Abstract
The herpes simplex virus 1 (HSV-1) UL6 portal protein forms a 12-subunit ring structure at a unique capsid vertex which functions as a conduit for the encapsidation of the viral genome. We have demonstrated previously that the leucine zipper region of UL6 is important for intersubunit interactions and stable ring formation (J. K. Nellissery, R. Szczepaniak, C. Lamberti, and S. K. Weller, J. Virol. 81:8868–8877, 2007). We now demonstrate that intersubunit disulfide bonds exist between monomeric subunits and contribute to portal ring formation and/or stability. Intersubunit disulfide bonds were detected in purified portal rings by SDS-PAGE under nonreducing conditions. Furthermore, the treatment of purified portal rings with dithiothreitol (DTT) resulted in the disruption of the rings, suggesting that disulfide bonds confer stability to this complex structure. The UL6 protein contains nine cysteines that were individually mutated to alanine. Two of these mutants, C166A and C254A, failed to complement a UL6 null mutant in a transient complementation assay. Furthermore, viral mutants bearing the C166A and C254A mutations failed to produce infectious progeny and were unable to cleave or package viral DNA. In cells infected with C166A or C254A, B capsids were produced which contained UL6 at reduced levels compared to those seen in wild-type capsids. In addition, C166A and C254A mutant proteins expressed in insect cells infected with recombinant baculovirus failed to form ring structures. Cysteines at positions 166 and 254 thus appear to be required for intersubunit disulfide bond formation. Taken together, these results indicate that disulfide bond formation is required for portal ring formation and/or stability and for the production of procapsids that are capable of encapsidation.
INTRODUCTION
The products of herpes simplex virus 1 (HSV-1) DNA replication are head-to-tail concatemers which are resolved into monomeric genomic units and packaged into a preformed capsid shell in the nucleus of the infected cell (reviewed in references 2, 6, and 10). The HSV-1 capsid shell is composed of the major capsid protein (VP5), two triplex proteins (VP19C and VP23), and VP26. Minor capsid proteins include UL6, UL15, UL17, UL25, UL28, and UL33. The process of cleavage and DNA packaging requires the six minor capsid proteins as well as UL32, which is not found associated with capsids (2, 6, 10, 21).
HSV capsid formation and genome encapsidation are reminiscent of the double-stranded DNA bacteriophages, in that a procapsid shell is preassembled around a scaffolding protein that is not present in the mature virion (3, 37, 38). Bacteriophage and herpesviruses share an important structural element, a dodecameric portal ring located at a unique capsid vertex (8, 9, 28, 40). During HSV genome encapsidation, the portal ring provides a docking site for the terminase, an ATP-driven molecular motor that facilitates the uptake of viral DNA (34, 42, 45, 46). Terminase is responsible not only for viral DNA uptake but also for the specific cleavage of viral genomes such that a monomeric unit of viral DNA is packaged in each capsid (1, 4, 18, 19, 33, 42, 44, 46, 47).
UL6 becomes incorporated into nascent HSV-1 capsids mediated by interaction with the UL26.5 major scaffold protein (15, 26, 29, 35). Procapsids can assemble in the absence of UL6 via an interaction between UL26.5 and VP5 (27); however, when UL6 is present at the initiation of assembly, UL6-containing capsids are formed, suggesting that the portal is incorporated at a very early step in assembly (26). These results also suggest that capsid assembly is regulated such that capsids lacking UL6 do not assemble efficiently in infected cells. UL6 is known to self assemble into a dodecameric ring in lysates from insect cells infected with recombinant UL6-expressing baculovirus (28). Interestingly, two UL6 mutant proteins, L429E L436E and D-LZ, bearing mutations in the leucine zipper region, are unable to produce rings and form polymorphic aggregates instead (25). Moreover, these mutant viruses assemble B capsids that are defective for virus growth and encapsidation. Thus, the ability to form a dodecameric portal ring appears to be essential for the formation of a procapsid that is competent for cleavage and packaging.
In this paper, we investigated another type of bonding interaction that contributes to ring formation and/or stability. UL6 portal rings from insect cells infected with recombinant baculovirus were disrupted when exposed to reducing agents. Although disulfide bonds have been reported previously between HSV-2 capsid proteins (48) and in HSV-1 scaffold proteins (43), this is the first report of disulfide linkages in the portal ring. The mutational analysis of UL6 identified cysteines 166 and 254 as essential for (i) intermolecular disulfide bond formation; (ii) the formation and/or stability of portal rings; and (iii) the production of procapsids that are capable of encapsidation.
MATERIALS AND METHODS
Viruses, cells, antibodies, and other reagents.
The KOS strain of herpes simplex virus 1 (HSV-1) was used as the wild-type (WT) virus and as the parental strain for the generation of recombinant viruses C166A and C254A. The UL6 null virus, hr74, contains an insertion of the E. coli lacZ gene under the control of the HSV-1 ICP6 promoter and was described previously (20). African green monkey kidney fibroblast cells (Vero) were obtained from the ATCC and used to propagate the WT type virus. The UL6 complementing cell line, UL6-31 (20, 25), was used to propagate hr74, C166A, and C254A recombinant viruses. Jay C. Brown (University of Virginia Health System) provided anti-UL6 monoclonal antibodies 1C9 and 4G9 (7). NEM (N-ethylmaleimide) was purchased from Sigma and dissolved in ethanol (0.5 M).
Recombinant baculoviruses and protein expression.
Wild-type protein and C166A and C254A recombinant proteins were expressed in Sf9 insect cells and purified as described previously (25). Briefly, 3.2 × 108 insect cells were infected with recombinant baculoviruses at a multiplicity of infection (MOI) of 5 PFU per cell and incubated at 28°C for 48 h. Inclusion bodies containing UL6 were isolated and resuspended in 0.5 ml of arginine (1.0 M, pH 7.5), kept on ice for 15 min with occasional mixing, and centrifuged at 30,000 × g in a Beckman TL-100 ultracentrifuge at 4°C for 15 min to remove insoluble UL6 as described in reference 25. The supernatant containing solubilized UL6 protein was used in all subsequent experiments.
Purification of UL6 rings.
UL6 rings were purified by sucrose gradient centrifugation. In short, the solubilized UL6 protein (0.25 ml) was loaded onto a 10 to 30% sucrose gradient containing 1.0 M arginine and spun at 48,000 rpm for 18 h at 4°C in an SW 55Ti rotor. Gradient fractions (250 μl each) were collected using a BIOCOMP Piston gradient fraction collector, stored on ice, and subjected to SDS-PAGE. UL6 was detected by silver staining with the following procedure: the gel was fixed for 1 h in a solution containing 30% ethanol and 10% acetic acid, washed twice in 20% ethanol (10 min each wash), and then washed once for 20 min with water. The gel was sensitized in 0.1% sodium thiosulfate for 1 min, briefly rinsed with water, and incubated in cold 0.1% silver nitrate for 30 min at room temperature. Silver nitrate solution was removed; the gel was washed twice in water (1 min each wash) and developed with 3% potassium bicarbonate and 0.05% formaldehyde. Once the protein bands were detected, staining was terminated by replacing the developing solution with 5% acetic acid solution for 10 min.
EM.
Purified rings in 1 M arginine were either untreated (control) or treated with 100 mM dithiothreitol (DTT), followed by the addition of 50 mM sodium bisulfite to react with the free sulfhydryl groups. For electron microscopy (EM) analysis, treated and untreated ring samples were adsorbed onto Formvar-carbon-coated grids, stained with uranyl acetate, and visualized in a Philips CM10 transmission electron microscope operating at 60 kV (52,000× magnification). Wild-type and mutant rings also were analyzed by EM as previously described (25).
Capsid isolation.
Confluent monolayers of Vero cells in T225 tissue culture flasks were infected with KOS wild-type or mutant virus at 37°C. At 18 to 20 h postinfection the medium was discarded, the monolayers were washed with phosphate-buffered saline (PBS), and the cells were scraped into 20 ml of PBS. Cells were pelleted at 200 × g for 15 min in a Beckman S4750 rotor, and the pellet containing 4 × 107 to 5 × 107 cells was resuspended in 5 ml of 20 mM Tris (pH 7.6) buffer followed by the addition of 5 ml of 2× lysis buffer (2% Triton, 20 mM Tris, pH 7.6, 1 M NaCl, 4 mM EDTA). Cell lysates were incubated on ice for 30 min, treated with MgCl2 and DNase at final concentrations of 20 mM and 0.1 mg/ml, respectively, for 15 to 20 min at 37°C, and briefly sonicated in a cup horn sonicator (three 20-s bursts at 50% power) to reduce viscosity. Insoluble material was removed by centrifugation at 10,000 × g for 15 min in a Beckman F6X100 rotor. The supernatants containing intracellular capsids were spun through a 1.5-ml cushion of 30% (wt/vol) sucrose in TNE buffer at 71,000 × g for 1 h in an SW41 rotor. Pellets containing capsids were resuspended in 700 μl of TNE, briefly sonicated in a cup horn sonicator to break up clumps, and layered over a continuous gradient of 20 to 50% (wt/vol) sucrose in TNE. Gradients were centrifuged at 71,000 × g for 1 h in an SW41 rotor and fractionated using a Biocomp piston gradient fraction collector. Collected fractions (250 μl each) were stored at −80°C for further analysis by SDS-PAGE and Western blotting. For capsids isolated in the presence of NEM, cell monolayers were incubated at 37°C in Dulbecco's modified essential medium (DMEM) containing 10 mM NEM for 10 min prior to harvesting. NEM at a final concentration of 10 mM also was added to all other buffers used during the process of capsid isolation. Total capsids were isolated as described above, except that instead of sucrose gradient centrifugation, capsids pelleted through a sucrose cushion were reconstituted in reducing or nonreducing SDS-PAGE loading buffer.
Infected cell lysates.
Infected cell lysates were prepared as follows. Vero cells in a 100-mm-diameter tissue culture dish (1 × 107 cells) were infected with wild-type or mutant virus stocks at an MOI of 10 PFU per cell at 37°C. Infected cells were incubated at 37°C in DMEM containing 10 mM NEM for 10 min prior to harvesting by scraping with a rubber spatula at 14 h postinfection. Cells were collected by centrifugation at 1,000 rpm for 10 min and rinsed twice with PBS containing 10 mM NEM. Cell pellets were resuspended in 1 ml of reducing or nonreducing SDS-PAGE loading buffer as described below and subjected to SDS-PAGE.
SDS-PAGE under reducing and nonreducing conditions.
Rings, capsids, and cell extracts were prepared either in reducing SDS buffer (50 mM Tris, pH 6.8, 10% glycerol, 2% SDS, 100 mM DTT, 5% [vol/vol] β-mercaptoethanol, 0.02% [wt/vol] bromophenol blue) or nonreducing SDS buffer (50 mM Tris, pH 6.8, 10% glycerol, 2% SDS, 20 mM NEM, 0.02% [wt/vol] bromophenol blue). Samples were heated at 95°C for 3 min, briefly sonicated, and resolved on a Tris-glycine gel cast according to instructions provided by Bio-Rad. Gels were silver stained or subjected to Western blotting.
Western blot analysis.
Proteins were electrotransferred to polyvinylidene difluoride (PVDF), blocked with 5% fat-free milk in TBST (150 mM NaCl, 20 mM Tris, pH 7.5, and 0.1% Tween), and incubated with antibodies against VP5 or UL6. Monoclonal antibody IC9 or 4G9 (anti-UL6) (25) was used at a dilution of 1:10,000 or 3E8 (anti-VP5) (5) at a dilution of 1:2,000. Membranes were washed with TBST and incubated with secondary antibodies conjugated to either horseradish peroxidase or alkaline phosphatase and washed again with TBST, and proteins were detected as described by the manufacturer's protocol.
Generation of cysteine substitution mutations.
Substitution mutagenesis was carried out as described previously (25). Sequences of the oligonucleotides used for mutagenesis are available upon request. HSV-1 genomic DNA was used as the template in a two-step PCR-based mutagenesis protocol (13). To generate the single and double cysteine substitutions, the PCR product was gel purified and cloned into the pENTR vector of the Gateway system (Invitrogen) and then moved to pDEST40 vector by following the manufacturer's protocol. The C166A and C254A mutations were moved to Gateway pDEST10 for transfer into recombinant baculovirus genomes as described previously (25). All plasmids containing mutant versions of UL6 were subjected to DNA sequencing to confirm the presence of the desired mutations.
Marker transfer.
Monolayers of the UL6-31 cell line (60-mm dishes at 70% confluence) were transfected with 1 μg mutant plasmid DNA and 1 μg hr74 virus DNA using the Lipofectamine Plus reagent (Invitrogen Corporation). Transfection was done in serum-free DMEM containing Plus reagent (8 μl) and Lipofectamine reagent (12 μl) in a total volume of 250 μl. Transfected cells were incubated in DMEM containing 5% fetal bovine serum for 24 h and were harvested along with the culture supernatant. The titers of viral stocks on the UL6-31 cell line were determined, and the stocks were replated at a suitable dilution to yield approximately 100 plaques on a 10-cm dish. The dishes were overlaid with 2% methyl cellulose in DMEM, and recombinant plaques were identified by overlaying with methyl cellulose solution containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (50 μg/ml) overnight. The parental hr74 plaques stain blue, while the recombinants that contain the mutant UL6 gene and hence have lost the lacZ gene are colorless. Colorless plaques were selected, and each mutant was plaque purified three times as described previously (25). The presence of the desired UL6 mutant was confirmed by sequence analysis following the PCR amplification of the region spanning the deletion.
Virus yield and plaque assays.
Virus yields were determined by infecting Vero or UL6-31 cells at an MOI of 0.5 PFU/cell, and samples were collected at 24 h postinfection. Serial dilutions were plated on the UL6-31 cell line, overlaid with methyl cellulose, incubated for 72 h, fixed in 4% formaldehyde, and stained with 1% crystal violet solution. Viral plaques were counted by eye.
Transient complementation assay.
The transient complementation assay was carried out as described previously (23). In brief, freshly trypsinized exponentially growing Vero cells were transfected with 1 μg of empty, wild-type, or mutant plasmid. At 18 h posttransfection, cells were superinfected with hr74 at an MOI of 3 PFU per cell. At 24 h postinfection, progeny viruses were harvested and assayed on the UL6-31 cell line for total yield. The percent complementation was calculated by dividing the titer obtained for the mutant plasmid by the titer of the WT plasmid and multiplying by 100. The background titers from the empty plasmid samples were subtracted.
Immunofluorescence microscopy.
Vero cells grown on coverslips (70% confluent) were transfected with 1 μg of plasmids bearing WT or mutant UL6 genes; at 16 h posttransfection the coverslips were washed with PBS and cells were fixed with 4% paraformaldehyde, washed again with PBS, permeabilized with 0.5% Triton X-100, and washed again with PBS. The coverslips then were incubated in 5% goat serum as blocking solution and treated with a 1:200 dilution of the 4G9 monoclonal antibody for 1 h. The coverslips were washed again in PBS and incubated with the secondary antibodies conjugated to Alexa Fluor 488 for 30 min. They then were washed and mounted on glass slides using glycerol-gelatin. The slides were examined using a Carl Zeiss Axiovert A410 confocal microscope, and images were collected. The images were exported to Photoshop to generate the final figures.
RESULTS
Purified UL6 rings are sensitive to reducing reagents.
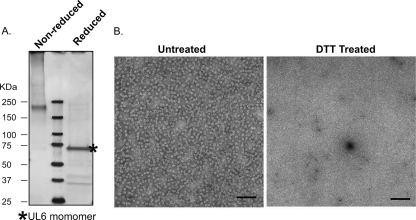
The UL6 protein plays multiple roles during viral assembly and encapsidation, including ring formation, interaction with scaffold and VP5 to nucleate the assembly of the procapsid shell, the docking of the terminase, and acting as a conduit for the uptake of viral DNA into capsids (8, 9, 20, 25, 26, 28, 31, 32, 40, 42, 46). It also is likely that the UL6 portal plays a role during subsequent infections. UL6 exists as a dodecameric ring, and the forces that promote UL6-UL6 intersubunit assembly are not well understood. In this paper, we tested the hypothesis that disulfide bonds contribute to UL6 ring formation and proper UL6 function. We first wanted to determine if purified UL6 protein contains disulfide bonds. To test this, UL6 was expressed in insect cells infected with recombinant baculovirus expressing wild-type protein as described previously (25). Portal rings were purified by sucrose gradient sedimentation as described in Materials and Methods. UL6-containing fractions were identified by SDS-PAGE followed by Western blot analysis using UL6 polyclonal antibody (data not shown). Purified UL6 rings were subjected to SDS-PAGE under reducing and nonreducing conditions (Fig. 1A). Under nonreducing conditions, slow-migrating forms of UL6 were observed, including a prominent band migrating faster than the 250-kDa molecular mass marker. On the other hand, in the presence of DTT, the major UL6 band migrated at a position expected for monomeric UL6 (74 kDa); the smaller bands in this lane react with UL6 antibody in immunoblots, suggesting that they are proteolytic fragments of UL6 (data not shown). The appearance of slow-migrating, cross-linked species that can be reduced to the monomeric form suggests the presence of disulfide bonds. Since no other viral proteins are expressed in this experiment, it is likely that the slow-migrating material represents intersubunit disulfide bonds; however, the oligomeric status of these forms is not clear. Although it is possible that slower-migrating material results from interactions of UL6 with insect cellular proteins such as chaperones, we think this is unlikely because the silver stained-reduced sample contained only UL6.
Fig. 1.
UL6 rings contain disulfide bonds and are destabilized in the presence of reducing agents. UL6 protein was expressed in insect cells and purified by sucrose gradient sedimentation as described in Materials and Methods. (A) Fractions containing UL6 were processed for SDS-PAGE under nonreducing and reducing conditions. Molecular mass markers are shown in the middle lane. The gel was silver stained, and the positions of molecular mass markers are shown on the left. (B) Electron micrographs show preparations of UL6 either untreated (left panel) or treated with DTT to reduce disulfide bonds, followed by the addition of sodium bisulfite to prevent free sulfhydryl groups from reforming disulfide bonds (right panel). The scale bar in each panel corresponds to 100 nm.
UL6 expressed in insect cells infected with recombinant baculovirus forms rings that can be observed by electron microscopy (25, 28). To address whether disulfide bonds contribute to ring stability, UL6 rings were treated with DTT followed by the addition of sodium bisulfite to prevent free sulfhydryl groups from reforming disulfide bonds. Figure 1B shows that in the untreated samples, portal rings were readily detected; however, in the DTT-treated samples, portal rings were not detected. These results suggest that disulfide bonds contribute to ring stability.
Disulfide bonds are observed in vivo.
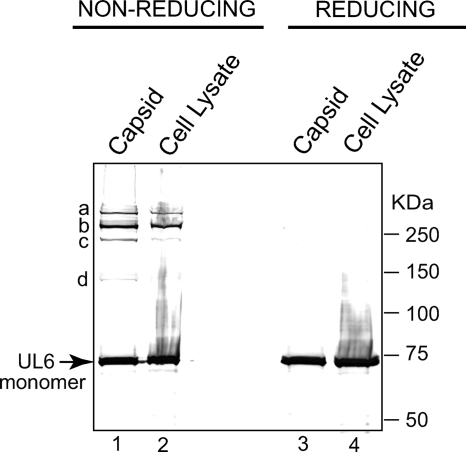
To determine whether disulfide bonds in the UL6 ring also could be detected in HSV-1-infected cells, cell lysates and capsids were harvested from infected cells at 24 h postinfection as described in Materials and Methods. Prior to harvest, N-ethylmaleimide (NEM) was added to bind and block any free sulfhydryl groups. The addition of NEM prior to lysis ensures that any disulfide bonds that are detected actually are present in the infected cell proteins in vivo rather than forming once the cells are lysed and the extracts are exposed to an oxidizing environment. Total capsids were harvested as described in Materials and Methods and compared to crude cell lysates. Capsids and lysates were subjected to SDS-PAGE under reducing and nonreducing conditions (Fig. 2). A prominent band corresponding to a monomeric unit of UL6 was detected in capsids and lysates under reducing conditions (Fig. 2, lanes 3 and 4). Under nonreducing conditions, four slower-migrating species (marked a to d) and monomeric UL6 were observed in capsids (Fig. 2, lane 1). The proportion of material in bands a to d and monomeric UL6 varies somewhat from preparation to preparation (data not shown), although band b is generally the most prominent of the slower-migrating bands. The variability likely is caused by the labile nature of disulfide bonds. Cell lysates contain bands a and b, a weaker signal for band c, no visible band d, and monomeric UL6 (Fig. 2, lane 2). Thus, UL6 protein appears to be linked through disulfide bonds in capsids and in cell lysates. The observation that even under nonreducing conditions UL6 monomer was detected indicates that not all of the subunits in the portal ring were cross-linked; however, since the amount of UL6 in various species is variable, it is not possible to quantify the proportion of cross-linked species in capsids and lysates.
Fig. 2.
UL6 protein exhibits disulfide linkages in vivo. Total capsids and crude cell lysates from infected cells were isolated and subjected to SDS-PAGE under nonreducing (lanes 1 and 2) and reducing (lanes 3 and 4) conditions as described in Materials and Methods. The slower-migrating species are labeled a to d. Band d is very faint in this figure but has been seen reproducibly in several experiments. The position of monomeric UL6 is marked with an arrow.
Cysteines 166 and 254 are essential for viral infectivity.
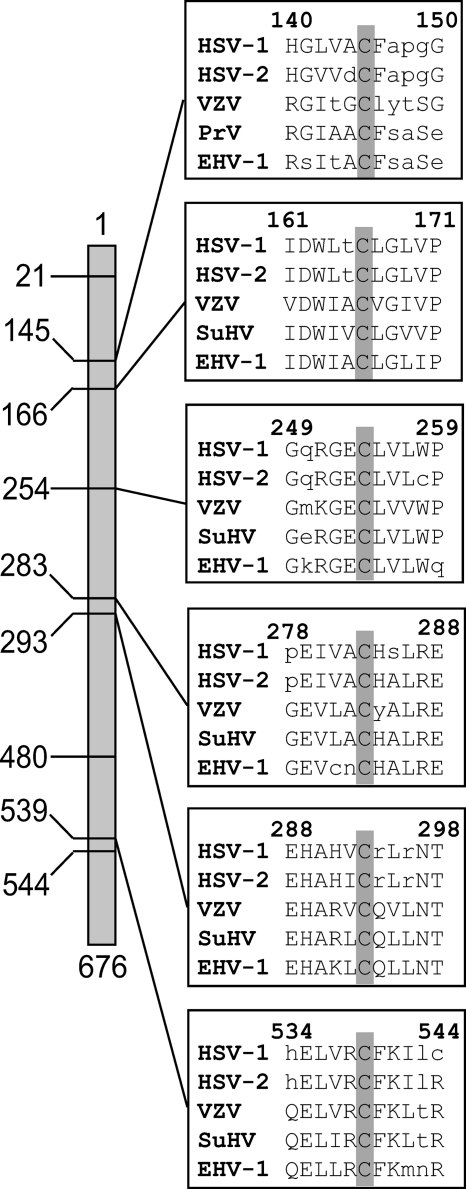
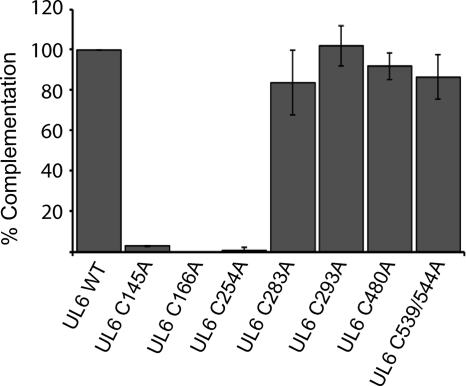
The predicted sequence of the HSV-1 UL6 gene reveals the presence of nine cysteines. UL6 genes from several alphaherpesviruses were analyzed using the Clustal W program (39), revealing that cysteines 145, 166, 254, 283, 293, and 539 are conserved in HSV-1, HSV-2, VZV, PrV, and EHV-1 (Fig. 3). To investigate which residues are important for the observed disulfide linkages in HSV-1 UL6, each cysteine was mutated to an alanine using site-directed mutagenesis as described in Materials and Methods; in addition, one double mutant was made. C-to-A mutants were tested for their ability to complement the growth of the UL6 null virus, hr74. Vero cells were transfected with plasmids bearing the wild-type or mutant UL6 genes and then infected with a UL6 null virus (hr74) at an MOI of 3 PFU/cell. After 24 h, virus was harvested and viral titers were determined on the UL6 complementing cell line UL6-31. Figure 4 shows that most of the substitution mutations were able to complement hr74 as well as the plasmid bearing the wild-type gene. An additional substitution mutant, C21A, also was able to complement hr74 at wild-type levels (data not shown). Three substitutions, C145A, C166A, and C254A, were unable to complement hr74.
Fig. 3.
Cysteines 145, 166, 254, 283, 293, and 539 are conserved in alphaherpesviruses. The positions of all nine cysteines in HSV-1 UL6 are shown on the left. Multiple sequence alignments of the portal proteins from human herpesvirus 1 (HHV-1, HSV-1) gp011, human herpesvirus 2 (HHV-2, HSV-1) gp08, human herpesvirus 3 (HHV-3, VZV) gp55, pseudorabies virus (SuHV-1) UL6, and equine herpesvirus 1 (EHV-1) orf56 reveal that cysteines 145, 166, 254, 283, 293, and 539 are conserved. The numbering reflects that of the HSV-1 sequence. The sequences were obtained from the NCBI viral genome database (accession numbers NC_001806, NC_001798, NC_001348, NC_006151, and NC_001491, respectively). Alignments were performed using Clustal W (39).
Fig. 4.
Cysteine-to-alanine substitutions at positions 145, 166, and 254 abolish complementation ability. Alanine substitution mutations were made at each of the cysteine residues. A transient complementation assay was used to determine the ability of the substitution mutants to complement the null virus hr74. Vero cells were transfected with plasmid DNAs bearing cysteine-to-alanine mutations and superinfected with hr74 virus, and the titer of the resulting progeny virus was determined on UL6-31 cells. This experiment was repeated three times. Error bars represent standard deviations.
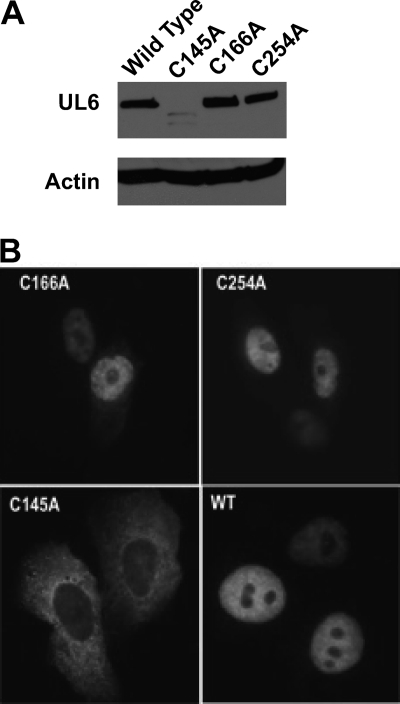
The lack of complementation indicates that cysteines 145, 166, and 254 are essential for UL6 function or, alternatively, that mutant proteins are globally or locally misfolded. To determine whether the noncomplementing mutants were able to synthesize stable UL6 protein, lysates from transfected cells were subjected to SDS-PAGE and Western blotting. C166A and C254A were able to synthesize wild-type levels of UL6; however, C145A was not, suggesting that this mutation caused misfolding leading to instability (Fig. 5A). To determine whether the mutant proteins could localize appropriately to the nucleus, immunofluorescence microscopy using anti-UL6 antibody was performed on cells transfected with plasmids expressing wild-type or mutant UL6 genes. Figure 5B shows that as previously reported (32), wild-type UL6 localizes to the nucleus of transfected cells even in the absence of other viral proteins. Cells transfected with plasmids expressing C166A and C254A mutant proteins exhibited nuclear fluorescence similar to that of the wild type; however, the C145A mutant protein signal was less intense and was detected only in the cytoplasm, which is consistent with the Western blot data. This suggests that the C145A mutant protein is globally misfolded and unable to adopt a conformation that can be recognized by the transport machinery.
Fig. 5.
Western blotting and immunofluorescence analysis of wild-type and mutant versions of UL6. (A) Vero cells were transfected with plasmids expressing wild-type or mutant UL6 proteins. Lysates were subjected to SDS-PAGE and blotted with anti-UL6 monoclonal antibody (1C9). Actin was included as an internal loading control. (B) Immunofluorescence images of Vero cells transfected with WT, C166A, C254A, and C145A plasmids stained with the 4G9 monoclonal antibody.
Marker transfer was used to introduce the C166A and C254A mutations into the HSV-1 genome as described in Materials and Methods. The global instability of the C145A mutant prevented the analysis of cysteine 145 in the context of infection. To determine whether cysteines 166 and 254 are required for viral growth, a viral yield experiment was performed on Vero and UL6-31 cells. Table 1 shows that in Vero cells infected with C166A and C254A mutant viruses, only background levels of infectious virus were produced. C166A and C254A mutants grew to wild-type levels on the UL6 complementing cell line UL6-31, indicating that the defect in virus growth was due to a mutation in the UL6 gene.
Table 1.
Growth of KOS, hr74, C166A, and C254A on Vero and UL6-31 cellsa
| Cell line | Virus growth (PFU/ml) |
|||
|---|---|---|---|---|
| KOS | hr74 | C166A | C254A | |
| Vero | 1.9 × 108 | 1.6 × 103 | 4.5 × 102 | 1.2 × 103 |
| UL6-31 | 1.0 × 108 | 7.5 × 107 | 1.6 × 107 | 3.1 × 107 |
Virus yields were determined by infecting Vero or UL6-31 cells at an MOI of 0.5 PFU/cell and collecting samples 24 h postinfection. Viral titers were determined on the permissive UL6-31 cell line.
C166A and C254A mutants produce B capsids that contain smaller amounts of UL6 than the wild type.
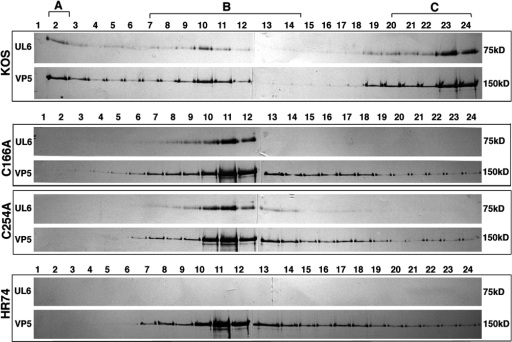
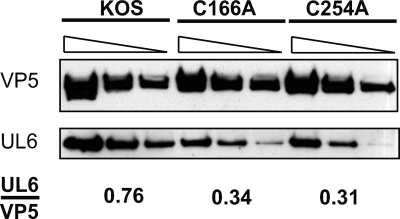
We next tested whether the C166A and C254A viral mutants could produce portal-containing capsids. Vero cells were infected with wild-type or mutant virus, and capsids were separated by sucrose gradient sedimentation. Fractions from across the gradient were subjected to SDS-PAGE and probed with monoclonal anti-UL6 and anti-VP5 antibodies. Figure 6 shows that cells infected with wild-type virus displayed the expected pattern of A, B, and C capsids and that UL6 specifically associated with capsid peaks. In contrast, cells infected with C166A, C254A, or the UL6 null virus (hr74) contained only B capsids, suggesting that packaging is defective (Fig. 6). Furthermore, mutant UL6 protein exhibited a specific association with viral capsids and was not distributed throughout the gradient. To compare the levels of UL6 in mutant and wild-type capsids, B capsids were recovered from sucrose gradients and analyzed by Western blotting with monoclonal anti-UL6 and anti-VP5 antibodies. Figure 7 shows serial dilutions of capsids isolated from cells infected with KOS, C166A, or C254A. Roughly similar amounts of VP5 were present in wild-type and mutant B capsids. On the other hand, UL6 was present in reduced amounts in capsids from the C166A or C254A mutant virus. The relative ratio of UL6 to VP5 was quantified and indicates that mutant capsids contain less UL6 than the wild type. The reduced amounts of UL6 in mutant capsids indicate that each capsid contains fewer than 12 molecules of UL6. Under this scenario, the ability of mutant UL6 protein to form aggregates may be sufficient to nucleate procapsid assembly, resulting in capsids each with diminished levels of UL6. Alternatively, it is possible that portal rings are able to form very inefficiently in cells infected with C166A or C254A mutants, resulting in a small number of capsids with an intact portal ring and a majority of capsids with none. Under this scenario, however, even if some capsids with an intact portal ring are produced, the absence of C capsids indicates that packaging is defective in mutant-infected cells. Additional experiments will be needed to determine the distribution of UL6 in capsids from mutant-infected cells. We can conclude, however, that cysteines 166 and 254 are essential for efficient portal ring formation and for the production of procapsids that are able to encapsidate viral genomes.
Fig. 6.
Substitution mutants C166A and C254A produce B, but not A or C, capsids. Vero cells were infected with KOS (WT), hr74 (null), C166A, or C254A as described in Materials and Methods. Infected cell lysates were subjected to sucrose density gradient sedimentation, and fractions (1, top; 24, bottom) were subjected to SDS-PAGE and immunoblot analysis. Blots were divided into strips and probed separately with anti-UL6 and anti-VP5 antibodies. The positions of A, B, and C capsids are shown.
Fig. 7.
Capsids isolated from cells infected with C166A and C254 mutant viruses contain reduced amounts of the UL6 protein compared to that of wild-type KOS. B capsids isolated from cells infected with KOS, C166A, or C254A were serially diluted (1:1, 1:2, and 1:4) and analyzed by Western blot analysis using anti-VP5 and anti-UL6 antibodies. The images were quantified using Image J software, and results are expressed as a ratio of the relative intensities of the UL6 and VP5 protein bands.
The production of B capsids only by cysteine substitution mutant viruses indicates that DNA packaging is defective. To verify this, DNA was isolated from cells infected with wild-type or mutant virus, digested with BamHI, and subjected to agarose gel electrophoresis and Southern blot hybridization. These experiments indicated that wild-type and mutant viruses are able to synthesize similar amounts of viral DNA, but that the terminal S and Q fragments indicative of cleavage were detected only in wild-type virus-infected cells (data not shown). Thus, C166A and C254A mutants are capable of DNA synthesis and procapsid assembly; however, procapsids are defective for cleavage and packaging and the production of infectious progeny.
Cysteines 166 and 254 are necessary for disulfide bond formation.
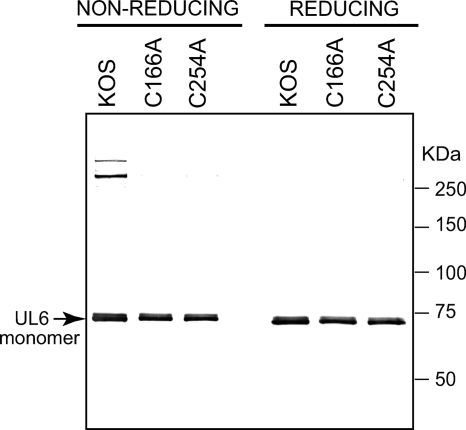
We next asked whether the C166A and C254A mutants displayed the same cross-linked protein bands as those seen in lysates and capsids from cells infected with wild-type virus (Fig. 2). Vero cells were infected with KOS, C166A, or C254A. After 24 h, cells were treated with NEM, and total intracellular capsids were isolated and subjected to SDS-PAGE under reducing and nonreducing conditions as described in Materials and Methods. While slower-migrating bands were observed in wild-type capsids under nonreducing conditions, no slower-migrating bands were seen in mutant capsids under the same conditions (Fig. 8). This result implies that cysteines 166 and 254 play a direct role in the formation of the disulfide bonds detected in UL6 in KOS capsids.
Fig. 8.
Cysteines 166 and 254 are essential for the formation of the slower-migrating species. Total capsids were prepared from cells infected with KOS, C166A, or C254A as described in Materials and Methods. For KOS, total capsids include A, B, and C capsids; however, the two mutants can only produce B capsids. Monomeric UL6 is labeled with an arrow.
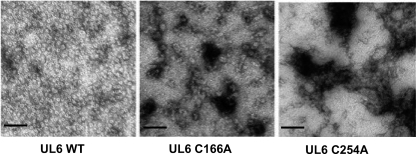
To investigate whether cysteines 166 and 254 are involved in ring formation, baculovirus recombinants bearing these mutations were constructed and used to infect insect cells. Solubilized UL6 was isolated from wild-type and mutant-infected cells and subjected to sucrose gradient centrifugation. Wild-type UL6 protein was able to form a ring structure that could be visualized by light scattering; however, no rings were observed in the expected position in gradients containing the mutant proteins (data not shown). Fractions sedimenting at the position of the wild-type rings were collected and analyzed by electron microscopy (Fig. 9). Rings were observed in the samples containing wild-type UL6 but not in samples from the mutant-infected cells. The mutant protein formed aggregates that exhibited various dimensions and did not contain a distinct central channel. These data demonstrate that cysteines 166 and 254 are necessary for ring formation and/or stability.
Fig. 9.
C166A and C254A mutant proteins do not form rings in vitro. Electron micrographs show purified preparations of mutant and wild-type UL6 protein expressed in insect cells using a recombinant baculovirus expression system. The photographs show negatively stained rings with dark central channels in the WT, and mutant proteins show amorphous particles of various dimensions and without a distinct central channel. The scale bar in each panel corresponds to 100 nm.
DISCUSSION
In this paper, we report that UL6 participates in disulfide bonds that are essential for portal ring integrity and for the formation of procapsids that are competent for the encapsidation of viral genomes. Slow-migrating UL6 bands were observed in purified portal rings that could be reduced to monomeric UL6 by the addition of reducing agents, indicating the likely presence of intersubunit disulfide cross-links. The observations that portal rings did not form in insect cells expressing cysteine substitution mutations (C166A and C254A) and that portal rings were destabilized in the presence of reducing agents are consistent with the suggestion that intersubunit disulfide bonds are present. Slow-migrating species of UL6 also were seen in cell lysates and capsids from cells infected with wild-type HSV but not in cells infected with mutants C166A and C254A, suggesting that these two cysteine residues are required for the disulfide linkages. Mutant C145A was misfolded, thus we cannot rule out that this residue also participates in disulfide bond formation. The inability of C166A and C254A to produce infectious virus and to package DNA is consistent with disulfide bond formation being essential for the production of procapsids that are competent for cleavage and packaging. Although it is likely that some of the slow-migrating species seen in cell lysates and capsids (Fig. 2 and 8) represent intersubunit interactions within the portal ring, it also is possible that some represent interactions between UL6 and other viral capsid proteins. We are intrigued by the possibility that during capsid formation, encapsidation, and the production of mature virus, the disulfide bond profile is dynamic. For instance, it is possible that intersubunit disulfide bonds play a role in ring formation and that later during capsid assembly some bonds shift such that interactions between UL6 and other capsid proteins form to stabilize the capsid. In fact, some differences were observed in the pattern of slow-migrating bands between B and C capsids and virions (unpublished results). Further experiments will be required to test this hypothesis and to identify relevant protein-protein interactions in this pathway.
Interestingly, although mutant C166A and C254A proteins cannot form portal rings, they form aggregates when expressed in insect cells. In cells infected with mutant viruses, B capsids were observed which, on average, contained UL6 in smaller amounts than those seen in wild-type B capsids. This behavior is reminiscent of our previous analysis of mutations in the leucine zipper region. UL6 protein expressed in insect cells infected with recombinant baculoviruses bearing mutations in this region were unable to form rings; instead, L429E L436E and D-LZ formed polymorphic aggregates. In the context of viral infection, leucine zipper mutants as well as C166A and C254A assemble B capsids that contain smaller amounts of UL6 than those seen in the wild type and are not capable of encapsidation (25 and this paper). This indicates that it is not necessary to form a complete portal ring for procapsid assembly to occur, and that aggregates of UL6 are able to interact with scaffold protein to initiate procapsid assembly. Alternatively, it is possible that a small minority of the B capsids seen in the mutant-infected cells contain a complete dodecameric ring and that the majority of procapsids contain little or no UL6. Thus, ring formation may be extremely inefficient, but the few rings that can form may nucleate procapsid formation, resulting in a small number of UL6-containing procapsids. Experiments are in progress to distinguish between these explanations. It also is possible that in the B capsids from mutant-infected cells, UL6 is not found at a unique vertex. In any case, it is clear that ring formation and/or stability is inefficient in both leucine zipper and cysteine substitution mutants. Furthermore, the addition of reducing agents to portal rings resulted in the disruption of the ring structure. Taken together, these results suggest that portal ring formation and/or stability requires at least two types of intersubunit interactions driven by disulfide bond formation and coiled-coil interactions typical of leucine zippers. It will be important to understand the assembly principles of this interesting structure, because an intact portal appears to be essential for all subsequent steps of capsid maturation. Portal assembly also provides a novel potential target for antiviral intervention.
We report that slower-migrating bands of UL6 were observed in capsids isolated from cells that had been pretreated with NEM. The presence of NEM prevents the formation of disulfide bonds that could occur upon cell lysis and exposure to oxidizing conditions. This result implies that the disulfide bonds in these capsids have already formed at the time of lysis. This may be surprising, since the nucleus generally is assumed to be a reducing environment (11, 16, 17). It is possible that the assembly pathway of the UL6 portal occurs in a microenvironment in the nucleus that allows disulfide bond formation. Several observations suggest that the conformation of UL6 is regulated in infected cells, and that the formation of portal rings takes place in an unusual environment. In infected cells, most of the UL6 has been reported to localize to replication compartments where procapsid assembly is believed to occur; however, we have reported that a subpopulation of UL6 could be observed in virus-induced chaperone-enriched (VICE) domains, which form adjacent to replication compartments during the earliest stages of HSV-1 infection (7, 22). VICE domains contain several components of the host protein quality control machinery, including molecular chaperones (e.g., Hsc70, the 20S proteasome, and ubiquitin) (7, 22). We have proposed that HSV-1 infection induces the formation of nuclear protein quality control centers to remodel or degrade aberrant nuclear proteins that otherwise would interfere with productive infection (22). Chaperone proteins in VICE domains may function to aid in the assembly of multimeric complexes. It is of interest that fluorescence recovery after photobleaching (FRAP) analysis indicates that the association of Hsc70 with VICE domains is dynamic (22), suggesting movement between VICE domains and replication compartments. It is possible that VICE domains provide a microenvironment conducive for disulfide bond formation and ring formation. According to this scenario, rings assembled in VICE domains would enter the replication compartments, where they would nucleate procapsid assembly. In any case, the presence of a subpopulation of UL6 in VICE domains suggests that its conformation is regulated by host cell chaperones.
The observation that the portal ring uses disulfide bond formation for stability is interesting in light of previous observations that HSV capsids contain disulfide bonds potentially involving VP5, triplex, and scaffold proteins (43, 48). We have confirmed the existence of disulfide bonds in capsids and virions and have demonstrated that these bonds contribute to overall capsid stability (38a). Viral capsids are dynamic structures that must protect viral genomes during the extracellular state while maintaining the ability to disassemble and release viral genomes during the next round of infection. Disulfide bonds may have several advantages for the creation of metastable capsids. In papilloma- and polyomaviruses, disulfide bonds have been shown to play an important role in the metastability of capsids (12, 14, 41). It is known that redox states are regulated differently in the cytoplasm, the nucleus, and the extracellular space (11, 16, 17). Once papilloma- and polyomaviruses enter the reducing environment of the cytoplasm, it appears that conformational changes occur which facilitate the uncoating and release of the viral genome to initiate infection. The observations that disulfide bonds are present in extracellular HSV capsids as well as in portal rings suggest a role for the redox state during HSV infection. Upon entry, capsids and tegument proteins are released into the cytoplasm, where rearrangements are possible (24). Capsids then are transported on microtubules and dock at the nuclear pore, where they release their genomes into the nucleus (30) (36).
In summary, we have now shown that intersubunit interactions of the portal ring of HSV-1 requires at least two types of protein-protein interactions: coiled-coil interactions and disulfide bond formation. UL6 plays several known roles in infection, including the nucleation of procapsid assembly, the docking of terminase, DNA encapsidation, and genome release during the next round of infection. It will be of interest to determine the precise role of disulfide bond cross-linking in portal ring formation, capsid assembly, and stabilization and genome release.
ACKNOWLEDGMENTS
We thank Greg Smith and the members of the Weller laboratory for helpful discussions and suggestions throughout the course of this study.
This work was supported by a grant from the NIH, AI37549, to S.K.W.
Footnotes
Published ahead of print on 18 May 2011.
REFERENCES
- 1. Adelman K., Salmon B., Baines J. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. U. S. A. 98:3086–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baines J., Weller S. K. 2005. Cleavage and packaging of herpes simplex virus 1 DNA. In Catalano C. (ed.), Viral genome packaging machines: genetics, structure and mechanism. Landes Bioscience, Austin, TX [Google Scholar]
- 3. Baker M., Jiang W., Rixon F., Chiu W. 2005. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 79:14967–14970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beard P., Taus N., Baines J. 2002. DNA cleavage and packaging proteins encoded by genes U(L)28, U(L)15, and U(L)33 of herpes simplex virus type 1 form a complex in infected cells. J. Virol. 76:4785–4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blewett E. L., Black D., Eberle R. 1996. Characterization of virus-specific and cross-reactive monoclonal antibodies to Herpesvirus simiae (B virus). J. Gen. Virol. 77:2787–2793 [DOI] [PubMed] [Google Scholar]
- 6. Brown J., McVoy M. A., Homa F. L. 2002. Packaging DNA into herpesvirus capsids, p. 111–155 In Bogner A. H. A. E. (ed.), Structure-function relationships of human pathogenic viruses. Kluwer Academic/Plenum Publishers, New York, NY [Google Scholar]
- 7. Burch A., Weller S. 2004. Nuclear sequestration of cellular chaperone and proteasomal machinery during HSV-1 infection. J. Virol. 78:7175–7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cardone G., et al. 2007. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology 361:426–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang J., Schmid M., Rixon F., Chiu W. 2007. Electron cryotomography reveals the portal in the herpesvirus capsid. J. Virol. 81:2065–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Conway J. F., Homa F. 2011. Nucleocapsid structure, assembly and DNA packaging of herpes simplex viru. In Weller S. K. (ed.), Alphaheresviruses: molecular virology. Caister Academic Press, Norwich, United Kingdom [Google Scholar]
- 11. Go Y.-M., Jones D. P. 2008. Redox compartmentalization in eukaryotic cells. Biochim. Biophys. Acta 1780:1273–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hare J., Chan J. 1968. Role of hydrogen and disulfide bonds in polyoma capsid structure. Virology 34:481–491 [DOI] [PubMed] [Google Scholar]
- 13. Higuchi R., Krummel B., Saiki R. K. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang E. S., Estes M. K., Pagano J. S. 1972. Structure and function of the polypeptides in simian virus 40. I. Existence of subviral deoxynucleoprotein complexes. J. Virol. 9:923–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huffman J., Newcomb W., Brown J., Homa F. 2008. Amino acids 143 to 150 of the herpes simplex virus type 1 scaffold protein are required for the formation of portal-containing capsids. J. Virol. 82:6778–6781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jones D. P., Go Y.-M. 2010. Redox compartmentalization and cellular stress. Diabetes Obes. Metab. 12(Suppl. 2):116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kietzmann T. 2010. Intracellular redox compartments: mechanisms and significances. Antioxid. Redox. Signal. 13:395–398 [DOI] [PubMed] [Google Scholar]
- 18. Koslowski K., et al. 1999. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J. Virol. 73:1704–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koslowski K., Shaver P., Wang X., Tenney D., Pederson N. 1997. The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J. Virol. 71:9118–9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lamberti C., Weller S. 1996. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology 226:403–407 [DOI] [PubMed] [Google Scholar]
- 21. Lamberti C., Weller S. K. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Livingston C. M., Ifrim M. F., Cowan A. E., Weller S. K. 2009. Virus-induced chaperone-enriched (VICE) domains function as nuclear protein quality control centers during HSV-1 infection. PLoS Pathog. 5:e1000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martinez R., Shao L., Weller S. K. 1992. The conserved helicase motifs of the herpes simplex virus type 1 origin-binding protein UL9 are important for function. J. Virol. 66:6735–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meckes D. G., Wills J. W. 2007. Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J. Virol. 81:13028–13036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nellissery J. K., Szczepaniak R., Lamberti C., Weller S. K. 2007. A putative leucine zipper within the herpes simplex virus type 1 UL6 protein is required for portal ring formation. J. Virol. 81:8868–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Newcomb W., Homa F., Brown J. 2005. Involvement of the portal at an early step in herpes simplex virus capsid assembly. J. Virol. 79:10540–10546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Newcomb W., Homa F., Thomsen D., Brown J. 2001. In vitro assembly of the herpes simplex virus procapsid: formation of small procapsids at reduced scaffolding protein concentration. J. Struct. Biol. 133:23–31 [DOI] [PubMed] [Google Scholar]
- 28. Newcomb W., et al. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923–10932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Newcomb W., Thomsen D., Homa F., Brown J. 2003. Assembly of the herpes simplex virus capsid: identification of soluble scaffold-portal complexes and their role in formation of portal-containing capsids. J. Virol. 77:9862–9871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ojala P. M., Sodeik B., Ebersold M. W., Kutay U., Helenius A. 2000. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 20:4922–4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Patel A., MacLean J. 1995. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology 206:465–478 [DOI] [PubMed] [Google Scholar]
- 32. Patel A., Rixon F., Cunningham C., Davison A. 1996. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology 217:111–123 [DOI] [PubMed] [Google Scholar]
- 33. Przech A., Yu D., Weller S. 2003. Point mutations in exon I of the herpes simplex virus putative terminase subunit, UL15, indicate that the most conserved residues are essential for cleavage and packaging. J. Virol. 77:9613–9621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sheaffer A., et al. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Singer G., Newcomb W., Thomsen D., Homa F., Brown J. 2005. Identification of a region in the herpes simplex virus scaffolding protein required for interaction with the portal. J. Virol. 79:132–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sodeik B., Ebersold M., Helenius A. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Steven A., et al. 1997. The making and breaking of symmetry in virus capsid assembly: glimpses of capsid biology from cryoelectron microscopy. FASEB J. 11:733–742 [DOI] [PubMed] [Google Scholar]
- 38. Steven A. C., Heymann J. B., Cheng N., Trus B. L., Conway J. F. 2005. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr. Opin. Struct. Biol. 15:227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a. Szczepaniak R., et al. 2011. Disulfide bond formation contributes to herpes simplex virus capsid stability and retention of pentons. J. Virol. 85:8625–8634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Trus B., et al. 2004. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78:12668–12671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walter G., Deppert W. 1975. Intermolecular disulfide bonds: an important structural feature of the polyoma virus capsid. Cold Spring Harb. Symp. Quant. Biol. 39:255–257 [DOI] [PubMed] [Google Scholar]
- 42. White C., Stow N., Patel A., Hughes M., Preston V. 2003. Herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J. Virol. 77:6351–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang C. C., Yang Y. Y., Lin K. L., Lin S. J. 2000. Different forms of HSV-1 VP22a within purified virion and infected cells. J. Microbiol. Immunol. Infect. 33:141–148 [PubMed] [Google Scholar]
- 44. Yang K., Baines J. 2006. The putative terminase subunit of herpes simplex virus 1 encoded by UL28 is necessary and sufficient to mediate interaction between pUL15 and pUL33. J. Virol. 80:5733–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang K., Homa F., Baines J. 2007. Putative terminase subunits of herpes simplex virus 1 form a complex in the cytoplasm and interact with portal protein in the nucleus. J. Virol. 81:6419–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu D., Weller S. 1998. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72:7428–7439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu D., Weller S. K. 1998. Genetic analysis of the UL15 gene locus for the putative terminase of herpes simplex virus type 1. Virol. 243:32–44 [DOI] [PubMed] [Google Scholar]
- 48. Zweig M., Heilman C. J., Hampar B. 1979. Identification of disulfide-linked protein complexes in the nucleocapsids of herpes simplex virus type 2. Virology 94:442–450 [DOI] [PubMed] [Google Scholar]