Abstract
DNA double strand breaks (DSBs) in repetitive sequences are a potent source of genomic instability, due to the possibility of non-allelic homologous recombination (NAHR). Repetitive sequences are especially at risk during meiosis, when numerous programmed DSBs are introduced into the genome to initiate meiotic recombination 1. Within the budding yeast repetitive ribosomal (r)DNA array, meiotic DSB formation is prevented in part through Sir2-dependent heterochromatin 2,3. Here, we demonstrate that the edges of the rDNA array are exceptionally susceptible to meiotic DSBs, revealing an inherent heterogeneity within the rDNA array. We find that this localised DSB susceptibility necessitates a border-specific protection system consisting of the meiotic ATPase Pch2 and the origin recognition complex subunit Orc1. Upon disruption of these factors, DSB formation and recombination specifically increased in the outermost rDNA repeats, leading to NAHR and rDNA instability. Strikingly, the Sir2-dependent heterochromatin of the rDNA itself was responsible for the induction of DSBs at the rDNA borders in pch2Δ cells. Thus, while Sir2 activity globally prevents meiotic DSBs within the rDNA, it creates a highly permissive environment for DSB formation at the heterochromatin/euchromatin junctions. Heterochromatinised repetitive DNA arrays are abundantly present in most eukaryotic genomes. Our data define the borders of such chromatin domains as distinct high-risk regions for meiotic NAHR, whose protection may be a universal requirement to prevent meiotic genome rearrangements associated with genomic diseases and birth defects.
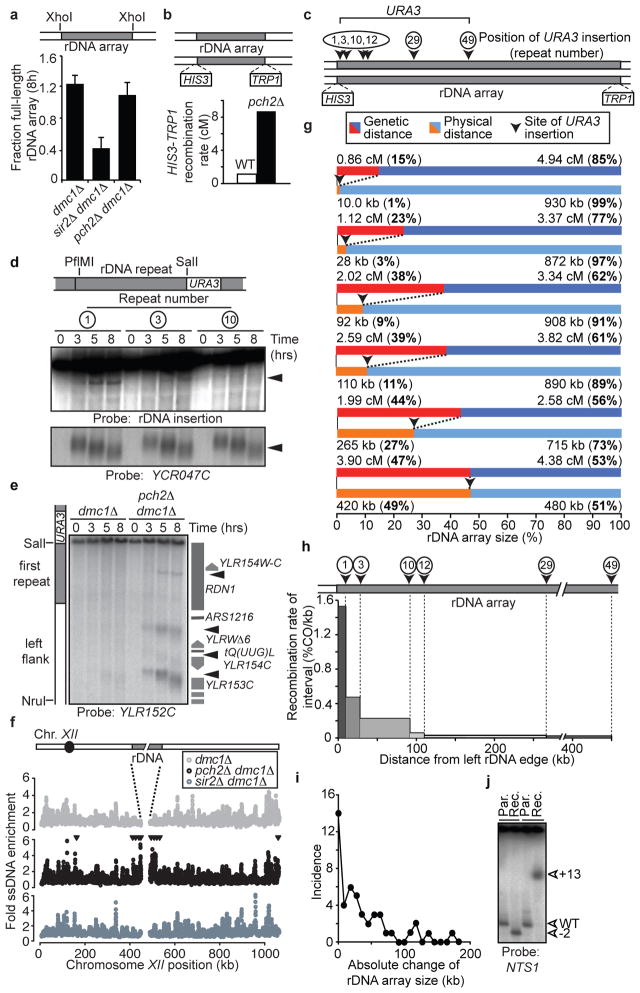
To better understand the mechanisms that protect repetitive DNA from meiotic NAHR, we analysed the single tandem rDNA array of budding yeast. Meiotic DSB formation and recombination within the rDNA are repressed by the histone deacetylase Sir2 2,3. Additionally, Pch2, a widely conserved meiosis-specific ATPase, suppresses meiotic recombination in the rDNA by an unknown mechanism 4,5. We used clamped-homogenous electric field (CHEF) electrophoresis and Southern blotting of excised rDNA arrays to address whether Pch2 regulates meiotic DSB formation in the rDNA. Consistent with previous results 2,3, the level of full-length rDNA arrays remaining 8 hours after meiotic induction was significantly reduced in sir2Δ mutants compared to wild-type cells, indicating increased DSB formation (Figures 1a, S1a). By contrast, no such reduction occurred in pch2Δ mutants, although we observed a 10-fold increase in crossover recombination across the rDNA array (Figure 1a,b). Because small changes in array length would not be detectable by the CHEF gel assay, we wondered whether DSB formation in pch2Δ mutants occurred specifically within the outermost rDNA repeats. To test this possibility, we generated pch2Δ strains carrying a URA3 insertion at defined positions in the rDNA array (Figure 1c) and analysed the rDNA repeat units directly flanking these insertions by Southern blotting. We observed a strong DSB site in repeat 1 and weak DSB formation in repeat 3, whereas no DSB formation was detectable in repeat 10 of the ~100 rDNA repeats (Figure 1d). Thus, pch2Δ cells experience increased meiotic DSB formation predominantly in the outermost rDNA repeats.
Figure 1. rDNA-associated DSB formation and recombination.
a, CHEF analysis of the rDNA of meiotic dmc1Δ (H5217), sir2Δ dmc1Δ (H2953), and pch2Δ dmc1Δ (H5216) cells. Schematic denotes the analysed XhoI restriction fragment. A dmc1Δ mutation was used to prevent DSB repair. Mean (+ s.e.m.) of five experiments is shown. Significance (one-tailed Student’s T-test): dmc1Δ vs. sir2Δ dmc1Δ, p-value=0.00122; dmc1Δ vs. pch2Δ dmc1Δ, p-value=0.254; pch2Δ dmc1Δ vs. sir2Δ dmc1Δ, p-value=0.00216. b, Schematic of markers inserted in unique single-copy sequences within 500 bp of the rDNA, and crossover rates in wild-type (H3026; n=467) and pch2Δ (H327; n=186 cells. c, Schematic indicating marker locations in the rDNA used in d and g. URA3 markers were inserted in the NTS1/2 region of indicated repeats. d, Southern blot for restriction fragments containing the indicated rDNA repeat units from pch2Δ dmc1Δ strains #1:H5622, #3:H5636, #10:H5706 (PflMI/SalI; probe: unique rDNA insertion). YCR047C (HindIII) was a positive control for DSB formation. e, Southern blot of left rDNA flank including the outermost rDNA repeat in dmc1Δ (H5583) and pch2Δ dmc1Δ (H5622) cells (SalI/NruI; probe: YLR152C). f, ssDNA enrichment profile of chromosome XII in dmc1Δ (H118, grey), pch2Δ dmc1Δ cells (H2629, black) and sir2Δ dmc1Δ cells (H2953, slate grey). Arrowheads indicate >2-fold elevated DSB formation in pch2Δ dmc1Δ compared to dmc1Δ. g, Tetrad analysis of pch2Δ URA3 rDNA insertions strains (#1:H4611, #3:H4613, #10:H3823, #12:H4612, #29:H3820, #49:H3821; Table S2). Recombination rates between URA3 and rDNA-flanking markers are shown in relation to the physical URA3 positions within the rDNA. h, Relative contribution of each measured interval indicated in g to total rDNA recombination (percent of crossovers/kb of interval). i, Incidence of changes in rDNA repeat number between URA3 insertion and the rDNA boundary in pch2Δ tetrads that had undergone crossover recombination. j, CHEF analysis of two tetrads that have undergone unequal recombination (rec.) with parental controls (par.; XhoI; probe: NTS1).
To determine whether PCH2 suppresses DSB formation only within the rDNA, or in other regions of the genome, we first analysed a chromosomal fragment spanning the single-copy/rDNA junction in a pch2Δ mutant by Southern blot. We observed additional strong DSB formation in the adjacent single-copy sequences (Figure 1e, S1b), which were previously shown to experience exceptionally low levels of meiotic DSBs in PCH2 cells 6,7 (Figure 1f). The observed break sites behaved similarly to known meiotic DSBs 8; they were induced during meiosis in dmc1Δ and DMC1 cells (Figures 1d,e, S1c), depended on the meiotic DSB machinery (Figure S1d) 9, promoted meiotic recombination (Figure S1e), and occurred in gene promoters (Figures 1e, S1b). Indeed, even the DSBs observed in repeat 1 mapped to the promoter of a gene (TAR1) that is encoded in every rDNA repeat 10 (Figure 1e). Genome-wide analysis of DSBs 6 in pch2Δ cells revealed that strong DSB induction occurred in 30–50 kb regions of single-copy sequence abutting both sides of the rDNA (Figure 1f). Mild increases in DSB formation were observed close to other heterochromatic regions (telomeres and HML), whereas the DSB landscape elsewhere in the genome was not significantly altered (Figures S1f,g, S2, Table S1). In contrast to pch2Δ mutants, loss of SIR2 did not lead to increased DSB formation adjacent to the rDNA array (Figure 1f). Thus, Pch2 represses recombination within the rDNA at the level of DSB formation, but in a manner distinct from Sir2.
We asked whether the increased DSB formation in the outermost rDNA repeats in pch2Δ mutants (Figure 1d) resulted in a local increase in rDNA recombination. We measured recombination rates using flanking markers to the left and right of the rDNA together with a collection of single URA3 insertions tiling inwards from the left side of the rDNA (Figure 1c). Analysis of a URA3 insertion in the centre of the rDNA (inserted next to repeat 49 of 99) indicated that recombination occurred in a symmetric pattern. Strikingly, ~80% of the recombination events in the left half of the rDNA occurred within the first 10 repeats from the left border (Figure 1g, Table S2), with ~30% taking place within repeat 1. Thus, there is a strong bias for recombination within the rDNA repeats very close to the array border.
Since recombination within repetitive DNA can lead to NAHR, we selected tetrads of pch2Δ mutants that had undergone recombination within the rDNA and determined the resulting rDNA repeat number between the URA3 insertion and the left rDNA boundary. In 70% (n=47) of the investigated tetrads from different URA3 integrants, we observed changes in repeat number, ranging from 1 to 19 repeats (Figures 1i, j, Table S2), demonstrating that rDNA crossovers in pch2Δ cells are frequently associated with NAHR. However, the prevalence of allelic recombinants (30%), together with the fact that repeat number changes were less than 20% of the ~100–110 rDNA repeats in our strains, imply that the homology search for DSBs in the outermost rDNA repeats in pch2Δ cells is restricted to close neighbours. Importantly, the distribution of repeat-number changes closely matched the pattern of crossover events (Figure 1h, j). This congruence indicates that the spread in the crossover distribution (Figure 1g, h) can largely be accounted for by non-allelic exchanges between rDNA repeats originating from DSBs in the outermost repeats of the array. Notably, although rDNA exchanges occurred at much lower frequency in wild-type cells, they were also associated with NAHR (Table S2), suggesting that Pch2 primarily acts to prevent NAHR by suppressing DSB formation. These results establish the rDNA borders as high-risk regions for meiotic NAHR.
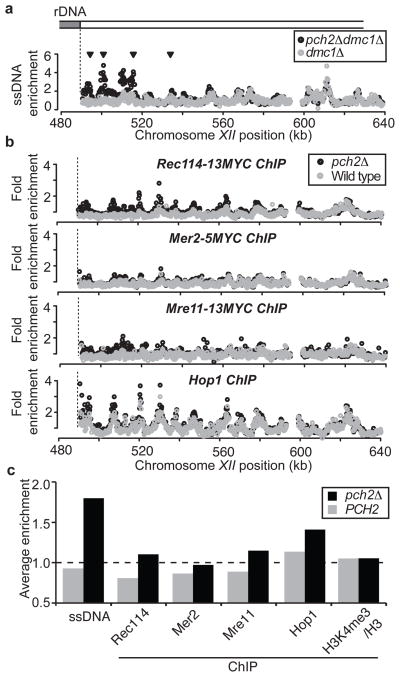
To determine how Pch2 suppresses DSB formation near the rDNA, we measured chromosome association of DSB-related factors at the time of DSB formation. Rec114, Mer2, and Mre11, the three essential DSB factors 11,12 we were able to analyse by chromatin immunoprecipitation (ChIP), were specifically enriched near the rDNA and HML in pch2Δ cells, mirroring the changes in DSB formation (Figures 2, S3). We wondered whether the regional exclusion of DSB factors could be explained by local depletion of the DSB-promoting chromosome axis protein Hop1, whose cytological distribution is affected by Pch2 4,13. Although Hop1 binding was slightly increased near the rDNA and HML in pch2Δ cells, it was abundant even in wild-type cells, suggesting Pch2 does not regulate the initial chromosomal recruitment of Hop1. Rather, the differences in Hop1 binding we observed might reflect an effect of Pch2 on chromosome structure 13. Finally, although DSBs are enriched in promoters containing histone H3 lysine 4 trimethylation 14, we observed no difference in the genome-wide levels of this modification with or without Pch2 (Figure 2c, S3d, S4), indicating Pch2 does not influence this chromatin modification. These findings suggest that Pch2 specifically blocks the stable recruitment of DSB factors to prevent local DSB formation.
Figure 2. Association of the meiotic DSB machinery near the rDNA.
a, ssDNA enrichment in region flanking the right border of the rDNA on chromosome XII (SGD coordinates) in dmc1Δ (H118, grey) and pch2Δ dmc1Δ (H2629, black) cells. Arrowheads indicate >2-fold elevated DSB formation in pch2Δ dmc1Δ compared to dmc1Δ. b, ChIP-chip analysis in same region as in a, for: Rec114-13Myc (first panel, wild type:H4890; pch2Δ:H4893), Mer2-5Myc (second panel, wild type:H5917; pch2Δ:H5916), Mre11-13Myc (third panel, wild type:H5547; pch2Δ:H5947) and Hop1 (fourth panel, wild type:H119; pch2Δ:H2817), in wild-type (grey) and pch2Δ cells (black). c, Average enrichment for the different PCH2 and pch2Δ ssDNA and ChIP-data sets (see a and b, Figure S4) within the 50 kb flanking the right rDNA border (see Methods for genomic coordinates). The genome-wide average is indicated by the dotted line.
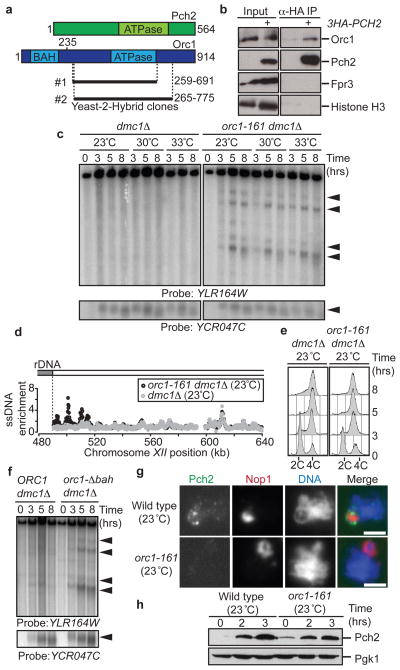
We sought to identify proteins that collaborate with Pch2 in preventing rDNA-proximal DSBs. A yeast-2-hybrid screen isolated a fragment of the Orc1 protein containing its ATPase domain as a Pch2 interactor (Figure 3a). This interaction was confirmed by co-immunoprecipitation (Figures 3b, S5a). Orc1 is a component of the conserved Origin Recognition Complex (ORC) that performs several important chromosomal functions, including the loading of the replicative helicase 15. Impairing Orc1 protein levels by a temperature-sensitive orc1-161 mutation 16 (Fig S5b) triggered DSB formation in the rDNA flanking regions similar to the loss of PCH2 (Fig 3c,d). DSB formation near the rDNA occurred even at a temperature (23 °C) that is permissive for pre-meiotic DNA replication and spore viability (Figures 3c–e, S5c,d). Similarly, we observed increased DSB levels near the rDNA in an orc1 mutant lacking the N-terminal bromo-adjacent homology (BAH) domain that is required for the chromatin-silencing function of Orc1 17, but dispensable for DNA replication (Figure 3f, S5e,f). These data indicate that the regulatory roles of Orc1 in DSB formation and bulk DNA replication are separable, although we cannot rule out that the analysed orc1 mutations locally affect rDNA replication. During meiosis, Pch2 concentrates within the nucleolus, the organelle assembled on the rDNA array 4. In orc1-161 cells the recruitment of Pch2 to the nucleolus was impaired, despite normal levels of cellular Pch2 (Figure 3g,h). Both Pch2 and Orc1 belong to the AAA+ family of ATPases that often function as multimeric complexes 18, and we found that the ATPase activity of Pch2 was required to prevent rDNA proximal DSBs (Figure S5g). These data define a role for Orc1 in the nucleolar recruitment and possible activation of Pch2 to prevent local DSB formation.
Figure 3. Orc1 and Pch2 collaborate to suppress DSB formation.
a, Schematic of Pch2 and Orc1 proteins indicating Orc1 clones identified by Yeast-2-Hybrid screen. b, Co-immunoprecipitation (IP) between 3HA-Pch2 and Orc1 in wild-type (H119) and 3HA-PCH2 (H3463) cells. Fpr3 and Histone H3 are controls for nucleolar and chromosomal fractions, respectively. c, Southern blot of right rDNA flank and YCR047C in meiotic dmc1Δ (H118) and orc1-161 dmc1Δ (H4952) cells grown at indicated temperatures d, ssDNA profiles of region flanking the right rDNA border in orc1-161 dmc1Δ (H5137, black) and dmc1Δ (H118, grey) cells grown at 23°C. e, DNA content analysis of meiotic dmc1Δ (H118) and orc1-161dmc1Δ (H5137) cells grown at 23°C. f, Southern blot of right rDNA flank and YCR047C in ORC1 dmc1Δ (H5838) and orc1-Δbah dmc1Δ (H5865) cells. g, Immunofluorescence of chromosome spreads stained for Pch2 (HA, green), Nop1 (nucleolar marker, red) and DNA (blue) in 3HA-PCH2 (H3463) and 3HA-PCH2 orc1-161 (H5033) cells at 3 hours after meiotic induction (at 23°C). Scale bar; 2 μm. h, Western blot analysis showing Pch2 (HA) expression in 3HA-PCH2 (H3463) and 3HA-PCH2 orc1-161 (H5033).
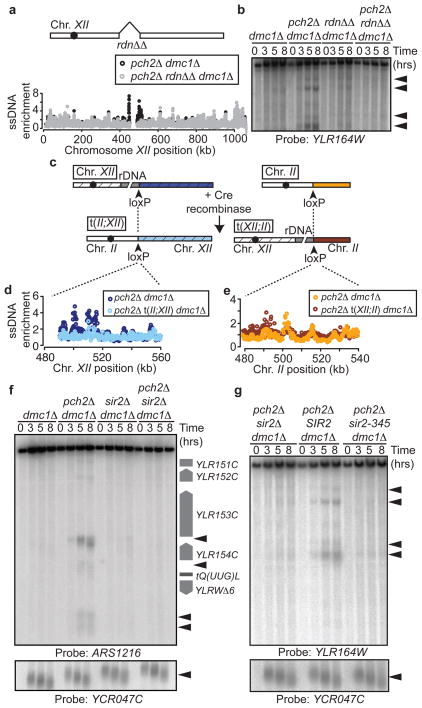
We wondered whether the specific DSB activity at the rDNA borders in pch2Δ mutants was linked to the presence of the rDNA itself. To test this possibility, we deleted the rDNA array from its genomic location. In these strains, loss of PCH2 no longer allowed DSB formation in the flanking regions (Figures 4a,b), demonstrating an intrinsic DSB-promoting activity in the rDNA. To ask whether the rDNA was sufficient to promote DSB formation, we created a translocation between chromosome II and XII that exchanged the rDNA-distal portion of Chromosome XII with a portion of Chromosome II (Figure 4c). In these strains, DSB levels were no longer increased in the former right flank of the rDNA (now flanked by Chromosome II sequences; Figure 4d), but importantly, increased DSB formation was observed on the chromosome II sequences that, after translocation, were flanking the rDNA (Figure 4e). Thus, in the absence of PCH2, the rDNA is necessary and sufficient to promote DSB formation.
Figure 4. rDNA chromatin promotes DSB formation.
a, Schematic of rdnΔΔ strain and ssDNA profiles of region flanking the right rDNA border in pch2Δ dmc1Δ (H2629, black) and pch2Δ rdnΔΔ dmc1Δ (H4737, grey) cells. b, Southern blot analysis of the right rDNA flank in dmc1Δ (H118), pch2Δ dmc1Δ (H2629), rdnΔΔ dmc1Δ (H4736), and pch2Δ rdnΔΔ dmc1Δ (H4737) cells. c, Strategy used to generate chromosomal translocations between chromosomes XII and II. d and e, ssDNA profiles of strains containing the XII;II translocation. In d, the depicted region is next to the rDNA in pch2Δ dmc1Δ (H2629) cells (dark blue) and next to the left arm of chromosome II in pch2Δ t(II;XII) dmc1Δ (H4798) cells (light blue). In e, the depicted region is located on chromosome II in pch2Δ dmc1Δ cells (orange) and next to rDNA in pch2Δ t(XII;II) dmc1Δ cells (dark red). f, Southern blot of left rDNA flank (HindIII; probe: ARS1216) and YCR047C in dmc1Δ (H118), pch2Δ dmc1Δ (H2629), sir2Δ dmc1Δ (H2953) and pch2Δ sir2Δ dmc1Δ (H3038) cells. g, Southern blot of the right rDNA flank and YCR047C in pch2Δ sir2Δ dmc1Δ (H3262), pch2Δ sir2Δ dmc1Δ leu2::SIR2 (H3261), and pch2Δ sir2Δ dmc1Δ leu2::sir2-345 (H3282) cells.
The rDNA is assembled into specialised, Sir2-dependent chromatin, and we asked how this chromatin state influenced DSB formation at the rDNA boundaries. Strikingly, loss of Sir2 protein or deacetylase activity 19 largely eliminated DSB formation in the rDNA flanking regions in pch2Δ mutants (Fig 4f,g). Thus, although Sir2-dependent heterochromatin suppresses meiotic DSBs within the rDNA array (2,3, Figure 1a), it has a profound DSB-promoting effect on the rDNA borders that is counteracted by Pch2 and Orc1. Intriguingly, Sir2 itself localises Pch2 to the nucleolus 4, reflecting an elegant coupling mechanism that maintains meiotic stability across the entire rDNA. The double dependence of Pch2 on Sir2 and Orc1 may promote Pch2 enrichment at the nucleolus/rDNA analogous to the bimodal recruitment mechanisms that restrict localisation of Aurora B and Shugoshin to centromeres 20.
Although repeat-associated chromatin marks differ substantially between organisms and even among individual loci 21, the assembly of heterochromatin on repetitive DNA arrays is a common strategy to protect the genome against destabilisation caused by errors in meiotic recombination 1. Our results establish heterochromatin/euchromatin borders as potential high-risk regions for meiotic DSB formation and NAHR, and reveal the existence of a secondary border-specific system that shields against these events. Buffer zones like those established by Pch2 and Orc1 may need to be broad, since even DSBs adjacent to repetitive DNA can trigger NAHR 22. Given the prominent presence of repetitive DNA arrays in genomes ranging from yeast to man 1, we propose that mechanisms that limit DSB activity around repetitive DNA might be a wide-spread phenomenon.
Methods Summary
Yeast strains were of the SK1 background and are listed in Table S3. Analysis of ssDNA profiles and ChIP-chip analysis were performed as described 6,23. These and additional standard techniques used are detailed in the Methods.
Methods
Yeast strains and 2-hybrid analysis
All yeast strains used in this study were constructed in the SK1 background and are listed in Table S3. Epitope tags and gene disruptions were introduced by standard PCR-based transformation. The orc1-Δbah mutant was generated using a plasmid containing a truncated version of Orc1 (amino acids 235-914; pSPB1.48, kind gift of Stephen P. Bell 24). To create URA3 insertions in the rDNA, cells were transformed with an SphI-linearised pRS306-NTS1/2 plasmid (this plasmid contains a 2341bp fragment harbouring the intergenic rDNA sequences, NTS1 and NTS2, ligated into the BamHI and EcoRI sites of pRS306). Insertion sites in the rDNA were mapped by CHEF gel analysis using a unique XhoI site in the inserted sequence (XhoI does not cut in the rDNA), and suitable clones were selected for further analysis. SK1 strains lacking the rDNA array were generated as described in 25. Briefly, cells were transformed with a very high-copy rDNA plasmid (pRDN-hyg::URA3::leu2-8) carrying a recessive point mutation that confers resistance to hygromycin 26. After selection on hygromycin, a clone was selected that had lost all but 3 repeats of the rDNA array through spontaneous deletion as determined by CHEF gel analysis. The remaining rDNA copies were subsequently deleted by conventional gene disruption using a HIS3 deletion cassette. Complete deletion of the rDNA array was confirmed by Southern blotting. Chromosomal translocations between chromosomes XII and II were generated essentially as described27. Briefly, plasmids containing a promoter-less ADE2 adjacent to a loxP site (loxP-ADE2::natMX4) and a GPD promoter (pGPD) with an adjacent loxP site (pGPD-loxP::hphMX4, both plasmids were kind gifts of Neil Hunter) were integrated at YLR162W-A and LYS2, respectively. After induction of Cre recombinase from a pGAL-Cre plasmid (Neil Hunter), cells were selected that had undergone translocation between LYS2 (chromosome II) and YLR162W-A (chromosome XII). Translocation was confirmed by Southern blot analysis. To identify interactors of Pch2 by 2-hybrid screen, full-length PCH2 was amplified from genomic DNA and the intron removed by site-directed mutagenesis. The Pch2 coding sequence was then cloned into pGBDU-C1, and the resulting bait plasmid was used to screen libraries in all 3 reading frames 28.
Synchronous meiosis
For synchronous meiosis, cells were grown for 24 hours in YPD at room temperature, then diluted in BYTA medium (1% yeast extract, 2% tryptone, 1% potassium acetate, 50 mM potassium phthalate) to OD600=0.3 (OD600=0.5 for orc1-161, rdnΔΔ and XII;II translocation strains), and grown for 16 hours at 30°C (or for 18 hours at room temperature in the case of temperature-sensitive strains). After 2 washes in water, cells were diluted into SPO medium (0.3% potassium acetate) at OD600=1.9 and incubated at 30°C unless otherwise stated.
ssDNA isolation
For ssDNA analysis 29,30, ~109 cells were fixed in 70% ethanol at −20°C at 0 and 5 hours after induction of meiosis. After spheroplasting in sorbitol buffer (1 M sorbitol, 1% betamercaptoethanol, 0.2 mg/ml zymolyase, and 0.1 M EDTA, pH 7.4), cells were lysed in NDS buffer (0.6% SDS, 300 mM EDTA, 10 mM Tris-HCl, pH 9.5). After treatment with proteinase K (0.25 mg/ml) and RNase A, the DNA was digested with EcoRI. ssDNA was enriched by adsorption to BND-cellulose, and eluted using 1.8% caffeine. This enriched ssDNA was subsequently used for microarray analysis. For this, 1.5 μg of the respective 0 and 5 hour ssDNA samples were labelled with Cy3-dUTP or Cy5-dUTP (GE Healthcare) by random priming without denaturation with 4 μg random primer (Integrated DNA Technologies) and 10 units of Klenow (New England Biolabs).
Western blotting and immunoprecipitation
For Western blotting, 5 ml of meiotic cells were harvested at the indicated time points and resuspended in 5% trichloroacetic acid (TCA). After incubation on ice for 10 minutes, samples were washed in acetone and dried overnight. Samples were lysed by bead beating in a FastPrep FP120 (Thermo Scientific) in TE lysis buffer (10 mM Tris pH 7.5, 1 mM EDTA, 2.75 mM dithiothreitol (DTT)). 3X SDS loading buffer was added and the pH of the sample was adjusted to neutral by addition of 1M Tris pH 8.0. Samples were separated by standard polyacrylamide gel electrophoresis. The following antibodies were used: α ORC (1108, 1:1000, gift of Stephen P. Bell), α Fpr3 (1:1000, gift of J. Thorner), α HA (3F10, 1:1000, Roche), α Pgk1 (1:1000, Invitrogen), and α Histone H3 (1:1000, Abcam). For immunoprecipitations, 50 ml of meiotic cells were harvested 3 hours after induction of meiosis. Cells were diluted in 2X lysis-buffer (20 mM HEPES, pH 7.5, 4 mM MgCl2, 0.6 M glutamic acid, 0.32 M sorbitol, 4 % glycerol, 0.5 % Triton X-100) containing protease inhibitors, and lysed by bead beating. Extracts were sonicated and cleared by centrifugation. After removal of one tenth of the extract for an input sample, extracts were immunoprecipitated with 2 μl α HA (3F10, Roche) for 3HA-Pch2, 2 μl α ORC (1108) and 2 μl of α Fpr3, in combination with 20μl of a 50% slurry of GammaBind-Sepharose beads (GE Healthcare) for 16 hours at 4°C. After 5 washes in 1X lysis buffer, 1X SDS-loading buffer was added to the beads, and samples were analysed by Western blotting with the indicated antibodies.
Chromatin immunoprecipitation
25 ml of meiotic cells were harvested (3 hours after induction of meiosis), and fixed for 15 minutes in 1% formaldehyde. The formaldehyde was quenched by addition of 125 mM glycine. Samples were processed as described 31. Before immunoprecipitation, one tenth of the sample was removed as input sample. The different antibodies used for immunoprecipitation were: 2 μl α Myc (9E11, Abcam; for Rec114-13Myc, Mer2-5Myc and Mre11-13Myc), 2 μl α Hop1 (gift of N. Hollingsworth), 2 μl anti-histone H3 (AB1791, Abcam) and 2 μl anti-trimethyl histone H3 lysine 4 (AB8580, Abcam), in combination with 20 μl of a 50% slurry of GammaBind-Sepharose (GE Healthcare) beads. Half of the ChIP sample and 1/10 of the input were labelled with Cy3-dUTP or Cy5-dUTP (GE Healthcare) as described in the ssDNA protocol, with the difference that the DNA was denatured for 5 minutes at 95°C prior to the extension reaction.
Microarray analysis
After removal of unincorporated dyes, Cy3 and Cy5-labeled samples were hybridised to custom 4X44K tiled genomic yeast microarrays (Agilent Technologies) for 16 hours at 65°C. Levels of Cy3 and Cy5 were calculated with the Agilent Feature Extractor CGH software. Background normalisation, log2 ratios for each experiment, and scale normalisations between experiments were calculated with the sma package in R (v2.1.0, http://www.r-project.org)29,30. Each data set is an average of two experiments. For comparison between isogenic wild-type and pch2Δ cells, data sets were scale normalised. To analyse the distribution of trimethyl histone H3 lysine 4, we normalised the enrichment to that of total histone H3 generated from the same extracts by subtracting the log2 ratios. To measure the average ssDNA or ChIP enrichment in different chromosomal regions, the following SGD coordinates were analysed, based on the position of available array features:
50 kb right of the rDNA: XII: 490,531-540,530
50 kb left of the rDNA: XII: 401,371-451,370.
Rest of Chromosome XII: XII, 1-401,370 and 540,531-1,078,177
First 100 kb of Chromosome III: III: 1-100,000
Rest of Chromosome III: III, 100,001-316,620
Chromosome VIII: VIII, 1-562,643
Chromosome spreads and immunofluorescence
Meiotic cells were spread as described 32. Cells were spheroplasted at 37°C in solution 1 (2% potassium acetate, 0.8 % sorbitol, 10 mM DTT, 130 mg/ml zymolyase 100T (Seikagaku)). Solution 2 (100 mM MES pH 6.4, 1 mM EDTA, 0.5 mM MgCl2, 1M sorbitol) was added to stop spheroplasting. Spheroplasted cells (15 μl) were fixed with 30μl of fixative solution (4% paraformaldehyde, 3.4 % sucrose) and lysed with 60 μl 1% lipsol. After addition of 60 μl fixative solution, cells were spread using a glass rod. After drying, the slides were blocked in blocking buffer (0.2 % gelatine, 0.5 % BSA in PBS) and stained with the following antibodies: α-HA (3F10, 1:500 dilution, Roche), and α Nop1 (1:500 dilution, Encor Biotechnology).
Clamped homogenous electric field gel electrophoresis (CHEF) and Southern blotting
Chromosome fragments for CHEF analysis were prepared by restriction digest in agarose plugs. Briefly, 20 ml of meiotic culture were killed by addition of sodium azide (0.1% w/v final), pelleted and stored on ice for the time of the time course. Cell pellets were washed twice in CHEF-TE (10 mM Tris-HCl, pH 7.5; 50 mM EDTA) and resuspended in 300μl CHEF-TE. Tubes were individually treated as follows: 4μl zymolyase T100 (10mg/ml) was added and the mix incubated at 42°C for 30s before addition of 500μl low-melting point agarose (1.5% SeaPlaque GTG, 125 mM EDTA) equilibrated at 42°C. 90 μl plugs were allowed to harden on ice in disposable plug molds (Bio-Rad) and incubated overnight at 37°C in 300 μl LET (10 mM Tris, pH 7.5; 500 mM EDTA) per plug. Plugs were deproteinised overnight at 50°C in 200μl NDS-PK (LET, 1% N-Lauroylsarcosine, 1mg/ml proteinase K (Amresco)) per plug. Proteinase K was inactivated by incubating plugs for 1 hour at 4°C in CHEF-TE containing 1mM PMSF, and plugs were washed another 3 times in CHEF-TE. Plugs were digested with XhoI and addition of 5 mM spermidine. To analyse the entire rDNA array, digested chromosomes were separated by CHEF in 1% agarose/0.5× TBE, 6V/cm, using 60s pulses for 15 hours and 90s pulses for 9 hours. For fine mapping of rDNA insertions and to analyse repeat number changes, digested chromosome fragments were separated using a 5–20s ramp over 20 hours. For conventional electrophoresis, DNA fragments were separated on 0.6% agarose/1X TBE and transferred onto Hybond-XL membranes (GE Healthcare) by alkaline transfer. Southern blotting was performed as described 33 and quantified with a Fujifilm BAS-2500 image reader V1.8 and Multi Gauge V2.2 software.
Probes for Southern analysis
Probe templates for non-rDNA sequences were generated by nested PCR and gel purification. The following probes (SGD coordinates) were used:
YLR164W: XII: 493,432-493,932.
YCR047C: III: 209,361-210,030.
ARS1216: XII: 450,407-451,150.
YLR152C: XII: 443,849-444,910.
NTS1: XhoI/XbaI-digest of pRS306-NTS1/2. This probe detects all NTS1/2 sequences (i.e. a pan-rDNA probe).
rDNA insertion: BciVI digest of pRS306-NTS1/2. This probe specifically detects the plasmid backbone of the URA3 insertion cassette.
TOM1/YDR457W: IV: 1,370,714-1,371,733
Flow cytometry
At the indicated time points, 150 μl of meiotic cells were fixed for 2 hours at 4°C after addition of 350 μl absolute ethanol. Cells were resuspended in 500 μl 50 mM sodium citrate containing 0.7 μl RNAse A (30 mg/ml, Sigma). Cells were incubated for 2 hours at 50°C. 10 μl proteinase K (Amresco) was added and cells were deproteinated for 2 hours at 50°C. 500 μl 50 mM sodium citrate containing 0.2 μl Sytox Green (Amersham) was added to the cells. Cells were briefly sonicated and analysed using a FACScalibur (Becton Dickinson) flow cytometer. DNA profiles were generated using CellQuest Software.
Recombination mapping
To determine crossover recombination rates, cells were sporulated for 24 hours in 3ml SPO and treated with zymolyase (1 mg/ml in 1M sorbitol) to remove ascus walls. Tetrads were dissected by micromanipulation and marker segregation was determined by replica plating on appropriate selective media. For mapping within the rDNA, only tetratypes were used to calculate recombination rates to avoid distortions originating from non-parental ditypes that were likely the result of prior mitotic recombination.
Supplementary Material
Acknowledgments
We thank Stephen P. Bell, Akira Shinohara, Neil Hunter, Nancy Hollingsworth, and Franz Klein for sharing (unpublished) reagents and data. We thank Iain Cheeseman, Mary Gehring and Viji Subramanian for helpful discussions and critically reading the manuscript. This work was supported by NIH grant GM088248 to A.H., and fellowships from the Netherlands Organisation for Scientific Research (NWO Rubicon-825.08.009 and NWO VENI-016.111.004) to G.V.; L.C. was supported by an HHMI Institutional Undergraduate Education Grant to MIT (Grant no. 52005879).
Footnotes
Author Contributions
G.V, H.G.B and A.H. designed and performed experiments and analysed the data. M.A.T. performed the yeast-2-hybrid analysis. J.E.F, L.C. and A.H. performed recombination mapping. G.V, H.G.B. and A.H wrote the paper.
Author Information
All data sets in this publication are available in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE30073. Reprints and permissions information are available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Sasaki M, Lange J, Keeney S. Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Biol. 11(3):182. doi: 10.1038/nrm2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56 (5):771. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 3.Mieczkowski PA, et al. Loss of a histone deacetylase dramatically alters the genomic distribution of Spo11p-catalyzed DNA breaks in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104 (10):3955. doi: 10.1073/pnas.0700412104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.San-Segundo PA, Roeder GS. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 1999;97 (3):313. doi: 10.1016/s0092-8674(00)80741-2. [DOI] [PubMed] [Google Scholar]
- 5.Wu HY, Burgess SM. Two distinct surveillance mechanisms monitor meiotic chromosome metabolism in budding yeast. Curr Biol. 2006;16 (24):2473. doi: 10.1016/j.cub.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blitzblau HG, et al. Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr Biol. 2007;17 (23):2003. doi: 10.1016/j.cub.2007.10.066. [DOI] [PubMed] [Google Scholar]
- 7.Gerton JL, et al. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97 (21):11383. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petes TD. Meiotic recombination hot spots and cold spots. Nat Rev Genet. 2001;2 (5):360. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- 9.Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 10.Coelho PS, et al. A novel mitochondrial protein, Tar1p, is encoded on the antisense strand of the nuclear 25S rDNA. Genes Dev. 2002;16 (21):2755. doi: 10.1101/gad.1035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arora C, Kee K, Maleki S, Keeney S. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol Cell. 2004;13 (4):549. doi: 10.1016/s1097-2765(04)00063-2. [DOI] [PubMed] [Google Scholar]
- 12.Keeney S, Neale MJ. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem Soc Trans. 2006;34 (Pt 4):523. doi: 10.1042/BST0340523. [DOI] [PubMed] [Google Scholar]
- 13.Borner GV, Barot A, Kleckner N. Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proc Natl Acad Sci U S A. 2008;105 (9):3327. doi: 10.1073/pnas.0711864105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borde V, et al. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28 (2):99. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell SP. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16 (6):659. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- 16.Gibson DG, Bell SP, Aparicio OM. Cell cycle execution point analysis of ORC function and characterization of the checkpoint response to ORC inactivation in Saccharomyces cerevisiae. Genes Cells. 2006;11 (6):557. doi: 10.1111/j.1365-2443.2006.00967.x. [DOI] [PubMed] [Google Scholar]
- 17.Bell SP, et al. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995;83 (4):563. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- 18.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6 (7):519. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 19.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403 (6771):795. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 20.Vader G, Lens SM. Chromosome segregation: taking the passenger seat. Curr Biol. 20(20):R879. doi: 10.1016/j.cub.2010.09.037. [DOI] [PubMed] [Google Scholar]
- 21.Moazed D. Common themes in mechanisms of gene silencing. Mol Cell. 2001;8 (3):489. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- 22.Hoang ML, et al. Competitive repair by naturally dispersed repetitive DNA during non-allelic homologous recombination. PLoS Genet. 6(12):e1001228. doi: 10.1371/journal.pgen.1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91 (1):59. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 24.Bell SP, et al. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995;83 (4):563. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi T. Strategies to maintain the stability of the ribosomal RNA gene repeats--collaboration of recombination, cohesion, and condensation. Genes Genet Syst. 2006;81 (3):155. doi: 10.1266/ggs.81.155. [DOI] [PubMed] [Google Scholar]
- 26.Chernoff YO, Vincent A, Liebman SW. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 1994;13 (4):906. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peoples TL, et al. Close, stable homolog juxtaposition during meiosis in budding yeast is dependent on meiotic recombination, occurs independently of synapsis, and is distinct from DSB-independent pairing contacts. Genes Dev. 2002;16 (13):1682. doi: 10.1101/gad.983802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144 (4):1425. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blitzblau HG, et al. Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr Biol. 2007;17 (23):2003. doi: 10.1016/j.cub.2007.10.066. [DOI] [PubMed] [Google Scholar]
- 30.Blitzblau HG, Hochwagen A. Genome-wide detection of meiotic DNA double-strand break hotspots using single-stranded DNA. Methods Mol Biol. 745:47. doi: 10.1007/978-1-61779-129-1_4. [DOI] [PubMed] [Google Scholar]
- 31.Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91 (1):59. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 32.Loidl J, Nairz K, Klein F. Meiotic chromosome synapsis in a haploid yeast. Chromosoma. 1991;100 (4):221. doi: 10.1007/BF00344155. [DOI] [PubMed] [Google Scholar]
- 33.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106 (1):59. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.