Abstract
Retina and anterior neural fold homeobox (Rax) gene encodes a transcription factor essential for vertebrate eye development. Recent microarray studies indicate that Rax is expressed in the adult rat pineal gland and retina. The present study reveals that Rax expression levels in the rat change significantly during retinal development with a peak occurring at embryonic day (E) 18, whereas Rax expression in the pineal is relatively delayed and not detectable until E20. In both tissues, Rax is expressed throughout postnatal development into adulthood. In the mature rat pineal gland, the abundance of Rax transcripts increases 2-fold during the light period with a peak occurring at dusk. These findings are consistent with the evidence that Rax is of functional importance in eye development and suggest a role of Rax in the developing pineal gland. In addition, it would appear possible that Rax contributes to phenotype maintenance in the mature retina and pineal gland and may facilitate 24-h changes in the pineal transcriptome.
Keywords: Rax, Rx, retina, pineal gland, developmental biology, circadian biology
Introduction
Formation of the CNS requires patterning of the neural tissue by expression of homeobox genes that encode highly conserved transcription factors (Lumsden and Krumlauf 1996). The homeobox gene Rax is first expressed broadly in the rostral part of the neural plate; expression becomes restricted later in fetal development to several brain regions (Furukawa et al. 1997a; Mathers et al. 1997). Rax is essential for eye development as indicated by the finding that Rax knockout mice do not specify the eye field or form the optic vesicle (Mathers et al. 1997; Zhang et al. 2000). Further, the Rax gene regulates the transcription factor network which controls the proliferative activity of the retinal progenitor cells of the developing eye (Alexiades and Cepko 1996; Furukawa et al. 1997a; Mathers et al. 1997; Bailey et al. 2004; Medina-Martinez et al. 2009), and mutations in the gene can cause anophthalmia and microphthalmia in humans (Voronina et al. 2004; Verma and Fitzpatrick 2007).
In mature photoreceptor cells, Rax protein transactivates photoreceptor cell-specific genes in cooperation with the cone-rod homeobox (Crx) protein (Kimura et al. 2000). Crx is specifically expressed in the retinal photoreceptors and the pineal gland and is known to transactivate genes involved in phototransduction and melatonin synthesis (Chen et al. 1997; Furukawa et al. 1997b; Furukawa et al. 1999; Rath et al. 2006; Rath et al. 2007).
The pineal gland and retina are linked in several ways. Functionally, both are integral parts of the circadian photoneuroendocrine system that conveys information about the environmental lighting condition to the mammalian organism by an increase in nocturnal pineal melatonin synthesis (Klein et al. 2010). Evolutionarily, it appears that both the vertebrate pinealocyte and retinal photoreceptor evolved from a common ancestral photoreceptive melatonin producing cell (Collin 1971; Klein 2004); this has traditionally been based on common structural features (Collin 1971), but additionally, significant similarity in the transcriptomes of both exists (Bailey et al. 2009). Further, the transcription factor cascades that control the development of the eye and the pineal gland appear to be similar (Ekström and Meissl 2003; Maronde and Stehle 2007). Consistent with the common origin hypothesis is the evidence that both structures develop from the diencephalic region of the neural tube (Rubenstein et al. 1998). An additional similarity shared by the pineal and retina is a 24-h pattern of activity, which in the retina reflects the light/dark cycle in photodetection and in the pineal gland reflects the 24-h rhythm in melatonin production and in the expression of many genes (Bailey et al. 2009), including homeobox genes (Rath et al. 2009a; Rath et al. 2009b). The rhythmic expression of these regulatory genes may contribute to the 24-h pattern of differential gene expression in the pineal gland and retina.
Rax has been found to be expressed in the adult pineal gland (Asbreuk et al. 2002; Bailey et al. 2009). In the current report this finding has been extended by analysis of the developmental pattern of appearance of Rax transcripts in the pineal gland and retina. We have also examined the abundance of Rax transcripts throughout the day. These issues are of special importance in defining the physiological role of Rax in regulating the pineal transcriptome. The results of these studies are presented here.
Materials and methods
Animals
Sprague-Dawley rats (Charles River, Sulzfeld, Germany) were used in all experiments. The animals were housed under a 12:12 light-dark schedule. For the constant darkness experiment, animals were shifted to a dark-dark schedule two days before they were euthanized.
For the radiochemical in situ hybridization developmental series, timed-pregnant mothers were anesthetized (500 mg/kg tribromethanol); the abdomen and uterus were opened and fetal animals were removed in utero. Postnatal animals were euthanized by decapitation. Euthanasia was done during daytime at Zeitgeber time (ZT) 6-8. Tissue was fixed by immersion in 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) for 2 days at 4°C, cryoprotected in 25% sucrose, and frozen on crushed solid CO2. For the pineal gland series, the whole fetus (E16-E17), head (E18-postnatal day (P) 2), or brain (P6-P30) was used, and for the retinal series, the fetus (E16-E17), head (E18-P6) or eyeball (P12-P30) was used.
For the 24-h tissue collections, adult male rats (200-350 g), were anesthetized (500 mg/kg tribromethanol) and euthanized by decapitation at eight time points throughout the 24-h period. Brains and eyeballs were immediately removed and frozen on crushed solid CO2. Tissue was prepared for radiochemical in situ hybridization as described below.
To collect tissues for western blot analysis, adult male rats (350-425 g), were anesthetized (500 mg/kg tribromethanol) and euthanized by decapitation at ZT12. The retina and selected parts of the CNS were immediately removed and frozen on crushed solid CO2.
Animal procedures were in accordance with the guidelines of EU Directive 86/609/EEC (approved by the Danish Council for Animal Experiments).
Radiochemical in situ hybridization
Cryostat sections (developmental series, 14 μm and adult animals, 12 μm) were prepared and mounted on glass slides. For detection of Rax mRNA, sections were hybridized (Rath et al. 2007) with a [35S]dATP-labelled 38 nucleotide antisense DNA probe: 5′–CGAGTCCTGCAGCTTCATGGACGACACTTCTAGTTTCT–3′ corresponding to residues 587 – 624 of the rat Rax mRNA (NM_053678.1) and a sense control probe. For detection of Aanat mRNA, a published probe sequence was used (Bailey et al. 2009).
Following hybridization and washing, the sections were exposed to an x-ray film for 2 weeks and the film was developed. Images of the x-ray film autoradiographs were digitized and the hybridization signal was quantified using ScionImage 1.42 software (Wayne Rasband, NIH, Bethesda, MD, USA). Optical densities were converted to dpm/mg tissue using simultaneously exposed 14C-standards calibrated against 35S brain-paste standards. Following x-ray film exposure, the sections were covered with a photographic emulsion (Amersham, Hillerød, Denmark) by immersion at 40°C and exposed for 4 weeks. The photographic emulsion was developed in amidol and fixed in sodium thiosulfate. The sections were counterstained in cresyl violet and photographed in a microscope in light and dark field mode.
Western blot analysis
Samples were homogenized in 2× Laemmli buffer containing 80% (v/v) 155 mM Tris/HCl buffer (pH 8.3), 10% (w/v) sodium dodecyl sulfate, 18 mM bromphenol blue and 20% (v/v) glycerol. Following homogenization, the samples were boiled at 95°C for 10 min and centrifuged for 1 h at 13 000 g at 4°C. The protein concentration of the supernatant was measured with Pierce BCA™ Protein Assay Kit (Thermo Scientific, Rockford, IL, USA) and the samples were further diluted in 2× Laemmli buffer including 5% (v/v) 2-mercaptoethanol. The protein samples were boiled for 5 min prior to loading and 100 μg protein per lane was run in a NuPAGE® 12% Bis-Tris Gel (Invitrogen, Taastrup, Denmark). After size-fractionation, the protein was transferred to nitrocellulose membrane (XCell SureLock™, Invitrogen). The nitrocellulose membrane was blocked with 2% (w/v) skim milk in phosphate-buffered saline (PBS) and incubated overnight at 4°C with polyclonal rabbit anti-Rax antibody (Antibodies-online GmbH, Aachen, Germany) diluted 1:500 in blocking solution. For the pre-absorption control, the diluted antibody was pre-incubated with 100 μg/mL blocking peptide (Antibodies-online GmbH) for 5 days at 4°C. The membrane was washed in PBS and incubated for 1 h at room temperature in biotinylated polyclonal swine anti-rabbit antibody (Dako, Glostrup, Denmark) diluted 1:500 in blocking solution and subsequently washed. The membrane was incubated for 45 min in avidin-biotin-peroxidase complex (Vector Laboratories, Burlingame, CA, USA), containing 0.5% (v/v) avidin solution and 0.5% (v/v) biotinylated-peroxidase solution in blocking solution, followed by washing for 3×5 min in PBS and 5 min in 0.05 M Tris/HCl buffer (pH 7.6). The membrane was incubated in a solution of chromogenic 1.4 mM diaminobenzidine, 0.01% (v/v) H2O2 in 0.05 M Tris/HCl buffer (pH 7.6) at room temperature until the protein bands became visible. The reaction was stopped by washing in deionized water. Precision Plus Protein Kaleidoscope™ Standards (Bio-Rad, Copenhagen, Denmark) were used as molecular weight markers.
Statistical analysis
Data were analyzed using GraphPad Prism® software. The densitometric quantification of in situ hybridization autoradiographs was analyzed with one- or two-way ANOVA and Tukey's multiple comparison post-test. A two-tailed p-value of <0.05 was considered to represent statistical significance.
Results
Expression of the Rax gene in the developing rat pineal gland
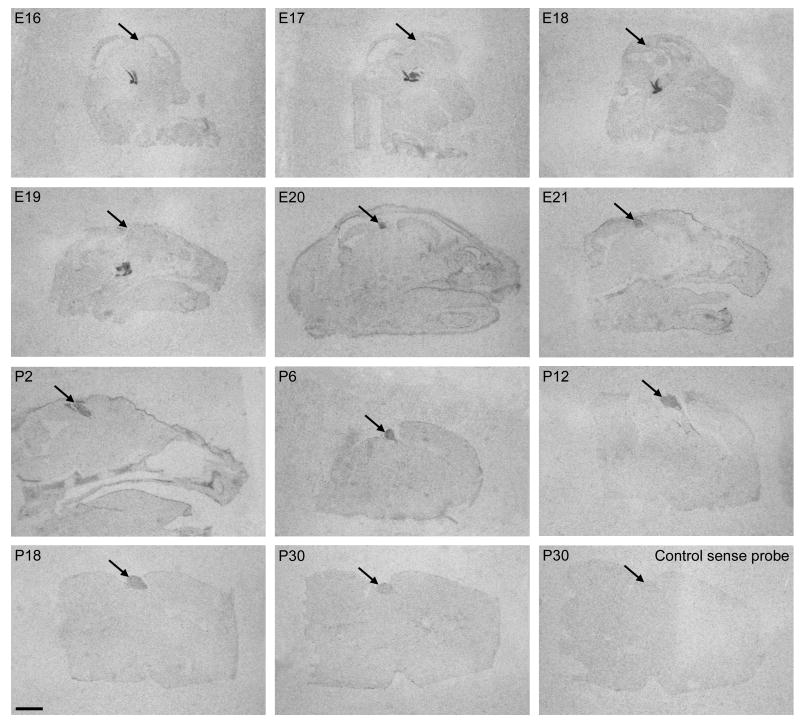
The Rax ontogenetic expression pattern was investigated in the developing pineal gland by use of in situ hybridization on median brain sections in a developmental series ranging from E16 to P30 (Fig. 1). At E16, when the pineal gland is present as a small tubular evagination of the diencephalic roof a Rax hybridization signal was not detected in this tissue (Fig. 1). However, expression was detected at developmental stages E16 to E19 in the hypothalamus and posterior pituitary on the same sections containing the pineal gland that did not generate a positive hybridization signal (Fig. 2). These observations confirm that Rax is not expressed in the pineal gland at these early stages at detectable levels. Hybridization with a sense control probe did not generate a signal (Fig. 1).
Figure 1. Rax transcripts in the developing rat pineal gland.
Autoradiographs of hybridized median brain sections in a developmental series ranging from E16 to P30 are displayed. A hybridization signal in the pineal gland is detectable from E20. The pineal gland is marked by arrows. The autoradiograph in the lower right corner shows a section hybridized with a sense control probe. Scale bar, 2 mm.
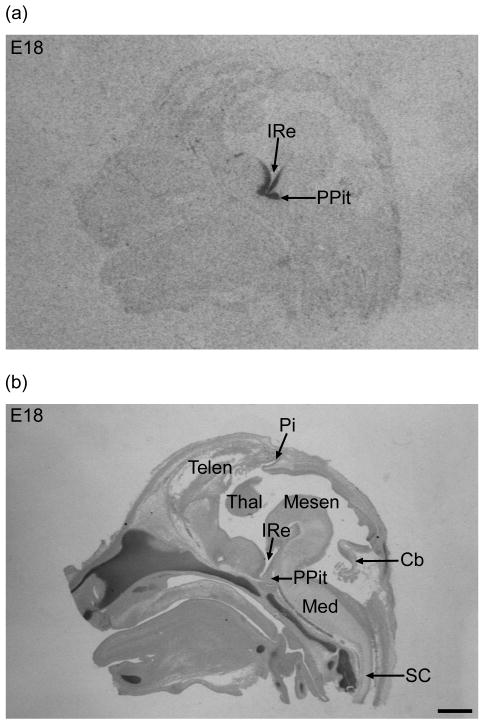
Figure 2. Rax mRNA in the brain and demonstration of the presence of the pineal “anlage” at E18.
(a) In situ hybridization autoradiograph for Rax mRNA detection in a median brain section exhibiting a signal around the infundibular recess of the 3rd ventricle (IRe) and in the posterior lobe of the pituitary (PPit). (b) The hybridized section was counterstained with cresyl violet after the exposure and the pineal gland (Pi) is seen in the diencephalic roof. The corresponding autoradiograph shows no pineal hybridization signal. PPit, posterior lobe of the pituitary; Ire, infundibular recess of the 3rd ventricle; Pi, pineal gland; Telen, telencephalon; Mesen, mesencephalon; Thal, thalamus; Cb, cerebellum; Med, medulla oblongata; SC, spinal cord. Scale bar, 1 mm.
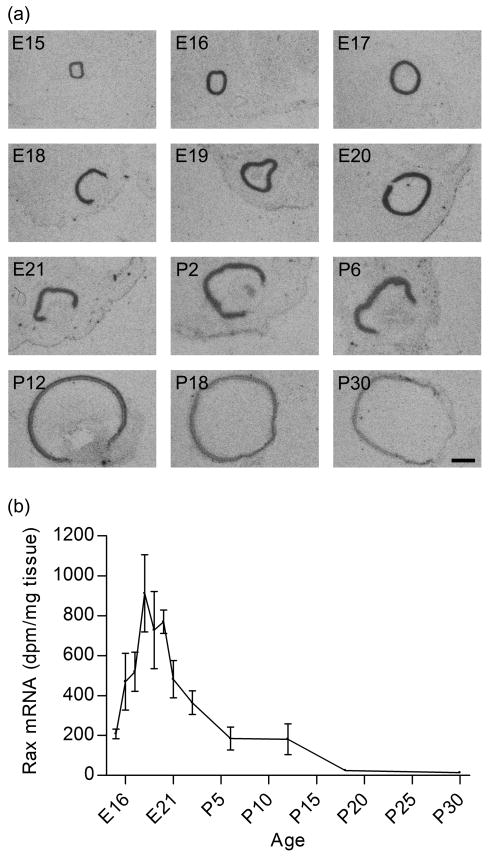
Pineal Rax mRNA was detected at developmental stage E20 at a high level, indicating that expression increased rapidly in the preceding day. Densitometric quantification revealed significant age-dependent differences in Rax expression levels during development (p<0.01, one-way ANOVA); following a peak at E20 values decreased (p-values <0.05, Tukey's multiple comparison test) (Fig. 3).
Figure 3. Quantification of in situ hybridization autoradiographs of ontogenetic Rax expression in the rat pineal gland.
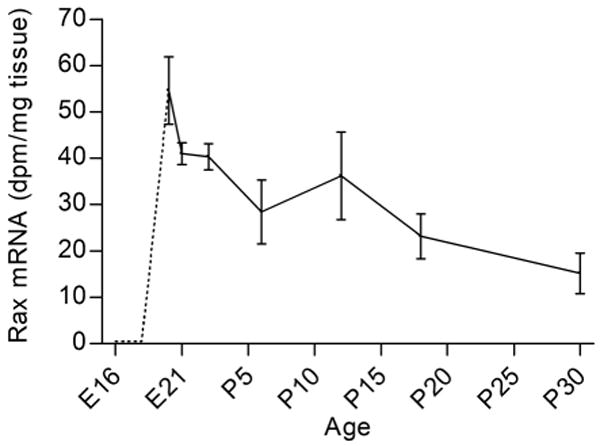
In the earliest stages (E16-E19), no hybridization signal was detected in the pineal gland (dotted line). Rax transcript levels varied significantly during the E20 to P30 developmental period (p<0.01, one-way ANOVA): Rax mRNA levels decreased from E20 to P18 (p<0.05, Tukey's multiple comparison test) and P30 (p<0.01, Tukey's multiple comparison test). The values on the graph represent the mean ± SEM of three animals at each developmental stage examined.
Examination of the distribution of label in the in situ hybridization images of the pineal gland revealed uniform distribution in the entire gland (Fig. 1). This pattern is consistent with Rax expression in the pinealocytes, which represent 95% of the cells in the mammalian pineal gland (Møller and Baeres 2002).
Expression of Rax in the developing rat retina
To compare the developmental expression pattern of Rax in the pineal gland to that in the retina, in situ hybridization was performed on cryostat sections of eyes in a developmental series ranging from E15 to P30 (Figs. 4 and 5). Rax mRNA was detected at all twelve stages examined (Fig. 4a). Densitometric quantification of the in situ hybridization autoradiographs revealed that Rax mRNA levels changed significantly during retinal development (p<0.0001, one-way ANOVA). Rax expression peaked around E18; the Rax mRNA level increased from E15 to E18 (p<0.01, Tukey's multiple comparison test) and decreased postnatally (E18 compared to P2-P30, p-values <0.05, Tukey's multiple comparison test) (Fig. 4b). Hybridization with a sense control probe generated no signal (data not shown).
Figure 4. Detection and quantification of Rax gene expression in the developing rat retina.
(a) Autoradiographs of in situ hybridization for detection of Rax mRNA in sections of embryonic and postnatal rat eyes. Developmental stages are indicated in the upper left corner of each photomicrograph (E15-P30). (b) Densitometric quantification of the retinal in situ hybridization signal. Differential expression levels of Rax during retinal development were detected (p<0.0001, one-way ANOVA). Retinal Rax mRNA levels peaked around E18; the expression increased from E15 to E18 (p<0.01, Tukey's multiple comparison test) and decreased gradually after birth (E18 to P2, p<0.05; E18 to P6, p<0.01; E18 to P12, p<0.01; E18 to P18, p<0.001; E18 to P30, p<0.001; Tukey's multiple comparison test). The values on the graph represent the mean ± SEM of three animals at each developmental stage examined. Scale bar, 1 mm.
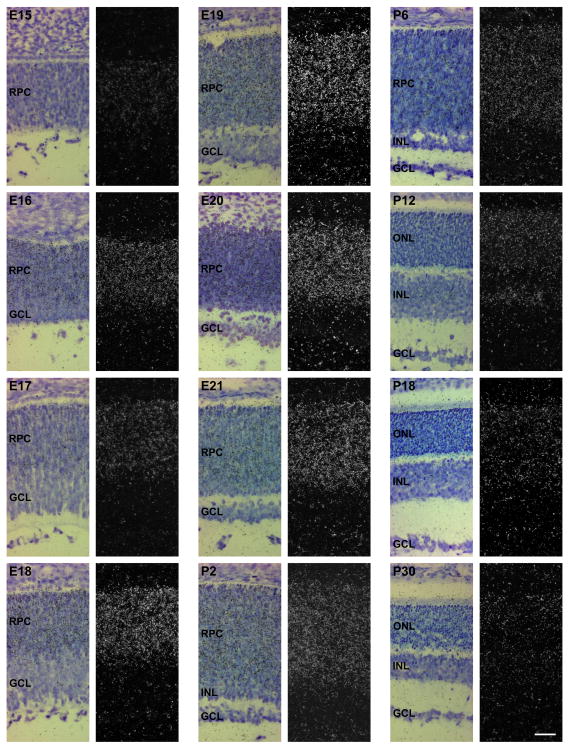
Figure 5. Localization of Rax transcripts in the developing rat retina.
Hybridized sections were dipped in a photographic emulsion, exposed, developed and counterstained. Developmental stages are indicated in the upper left corner of each photomicrograph (E15-P30). Left columns: bright field photomicrographs. Right columns: dark field photomicrographs of the same parts of the hybridized sections. RPC, retinal progenitor cells; GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. Scale bar, 40 μm.
The Rax expression pattern in the developing rat retina was examined at a higher spatial resolution by applying a photographic emulsion directly to the sections (Fig. 5). At E15, the Rax transcript was detected throughout neural retina. Rax gene expression was not observed in the pigment epithelium at any developmental stages examined. The presumptive ganglion cell layer becomes distinguishable at E16; however, Rax expression was at this stage not observed in the ganglion cells, whereas persistent expression was detected in the retinal progenitor cells. This spatial pattern of expression was maintained from E17 through E21. The ganglion cell layer starts to separate from the rest of the developing neural retina at E19, and at P2, the presumptive inner nuclear layer begins to form at the inner part of the retinal progenitor cells. At P2 and P6, Rax expression was found to be localized to the retinal progenitor cells and the outer part of the presumptive inner nuclear layer, whereas Rax transcripts were not detected in the inner part of the presumptive inner nuclear layer, the inner plexiform layer, and the ganglion cell layer. At P12, the mature layers of the neural retina were fully distinguishable; at this stage, the Rax transcript was detected in the inner segments of the photoreceptors, the outer nuclear layer, and the outer part of the inner nuclear layer. This expression pattern persisted into adulthood (Fig. 5).
Diurnal rhythm of Rax expression in the adult rat pineal gland and retina
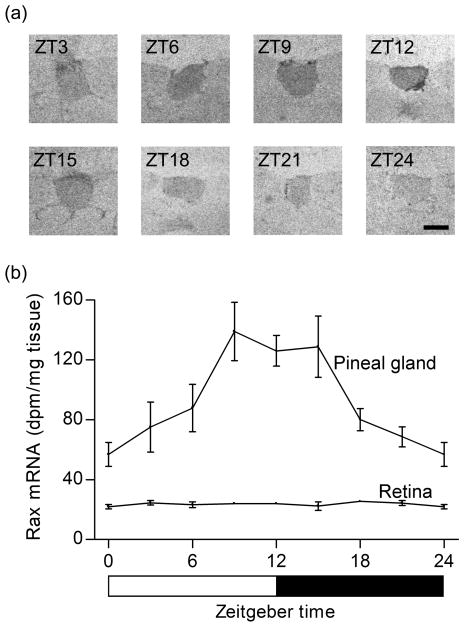
In the adult pineal gland, Rax mRNA was detected at eight time points throughout the 24-h period on in situ hybridization autoradiographs (Fig. 6a). Densitometric quantification revealed that Rax mRNA levels changed significantly during the 24-h period (p<0.01, one-way ANOVA); the Rax mRNA level significantly increased during the light period and gradually decreased during the dark period (p<0.05, Tukey's multiple comparison test) (Fig. 6b). The ZT12 / ZT24-ratio was 2.2 ± 0.4 (Fig. 6b). The pineal expression of Rax was not affected by changing the 12:12 light-dark schedule to constant darkness (p=0.24, two-way ANOVA) indicating that the expression is circadian (Supplementary data, Fig. S1).
Figure 6. Rax expression in the adult rat pineal gland exhibits a daily rhythm.
(a) In situ hybridization autoradiographs of coronal brain sections through the pineal gland of rats sacrificed in 3 h intervals at the indicated time points. (b) Densitometric quantification of pineal and retinal in situ hybridization autoradiographs. In the pineal gland, a significant Rax mRNA diurnal rhythm (p<0.01, one-way ANOVA) was detected; Rax mRNA levels increased during the light period and declined during the dark period (p<0.05, Tukey's multiple comparison test). In the retina, daily changes in Rax expression were not detected (p = 0.76, one-way ANOVA). Rats were housed under a 12:12 light-dark schedule. Values on the Rax mRNA graphs represent the mean ± SEM of three animals at each time point examined. Scale bar, 1 mm.
In the retina, Rax mRNA was detected at eight time points throughout the 24-h period on in situ hybridization autoradiographs (Supplementary data, Fig. S2). However, in contrast to the pineal gland, a daily change in retinal Rax expression was not detected (p = 0.76, one-way ANOVA) (Fig. 6b).
Rax protein is detectable in the adult rat pineal gland and retina
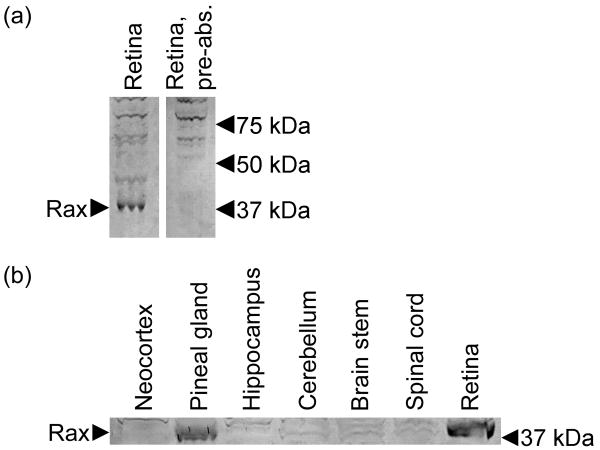
Western blot analysis using a commercially available antiserum detected a band of ∼37 kDa (Fig. 7a); pre-absorption of the antiserum specifically eliminated this band, thereby confirming that the band represented Rax protein, which has a predicted mass of 36.3 kDa. Rax protein was detected in pineal and retinal extracts, whereas it was not detected in several brain regions (Fig. 7b).
Figure 7. Rax protein in the adult rat pineal gland and retina.
(a) Elimination of a 37 kDa immunopositive band by pre-absorption with Rax peptide, indicating that the detected band represents Rax protein (NP_446130.1; predicted molecular weight = 36.3 kDa). (b) A 37 kDa band is detected specifically in the extracts of the pineal gland and retina, but not in other tissues examined. Tissues were collected from rats euthanized at ZT12. Arrows indicate the position of the Rax band and molecular weight markers.
Discussion
The results presented here provide a detailed description of the expression of the Rax homeobox gene in the developing and adult pineal gland and retina of the rat. In both tissues, Rax is expressed from intrauterine stages into adulthood, but with a distinct lag of onset in the pineal gland as compared to the retina. The expression changes dynamically during development and on a daily basis in the adult rat pineal gland.
Our results on Rax expression in the developing rat retina extend previous murine studies (Furukawa et al. 1997a; Mathers et al. 1997) by adding information on the distribution of Rax transcripts and a quantitative analysis of gene expression levels. Rax is continuously expressed in the mitotic retinal progenitor cells of the neural retina during the last fetal week and the first postnatal week. The decrease in retinal Rax expression during the first postnatal week correlates in a timely manner to a previously reported decline in the percentage of mitotic cells (Alexiades and Cepko 1996), thus supporting a role of Rax as a transcription factor that maintains the pool of retinal progenitor cells by promoting proliferation and inhibiting differentiation (Bailey et al. 2004; Medina-Martinez et al. 2009). However, in addition to the developmental role of Rax, the persistent expression of Rax into adulthood in the outer nuclear layer suggests a role in normal photoreceptor cell function; this is in accordance with the reported ability of the Rax protein to interact with photoreceptor cell-specific gene promoters and stimulate expression of genes involved in phototransduction, e.g. rhodopsin and arrestin (Kimura et al. 2000; Pan et al. 2010). These genes are also activated by Crx (Furukawa et al. 1997b), which has been shown in vitro to at least partly exert its function via cooperation with Rax to generate high expression levels of photoreceptor cell-specific genes (Kimura et al. 2000).
As indicated above, it is known that Rax is expressed in the mature rodent pineal gland (Asbreuk et al. 2002; Bailey et al. 2009). Our developmental analysis demonstrate Rax expression is not detectable in the gland prior to E20, when there is a clear expression that continues thereafter. When Rax expression is initiated in the pineal gland, cell division is decreasing and genetic markers of pinealocytes start to appear (Ellison et al. 1972; Quay 1974; Sugden and Klein 1983; Babila et al. 1992; Pfeffer and Stehle 1998; Ribelayga et al. 1998; Calvo et al. 2004). Accordingly, the temporal expression pattern of Rax is inconsistent with a primary role of the Rax gene in pinealocyte progenitor cells, in contrast to homeobox genes essential for pineal gland development, e.g. Otx2 and Pax6 (Estivill-Torrus et al. 2001; Nishida et al. 2003), which are highly expressed in the pineal gland when progenitor cells are dividing (Rath et al. 2006; Rath et al. 2009b) (Fig. 8). Rather, the continuous expression into adulthood is consistent with a role of Rax in cell fate determination and maintenance of the pinealocyte phenotype.
Figure 8. Developmental appearance of Rax, Otx2 and Crx homeobox gene expression in the rat pineal gland and establishment of pineal function.
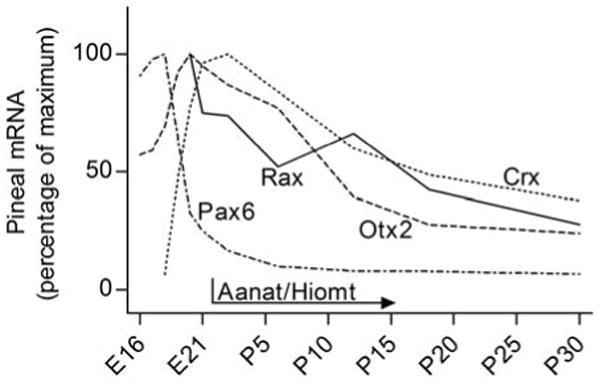
Otx2 is expressed in the developing rat pineal gland with a peak at E20 (Rath et al. 2006), at which time the expression of Crx (Rath et al. 2006) and Rax (the present study) start and peak, respectively. This is consistent with the proposal that Otx2 is involved in the transactivation of Rax in the mammalian pineal gland as reported in submammalian vertebrates. Notably, Pax6 is expressed very early in pineal development (Rath et al. 2009b). Rax and Crx expression peak immediately prior to birth, at which time the genes encoding the penultimate enzymes in melatonin synthesis Aanat and Hiomt are turned on (Pfeffer and Stehle 1998; Ribelayga et al. 1998). This supports the hypothesis that Rax and Crx exert their function in concert to positively influence the development of pineal physiology.
Several studies link Rax at the molecular level to other homeobox genes regulating pineal development and adult function. Data from Xenopus studies show that Rax is a downstream target of the Otx2 transcription factor, which binds and transactivates the Rax gene promoter (Danno et al. 2008). Otx2 is expressed in the rat pineal gland from E16 into adulthood with a peak at E20 (Rath et al. 2006), at which time Rax expression is initiated, thus making it reasonable to suspect that a similar transactivating function of Otx2 is present in the mammalian pineal gland (Fig. 8). Besides the role of the Crx transcription factor in the retinal photoreceptors, Crx also transactivates genes involved in melatonin synthesis, e.g. arylalkylamine N-acetyltransferase (Aanat) and hydroxyindole-O-methyltransferase (Hiomt) (Li et al. 1998; Bernard et al. 2001). The peak in expression of both Rax and Crx just before birth in the rat pineal gland (Rath et al. 2006), at which time Aanat and Hiomt expression start to increase (Ellison et al. 1972; Sugden and Klein 1983; Pfeffer and Stehle 1998; Ribelayga et al. 1998) (Fig. 8), in addition to the synergistic relation between Rax and Crx discussed above, establishes these transcription factors as candidates for a role in controlling development of the central physiological function of the pineal gland, melatonin synthesis.
The presence of Rax mRNA and the corresponding protein in both the pineal gland and retina supports the previously proposed common ancestral origin of the pinealocyte and retinal photoreceptor as suggested by morphology (Collin 1971; Korf 1999) and molecular data (Klein 2004; Bailey et al. 2009). However, the inverse ontogenetic expression patterns of Rax in the pineal gland and retina are in marked contrast to the similar onset and peak in expression of other homeobox genes, e.g. Otx2, Crx, Pax4 and Pax6, in these tissues (Rath et al. 2006; Rath et al. 2007; Rath et al. 2009a; Rath et al. 2009b). The distinct difference in onset of Rax expression in the pineal gland and retina suggests a profound role of the Rax transcription factor in determining the differential development of these related neuronal structures
The investigation of the daily expression pattern of the Rax gene in the adult rat pineal gland has revealed that Rax mRNA levels increase 2-fold during the light period and decrease gradually during the dark period, which is clearly out of phase with the pineal melatonin release, which occurs at night (Møller and Baeres 2002; Klein 2007). It seems reasonable to suspect that the diurnal rhythm of Rax expression may contribute to the regulation of the 24-h dynamics of the pineal gland by controlling rhythmic expression of other genes.
Supplementary Material
Acknowledgments
This study was supported by the Novo Nordisk Foundation, the Danish Medical Research Council, the Lundbeck Foundation and the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health. We wish to thank Ms Tine Thorup Mellergaard for expert histological assistance. The authors declare no conflicts of interest.
Abbreviations
- Aanat
arylalkylamine N-acetyltransferase
- Crx
cone-rod homeobox
- E
embryonic day
- Hiomt
hydroxyindole-O-methyltransferase
- P
postnatal day
- PBS
phosphate-buffered saline
- Rax
retina and anterior neural fold homeobox
- ZT
Zeitgeber time
References
- Alexiades MR, Cepko C. Quantitative analysis of proliferation and cell cycle length during development of the rat retina. Dev Dyn. 1996;205:293–307. doi: 10.1002/(SICI)1097-0177(199603)205:3<293::AID-AJA9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Asbreuk CH, van Schaick HS, Cox JJ, Smidt MP, Burbach JP. Survey for paired-like homeodomain gene expression in the hypothalamus: restricted expression patterns of Rx, Alx4 and goosecoid. Neuroscience. 2002;114:883–889. doi: 10.1016/s0306-4522(02)00325-1. [DOI] [PubMed] [Google Scholar]
- Babila T, Schaad NC, Simonds WF, Shinohara T, Klein DC. Development of MEKA (phosducin), G beta, G gamma and S-antigen in the rat pineal gland and retina. Brain Res. 1992;585:141–148. doi: 10.1016/0006-8993(92)91199-o. [DOI] [PubMed] [Google Scholar]
- Bailey MJ, Coon SL, Carter DA, Humphries A, Kim JS, Shi Q, Gaildrat P, Morin F, Ganguly S, Hogenesch JB, Weller JL, Rath MF, Møller M, Baler R, Sugden D, Rangel ZG, Munson PJ, Klein DC. Night/day changes in pineal expression of >600 genes: central role of adrenergic/cAMP signaling. J Biol Chem. 2009;284:7606–7622. doi: 10.1074/jbc.M808394200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TJ, El-Hodiri H, Zhang L, Shah R, Mathers PH, Jamrich M. Regulation of vertebrate eye development by Rx genes. Int J Dev Biol. 2004;48:761–770. doi: 10.1387/ijdb.041878tb. [DOI] [PubMed] [Google Scholar]
- Bernard M, Dinet V, Voisin P. Transcriptional regulation of the chicken hydroxyindole-O-methyltransferase gene by the cone-rod homeobox-containing protein. J Neurochem. 2001;79:248–257. doi: 10.1046/j.1471-4159.2001.00555.x. [DOI] [PubMed] [Google Scholar]
- Calvo JL, Boya J, Carbonell AL, Garcia-Maurino JE. Time of origin of the rat pineal gland cells. A bromodeoxyuridine immunohistochemical study. Histol Histopathol. 2004;19:137–142. doi: 10.14670/HH-19.137. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- Collin JP. Differentiation and regression of the cells of the sensory line in the epiphysis cerebri. In: Wolstenholme GEW, Knight J, editors. The pineal gland. Churchill-Livingstone; Edinburgh: 1971. pp. 79–125. [Google Scholar]
- Danno H, Michiue T, Hitachi K, Yukita A, Ishiura S, Asashima M. Molecular links among the causative genes for ocular malformation: Otx2 and Sox2 coregulate Rax expression. Proc Natl Acad Sci U S A. 2008;105:5408–5413. doi: 10.1073/pnas.0710954105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekström P, Meissl H. Evolution of photosensory pineal organs in new light: the fate of neuroendocrine photoreceptors. Philos Trans R Soc Lond B Biol Sci. 2003;358:1679–1700. doi: 10.1098/rstb.2003.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison N, Weller JL, Klein DC. Development of a circadian rhythm in the activity of pineal serotonin N-acetyltransferase. J Neurochem. 1972;19:1335–1341. doi: 10.1111/j.1471-4159.1972.tb01458.x. [DOI] [PubMed] [Google Scholar]
- Estivill-Torrus G, Vitalis T, Fernandez-Llebrez P, Price DJ. The transcription factor Pax6 is required for development of the diencephalic dorsal midline secretory radial glia that form the subcommissural organ. Mech Dev. 2001;109:215–224. doi: 10.1016/s0925-4773(01)00527-5. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Kozak CA, Cepko CL. rax, a novel paired-type homeobox gene, shows expression in the anterior neural fold and developing retina. Proc Natl Acad Sci U S A. 1997a;94:3088–3093. doi: 10.1073/pnas.94.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997b;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- Kimura A, Singh D, Wawrousek EF, Kikuchi M, Nakamura M, Shinohara T. Both PCE-1/RX and OTX/CRX interactions are necessary for photoreceptor-specific gene expression. J Biol Chem. 2000;275:1152–1160. doi: 10.1074/jbc.275.2.1152. [DOI] [PubMed] [Google Scholar]
- Klein DC. The 2004 Aschoff/Pittendrigh lecture: Theory of the origin of the pineal gland--a tale of conflict and resolution. J Biol Rhythms. 2004;19:264–279. doi: 10.1177/0748730404267340. [DOI] [PubMed] [Google Scholar]
- Klein DC. Arylalkylamine N-acetyltransferase: “the Timezyme”. J Biol Chem. 2007;282:4233–4237. doi: 10.1074/jbc.R600036200. [DOI] [PubMed] [Google Scholar]
- Klein DC, Bailey MJ, Carter DA, Kim JS, Shi Q, Ho AK, Chik CL, Gaildrat P, Morin F, Ganguly S, Rath MF, Møller M, Sugden D, Rangel ZG, Munson PJ, Weller JL, Coon SL. Pineal function: impact of microarray analysis. Mol Cell Endocrinol. 2010;314:170–183. doi: 10.1016/j.mce.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf HW. Evolution of melatonin-producing pinealocytes. Adv Exp Med Biol. 1999;460:17–29. doi: 10.1007/0-306-46814-x_3. [DOI] [PubMed] [Google Scholar]
- Li X, Chen S, Wang Q, Zack DJ, Snyder SH, Borjigin J. A pineal regulatory element (PIRE) mediates transactivation by the pineal/retina-specific transcription factor CRX. Proc Natl Acad Sci U S A. 1998;95:1876–1881. doi: 10.1073/pnas.95.4.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- Maronde E, Stehle JH. The mammalian pineal gland: known facts, unknown facets. Trends Endocrinol Metab. 2007;18:142–149. doi: 10.1016/j.tem.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Mathers PH, Grinberg A, Mahon KA, Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- Medina-Martinez O, Amaya-Manzanares F, Liu C, Mendoza M, Shah R, Zhang L, Behringer RR, Mahon KA, Jamrich M. Cell-autonomous requirement for rx function in the mammalian retina and posterior pituitary. PLoS One. 2009;4:e4513. doi: 10.1371/journal.pone.0004513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller M, Baeres FM. The anatomy and innervation of the mammalian pineal gland. Cell Tissue Res. 2002;309:139–150. doi: 10.1007/s00441-002-0580-5. [DOI] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- Pan Y, Martinez-De Luna RI, Lou CH, Nekkalapudi S, Kelly LE, Sater AK, El-Hodiri HM. Regulation of photoreceptor gene expression by the retinal homeobox (Rx) gene product. Dev Biol. 2010;339:494–506. doi: 10.1016/j.ydbio.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer M, Stehle JH. Ontogeny of a diurnal rhythm in arylalkylamine-N-acetyltransferase mRNA in rat pineal gland. Neurosci Lett. 1998;248:163–166. doi: 10.1016/s0304-3940(98)00356-5. [DOI] [PubMed] [Google Scholar]
- Quay WB. Pineal chemistry in cellular and physiological mechanisms. Charles C. Thomas; Springfield: 1974. [Google Scholar]
- Rath MF, Bailey MJ, Kim JS, Coon SL, Klein DC, Møller M. Developmental and daily expression of the Pax4 and Pax6 homeobox genes in the rat retina: localization of Pax4 in photoreceptor cells. J Neurochem. 2009a;108:285–294. doi: 10.1111/j.1471-4159.2008.05765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath MF, Bailey MJ, Kim JS, Ho AK, Gaildrat P, Coon SL, Møller M, Klein DC. Developmental and diurnal dynamics of Pax4 expression in the mammalian pineal gland: nocturnal down-regulation is mediated by adrenergic-cyclic adenosine 3′,5′-monophosphate signaling. Endocrinology. 2009b;150:803–811. doi: 10.1210/en.2008-0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath MF, Morin F, Shi Q, Klein DC, Møller M. Ontogenetic expression of the Otx2 and Crx homeobox genes in the retina of the rat. Exp Eye Res. 2007;85:65–73. doi: 10.1016/j.exer.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Rath MF, Munoz E, Ganguly S, Morin F, Shi Q, Klein DC, Møller M. Expression of the Otx2 homeobox gene in the developing mammalian brain: embryonic and adult expression in the pineal gland. J Neurochem. 2006;97:556–566. doi: 10.1111/j.1471-4159.2006.03773.x. [DOI] [PubMed] [Google Scholar]
- Ribelayga C, Gauer F, Pevet P, Simonneaux V. Ontogenesis of hydroxyindole-O-methyltransferase gene expression and activity in the rat pineal gland. Brain Res Dev Brain Res. 1998;110:235–239. doi: 10.1016/s0165-3806(98)00114-x. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL, Shimamura K, Martinez S, Puelles L. Regionalization of the prosencephalic neural plate. Annu Rev Neurosci. 1998;21:445–477. doi: 10.1146/annurev.neuro.21.1.445. [DOI] [PubMed] [Google Scholar]
- Sugden D, Klein DC. Regulation of rat pineal hydroxyindole-O-methyltransferase in neonatal and adult rats. J Neurochem. 1983;40:1647–1653. doi: 10.1111/j.1471-4159.1983.tb08138.x. [DOI] [PubMed] [Google Scholar]
- Verma AS, Fitzpatrick DR. Anophthalmia and microphthalmia. Orphanet J Rare Dis. 2007;2:47. doi: 10.1186/1750-1172-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina VA, Kozhemyakina EA, O'Kernick CM, Kahn ND, Wenger SL, Linberg JV, Schneider AS, Mathers PH. Mutations in the human RAX homeobox gene in a patient with anophthalmia and sclerocornea. Hum Mol Genet. 2004;13:315–322. doi: 10.1093/hmg/ddh025. [DOI] [PubMed] [Google Scholar]
- Zhang L, Mathers PH, Jamrich M. Function of Rx, but not Pax6, is essential for the formation of retinal progenitor cells in mice. Genesis. 2000;28:135–142. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.