Abstract
We report here an unusual mechanism for enzyme regulation: the disassembly of all three subunits of RecBCD enzyme after its interaction with a Chi recombination hot spot. The enzyme, which is essential for the major pathway of recombination in Escherichia coli, acts on linear double-stranded DNA bearing a Chi site to produce single-stranded DNA substrates for strand exchange by RecA protein. We show that after reaction with DNA bearing Chi sites, RecBCD enzyme is inactivated and the three subunits migrate as separate species during glycerol gradient ultracentrifugation or native gel electrophoresis. This Chi-mediated inactivation and disassembly of purified RecBCD enzyme can account for the previously reported Chi-dependent loss of Chi activity in E. coli cells containing broken DNA. Our results support a model of recombination in which Chi regulates one RecBCD enzyme molecule to make a single recombinational exchange (‘one enzyme-one exchange’ hypothesis).
Keywords: RecBCD enzyme, Chi sites, Escherichia coli, genetic recombination, disassembly
Biological processes are frequently controlled by regulation of the activity of the first enzyme in the process. For example, many of the first enzymes in pathways of small molecule biosynthesis are subject to feedback inhibition or activation. DNA replication is likewise controlled at the initial stage, activation of the origin of replication. Here, we report experiments on the regulation of RecBCD enzyme, the first enzyme acting in the major pathway of homologous recombination in Escherichia coli.
Homologous recombination is a multistep process, involving many proteins, that can repair double-strand (ds) DNA breaks and generate new combinations of alleles. In E. coli there are multiple pathways of recombination; in wild-type cells the major (RecBCD) pathway depends upon the RecBCD enzyme (for review, see Smith 1989, 1998; Kowalczykowski et al. 1994). This enzyme contains three subunits, encoded by the recB, recC, and recD genes, with a composite mass of 330 kD; the active form of the enzyme contains one copy of each polypeptide (Taylor and Smith 1995a). The enzyme is inactive on circular dsDNA but binds tightly to a dsDNA end with a Kd of ∼0.1 nm, or ∼1/10 the concentration of one dsDNA end per E. coli cell (Taylor and Smith 1995a). In the presence of the essential cofactors ATP and Mg2+ the enzyme rapidly unwinds the DNA at ∼350 bp/sec (Taylor and Smith 1980). Upon encountering and acting at a properly oriented Chi site (5′-GCTGGTGG-3′) and continuing its unwinding of DNA, RecBCD enzyme makes single-stranded (ss) DNA with an end near Chi. When [ATP] > [Mg2+], this ssDNA end results from RecBCD enzyme nicking one DNA strand near Chi, followed by continued unwinding by the enzyme (Ponticelli et al. 1985; Taylor et al. 1985). When [Mg2+] > [ATP], RecBCD enzyme acts as a potent exonuclease (Wright et al. 1971) whose 3′ → 5′ exonuclease activity is suppressed on encountering a Chi site (Dixon and Kowalczykowski 1993). Continued unwinding or the derepression of a 5′ → 3′ exonuclease activity at Chi (Anderson and Kowalczykowski 1997a) provides a ssDNA end extending from Chi. This ssDNA end is a potent substrate for homologous DNA strand exchange by RecA protein, a reaction aided by the Chi- and RecBCD enzyme-mediated loading of RecA protein onto the ssDNA end (Anderson and Kowalczykowski 1997b). The joint DNA molecules thereby produced appear to be resolved into recombinant or repaired DNA molecules by some combination of the RuvABC, RecG, and other proteins (for review, see Taylor 1992; West 1996). Recombination and repair also appear to involve DNA replication (for review, see Smith 1991; Kogoma 1997).
In addition to RecBCD enzyme acting on the DNA at Chi, Chi changes RecBCD enzyme. After nicking the DNA at Chi, RecBCD enzyme loses its ability to nick the DNA at a properly oriented Chi site encountered subsequently on the same DNA molecule (Taylor and Smith 1992). Although the enzyme continues to unwind this DNA, it loses the ability to unwind a subsequently encountered DNA molecule or to nick at a Chi site on it. A parallel change is also seen in vivo: Chi on a linear DNA molecule reduces or abolishes the activity of Chi on another DNA molecule via a change in RecBCD enzyme (Köppen et al. 1995; Myers et al. 1995; see Discussion). These alterations of enzymatic activity regulate RecBCD enzyme and hence homologous recombination. We report here a physical basis for the Chi-mediated loss of activity on subsequently encountered DNA.
Results
Chi-dependent loss of three activities of RecBCD enzyme
As RecBCD enzyme has <40% probability of nicking DNA at Chi when it passes a single, correctly oriented Chi site (Taylor and Smith 1992), we used a 6- to 10-fold molar excess of a 345-bp DNA fragment, denoted Chi+, containing three tandem Chi sites to inactivate RecBCD enzyme. In this way enzyme that was not inactivated during the first passage could be inactivated during subsequent passages through the Chi+ DNA. As a control we used a similar DNA fragment, denoted Chi0, lacking the Chi inserts. After an initial incubation with unlabeled Chi+ or Chi0 DNA, RecBCD enzyme activities were assayed by incubation with non-homologous [3H] DNA to measure dsDNA exonuclease activity (Figs. 1 and 2) or with nonhomologous 32P-labeled DNA with or without Chi to measure DNA-unwinding and Chi-nicking activities (Fig. 2).
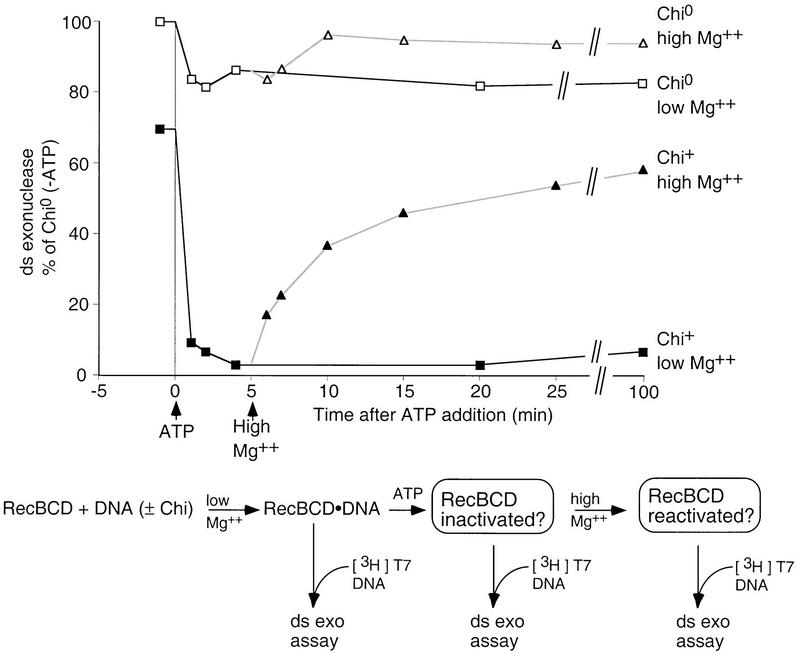
Figure 1.
Rapid inactivation and slower reactivation of RecBCD enzyme. RecBCD enzyme was incubated with a 10-fold molar excess of unlabeled Chi0 (□, ▵) or Chi+ (█, ▴) DNA for 5 min under standard reaction conditions (but without ATP) to allow binding of enzyme to DNA ends, and reactions started by addition of ATP to 5 mm (low Mg2+, □, █). After 5 min reaction, excess Mg2+ (total 13 mm) was added to a portion of each reaction (high Mg2+, ▵, ▴). Samples taken at the indicated times were assayed for dsDNA exonuclease activity (Eichler and Lehman 1977; to assure linearity, <30% of the substrate was made acid-soluble in any assay). Activity is expressed as a percentage of that measured for enzyme incubated with Chi0 DNA, prior to addition of ATP.
Figure 2.
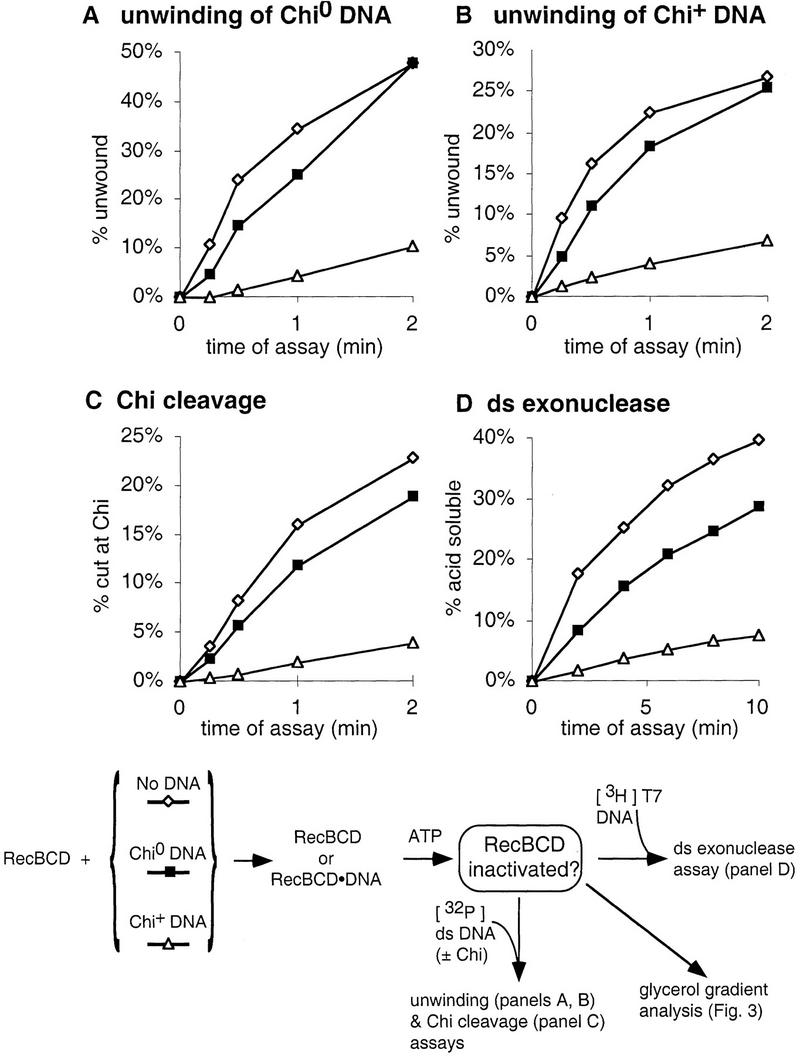
Chi-dependent loss of RecBCD enzyme activities. RecBCD enzyme was reacted without DNA (⋄) or with Chi0 (█) or Chi+ DNA (▵) for 2.5 min and the reactions were stopped by addition of EDTA to 5 mm. Samples of the reacted enzyme were added to radioactive pBR322 DNA (χ0 or χ+F χ+H) for Chi cleavage and unwinding assays, or to radioactive T7 DNA for dsDNA exonuclease assay under high Mg2+ conditions (Eichler and Lehman 1977). The fraction of the radioactive DNA that was cleaved at Chi sites, unwound or rendered acid-soluble is plotted vs. the duration of the assay.
Reaction with a 10-fold molar excess of Chi+ DNA produced a rapid, almost total loss (>30-fold reduction) of ds exonuclease activity after reaction with Chi+ DNA, but <20% loss after reaction with Chi0 DNA (Fig. 1). DNA bearing a single Chi site gave a similar, but less extensive (15- vs. 30-fold reduction) reduction (data not shown). Most of the inactivation seen in Figure 1 occurred within the first minute of reaction, persisted for 100 min in this experiment, and persisted apparently indefinitely if ATP concentrations were maintained above the Mg2+ concentration (by the use of an ATP-regenerating system; data not shown). The slight (30%) loss of activity before the addition of ATP to the Chi+ substrate probably reflects occasional inactivation of the enzyme as it traversed the already bound Chi+ DNA before attacking the 3H assay substrate. After reaction with Chi0 DNA, all of the DNA was unwound (data not shown), yet the enzyme was not inactivated (Fig. 1). Hence, the inhibition of RecBCD enzyme by ssDNA reaction products, seen under some reaction conditions (Anderson and Kowalczykowski 1998), does not occur here.
Enzyme inactivated by reaction with Chi+ DNA can be reactivated by subsequent addition of Mg2+ in excess over ATP (Dixon et al. 1994; Fig. 1). About 50% of the initial dsDNA exonuclease activity was recovered in 15 min after addition of excess Mg2+. Unwinding and Chi-cleavage activities were also recovered (data not shown). The slight loss of activity seen upon incubation with Chi0 DNA was also recovered upon addition of excess Mg2+, suggesting that the loss resulted from a mechanism similar to that of Chi-mediated inactivation. Little or no loss of unwinding, Chi cleavage or dsDNA exonuclease activity was seen if Mg2+ levels [10 mm] were always higher than the ATP level (data not shown).
All RecBCD enzyme activities assayed were inactivated to a similar extent by Chi-containing DNA (Fig. 2). This extensive loss of activities after reaction with Chi-containing DNA has been reported previously (Taylor and Smith 1992; Dixon et al. 1994). The initial reaction rates (Table 1A) show that, in this experiment, all activities were reduced six- to eightfold by prior incubation with Chi+ DNA, but less than twofold by incubation with Chi0 DNA. Chi-dependent inactivation in this experiment was less drastic than that in Figure 1, perhaps because of the shorter incubation and smaller excess of DNA over enzyme (6-fold rather than 10-fold). Although RecBCD enzyme activity can be recovered by addition of excess Mg2+ (13 mm Mg2+ and 5 mm ATP; Fig. 1), such recovery does not occur during the ds exonuclease assays, which used excess Mg2+; this is seen both by comparing the dsDNA exonuclease assays (Fig. 2D) with the other assays in Figure 2D, which used excess ATP, and by noting the high level of inactivation observed by dsDNA exonuclease assay in Figure 1. Reactivation may have been prevented by the low enzyme concentration in the assay or by the reaction conditions.
Table 1.
RecBCD enzyme inactivation and subunit disassembly
| A. RecBCD enzyme activities after prior incubationwith Chi+ or Chi0 DNA | ||
|---|---|---|
|
Enzyme activity
|
Initial rate (% of no DNA) after incubation with
|
|
| Chi0 DNA | Chi+ DNA | |
| Unwinding of Chi0 DNA | 52 | 12 |
| Unwinding of Chi+ DNA | 64 | 13 |
| Chi cleavage | 73 | 12 |
| Double-strand exonuclease | 67 | 17 |
| B. Distribution of RecBCD polypeptides after prior incubation with Chi+ or Chi0 DNA | ||||
|---|---|---|---|---|
|
DNA
|
Species
|
Polypeptides (%)
|
||
| RecB | RecC | RecD | ||
| None | free subunits | 6 | 1 | 4 |
| RecBCD | 77 | 73 | 91 | |
| RecBC | 17 | 20 | ||
| Chi0 | free subunits | 13 | 8 | 5 |
| RecBCD | 77 | 84 | 91 | |
| RecBC | 6 | 7 | ||
| Chi+ | free subunits | 68 | 66 | 67 |
| RecBCD | 21 | 21 | 28 | |
| RecBC | 8 | 8 | ||
(A) After prior incubation of RecBCD enzyme with unlabeled Chi+ or Chi0 DNA, several RecBCD enzyme activities were assayed, using radioactive DNA substrates, all described in Fig. 2. Reaction rates were estimated by linear regression from the initial, linear portions of the data in Fig. 2 and are expressed as a percentage of the rate obtained after prior incubation without DNA.
(B) Samples of the incubations in A were separated by glycerol gradient centrifugation, and fractions assayed for RecBCD polypeptides. The recovery of each species was calculated from the data in Fig. 4 and is expressed as a percentage of the total recovery of each polypeptide (fractions 5–50). For the reaction without DNA the fractions summed were 29–33 (RecB), 35–41 (RecC), 26–30 (RecD), 16–24 (RecBCD), and 25–28 (RecBD). For reactions with Chi0 DNA the fractions summed were 30–35 (RecB), 35–41 (RecC), 24–30 (RecD), 5–23 (RecBCD), and 24–28 (RecBC). The Chi+ data used the same fractions as the Chi0 data, except for RecB (fractions 28–33) and RecC (fractions 34–41).
We have thus shown that Chi-mediated inactivation of RecBCD enzyme is rapid, results in the loss of all RecBCD enzyme activities tested, and persists indefinitely in the absence of excess Mg2+. We next investigated the nature of the change to the enzyme that results in these effects.
Disassembly of the three RecBCD enzyme subunits: glycerol-gradient analysis
Based on the properties of Chi and of recD mutants of E. coli, Thaler et al. (1988) hypothesized that the RecD subunit of RecBCD enzyme is ejected when the enzyme encounters Chi. To determine whether ejection of RecD, or any other subunits, was responsible for the Chi-dependent inactivation reported above, we first used glycerol-gradient ultracentrifugation to assess the state of assembly of the RecBCD enzyme subunits after inactivation by Chi. The results showed, to our surprise, that all three subunits were disassembled.
To examine the Chi-inactivated RecBCD enzyme physically, RecBCD enzyme was incubated with Chi+ DNA and then centrifuged through a glycerol gradient (Figs. 3 and 4). Whereas the three subunits of mock-reacted RecBCD enzyme cosedimented (Fig. 3A), very little intact enzyme was observed after reaction with Chi+ DNA (Fig. 3C), as expected from the low dsDNA exonuclease activity (17% of the input) of the reaction products. The RecB and RecC polypeptides were recovered in good yield (58% and 61%) but, unexpectedly, sedimented at very different rates in the glycerol gradient. Whereas RecC sedimented as expected for the free polypeptide, RecB sedimented faster than free RecB, but slower than RecBC, as shown by their sedimentation positions in separate gradients (marked below Fig. 3C). As RecB and RecC have very similar molecular masses (134 and 129 kD; Finch et al. 1986a,b) and sedimentation rates (data not shown), a dimer of RecB would be expected to sediment at about the position of RecBC. The faster sedimentation of RecB thus cannot be caused either by homodimerization or by formation of a RecB–RecD complex, as the majority of RecD did not cosediment with RecB (Fig. 4C). The most plausible explanation for the faster sedimentation of RecB, a RecB–DNA complex, is supported by experiments described below.
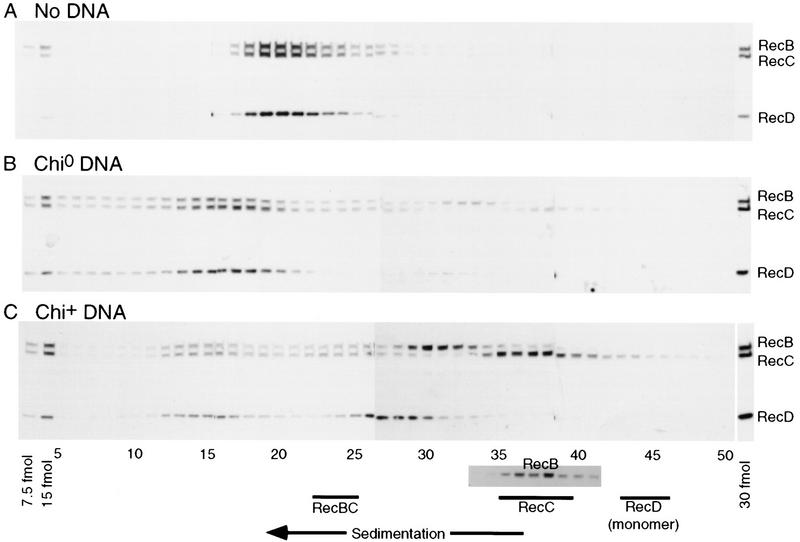
Figure 3.
RecBCD enzyme subunits disassemble after reaction with Chi-containing DNA. RecBCD enzyme was reacted for 2.5 minutes with no DNA (A), Chi0 DNA (B) or Chi+ DNA (C) and assayed for enzyme activities (described in Fig. 2 and Table 1A). The reaction products were fractionated by ultracentrifugation on low-salt glycerol gradients. The RecBCD polypeptides in each fraction were separated on SDS–polyacrylamide gels, transferred to membranes and detected by incubation with mouse anti-RecB, RecC, and RecD monoclonal antibodies. Each panel shows fractions 5–50 of the 68 fractions collected, flanked by known amounts of RecBCD enzyme. The sedimentation positions of uncomplexed RecB (SDS gel data in inset), RecC and RecBC (black lines) are shown below C, determined from similar gradients loaded with the individual polypeptides or a mix of the two. The expected position of monomeric RecD (black line) was inferred from the sedimentation position of BSA (whose molecular mass is similar to that of RecD) included in each sample.
Figure 4.
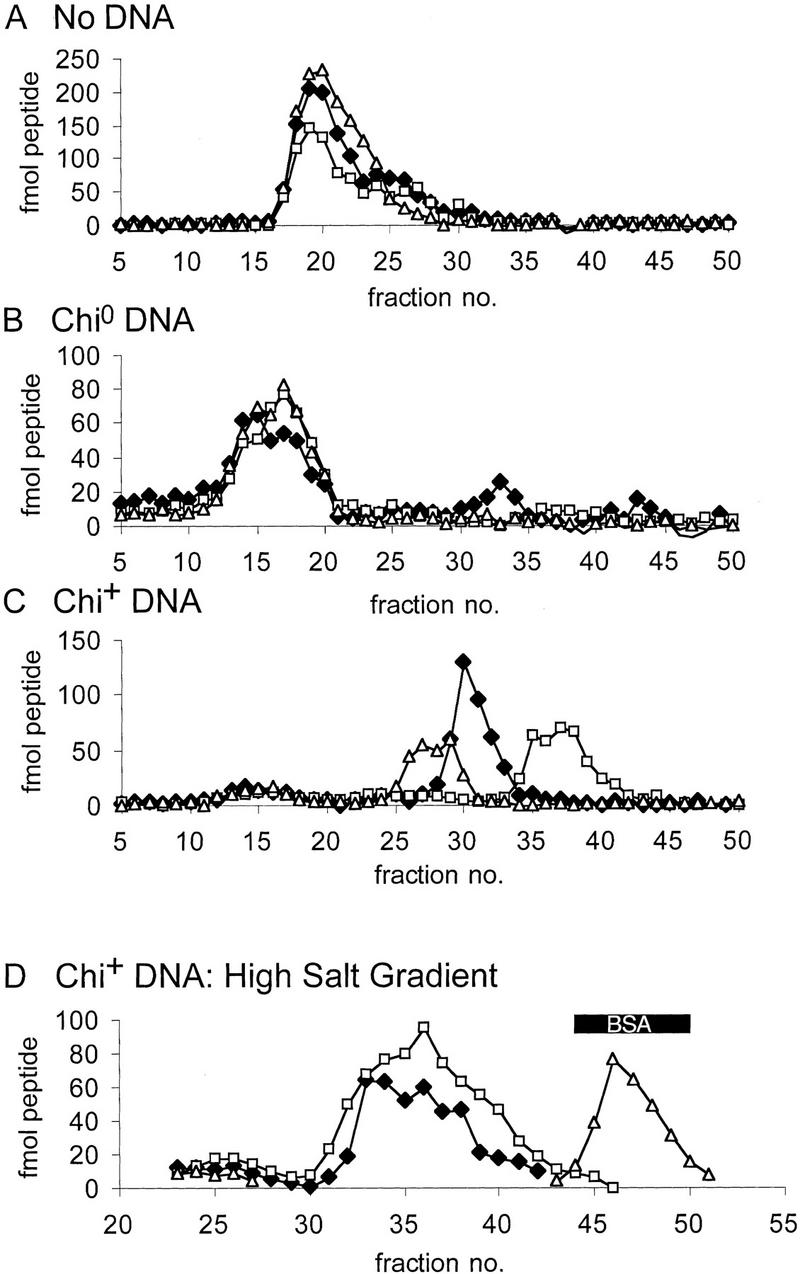
Quantitation of RecBCD enzyme subunit disassembly. Total recovery, per gradient fraction, of each polypeptide is plotted against gradient fraction for all the fractions analyzed in Fig. 3 and for a similar reaction, using Chi+ DNA, analyzed on a high salt (500 mm NaCl) gradient. Recoveries of RecB (♦), RecC (□) and RecD (▵), respectively, were 86%, 67%, and 69% for A; 55%, 56%, and 33% for B; 58%, 61%, and 35% for C, and 68%, 100%, and 57% for D.
After reaction with Chi+ DNA, RecD was separated physically from the other subunits. As shown in Figures 3 and 4, RecD did not cosediment with RecB or RecC or at the position expected (based on the almost identical molecular masses of bovine serum albumin and RecD) for monomeric RecD. Its position (faster than RecC but slower than RecBC) suggested that it existed primarily as a trimeric species. Overexpressed RecD aggregates during purification (Masterson et al. 1992), and disassembled RecD also apparently aggregates. Whereas we cannot eliminate the possibility that the fast sedimentation of RecD is caused by its binding DNA, this seems unlikely, as purified RecD binds DNA only very weakly (Chen et al. 1997). We infer that after reaction with Chi+ DNA RecD dissociated from RecB and RecC and aggregated.
To further investigate the RecD subunit and the faster-than-expected sedimentation of RecB, RecBCD enzyme was reacted with Chi+ DNA and sedimented through a high-salt glycerol gradient (0.5 m NaCl). Native RecBCD enzyme is stable at this salt concentration (Lieberman and Oishi 1973; data not shown). RecD was recovered in high yield from the high-salt gradient and sedimented as a monomer, as shown by its cosedimentation with bovine serum albumin (Fig. 4D). The poor recovery of RecD from low salt gradients (50 mm; Fig. 3) presumably resulted either from rapidly sedimenting multimeric RecD or from the (hydrophobic) RecD being lost from solution. RecB and RecC cosedimented under these high-salt conditions, presumably because of displacement of the DNA bound to RecB. Thus, high salt disrupts the RecD aggregate and the putative RecB–DNA complex. The results showed that after reaction with Chi+ DNA the three subunits of RecBCD enzyme were physically separate, with RecB apparently complexed with DNA. Further evidence for a RecB–DNA complex after reaction with Chi+ DNA is presented below.
The state of association of the subunits of RecBCD enzyme after reaction with Chi+ or Chi0 DNA quantitatively reflects the degree of inactivation of the enzyme (Fig.4, Table 1). After reaction with Chi+ DNA 66%–68% of each polypeptide was free, and 21%–28% was in native RecBCD enzyme (Table 1B), in good agreement with the observed Chi-dependent loss of enzyme activities (12%–17% remaining, Table 1A). The modest loss of activity after incubation with Chi0 DNA is consistent with the minor release of free RecB, RecC, and RecD subunits (5%–13% of total, Table 1B) seen in Fig. 3B. Intact enzyme remaining after reaction with DNA (Figs. 3, B and C) was presumably bound to DNA fragments and hence sedimented faster than free enzyme (Fig. 3A). Enzyme that had been incubated with the 345-bp DNA substrate in the absence of ATP, an essential cofactor for RecBCD enzyme reactions, sedimented even faster than that in Figure 3, B and C (data not shown), in accord with its high binding affinity under that condition (Taylor and Smith 1995a).
The data in Figures 3 and 4 thus show that the extensive disassembly of RecBCD enzyme into its three subunits is dependent on the presence of Chi sites on the DNA substrate. Similar results were obtained in many independent glycerol-gradient separations using these and other detection methods (data not shown) and in the native gel analyses described next.
Disassembly of the three RecBCD enzyme subunits: native gel analysis
The glycerol-gradient separations described above enabled us to monitor the fate of all three subunits of RecBCD enzyme but were cumbersome and of limited resolution. To confirm and extend the above observations, we turned to native PAGE (Taylor and Smith 1995a). We detected species containing RecB or RecC by Western analysis using anti-RecB and anti-RecC monoclonal antibodies and confirmed their identities by comparison to the migration of individual RecB and RecC polypeptides, RecBC and RecBCD enzyme on the gels (Fig. 5). The isolated RecB and RecC subunits were well separated from RecBC and RecBCD enzyme (Fig. 5A), the order of their migration corresponding to the calculated ratio of the charge to the mass of each complex (A.F. Taylor, unpubl.). Isolated RecD has a slight positive charge at the pH of the gel (7.0) and migrated in the direction opposite to that of the other species and hence was not detected in these experiments (data not shown). As noted previously (Masterson et al. 1992; Taylor and Smith 1995a), purified RecBCD enzyme preparations typically contain a small amount of RecBC enzyme, which remained at comparable levels throughout these experiments.
Figure 5.
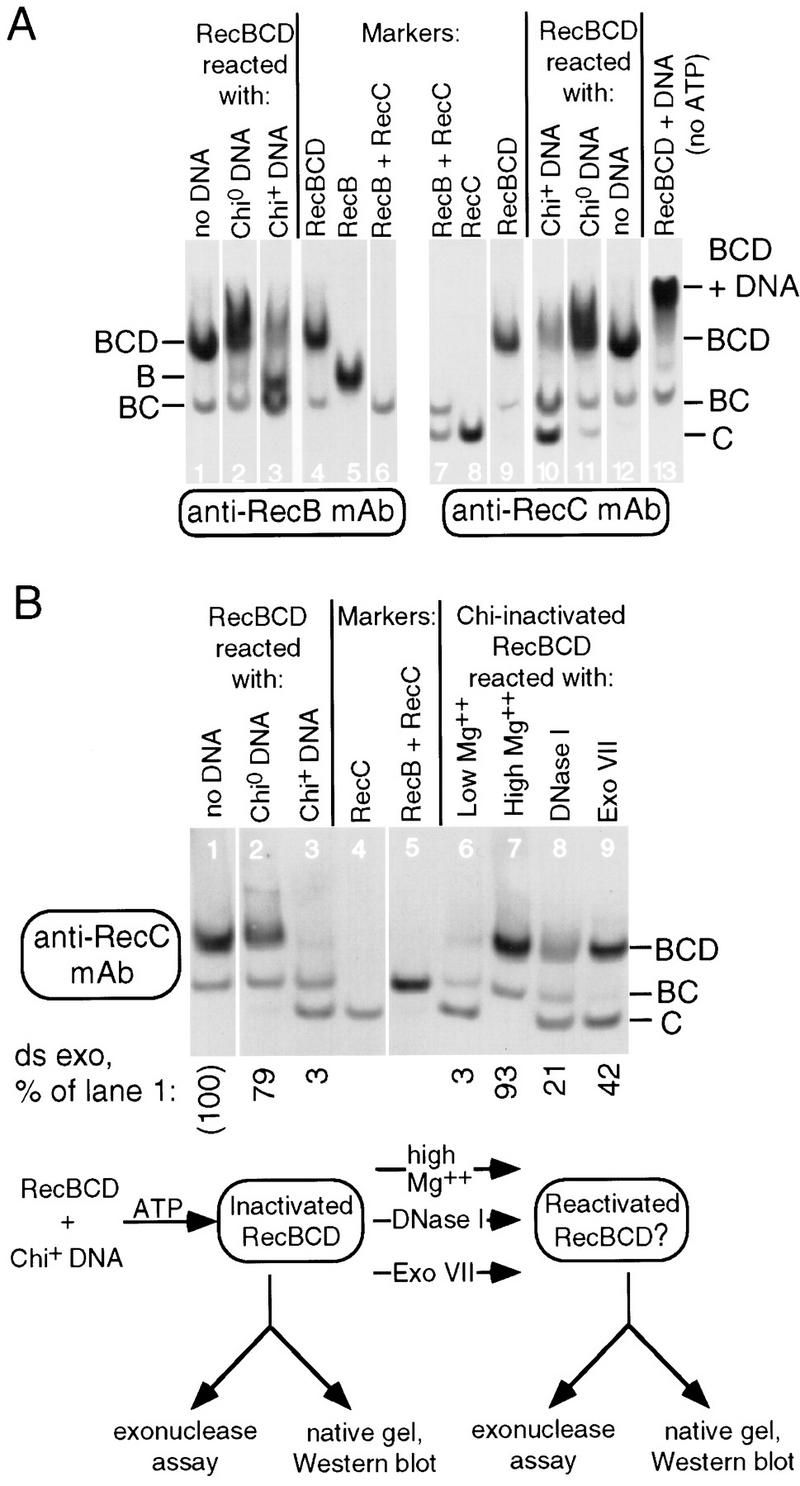
Disassembly and reassembly of RecBCD enzyme. (A) Chi-dependent disassembly of RecBCD enzyme. RecBCD enzyme was reacted for 1 min without DNA or with Chi0 or Chi+ DNA, and the products (from 25 fmoles of RecBCD enzyme) were separated on native polyacrylamide gels and detected by Western analysis. RecBCD (20 fmoles), RecB (50 fmoles), RecC (30 fmoles), or RecB plus RecC (15 fmoles each) were run as markers. (B) Nuclease and Mg2+-dependent reassembly and reactivation of RecBCD enzyme. RecBCD enzyme was reacted for 10 min without DNA or with Chi+ or Chi0 DNA. Samples of the Chi+ DNA-inactivated RecBCD enzyme were incubated for a further 120 minutes with low Mg2+ (3 mm), high Mg2+ (13 mm), DNase I or exonuclease VII. Samples were assayed for dsDNA exonuclease activity and were separated on native polyacrylamide gels, together with RecC and RecB plus RecC markers (10 fmoles of each polypeptide marker or reaction product), and detected with anti-RecC monoclonal antibodies.
Native gel analysis of RecBCD enzyme that had reacted with Chi0 or Chi+ DNA mirrored the results obtained with glycerol gradients. Reaction of RecBCD enzyme with Chi+ DNA caused most of the RecB and RecC to be released as free subunits (Fig. 5 A, lanes 3 and 10, and B, lane 3), whereas reaction of Chi0 DNA with RecBCD enzyme left most of the enzyme unchanged (Fig. 5, A, lanes 2 and 11, and B, lane 2). Small amounts of RecB and RecC were released after reaction with Chi0 DNA, but the majority of the polypeptides migrated as free RecBCD (cf. Fig. 5A, lanes 1 and 12) or DNA-bound RecBCD (cf. lane 13). Reconstruction experiments (not shown) revealed that a RecB–DNA complex migrated at the same rate as free RecB in these gels, preventing us from examining, by gel analysis, the existence of such a complex after Chi-mediated inactivation of RecBCD enzyme.
Glycerol-gradient and native gel analyses thus both reveal that RecB and RecC are released as free subunits, uncomplexed with any other subunit of the enzyme, as a result of RecBCD enzyme’s reaction with, and inactivation by, Chi+ DNA.
Reactivation and reassembly of Chi-inactivated RecBCD enzyme by nucleases
After inactivation of RecBCD enzyme by Chi the RecB subunit was not complexed with RecC or RecD but nonetheless sedimented faster than free RecB (Fig. 3C). We hypothesized that RecB remained bound to a DNA fragment whose mass (∼60 kD) and high density (1.7) would increase the sedimentation velocity of RecB. Furthermore, a DNA fragment bound to RecB might prevent the reassociation, and hence reactivation, of RecBCD enzyme. We therefore tested the ability of nucleases to reactivate Chi-inactivated RecBCD enzyme, as outlined in Figure 5.
We found that either E. coli exonuclease VII or bovine pancreatic DNase I reactivated RecBCD enzyme, as seen by ds exonuclease assays, and caused the reappearance of intact RecBCD enzyme, as seen by native gel analysis (Fig. 5B). RecBCD enzyme was inactivated by incubation with Chi+ DNA; in this experiment 3% of the initial ds exonuclease activity remained. Samples further incubated with exonuclease VII or DNase I regained 42% or 21% of the initial activity in the subsequent assay for ds exonuclease. The small amount of exonuclease VII or DNase I added to reactivate RecBCD enzyme contributed very little (1.4% and 0.2%, respectively) to the exonuclease activity measured in the subsequent assay. Incubation with heat-inactivated nucleases did not reactivate RecBCD enzyme (data not shown). For comparison, Chi-inactivated RecBCD enzyme further incubated with excess Mg2+ regained 93% of the initial activity. The action of exonuclease VII and DNase I, which specifically degrade DNA (Chase and Richardson 1974; Moore 1981), indicates that DNA blocks the reactivation and reassembly of Chi-inactivated RecBCD enzyme. These observations support the proposal stated above that after reaction at Chi RecB remains bound to a DNA fragment.
Discussion
We have shown that the permanent inactivation of RecBCD enzyme by Chi sites in duplex DNA occurs by the disassembly of the enzyme into its three constituent subunits. We hypothesize that this inactivation occurs in two distinct steps. Upon encountering a Chi sequence, RecBCD enzyme undergoes its first change: it retains its ability to travel along the DNA and to cut a hairpin DNA structure at the distal end of the DNA but loses its ability to nick at subsequently encountered Chi sites on the same DNA molecule (Taylor and Smith 1992). The second change, the disassembly of the enzyme into three inactive subunits, may occur either during continued unwinding beyond Chi or upon reaching the end of the DNA.
We observed distinct fates for the three disassembled subunits of RecBCD enzyme. The RecC subunit was released free into solution, whereas the RecB subunit appeared to remain in a noncovalent complex with ssDNA. RecD was recovered as an oligomer separate from either RecB or RecC (Figs. 3 and 4), but it was recovered as a free monomer after treatment with high salt (see Results), consistent with its tendency to self-associate (Masterson et al. 1992). Under appropriate conditions the RecD subunit is able to reassemble with the other enzyme subunits to recreate active RecBCD enzyme (Amundsen et al. 1986).
Inactivation of RecBCD enzyme by disassembly of all three subunits is an unusual mechanism of regulation of enzyme activity, but we are aware of related examples. In E. coli the σ factor of RNA polymerase dissociates after the initiation of transcription (Helmann and Chamberlin 1988), although in that case only one of the five subunits dissociates, and it reassociates and restores promoter-recognition to the enzyme after the termination of transcription. In Salmonella typhimurium the FlgM factor regulates transcription by dissociating the flagellar-gene-specific σ factor from RNA polymerase (Chadsey et al. 1998). Other multiprotein complexes, such as ribosomes and spliceosomes, may also be regulated by subunit disassembly (Moore et al. 1993; Merrick and Hershey 1996).
We discuss below a model for the two-step inactivation of RecBCD enzyme, evidence for its occurrence in E. coli cells, and the implications of this inactivation for the regulation of homologous recombination.
A two-step model for Chi-mediated inactivation of RecBCD enzyme
RecBCD enzyme binds to the end of duplex DNA (Taylor and Smith 1995a) to form a stable initiation complex (Fig. 6A) in which the RecB subunit contacts the 3′-terminated strand and the RecC and RecD subunits contact the 5′-terminated strand (Ganesan and Smith 1992). Upon addition of ATP (with [ATP] > [Mg2+]), the enzyme travels along the DNA and unwinds it via a loop–tail intermediate (Fig. B; Taylor and Smith 1980). The loop and associated short tail are on the strand at whose 3′ terminus the enzyme entered the DNA (Braedt and Smith 1989) and on which Chi is recognized (Bianco and Kowalczykowski 1997). This suggests an interaction between RecB and the loop structure, consistent with the ssDNA-dependent ATP hydrolysis activity (Boehmer and Emmerson 1992) and limited helicase activity of isolated RecB (Boehmer and Emmerson 1992; Phillips et al. 1997).
Figure 6.
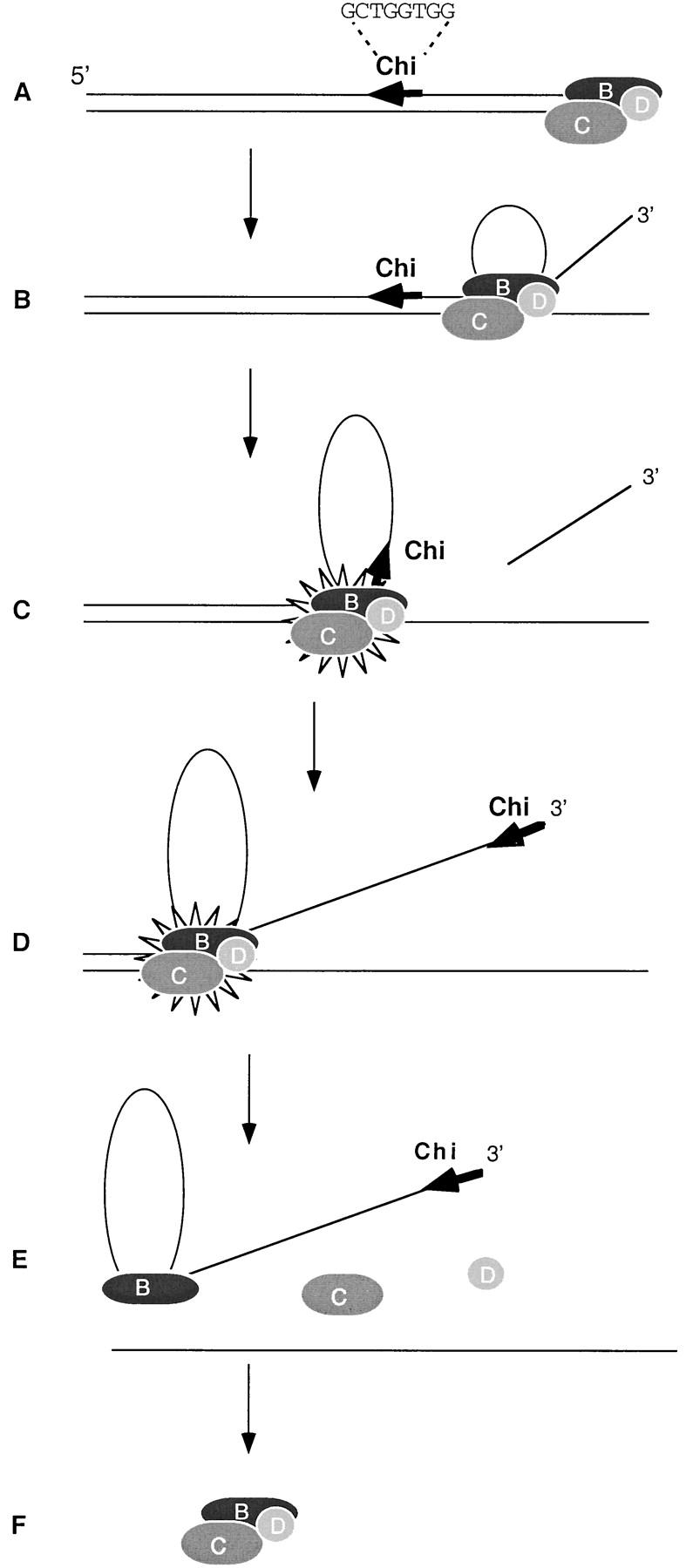
A model for the two-step inactivation of RecBCD enzyme by Chi. RecBCD enzyme binds to a dsDNA end (A) and unwinds the DNA with the production of an ssDNA loop and tail (B). At Chi it produces a 3′ end (C); continued unwinding produces the 3′ Chi tail (D) onto which RecA protein is loaded (not shown). After encountering Chi, RecBCD enzyme is depicted in an altered form (C and D; the first change). During continued unwinding or upon reaching the end of the DNA the enzyme disassembles into separate subunits (E; the second change). Reassembly is prevented by the DNA entrapped within RecB (E). Removal of this DNA, via nuclease treatment or incubation with excess Mg2+, allows reassembly and reactivation of the enzyme (F). (See text for details).
The first step of the inactivation of RecBCD enzyme occurs upon the enzyme’s encountering a Chi site (Fig. 6C). At this point two events occur, perhaps simultaneously. The “upper” strand of the DNA is nicked a few nucleotides to the 3′ side of the Chi octamer (Taylor et al. 1985; Taylor and Smith 1995b), and the enzyme loses its ability to act at a subsequently encountered Chi site on the same DNA molecule (Taylor and Smith 1992). The Chi-modified enzyme continues to travel along and unwind the initial DNA molecule (Fig. 6D; Taylor et al. 1985; Taylor and Smith 1992). The nature of the first change in the enzyme at Chi is still unknown. It has been hypothesized to be the simple release of RecD (Thaler et al. 1988), implying that the active species remaining on the initial DNA molecule is RecBC enzyme, although some evidence suggests otherwise (Taylor and Smith 1992; Anderson et al. 1997). Under some conditions RecBC enzyme can unwind dsDNA at ∼25% the rate of RecBCD enzyme (Korangy and Julin 1994), but it is inactive under the conditions used here (Palas and Kushner 1990; Dixon et al. 1994). The active species after the first change may, however, be RecBC enzyme, as the topology and hence the activities of RecBC enzyme binding afresh to the ends of a duplex DNA may differ from that of RecBC enzyme generated by the (hypothesized) ejection of RecD during unwinding. A conformational change in the RecB subunit of the enzyme has been proposed as an alternative mechanism (Yu et al. 1998). In the absence of definitive information we merely depict the enzyme as altered upon its encounter with Chi (Fig. 6C,D).
The second step in the inactivation of RecBCD enzyme occurs during or after continued unwinding beyond Chi. As a result of the first change at Chi, the enzyme eventually undergoes a second change: It dissociates into its separate subunits (Fig. 6E) and hence loses all of its activities on subsequently encountered dsDNA molecules (Masterson et al. 1992). This disassembly may occur either during continued unwinding beyond Chi or when the enzyme reaches the distal end of the DNA. Release during unwinding may reflect reduced processivity of unwinding by RecBCD enzyme after the first change to the enzyme; processivity is reduced under certain reaction conditions (Roman et al. 1992) or by mutation of the ATP-binding site in RecD (Korangy and Julin 1992), showing it to be sensitive to subtle changes in the subunits of the enzyme. The RecC subunit (and the RecD subunit, if it is still present) is released free into solution. If the enzyme has reached the end of the DNA, RecB remains trapped within the remaining loop and/or tail of ssDNA on the upper DNA strand. If the enzyme has not reached the end, RecB is trapped within a partially unwound structure resembling a loop–tail unwinding structure (Taylor and Smith 1980, as in Fig. 6D). Free RecB and RecC polypeptides can rapidly associate to form RecBC (Masterson et al. 1992; Korangy and Julin 1993): we therefore hypothesize that DNA bound to RecB prevents reassociation with RecC, perhaps via steric hindrance or a conformational change (Phillips et al. 1997; Yu et al. 1998). In the absence of further treatment the enzyme remains inactive for >1 hr (Fig. 1).
The Chi-inactivated enzyme can be reactivated by treatment with DNases or excess Mg2+ (Figs. 1 and 5). Free Mg2+ may stimulate the dsDNA exonuclease activity of the residual active RecBCD enzyme (Eggleston and Kowalczykowski 1993), allowing it, like exogenous DNase, to digest the ssDNA bound to RecB. Alternatively, excess Mg2+ may stimulate the helicase activity of RecB (Boehmer and Emmerson 1992; Phillips et al. 1997), allowing it to roll off the end of the DNA. The three enzyme subunits, once free in solution (Fig. 6F), then reassemble rapidly to form fully active RecBCD enzyme (Lieberman and Oishi 1974; Amundsen et al. 1986; Masterson et al. 1992).
In vivo evidence for the second step of Chi-mediated inactivation
Two types of experiments in E. coli have shown that Chi on a linear DNA molecule blocks the activity of Chi on another DNA molecule, via the second change of RecBCD enzyme. (1) The induction of bacteriophage λ terminase in vivo linearizes a plasmid carrying a λ cos site and allows entry of RecBCD enzyme. When the linearized plasmid carries Chi sites, it protects a separate linearized Chi0 plasmid from degradation (Kuzminov et al. 1994) and reduces, but does not abolish, the hot spot activity of Chi on injected, nonreplicating λ DNA (Myers et al. 1995). (2) After bleomycin treatment, which presumably makes multiple double-strand breaks in the E. coli chromosome and allows RecBCD enzyme access to the 1009 Chi sites on the chromosome (Burland et al. 1997), hot spot activity of Chi on infecting λ DNA is reduced strongly for at least 2 hr (Köppen et al. 1995). The dsDNA exonuclease activity of RecBCD enzyme, as measured in extracts or by the ability of phage T4 gene 2− mutants to grow, is also strongly reduced by bleomycin treatment.
The RecBCD enzyme-specificity of Chi hot spot activity and of the assays for ATP-dependent dsDNA exonuclease indicates that these effects of Chi are via an effect on RecBCD enzyme. The presence of a plasmid expressing recD reverses the effects of Chi partially or completely (Köppen et al. 1995; Myers et al. 1995). Where tested (Köppen et al. 1995), the recovery of RecBCD enzyme activity occurred 30–120 min after induction of recD expression. The eventual recovery of Chi activity may result from synthesis and assembly of new RecBCD enzyme molecules, which may be limited by the availability of RecD.
This Chi-mediated loss of RecBCD enzyme activity also persists for at least 1 hr in vitro with [ATP] > [Mg2+] (Fig. 1). This effect of Chi on purified RecBCD enzyme was barely detectable with [Mg2+] > [ATP] (data not shown). The similarity between the long-lasting effect of Chi in vitro at low [Mg2+] and the effects seen in vivo suggests that reaction conditions with low [Mg2+] better approximate those in E. coli cells than do those with excess [Mg2+]. A corollary is that the nicking of one DNA strand at Chi during DNA unwinding seen in vitro with excess ATP (Ponticelli et al. 1985; Taylor et al. 1985) may also occur in vivo. Examination of the oligomeric state of RecBCD enzyme after interaction with Chi sites in vivo may help test this hypothesis.
Regulation of homologous recombination
Current evidence indicates that Chi, via its two changes of RecBCD enzyme, regulates homologous recombination in two ways. Chi stimulates recombination at and to one side of itself on the DNA molecule on which it resides (e.g., Stahl et al. 1975; Dabert and Smith 1997). RecBCD enzyme makes ssDNA with Chi near its 3′ end (the ‘Chi tail’; Taylor et al. 1985; Fig. 6). The generation of additional 3′ ssDNA ends “downstream” of the initial one is precluded by the first change of RecBCD enzyme by Chi (Taylor and Smith 1992). The localized stimulation of recombination by Chi is accounted for adequately by the production of ssDNA with a 3′ end near Chi (Taylor et al. 1985; Dixon and Kowalczykowski 1993; Fig. 6), the loading of RecA protein preferentially onto this ssDNA by RecBCD enzyme (Anderson and Kowalczykowski 1997b), and the synapsis of this RecA protein–ssDNA complex with homologous dsDNA and subsequent strand exchange (West 1992).
The first change of RecBCD enzyme at Chi can account for the prevalence of single recombinational exchanges near a DNA end in E. coli recombination (Smith 1991). The linear invading dsDNA fragment that recombines with the circular chromosome in E. coli transduction or conjugation typically has between 10 and 100 Chi sites. The initial action of Chi on RecBCD enzyme assures a single exchange near a Chi site near each end of the linear fragment. The resultant two exchanges are the minimum required to maintain circularity of the chromosome and viability of the cell. Odd numbers of exchanges, which would often occur if there were uncoordinated multiple exchanges, would be lethal. Repair of a dsDNA break by homologous recombination with an intact sister chromosome would also occur with just two exchanges, the minimum number required. The occurrence of a single exchange near each end of the linear fragment would result in positive interference of genetic exchanges; such interference is difficult to measure in E. coli crosses, but is well documented in most eukaryotes (Smith 1991; Foss et al. 1993).
The second change of RecBCD enzyme by Chi results in complete inactivation of the enzyme. This seemingly permanent inactivation implies that one RecBCD enzyme molecule promotes only one recombinational event (‘one enzyme-one exchange‘ hypothesis). With respect to this overall reaction RecBCD enzyme may act stoichiometrically.
As wild-type E. coli contains only ∼10 RecBCD enzyme molecules per cell (Taylor and Smith 1980; A.F. Taylor, unpubl.), the consequences of the inactivation of RecBCD enzyme by Chi may depend on the number of dsDNA breaks per cell. The first step of inactivation by Chi may be most important when there are few dsDNA breaks in a cell, as in conjugation or transduction. This inactivation would limit the number of exchanges to one per DNA end (Smith 1991) but would not inactivate other RecBCD enzyme molecules in the cell. Complete inactivation of RecBCD enzyme by Chi, the second step, may be most important when there are many breaks, as after extensive DNA damage. Additional breaks may be repaired by other, Chi-independent factors activated by the SOS-inducing function of RecBCD enzyme after extensive DNA damage (McPartland et al. 1980; Rinken and Wackernagel 1992). Such induced factors and the possible titration of RecBCD enzyme on broken DNA complicate inferences from studies of whole cells. Further studies of purified RecBCD enzyme and its interaction with Chi may reveal additional features of the regulation of homologous recombination.
Materials and methods
Enzymes
RecBCD enzyme was purified from IPTG-induced E. coli strain V2445 [Δ (pro-lac) ara thi (F′ traD36 proAB lacIq lacZΔM15)], containing plasmids pB520 and pB800 (Boehmer and Emmerson 1991). The enzyme was purified to apparent homogeneity using HiTrap Q, HiTrap Heparin, and HiPrep Sephacryl S-300 columns (Pharmacia Biotech). Lysis conditions and buffers were as used previously (Taylor and Smith 1995a). Protein concentration was determined from its A280 (Roman and Kowalczykowski 1989). Native gel electrophoresis of RecBCD enzyme, and the glycerol-gradient experiments reported here showed that purified enzyme typically contained 10%–20% RecBC (Taylor and Smith 1995a; Table 1B).
Exonuclease VII (GIBCO-BRL) was used at 0.13 U/μl. DNase I (GIBCO-BRL) was diluted to 0.5 U/μl in 10 mm magnesium acetate and used at a final concentration of 0.05 U/μl. Other enzymes were from GIBCO-BRL or New England Biolabs and were used as suggested by the manufacturer.
DNA substrates
Plasmids pUC19 (Yanisch-Perron et al. 1985) and a derivative bearing three Chi sequences (pChi3-A2, from Andrew Eisen, Albert Einstein College of Medicine, New York, NY) were used to produce, respectively, the Chi0 and Chi+ fragments used as substrates for RecBCD enzyme. To construct pChi3-A2, oligonucleotides AE-6 and AE-7 (below) were annealed, filled-in using the Klenow fragment of DNA polymerase I and dNTPs, cut with BamHI and XbaI, and ligated directionally into similarly cut pUC19.
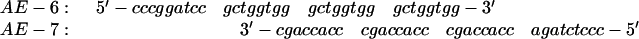 |
Plasmid DNAs were purified twice by cesium chloride/ethidium bromide density-gradient centrifugation (Sambrook et al. 1989) and cut with AseI and FspI. The 321-bp fragment from pUC19 and the 345-bp fragment from pChi3-A2 bearing three centrally located Chi sequences were isolated and purified by electrophoresis through 4% polyacrylamide gels in TAE buffer (Sambrook et al. 1989), followed by electroelution and purification with Geneclean (Bio101, Inc.). After phenol extraction and ethanol precipitation the DNA was dissolved in ME buffer (20 mm MOPS-KOH, 0.1 mm EDTA at pH 7.0) and its concentration determined by its absorbance at 260 nm. All DNA and protein concentrations are given as molarities of molecules.
Radioactive DNA substrates for Chi cleavage and unwinding assays were made by linearizing plasmid pBR322, bearing either no Chi sites (χ0) or one Chi site facing each direction (χ+F χ+H; Dixon and Kowalczykowski 1991), with EcoRI, followed by 5′-end labeling with 32P.
Reaction conditions
Standard reaction mixtures contained 20 mm MOPS-KOH at pH 7.0, 5 mm ATP, 3 mm magnesium acetate, 0.5 mg/ml BSA (Boehringer Mannheim), 20 mm DTT, 100 μg/ml polyvinylpyrrolidone (PVP; average molecular mass 40,000; Sigma), DNA and RecBCD enzyme and were incubated at 23°C. The DNA and RecBCD concentrations were, respectively, 100 and 10 nm in the reactions in Figures 1 and 5B, 60 and 10 nm in Figures 2–4 , and 15 and 2.5 nm in Figure 5A. Reactions were synchronized by prior incubation without ATP. dsDNA exonuclease activity was assayed as described (Eichler and Lehman 1977, but with 50 μm ATP), using 200 pm 3H-labeled T7 DNA and <100 pm RecBCD enzyme. Chi nicking and unwinding were assayed, using 5′ 32P-end-labeled EcoRI-digested plasmid pBR322 χ+F χ+H, or χ0 DNA, under low Mg2+ conditions (Taylor and Smith 1995b, equivalent to standard conditions but with 1 mm DTT and lacking BSA), using 0.9 nm RecBCD enzyme and 0.45 nm DNA. After incubation at 23°C, the reactions were stopped by addition of EDTA to 10 mm, sucrose to 10%, and tracking dyes to 0.04%. Reaction products were separated on 1.2% agarose gels in TAE buffer (Sambrook et al. 1989) and quantitated by PhosphorImager analysis of the dried gel.
Glycerol-gradient centrifugation and analysis
Reaction samples (200 μl) were layered onto 5 ml of 20%–40% (vol/vol) glycerol gradients in siliconized (Sigmacote, Sigma) polyallomer tubes and centrifuged for 17 hr at 55,000 rpm in a Beckman SW55Ti rotor at 4°C. Gradients contained 20 mm potassium phosphate at pH 6.8, 50 mm NaCl, 0.1 mm EDTA, 20 mm DTT, and 100 μg/ml PVP. The high salt gradient contained 0.5 m NaCl. Fractions (68 one-drop fractions per gradient) were collected by bottom puncture in siliconized microtiter trays. Samples (20 μl) of each fraction were electrophoresed on 6% polyacrylamide Tris-glycine SDS minigels (Novex), together with samples of each reaction mixture and of known amounts of RecBCD enzyme (7.5–120 fmoles).
Proteins from the four gels used to analyze each gradient were transferred to a single PVDF membrane (Immobilon-P, Millipore) and processed together. The blots were probed with mouse monoclonal antibodies specific for RecB, RecC, and RecD, and the antibodies visualized with horseradish peroxidase-linked horse anti-mouse IgG and a Phototope-HRP detection kit (New England Biolabs). Films were scanned on a Sharp JX-325 scanner and quantitated using Molecular Dynamics ImageQuant Software. Linear regression (Microsoft Excel) of the data from the standards was used to estimate the amount of RecB, RecC, or RecD polypeptide present in each gel lane. Similar quantitation of a sample of each reaction mixture was used to calculate the recovery of each polypeptide. The migration position of BSA was visualized by Amido Black staining of the membranes.
Native-polyacrylamide gel electrophoresis
Polyacrylamide gels (5% polyacrylamide, 37.5:1 acrylamide:bis) in 50 mm MOPS-KOH at pH 7.0 and 3 mm magnesium acetate were poured in 1-mm Novex gel cassettes. Gels were prerun for 1 hr and the buffer was changed before the addition of samples. Samples were mixed with one-fifth volume of loading solution (50% glycerol, 0.2% bromophenol blue) and run at 100 V for 2–3.5 hr at 4°C prior to transfer to membranes and antibody detection as described above.
Acknowledgments
We are grateful to Douglas Julin (University of Maryland) for samples of RecB and RecC subunits, Paul Boehmer (New Jersey Medical School) for plasmids expressing the recB, recC, and recD genes, Andrew Eisen (Albert Einstein College of Medicine) for plasmids bearing multiple Chi sites, and Elizabeth Wayner (Fred Hutchinson Cancer Research Center Hybridoma Facility) for preparation of monoclonal antibodies. We thank our colleagues in the Smith laboratory and Jim Roberts, Mark Roth, and Meng-Chao Yao for helpful comments on the manuscript and Karen Brighton for help in preparing it. This work was supported by grants GM31693 and GM32194 from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gsmith@fhcrc.org; FAX (206) 667-6497.
References
- Amundsen SK, Taylor AF, Chaudhury AM, Smith GR. recD: The gene for an essential third subunit of exonuclease V. Proc Natl Acad Sci. 1986;83:5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DG, Kowalczykowski SC. The recombination hot spot χ is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes & Dev. 1997a;11:571–581. doi: 10.1101/gad.11.5.571. [DOI] [PubMed] [Google Scholar]
- ————— The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell. 1997b;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- ————— SSB protein controls RecBCD enzyme nuclease activity during unwinding: A new role for looped intermediates. J Mol Biol. 1998;282:275–285. doi: 10.1006/jmbi.1998.2013. [DOI] [PubMed] [Google Scholar]
- Anderson DG, Churchill JJ, Kowalczykowski SC. Chi-activated RecBCD enzyme possesses 5′ → 3′ nucleolytic activity, but RecBC enzyme does not: Evidence suggesting that the alteration induced by Chi is not simply ejection of the RecD subunit. Genes Cells. 1997;2:117–128. doi: 10.1046/j.1365-2443.1997.1130311.x. [DOI] [PubMed] [Google Scholar]
- Bianco PR, Kowalczykowski SC. The recombination hot spot χ is recognized by the translocating RecBCD enzyme as the single strand of DNA containing the sequence 5′-GCTGGTGG-3′. Proc Natl Acad Sci. 1997;94:6706–6711. doi: 10.1073/pnas.94.13.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer PE, Emmerson PT. Escherichia coli RecBCD enzyme: Inducible overproduction and reconstitution of the ATP-dependent deoxyribonuclease from purified subunits. Gene. 1991;102:1–6. doi: 10.1016/0378-1119(91)90529-k. [DOI] [PubMed] [Google Scholar]
- ————— The RecB subunit of the Escherichia coli RecBCD enzyme couples ATP hydrolysis to DNA unwinding. J Biol Chem. 1992;267:4981–4987. [PubMed] [Google Scholar]
- Braedt G, Smith GR. Strand specificity of DNA unwinding by RecBCD enzyme. Proc Natl Acad Sci. 1989;86:871–875. doi: 10.1073/pnas.86.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Chadsey MS, Karlinsey JE, Hughes KT. The flagellar anti-σ factor FlgM actively dissociates Salmonella typhimurium σ28 RNA polymerase holoenzyme. Genes & Dev. 1998;12:3123–3136. doi: 10.1101/gad.12.19.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase JW, Richardson CC. Exonuclease VII of Escherichia coli. Mechanism of action. J Biol Chem. 1974;249:4553–4561. [PubMed] [Google Scholar]
- Chen H-W, Ruan B, Yu M, Wang J-d, Julin DA. The RecD subunit of the RecBCD enzyme from Escherichia coli is a single-stranded DNA dependent ATPase. J Biol Chem. 1997;272:10072–10079. doi: 10.1074/jbc.272.15.10072. [DOI] [PubMed] [Google Scholar]
- Dabert P, Smith GR. Gene replacement in wild-type Escherichia coli: Enhancement by Chi sites. Genetics. 1997;145:877–889. doi: 10.1093/genetics/145.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DA, Kowalczykowski SC. Homologous pairing in vitro stimulated by the recombination hot spot, Chi. Cell. 1991;66:361–371. doi: 10.1016/0092-8674(91)90625-9. [DOI] [PubMed] [Google Scholar]
- ————— The recombination hot spot χ is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell. 1993;73:87–96. doi: 10.1016/0092-8674(93)90162-j. [DOI] [PubMed] [Google Scholar]
- Dixon DA, Churchill JJ, Kowalczykowski SC. Reversible inactivation of the Escherichia coli RecBCD enzyme by the recombination hot spot χ in vitro: Evidence for functional inactivation or loss of the RecD subunit. Proc Natl Acad Sci. 1994;91:2980–2984. doi: 10.1073/pnas.91.8.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggleston AK, Kowalczykowski SC. Biochemical characterization of a mutant recBCD enzyme, the recB2109CD enzyme, which lacks χ-specific, but not non-specific, nuclease activity. J Mol Biol. 1993;231:605–620. doi: 10.1006/jmbi.1993.1313. [DOI] [PubMed] [Google Scholar]
- Eichler DC, Lehman IR. On the role of ATP in phosphodiester bond hydrolysis catalyzed by the RecBC deoxyribonuclease of Escherichia coli. J Biol Chem. 1977;252:499–503. [PubMed] [Google Scholar]
- Finch PW, Storey A, Chapman KE, Brown K, Hickson ID, Emmerson PT. Complete nucleotide sequence of the Escherichia coli recB gene. Nucleic Acids Res. 1986a;14:8573–8582. doi: 10.1093/nar/14.21.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch PW, Wilson RE, Brown K, Hickson ID, Thompkinson AE, Emmerson PT. Complete nucleotide sequence of the Escherichia coli recC gene and of the thyA recC intergenic region. Nucleic Acids Res. 1986b;14:4437–4451. doi: 10.1093/nar/14.11.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss E, Lande R, Stahl FW, Steinberg CM. Chiasma interference as a function of genetic distance. Genetics. 1993;133:681–691. doi: 10.1093/genetics/133.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan S, Smith GR. Strand-specific binding to duplex DNA ends by the subunits of Escherichia coli RecBCD enzyme. J Mol Biol. 1992;229:67–78. doi: 10.1006/jmbi.1993.1008. [DOI] [PubMed] [Google Scholar]
- Helmann JD, Chamberlin MJ. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Kogoma T. Stable DNA replication: Interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köppen A, Krobitsch S, Thoms B, Wackernagel W. Interaction with the recombination hot spot χ in vivo converts the RecBCD enzyme of Escherichia coli into a χ-independent recombinase by inactivation of the RecD subunit. Proc Natl Acad Sci. 1995;92:6249–6253. doi: 10.1073/pnas.92.14.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korangy F, Julin DA. A mutation in the consensus ATP-binding sequence of the RecD subunit reduces the processivity of the RecBCD enzyme from Escherichia coli. J Biol Chem. 1992;267:3088–3095. [PubMed] [Google Scholar]
- ————— Kinetics and processivity of ATP hydrolysis and DNA unwinding by the RecBC enzyme from Escherichia coli. Biochemistry. 1993;32:4873–4880. doi: 10.1021/bi00069a024. [DOI] [PubMed] [Google Scholar]
- ————— Efficiency of ATP hydrolysis and DNA unwinding by the RecBC enzyme from Escherichia coli. Biochemistry. 1994;33:9552–9560. doi: 10.1021/bi00198a022. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A, Schabtach E, Stahl FW. Chi sites in combination with RecA protein increase the survival of linear DNA in Escherichia coli by inactivating exoV activity of RecBCD nuclease. EMBO J. 1994;13:2764–2776. doi: 10.1002/j.1460-2075.1994.tb06570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman RP, Oishi M. Formation of the recB–recC DNase by in vitro complementation and evidence concerning its subunit nature. Nat New Biol. 1973;243:75–77. [PubMed] [Google Scholar]
- ————— The recBC deoxyribonuclease of Escherichia coli: Isolation and characterization of the subunit proteins and reconstitution of the enzyme. Proc Natl Acad Sci. 1974;71:4816–4820. doi: 10.1073/pnas.71.12.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson C, Boehmer PE, McDonald F, Chaudhuri S, Hickson ID, Emmerson PT. Reconstitution of the activities of the RecBCD holoenzyme of Escherichia coli from the purified subunits. J Biol Chem. 1992;267:13564–13572. [PubMed] [Google Scholar]
- McPartland A, Green L, Echols H. Control of recA gene RNA in E. coli: Regulatory and signal genes. Cell. 1980;20:731–737. doi: 10.1016/0092-8674(80)90319-0. [DOI] [PubMed] [Google Scholar]
- Merrick WC, Hershey JWB. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey JWB, Mathews MB, Sonenberg N, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 31–69. [Google Scholar]
- Moore S. Pancreatic DNase. In: Boyer PD, editor. The enzymes. XIV. New York, NY: Academic Press, Inc; 1981. pp. 281–296. [Google Scholar]
- Moore MJ, Query CC, Sharp PA. Splicing of precursors to mRNA by the spliceosome. In: Gesteland RF, Atkins JF, editors. The RNA world. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- Myers RS, Kuzminov A, Stahl FW. The recombination hot spot χ activates RecBCD recombination by converting Escherichia coli to a recD mutant phenocopy. Proc Natl Acad Sci. 1995;92:6244–6248. doi: 10.1073/pnas.92.14.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palas KM, Kushner SR. Biochemical and physical characterization of exonuclease V from Escherichia coli. J Biol Chem. 1990;265:3447–3454. [PubMed] [Google Scholar]
- Phillips RJ, Hickelton DC, Boehmer PE, Emmerson PT. The RecB protein of Escherichia coli translocates along single-stranded DNA in the 3′ to 5′ direction: A proposed ratchet mechanism. Mol & Gen Genet. 1997;254:319–329. doi: 10.1007/pl00008605. [DOI] [PubMed] [Google Scholar]
- Ponticelli AS, Schultz DW, Taylor AF, Smith GR. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell. 1985;41:145–151. doi: 10.1016/0092-8674(85)90069-8. [DOI] [PubMed] [Google Scholar]
- Rinken R, Wackernagel W. Inhibition of the recBCD-dependent action of Chi recombinational hot spots in SOS-induced cells of Escherichia coli. J Bacteriol. 1992;174:1172–1178. doi: 10.1128/jb.174.4.1172-1178.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman LJ, Kowalczykowski SC. Characterization of the helicase activity of Escherichia coli RecBCD enzyme using a novel helicase assay. Biochemistry. 1989;28:2863–2873. doi: 10.1021/bi00433a018. [DOI] [PubMed] [Google Scholar]
- Roman LJ, Eggleston AK, Kowalczykowski SC. Processivity of the DNA helicase activity of Escherichia coli recBCD enzyme. J Biol Chem. 1992;267:4207–4214. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Smith GR. Homologous recombination in E. coli: Multiple pathways for multiple reasons. Cell. 1989;58:807–809. doi: 10.1016/0092-8674(89)90929-x. [DOI] [PubMed] [Google Scholar]
- ————— Conjugational recombination in E. coli: Myths and mechanisms. Cell. 1991;64:19–27. doi: 10.1016/0092-8674(91)90205-d. [DOI] [PubMed] [Google Scholar]
- ————— . DNA double-strand break repair and recombination in Escherichia coli. In: Nickoloff JA, Hoekstra MF, editors. DNA damage and repair. I. DNA repair in prokaryotes and lower eukaryotes. Totowa, NJ: Humana Press; 1998. pp. 135–162. [Google Scholar]
- Stahl FW, Crasemann JM, Stahl MM. Rec-mediated recombinational hot spot activity in bacteriophage λ III. Chi mutations are site-mutations stimulating Rec-mediated recombination. J Mol Biol. 1975;94:203–212. doi: 10.1016/0022-2836(75)90078-9. [DOI] [PubMed] [Google Scholar]
- Taylor AF. Movement and resolution of Holliday junctions by enzymes from E. coli. Cell. 1992;69:1063–1065. doi: 10.1016/0092-8674(92)90626-n. [DOI] [PubMed] [Google Scholar]
- Taylor A, Smith GR. Unwinding and rewinding of DNA by the RecBC enzyme. Cell. 1980;22:447–457. doi: 10.1016/0092-8674(80)90355-4. [DOI] [PubMed] [Google Scholar]
- ————— RecBCD enzyme is altered upon cutting DNA at a Chi recombination hot spot. Proc Natl Acad Sci. 1992;89:5226–5230. doi: 10.1073/pnas.89.12.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Monomeric RecBCD enzyme binds and unwinds DNA. J Biol Chem. 1995a;270:24451–24458. doi: 10.1074/jbc.270.41.24451. [DOI] [PubMed] [Google Scholar]
- ————— Strand specificity of nicking of DNA at Chi sites by RecBCD enzyme: Modulation by ATP and magnesium levels. J Biol Chem. 1995b;270:24459–24467. doi: 10.1074/jbc.270.41.24459. [DOI] [PubMed] [Google Scholar]
- Taylor AF, Schultz DW, Ponticelli AS, Smith GR. RecBC enzyme nicking at Chi sites during DNA unwinding: Location and orientation dependence of the cutting. Cell. 1985;41:153–163. doi: 10.1016/0092-8674(85)90070-4. [DOI] [PubMed] [Google Scholar]
- Thaler DS, Sampson E, Siddiqi I, Rosenberg SM, Stahl FW, Stahl M. A hypothesis: Chi-activation of RecBCD enzyme involves removal of the RecD subunit. In: Friedberg E, Hanawalt P, editors. Mechanisms and consequences of DNA damage processing. New York, NY: Alan R. Liss; 1988. pp. 413–422. [Google Scholar]
- West SC. Enzymes and molecular mechanisms of genetic recombination. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]
- ————— The RuvABC proteins and Holliday junction processing in Escherichia coli. J Bacteriol. 1996;178:1237–1241. doi: 10.1128/jb.178.5.1237-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M, Buttin G, Hurwitz J. The isolation and characterization from Escherichia coli of an adenosine triphosphate-dependent deoxyribonuclease directed by rec B, C genes. J Biol Chem. 1971;246:6543–6555. [PubMed] [Google Scholar]
- Yanisch-Perron C, Vièira J, Messing J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13 mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yu M, Souaya J, Julin DA. The 30-kDa C-terminal domain of the RecB protein is critical for the nuclease activity, but not the helicase activity, of the RecBCD enzyme from Escherichia coli. Proc Natl Acad Sci. 1998;95:981–986. doi: 10.1073/pnas.95.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]