cellular and viral proteases to generate three structural (core, E1, and E2) and seven nonstructural (p7; NS2 to NS5B) proteins. Although HCV is a highly prevalent pathogen, no protective vaccine is available yet. Current standard therapy is pegylated alpha interferon (IFN-α) combined with ribavirin. However, this therapy shows some side effects and results in a sustained virological response in only a small portion of patients. Thus, there is an urgent need to develop more-effective therapeutic strategies for HCV-associated chronic hepatitis.
Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (Pin1) was first discovered in a screen for molecules regulating mitosis (34). Pin1 consists of 163 amino acids and contains two functional domains: the N-terminal WW binding domain and the C-terminal peptidyl-prolyl isomerase domain (12, 13, 32, 33). The N-terminal WW binding domain is responsible for binding to specific proteins that are phosphorylated at Ser/Thr-Pro motifs, whereas the C-terminal isomerase domain promotes the cis-trans isomerization of the bound peptide. Such conformational changes have significant effects on the phosphorylation status, subcellular localization, protein stability, and functions of many Pin1 substrates (12, 13, 32, 33). Accordingly, Pin1 plays important roles in many cellular events, including cell cycle progression, cell proliferation, transcriptional regulation, and neoplastic transformation. This protein has also been linked to several diseases, such as cancer, Alzheimer's disease, and asthma. Pin1 is overexpressed in many human cancers, including HCC (11); it has been found to be overexpressed in more than 50% of HCCs. All cases with Pin1 overexpression also showed β-catenin accumulation, and 68% of cases showed concomitant β-catenin and cyclin D1 accumulation (16). Furthermore, overexpression of Pin1 in a nontransformed human liver cell line leads to hepatocyte transformation, and inhibition of Pin1 expression suppresses HCC tumorigenesis (18). It has been reported recently that Pin1 interacts with a specific serine-proline motif of hepatitis B virus (HBV) X protein (HBx) to enhance hepatocarcinogenesis in HBV patients (17). In the present study, we demonstrate for the first time that Pin1 interacts directly with the HCV NS5A and NS5B (NS5A/5B) proteins and plays unique roles in HCV replication. In addition, juglone (5-hydroxy-1,4-naphthoquinone), a natural inhibitor of Pin1, impairs the interaction between Pin1 and the HCV NS5A/5B proteins and inhibits HCV propagation. Therefore, Pin1 may be a potential target for HCV treatment.
MATERIALS AND METHODS
Plasmids and DNA transfection.
Plasmids expressing Myc-tagged NS4B, Myc-tagged NS5A, and Myc-tagged NS5B have been described previously (3, 19). Full-length human Pin1 cDNA was amplified from the pCNS-D2-Pin1 plasmid (21C Frontier Human Gene Bank) and was subcloned into the pGEX-4T1 (Amersham Biosciences) and p3×Flag-CMV10 (Sigma-Aldrich) vectors to generate the GST-Pin1 and Flag-Pin1 expression plasmids, respectively. Pin1 mutants were generated by site-directed mutagenesis (Stratagene) using the primers listed in Table 1 according to the manufacturer's instructions. Small interfering RNA (siRNA)-resistant mutant Pin1 contains two silent mutations in the siRNA binding site. To generate siRNA-resistant binding-defective mutant Pin1 (13) and siRNA-resistant isomerase-inactive mutant Pin1 (31), the substitution mutations S16A and C113A, respectively, were introduced into siRNA-resistant mutant Pin1. For the cloning of human cyclophilin A (CypA) and CypB (mature form), total RNAs were extracted from Huh7.5 cells and were used for reverse transcription-PCR (RT-PCR) with the primer sets CypA-F/CypA-R and CypB-F/CypB-R (Table 1). PCR products were inserted into the EcoRI and BamHI sites of plasmid p3×Flag-CMV10. For DNA transfection, cells were transfected with the expression plasmid by using a polyethyleneimine reagent (Sigma-Aldrich) as we described previously (19).
Table 1.
List of primers used in this study
| Primer | Sequence | Purpose |
|---|---|---|
| GST-Pin1-F | CGCGAATTCATGGCGGACGAG | Cloning of Pin1 into the pGEX4T1 vector |
| GST-Pin1-R | TCCCTCGAGTCACTCAGTGCG | |
| Flag-Pin1-F | GGAGAATTCAATGGCGGACGAG | Cloning of Pin1 into the p3×Flag-CMV10 vector |
| Flag-Pin1-R | CCGGGATCCTCACTCAGTGCG | |
| AD-Pin1-F | AAGGATCCATGGCGGACGAGGA | Cloning of Pin1 into the pCMV-AD vector |
| AD-Pin1-R | CCGAATTCTCACTCAGTGCGGAG | |
| BD-NS4B-F | GGGATCCGCCTCACAACTTCCTTAC | Cloning of NS4B into the pCMV-BD vector |
| BD-NS4B-R | GCGAATTCTTAGCATGGCGTAGAGCAG | |
| BD-NS5A-F | GGAAGCTTGTCCGGCTCGTGGCTAA | Cloning of NS5A into the pCMV-BD vector |
| BD-NS5A-R | AGTCTAGACTAGCAGCAGACGACATCC | |
| BD-NS5B-F | GCGAATTCTCAATGTCCTATACGTG | Cloning of NS5B into the pCMV-BD vector |
| BD-NS5B-R | AGTCTAGACTATCGGTTGGGGAGCA | |
| 5′NTR-F | TGAGTGTCGTACAGCCTCCA | Quantitative real-time PCR |
| 5′NTR-R | ACGCTACTCGGCTAGCAGTC | |
| qPin1-F | ATCAAGTCGGGAGAGGAG | Quantitative real-time PCR |
| qPin1-R | GCGTCTTCAAATGGCTTCT | |
| qGAPDH-F | CGCTCTCTGCTCCTCCTGTTC | Quantitative real-time PCR |
| qGAPDH-R | CGCCCAATACGACCAAATCCG | |
| CypA-F | GCG AAT TCA ATG GTC AAC CCC A | Cloning of human cyclophilin A into the p3×Flag-CMV10 vector |
| CypA-R | CGG GAT CCT TAT TCG AGT TGT CCA | |
| CypB-F | GCG AAT TCA CTG CTG CCG GGA CCT T | Cloning of human cyclophilin B into the p3×Flag-CMV10 vector |
| CypB-R | CGG GAT CCC TAC TCC TTG GCG ATG | |
| Pin1-SR-F | TGAGCCGCAGCTCAGGGAGAGTGTACTACTTCAAC | Generation of siRNA-resistant mutant of Pin1 |
| Pin1-SR-R | GTTGAAGTAGTACACTCTCCCTGAGCTGCGGCTCA | |
| Pin1-S16A-F | GGAGAAGCGCATGGCCCGCAGCTCAGGC | Generation of binding-defective mutant of Pin1 |
| Pin1-S16A-R | GCCTGAGCTGCGGGCCATGCGCTTCTCC | |
| Pin1-C113A-F | TCACAGTTCAGCGACGCCAGCTCAGCCAAGGC | Generation of isomerase-inactive mutant of Pin1 |
| Pin1-C113A-R | GCCTTGGCTGAGCTGGCGTCGCTGAACTGTGA |
Cell culture.
All cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum-1% penicillin-streptomycin under 5% CO2 at 37°C. HCV subgenomic replicon (genotype 1b) cells and cells stably expressing NS5A were grown as reported previously (3).
Chemicals and antibodies.
Juglone, a natural inhibitor of Pin1, was purchased from Calbiochem. Antibodies were purchased from the following sources: anti-Pin1 antibodies from Cell Signaling and Santa Cruz; anti-Flag antibodies from Sigma-Aldrich; and antibodies against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Myc from Santa Cruz. Both the anti-NS5A and anti-NS5B antibodies have been described elsewhere (3). Both the rabbit anti-core and anti-NS3 antibodies were obtained from Byung-Yoon Ahn (Korea University).
Generation of HCV Jc1 and virus infection.
Infectious HCVs were generated as described previously (8, 21) with minor modifications. Briefly, the pFK-Jc1 plasmid was linearized at the 3′ end of the Jc1 genome by MluI digestion and was purified by phenol-chloroform extraction. RNA transcripts were generated using Ambion MEGAscript T7 kit and were purified with Trizol LS (Invitrogen) according to the manufacturer's protocols. Ten micrograms of in vitro-transcribed genomic Jc1 RNA was mixed with 7.5 × 106 Huh7.5 cells in a 0.4-cm-gap cuvette, and the mixture was electroporated at 300 V and 975 μF using a Bio-Rad GenePulser II electroporator. Cells were gently transferred to complete medium (low-glucose DMEM containing 10% fetal bovine serum [FBS], 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine, 1 mM nonessential amino acids [NEAA], and 10 mM HEPES) and were plated on a 150-mm dish. Twenty-four hours later, the medium was replaced with fresh complete medium to remove dead cells. At 4 days after electroporation, the culture medium was collected, filtered through a 0.45-mm syringe-top filter unit (Millipore), and kept as a virus stock. The Jc1/GND mutant, containing an active-site mutation in NS5B that destroys RNA-dependent RNA polymerase activity, was generated in parallel. The culture supernatant harvested from cells transfected with Jc1/GND mutant RNA was prepared as described above and was used for mock infection. For virus infection experiments, cells were washed twice with phosphate-buffered saline (PBS) and were incubated for 4 h with cell culture-grown HCV (HCVcc). HCVcc-infected cells were washed again with PBS and were further grown in complete medium.
RNA interference.
A human cell cycle regulation siRNA library (ON-TARGETplus) was purchased from Dharmacon (Lafayette, CO). This library contains 131 siRNA pools targeting genes whose functions are crucial for controlling the cell cycle. Each pool consists of 4 siRNA duplexes designed to target different regions of the same target mRNA sequence. A SMARTpool siRNA, when transfected at a concentration of 100 nM, was guaranteed by Dharmacon to silence at least 75% of the expression of the target gene at the mRNA level. siRNAs targeting Pin1 (5′-GCU CAG GCC GAG UGU ACU A-3′) and the universal negative-control siRNA were also purchased from Dharmacon. A siRNA targeting the 5′ nontranslated region (NTR) of Jc1 (5′-CCU CAA AGA AAA ACC AAA CUU-3′) was used as a positive control (22). siRNA transfection was performed using the Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA).
Quantification of HCV RNA.
Both intracellular and extracellular RNAs were isolated from HCVcc-infected cells, cell culture media, or replicon cells using the Trizol LS reagent (Invitrogen, Carlsbad, CA) and were reverse transcribed using the SuperScript Vilo cDNA synthesis kit (Invitrogen, Carlsbad, CA). Quantitative real-time PCR (qRT-PCR) experiments were carried out using an iQ5 multicolor real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA) under the following conditions: 15 min at 95°C, followed by 40 cycles of 95°C for 20 s, 55°C for 20 s, and 72°C for 20 s. Seventy one cycles of 10 s, with 0.5°C temperature increments from 60 to 95°C, were used for the melting curves. The HCV 5′ NTR sequence was cloned into the T-A vector (RBC) and was used as a standard for qRT-PCR. The standards were run in parallel with the samples, and the absolute RNA copy number was determined based on the standard curve.
Determination of virus titers.
Naïve Huh7.5 cells were seeded at 2 × 104/well in 8-well chamber culture slides (Becton Dickinson/Falcon, Franklin Lakes, NJ) with 200 μl of culture medium. Following 24 h of incubation, Huh7.5 cells were inoculated with serial dilutions of cell culture medium harvested from HCVcc-infected cells. At 2 days after inoculation, indirect immunofluorescence was performed for the presence of intracellular core antigen in order to determine the number of focus-forming units (FFU). Each cluster of infected cells identified by staining for NS5A was considered to be a single infectious focus. Virus titers were calculated as focus-forming units per milliliter.
MTT assay.
Host cell viability was determined by using a 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) cell proliferation assay kit (Roche) according to the manufacturer's protocol.
Generation of Pin1-stable cells.
To make cell lines stably expressing Flag-Pin1 (Pin1-stable cell lines), Huh7 cells were transfected with the p3×Flag-CMV10-Pin1 expression plasmid and were cultured for 4 weeks in the presence of 300 μg/ml of G418. Single positive clones were selected by immunoblot analysis using an anti-Flag monoclonal antibody. Huh7 cells transfected with an empty vector (p3×Flag-CMV10 only) were selected as described above and were used as a control.
Immunoblot analysis.
Cell lysates were prepared and immunoblotted as we described previously (3). Briefly, cells were lysed in a cell lysis buffer containing 50 mmol/liter Tris-HCl (pH 7.5), 150 mmol/liter NaCl, 1 mmol/liter EDTA, 1% Nonidet P-40, 10% glycerol, and a protease inhibitor cocktail (Roche) for 20 min on ice. The protein concentration was determined by the Bradford assay (Bio-Rad). Equal amounts of proteins were subjected to either 10% or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were electrotransferred to a nitrocellulose membrane. The membrane was blocked in PBS containing 5% nonfat dry milk for 1 h and was then incubated for 2 h at room temperature with the indicated antibodies in Tris-buffered saline-Tween (20 mmol/liter Tris-HCl [pH 7.5], 500 mmol/liter NaCl, and 0.05% Tween 20). Following two washes in Tris-buffered saline-Tween, the membrane was incubated either with a horseradish peroxidase-conjugated goat anti-rabbit antibody or with a goat anti-mouse antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) in Tris-buffered saline-Tween for 1 h at room temperature. Proteins were detected using an ECL kit (Amersham Biosciences).
GST pulldown assay and coimmunoprecipitation.
The glutathione S-transferase (GST)-Pin1 fusion protein was expressed and purified as we described previously (3). Both in vitro binding and coimmunoprecipitation assays were performed as we described elsewhere (4).
Confocal microscopy.
Either Huh7 or Huh7.5 cells cultured on glass slides were fixed with 4% paraformaldehyde in PBS at room temperature for 30 min. After two washes with PBS, fixed cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min and were incubated with a mouse anti-Pin1 antibody, a mouse anti-Flag antibody, a rabbit anti-NS5A antibody, or a rabbit anti-NS5B antibody for 2 h. After three washes with PBS, cells were further incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse or anti-rabbit IgG (American Qualex, San Clemente, CA) and with tetramethylrhodamine isothiocyanate (TRITC)-conjugated donkey anti-rabbit or anti-mouse IgG (Jackson Immunoresearch Laboratories) for 1 h at room temperature. After two washes with 0.1% Triton X-100 in PBS and three washes with PBS, cells were analyzed using the Zeiss LSM 700 laser confocal microscopy system (Carl Zeiss, Inc., Thornwood, NY). To quantify the colocalization of Pin1 with NS5A and NS5B, immunofluorescence data were analyzed using the Zeiss LSM 700 software tool. The image threshold was obtained for both channels, and the extent of true colocalization in the selected region of interest (yellow pixels in the merged image) was analyzed using the overlap coefficient (colocalization module; Carl Zeiss), with a coefficient of zero indicating no colocalization and a coefficient of 1 indicating complete colocalization. More than 10 cells were analyzed for each experiment.
Luciferase replication assay.
Huh7.5 cells were electroporated with 10 μg of in vitro-transcribed full-length JFH-Luc RNA. At 24 h after electroporation, cells were transfected with 20 nM negative-control, positive-control, or Pin1 siRNA. Luciferase assays were performed as described previously (3) using cells harvested 24 h, 48 h, and 72 h after siRNA transfection.
JEV infection.
BHK-21 cells were either mock infected or infected with Japanese encephalitis virus (JEV) as described previously (36). Equal amounts of total-cell lysates harvested at either 24 h or 48 h were analyzed for JEV protein expression by immunoblotting with the rabbit anti-JEV NS1 antibody as described above.
Statistical analysis.
Data are presented as means ± standard deviations (SD). Student's t test was used for statistical analysis. A P value of <0.05 was considered statistically significant.
RESULTS
Pin1 is a cellular factor required for HCV replication.
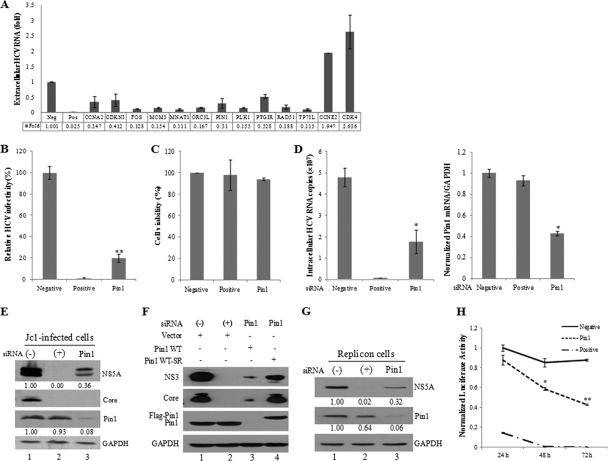
HCV replication and translation depend strongly on the host cell cycle (26). HCV interferes with the host cell cycle and exploits host proteins to complete its life cycle. To identify the cellular proteins required for HCV replication, we screened a siRNA library targeting 131 genes that control the cell cycle in HCVcc-infected cells. Huh7.5 cells were transfected with 20 nM siRNA for 48 h and were then infected with HCV Jc1 for 4 h. At 2 days postinfection, culture supernatants were collected, and extracellular HCV RNAs were quantified by qPCR. Experiments were carried out in duplicate. siRNAs that altered HCV production by an average 2-fold increase or decrease relative to the SD compared with the effect of a universal negative-control siRNA and that exerted no cytotoxicity were selected as hits. From 131 siRNA pools, 13 pools were identified as candidate hits (Fig. 1A). These pools were retested in a secondary screen. Cell viability was measured by the MTT assay. A siRNA targeting the 5′ NTR of HCV was used as a positive control.
Fig. 1.
Pin1 is essential for HCV replication. (A) Huh7.5 cells were transfected with 20 nM siRNAs for 48 h and were then infected with HCV Jc1 for 4 h. At 2 days postinfection, culture supernatants were collected, and extracellular HCV RNAs were quantified by qPCR. From the siRNA library targeting 131 human cell cycle genes, 13 siRNAs that altered (increased or decreased) the level of HCV production 2-fold from that for the negative control were selected. (B) Huh7.5 cells were transfected with 20 nM negative (−), positive (+), or Pin1 siRNA duplexes for 48 h, followed by infection with Jc1 for 4 h. At 2 days postinfection, HCV infectivity was determined by a focus-forming assay. (C) Huh7.5 cells were transfected with the indicated siRNAs. At 96 h after transfection, cell viability was assessed by an MTT assay. (D) Both intracellular RNA (left) and Pin1 mRNA (right) isolated from cells treated as described in the legend to panel B were analyzed by qPCR. (E) Total-cell lysates harvested from cells treated as described in the legend to panel B were immunoblotted with the indicated antibodies. (F) Huh7.5 cells were transfected with the indicated siRNA. At 2 days after siRNA transfection, cells were infected with Jc1 for 4 h and were then transfected with 3 μg of a vector or plasmid expressing wild-type Pin1 (Flag-Pin1 WT) or siRNA-resistant Pin1 (Flag-Pin1 WT-SR). Protein expression was determined by immunoblot analysis at 2 days after transfection. (−), the universal negative-control siRNA; (+), siRNA targeting the 5′ NTR of Jc1 as a positive control. (G) Subgenomic replicon cells were transfected with the indicated siRNAs for 96 h. Protein expression was determined by immunoblot analysis. The band intensity was quantified using ImageJ software. (H) Huh7.5 cells were transfected with in vitro-transcribed HCV-Luc RNA. At 2 days after transfection, cells were transfected with a negative-control, positive-control, or Pin1 siRNA. Luciferase activity was measured at the indicated times. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01) from activity for the negative control.
Among candidate genes, we selected and characterized the gene encoding Pin1, because it has been implicated in many viral infections, including human immunodeficiency virus (HIV) replication (29), HBV-mediated hepatocarcinogenesis (17), and human T cell leukemia virus type 1 (HTLV-1) malignant transformation (7, 20). Silencing of Pin1 expression led to strong inhibition of HCV production (Fig. 1B) with minimal toxicity (Fig. 1C). To determine which step in the HCV life cycle required Pin1, we quantified intracellular viral RNA in Pin1 knockdown cells. As shown in Fig. 1D, the Pin1 mRNA level was significantly suppressed by siRNA treatment. As a result, the intracellular HCV RNA level was also significantly reduced in Pin1 knockdown cells. These results indicate that Pin1 may be involved in HCV replication. To further confirm the effect of Pin1 on HCV replication, we investigated viral protein expression levels in Pin1 knockdown cells. The positive-control siRNA completely blocked NS5A and core expression (Fig. 1E, lane 2). Similarly, silencing of Pin1 impaired HCV protein expression (Fig. 1E, lane 3). To rule out the off-target effect of Pin1 siRNA, we generated a siRNA-resistant Pin1 mutant. As shown in Fig. 1F, exogenous expression of a siRNA-resistant Pin1 mutant, but not of wild-type Pin1, rescued HCV protein expression (lane 4). The role of Pin1 in HCV replication was also examined in Huh7 cells harboring HCV subgenomic replicons. Figure 1G shows that knockdown of Pin1 in HCV replicon cells significantly inhibited NS5A protein expression. It was noteworthy that the Pin1 level in replicon cells was also decreased by positive siRNA treatment compared to negative siRNA treatment. In order to measure viral replication more accurately, the effect of Pin1 siRNA on HCV replication was determined by the JFH-Luc reporter system. As shown in Fig. 1H, luciferase activity was decreased slightly at 24 h after Pin1 siRNA treatment, and was dramatically reduced at 48 to 72 h, in cells transfected with Pin1 siRNA compared to those transfected with the negative-control siRNA. Taken together, these data suggest that Pin1 is required for HCV propagation.
Overexpression of Pin1 enhances HCV propagation.
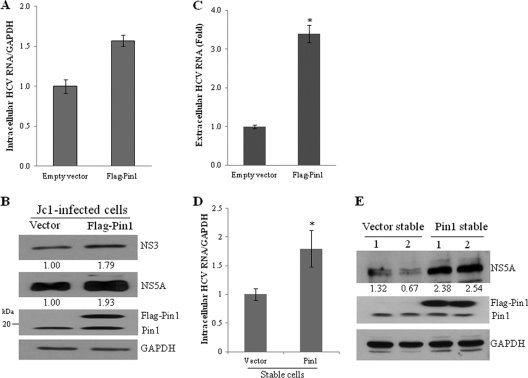
Because silencing of Pin1 suppressed HCV replication, we speculated that overexpression of Pin1 might have a positive effect on HCV replication. To investigate this possibility, Huh7.5 cells infected with Jc1 for 48 h were transiently transfected with Flag-tagged Pin1 and were then harvested at 48 h after transfection. We showed that overexpression of Pin1 increased the levels of intracellular HCV RNA (Fig. 2A), intracellular viral proteins (Fig. 2B), and extracellular HCV RNA (Fig. 2C). We further demonstrated that both intracellular HCV RNA (Fig. 2D) and protein (Fig. 2E) levels were also higher in Jc1-infected Pin1-stable cells than in vector-stable cells. We further found that both HCV RNA and protein levels were increased in other clones of Pin1-stable cells (data not shown), confirming that this phenomenon was not caused by clonal selection of Pin1-stable cells.
Fig. 2.
Overexpression of Pin1 increases HCV propagation. (A) Huh7.5 cells were infected with Jc1 for 48 h and were then transiently transfected with 2 μg of either a vector or the Flag-Pin1 plasmid. At 48 h after transfection, intracellular HCV RNAs were analyzed by qPCR. (B) Total-cell lysates harvested from these cells were immunoblotted with the indicated antibodies. (C) Extracellular RNAs isolated from the culture supernatant were quantified by qPCR. Experiments were carried out in duplicate. Error bars indicate standard deviations. (D) Vector-stable and Pin1-stable cells were infected with Jc1. At 48 h postinfection, intracellular HCV RNAs were quantified by qPCR. The asterisk indicates a significant difference (*, P < 0.05) from the value for the vector control. (E) Two clones each of vector-stable and Pin1-stable cells were infected with Jc1. At 48 h postinfection, total-cell lysates were immunoblotted with the indicated antibodies. The band intensity was quantified using ImageJ software.
Pin1 interacts with HCV NS5A and NS5B proteins.
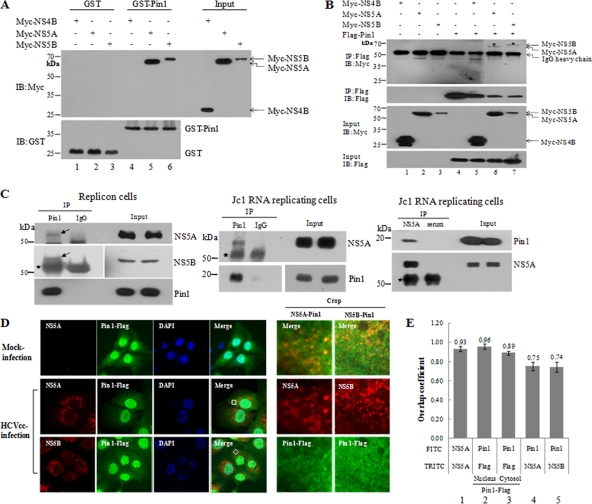
Pin1 is a member of the parvulin family of peptidyl prolyl cis-trans isomerases (PPIases). Among the PPIases that have been studied to date, cyclophilin A (CypA) and CypB have been reported to play essential roles in HCV replication (30, 35). Like other PPIases, Pin1 interacts with phosphorylated proteins through the serine/threonine-proline motif. Because both CypA and CypB interact with the HCV NS5A and NS5B proteins (2, 5, 10, 30, 35), we speculated that Pin1 might upregulate HCV propagation through interaction with either NS5A or NS5B protein. Both NS5A and NS5B are phosphoproteins that contain various phosphorylated serine/threonine-proline motifs; thus, both proteins could be potential substrates of PPIases. We first examined this possibility by in vitro GST pulldown assays using GST and GST-Pin1 fusion proteins expressed in Escherichia coli and Myc-NS4B, Myc-NS5A, and Myc-NS5B proteins expressed individually in HEK293T cells. HEK293T cell extracts containing Myc-tagged NS4B, NS5A, or NS5B were incubated with either purified GST or GST-Pin1. Complexes were precipitated with glutathione beads, and bound proteins were analyzed by immunoblot analysis using anti-Myc antibodies. As shown in Fig. 3A, both the NS5A and NS5B proteins were selectively bound to Pin1 (lanes 5 and 6, respectively), whereas neither GST nor NS4B interacted with Pin1. To confirm the in vitro result, we performed a coimmunoprecipitation assay. Myc-tagged NS4B, NS5A, or NS5B was coexpressed with Flag-tagged Pin1 in HEK293 cells. Cell lysates were immunoprecipitated with an anti-Flag monoclonal antibody, and coprecipitated protein was then detected by immunoblot analysis using an anti-Myc monoclonal antibody. Figure 3B shows that both the NS5A and NS5B proteins interacted specifically with Pin1 in vivo (lanes 6 and 7). We confirmed these results by a mammalian two-hybrid assay (data not shown), which also showed that both NS5A and NS5B interacted specifically with Pin1.
Fig. 3.
Pin1 interacts directly with both the HCV NS5A and NS5B proteins. (A) HEK293T cells were transfected with Myc-NS4B, Myc-NS5A, or Myc-NS5B. At 36 h after transfection, total-cell lysates were harvested and were incubated with either purified GST or GST-Pin1. (Top) Complexes were precipitated with glutathione beads, and bound proteins were analyzed by immunoblotting (IB) using an anti-Myc antibody. (Bottom) The expression of the GST and GST-Pin1 proteins was confirmed with an anti-GST antibody. (B) HEK293T cells were transfected with the indicated combinations of expression plasmids. (Top panel) At 36 h after transfection, cell lysates were immunoprecipitated (IP) with an anti-Flag monoclonal antibody, and bound proteins were then detected by IB analysis. (Lower panels) Protein expression of Myc-NS4B, Myc-NS5A, Myc-NS5B, and Flag-Pin1 in the same cell lysates was also verified by immunoblot analysis. (C) (Left) Total-cell lysates harvested from HCV replicon cells were immunoprecipitated with either an anti-Pin1 antibody or IgG. Bound protein was detected by immunoblotting with an antibody against either NS5A or NS5B. (Center) Huh7.5 cells were electroporated with 10 μg Jc1 RNA and were incubated for 3 days. Cell lysates were immunoprecipitated with either an anti-Pin1 antibody or IgG. Bound protein was immunoblotted with an anti-NS5A antibody. (Right) Reciprocally, the same cell lysates were immunoprecipitated with either a rabbit anti-NS5A antibody or a rabbit control serum. Bound protein was immunoblotted with an anti-Pin1 antibody. The asterisk indicates a heavy chain. (D) Huh7.5 cells were infected with HCV Jc1 for 4 h. At 2 days postinfection, cells were transfected with a plasmid expressing Flag-tagged Pin1. At 24 h after transfection, cells were fixed in 4% paraformaldehyde, and immunofluorescence staining was performed by using an anti-Pin1 monoclonal antibody and fluorescein isothiocyanate-conjugated goat anti-mouse IgG to detect Pin1 (green) and a rabbit anti-NS5A or anti-NS5B antibody and TRITC-conjugated goat anti-rabbit IgG to detect NS5A (red) or NS5B (red). Dual staining showed colocalization of Pin1 and NS5A as yellow fluorescence in the merged image. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to label nuclei. The boxed area in the merged image is enlarged on the right (crop). (E) Huh7.5 cells infected with HCV Jc1 were fixed, incubated with a rabbit anti-NS5A antibody, and further incubated with either FITC- or TRITC-conjugated anti-rabbit antibodies (bar 1). Flag-Pin1-stable cells were fixed, incubated with both a mouse anti-Pin1 and a rabbit anti-Flag antibody, and further incubated with both TRITC-conjugated anti-mouse and FITC-conjugated anti-rabbit antibodies (bars 2 and 3). Colocalization of NS5A (bar 4) and NS5B (bar 5) with Pin1 was quantified by Manders' overlap coefficient as described in Materials and Methods.
To further demonstrate protein interplay between Pin1 and NS5A/NS5B in a more authentic system, we performed immunoprecipitation using HCV replicon cells and HCVcc-infected cells. As shown in Fig. 3C, Pin1 interacted with NS5A/NS5B in HCV replicon cells (left). We further demonstrated this interaction in HCVcc-infected Huh7.5 cells by reciprocal immunoprecipitation assays (Fig. 3C, center and right). Both in vitro and in vivo protein-protein interaction data suggest that Pin1 may colocalize with NS5A/NS5B. To investigate this possibility, HCVcc-infected cells were examined for the subcellular localization of Pin1 and HCV proteins by confocal microscopy. Figure 3D shows that Pin1 was localized mainly in the nucleus and partly in the cytoplasm in hepatoma cells and that the cellular distribution of Pin1 was not affected by viral proteins. Both the NS5A and NS5B proteins were localized in the cytoplasm, and dual staining showed colocalization of NS5A and NS5B with Pin1 in the cytoplasm as yellow fluorescence in HCVcc-infected cells. To verify this result, we quantified the colocalization of Pin1 with HCV proteins by Manders' overlap coefficient as reported previously (14). By use of this method and labeling of NS5A with two different probes (FITC and TRITC), the overlap coefficient of NS5A is ∼0.93 (Fig. 3E, bar 1). Because Pin1 is localized primarily in the nucleus and partly in the cytoplasm, we determined the overlap coefficients of Pin1 in two different cellular compartments. Dual-staining data showed that the overlap coefficient of Pin1 in the nucleus is 0.96 ± 0.27 (Fig. 3E, bar 2), while that of Pin1 in the cytoplasm is 0.89 ± 0.20 (Fig. 3E, bar 3). The overlap coefficients of Pin1/NS5A and Pin1/NS5B were 0.75 and 0.74, respectively, indicating that these proteins were colocalized in the cytoplasm (Fig. 3E, bars 4 and 5). Taken together, these data indicate that Pin1 specifically interacts with NS5A and NS5B both in vitro and in vivo.
NS5B but not NS5A increases Pin1 expression.
It has been reported previously that Pin1 is overexpressed in many types of cancer, including HCC (11). To investigate whether Pin1 could be upregulated by HCV infection, we examined the expression level of Pin1 protein in HCV-infected cells. As shown in Fig. 4A (left), the level of Pin1 protein expression was elevated in wild-type HCV-infected cells at 4 days postinfection. This was confirmed by scanning of the blot images and measurement of the protein pixel intensity (Fig. 4A, center). We also showed that the Pin1 mRNA level was significantly increased in wild-type virus-infected cells (Fig. 4A, right). To confirm these results, we further examined Pin1 expression levels in HCV subgenomic replicon cells. Indeed, both the protein and mRNA levels of Pin1 were significantly higher in replicon cells than in IFN-cured cells (Fig. 4B). These results suggested that HCV nonstructural proteins might induce Pin1 expression. Since Pin1 interacted directly with NS5A and NS5B proteins, we speculated that NS5A and NS5B proteins might be responsible for Pin1 induction. To determine which protein is responsible for the Pin1 increase, Huh7 cells were transfected with either a Myc-tagged NS5A or a Myc-tagged NS5B expression plasmid, and the Pin1 level was then analyzed by immunoblot analysis. As shown in Fig. 4C, the Pin1 protein level was significantly increased in cells expressing NS5B protein (lane 3). Although the protein expression level of NS5A was higher than that of NS5B (Fig. 4C, lane 2 versus lane 3), the Pin1 protein level was not changed by NS5A protein. To further examine whether another positive-strand RNA virus could increase Pin1 expression, we investigated JEV-infected BHK cells. The JEV infection was confirmed by immunoblot analysis using a rabbit anti-JEV NS1 antibody (36). However, JEV infection did not increase Pin1 expression (data not shown), indicating that this phenomenon was HCV specific.
Fig. 4.
NS5B but not NS5A increases Pin1 expression. (A) (Left) Huh7.5 cells were either mock infected or infected with HCV Jc1 for 4 h. Total-cell lysates harvested at 4 days postinfection were immunoblotted with the indicated antibodies. (Center) Blot images were scanned, and protein pixel intensity was measured using ImageJ software. (Right) Total RNAs were extracted, and qPCR was performed to quantify the Pin1 mRNA level. Data from two independent experiments were quantified. The asterisk indicates a significant difference (P < 0.05) between mock-infected and HCV Jc1-infected cells. (B) (Left) Equal amounts of cell lysates harvested from either IFN-cured or subgenomic replicon cells were immunoblotted with the indicated antibodies. (Center) Blot images were scanned, and the protein pixel intensity was measured using ImageJ software. (Right) Total RNAs were extracted, and qPCR was performed to quantify the Pin1 mRNA level. Data from two independent experiments were quantified. Asterisks indicate a significant difference (P < 0.05) between IFN-cured and replicon cells. (C) Huh7 cells were transfected with either a vector, Myc-tagged NS5A, or Myc-tagged NS5B. At 48 h after transfection, protein expression was determined by immunoblot analysis.
Both the binding and isomerase activities of Pin1 are essential for HCV replication.
Our data raised the important question of whether the interactions between Pin1 and HCV proteins play any role in HCV replication. To answer this question, we constructed siRNA-resistant mutant Pin1 (Pin1 WT-SR), siRNA-resistant binding-defective mutant Pin1 (Pin1 S16A-SR), and siRNA-resistant isomerase-inactive mutant Pin1 (Pin1 C113A-SR). We first investigated whether these Pin1 mutants could interact with the NS5A and NS5B proteins. Huh7.5 cells were transfected with the combinations of expression plasmids shown in Fig. 5. At 48 h after transfection, cell lysates were immunoprecipitated with an anti-Flag antibody, and bound proteins were then detected by immunoblot analysis using an anti-Myc antibody. As shown in Fig. 5A, Pin1 S16A-SR no longer interacted with the NS5A and NS5B proteins (lanes 4 and 5), whereas Pin1 C113A-SR retained binding capability (lanes 6 and 7). To investigate the effects of the binding and isomerase activities of Pin1 on HCV propagation, Huh7.5 Pin1 knockdown cells were infected with Jc1 and were then transfected with a plasmid expressing either wild-type Pin1, Pin1 S16A-SR, Pin1 C113A-SR, or Pin1 WT-SR. Overexpression of Pin1 WT-SR in HCV-infected cells rescued intracellular HCV RNA expression (Fig. 5B), HCV protein expression (Fig. 5C, lane 6), and virus titers (Fig. 5D) in Pin1 knockdown cells. However, neither Pin1 S16A-SR nor Pin1 C113A-SR overexpression in Pin1 knockdown cells was able to rescue viral RNA or protein expression (Fig. 5B to D), indicating that both the binding and isomerase activities of Pin1 are required for HCV propagation.
Fig. 5.
Both the binding and isomerase activities of Pin1 are essential for HCV replication. (A) Huh7.5 cells were transfected with the indicated combinations of expression plasmids. (Top panel) At 48 h after transfection, cell lysates were immunoprecipitated (IP) with an anti-Flag monoclonal antibody, and bound proteins were then detected by an immunoblot (IB) assay. (Lower panels) Protein expression in the same cell lysates was verified with the indicated antibodies. Pin1 WT-SR, siRNA-resistant mutant Pin1; Pin1 S16A-SR, siRNA-resistant binding-defective mutant Pin1; Pin1 C113A-SR, siRNA-resistant isomerase-inactive mutant Pin1. The asterisk indicates the IgG heavy chain. (B) Huh7.5 cells were transfected with the indicated siRNA. At 2 days after siRNA transfection, cells were infected with Jc1 for 4 h and were then transfected with 2 μg of a plasmid expressing wild-type Pin1 (Pin1 WT), Pin1 S16A-SR, Pin1 C113A-SR, or Pin1 WT-SR. Intracellular RNAs isolated at 2 days after Pin1 transfection were analyzed by qPCR. (C) Protein expression levels of both HCV and host cells were determined by immunoblot analysis using the indicated antibodies. (D) The effects of Pin1 mutants on HCV infectivity were determined by focus-forming assays. Experiments were carried out in duplicate. Error bars indicate standard deviations.
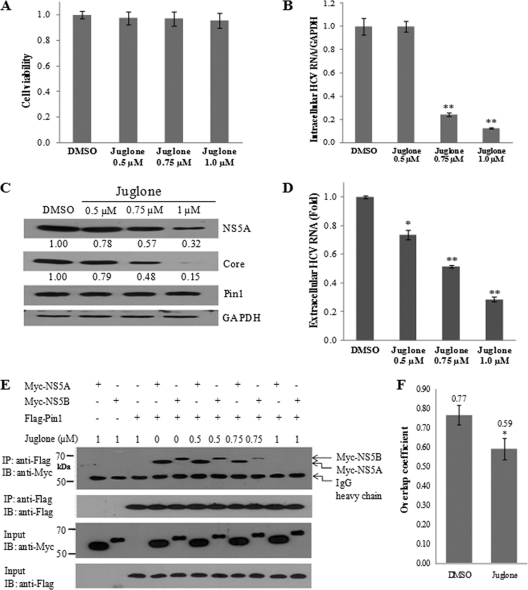
Juglone inhibits HCV replication by disrupting protein interactions between Pin1 and NS5A/NS5B.
Juglone (5-hydroxy-1,4-naphthoquinone) is a natural inhibitor of Pin1 with anti-inflammatory and antifibrotic properties (23). It binds irreversibly to Pin1. At high concentrations, juglone inhibits the activities of RNA polymerases I, II, and III and shows cytotoxic effects (1). We demonstrated that treatment of Huh7.5 cells with juglone at concentrations as high as 1 μM induced no cell toxicity as measured by an MTT assay (Fig. 6A). We also showed that 1 μM juglone efficiently inhibited HCV propagation at the levels of intracellular HCV RNA (Fig. 6B), protein (Fig. 6C), and extracellular HCV RNA (Fig. 6D) in Huh7.5 cells and that inhibition occurred in a dose-dependent manner. However, the levels of Pin1 and GAPDH protein expression were unaffected by juglone. To gain more insight into the role of juglone in HCV inhibition, we investigated the possible effect of juglone on the interaction between the Pin1 and NS5A/NS5B proteins. As shown in Fig. 6E, juglone inhibited the protein interplay between Pin1 and NS5A and between Pin1 and NS5B in a dose-dependent manner. It was noteworthy that 1 μM juglone completely blocked the interaction between Pin1 and HCV proteins. To further confirm the inhibitory role of juglone, Huh7.5 cells infected with Jc1 were transiently transfected with Flag-tagged Pin1 and were treated with either dimethyl sulfoxide (DMSO) or 1 μM of juglone. Samples were fixed and were incubated with both anti-NS5A and anti-Flag antibodies. The level of colocalization of Pin1 with NS5A was quantified by Manders' overlap coefficient (14). Figure 6F shows that the colocalization level of Pin1 with NS5A was prominently decreased by juglone. These results further demonstrate that protein interactions between cellular Pin1 and viral NS5A/5B play a crucial role in HCV replication.
Fig. 6.
Juglone represses HCV replication by inhibiting the interplay between Pin1 and HCV proteins. (A) Huh7.5 cells were infected with Jc1. One day after HCV infection, cells were treated with different concentrations of juglone for 1 day. Cell viability was assessed by the MTT assay. (B) Intracellular RNAs isolated from these cells were analyzed by qPCR. (C) Protein expression in these cells was analyzed by immunoblotting (IB). (D) Extracellular RNAs isolated from these cells were quantified by qPCR. Experiments were carried out in duplicate. Asterisks indicate significant differences (*, P < 0.05; **, P < 0.01) from results with DMSO (vehicle). (E) HEK293T cells were transfected with the indicated combinations of expression plasmids. (Top panel) At 24 h after transfection, cell lysates were immunoprecipitated (IP) with an anti-Flag monoclonal antibody, and bound proteins were then detected by immunoblot analysis using an anti-Myc antibody. (Lower panels) The expression of the NS5A, NS5B, and Pin1 proteins in the same cell lysates was confirmed by immunoblot analysis using the indicated antibodies. (F) Huh7.5 cells infected with HCV Jc1 were transiently transfected with a plasmid expressing Flag-tagged Pin1 at 24 h postinfection. Cells were then treated with either DMSO (vehicle) or 1 μM of juglone at 24 h after transfection. Samples were fixed and were incubated with both anti-NS5A and anti-Flag antibodies. The overlap coefficient was determined as described in Materials and Methods using immunofluorescence data. *, P < 0.05.
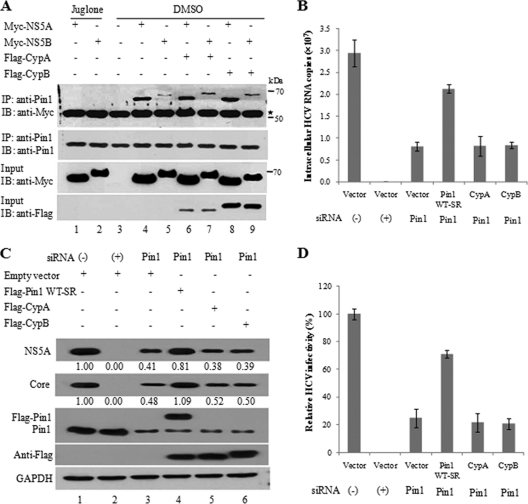
Pin1 and CypA/CypB are differentially involved in HCV proliferation.
Since CypA and CypB, which are also members of the PPIases, also interact with the NS5A and NS5B proteins and play important roles in HCV replication (2, 5, 10), we explored the possible competition between Pin1 and CypA/CypB for interaction with HCV proteins. HEK293T cells were transfected with Myc-tagged NS5A or NS5B and with a Flag-tagged CypA or CypB expression plasmid as indicated in Fig. 7. The transfected cells were then treated with either juglone or a vehicle (DMSO). Cell lysates were immunoprecipitated with an anti-Pin1 antibody, and bound proteins were then detected by immunoblot analysis using an anti-Myc antibody. As shown in Fig. 7A, juglone completely inhibited protein interactions between endogenous Pin1 and NS5A/NS5B (lanes 1 and 2). However, endogenous Pin1 specifically interacted with NS5A and NS5B in the absence of juglone (lanes 4 and 5). Nevertheless, the protein interactions between Pin1 and NS5A and between Pin1 and NS5B were not disturbed by exogenous expression of CypA (Fig. 7A, lanes 6 and 7) or CypB (Fig. 7A, lanes 8 and 9). We then examined whether CypA/CypB could complement the functional loss of HCV propagation in Pin1 knockdown cells. Huh7.5 cells transfected with the siRNAs indicated in Fig. 7B were infected with the Jc1 virus and were then transfected with a plasmid expressing Pin1 WT-SR, CypA, or CypB. Figure 7B shows that neither CypA nor CypB was able to rescue HCV RNA replication in Pin1 knockdown cells. This was further confirmed at the protein level (Fig. 7C, lanes 5 and 6) and the level of infectivity (Fig. 7D). These data suggest that Pin1 may be distinctively involved in HCV propagation.
Fig. 7.
Pin1, CypA, and CypB are distinctively involved in HCV replication. (A) HEK293T cells were transfected with the indicated combinations of expression plasmids. At 24 h after transfection, cells were treated with either 1 μM juglone or a vehicle (DMSO) for 24 h. (Top panel) Cell lysates were immunoprecipitated (IP) with an anti-Pin1 polyclonal antibody, and bound proteins were then detected by immunoblot (IB) analysis. (Lower panels) The expression of the NS5A, NS5B, Pin1, CypA, and CypB proteins in the same cell lysates was confirmed by immunoblot analysis using the indicated antibodies. (B) Huh7.5 cells were transfected with the indicated siRNA. At 2 days after siRNA transfection, cells were infected with Jc1 for 4 h and were then transfected with 2 μg of an empty vector or a plasmid expressing WT-SR, CypA, or CypB. Intracellular RNAs were isolated and were analyzed by qPCR. (C) Both viral and cellular proteins isolated from these cells were analyzed by immunoblot analysis using the indicated antibodies. The band intensity was quantified as described for Fig. 1. (D) Virus titers were determined by focus-forming assays. Experiments were carried out in duplicate.
DISCUSSION
The HCV life cycle is highly dependent on cellular factors. Identification of host factors involved in HCV replication not only facilitates understanding of the HCV life cycle but also aids in the development of novel anti-HCV therapies. Since the mutation rates in cellular genes are substantially lower than those in viral genomes, targeting of host factors may provide a higher barrier to the generation of drug resistance. To identify the cellular factors involved in HCV replication, we screened a siRNA library targeting the cell cycle in HCVcc-infected cells. Out of 11 candidate hits that decreased extracellular HCV RNA levels, we selected and characterized the gene encoding Pin1. Pin1 catalyzes a conformational change in the bound substrate in a phosphorylation-dependent manner. This protein has been shown to regulate the stability and localization of its substrates during transcriptional activation, cell cycle progression, and cell death. Pin1 has also been found to be involved in infection by many viruses. It interacts with hepatitis B virus X protein to enhance hepatocarcinogenesis (17). Pin1 stabilizes human T cell leukemia virus type 1 (HTLV-1) Tax oncoprotein and promotes malignant transformation (7, 20). Pin1 also modulates the replication of human immunodeficiency virus type 1 (HIV-1) via interaction with APOBEC3G protein (29). In the present study, we showed that knockdown of Pin1 significantly reduced extracellular HCV RNA expression as well as intracellular HCV RNA and protein expression, suggesting that Pin1 is required for HCV replication.
Pin1 interacted directly with both the HCV NS5A and NS5B proteins, as demonstrated by a GST pulldown assay. Protein interactions between Pin1 and NS5A/NS5B were confirmed by coimmunoprecipitation analysis and confocal microscopy. Neither binding-defective mutant Pin1 nor isomerase inactive mutant Pin1 was able to rescue HCV replication in Pin1 knockdown cells. These data suggested that the protein interaction and isomerase activities of Pin1 are essential for HCV propagation. Pin1 is upregulated in many cancers, including HCC (11, 24). In this study, we found that Pin1 mRNA and protein expression levels were elevated in both subgenomic replicon cells and HCV-infected cells. Nevertheless, intriguingly, NS5B but not NS5A increased Pin1 expression, although Pin1 interacts with both these proteins. Therefore, further studies are required in order to demonstrate the molecular mechanism that causes NS5B to increase Pin1 expression. Because Pin1 overexpression significantly increases HCV replication, HCV may hijack cellular Pin1 for its own propagation.
We used juglone, a naturally occurring Pin1 inhibitor, to evaluate the effects of Pin1 inhibition on HCV propagation. Juglone is known to inhibit the activities of RNA polymerases I, II, and III (1). In the present study, juglone significantly suppressed HCV propagation. Furthermore, juglone inhibited the protein interplay between Pin1 and NS5A and between Pin1 and NS5B. These data suggest that juglone may inhibit the RNA-dependent RNA polymerase activity of NS5B. However, further studies are required to demonstrate the molecular mechanisms that regulate the inhibitory effects of juglone on HCV replication. Juglone may represent a potent anti-HCV therapeutic agent.
Previous studies have demonstrated that both CypA and CypB interact with the NS5A and NS5B proteins and thus modulate HCV replication (2, 5, 10). We wondered whether these PPIases competed for interaction with HCV proteins. We demonstrated that protein interactions between Pin1 and NS5A and between Pin1 and NS5B were not altered by CypA or CypB. Furthermore, the functional loss of Pin1 in HCV propagation could not be rescued by either CypA or CypB, suggesting that each of these PPIases may have a distinctive role in HCV propagation.
It has been reported previously that Pin1 negatively regulates innate IFN-regulatory factor 3 (IRF3)-dependent antiviral responses (25). However, the phenotypes observed here may not be due to the regulation of the IRF3 signaling pathway by Pin1. Because Pin1 regulates IRF3 through interaction with the phosphorylated form of IRF3, retinoic acid-inducible gene I (RIG-I)-mediated IRF3 signaling modulation by Pin1 is blocked in Huh7.5 cells in which RIG-I is defective (27). HCV has the potential to encounter host immune responses in many ways. Proteolytic processing of components of both the RIG-I and Toll-like receptor 3 (TLR3) pathways by HCV NS3/NS4A leads to abrogation of the cascade that activates IRF3 and NF-κB. In addition, the HCV core, E2, and NS5A proteins block the protein kinase R (PKR) and JAK/STAT pathways. In our study, Pin1 interacted with NS5A/NS5B. NS5B increased Pin1 expression, which resulted in an increase in HCV propagation. Because there are multiple pathways through which IRF3 could be activated, it is also possible that Pin1 may mediate HCV propagation indirectly, probably via its negative regulation of IRF3. Taking these findings together, our study has revealed, for the first time, the essential roles of Pin1 in HCV propagation via its interactions with NS5A and NS5B proteins. Moreover, juglone, a natural inhibitor of Pin1, impaired the interaction between Pin1 and the HCV NS5A/5B proteins and thus inhibited HCV propagation. Our study may provide a new strategy for anti-HCV therapy.
ACKNOWLEDGMENTS
We thank Young-Min Lee (Chungbuk National University, South Korea) for JEV, the rabbit anti-JEV NS1 antibody, and BHK cells and Moon Jung Song (Korea University) for the anti-phospho-IRF3 antibody. JFH-Luc was kindly provided by Ralf Bartenschlager (University of Heidelberg, Heidelberg, Germany).
This work was supported by the National R&D Program for Cancer Control, Ministry for Health and Welfare, South Korea (1020290), and by the National Research Laboratory Grant Program (ROA-2007-0056700) and Biotechnology Development (2008-2004100) from the Ministry of Education, Science and Technology, South Korea.
No conflicts of interest exist.
Footnotes
Published ahead of print on 15 June 2011.
REFERENCES
- 1. Chao S. H., Greenleaf A. L., Price D. H. 2001. Juglone, an inhibitor of the peptidyl-prolyl isomerase Pin1, also directly blocks transcription. Nucleic Acids Res. 29:767–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chatterji U., et al. 2010. HCV resistance to cyclosporin A does not correlate with a resistance of the NS5A-cyclophilin A interaction to cyclophilin inhibitors. J. Hepatol. 53:50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Choi S. H., Hwang S. B. 2006. Modulation of the transforming growth factor-beta signal transduction pathway by hepatitis C virus nonstructural 5A protein. J. Biol. Chem. 281:7468–7478 [DOI] [PubMed] [Google Scholar]
- 4. Choi S. H., Jeong S. H., Hwang S. B. 2007. Large hepatitis delta antigen modulates transforming growth factor-beta signaling cascades: implication of hepatitis delta virus-induced liver fibrosis. Gastroenterology 132:343–357 [DOI] [PubMed] [Google Scholar]
- 5. Fernandes F., Ansari I. U., Striker R. 2010. Cyclosporine inhibits a direct interaction between cyclophilins and hepatitis C NS5A. PLoS One 5:e9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Giannini C., Brechot C. 2003. Hepatitis C virus biology. Cell Death Differ. 10(Suppl. 1):S27–S38 [DOI] [PubMed] [Google Scholar]
- 7. Jeong S. J., Ryo A., Yamamoto N. 2009. The prolyl isomerase Pin1 stabilizes the human T-cell leukemia virus type 1 (HTLV-1) Tax oncoprotein and promotes malignant transformation. Biochem. Biophys. Res. Commun. 381:294–299 [DOI] [PubMed] [Google Scholar]
- 8. Kato T., et al. 2006. Cell culture and infection system for hepatitis C virus. Nat. Protoc. 1:2334–2339 [DOI] [PubMed] [Google Scholar]
- 9. Lindenbach B. D., Rice C. M. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436:933–938 [DOI] [PubMed] [Google Scholar]
- 10. Liu Z., Yang F., Robotham J. M., Tang H. 2009. Critical role of cyclophilin A and its prolyl-peptidyl isomerase activity in the structure and function of the hepatitis C virus replication complex. J. Virol. 83:6554–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu K. P. 2003. Prolyl isomerase Pin1 as a molecular target for cancer diagnostics and therapeutics. Cancer Cell 4:175–180 [DOI] [PubMed] [Google Scholar]
- 12. Lu K. P., Zhou X. Z. 2007. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat. Rev. Mol. Cell Biol. 8:904–916 [DOI] [PubMed] [Google Scholar]
- 13. Lu P. J., Zhou X. Z., Liou Y. C., Noel J. P., Lu K. P. 2002. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J. Biol. Chem. 277:2381–2384 [DOI] [PubMed] [Google Scholar]
- 14. Manders E. E. M., Verbeek F. J., Aten J. A. 1993. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169:375–382 [DOI] [PubMed] [Google Scholar]
- 15. Moradpour D., Penin F., Rice C. M. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453–463 [DOI] [PubMed] [Google Scholar]
- 16. Pang R., et al. 2004. Pin1 overexpression and β-catenin gene mutations are distinct oncogenic events in human hepatocellular carcinoma. Oncogene 23:4182–4186 [DOI] [PubMed] [Google Scholar]
- 17. Pang R., et al. 2007. Pin1 interacts with a specific serine-proline motif of hepatitis B virus X-protein to enhance hepatocarcinogenesis. Gastroenterology 132:1088–1103 [DOI] [PubMed] [Google Scholar]
- 18. Pang R. W., et al. 2006. Pin1 expression contributes to hepatic carcinogenesis. J. Pathol. 210:19–25 [DOI] [PubMed] [Google Scholar]
- 19. Park C. Y., Choi S. H., Hwang S. B. 2009. Nonstructural 5A protein activates β-catenin signaling cascades: implication of hepatitis C virus-induced liver pathogenesis. J. Hepatol. 51:853–864 [DOI] [PubMed] [Google Scholar]
- 20. Peloponese J. M., Jr., Yasunaga J., Kinjo T., Watashi K., Jeang K. T. 2009. Peptidyl proline cis-trans-isomerase Pin1 interacts with human T-cell leukemia virus type 1 Tax and modulates its activation of NF-κB. J. Virol. 83:3238–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pietschmann T., et al. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. U. S. A. 103:7408–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Randall G., et al. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U. S. A. 104:12884–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reese S., et al. 2010. The Pin 1 inhibitor juglone attenuates kidney fibrogenesis via Pin 1-independent mechanisms in the unilateral ureteral occlusion model. Fibrogenesis Tissue Repair 3:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rippmann J. F., et al. 2000. Phosphorylation-dependent proline isomerization catalyzed by Pin1 is essential for tumor cell survival and entry into mitosis. Cell Growth Differ 11:409–416 [PubMed] [Google Scholar]
- 25. Saitoh T., et al. 2006. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 7:598–605 [DOI] [PubMed] [Google Scholar]
- 26. Scholle F., et al. 2004. Virus-host cell interactions during hepatitis C virus RNA replication: impact of polyprotein expression on the cellular transcriptome and cell cycle association with viral RNA synthesis. J. Virol. 78:1513–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sumpter R., Jr., et al. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suzuki T., Ishii K., Aizaki H., Wakita T. 2007. Hepatitis C viral life cycle. Adv. Drug Deliv. Rev. 59:1200–1212 [DOI] [PubMed] [Google Scholar]
- 29. Watashi K., et al. 2008. Human immunodeficiency virus type 1 replication and regulation of APOBEC3G by peptidyl prolyl isomerase Pin1. J. Virol. 82:9928–9936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Watashi K., et al. 2005. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 19:111–122 [DOI] [PubMed] [Google Scholar]
- 31. Winkler K. E., Swenson K. I., Kornbluth S., Means A. R. 2000. Requirement of the prolyl isomerase Pin1 for the replication checkpoint. Science 287:1644–1647 [DOI] [PubMed] [Google Scholar]
- 32. Wulf G., Finn G., Suizu F., Lu K. P. 2005. Phosphorylation-specific prolyl isomerization: is there an underlying theme? Nat. Cell Biol. 7:435–441 [DOI] [PubMed] [Google Scholar]
- 33. Wulf G. M., et al. 2001. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 20:3459–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yaffe M. B., et al. 1997. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 278:1957–1960 [DOI] [PubMed] [Google Scholar]
- 35. Yang F., et al. 2008. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J. Virol. 82:5269–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yun S. I., Choi Y. J., Song B. H., Lee Y. M. 2009. 3′ cis-acting elements that contribute to the competence and efficiency of Japanese encephalitis virus genome replication: functional importance of sequence duplications, deletions, and substitutions. J. Virol. 83:7909–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]