Abstract
Signals from TGF-β superfamily receptors are transduced to the nucleus by Smad proteins, which transcriptionally activate target genes. In Caenorhabditis elegans, defects in a TGF-β-related pathway cause a reversible developmental arrest and metabolic shift at the dauer larval stage. Null mutations in daf-3 suppress mutations in genes encoding this TGF-β signal, its receptors, and associated Smad signal transduction proteins. daf-3 encodes a Smad protein that is most closely related to mammalian DPC4, and is expressed throughout development in many of the tissues that are remodeled during dauer development. DAF-4, the type II TGF-β receptor in this pathway, is also expressed in remodeled tissues. These data suggest that the DAF-7 signal from sensory neurons acts as a neuroendocrine signal throughout the body to directly regulate developmental and metabolic shifts in tissues that are remodeled during dauer formation. A full-length functional DAF-3/GFP fusion protein is predominantly cytoplasmic, and this localization is independent of activity of the upstream TGF-β-related pathway. However, this fusion protein is associated with chromosomes in mitotic cells, suggesting that DAF-3 binds DNA directly or indirectly. DAF-3 transgenes also interfere with dauer formation, perhaps attributable to a dosage effect. A truncated DAF-3/GFP fusion protein that is predominantly nuclear interferes with dauer formation, implying a role for DAF-3 in the nucleus. These data suggest that DAF-7 signal transduction antagonizes or modifies DAF-3 Smad activity in the nucleus to induce reproductive development; when DAF-7 signals are disabled, unmodified DAF-3 Smad activity mediates dauer arrest and its associated metabolic shift. Therefore, daf-3 is unique in that it is antagonized, rather than activated, by a TGF-β pathway.
Keywords: diapause, neuroendocrine, TGF-β signaling
In response to environmental signals, Caenorhabditis elegans has two distinct life-cycles—reproductive, in which the animals rapidly grow to the adult reproductive stage, and dauer, in which the animals arrest development at the anatomically and metabolically distinctive third-larval dauer stage (Riddle and Albert 1997). Environmental conditions are sensed in chemosensory neurons (Bargmann and Horvitz 1991), which couple to a transforming growth factor β (TGF-β) signaling pathway (Georgi et al. 1990; Estevez et al. 1993; Ren et al. 1996) as well as an insulin-related signaling pathway (Morris et al. 1996; Kimura et al. 1997) to trigger changes in the development of the many tissues remodeled in dauer larvae (Riddle and Albert 1997). Mutations in daf-7 [a TGF-β homolog (Ren et al. 1996)], daf-4 [a type II TGF-β receptor (Estevez et al. 1993)], daf-1 [a type I TGF-β receptor (Georgi et al. 1990)], daf-8, and daf-14 [Smad (for small and mad) homologs (A. Estevez and D.L. Riddle, pers. comm.; T. Inoue and J. Thomas, pers. comm.)] cause constitutive arrest at the dauer stage even in conditions that promote reproductive growth. These genes constitute a neuronal signaling pathway that is active during reproductive development. The DAF-7 TGF-β signal is produced by the sensory neuron ASI during reproductive development, whereas daf-7 expression in this neuron is inhibited during dauer-inducing conditions (Ren et al. 1996; Schackwitz et al. 1996). DAF-7 binding to the DAF-1/DAF-4 receptors has been suggested to activate the DAF-8/DAF-14 Smads to promote a commitment to reproductive growth (Ren et al. 1996; A. Estevez and D.L. Riddle, pers. comm.; T. Inoue and J. Thomas, pers. comm.).
A model for signaling by TGF-β and related receptors has emerged recently. Binding of ligand induces dimerization of receptors to produce an active signaling kinase (Attisano et al. 1994; Miyazano et al. 1994). This complex phosphorylates cytoplasmic Smad proteins and causes them to relocate to the nucleus, where they act as transcription factors (Chen et al. 1996; Liu et al. 1996; Mascias-Silva et al. 1996; Chen et al. 1997; Kim et al. 1997; Kretzschmar et al. 1997). The Smad protein phosphorylated is determined by the identity of the complex, for example, BMP receptors phosphorylate Smad1 (Kretzschmar et al. 1997), whereas TGF-β and activin receptors target Smad2 or Smad3 (Chen et al. 1996; Chen et al. 1996; Lagna et al. 1996; Mascias-Silva et al. 1996; Zhang et al. 1996, 1997). All of the above Smads interact with DPC4, a member of a second class of Smad proteins, and all three pathways are potentiated by DPC4 (Lagna et al. 1996; Mascias-Silva et al. 1996; Zhang et al. 1996, 1997; Kretzschmar et al. 1997); therefore, DPC4 is thought to be a general cofactor for pathway-specific Smads such as Smad1, Smad2, and Smad3. The small TGF-β-related pathway in C. elegans also requires both a DPC4-like Smad (sma-4) and two other Smads more closely related to Smad-1 (sma-2 and sma-3) (Savage et al. 1996). Drosophila melanogaster also has Mad, which resembles Smad1, and a DPC4 homolog as well (P. Das and R. Padgett, pers. comm.). The requirement for these two basic types of Smads may be conserved in TGF-β related-pathways across metazoan phylogeny.
Because Smad proteins are positive transducers of signals, the biological effect of overexpressing Smads is the same as overexpressing or activating receptors (Baker and Harland 1996; Graff et al. 1996), and the effect of Smad mutations mimics the effect of loss-of-function mutations in the receptors (Raftery et al. 1995; Sekelsky et al. 1995; Savage et al. 1996). And, although screens for enhancers of TGF-β pathway mutations have been successful (Raftery et al. 1995), suppressors of TGF-β pathway mutants have not been identified, except in the C. elegans dauer pathway (Riddle et al. 1981; Thomas et al. 1993).
daf-3 is a unique TGF-β pathway gene in that it is antagonized, rather than activated by the daf-7 TGF-β-related pathway. The dauer-constitutive phenotypes of mutations in the DAF-7 signal transduction pathway genes (including putative null mutations, see below) are suppressed fully by mutations in daf-3 (Riddle et al. 1981; Vowels and Thomas 1992). These genetic data indicate that in the absence of DAF-7 signaling, the lack of antagonism releases DAF-3 activity to induce dauer arrest. In this report, we show that daf-3 encodes a novel Smad, which is antagonized rather than activated by a TGF-β-related pathway. We show that DAF-3 and the TGF-β receptor DAF-4 are expressed widely in tissues that remodel during dauer development, and therefore, may transduce DAF-7 neuroendocrine signals throughout the body to regulate tissue metabolism and development. We show that a DAF-3/GFP fusion protein is nuclear localized and perturbs dauer arrest when mutations are directed to a conserved domain in the carboxy-terminal region, and that DAF-3/GFP fusion proteins are associated with mitotic chromosomes, indicating that DAF-3 functions in the nucleus. The DAF-3 expression level and subcellular localization is independent of upstream DAF-7 signal transduction activity, indicating that the regulation of DAF-3 by DAF-7 signaling may occur downstream of nuclear localization. We propose a model for regulation of DAF-3 activity by the DAF-8/DAF-14 Smads.
Results
daf-3 encodes a novel Smad
daf-3 mutations suppress the dauer constitutive phenotype induced by mutations in daf-7 and its associated receptors and Smad proteins (Riddle et al. 1981; Vowels and Thomas 1992; Thomas et al. 1993). To understand the role of wild-type daf-3 in this pathway, it is important to determine if this suppression is a loss-of-function or gain-of-function phenotype, and whether it is the null phenotype. We isolated a daf-3 null allele, which is missing the entire daf-3-coding region (see Materials and Methods; Fig. 1A). This allele completely suppresses the dauer constitutive phenotype of mutants in the daf-7 pathway (Table 1). Therefore, dauer arrest in a daf-7 mutant depends on wild-type DAF-3 activity, suggesting that DAF-7 signaling normally negatively regulates or modifies DAF-3 gene activity to allow reproductive development.
Figure 1.
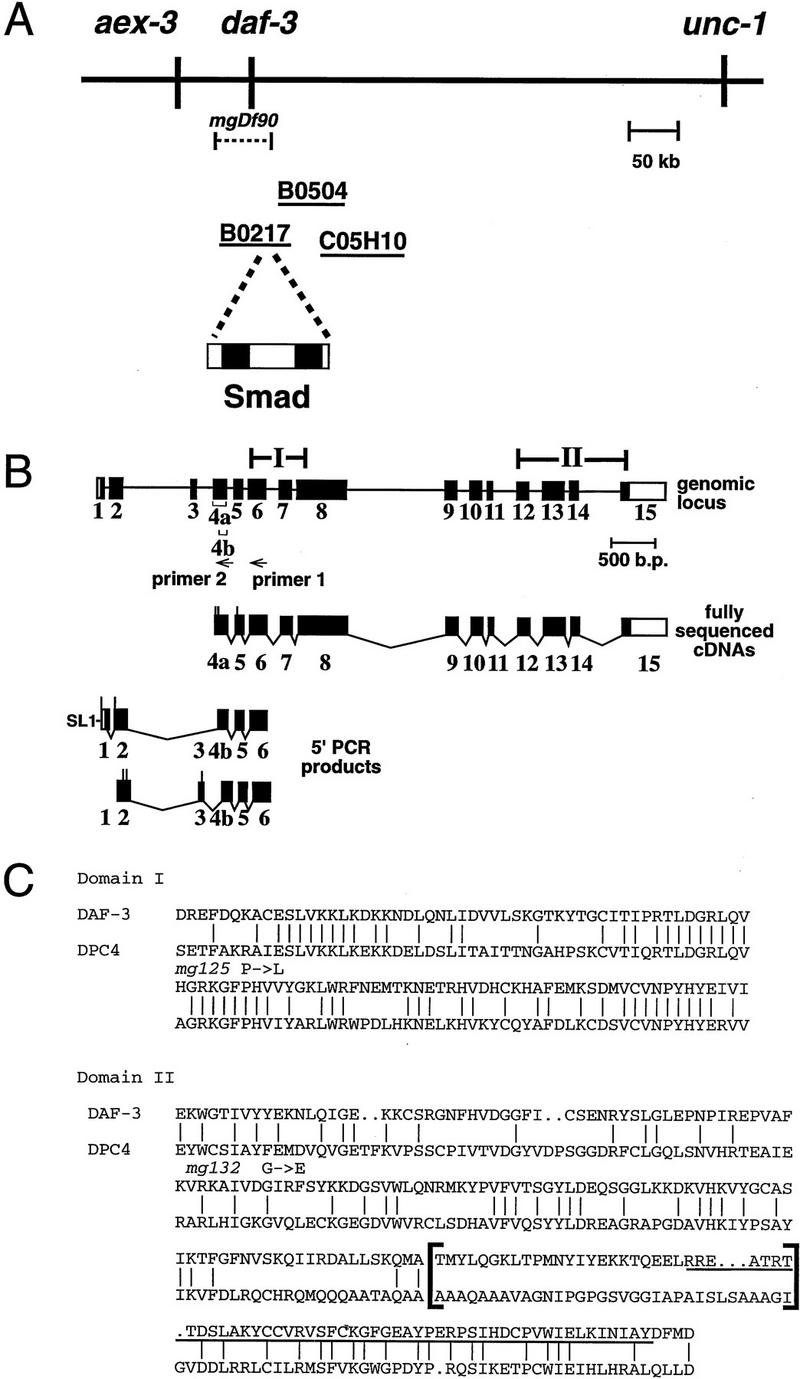
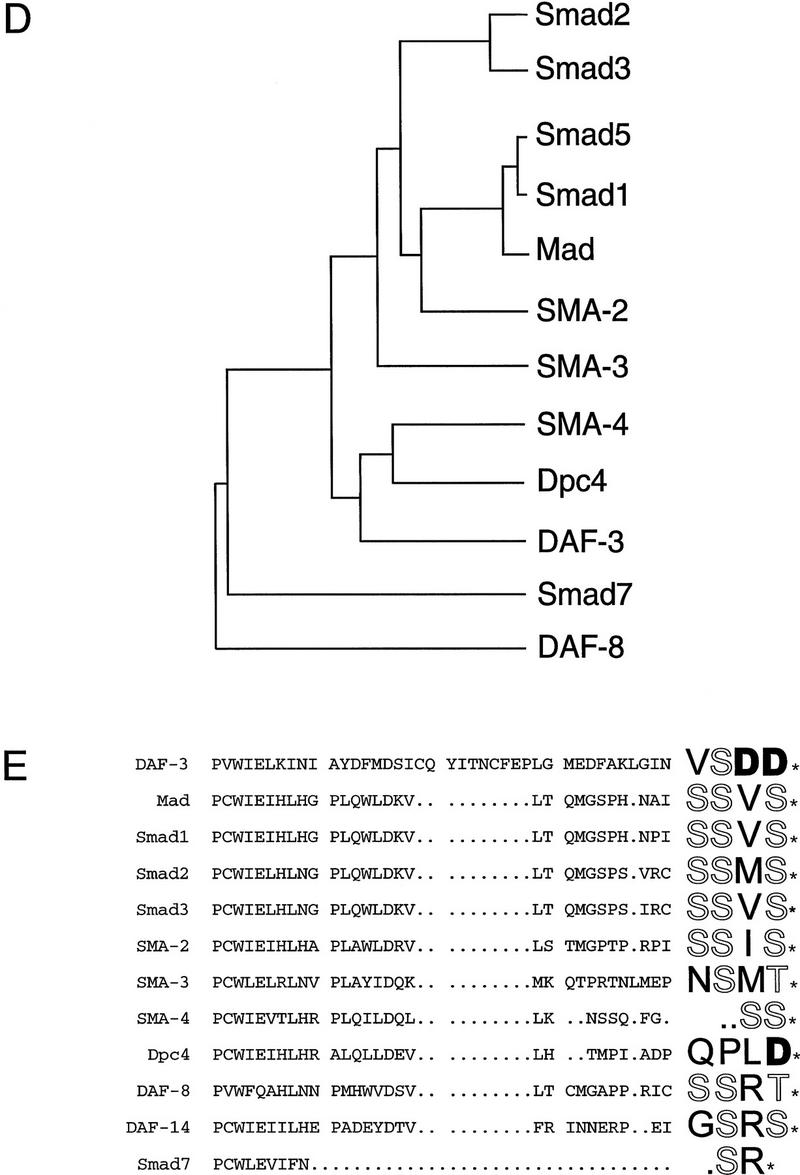
daf-3 encodes a Smad protein most closely related to DPC4. (A) daf-3 cloning. daf-3 was genetically mapped to a region on the X chromosome between aex-3 and unc-1 (A. Koweek and G. Patterson, unpubl.). B0217 complements daf-3 mutants in transformation rescue experiments; B0504 and C05H10 do not. mgDf90 is a deletion that removes all of daf-3. (B) Structure of daf-3-coding region. At the top is the exon/intron structure of daf-3; coding exons are solid boxes, noncoding regions are open boxes, and lines are introns. Conserved Smad domains I and II are indicated. The three classes of cDNA are shown, and 5′ ends are indicated by vertical lines. The accession numbers for the cDNAs shown are, top to bottom: AF005205, AF005206, and AF005207. (C) Protein sequence alignment of C. elegans DAF-3 and its closest homolog, human DPC4, in the Smad conserved domains I and II. Dots indicate gaps introduced to maximize alignment. The Smad mutational hot spot is underscored, and the insertion in DAF-3 and DPC4 relative to other Smads is bracketed. In addition to mg125 and mg132, seven other daf-3 alleles were sequenced in the hot spot; none of them contains a mutation. Alleles sequenced were mg91, mg93, mg105, mg121, mg126, mg133 (isolated by A. Koweek and G. Patterson, unpubl.) and sa205 (Thomas et al. 1993). (D) Relationship of DAF-3 domain I to other Smads. Lineup was performed with the program pileup (Genetics Computer Group 1994) using amino acids 137–245 of DAF-3 (GenBank accession no. AF005205) and corresponding residues of the other Smads. (E) Comparison of carboxyl termini of Smads. The final 28–44 residues are shown. Residues that are phosphorylated by receptor (in Smad1 and Smad2) or similar residues in similar positions (in other Smads) are shown in outline. Aspartates in similar positions are shown in boldface type.
Table 1.
Suppression of dauer-constitutive mutants by a null daf-3 allele
| Dauer constitutive mutation
|
Percent dauer arrest (no.)
|
|
|---|---|---|
|
daf-3(+)
|
daf-3(mgDf90)
|
|
| + | 0 (434) | 0 (251) |
| daf-7(e1372) | 100 (208) | 0 (286) |
| daf-7(m62) | 81 (211) | 0 (291) |
| daf-8(sa233) | 85 (267) | 0 (285) |
| daf-14(m77) | 76 (84) | 0 (271) |
| daf-8(sa233); daf-14(m77) | 100 (383) | 0 (600) |
To discern the molecular basis of DAF-3 function in this pathway, we determined the sequence and expression pattern of daf-3. Cosmids in the daf-3 genetic region were assayed for gene activity by transformation. Cosmid B0217 partially complements a daf-3 mutation, whereas other cosmids from the region do not (Fig. 1A). Examination of sequence provided by the C. elegans Sequencing Consortium revealed a Smad-homologous gene on B0217. A subclone of B0217 containing only a Smad homolog, but no other predicted coding regions, also rescues daf-3. Our detection of mutations in the Smad homolog (see below) confirmed its assignment to daf-3. Analysis of daf-3 cDNAs reveals that the gene is transcribed from 15 exons and is alternatively spliced. Four daf-3 cDNAs were isolated and sequenced (Fig. 1B). These cDNAs contain a 400-bp 3′-untranslated region (UTR), but no poly(A) tail; a C. elegans consensus polyadenylation sequence is found 12 bp beyond the 3′ end of the cDNAs. The longest of these four is likely to be nearly full-length, as it contains a methionine codon and the genomic sequence contains no other methionine codon and no putative splice sites upstream before in-frame stop codons. To characterize further the 5′ end of daf-3, PCR products from libraries or individual daf-3 cDNAs were sequenced. Two alternate spliced forms were identified. Note that the first has the trans-spliced leader SL1 that is found at the 5′ end of many C. elegans cDNAs; therefore, this cDNA represents a probable daf-3 5′ end. The alternative splicing affects regions upstream of the conserved Smad protein domains. No genetic data support distinct functions for these distinct DAF-3 isoforms.
The DAF-3 Smad protein is most closely related to vertebrate DPC4, which is a likely cofactor for Smad1, Smad2, and Smad3 (Lagna et al. 1996; Mascias-Silva et al. 1996; Zhang et al. 1996, 1997; Chen et al. 1997; Kretzschmar et al. 1997). Smads have two conserved domains (Sekelsky et al. 1995; Massague 1996; Wrana and Attisano 1996) and DAF-3 has 55% amino acid identity in the probable DNA-binding domain I to DPC4 (Fig. 1C) and 31% identity in the probable receptor interacting domain II. DPC4 and SMA-4 are a distinct subclass of Smads based on sequence and function (Hahn et al. 1996; Lagna et al. 1996; Mascias-Silva et al. 1996; Savage et al. 1996; Zhang et al. 1996, 1997; Kretzschmar et al. 1997). DAF-3, SMA-4, and DPC4 all have a small insertion in the same location in domain II relative to Smad1, Smad2, and Smad3 (shown in brackets in Fig. 1C). When DAF-3 is compared with other Smads, domain I of DAF-3 falls into a subfamily with DPC4 and Sma-4 (Fig. 1D). Domain II of DAF-3 does not fall into any Smad subfamily (data not shown).
The predicted DAF-3 protein has a unique sequence feature at the carboxyl terminus. Most Smads have the sequence SSXS at the carboxyl terminus, or a closely related sequence (Fig. 1E). This sequence has been shown to be phosphorylated by the upstream TGF-β-related receptors in the case of Smad1 and Smad2, and this phosphorylation event activates nuclear entry and transcriptional activation by the Smads (Mascias-Silva et al. 1996; Kretzschmar et al. 1997). At this same location, daf-3 has one serine and two aspartate residues. This change is interesting because, in many cases, a serine to aspartate change has the same effect as phosphorylated serine—the acidic amino acid simulates a negatively charged phosphate group (e.g., see Weiser et al. 1995).
We identified three mutations in daf-3, all of which were isolated as suppressors of daf-7(e1372). mgDf90 is a homozygous viable deletion of 15–90 kb that removes the entire Smad gene (Fig. 1A). This allele efficiently suppresses daf-7 mutants (Table 1). Therefore, suppression of the daf-7 dauer-constitutive phenotype is the daf-3 null phenotype, demonstrating that wild-type DAF-7 TGF-β pathway signaling acts to antagonize DAF-3. The daf-3(mgDf90) strain does not have any apparent pleiotropies—the animals are healthy, move normally, progress from egg to adult at the same rate as wild type, and >99% of the eggs hatch, similar to wild type. daf-3(mg125) and daf-3(mg132) are missense mutations that alter conserved residues in domains 1 and 2, respectively (Fig. 1C). A majority of the mutations detected in other Smads localize to a 45-amino-acid segment of domain II (Massague 1996; Wrana and Attisano 1996). Clustering of mutations is observed even in DPC4, for which homozygous null mutations have been identified (Hahn et al. 1996), so the clustering is unlikely to be attributable to selection for non-null mutations. This hotspot region was sequenced in nine daf-3 alleles, and no mutations were detected. This difference in mutation location may be a simple statistical fluke, or may indicate functional differences between DAF-3 and other Smad proteins, consistent with the fact that DAF-3 is antagonized, rather than activated, by upstream TGF-β signaling.
DAF-3 Smad protein activity functions antagonistically to Smad signaling downstream of DAF-7 TGF-β-like signals
daf-3 mutations suppress the dauer constitutive phenotype of mutations in five genes, daf-1, daf-4, daf-7, daf-8, and daf-14 (Riddle et al. 1981; Thomas et al. 1993). daf-7 encodes the probable TGF-β ligand (Ren et al. 1996), daf-1 and daf-4 encode the probable DAF-7 receptors (Georgi et al. 1990; Estevez et al. 1993), and daf-8 and daf-14 encode Smad proteins that are likely to couple to those receptors (A. Estevez and D.L. Riddle, pers. comm.; T. Inoue and J. Thomas, pers. comm.). The daf-3(mgDf90) null allele also suppresses completely the dauer constitutive phenotypes of mutations in these probable upstream TGF-β signaling components (Table 1). The suppression of mutations in the probable DAF-7 receptors as well as in the probable downstream DAF-8 and DAF-14 Smad proteins by daf-3(mgDf90) argues strongly that DAF-3 does not antagonize this pathway by, for example, competing with the DAF-8 and DAF-14 Smad proteins for coupling to the receptor kinases, as has been observed for Smad7 (Hayashi et al. 1997; Topper et al. 1997). Given that Smad proteins have been shown to interact physically with each other and with upstream receptor kinases, we tested the model that for example unphosphorylated DAF-8 or DAF-14 Smad proteins may be necessary for DAF-3-mediated dauer arrest. We found that a daf-8(sa233); daf-14(m77); daf-3(mgDf90) triple mutant is not dauer-constitutive, unlike the daf-8(sa233); daf-14(m77) double mutant or the daf-8(sa233) or daf-14(m77) single mutants (Table 1). Therefore, when daf-3 is absent, daf-8 and daf-14 are not required for reproductive growth. This suggests that engagement of the DAF-1 and DAF-4 receptors by the DAF-7 ligand during reproductive growth induces DAF-8 and DAF-14 Smad proteins to antagonize DAF-3 activity, and that DAF-8 and DAF-14 have no function in the induction of dauer arrest by the DAF-3 activity.
daf-3 and daf-4 are expressed in tissues remodeled during dauer formation
To determine where DAF-3 may function to control dauer formation, we examined the expression pattern of a DAF-3/green fluorescent protein (GFP) fusion gene, which partially rescues a daf-3 mutant (Fig. 3, below). DAF-7 signaling from the ASI neuron begins during the L1 stage (Ren et al. 1996; Schackwitz et al. 1996), and neuron ablations and dauer-formation assays in various environmental conditions indicate that the signal for dauer formation is also received during the first two larval stages (Swanson and Riddle 1981; Golden and Riddle 1984; Bargmann and Horvitz 1991). Therefore, we carefully examined L1 larvae. However, this pathway is not only active during the dauer decision in L1 and L2. daf-7 and the other TGF-β-pathway-related genes affect dauer recovery in the L3 stage, and egg-laying and locomotory behavior in adults (Thomas et al. 1993; Trent et al. 1983). In addition, these genes affect fat metabolism throughout larval and adult development (Thomas et al. 1993; Kimura et al. 1997; G. Ruvkun, unpubl.). daf-3 mutations suppress these phenotypes efficiently, therefore, we also examined daf-3 expression at later stages. We find that the expression of DAF-3/GFP is similar in the four larval stages and adults (see below).
Figure 3.
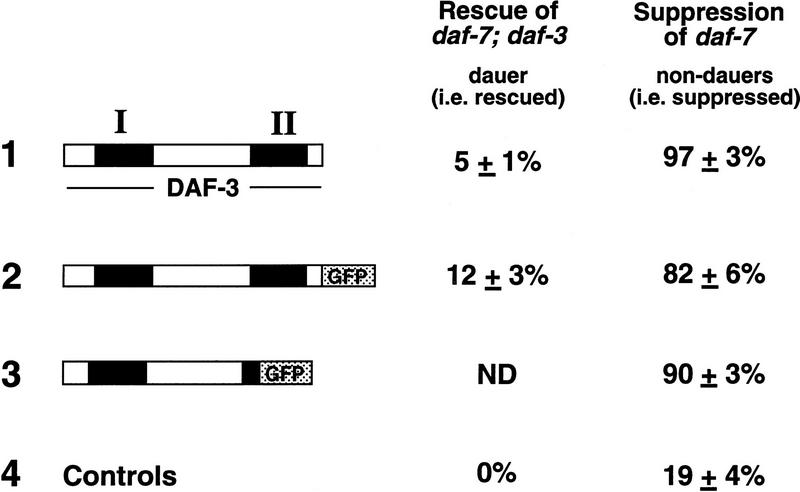
Rescuing ability and suppression of daf-7 by daf-3 plasmids. The solid boxes represent the Smad conserved domains I and II of daf-3; the stippled boxes represent GFP. For all experiments shown, daf-3 plasmids were injected at a concentration of 10 ng/μl and the pRF4 injection marker was injected at a concentration of 90 ng/μl. To score dauer formation, synchronous broods of transgenic animals were scored after two days at 25°C. The rescue experiment shows the rescue of daf-7(m62); daf-3(e1376) by each of the fusion proteins. The control is a transgenic array with the pRF4 transformation marker and a nonrescuing cosmid. For each construct, four or more lines were measured in two separate experiments. To measure suppression of daf-7, transgenic arrays were crossed to a daf-7 strain (for plasmids 1 and 3), or produced by injecting directly into daf-7 (plasmid 2). The controls are two transgenic strains with the pRF4 marker and an unrelated GFP-expressing transgene.
Almost every transgenic animal shows strong DAF-3/GFP expression in many, but not all, head neurons (Fig. 2A), the ventral nerve cord (both cell bodies and processes; Fig. 2B), the intestinal cells (Fig. 2C), especially the membrane adjacent to the intestinal lumen, and tail hypodermal cells and neurons. Weak expression in the pharynx, hypodermal V blast cells, P blast cells and hyp 7 hypodermal cells is observed in about half of the transgenic animals (data not shown). Expression in the tail hypodermal cells hyp 9, hyp 10, and hyp 11 is clearly seen in nearly every animal. This apparent difference between tail hypodermal expression and main body expression may be a consequence of the anatomy of the animal. The main body hypodermis is underlain by bright GFP in the intestine and ventral nerve cord, so weak expression in the hypodermis is hard to see against this background. In the region of hyp 9, hyp 10, and hyp 11, the body is solid hypodermis with no underlying tissue (Sulston et al. 1983). Expression is rarely detected in dorsal body wall muscle. DAF-3/GFP is expressed in the distal tip cells and in their precursors, Z1.a and Z4.p, throughout development (Figs. 2D and 4, below). The distal tip cells have a key role in development of the somatic gonad and germ line (Kimble 1981; Kimble and White 1981); expression of DAF-3/GFP there may indicate a role for daf-3 in gonad developmental arrest. DAF-3/GFP is also expressed strongly in unidentified vulval cells in adults. In wild-type embryos of 200–400 cells, DAF-3/GFP is expressed uniformly thoughout the embryo (Fig. 2E). Under the conditions of the experiment, which promote reproductive growth, the subcellular localization of the DAF-3/GFP protein is mainly cytoplasmic (Fig. 2B–E, and see below).
Figure 2.
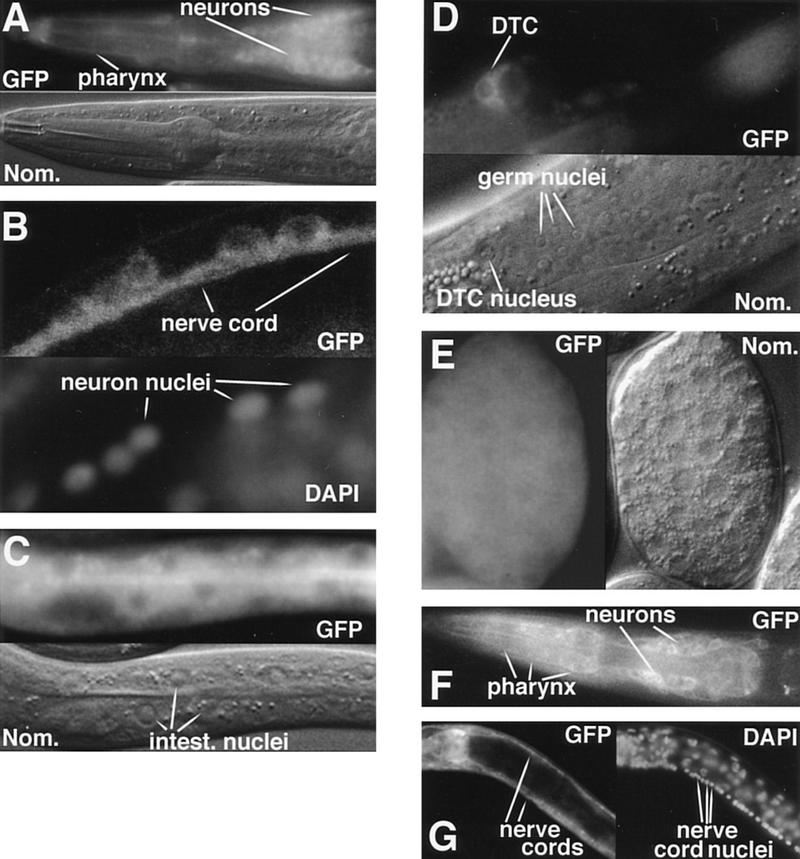
DAF-3 and DAF-4 expression. All DAF-3 photographs show animals with the full-length functional DAF-3/GFP fusion gene shown in Fig. 3. In A and C–F, intrinsic fluorescence is used to visualize GFP; in B and G, anti-GFP antibodies are used. (A) DAF-3/GFP head expression in an L1 animal. Weak DAF-3/GFP expression in the pharynx impedes cell identification, but the main body of the pharynx is filled, implying expression in pharyngeal muscle. (B) DAF-3/GFP expression in the ventral nerve cord of an adult animal. L1 animals look similar. (C) DAF-3/GFP expression in the intestine of an L1 animal. (D) DAF-3/GFP expression in the distal tip cell of an L4 animal. (E) DAF-3/GFP expression in an embryo with ∼200 nuclei. (F) DAF-4/GFP expression in the head of an L1 animal. (G) DAF-4/GFP expression in the dorsal nerve cord and ventral nerve cord of an L4 animal. Although the intestine can be seen in this picture, the DAF-4/GFP is not visible, because in this focal plane, the intestinal cell membrane is obscured by the bright fluorescence in the nerve cords.
Figure 4.
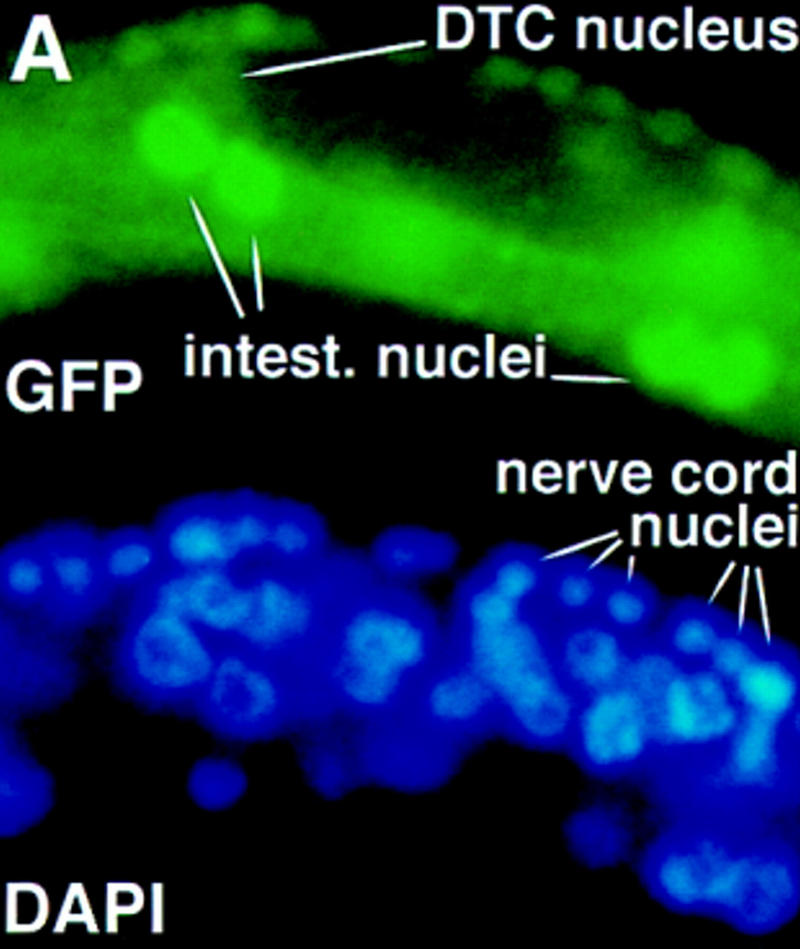
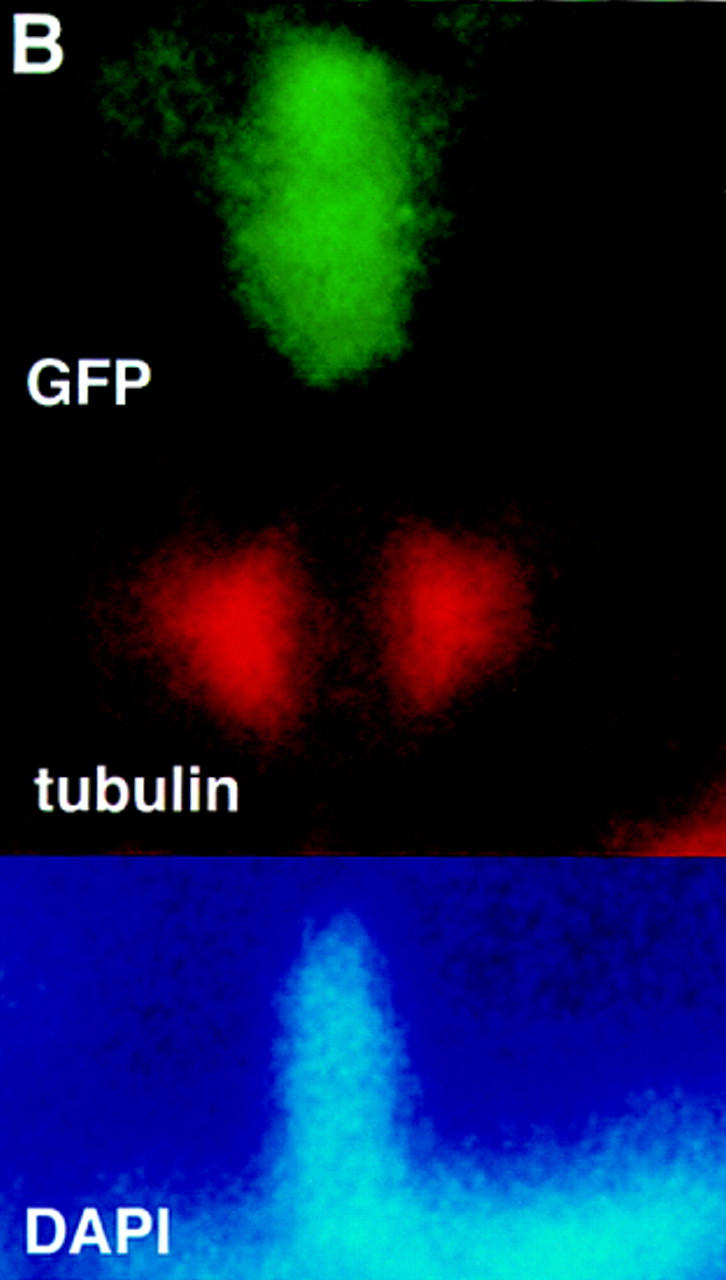
(A) A truncated DAF-3/GFP protein is predominantly nuclear. Wild-type animals were injected with the truncated construct shown in Fig. 3 at a concentration of 10 ng/ml. The pRF4 transformation marker was injected at 100 ng/ml. The photograph shows an early L2 animal, and DAF-3 is predominantly nuclear. The clear spot in the center of some of the nuclei is the nucleolus. All cells in these animals have predominantly nuclear DAF-3/GFP, including the ventral cord neurons, intestinal cells and distal tip cell (all shown), as well as head and tail neurons and hypodermal cells. (B) DAF-3/GFP is associated with metaphase chromosomes. Fixed L1 animals were immunostained with anti-GFP antibody (Clontech) and anti-α-tubulin antibody (Amersham). DNA was visualized with DAPI.
The expression pattern of DAF-4 supports the model that DAF-4 and DAF-3 transduce DAF-7 signals in tissues that are remodelled during dauer development. We examined the expression of a DAF-4/GFP fusion gene that complements a daf-4 mutant (Fig. 2F–G; see Materials and Methods). The pattern of DAF-4/GFP expression is similar to that of DAF-3/GFP, except that DAF-4/GFP is localized to membranes, consistent with its role as a receptor. As in the case of DAF-3/GFP, almost every DAF-4/GFP transgenic animal shows strong GFP fluorescence in many, but not all, head neurons (Fig. 2F), the ventral nerve cord (both cell bodies and processes, see Fig. 2G), intestinal cells, and tail neurons (data not shown). DAF-4/GFP is expressed more strongly than DAF-3/GFP in the pharynx (Fig. 2F,G), and more weakly in the ventral nerve cord cell bodies. DAF-4/GFP is expressed rarely in main body hypodermis, but, like DAF-3, is expressed in tail hypodermis. As described in the previous paragraph, this apparent difference in the tail versus the main body may be attributable to effects of the anatomy on scoring. DAF-4/GFP is not localized to the membrane surrounding the intestinal lumen, nor is it expressed in the distal tip cell or its precursors, unlike DAF-3/GFP. As with DAF-3, expression of DAF-4/GFP is similar in larval stages and adults. Expression of DAF-4/GFP in wild-type animals is detected later than DAF-3/GFP; DAF-4/GFP is first detectable at late embryogenesis when the embryo resembles an L1 larva. Using anti-GFP antibodies, we verified the difference in embryonic expression of DAF-4/GFP and DAF-3/GFP. We found that the antibodies recapitulated the results with direct GFP fluorescence: DAF-3/GFP is expressed in early embryos; DAF-4/GFP is not.
Smad1 and Smad2 relocalize to become predominantly nuclear when the upstream TGF-β signaling pathways are activated (Baker and Harland 1996; Hoodless et al. 1996; Liu et al. 1996; Macias-Silva et al. 1996; Kretzschmar et al. 1997). In wild type, DAF-3/GFP is primarily, although not exclusively, cytoplasmic. DAF-3/GFP subcellular distribution was examined in head neurons in the vicinity of ASI (the cell that produces the DAF-7 signal), as well as in intestinal cells. DAF-3/GFP was predominantly cytoplasmic in all animals. However, in all animals, dim GFP fluorescence was seen in the nucleus of some of the cells with bright cytoplasmic fluorescence, and in ∼25% of the animals, equivalent DAF-3/GFP levels in the nucleus and cytoplasm was observed in one or more cells.
Because DAF-3 activity is antagonized by DAF-7 TGF-β pathway signaling, we expect that DAF-3 is most active (and perhaps localized to the nucleus) when these genes are inactive. We therefore observed the subcellular localization of the full-length DAF-3/GFP fusion protein in the head neurons, tail neurons, and intestine of dauer-constitutive mutant L1 worms, when DAF-3 gene activity is predicted to be highest. In daf-1(m402), daf-4(m72), daf-7(m62), daf-8(sa233), and daf-14(m77) mutants, DAF-3/GFP was predominantly cytoplasmic, although, as in wild type, nuclear GFP was observed in a subset of cells. Even in the neurons nearest to ASI, where the DAF-7 signal should be strongest, we detected no change in DAF-3/GFP subcellular localization in these mutant animals. The DAF-3/GFP fusion protein is also predominantly cytoplasmic in L1 and L2 stages of larvae induced to form dauers by environmental conditions or by mutations in the insulin receptor pathway gene daf-2, rather than by mutations in the DAF-7 signaling pathway mutants (data not shown). The tissue-specific expression pattern of DAF-3/GFP was unaltered in these mutant backgrounds (data not shown). Therefore, we see no evidence that DAF-3 localization is regulated by these upstream receptors or Smads.
A truncated DAF-3/GFP is nuclear localized and full-length DAF-3/GFP associates with mitotic chromosomes
The homology of DAF-3 domain I to the DNA-binding domain of Drosophila MAD indicates that DAF-3 functions in the nucleus. This hypothesis is supported by two additional observations. First, a truncated DAF-3/GFP fusion that is missing most of conserved domain II is predominantly nuclear (Fig. 4A, below). This fusion protein interferes with dauer induction; like a daf-3 loss-of-function mutant, it suppresses mutations in daf-7 (Fig. 3). Because this truncated protein is predominantly nuclear, it may repress dauer formation by acting in the nucleus. This result suggests that wild-type DAF-3 also has a function in the nucleus. However, we find that a full-length DAF-3/GFP fusion also suppresses mutations in daf-7, as does a full-length DAF-3 transgene without GFP (Fig. 3). These constructs are likely to contain complete regulatory sequences for daf-3, as each contains DNA extending to the next predicted gene both left and right of daf-3; therefore, we suspect that the suppression is attributable to overexpression, rather than misexpression, of DAF-3. This suppression indicates that overexpression of DAF-3 has dominant-negative activity, perhaps attributable to interference with DAF-3 interactions with receptors or cofactors such as other Smads.
A second indication that DAF-3 acts in the nucleus is that DAF-3/GFP is associated with chromosomes in intestinal cells during mitosis. These cells divide at the end of the L1 stage, and antibody staining with anti-GFP antibodies and anti-α-tubulin antibodies reveals that DAF-3/GFP associates with chromosomes between the spindles during mitosis (Fig. 4B). DAF-3/GFP was seen between spindles (190 spindles were examined; 130 showed GFP fluorescence between the spindles, coincident with DAPI). Two control GFP fusion genes (see Materials and Methods) did not show such mitotic localization (0/91 spindles). We see DAF-3 GFP co-localized with DAPI from prophase to late anaphase. DAF-3/GFP was associated with nuclei in prophase by the following criteria. The spindles were present on either side of the nucleus, but the nucleus had not broken down completely. In particular, an indistinct nucleolus was present. In some cases, we see the anaphase or telophase chromosomes—two spots in which GFP and DAPI co-localize can be seen, flanked by spindles. At this point in mitosis, DAF-3/GFP fades and becomes undetectable as the nuclei reform the nuclear envelope and nucleolus. Therefore, DAF-3 can associate with chromosomes, consistent with the hypothesis that it is a transcriptional activator that acts in the nucleus. DAF-3 is not predicted from its mutant phenotype to have a role in mitosis, and we have found that the nuclear divisions in the intestine occur normally in daf-3 null mutants (data not shown). It is possible that the brighter GFP on mitotic chromosomes is attributable to increased access to DNA caused by the breakdown of the nuclear envelope.
The DAF-3/GFP fusion that has an intact domain 1 but a truncated domain 2 is also associated with mitotic chromosomes (62/107 spindles). This result is consistent with the finding that Drosophila MAD domain 1 binds DNA (Kim et al. 1997), whereas domain 2 of Smad3 and Smad4 serve to allow interactions with other Smads (Wu et al. 1997). This DAF-3/GFP fusion protein, with its homologous domain 1 intact, and most of domain 2 missing, interacts with metaphase chromosomes to the same extent as full-length DAF-3/GFP. In addition, this protein can interfere with wild-type DAF-3 to the same extent as high-copy DAF-3 (Fig. 3). These results are consistent with DAF-3 domain 1 mediating DAF-3-binding to DNA.
Discussion
The DAF-3 Smad protein is antagonized by, rather than activated by, the DAF-7 TGF-β-related signaling pathway. DAF-3 and DAF-4, the type II TGF-β receptor in this signaling pathway, are expressed widely in the animal, and therefore may constitute a signal transduction pathway that controls the morphogenetic and metabolic changes in tissues that are remodelled during dauer formation. A DAF-3/GFP protein from a rescuing expression construct localizes to both the cytoplasm and the nucleus in the many cells that express the protein. The expression level and subcellular localization of DAF-3 is not coupled to environmental conditions or mutations in pathways that regulate dauer formation, suggesting that the upstream TGF-β-signaling pathway genes regulate DAF-3 activity at a step other than subcellular localization.
The combination of the DAF-3 and DAF-4 expression patterns suggests that these genes act to transduce DAF-7 neuroendocrine signals in tissues that are remodelled during dauer development. A neuroendocrine mode of action is similar to that of the TGF-β homolog activin, which was discovered as a neuroendocrine factor that acts in gonadotropin signaling in humans (Vale et al. 1990). The early expression of DAF-3 in embryos is also consistent with a model that DAF-3 acts during embryonic development, for example, to mediate the development of neuronal pathways that emit neuroendocrine signals to antagonize DAF-7 TGF-β signaling during the L1 stage. However, our data favor a function for DAF-3 in transduction of environmental signals during the L1 and L2 stages (1) DAF-7 TGF-β signal from ASI neurons occurs during the L1 and L2 stages and is repressed by dauer-inducing environmental conditions (Ren et al. 1996; Schackwitz et al. 1996). Genetic analysis of daf-3 shows that its activity is regulated by DAF-7 (Table 1). (2) Expression of the DAF-4 type II receptor begins in very late embryogenesis. (3) The expression patterns of DAF-3 and DAF-4 are coincident in most of the tissues remodeled during dauer morphogenesis. For example, DAF-3 and DAF-4 are both expressed in the hypodermis that secretes a modified dauer cuticle and in the pharynx that is slimmed. In addition, DAF-4 and DAF-3 are expressed in the intestine and hypodermis of dauer larvae, which contain large fat stores indicative of a metabolic shift to fat storage (Kimura et al. 1997; Riddle and Albert 1997). The expression of both the DAF-4 TGF-β family receptor kinase and the DAF-3 Smad protein in these target tissues is consistent with a model that the DAF-7 neuroendocrine signal from the ASI neuron is received directly by these tissues during reproductive development. In addition, the observation that DAF-4 and DAF-3 are expressed in many of the same cells is consistent with a model that DAF-4/DAF-1 signaling to downstream Smads (DAF-8 and DAF-14 are likely candidates) directly regulates DAF-3 gene activity. One exception to this coexpression is the distal tip cells of the gonad, which control gonad growth and development during the C. elegans reproductive life cycle (Kimble 1981; Kimble and White 1981). Gonad development is arrested in animals that enter the dauer stages, so expression of DAF-3/GFP in the distal tip cells is consistent with the control of gonad development by DAF-3. However, we do not observe expression of DAF-4/GFP in the distal tip cells. It is possible that this cell does not have a key role in gonad developmental arrest at the dauer stage, or that DAF-4/GFP is below the detection threshold in these cells.
Both DAF-3/GFP and DAF-4/GFP have a similar expression pattern in larval and adult stages. This result is not surprising given that daf-7 and the other TGF-β-pathway-related genes affect fat metabolism and egg-laying and locomotory behavior in adults (Trent et al. 1983; Thomas et al. 1993; Kimura et al. 1997; G. Ruvkun unpubl.). daf-3 mutations act to suppress all of these phenotypes. It is possible that daf-7 continues to have a role in transducing environmental signals in adults to affect these phenotypes and that continued expression of DAF-3 and DAF-4 is required for appropriate expression of this signal.
We find that a functional DAF-3/GFP fusion protein is localized to both the cytoplasm and nucleus (Fig. 2), and that DAF-3/GFP subcellular localization is not apparently coupled to DAF-7 signaling pathway activity. Even though we detect no change in DAF-3/GFP subcellular localization in these mutants, we do detect some DAF-3/GFP in nuclei. A change in DAF-3 activity attributable to phosphorylation state or association with DAF-8 and DAF-14 may couple DAF-3 to DAF-7 signaling. However, it is possible that DAF-3 localization is coupled to the receptors in a subtle way. In fact, the subcellular localization of Drosophila MAD protein is not detectably altered in wild type when receptor signaling to MAD occurs; relocalization is seen only if the DPP ligand is overexpressed drastically (Newfeld et al. 1997). It is unlikely that a set of undiscovered TGF-β receptors regulates DAF-3. The genome sequencing project has sequenced in excess of 85% of the genes in C. elegans, and there is only one candidate TGF-β receptor gene other than daf-1 and daf-4. If this receptor were a positive regulator of DAF-3, mutants would be expected to, like daf-3 mutants, suppress daf-7 mutants. This receptor acts in a signaling pathway distinct from DAF-3, and it is not a suppressor of daf-7 (S. Krishna and R. Padgett, pers. comm.). We favor the model that phosphorylated DAF-8 and DAF-14 Smad proteins antagonize DAF-3 action in the nucleus (see below) but do not affect nuclear localization of DAF-3.
Features of the DAF-3 sequence support the model that DAF-3 activity may be independent of upstream receptor signaling. DAF-3 has two aspartates at the carboxyl terminus where Smad1 and Smad2 have three serines that are phosphorylated by a receptor kinase. In many cases, a serine to aspartate substitution has the same effect as phosphorylated serine because the acidic amino acid simulates a negatively charged phosphate group (e.g., see Weiser et al. 1995). Therefore, we propose that DAF-3 may be a constitutively active Smad because of these aspartates. However, it should be borne in mind that a serine residue precedes these acidic residues. Therefore DAF-3 activity could be additionally modulated by phosphorylation by an unknown kinase. However, phosphorylation of DAF-3 by DAF-1 and DAF-4 is not required for DAF-3 activity because genetic analysis suggests that DAF-3 is active in the absence of DAF-1 and DAF-4 activity (Table 1; Riddle et al. 1981; Thomas et al. 1993).
Other features of the DAF-3 sequence and observations of DAF-3/GFP subcellular localization support a nuclear function. DAF-3 has a well-conserved conserved Smad domain 1 (Fig. 1D), which has been shown to be a DNA-binding domain of Drosophila MAD (Kim et al. 1997), implying that DAF-3 retains this function. DAF-3 associates with mitotic chromosomes in the intestinal cells (Fig. 4), consistent with a role in the nucleus. In addition, we find that both a full-length DAF-3/GFP, which is primarily cytoplasmic, and a truncated version of DAF-3/GFP, which is primarily nuclear, interfere with dauer formation in a daf-7 mutant. Therefore, we suggest that DAF-3/GFP can interfere with wild-type DAF-3 function both in the cytoplasm and the nucleus, implying a functional role in each compartment. However, it is possible that these transgenes interfere with dauer formation in some other way, in which case, the subcellular localization of the transgene may not reflect the site of action of the normal gene.
The constitutive nuclear localization of a truncated DAF-3/GFP fusion gene missing part of domain II is consistent with the reports that the Drosophila MAD amino-terminal conserved domain mediates DNA binding (Kim et al. 1997) and that the Smad2 carboxy-terminal conserved domain mediates receptor coupling in the cytoplasm (Mascias-Silva et al. 1996; Kretzschmar et al. 1997). It is inconsistent with the finding that a Smad2 construct containing only conserved domain II is localized constitutively to the nucleus (Baker and Harland 1996), or that a DPC4 construct in which domain 2 is truncated is cytoplasmic (Zhang et al. 1997). It is unclear at this point whether these differences between nuclear and cytoplasmic localization sequences are attributable to distinctions between different Smad proteins or methods of analysis.
Recently, Smad7 has been reported as an antagonistic Smad (Hayashi et al. 1997; Topper et al. 1997). Smad7 antagonizes TGF-β signaling by stably associating with receptors and thereby blocking the phosphorylation and concomitant activation of Smad2. DAF-3, like Smad7, lacks the SSXS motif that is the phosphorylation target of the receptor. However, DAF-3 is unlikely to function like Smad7. Smad7 blocks the receptors from signaling; daf-3, on the other hand, is active in the absence of signaling from receptors and associated Smads (Table 1; Riddle et al. 1981; Thomas et al. 1993). This genetic result suggests that DAF-3 does not block the receptors; instead, the receptors and associated Smads block DAF-3 from inducing dauer.
We propose a model for the TGF-β pathway in dauer formation (Fig. 5). We suggest that in the absence of DAF-7 signaling, DAF-3 forms homodimers or higher order oligomers that act in the nucleus to induce the expression of genes that mediate the metabolic and morphological changes during dauer arrest (Fig. 5A). In conditions that promote reproductive growth, DAF-7 TGF-β ligand from the ASI sensory neuron (Ren et al. 1996; Schackwitz et al. 1996) binds to the DAF-1/DAF-4 receptor kinases, which then phosphorylate the Smads DAF-8 and/or DAF-14, analogous to the phosphorylation and activation of Smad1, Smad2, and Smad3 (Hoodless et al. 1996; Lagna et al. 1996; Macias-Silva et al. 1996; Zhang et al. 1996; Kretzschmar et al. 1997). We propose that under these conditions DAF-3 functions like its closest homolog, DPC4, which dimerizes with phosphorylated Smad1 and Smad2 even under conditions that do not lead to detectable DPC4 phosphorylation (Lagna et al. 1996). Therefore, we suggest that the dauer promoting DAF-3 homo-oligomers are disrupted when DAF-3 hetero-oligomerizes with a phosphorylated DAF-8 and/or DAF-14 (Fig. 5B). We show the heterodimers in the nucleus because we saw no indication of relocalization of DAF-3 in daf-8 or daf-14 mutants. The complete suppression of the dauer constitutive phenotype of a daf-8; daf-14 double mutant by the daf-3(mg90) null allele (Table 1) strongly supports the model that the major function of the DAF-8 and DAF-14 Smad proteins in reproductive development is to antagonize DAF-3 activity.
Figure 5.
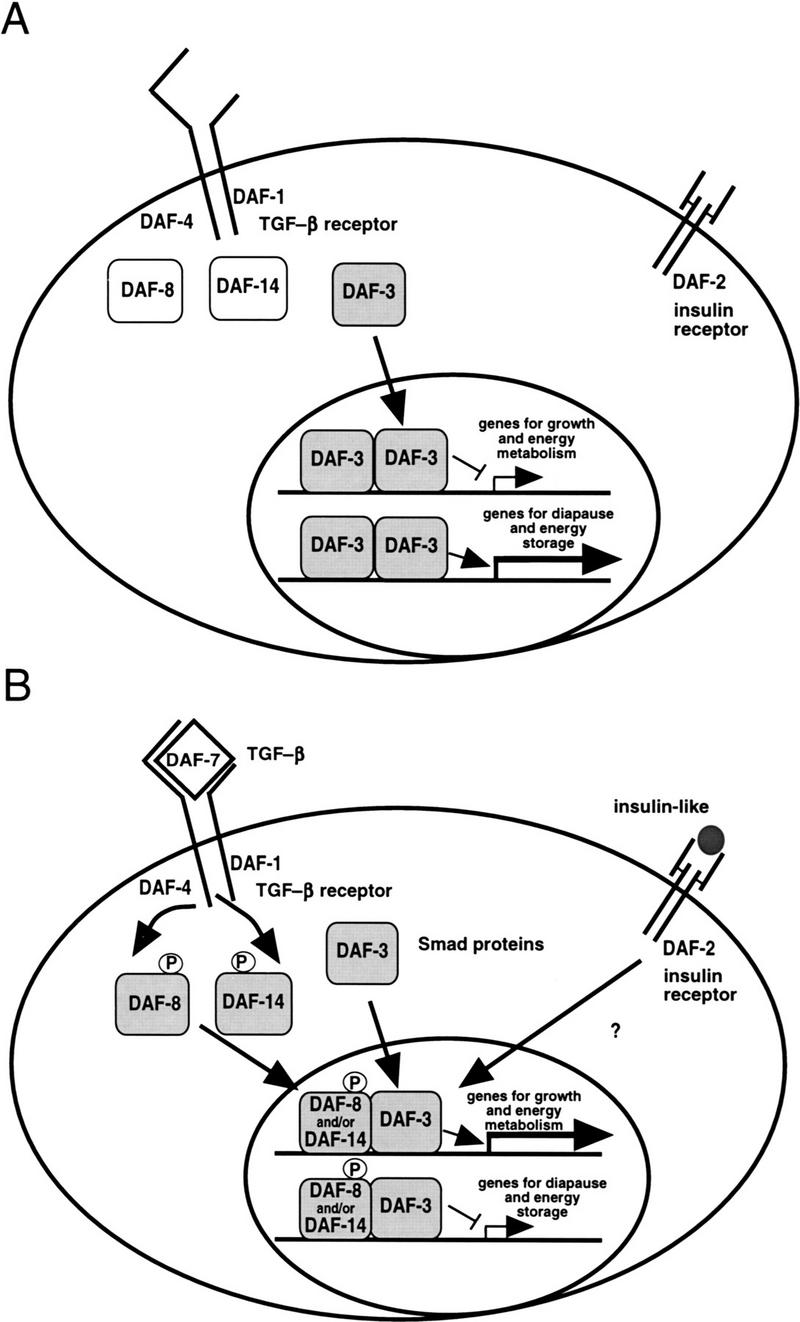
A model for the role of the DAF-3/DAF-8/DAF-14 Smads in dauer formation, as described in text. (A) Dauer growth induction; (B) reproductive growth induction.
Because daf-8 and daf-14 are only partially redundant (Riddle et al. 1981; Vowels and Thomas 1992; T. Inoue and J. Thomas, pers. comm.), each is likely to perform a unique function in reproductive development. Therefore, DAF-3/DAF-8 dimers may have different activity from DAF-3/DAF-14. Perhaps each activates a subset of genes required for reproductive development and metabolism. We show in the model that the DAF-8/DAF-3 and DAF-14/DAF-3 homodimers activate genes involved in reproductive growth. However, it is important to note that this complex cannot be absolutely required, as reproductive growth occurs even in a daf-8; daf-14; daf-3 triple mutant (Table 1). Therefore, the regulation of DAF-3 activity by upstream Smads is capable of blocking dauer formation and associated metabolic changes by DAF-3; however, the parallel daf-2 insulin-related pathway (Kimura et al. 1997) and other pathways (Thomas et al. 1993; Hobert et al. 1997) also regulate metabolic genes. The convergence of TGF-β-related and insulin-related pathways in C. elegans metabolic control has implications for human disease in that defects of human homologs of these TGF-β related genes may cause diabetes in some populations.
Materials and methods
Cosmid rescue and scoring of dauer-constitutive phenotypes
Cosmids were injected at 5 ng/μl each, with pRF4 as an injection marker to bring the total DNA concentration to 100 ng/μl as described by Mello et al. (1991). Injections were into the strain daf-7(e1372) daf-3(e1376) and transgenic lines were scored for rescue at 25°C; rescued animals were dauers (indicating return to the daf-7(e1372) phenotype). Cosmid B0217 rescued daf-3 (18/19 lines). Cosmids C05H10 and B0504 did not (0/10 and 0/7 lines respectively). Synchronized broods were scored at 24.5°C ± 0.5°C to generate the data in Table 1. Two plates were scored for each genotype. In a second experiment performed at 25.5°C, daf-3 also completely suppressed the dauer-constitutive phenotype of all the mutants shown.
cDNA characterization
All cDNAs were from libraries λACT–RB1 (oligo-dT primed) and λACT–RB2 (random primed) provided by R. Barstead (Oklahoma Medical Research Foundation, Oklahoma City). Libraries were probed with a PCR product generated from genomic DNA with the primers daf 3-1 (ctggcagtcactaacacacg) and daf 3-4 (accctcatgcctactgtcag); this product spans exons 4–8. Three cDNAs isolated in this manner were sequenced fully. A fourth cDNA isolated and sequenced by J. Thatcher and P. Okkema (University of Illinois, Chicago) was also sequenced and verified the gene structure. To examine the 5′ end two strategies were used. First, a DNA prep from the library RB2 (F. Slack, unpubl.) was PCR-amplified with the primers SL1 (ggactagttttaattacccaagtttga) and daf 3-8 (ttcaccagggactcgcaagc; this is primer 1 in Fig. 1B). This PCR product was sequenced and produced the SL1-spliced product shown in Figure 1B. The second strategy was to PCR-amplify from individual plaques that hybridized to the PCR product amplified with daf 3-1 and daf-3-4. These were amplified with a primer to the vector (OFS30:cgatgatgaagatacccc or OFS31: cagttgaagtgaacttgcgg) and primer daf 3-8. These cDNAs ended at locations shown as vertical lines on the 5′ PCR products shown in Figure 1B.
Scoring of transgenic animals for GFP
All DAF-3/GFP transgenic lines were produced by the method of Mello et al. (1991) using 10 ng/μl of GFP plasmid and 90 ng/μl or pRF4 as a transformation marker. DAF-4/GFP transgenic lines were injected with 50 ng/μl of GFP plasmid and 70 ng/μl of pRF4. For all GFP scoring, animals were grown at 25–26°C. For scoring of DAF-3/GFP in wild-type and in dauer-constitutive mutant backgrounds, three or more lines were scored in each case. A large number of animals were surveyed to determine the expression pattern, and at least 30 animals were scored head-to-tail, and expression was tallied for each tissue. In three daf-4(m72) mutant lines, DAF-3/GFP was localized to the nucleus more than in wild type lines. When these strains were crossed to wild-type, the increased nuclear localization was seen in both the daf-4 and wild-type segregants. Therefore the increased nuclear GFP was a property of the array, rather than of daf-4.
When GFP was scored by antibody, polyclonal antibodies from Clontech were used with the protocol of Finney and Ruvkun (1990). For scoring of DAF-3/GFP association with chromosomes in mitotic cells, the animals were prepared by hypochlorite treatment followed by incubation without food to generate synchronized L1 larvae, followed by growth at 20°C for 14–15 hr (until animals were at the end of the L1 stage, at which time intestinal nuclei divide). Fixed animals were stained with the anti-GFP antibody and a monoclonal anti-α-tubulin antibody (Amersham). The control strains for this experiment contained one of two GFP constructs expressed in the intestine. The first was a GFP fusion to the first exon of daf-16, which is expressed in the intestine throughout larval development (S. Ogg, pers. comm.). The second is a construct that provides intestinal expression of a GFP with a fused SV40 nuclear localization sequence (S. Marshall and J. McGhee, pers. comm.). This construct is nuclear in interphase, but no GFP is seen associated with chromosomes during metaphase.
Subcloning
pGP8 is a 13-kb EagI–SacI restriction fragment from cosmid B0217 cloned into pBluescript SK(−) (Stratagene). This clone goes from 5 kb upstream of exon 1 to 500 bp downstream of the putative poly-A addition site. To make the full-length rescuing DAF-3/GFP, an AvrII–SacI fragment containing exon 15 and DNA from the preceding intron and following 3′ untranslated and flanking DNA was removed from pGP8. A PCR product was generated from pGP8 with the primers daf 3-34(ctacgagaagaagactcagg) and daf3poly1 (tgcatggagctcttaagctagctggccggccggcgcgccgtcatcactgacgttgattcc), cut with AvrII and SacI and used to replace the AvrII–SacI fragment from pGP8. This maneuver replaces the exon 15 with identical coding sequence (verified by sequencing) except that several restriction sites are placed between the last codon and the stop codon. A GFP/unc-54 3′ end PCR product from pPD95.81 (A. Fire, pers. comm.) was PCR-amplified and cloned into the AscI and FseI restriction sites to produce pGP19.
The truncated construct pGP7 consists of 8 kb of daf-3 fused to GFP. An 8-kb EcoRI fragment from B0217 was cloned into the EcoRI site of pBluescript SK(−). A PvuI–SalI fragment of this subclone was ligated to a PvuI–SalI fragment from the GFP vector pPD95.81. The resulting plasmid contains ∼2.5 kb of sequence upstream of exon 1 of daf-3 and coding region through the first 58 amino acid residues of domain II. The remaining 175 amino acids of daf-3 and the 3′ noncoding region are replaced with GFP and the unc-54 3′ end. Three transgenic lines were isolated, and all had a similar phenotype.
To make a rescuing DAF-4/GFP construct, a 10-kb SalI fragment from cosmid C05D2, which contains 3 kb of sequence upstream of the daf-4 transcriptional start, and all of the daf-4 coding region except codons for the last 14 residues of daf-4, was used. This fragment was subcloned into the SalI site of the GFP plasmid TU#61 (Chalfie et al. 1994) to create a nearly full-length daf-4-coding region, followed by GFP and the unc-54 3′ end. This plasmid was injected into a daf-4(m72) strain to test the fusion for DAF-4 activity. More than 95% of transgenic animals were rescued for the dauer-constitutive and small phenotypes of daf-4(m72), indicating that the fusion has robust DAF-4 activity.
Isolation and characterization of mutant alleles
mgDf90 was identified as a spontaneous mutation that suppressed daf-7 in the strain daf-7(e1372) III; mut-6(st702) unc-22(st192)IV (G. Patterson, unpubl.). daf-3(mg125) and daf-3(mg132) were isolated as EMS-induced suppressors of daf-7(e1372) (A. Koweek, unpubl.). Genomic DNA from mg125 and mg132 animals was PCR-amplified for sequencing, and sequencing was performed directly on the PCR products. The mgDf90 deletion was mapped by Southern blotting. Genomic DNA from an mgDf90 strain was digested with EcoRI or SpeI and compared with similarly digested DNA from N2 (wild type). All bands corresponding to the daf-3 protein-coding region were missing from the mgDf90 digest.
Acknowledgments
We thank A. Estevez, D. Riddle, T. Inoue, J. Thomas, S. Newfeld, B. Gelbart, J. Thatcher, P. Okkema, and the C. elegans Genome Sequencing Consortium for communicating results before publication. We also thank J. Thatcher and P. Okkema for isolating and sequencing one of the daf-3 cDNAs; R. Barstead and A. Fire for providing clones, and members of the Ruvkun lab for comments on the manuscript. We thank S. Marshall, J. McGhee, O. Hobert, and S. Ogg for providing GFP-expressing strains. Some of the strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Center for Research Resources. G.I.P. is an Amersham/US Biochemicals Fellow of the Life Sciences Research Foundation. This work was supported by a Hoechst Grant to the Department of Molecular Biology at Massachusetts General Hospital, and an NIH grant to G.R. (R01 AG14161).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Ruvkun@frodo.mgh.harvard.edu; FAX (617) 726-6893.
References
- Attisano L, Wrana JL, Montalvo E, Massague J. TGF-β receptors and actions. Biochim Biophys Acta. 1994;1222:71–80. doi: 10.1016/0167-4889(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Baker JC, Harland RM. A novel mesoderm inducer, Madr2, functions in the activin signal transduction pathway. Genes & Dev. 1996;10:1880–1889. doi: 10.1101/gad.10.15.1880. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Horvitz HR. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science. 1991;251:1243–1246. doi: 10.1126/science.2006412. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Chen X, Rubrock M, Whitman M. A transcriptional partner for MAD proteins in TGF-β signaling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- Chen, X., E. Weisberg, V. Fridmacher, M. Watanabe, G. Naco, and M. Whitman. 1997. Role of Smad4 and FAST-1 in the assembly of activin responsive factor. Nature (in press). [DOI] [PubMed]
- Chen Y, Lebrun J, Vale W. Regulation of transforming growth factor-β and activin-induced transcription by mammalian Mad proteins. Proc Natl Acad Sci. 1996;93:12992–12997. doi: 10.1073/pnas.93.23.12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez M, Attisano L, Wrana JL, Albert PS, Massague J, Riddle DL. The daf-4 gene encodes a bone morphogenetic protein receptor controlling C. elegans dauer larva development. Nature. 1993;365:644–649. doi: 10.1038/365644a0. [DOI] [PubMed] [Google Scholar]
- Finney M, Ruvkun GB. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Genetics Computer Group. 1994. Program manual for the Wisconsin package, version 8. Madison, WI.
- Georgi LL, Albert PS, Riddle DL. daf-1, a C. elegans gene controlling dauer larva development, encodes a novel receptor protein kinase. Cell. 1990;61:635–645. doi: 10.1016/0092-8674(90)90475-t. [DOI] [PubMed] [Google Scholar]
- Golden JW, Riddle DL. The Caenorhabditis elegans dauer larva: Developmental effects of pheromone, food, and temperature. Dev Biol. 1984;102:368–378. doi: 10.1016/0012-1606(84)90201-x. [DOI] [PubMed] [Google Scholar]
- Graff JM, Bansal A, Melton DA. Xenopus mad proteins transduce distinct subsets of signals for the TGF-β superfamily. Cell. 1996;85:479–485. doi: 10.1016/s0092-8674(00)81249-0. [DOI] [PubMed] [Google Scholar]
- Hahn SA, Schutte M, Shamsul Hoque ATM, Moskaluk CA, da Costa LT, Rosenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Abdollah S, Qiu Y, Cai J, Xu Y, Grinnell BW, Richardson MA, Topper JN, Gimbrone MA, Wrana JL, Falb D. The MAD related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- Hobert O, Mori I, Yamashita Y, Honda H, Ohshima Y, Liu Y, Ruvkun G. Regulation of interneuron function in the C. elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron. 1997;19:345–357. doi: 10.1016/s0896-6273(00)80944-7. [DOI] [PubMed] [Google Scholar]
- Hoodless PA, Haerry T, Abdollah S, Stapleton M, O’Conner MB, Attisano L, Wrana JL. MADR1, a MAD-related protein that functions in BMP2 signaling pathways. Cell. 1996;85:489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- Kim J, Johnson K, Chen HJ, Carroll S, Laughon A. Mad binds to DNA and directly mediates activation of vestigial by DPP. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- Kimble J. Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev Biol. 1981;87:286–300. doi: 10.1016/0012-1606(81)90152-4. [DOI] [PubMed] [Google Scholar]
- Kimble J, White JG. On the control of germ cell development in Caenorhabditis elegans. Dev Biol. 1981;81:208–219. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin-receptor-like gene that regulates longevity and diapause in C. elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Liu F, Hata A, Doody J, Massague J. The TGF-β family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes & Dev. 1997;11:984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Partnership between DPC4 and SMAD proteins in TGF-β signaling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- Liu F, Hata A, Baker JC, Doody J, Carcamo J, Harland RM, Massague J. A human mad protein acting as a BMP-regulated transcriptional activator. Nature. 1996;381:620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]
- Macias-Silva M, Abdollah S, Hoodless PA, Pirone R, Attisano L, Wrana JL. MADR2 is a substrate of the TGF-β receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- Massague J. TGF-β signaling: Receptors, transducers and mad proteins. Cell. 1996;85:947–950. doi: 10.1016/s0092-8674(00)81296-9. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazano K, ten Dijke P, Ichijo H, Heldin C-H. Receptors for transforming-growth-factor-β. Adv Immunol. 1994;55:181–220. [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Newfeld SJ, Mehra A, Singer MA, Wrana JL, Attisano L, Gelbart WM. Mothers against dpp participates in a DPP/TGF-β responsive serine-threonine kinase signal transduction cascade. Development. 1997;124:3167–3176. doi: 10.1242/dev.124.16.3167. [DOI] [PubMed] [Google Scholar]
- Raftery LA, Twombly V, Wharton K, Gelbart WM. Genetic screens to identify elements of the decapentaplegic signaling pathway in Drosophila. Genetics. 1995;139:241–254. doi: 10.1093/genetics/139.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren P, Lim C, Johnsen R, Albert PS, Pilgrim D, Riddle DL. Control of C. elegans larval development by neuronal expression of a TGF-β homolog. Science. 1996;274:1389–1391. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Albert PS. Genetic and environmental regulation of dauer larva development. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 739–768. [PubMed] [Google Scholar]
- Riddle DL, Swanson MM, Albert PS. Interacting genes in dauer larva formation. Nature. 1981;290:668–671. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- Savage C, Das P, Finelli AL, Townsend SR, Sun C, Baird SE, Padgett RW. Caenorhabditis elegans genes sma-2, sma-3, and sma-4 define a conserved family of transforming growth factor-β pathway components. Proc Natl Acad Sci. 1996;93:790–794. doi: 10.1073/pnas.93.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schackwitz WS, Inoue T, Thomas JH. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron. 1996;17:719–728. doi: 10.1016/s0896-6273(00)80203-2. [DOI] [PubMed] [Google Scholar]
- Sekelsky JJ, Newfeld SJ, Raftery LA, Chartoff EH, Gelbart WM. Genetic characterization and cloning of Mothers against Dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics. 1995;139:1347–1358. doi: 10.1093/genetics/139.3.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Swanson MM, Riddle DL. Critical periods in the development of the Caenorhabditis elegans dauer larva. Dev Biol. 1981;84:27–40. doi: 10.1016/0012-1606(81)90367-5. [DOI] [PubMed] [Google Scholar]
- Thomas JT, Birnby DA, Vowels JJ. Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans. Genetics. 1993;134:1105–1117. doi: 10.1093/genetics/134.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent C, Tsung N, Horvitz HR. Egg laying defective mutants of the nematode Caenorhabditis elegans. Genetics. 1983;104:619–647. doi: 10.1093/genetics/104.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper JN, Cai J, Qui Y, Anderson KR, Xu Y, Deeds J, Feeley R, Wolf B, Gimeno C, Sampson BA, et al. Vascular MADs: Two novel MAD related genes selectively inducible by flow in human vascular endothelium. Proc Natl Acad Sci. 1997;94:9314–9319. doi: 10.1073/pnas.94.17.9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale W, Hsueh A, Rivier C, Yu J. The inhibin/activin family of hormones and growth factors. In: Sporn MB, Roberts AB, editors. Peptide growth factors and their receptors. Heidelberg, Germany: Springer-Verlag; 1990. pp. 211–248. [Google Scholar]
- Vowels JJ, Thomas JH. Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics. 1992;130:105–123. doi: 10.1093/genetics/130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser R, Wrana JL, Massague J. GS domain mutations that constitutively activate TβR-1, the downstream signaling component in the TGF-β receptor complex. EMBO J. 1995;14:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrana JL, Attisano L. MAD-related proteins in TGF-β signaling. Trends Genet. 1996;12:493–496. doi: 10.1016/s0168-9525(96)30109-1. [DOI] [PubMed] [Google Scholar]
- Wu R-Y, Zhang Y, Feng X-H, Derynck R. Heteromeric and homomeric interactions correlate with signaling activity and functional cooperativity of Smad3 and Smad4/DPC4. Mol Cell Biol. 1997;17:2521–2528. doi: 10.1128/mcb.17.5.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng X, Wu R, Derynck R. Receptor-associated Mad homologs synergize as effectors of the TGF-β response. Nature. 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Musci T, Derynck R. The tumor suppressor Smad4/DPC 4 as a central mediator of Smad function. Curr Biol. 1997;7:270–276. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]