Abstract
Protein misfolding and assembly into ordered, self-templating aggregates (amyloid) has emerged as a novel mechanism for regulating protein function. For a subclass of amyloidogenic proteins known as prions, this process induces transmissible changes in normal cellular physiology, ranging from neurodegenerative disease in animals and humans to new traits in fungi. The severity and stability of these altered phenotypic states can be attenuated by the conformation or amino-acid sequence of the prion, but in most of these cases, the protein retains the ability to form amyloid in vitro. Thus, our ability to link amyloid formation in vitro with its biological consequences in vivo remains a challenge. In two recent studies, we have begun to address this disconnect by assessing the effects of the cellular environment on traits associated with the misfolding of the yeast prion Sup35. Remarkably, the effects of quality control pathways and of limitations on protein transfer in vivo amplify the effects of even slight differences in the efficiency of Sup35 misfolding, leading to dramatic changes in the associated phenotype. Together, our studies suggest that the interplay between protein misfolding pathways and their cellular context is a crucial contributor to prion biology.
Key words: prion, protein misfolding, chaperones, amyloid, ordered aggregates, transmission, aggregate size, Sup35, Hsp104
Prion Propagation In Vivo
According to the prion hypothesis, an alternative conformation of a normal, host-encoded protein, known as a prion, can function as an epigenetic determinant of transmissible phenotypic states in vivo. This idea was originally proposed as a disease mechanism for the Transmissible Spongiform Encephalopathies (TSEs), a group of severe mammalian neurodegenerative disorders associated with the misfolding of the prion PrP.1,2 More recently, the prion hypothesis has provided a novel framework for understanding other enigmatic biological phenomena, including a group of metastable traits in fungi that are inherited through a non-Mendelian route.3,4 Although once met with skepticism, this protein-only mechanism has now been proven in multiple studies, where new transmissible phenotypes were induced in experimental organisms simply by introducing particular conformational variants (conformers) of prions.5–11
Prion-associated phenotypes are thought to arise because conformational conversion alters the activity of the protein.4 However, this link between discrete physical and functional states of a protein only provides a steady-state snapshot of the endpoints of a dynamic process. The appearance, spread and reversal of prion-associated phenotypes necessarily involve changes in protein physical state.12 However, these transitions are only effective if the alternative conformer is preferentially amplified, at the expense of all others, to a level that can alter cellular physiology.
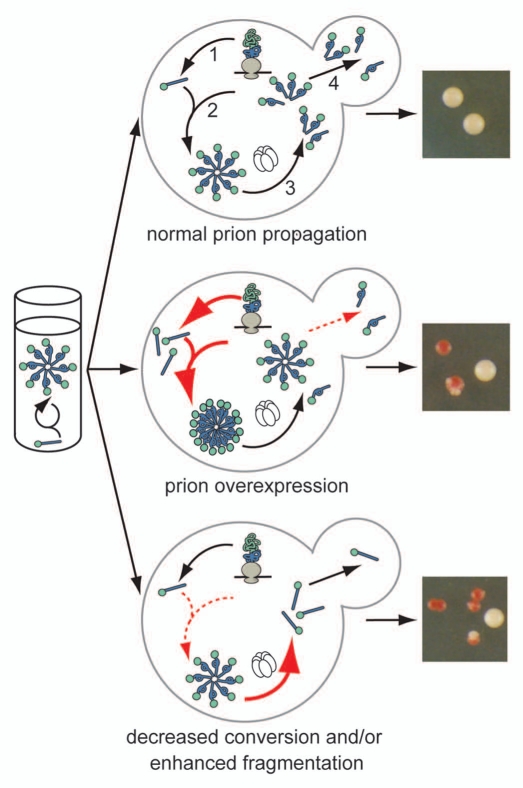
Much of our understanding of this conformational selection and amplification results from studies in vitro, where the assembly of prion conformers into amyloid aggregates directs the continued misfolding of the protein by providing a template for this conversion.13–19 However, studies on the yeast prion Sup35 indicate that the process of conformational self-replication is more complex in vivo (Fig. 1, top), and a similar pathway has been proposed for prion propagation in mammals.20 Based on this work, aggregates or their component prion-state protein must act in three roles to sustain the prion-associated phenotype in a population. As is the case in vitro, these aggregates template the conversion of newly made Sup35 to the prion conformer;21,22 however, in vivo, simple growth of the aggregates will not allow the spread of prion-associated phenotypes in a dividing culture. Thus, these aggregates also must be continually fragmented to create new conversion surfaces23 and transmitted to daughter cells (Fig. 1, top).23,24
Figure 1.
The cellular environment modulates prion misfolding pathways to create protein-based traits. (top) Self-replicating protein conformations create stable phenotypes (white colonies) in vivo when the processes of synthesis (1), conversion (2), fragmentation (3) by Hsp104 (hexamer) and transmission (4) are balanced to allow aggregates of prion proteins to persist in vivo. (middle) Overexpression of a prion protein promotes the conversion reaction (red arrows), leading to the accumulation of large aggregates that are inefficiently transmitted to daughter cells (dotted red arrow) and loss of the prion-associated phenotype (red colonies). (bottom) Dominant inhibition of prion propagation by mutants that decrease conversion efficiency (dotted red arrow) or enhance fragmentation efficiency (solid red arrow) promote aggregate disassembly (ball and stick) and induce prion loss (red colonies).
Prion Variants
While the pathway of prion propagation in vivo explains how the appearance of a misfolded conformer can be amplified to induce and maintain a new phenotype, this scheme alone is not sufficient to explain the diversity of prion-associated phenotypes. Remarkably, each prion has the capacity to create a spectrum of phenotypic states, known as strains or variants, which retain their characteristics upon serial passage in the same host.25–30 In mammals, variants of PrP are characterized by differences in host-specific incubation periods between initial infection and clinical disease,31–34 the distribution of pathological changes in the brain,25,33,35 clinical symptoms,36 sensitivity to thermal inactivation37–40 and the interspecies transmission of disease.32,38,41–43 In contrast, variants of the fungal prions do not specify new phenotypes but rather alter the severity of the prototypical prion-associated phenotype to generate a continuum of similar traits.26,27,29
The existence of variants was once considered incompatible with a protein-only mechanism, but studies in both mammals and yeast have linked this phenotypic diversity to a parallel collection of unique conformers for each prion, which are characterized by changes in protease sensitivity, thermodynamic stability, and the extent of the core amyloid structure.7,25,28,44–49 For both mammalian and fungal prions, these conformers are all self-replicated through the same pathway, but the physical differences between them affect the activities of aggregates and presumably their phenotypic consequences.6,7,50 Aggregates composed of distinct prion conformers differ in their templating activity49–51 and in their thermodynamic stabilities, which likely affect both their ability to be fragmented and to be cleared by quality control pathways in vivo.49,52,53 What has emerged from these analyses is that unique combinations of conversion and fragmentation efficiencies define prion variants, but how these biochemical parameters created distinct biological outcomes remained a major unanswered question.
Connecting Prion Misfolding to Phenotype
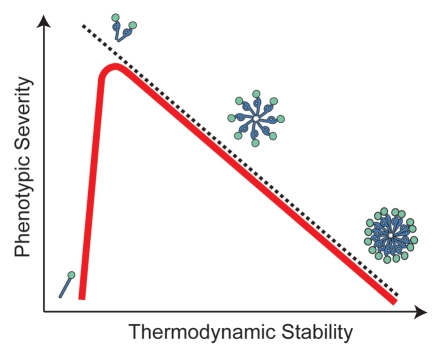
In general, the thermodynamic stability of mouse, both natural and synthetic (i.e., in vitro generated), and yeast prion aggregates is inversely correlated with the severity of their associated phenotypes (Fig. 2), most typically scored as incubation time in mammals and extent of aggregation in fungi.49,52,53 Based on these observations, it was suggested that the efficiency of aggregate fragmentation in vivo, which should be a function of aggregate thermodynamic stability, has a profound effect on prion biology. If the system has reached steady-state, modest changes in fragmentation efficiency are predicted to inversely affect the size of aggregates and directly affect their accumulation, as prion synthesis is on-going,54 and either (or both) of these variables could impact the creation of unique, conformation-based phenotypes. To distinguish between these size and abundance-based models of prion variants, we focused on the yeast protein Sup35, a component of the translation termination complex whose function is modulated by a prion cycle.55–57 In the non-prion [(psi−)]conformer, Sup35 is soluble and facilitates translation termination; however, in its prion [(PSI+)] conformers, Sup35 aggregates to different extents, establishing a corresponding range of decreased translation termination efficiencies.21,58,59
Figure 2.
Relating prion phenotypic severity to aggregate thermodynamic stability. For many prion variants, there is a linear but inverse relationship between aggregate thermodynamic stability and phenotypic severity (black dotted line), but this trend cannot explain the phenotypes associated with all prion variants or the effects of dominant-negative prion mutants (see text for details). Our studies in vivo on the yeast prion [PSI+] suggest that thermodynamic stability poses a limit on prion persistence at both extremes (red line) by impacting aggregate size and accumulation (shown schematically). The least thermodynamically stable aggregates are efficiently resolubilized, while the most thermodynamically stable aggregates are inefficiently transmitted.
Given the multiple roles of aggregates in prion propagation in vivo, we employed a computational approach to guide our experiments on the in vivo pathway through which prion variants are created. TSE propagation can be mathematically described in terms of PrP synthesis, conversion, fragmentation and decay rates,20 and by substituting prion dilution during cell division for prion decay, the same formulation accurately captures the differences in soluble Sup35 levels found in two [PSI+] variants, known as weak and strong.27,49 However, this model cannot accurately predict other defining characteristics of yeast prion variants, including differences in the size distribution of aggregates,60,61 the spontaneous frequency of prion loss,27,62 and the elevated frequency of prion loss observed upon Sup35 overexpression,63,64 a phenomenon that is also observed for another yeast prion.65–67
Each of these characteristics is specific to prion propagation in vivo; therefore, we formulated a new model that incorporated aspects of the cellular environment that were known to impact prion propagation: the transmission of Sup35 protein to daughter cells23,68 and the limitation on fragmentation efficiency imposed by the concentration of the molecular chaperone Hsp104, the catalyst for this reaction.23,69,70 With this reformulation, we simulated [PSI+] propagation via a transmission model that was either based on aggregate abundance or size and assessed their accuracy in computationally recapitulating all of the characteristics of the [PSI+]weak and [PSI+]strong variants described above. In contrast to in vivo observations, the prion state was completely stable under all conditions in the abundance-based model because aggregates, if they accumulated to any extent, were efficiently transferred to daughter cells, as had been previously predicted.62 However, the severities, stabilities and aggregate size distributions of [PSI+]weak and [PSI+]strong variants were accurately recapitulated if a size threshold for aggregate transmission was imposed, but only if this limitation fell within the size range of SDS-resistant aggregates that distinguished the two variants.60
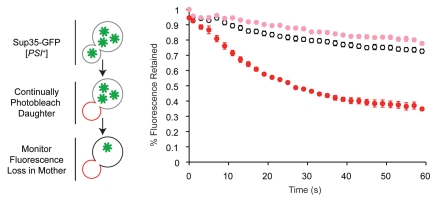
Our modeling suggested that the heritable prion species, known as a propagon,71 represented a subset of aggregates that was distinguished simply by their size and that variants differed in their accumulation of these species. If true, the stability of variant-associated phenotypes should correlate with the proportion of Sup35 that was transmissible. To directly test this idea, we developed a fluorescence loss in photobleaching (FLIP) assay, in which daughter cells of yeast strains expressing Sup35-GFP are continually bleached prior to cytokinesis and fluorescence loss in the mother is monitored over time as a proxy for transmission (Fig. 3). Indeed, a [PSI+] weak variant transmitted 50% less Sup35-GFP and accumulated 50% fewer propagons than a [PSI+]strong variant, and these differences correlated with a three order of magnitude increase in prion loss during cell division (mitotic instability) for the [PSI+]weak variant.27
Figure 3.
Transmission of Sup35 protein to daughter cells is conformation-dependent. (left) Schematic of fluorescence loss in photobleaching assay (FLIP) for Sup35 transmission to daughter cells. Bleached daughter (red) and monitored mother (black) are indicated. (right) Fluorescence retention in mother cells expressing Sup35-GFP in the [PSI+]strong (white), [PSI+]weak (pink) or [psi−] (red) conformation.
If aggregate size alone is responsible for these effects, shifting this distribution, in the absence of conformational changes, should also impact both the transmissibility of Sup35 and the stability of the associated phenotype. In vivo, the steady-state size of aggregates is a function of both the conversion and fragmentation reactions; therefore, we varied Sup35 expression level to modulate the conversion rate. The size of SDS-resistant aggregates was increased by mild overexpression of Sup35 (∼5-fold) and dramatically decreased by repression of Sup35 synthesis for even one generation. By FLIP, conditions associated with the accumulation of larger aggregates resulted in a decrease in Sup35-GFP transmission (Fig. 1, middle), while those associated with smaller aggregates resulted in an increase in Sup35-GFP transmission. As was the case for different variants, these differences in transmission correlated directly with the number of propagons and indirectly with the mitotic stability of the prion-associated phenotype. Thus, our studies suggest that there is no inherent functional difference between aggregates of the same conformer but rather that their behavior in vivo is modulated by differential interactions with the cellular environment.
Conformation-dependent differences in prion-associated phenotypes were previously thought to arise from distinct equilibria between soluble and aggregated protein (i.e., abundance-based model) in all cells in a population.49 However, our size-based model suggested that prion-associated phenotypes arose through a population-based mechanism. Specifically, the size threshold for aggregate transmission is predicted to create heterogeneity in the population, with mother cells accumulating more and larger aggregates than their daughters. By assessing Sup35 physical state in either mother or daughter cells by FLIP or a gel-based assay following centrifugal elutriation, we provided experimental support for this prediction. These age-dependent differences and the fact that prion variants produce phenotypically homogeneous colonies suggested that the aggregate complement and associated phenotype must change over time. Our modeling indeed predicted that the fraction of Sup35 in aggregates, which determines translation termination activity,72 and their size both increased with replicative age. Consistent with this prediction, we demonstrated that the number of propagons transmitted to daughters increases with the replicative age of the mother and that the fidelity of translation termination, as monitored by a fluorescent reporter, decreases with replicative age. Thus, prion-associated phenotypes arise through a dynamic pathway of prion biogenesis rather than a simple equilibrium between the prion and non-prion conformers.
The size-dependent transmission of aggregates provides a framework for understanding prion-associated phenotypes in a dividing yeast culture, but can differences in aggregate size also provide insight into prion biology in mammalian post-mitotic neurons? Mathematical models have suggested that aggregate size as well as abundance could be important contributors to prion propagation in mammals.20 Indeed, biochemical analyses have linked small aggregates to high infectivity and short incubation times.73,74 Mechanistically, aggregate size appears to impact converting activity,73,75 but others have suggested an effect on transmission as well.20,76 Consistent with this idea, more thermodynamically stable and presumably larger, aggregates are associated with localized pathology while less thermodynamically stable aggregates are associated with more widespread pathology.77 Moreover, mammalian prion biology is altered by changes in expression of the prion protein, as is the case for yeast prion biology.24,63–67 For example, the incubation time for clinical disease, but not for the generation of prion infectivity, is highly dependent on PrP expression level.78,79 While other mechanisms are possible,78,80,81 an intriguing model to explain these observations is that infectivity increases with aggregate abundance until reaching a plateau, which represents a limitation on the system. If that limitation is fragmentation activity, aggregates will increase in size but not abundance, and this process would proceed more rapidly at higher PrP expression levels. In this scenario, prion toxicity, corresponding to clinical disease, could result from the titration of other cellular factors through their association with aggregates, a mechanism that has been proven for the toxicity of overexpressed Sup35 in [PSI+] yeast strains.82 Thus, aggregate size may be an underappreciated contributor to the phenotypic outcomes of prion variants in mammals, as well.
Balancing Aggregate Assembly and Disassembly Pathways
While fragmentation efficiency appears to be a limitation on prion propagation in vivo, there may also be an upper boundary on the level of activity that is compatible with the prion state (Fig. 2). Several variants of hamster prion assemble into aggregates of lower thermodynamic stability than most other prion variants,25,32,83,84 but their incubation periods are significantly longer.28,49,52 One potential explanation for these disconnects is that the rate of aggregate disassembly might approach that of aggregate assembly for these variants, leading to clearance.50,51,84,85 Consistent with this idea that aggregate assembly and disassembly pathways compete with one another in vivo, infectious particles delivered by direct inoculation, which presumably do not represent a load on the system, are largely cleared from experimental organisms,78,86–90 and even established prion infections are reversed in vivo upon repression of new PrP synthesis.91–95
Other examples of the importance of competition between aggregate assembly and disassembly pathways in prion phenotypic outcomes may also be found in nature. Individuals with PrP polymorphisms are genetically less susceptible to prion disease.96–105 These polymorphisms clearly restrict the range of conformations accessible to the prion and therefore its ability to replicate certain variants.31,43,106–110 However, many of the same sequence changes also function as dominant inhibitors of prion propagation in vivo,105,111–118 and a similar effect occurs upon co-expression of PrP homologues from different species.79,119–124 Thus, these sequence variants must target crucial events in prion propagation by the wild-type protein, and elucidating their mechanisms of action could be instructive for developing therapeutic interventions for these diseases.114–116
Early models suggested that PrP dominant-negative mutants acted by titrating away a host-encoded cofactor (protein X) required for the conversion reaction;122 however, these sequence variants also inhibit prion propagation in cell-free systems, suggesting that their effects are mediated directly through prion-prion interactions.125–127 In this scenario, PrP sequence variants would interact with the templating surface on an aggregate and either slow, as has been proposed for a Q219K variant, or block, as has been proposed for a Q172R variant, conversion of wild-type PrP, depending on their affinities for one another.125,126 Potentially supporting this affinity-based model, PrP dominant-negative mutants differ in their effective inhibitory ratios relative to wild-type protein,111,125–129 but the same observation is also consistent with a model in which these mutants target different events in prion propagation in vivo. According to this latter idea, mutants that act at the templating surface would be effective at lower doses than those affecting fragmentation, which occurs along the length of the aggregate.130
Propagation of the yeast prion [PSI+] is also disrupted by co-expression of Sup35 mutants, such as Q24R and G58D,131–133 and we used this system to explore the mechanisms underlying prion dominant inhibition in vivo.134 We found that Q24R, like PrP Q172R, acted at sub-stochiometric ratios while G58D, like Q219K, interfered only at higher ratios relative to wild-type protein.113,125,135 Using a series of in vivo analyses, we linked these differences to defects in discrete events in the prion propagation pathway. While we detected no defect in prion transmission by FLIP, we determined that G58D, but not Q24R, could efficiently join wild-type complexes by monitoring conversion by a translation termination defect that appears upon Sup35 incorporation into aggregates,22 as had been previously suggested.64,131,136,137 In contrast, G58D, but not Q24R, incorporation into aggregates led to their destabilization, as measured by their sensitivity to disruption in SDS at increasing temperatures. Thus, Q24R expression decreases conversion efficiency while G58D expression likely increases fragmentation efficiency. In addition to revealing the molecular basis of the differences in effective inhibitory ratios, these effects on Sup35 biogenesis also explain the differential dominant interactions of Q24R and G58D with the [PSI+]weak variant, which is characterized by a reduced accumulation of aggregates due to inefficient fragmentation.49 Q24R expression, which limits the conversion reaction and would further diminish aggregate accumulation, is completely incompatible with the [PSI+]weak variant (our unpublished observations), while G58D expression, which enhances fragmentation and would increase aggregate accumulation, promotes [PSI+]weak mitotic stability.64
Consistent with these predictions, propagon accumulation in [PSI+]strong haploid strains decreased with Q24R expression but increased with G58D expression. However, these effects were modulated by the cellular environment: in [PSI+] strong diploid strains, expression of either mutant decreased propagon accumulation. We hypothesized that these observations could be explained by the relative doses of Sup35 and Hsp104 under the two conditions (2:1 vs. 1:1, respectively). If true, modulating chaperone levels in diploids should affect the severity of the inhibition. Indeed, the dominant-negative effects of both Q24R and G58D were partially or completely reversed, respectively, by lowering Hsp104 levels and were enhanced by overexpression of Hsp104. Moreover, we found differences in the Sup35:Hsp104 ratio in yeast strains of different genetic backgrounds, providing a molecular explanation for known variations in G58D efficacy.64,132
The dependency of prion inhibition on chaperone level suggested that the disassembly pathway was competing more efficiently with the assembly pathway in the presence of the dominant-negative mutants. To determine if aggregates were actually being resolubilized, we monitored the fate of existing Sup35 after treating [PSI+]strong cultures with cycloheximide. In these experiments, Sup35 transitioned from an SDS-resistant to an SDS-sensitive form in yeast strains expressing both wild-type and dominant-negative mutants but not in those expressing wild-type protein alone. Mechanistically, we predict that Q24R and G58D induce this same outcome through different pathways: the conversion defect of Q24R would increase the ratio of enzyme (Hsp104) to substrate (aggregates) for the fragmentation reaction, while the efficiency of that reaction would increase in the presence of G58D (Fig. 1, bottom). Thus, in either case, expression of dominant-negative mutants skews the competition between aggregate assembly and disassembly pathways toward the latter, allowing the effective clearance of an established prion conformer in vivo (Fig. 2). Given the heterogeneity in aggregate accumulation that we have observed in individual wild-type yeast cells, the balance between these opposing forces may also vary among cells in the population, contributing a currently unappreciated influence on the severity and stability of prion-associated phenotypes under normal conditions.
Strikingly, there are many parallels between Sup35 Q24R and PrP Q172R and between Sup35 G58D and PrP Q219K. In addition to displaying similar effective inhibitory ratios, PrP Q172R, like Sup35 Q24R, is conversion defective and is incompatible with prion variants of different thermodynamic stabilities.28,117,125,135 In contrast, PrP Q219K, like, Sup35 G58D, converts to the prion conformer efficiently, destabilizes aggregates and is incompatible only with prion variants of reduced thermodynamic stability.28,105,117,125,126 Thus, despite the distinct cellular environments in which mammalian and yeast prions propagate, the pathways of dominant inhibition that we have uncovered for Sup35 mutants may be applicable to PrP mutants and may provide mechanistic insight into the phenotypic outcomes of prion infections in that system.
Conclusions
Despite our detailed understanding of amyloidogenesis in vitro, our ability to mechanistically link this process to its biological consequences in vivo lags far behind.138,139 Collectively, our studies on the yeast prion [PSI+] have begun to bridge this gap by demonstrating that the cellular environment dramatically influences the physiological consequences of Sup35 misfolding. Notably, conformation-based phenotypes are created through the interplay among Sup35 folding, protein quality control and other aspects of yeast cell biology,24 and imbalances in these forces lead to the dominant inhibition of prion propagation in vivo.134 Given the many parallels between prion propagation in yeast and in mammals, the insight gained through these studies may provide a new framework for connecting protein misfolding pathways in vitro to disease mechanisms in vivo.
Acknowledgments
We thank A. Derdowski for the FLIP image and members of the Serio lab for helpful discussions and comments on the manuscript. This work was supported by awards from the National Institutes of Health to T.R.S. (GM069802) and to S.D. (AG032818).
Note Added in Proof
Recent studies by the Weissman group have determined that amyloid fibers composed of G58D alone, when present in a particular conformation, exhibit the same conversion and thermodynamic stability differences in comparison with wild-type fibers in vitro as we have identified for mixed G58D-wild-type prion aggregates in vivo.140
References
- 1.Griffith JS. Self-replication and scrapie. Nature. 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 3.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 4.Tuite MF, Serio TR. The prion hypothesis: from biological anomaly to basic regulatory mechanism. Nat Rev Mol Cell Biol. 2010;11:823–833. doi: 10.1038/nrm3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brachmann A, Baxa U, Wickner RB. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 8.Patel BK, Liebman SW. “Prion-proof” for [PIN+]: Infection with in vitro-made amyloid aggregates of Rnq1p-(132–405) induces [PIN+] J Mol Biol. 2007;365:773–782. doi: 10.1016/j.jmb.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ. Amyloid aggregates of the HET-s prion protein are infectious. Proc Natl Acad Sci USA. 2002;99:7402–7407. doi: 10.1073/pnas.072199199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JI, Cali I, Surewicz K, Kong Q, Raymond GJ, Atarashi R, et al. Mammalian prions generated from bacterially expressed prion protein in the absence of any mammalian cofactors. J Biol Chem. 2010;285:14083–14087. doi: 10.1074/jbc.C110.113464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, Wang X, Yuan CG, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science. 2010;327:1132–1135. doi: 10.1126/science.1183748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pezza JA, Serio TR. Prion propagation: The role of protein dynamics. Prion. 2007;1:36–43. doi: 10.4161/pri.1.1.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 14.Kocisko DA, Come JH, Priola SA, Chesebro B, Raymond GJ, Lansbury PT, et al. Cell-free formation of protease-resistant prion protein. Nature. 1994;370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 15.Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 16.King CY, Tittmann P, Gross H, Gebert R, Aebi M, Wuthrich K. Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci USA. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor KL, Cheng N, Williams RW, Steven AC, Wickner RB. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- 18.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411:810–813. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- 19.Dos Reis S, Coulary-Salin B, Forge V, Lascu I, Begueret J, Saupe SJ. The HET-s prion protein of the filamentous fungus Podospora anserina aggregates in vitro into amyloid-like fibrils. J Biol Chem. 2002;277:5703–5706. doi: 10.1074/jbc.M110183200. [DOI] [PubMed] [Google Scholar]
- 20.Masel J, Jansen VA, Nowak MA. Quantifying the kinetic parameters of prion replication. Biophys Chem. 1999;77:139–152. doi: 10.1016/s0301-4622(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 21.Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 22.Satpute-Krishnan P, Serio TR. Prion protein remodelling confers an immediate phenotypic switch. Nature. 2005;437:262–265. doi: 10.1038/nature03981. [DOI] [PubMed] [Google Scholar]
- 23.Satpute-Krishnan P, Langseth SX, Serio TR. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol. 2007;5:24. doi: 10.1371/journal.pbio.0050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derdowski A, Sindi SS, Klaips CL, DiSalvo S, Serio TR. A size threshold limits prion transmission and establishes phenotypic diversity. Science. 2010;330:680–683. doi: 10.1126/science.1197785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bessen RA, Marsh RF. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol. 1994;68:7859–7868. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci USA. 2002;99:16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, et al. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 29.Schlumpberger M, Prusiner SB, Herskowitz I. Induction of distinct [URE3] yeast prion strains. Mol Cell Biol. 2001;21:7035–7046. doi: 10.1128/MCB.21.20.7035-7046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wickner RB, Edskes HK, Shewmaker F, Kryndushkin D, Nemecek J. Prion variants, species barriers, generation and propagation. J Biol. 2009;8:47. doi: 10.1186/jbiol148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dickinson AG, Meikle VM. Host-genotype and agent effects in scrapie incubation: change in allelic interaction with different strains of agent. Mol Gen Genet. 1971;112:73–79. doi: 10.1007/BF00266934. [DOI] [PubMed] [Google Scholar]
- 32.Bessen RA, Marsh RF. Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. J Gen Virol. 1992;73:329–334. doi: 10.1099/0022-1317-73-2-329. [DOI] [PubMed] [Google Scholar]
- 33.Bruce ME, McConnell I, Fraser H, Dickinson AG. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J Gen Virol. 1991;72:595–603. doi: 10.1099/0022-1317-72-3-595. [DOI] [PubMed] [Google Scholar]
- 34.Pattison IH, Millson GC. Scrapie produced experimentally in goats with special reference to the clinical syndrome. J Comp Path. 1961;71:101–109. doi: 10.1016/s0368-1742(61)80013-1. [DOI] [PubMed] [Google Scholar]
- 35.Fraser H, Dickinson AG. Scrapie in mice. Agent-strain differences in the distribution and intensity of grey matter vacuolation. J Comp Path. 1973;83:29–40. doi: 10.1016/0021-9975(73)90024-8. [DOI] [PubMed] [Google Scholar]
- 36.Carp RI, Callahan SM, Sersen EA, Moretz RC. Preclinical changes in weight of scrapie-infected mice as a function of scrapie agent-mouse strain combination. Intervirology. 1984;21:61–69. doi: 10.1159/000149503. [DOI] [PubMed] [Google Scholar]
- 37.Dickinson AG, Taylor DM. Resistance of scrapie agent to decontamination. N Engl J Med. 1978;299:1413–1414. doi: 10.1056/NEJM197812212992512. [DOI] [PubMed] [Google Scholar]
- 38.Kimberlin RH, Walker CA. Evidence that the transmission of one source of scrapie agent to hamsters involves separation of agent strains from a mixture. J Gen Virol. 1978;39:487–496. doi: 10.1099/0022-1317-39-3-487. [DOI] [PubMed] [Google Scholar]
- 39.Kimberlin RH, Walker CA, Millson GC, Taylor DM, Robertson PA, Tomlinson AH, et al. Disinfection studies with two strains of mouse-passaged scrapie agent. Guidelines for Creutzfeldt-Jakob and related agents. J Neurol Sci. 1983;59:355–369. doi: 10.1016/0022-510x(83)90021-7. [DOI] [PubMed] [Google Scholar]
- 40.Taylor DM, Fernie K, McConnell I, Steele PJ. Observations on thermostable subpopulations of the unconventional agents that cause transmissible degenerative encephalopathies. Vet Microbiol. 1998;64:33–38. doi: 10.1016/s0378-1135(98)00257-0. [DOI] [PubMed] [Google Scholar]
- 41.Kimberlin RH, Cole S, Walker CA. Temporary and permanent modifications to a single strain of mouse scrapie on transmission to rats and hamsters. J Gen Biol. 1987;68:1875–1881. doi: 10.1099/0022-1317-68-7-1875. [DOI] [PubMed] [Google Scholar]
- 42.Asante EA, Linehan JM, Desbruslais M, Joiner S, Gowland I, Wood AL, et al. BSE prions propagate as either variant CJD-like or sporadic CJD-like prion strains in transgenic mice expressing human prion protein. EMBO J. 2002;21:6358–6366. doi: 10.1093/emboj/cdf653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wadsworth JD, Asante EA, Desbruslais M, Linehan JM, Joiner S, Gowland I, et al. Human prion protein with valine 129 prevents expression of variant CJD phenotype. Science. 2004;306:1793–1796. doi: 10.1126/science.1103932. [DOI] [PubMed] [Google Scholar]
- 44.Caughey B, Raymond GJ, Bessen RA. Strain-dependent differences in beta-sheet conformations of abnormal prion protein. J Biol Chem. 1998;273:32230–32235. doi: 10.1074/jbc.273.48.32230. [DOI] [PubMed] [Google Scholar]
- 45.Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 46.Peretz D, Williamson RA, Legname G, Matsunaga Y, Vergara J, Burton DR, et al. A change in the conformation of prions accompanies the emergence of a new prion strain. Neuron. 2002;34:921–932. doi: 10.1016/s0896-6273(02)00726-2. [DOI] [PubMed] [Google Scholar]
- 47.Telling GC, Parchi P, DeArmond SJ, Cortelli P, Montagna P, Gabizon R, et al. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 48.Peretz D, Scott MR, Groth D, Williamson RA, Burton DR, Cohen FE, et al. Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci. 2001;10:854–863. doi: 10.1110/ps.39201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 50.Bessen RA, Kocisko DA, Raymond GJ, Nandan S, Lansbury PT, Caughey B. Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature. 1995;375:698–700. doi: 10.1038/375698a0. [DOI] [PubMed] [Google Scholar]
- 51.Mulcahy ER, Bessen RA. Strain-specific kinetics of prion protein formation in vitro and in vivo. J Biol Chem. 2004;279:1643–1649. doi: 10.1074/jbc.M307844200. [DOI] [PubMed] [Google Scholar]
- 52.Legname G, Nguyen HO, Peretz D, Cohen FE, DeArmond SJ, Prusiner SB. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc Natl Acad Sci USA. 2006;103:19105–19110. doi: 10.1073/pnas.0608970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prusiner SB, Scott MR, DeArmond SJ, Cohen FE. Prion protein biology. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 54.Sindi SS, Serio TR. Prion dynamics and the quest for the genetic determinant in protein-only inheritance. Curr Opin Microbiol. 2009;12:623–630. doi: 10.1016/j.mib.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Serio TR, Lindquist SL. [PSI+]: an epigenetic modulator of translation termination efficiency. Annu Rev Cell Dev Biol. 1999;15:661–703. doi: 10.1146/annurev.cellbio.15.1.661. [DOI] [PubMed] [Google Scholar]
- 56.Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, et al. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, et al. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cox B. [PSI], a cytoplasmic suppressor of super-suppression in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- 59.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 60.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 61.Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Meth Enz. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
- 62.Byrne LJ, Cole DJ, Cox BS, Ridout MS, Morgan BJ, Tuite MF. The number and transmission of [PSI] prion seeds (Propagons) in the yeast Saccharomyces cerevisiae. PLoS One. 2009;4:4670. doi: 10.1371/journal.pone.0004670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, et al. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227–1242. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Derkatch IL, Bradley ME, Zhou P, Liebman SW. The PNM2 mutation in the prion protein domain of SUP35 has distinct effects on different variants of the [PSI+] prion in yeast. Curr Genet. 1999;35:59–67. doi: 10.1007/s002940050433. [DOI] [PubMed] [Google Scholar]
- 65.Crapeau M, Marchal C, Cullin C, Maillet L. The cellular concentration of the yeast Ure2p prion protein affects its propagation as a prion. Mol Biol Cell. 2009;20:2286–2296. doi: 10.1091/mbc.E08-11-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edskes HK, Gray VT, Wickner RB. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci USA. 1999;96:1498–1503. doi: 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baudin-Baillieu A, Fernandez-Bellot E, Reine F, Coissac E, Cullin C. Conservation of the prion properties of Ure2p through evolution. Mol Biol Cell. 2003;14:3449–3458. doi: 10.1091/mbc.E03-01-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kawai-Noma S, Pack CG, Tsuji T, Kinjo M, Taguchi H. Single mother-daughter pair analysis to clarify the diffusion properties of yeast prion Sup35 in guanidine-HCl-treated [PSI] cells. Genes Cells. 2009;4:1045–1054. doi: 10.1111/j.1365-2443.2009.01333.x. [DOI] [PubMed] [Google Scholar]
- 69.Haslberger T, Bukau B, Mogk A. Towards a unifying mechanism for ClpB/Hsp104-mediated protein disaggregation and prion propagation. Biochem Cell Biol. 2010;88:63–75. doi: 10.1139/o09-118. [DOI] [PubMed] [Google Scholar]
- 70.Ness F, Ferreira P, Cox BS, Tuite MF. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol Cell Biol. 2002;22:5593–5605. doi: 10.1128/MCB.22.15.5593-5605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cox B, Ness F, Tuite M. Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou P, Derkatch IL, Uptain SM, Patino MM, Lindquist S, Liebman SW. The yeast non-Mendelian factor [ETA+] is a variant of [PSI+], a prion-like form of release factor eRF3. EMBO J. 1999;18:1182–1191. doi: 10.1093/emboj/18.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, et al. The most infectious prion protein particles. Nature. 2005;437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tixador P, Herzog L, Reine F, Jaumain E, Chapuis J, Le Dur A, et al. The physical relationship between infectivity and prion protein aggregates is strain-dependent. PLoS Path. 2010;6:1000859. doi: 10.1371/journal.ppat.1000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Calvez V, Lenuzza N, Oelz D, Deslys JP, Laurent P, Mouthon F, et al. Size distribution dependence of prion aggregates infectivity. Math Biosci. 2009;217:88–99. doi: 10.1016/j.mbs.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 76.Zampieri M, Legname G, Altafini C. Investigating the conformational stability of prion strains through a kinetic replication model. PLoS Comput Biol. 2009;5:1000420. doi: 10.1371/journal.pcbi.1000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Legname G, Nguyen HO, Baskakov IV, Cohen FE, Dearmond SJ, Prusiner SB. Strain-specified characteristics of mouse synthetic prions. Proc Natl Acad Sci USA. 2005;102:2168–2173. doi: 10.1073/pnas.0409079102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sandberg MK, Al-Doujaily H, Sharps B, Clarke AR, Collinge J. Prion propagation and toxicity in vivo occur in two distinct mechanistic phases. Nature. 2011;470:540–542. doi: 10.1038/nature09768. [DOI] [PubMed] [Google Scholar]
- 79.Scott M, Foster D, Mirenda C, Serban D, Coufal F, Walchli M, et al. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell. 1989;59:847–857. doi: 10.1016/0092-8674(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 80.Wickner RB. Prion diseases: Infectivity versus toxicity. Nature. 2011;470:470–471. doi: 10.1038/470470a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ghaemmaghami S, Phuan PW, Perkins B, Ullman J, May BC, Cohen FE, et al. Cell division modulates prion accumulation in cultured cells. Proc Natl Acad Sci USA. 2007;104:17971–17976. doi: 10.1073/pnas.0708372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vishveshwara N, Bradley ME, Liebman SW. Sequestration of essential proteins causes prion associated toxicity in yeast. Mol Microbiol. 2009;73:1101–1114. doi: 10.1111/j.1365-2958.2009.06836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bessen RA, Marsh RF. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J Virol. 1992;66:2096–2101. doi: 10.1128/jvi.66.4.2096-2101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ayers JI, Schutt CR, Shikiya RA, Aguzzi A, Kincaid AE, Bartz JC. The strain-encoded relationship between PrP replication, stability and processing in neurons is predictive of the incubation period of disease. PLoS Path. 2011;7:1001317. doi: 10.1371/journal.ppat.1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shikiya RA, Ayers JI, Schutt CR, Kincaid AE, Bartz JC. Coinfecting prion strains compete for a limiting cellular resource. J Virol. 2010;84:5706–5714. doi: 10.1128/JVI.00243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 87.Dickinson AG, Fraser H, Outram GW. Scrapie incubation time can exceed natural lifespan. Nature. 1975;256:732–733. doi: 10.1038/256732a0. [DOI] [PubMed] [Google Scholar]
- 88.Race R, Raines A, Raymond GJ, Caughey B, Chesebro B. Long-term subclinical carrier state precedes scrapie replication and adaptation in a resistant species: analogies to bovine spongiform encephalopathy and variant Creutzfeldt-Jakob disease in humans. J Virol. 2001;75:10106–10112. doi: 10.1128/JVI.75.21.10106-10112.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Safar JG, Kellings K, Serban A, Groth D, Cleaver JE, Prusiner SB, et al. Search for a prion-specific nucleic acid. J Virol. 2005;79:10796–10806. doi: 10.1128/JVI.79.16.10796-10806.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Outram GW. The pathogenesis of scrapie in mice. In: Kimberlin RH, editor. Slow Virus Diseases of Animals and Man. Amsterdam: North-Holland; 1976. pp. 325–357. [Google Scholar]
- 91.Safar JG, DeArmond SJ, Kociuba K, Deering C, Didorenko S, Bouzamondo-Bernstein E, et al. Prion clearance in bigenic mice. J Gen Virol. 2005;86:2913–2923. doi: 10.1099/vir.0.80947-0. [DOI] [PubMed] [Google Scholar]
- 92.Mallucci G, Dickinson A, Linehan J, Klohn PC, Brandner S, Collinge J. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 93.Mallucci GR, White MD, Farmer M, Dickinson A, Khatun H, Powell AD, et al. Targeting cellular prion protein reverses early cognitive deficits and neurophysiological dysfunction in prion-infected mice. Neuron. 2007;53:325–335. doi: 10.1016/j.neuron.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 94.Pfeifer A, Eigenbrod S, Al-Khadra S, Hofmann A, Mitteregger G, Moser M, et al. Lentivector-mediated RNAi efficiently suppresses prion protein and prolongs survival of scrapie-infected mice. J Clin Invest. 2006;116:3204–3210. doi: 10.1172/JCI29236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.White MD, Farmer M, Mirabile I, Brandner S, Collinge J, Mallucci GR. Single treatment with RNAi against prion protein rescues early neuronal dysfunction and prolongs survival in mice with prion disease. Proc Natl Acad Sci USA. 2008;105:10238–10243. doi: 10.1073/pnas.0802759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goldmann W, Chong A, Foster J, Hope J, Hunter N. The shortest known prion protein gene allele occurs in goats, has only three octapeptide repeats and is non-pathogenic. J Gen Virol. 1998;79:3173–3176. doi: 10.1099/0022-1317-79-12-3173. [DOI] [PubMed] [Google Scholar]
- 97.Dickinson AG, Stamp JT, Renwick CC, Rennie JC. Some factors controlling the incidence of scrapie in Cheviot sheep injected with a Cheviot-passaged scrapie agent. J Comp Path. 1968;78:313–321. doi: 10.1016/0021-9975(68)90007-8. [DOI] [PubMed] [Google Scholar]
- 98.Hunter N, Moore L, Hosie BD, Dingwall WS, Greig A. Association between natural scrapie and PrP genotype in a flock of Suffolk sheep in Scotland. Vet Rec. 1997;140:59–63. doi: 10.1136/vr.140.3.59. [DOI] [PubMed] [Google Scholar]
- 99.Laplanche JL, Delasnerie-Laupretre N, Brandel JP, Chatelain J, Beaudry P, Alperovitch A, et al. Molecular genetics of prion diseases in France. French research group on epidemiology of human spongiform encephalopathies. Neurology. 1994;44:2347–2351. doi: 10.1212/wnl.44.12.2347. [DOI] [PubMed] [Google Scholar]
- 100.Belt PB, Muileman IH, Schreuder BE, Bos-de Ruijter J, Gielkens AL, Smits MA. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. J Gen Virol. 1995;76:509–517. doi: 10.1099/0022-1317-76-3-509. [DOI] [PubMed] [Google Scholar]
- 101.Maciulis A, Hunter N, Wang S, Goldmann W, Hope J, Foote WC. Polymorphisms of a scrapie-associated fibril protein (PrP) gene and their association with susceptibility to experimentally induced scrapie in Cheviot sheep in the United States. Am J Vet Res. 1992;53:1957–1960. [PubMed] [Google Scholar]
- 102.Dickenson AG, Outram GW. Genetic aspects of unconventional virus infections: the basis fo the virino hypothesis. Ciba Foundation Symposium. 1988;135:63–83. doi: 10.1002/9780470513613.ch5. [DOI] [PubMed] [Google Scholar]
- 103.Bossers A, de Vries R, Smits MA. Susceptibility of sheep for scrapie as assessed by in vitro conversion of nine naturally occurring variants of PrP. J Virol. 2000;74:1407–1414. doi: 10.1128/jvi.74.3.1407-1414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shibuya S, Higuchi J, Shin RW, Tateishi J, Kitamoto T. Codon 219 Lys allele of PRNP is not found in sporadic Creutzfeldt-Jakob disease. Ann Neurol. 1998;43:826–828. doi: 10.1002/ana.410430618. [DOI] [PubMed] [Google Scholar]
- 105.Hizume M, Kobayashi A, Teruya K, Ohashi H, Ironside JW, Mohri S, et al. Human prion protein (PrP) 219K is converted to PrPSc but shows heterozygous inhibition in variant Creutzfeldt-Jakob disease infection. J Biol Chem. 2009;284:3603–3609. doi: 10.1074/jbc.M809254200. [DOI] [PubMed] [Google Scholar]
- 106.Parchi P, Castellani R, Capellari S, Ghetti B, Young K, Chen SG, et al. Molecular basis of phenotypic variability in sporadic Creutzfeldt-Jakob disease. Ann Neurol. 1996;39:767–778. doi: 10.1002/ana.410390613. [DOI] [PubMed] [Google Scholar]
- 107.Parchi P, Giese A, Capellari S, Brown P, Schulz-Schaeffer W, Windl O, et al. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann Neurol. 1999;46:224–233. [PubMed] [Google Scholar]
- 108.Collinge J, Sidle KC, Meads J, Ironside J, Hill AF. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature. 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 109.Hill AF, Joiner S, Wadsworth JD, Sidle KC, Bell JE, Budka H, et al. Molecular classification of sporadic Creutzfeldt-Jakob disease. Brain. 2003;126:1333–1346. doi: 10.1093/brain/awg125. [DOI] [PubMed] [Google Scholar]
- 110.Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 111.Kaneko K, Zulianello L, Scott M, Cooper CM, Wallace AC, James TL, et al. Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc Natl Acad Sci USA. 1997;94:10069–10074. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zulianello L, Kaneko K, Scott M, Erpel S, Han D, Cohen FE, et al. Dominant-negative inhibition of prion formation diminished by deletion mutagenesis of the prion protein. J Virol. 2000;74:4351–4360. doi: 10.1128/jvi.74.9.4351-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Perrier V, Kaneko K, Safar J, Vergara J, Tremblay P, DeArmond SJ, et al. Dominant-negative inhibition of prion replication in transgenic mice. Proc Natl Acad Sci USA. 2002;99:13079–13084. doi: 10.1073/pnas.182425299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Crozet C, Lin Y, Mettling C, Mourton-Gilles C, Corbeau P, Lehmann S, et al. Inhibition of PrPSc formation by lentiviral gene transfer of PrP containing dominant negative mutants. J Cell Sci. 2004;117:5591–5597. doi: 10.1242/jcs.01484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Furuya K, Kawahara N, Yamakawa Y, Kishida H, Hachiya NS, Nishijima M, et al. Intracerebroventricular delivery of dominant negative prion protein in a mouse model of iatrogenic Creutzfeldt-Jakob disease after dura graft transplantation. Neurosci Lett. 2006;402:222–226. doi: 10.1016/j.neulet.2006.03.062. [DOI] [PubMed] [Google Scholar]
- 116.Toupet K, Compan V, Crozet C, Mourton-Gilles C, Mestre-Frances N, Ibos F, et al. Effective gene therapy in a mouse model of prion diseases. PLoS One. 2008;3:2773. doi: 10.1371/journal.pone.0002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Atarashi R, Sim VL, Nishida N, Caughey B, Katamine S. Prion strain-dependent differences in conversion of mutant prion proteins in cell culture. J Virol. 2006;80:7854–7862. doi: 10.1128/JVI.00424-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ott D, Taraborrelli C, Aguzzi A. Novel dominant-negative prion protein mutants identified from a randomized library. Protein Eng Des Sel. 2008;21:623–629. doi: 10.1093/protein/gzn042. [DOI] [PubMed] [Google Scholar]
- 119.Prusiner S, Scott M, Foster D, Pan K, Groth D, Mirenda C, et al. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 120.Scott MR, Kohler R, Foster D, Prusiner SB. Chimeric prion protein expression in cultured cells and transgenic mice. Protein Sci. 1992;1:986–997. doi: 10.1002/pro.5560010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Prusiner SB, Groth D, Serban A, Koehler R, Foster D, Torchia M, et al. Ablation of the prion protein (PrP) gene in mice prevents scrapie and facilitates production of anti-PrP antibodies. Proc Natl Acad Sci USA. 1993;90:10608–10612. doi: 10.1073/pnas.90.22.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Telling GC, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen FE, et al. Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 123.Telling GC, Scott M, Hsiao KK, Foster D, Yang SL, Torchia M, et al. Transmission of Creutzfeldt-Jakob disease from humans to transgenic mice expressing chimeric human-mouse prion protein. Proc Natl Acad Sci USA. 1994;91:9936–9940. doi: 10.1073/pnas.91.21.9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Korth C, Kaneko K, Groth D, Heye N, Telling G, Mastrianni J, et al. Abbreviated incubation times for human prions in mice expressing a chimeric mouse-human prion protein transgene. Proc Natl Acad Sci USA. 2003;100:4784–4789. doi: 10.1073/pnas.2627989100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Geoghegan JC, Miller MB, Kwak AH, Harris BT, Supattapone S. Trans-dominant inhibition of prion propagation in vitro is not mediated by an accessory cofactor. PLoS Path. 2009;5:1000535. doi: 10.1371/journal.ppat.1000535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lee CI, Yang Q, Perrier V, Baskakov IV. The dominant-negative effect of the Q218K variant of the prion protein does not require protein X. Protein Sci. 2007;16:2166–2173. doi: 10.1110/ps.072954607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Horiuchi M, Priola SA, Chabry J, Caughey B. Interactions between heterologous forms of prion protein: binding, inhibition of conversion and species barriers. Proc Natl Acad Sci USA. 2000;97:5836–5841. doi: 10.1073/pnas.110523897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Priola SA. Prion protein and species barriers in the transmissible spongiform encephalopathies. Biomed Pharmacother. 1999;53:27–33. doi: 10.1016/s0753-3322(99)80057-2. [DOI] [PubMed] [Google Scholar]
- 129.Priola SA, Caughey B, Race RE, Chesebro B. Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J Virol. 1994;68:4873–4878. doi: 10.1128/jvi.68.8.4873-4878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Masel J, Jansen VA. Designing drugs to stop the formation of prion aggregates and other amyloids. Biophys Chem. 2000;88:47–59. doi: 10.1016/s0301-4622(00)00197-6. [DOI] [PubMed] [Google Scholar]
- 131.DePace AH, Santoso A, Hillner P, Weissman JS. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 132.Doel SM, McCready SJ, Nierras CR, Cox BS. The dominant PNM2-mutation which eliminates the psi factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics. 1994;137:659–670. doi: 10.1093/genetics/137.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Young C, Cox B. Extrachromosomal elements in a super-suppression system of yeast I. A nuclear gene controlling the inheritance of the extrachromosomal elements. Heredity. 1971;26:413–422. [Google Scholar]
- 134.DiSalvo S, Derdowski A, Pezza JA, Serio TR. Dominant-negative prion mutants induce curing through distinct pathways that promote chaperone-mediated aggregate disassembly. Nature Struct Mol Biol. 2011;18:486–492. doi: 10.1038/nsmb.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bossers A, Belt P, Raymond GJ, Caughey B, de Vries R, Smits MA. Scrapie susceptibility-linked polymorphisms modulate the in vitro conversion of sheep prion protein to protease-resistant forms. Proc Natl Acad Sci USA. 1997;94:4931–4936. doi: 10.1073/pnas.94.10.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kochneva-Pervukhova NV, Paushkin SV, Kushnirov VV, Cox BS, Tuite MF, Ter-Avanesyan MD. Mechanism of inhibition of [PSI+] prion determinant propagation by a mutation of the N-terminus of the yeast Sup35 protein. EMBO J. 1998;17:5805–5810. doi: 10.1093/emboj/17.19.5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Osherovich LZ, Cox BS, Tuite MF, Weissman JS. Dissection and design of yeast prions. PLoS Biol. 2004;2:86. doi: 10.1371/journal.pbio.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Luheshi LM, Dobson CM. Bridging the gap: from protein misfolding to protein misfolding diseases. FEBS Lett. 2009;583:2581–2586. doi: 10.1016/j.febslet.2009.06.030. [DOI] [PubMed] [Google Scholar]
- 139.Bellotti V, Chiti F. Amyloidogenesis in its biological environment: challenging a fundamental issue in protein misfolding diseases. Curr Opin Struct Biol. 2008;18:771–779. doi: 10.1016/j.sbi.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 140.Verges KJ, Smith MH, Toyama BH, Weissman JS. Strain conformation, primary structure and the propagation of the yeast prion [PSI+] Nat Struct Mol Biol. 2011;18:493–499. doi: 10.1038/nsmb.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]