Abstract
Two symmetrical cyanine dyes based on benzothiazole heterocycles and a trimethine bridge were found to bind to a parallel-stranded DNA guanine quadruplex based on the MYC oncogene promoter sequence with high nanomolar affinity and 1:1 stoichiometry. The dyes exhibited substantial fluorescence enhancements upon binding. In the presence of homologous guanine-rich peptide nucleic acid oligomers, PNA-DNA heteroquadruplexes were formed. The dyes retained their ability to bind to the heteroquadruplexes at low micromolar concentrations and with varying fluorescence enhancements, although indeterminate stoichiometries preclude quantitative comparison of the affinities with the DNA homoquadruplex precursor. The difference in fluorescence enhancement between DNA homoquadruplex and PNA-DNA heteroquadruplex allows the dyes to be used as fluorogenic indicators of hybridization in a facile method for determining PNA-DNA stoichiometry.
Key words: PNA-DNA heteroquadruplex, cyanine dyes, hybridization, small molecule-quadruplex recognition, fluorescence enhancement
Introduction
1The growing recognition of the important roles of guanine quadruplex structures in regulating gene expression and telomere biology have motivated considerable efforts to design or discover synthetic molecules that can bind to quadruplexes with high affinity and selectivity.1–3 Quadruplex-binding ligands might find applications in chemical biology, where they could report the presence or perturb the biological function of a quadruplex.4,5 Clinical applications of drug-like small molecules targeted to G-quadruplexes are also envisioned.6
Small molecules bind to G-quadruplexes primarily through shape-based recognition. An alternative strategy relies on sequence-based recognition, where complementary C-rich oligonucleotides bind via Watson-Crick base pairing to form heteroduplexes7–10 or homologous G-rich oligonucleotides bind via Hoogsteen-based G-tetrad formation to yield heteroquadruplexes.11–15 Peptide nucleic acid (PNA) probes can bind to G-quadruplexes by either of these mechanisms, leading to stable hybrid structures.
Our recent focus has been on PNA-DNA/RNA heteroquadruplex formation, based on the high affinity (low nanomolar Kds) of the PNA for its homologous DNA11,13,14 and RNA12,16 targets, as well as excellent sequence discrimination for homologous versus complementary targets that can be achieved through PNA backbone modification.14 Short PNAs consisting of two G2 or G3 tracts and various nucleobase or abasic loops successfully invade G-quadruplex targets under physiological conditions to form stable heteroquadruplex structures. The heteroquadruplex stoichiometries are typically 2 PNA: 1 DNA or RNA. Hence, the short PNAs disrupt the folded homoquadruplex then hybridize such that each of two PNA strands recognizes two of the four G-tracts that make up the quadruplex target.
The current report describes our first steps toward combining sequence- and shape-based recognition of quadruplexes through the simultaneous application of PNAs and small molecules. In our prior work, we used a covalently conjugated fluorogenic cyanine dye to provide a fluorescence response to PNA binding, as in the “light-up” probes originally reported by Ishiguro and coworkers for DNA17 and subsequently by Svanvik and coworkers for PNA.18,19 Recently, Ladame and coworkers15 combined a PNA G3 tract that could form one edge of a heteroquadruplex with an acridone stacking ligand previously shown to end-stack on G-tetrads.20
Although the examples cited above clearly illustrate that a covalently conjugated dye can interact with a PNA-DNA heteroquadruplex, it was unclear to what extent the dye would interact with the heteroquadruplex if not for the covalent linkage to the PNA terminus. While still composed of G-tetrads found in DNA homoquadruplexes, PNA-DNA heteroquadruplexes offer unique structural characteristics that could be useful for small molecule recognition. For example, the negative charge density in a heteroquadruplex is less than half that of a homoquadruplex, due to the uncharged nature of the backbone and the presence of cationic termini on the PNA. Thus, electrostatic interactions offered by the two types of quadruplex will be quite different. Similarly, the van der Waals surfaces of the quadruplex grooves should vary considerably between homo- and heteroquadruplexes.
Cyanine dyes have found numerous applications in nucleic acid recognition,21,22 with various reports on intercalation, groove binding and end-stacking on duplexes,23 triplexes24,25 and quadruplexes.25–30 The wide range of absorption/emission wavelengths accessible to the cyanines, the ease with which the net charge on the molecule can be varied through substituents, and the versatility of the dyes, in terms of either having environmentally sensitive or insensitive quantum yields, have been exploited in these diverse studies. In this report, we describe two carbocyanine dyes that bind noncovalently to PNA-DNA heteroquadruplexes. These dyes can exhibit variable fluorescence enhancements upon binding, depending on the loop composition of the PNA strand. These results open the door to using cyanine dyes in combination with G-rich PNAs to form and detect heteroquadruplex structures.
Results and Discussion
Cyanine Dye-DNA quadruplex interactions.
The carbocyanine dyes DiSC1(3) and β-Me-DiSC1(3) (Scheme 1; these dyes are also known as Cyan 46 and Cyan 2, respectively31) have been shown to exhibit substantial increases in fluorescence in the presence of quadruplex DNA.25,30 The quadruplexes studied in the prior reports were modeled on telomeric DNA sequences and other G-rich motifs. For our experiments, we focused on a DNA quadruplex formed by a 19-base sequence found in the promoter region of the MYC oncogene32 (Myc19, Scheme 1). This sequence is known to fold into a stable intramolecular G quadruplex with a parallel arrangement of the four strands.33
Scheme 1.
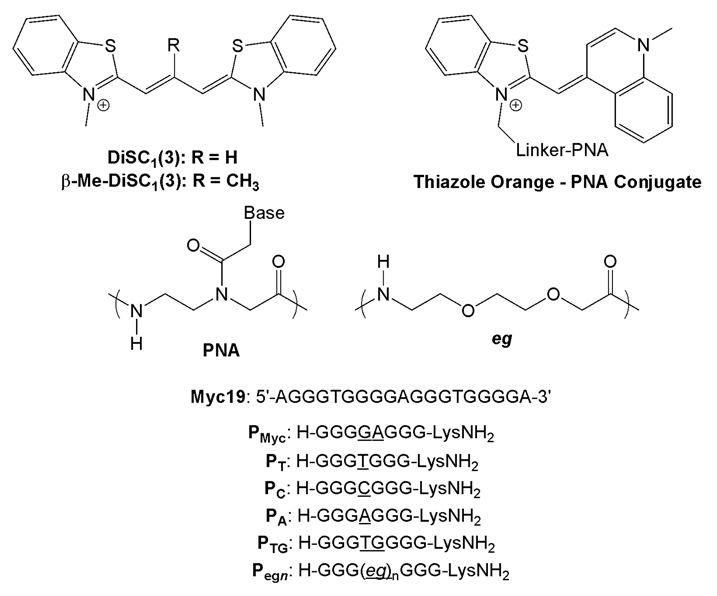
Cyanine dye structures and DNA target and PNA probe sequences. Differences among PNA probes are underlined; “eg” = 8-amino-3,6-dioxa-octanoic acid.
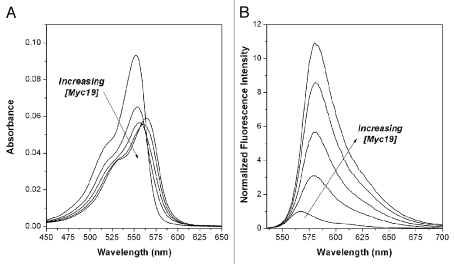
The interaction of DiSC1(3) with Myc19 is illustrated in Figure 1, where UV-vis (A) and fluorescence (B) spectra of the dye undergo significant perturbations in the presence of the quadruplex. As the DNA is titrated into the dye, the absorption spectrum exhibits batho- and hypochromism, while the fluorescence spectrum also exhibits a bathochromic shift and enhanced fluorescence. These observations are consistent with a binding interaction between the dye and the DNA quadruplex.
Figure 1.
Effect of Myc19 quadruplex DNA on UV-vis (A) and fluorescence (B) spectra of DiSC1(3). Samples contained 1.0 µM DiSC1(3) and 0, 0.25, 0.5, 1.0 and 2.0 µM DNA. Fluorescence spectra were excited at λ = 520 nm.
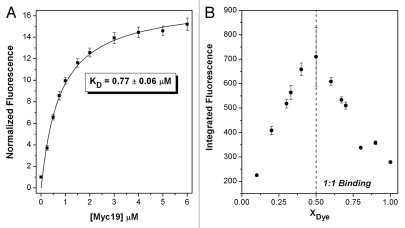
Data from a full fluorescence titration experiment were fit adequately by a simple 1:1 binding model, yielding KD = 0.77 ± 0.06 µM (Fig. 2A). The stoichiometry was further supported by a continuous variations experiment (Fig. 2B). Thus, the carbocyanine dye DiSC1(3) binds with high nanomolar affinity to the Myc19 quadruplex to form a stable 1:1 complex. Circular dichroism experiments indicated that the parallel morphology of the quadruplex (positive peak at 260 nm) is maintained in the presence of the dye (data not shown).
Figure 2.
Fluorescence titration (A) and continuous variations (B) experiments to determine KD and stoichiometry of DiSC1(3) binding to Myc19 quadruplex. For (A), DNA was titrated into a solution of 1.0 µM dye. For (B), a total concentration of dye + DNA = 1.0 µM was used.
The binding site for the cyanine dye on the DNA quadruplex is unknown. The parallel structure of the Myc19 quadruplex features three double-chain reversal loops, in which single- or double-nucleotide loops fold back across the groove to allow adjacent G-tracts to align in parallel orientations. This leaves one groove unblocked and potentially accessible to the cyanine dye. However, we cannot rule out the possibility that the dye stacks preferentially on one end of the quadruplex.
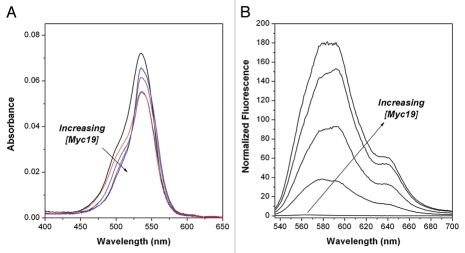
Similar experiments were undertaken with the bridge-substituted cyanine dye β-Me-DiSC1(3). Absorption and fluorescence spectra recorded as a function of Myc19 concentration are shown in Figure 3. The shape of the absorption spectrum changes with DNA concentration, although the peak does not shift (Fig. 3A). Meanwhile, the fluorescence of the dye in solution is much lower than the non-bridge substituted analogue DiSC1(3). Bridge-substitution of cyanine dyes is known to suppress fluorescence, possibly due to inducing a conformational change to a structure34 with a lower quantum yield. The fluorescence of β-Me-DiSC1(3) is strongly enhanced in the presence of the Myc19 quadruplex (Fig. 3B) similar to prior work by Yarmoluk and coworkers with duplex and quadruplex DNAs.25,31,35 The spectra also exhibit minor variation in shape with DNA concentration, perhaps reflecting the presence of multiple binding modes. The shoulder observed at 640 nm in these spectra is also unexpected and is not observed if the KCl concentration is reduced from 100 mM to 1 mM (Sup. Fig. 1). This suggests that the shoulder is due to aggregation of the dye, since dimerization/aggregation of cyanines is known to be favored at elevated ionic strength.36
Figure 3.
Effect of Myc19 quadruplex DNA on UV-vis (A) and fluorescence (B) spectra of β-Me-DiSC1(3). Samples contained 1.0 µM β-Me-DiSC1(3) and 0, 0.25, 0.5, 1.0 and 2.0 µM DNA. Fluorescence spectra were excited at λ = 520 nm.
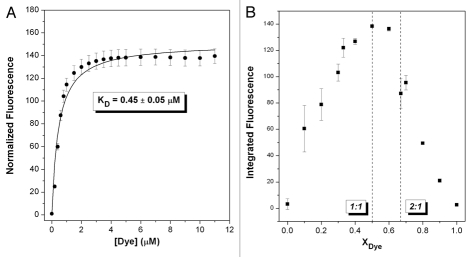
A continuous variations experiment indicated that a 1:1 dye:DNA complex is formed (Fig. 4B), although the turnover point is not as well resolved as for DiSC1(3). Fitting the titration data to a 1:1 model as shown in Figure 4A indicates that this dye binds slightly stronger to the quadruplex than does the unsubstituted analogue DiSC1(3). Although both DiSC1(3) and β-Me-DiSC1(3) have similar quantum yields when fully bound to the quadruplex, the lower quantum yield for unbound β-Me-DiSC1(3) leads to a much stronger enhancement.
Figure 4.
Fluorescence titration (A) and continuous variations (B) experiments to determine KD and stoichiometry of β-Me-DiSC1(3) binding to Myc19 quadruplex. For (A), DNA was titrated into a solution of 1.0 µM dye. For (B), a total concentration of dye + DNA = 1.0 µM was used.
Cyanine Dye-PNA/DNA heteroquadruplex interactions.
We previously demonstrated the ability of G-rich peptide nucleic acid (PNA) probes to form heteroquadruplexes with homologous DNA targets such as Myc19. For example, the PNA probe PMyc forms a 2:1 heteroquadruplex with Myc19 (KD = 5 nM).13 Similar to prior reports in which fluorogenic cyanine dyes were used as reporters for heteroduplex formation, the fluorogenic dye thiazole orange was a useful reporter molecule for PNA-DNA heteroquadruplex formation. (This dye has been used as the basis for a fluorescent displacement assay to screen quadruplex-binding ligands).27 When conjugated to the PNA N-terminus (Scheme 1), the dye exhibits a substantial fluorescence enhancement upon heteroquadruplex formation, allowing determination of the stoichiometry and affinity. Thus, we were interested to determine whether the fluorogenic trimethine dyes DiSC1(3) and β-Me-DiSC1(3) would bind noncovalently to PNA-DNA heteroquadruplexes.
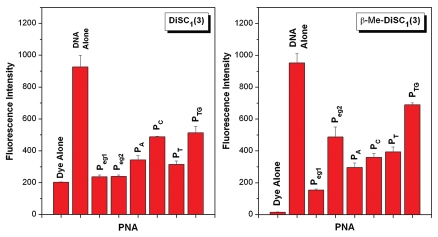
UV-vis and fluorescence titrations of the dyes with various PNA-DNA heteroquadruplexes did not produce well-defined stoichiometries, precluding determination of binding affinities and molecularities (data not shown). Nevertheless, UV-vis spectral perturbations and fluorescence enhancements were evident in many cases. Figure 5 shows the fluorescence intensities of the dyes alone, in the presence of the Myc19 DNA homoquadruplex and with a variety of PNA-DNA heteroquadruplexes at constant dye, DNA and PNA concentrations. Modest fluorescence enhancements, relative to unbound dye, are observed for DiSC1(3) and the PNA-DNA heteroquadruplexes, with slightly larger enhancements observed for those PNAs having bases in their loops (PA, PC, PT and PTG) compared to the PNAs having abasic loops (Peg1 and Peg2). As a result of its lower background fluorescence, β-Me-DiSC1(3) exhibits considerably larger fluorescence enhancements for all PNA-DNA heteroquadruplexes. In this case, the strongest fluorescence for the heteroquadruplexes is observed for those PNAs having the capacity to form longer loops when bound to DNA (Peg2 and PTG).
Figure 5.
Fluorescence of cyanine dyes DiSC1(3) (left) and β-Me-DiSC1(3) (right) in presence of various DNA and PNA-DNA quadruplexes. Samples contained 1.0 µM dye, 1.0 µM DNA and 2.0 µM PNA.
It is difficult to interpret the results of Figure 5 due to uncertainty in the affinities and binding modes for the two dyes. For example, differences in fluorescence intensity could arise from differences in affinity and/or quantum yield for the bound dye. Nevertheless, the fact that the intensity is substantially higher for the DNA homoquadruplex relative to the PNA-DNA heteroquadruplexes allows a useful experiment to be performed to determine the stoichiometry of the PNA-DNA quadruplexes. PNA and DNA are mixed in various ratios but constant total concentration. A constant amount of cyanine dye is also present in solution. The fluorescence of the dye is determined for each PNA:DNA ratio and is plotted versus mole fraction of PNA. As shown in Figure 6, the results of this continuous variations experiment reveal a 2:1 PNA:DNA stoichiometry for Peg2-Myc19, consistent with our prior investigations of this heteroquadruplex.
Figure 6.
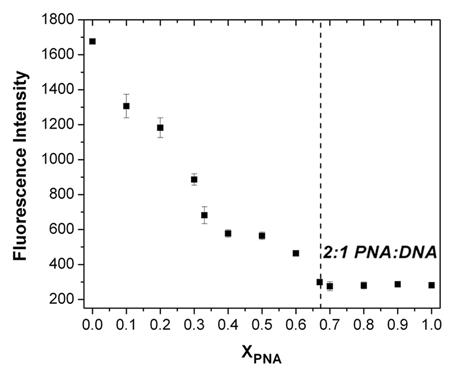
Job plot constructed by mixing together Peg2 and Myc19 in varying ratios and 1.0 µM total concentration in the presence of 1.0 µM DiSC1(3). Fluorescence was recorded with excitation at 520 nm.
We previously used two methods to determine PNA-DNA stoichiometry. In the first, a fluorogenic TO dye was conjugated to the end of the PNA and the difference in fluorescence intensity in the free versus hybridized states was used to construct a Job plot that revealed 2:1 binding. Alternatively, surface plasmon resonance experiments in which the total amount of immobilized DNA and captured PNA are known can provide the stoichiometry directly.13 Use of a noncovalent reporter dye such as the cyanines described herein provides an appealing alternative, since this bypasses synthesis of a separate PNA-dye conjugate for each system or the need for a specialized and costly SPR instrument, although, of course other methods such as electrophoretic mobility shift assay could provide the same information.
Materials and Methods
PNAs were synthesized using t-Boc-protected monomers (ASM chemicals) and solid phase methods, as reported previously in reference 38 and 39. Crude oligomers were purified by reverse-phase HPLC and characterized by MALDI-TOF mass spectrometry. PNAs Pmyc, Peg1 and Peg2 were reported previously. (Expected m/z for PA, 2168.1, found, 2171.2; expected m/z for PC, 2144.1, found, 2147.2; expected m/z for PT, 2159.1, found, 2157.9; and the expected m/z for PTG, 2450.3, found, 2454.1). DNA oligonucleotides were purchased from Integrated DNA Technologies (www.idtdna.com) and used as received. Cyanine dyes DiSC1(3) and β-Me-DiSC1(3) are known compounds and were synthesized by Dr. Gloria Silva. Products were characterized using ESI mass spectrometry (Thermo-Fisher LCQ ESI/APCI Ion Trap). (β-Me-DiSC1(3), expected m/z for M+, 351.5; found, 351.3. DiSC1(3), expected m/z for M+, 337.4; found, 337.3). UV-vis spectra for the compounds matched those from the literature.31
All DNA and PNA concentrations were determined by measuring the absorbance at 260 nm at 85°C on a Cary 300 Bio UV-visible spectrophotometer. At high temperatures, the bases are assumed to be unstacked and the extinction coefficient of the oligomers is estimated as the sum of the extinction coefficients of the individual bases at that temperature. For the DNA oligomers, the extinction coefficients were used as reported in the literature.40 The individual nucleobase extinction coefficients used to calculate concentrations were 13,700 M−1cm−1 (A), 11,700 M−1cm−1 (G), 6,600 M−1cm−1 (C) and 8,600 M−1cm−1 (T).
Absorption and emission spectroscopy.
Absorption spectra of pre-annealed (i.e., heated to 95°C for 5 min before slow cooling to room temperature) samples were collected using Varian Cary 300 Bio UV-Visible Spectrophotometer. Corresponding baselines corrections were made prior to absorbance measurement. Emission (fluorescence) spectra of pre-annealed samples were collected using a Varian Cary Eclipse Fluorescence Spectrophotometer. Samples containing DNA, PNA and dye were prepared in a buffer solution containing 10 mM Tris-HCl (pH 7), 0.1 mM Na2EDTA and 100 mM KCl. (This buffer was used for all spectroscopic experiments).
Two types of continuous variations experiments were performed. For DNA-dye stoichiometries, DNA and dye were mixed at varying ratios but constant total concentration of 1.0 µM. Fluorescence spectra were recorded with excitation at 520 nm and the integrated fluorescence intensity was plotted versus dye mole fraction to determine the empirical stoichiometry of the DNA-dye complex. For PNA-DNA stoichiometries, PNA and DNA were mixed at varying ratios but constant total concentration of 1.0 µM. Dye was also present at 1.0 µM concentration. Fluorescence spectra were recorded with excitation at 520 nm and the integrated fluorescence intensity was plotted versus dye mole fraction to determine the empirical stoichiometry of the PNA-DNA complex.
Circular dichroism (CD) spectropolarimetry.
CD measurements were performed using a Jasco 715 CD spectropolarimeter, equipped with a water-circulating temperature controller. Samples were annealed prior to CD recording. Each CD spectrum represents an average of six scans, collected at a rate of 100 nm/min.
Conclusion
The cyanine dyes DiSC1(3) and β-Me-DiSC1(3) bind to the DNA quadruplex Myc19 in 1:1 stoichiometries and with high nanomolar affinities. While these dyes are unlikely to exhibit selective binding to a particular DNA quadruplex relative to other quadruplexes, double-stranded DNA or RNA, the cyanine scaffold is readily modified with various substituents on the heterocycle periphery, the (poly)methine bridge or the heterocycle nitrogens. Thus, libraries of cyanine dyes could be screened in order to identify substitution patterns that favor selective binding, as has been done with other scaffolds.27,30,37 The low background fluorescence of β-Me-DiSC1(3) makes it particularly attractive for further study since dyes based on this structure might exhibit selective staining of quadruplex structures in cells without having to wash out unbound dye.
In addition to the dye-DNA results presented above, this report provides the first examples of noncovalent binding of small molecules to PNA-DNA heteroquadruplexes. The variation in fluorescence intensities for the different heteroquadruplexes evident in Figure 5 indicates that the dyes sense subtle structural variations and this impacts the affinities and/or quantum yields of the bound dyes. These dyes are useful in the shortterm for determining PNA-DNA stoichiometries; in the longer view, we hope to identify dyes that will bind selectively to PNA-containing heteroquadruplexes over DNA or RNA homoquadruplexes, duplexes or other structures. Such dyes would provide sensitive indicators of successful PNA hybridization in complex biological samples, including within cells. Alternatively, covalent conjugation of the dyes to PNA could provide alternatives to the TO conjugates used previously, with longer wavelength emission and potentially larger fluorescence enhancements, particularly in the case of β-Me-DiSC1(3).
Acknowledgments
We are grateful to Dr. Gloria Silva for synthesizing the cyanine dyes used in these studies. This research was supported by the National Institutes of Health grant R01 GM58547. Mass spectra were recorded in the Center for Molecular Analysis at Carnegie Mellon, supported by NSF CHE-9808188 and DBI-9729351.
Supplementary Material
References
- 1.Neidle S, Balasubramanian S, editors. Quadruplex nucleic acids. Cambridge, UK: Royal Society of Chemistry; 2006. [Google Scholar]
- 2.Huppert J. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem Soc Rev. 2008;37:1375–1384. doi: 10.1039/b702491f. [DOI] [PubMed] [Google Scholar]
- 3.Qin Y, Hurley LH. Structures, folding patterns and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monchaud D, Teulade-Fichou MP. A hitchhiker's guide to G-quadruplex ligands. Org Biomol Chem. 2008;6:627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- 5.Luedtke NW. Targeting G-quadruplex DNA with small molecules. Chimia. 2009;63:134–139. [Google Scholar]
- 6.Balasubramanian S, Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr Opin Chem Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datta B, Armitage BA. Hybridization of PNA to structured DNA targets: quadruplex invasion and the overhang effect. J Am Chem Soc. 2001;123:9612–9619. doi: 10.1021/ja016204c. [DOI] [PubMed] [Google Scholar]
- 8.Green JJ, Ying L, Klenerman D, Balasubramanian S. Kinetics of unfolding the human telomeric DNA quadruplex using a PNA trap. J Am Chem Soc. 2003;125:3763–3767. doi: 10.1021/ja029149w. [DOI] [PubMed] [Google Scholar]
- 9.Kumar N, Patowary A, Sivasubbu S, Petersen M, Maiti S. Silencing c-MYC expression by targeting quadruplex in P1 promoter using locked nucleic acid trap. Biochemistry. 2008;47:13179–13188. doi: 10.1021/bi801064j. [DOI] [PubMed] [Google Scholar]
- 10.Amato J, Oliviero G, De Pauw E, Gabelica V. Hybridization of short complementary PNAs to G-quadruplex forming oligonucleotides: an electrospray mass spectrometry study. Biopolymers. 2009;91:244–255. doi: 10.1002/bip.21124. [DOI] [PubMed] [Google Scholar]
- 11.Datta B, Schmitt C, Armitage BA. Formation of a PNA2-DNA2 hybrid quadruplex. J Am Chem Soc. 2003;125:4111–4118. doi: 10.1021/ja028323d. [DOI] [PubMed] [Google Scholar]
- 12.Marin VL, Armitage BA. Hybridization of complementary and homologous peptide nucleic acid oligomers to a guanine quadruplex-forming RNA. Biochemistry. 2006;45:1745–1754. doi: 10.1021/bi051831q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy S, Tanious F, Wilson WD, Ly DH, Armitage BA. High affinity homologous peptide nucleic acid probes for targeting a quadruplex forming sequence from a MYC promoter element. Biochemistry. 2007;46:10433–10443. doi: 10.1021/bi700854r. [DOI] [PubMed] [Google Scholar]
- 14.Lusvarghi S, Murphy CT, Roy S, Tanious FA, Sacui I, Wilson WD, et al. Loop and backbone modifications of PNA improve G quadruplex binding selectivity. J Am Chem Soc. 2009;131:18415–18424. doi: 10.1021/ja907250j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul A, Sengupta P, Krishnan Y, Ladame S. Combining G-quadruplex targeting motifs on a single peptide nucleic acid scaffold: a hybrid (3 + 1) PNA-DNA bimolecular quadruplex. Chem Eur J. 2008;14:8682–8689. doi: 10.1002/chem.200800605. [DOI] [PubMed] [Google Scholar]
- 16.Marin VL, Armitage BA. RNA guanine quadruplex invasion by complementary and homologous PNA probes. J Am Chem Soc. 2005;127:8032–8033. doi: 10.1021/ja051102y. [DOI] [PubMed] [Google Scholar]
- 17.Ishiguro T, Saitoh J, Yawata H, Otsuka M, Inoue T, Sugiura Y. Fluorescence detection of specific sequence of nucleic acids by oxazole yellow-linked oligonucleotides. Homogeneous quantitative monitoring of in vivo transcription. Nucleic Acids Res. 1996;24:4992–4997. doi: 10.1093/nar/24.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svanvik N, Westman G, Wang D, Kubista M. Light-up probes: thiazole orange-conjugated peptide nucleic acid for detection of target nucleic acid in homogeneous solution. Anal Biochem. 2000;281:26–35. doi: 10.1006/abio.2000.4534. [DOI] [PubMed] [Google Scholar]
- 19.Svanvik N, Nygren J, Westman G, Kubista M. Free-Probe Fluorescence of light-up probes. J Am Chem Soc. 2001;123:803–809. doi: 10.1021/ja002294u. [DOI] [PubMed] [Google Scholar]
- 20.Harrison RJ, Reszka AP, Haider SM, Romagnoli B, Morrell J, Read MA, et al. Evaluation of disubstituted acridone derivatives as telomerase inhibitors: the importance of G-quadruplex binding. Bioorg Med Chem Lett. 2004;14:5845–5849. doi: 10.1016/j.bmcl.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 21.Armitage BA. Cyanine dye-DNA interactions: intercalation, groove binding and aggregation. Top Curr Chem. 2005;253:55–76. [Google Scholar]
- 22.Ihmels H, Otto D. Intercalation of organic dye molecules into double-stranded DNA—general principles and recent developments. Top Curr Chem. 2005;258:161–204. [Google Scholar]
- 23.Armitage BA. Cyanine dye-nucleic acid interactions. Top Heterocyc Chem. 2008;14:11–29. [Google Scholar]
- 24.Cao R, Venezia CF, Armitage BA. Investigation of DNA binding modes for a symmetrical cyanine dye trication: effect of DNA sequence and structure. J Biomol Struct Dyn. 2001;18:844–856. doi: 10.1080/07391102.2001.10506712. [DOI] [PubMed] [Google Scholar]
- 25.Kovalska VB, Losytskyy MY, Yarmoluk SM, Lubitz I, Kotlyar AB. Mono and trimethine cyanines Cyan 40 and Cyan 2 as probes for highly selective fluorescent detection of non-canonical structures. J Fluoresc. 2011;21:223–230. doi: 10.1007/s10895-010-0709-y. [DOI] [PubMed] [Google Scholar]
- 26.Chen Q, Kuntz ID, Shafer RH. Spectroscopic recognition of guanine dimeric hairpin quadruplexes by a carbocyanine dye. Proc Natl Acad Sci USA. 1996;93:2635–2639. doi: 10.1073/pnas.93.7.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monchaud D, Allain C, Bertrand H, Smargiasso N, Rosu F, Gabelica V, et al. Ligands playing musical chairs with quadruplex DNA: a rapid and simple displacement assay for identifying selective G-quadruplex binders. Biochimie. 2008;90:1207–1223. doi: 10.1016/j.biochi.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 28.Kerwin SM, Sun D, Kern JT, Rangan A, Thomas PW. G-quadruplex DNA binding by a series of carbocyanine dyes. Bioorg Med Chem Lett. 2001;11:2411–2414. doi: 10.1016/s0960-894x(01)00490-5. [DOI] [PubMed] [Google Scholar]
- 29.Yang Q, Xiang J, Yang S, Zhou Q, Li Q, Tang Y, et al. Verification of specific G-quadruplex structure by using a novel cyanine dye supramolecular assembly: I. Recognizing mixed G-quadruplex in human telomeres. Chem Commun. 2009:1103–1105. doi: 10.1039/b820101c. [DOI] [PubMed] [Google Scholar]
- 30.Paramasivan S, Bolton PH. Mix and measure fluorescence screening for selective quadruplex binders. Nucleic Acids Res. 2008;36:106. doi: 10.1093/nar/gkn487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yarmoluk SM, Kovalska VB, Lukashov SS, Slominskii YL. Interaction of cyanine dyes with nucleic acids. XII. β-Substituted carbocyanines as possible fluorescent probes for nucleic acids detection. Bioorg Med Chem Lett. 1999;9:1677–1678. doi: 10.1016/s0960-894x(99)00253-x. [DOI] [PubMed] [Google Scholar]
- 32.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 34.West W, Pearce S, Grum F. Stereoisomerism in cyanine dyes—meso-substituted thiacarbocyanines. J Phys Chem. 1967;71:1316–1326. [Google Scholar]
- 35.Yarmoluk SM, Lukashov SS, Losytskyy MY, Akerman B, Kornyushyna OS. Interaction of cyanine dyes with nucleic acids. XXVI. Intercalation of the trimethine cyanine dye Cyan 2 into double-stranded DNA: study by spectral luminescence methods. Spectrochimica Acta A. 2002;58:3223–3232. doi: 10.1016/s1386-1425(02)00100-2. [DOI] [PubMed] [Google Scholar]
- 36.Herz AH. Dye-dye interactions in solution and at AgBr surfaces. Photograph Sci Eng. 1974;18:323–335. [Google Scholar]
- 37.Redman JE, Ladame S, Reszka AP, Neidle S, Balasubramanian S. Discovery of G-quadruplex stabilizing ligands through direct ELISA of a one-bead-one-compound library. Org Biomol Chem. 2006;4:4364–4369. doi: 10.1039/b611716c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christensen L, Fitzpatrick R, Gildea B, Petersen KH, Hansen HF, Koch T, et al. Solid-phase synthesis of peptide nucleic acids. J Pept Sci. 1995;3:175–183. doi: 10.1002/psc.310010304. [DOI] [PubMed] [Google Scholar]
- 39.Koch T. PNA synthesis by Boc chemistry. In: Nielsen PE, editor. Peptide Nucleic Acids: Protocols and Applications. Norfolk: Horizon Bioscience; 2004. pp. 37–60. [Google Scholar]
- 40.Dawson RMC, Elliott DC, Elliott WH, Jones KM. Data for Biochemical Research. New York: Oxford University Press; 1986. pp. 103–114. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.