Abstract
Highly pathogenic avian influenza (HPAI) H5N1 virus persists in Asia, posing a threat to poultry, wild birds, and humans. Previous work in Southeast Asia demonstrated that HPAI H5N1 risk is related to domestic ducks and people. Other studies discussed the role of migratory birds in the long distance spread of HPAI H5N1. However, the interplay between local persistence and long-distance dispersal has never been studied. We expand previous geospatial risk analysis to include South and Southeast Asia, and integrate the analysis with migration data of satellite-tracked wild waterfowl along the Central Asia flyway. We find that the population of domestic duck is the main factor delineating areas at risk of HPAI H5N1 spread in domestic poultry in South Asia, and that other risk factors, such as human population and chicken density, are associated with HPAI H5N1 risk within those areas. We also find that satellite tracked birds (Ruddy Shelduck and two Bar-headed Geese) reveal a direct spatio-temporal link between the HPAI H5N1 hot-spots identified in India and Bangladesh through our risk model, and the wild bird outbreaks in May–June–July 2009 in China (Qinghai Lake), Mongolia, and Russia. This suggests that the continental-scale dynamics of HPAI H5N1 are structured as a number of persistence areas delineated by domestic ducks, connected by rare transmission through migratory waterfowl.
Electronic supplementary material
The online version of this article (doi:10.1007/s10393-010-0672-8) contains supplementary material, which is available to authorized users.
Keywords: avian influenza, epidemiology, disease ecology, migration
Introduction
Since 2003, highly pathogenic avian influenza (HPAI) H5N1 has caused outbreaks in poultry and wild birds across Asia, Europe, and Africa, and persisted over several years in many countries (Liu et al., 2005; Alexander, 2007). Compared to epizootics caused by other subtypes, HPAI H5N1 showed a capacity to spread on continental and subcontinental scales. The success of the HPAI H5N1 virus, when compared to other HPAI subtypes, may result from its capacity to persist and evolve in localized areas (Li et al., 2004) and to be transmitted over long distances.
Several conditions that promote long-term, local persistence have been identified. First, controlling HPAI H5N1 virus is difficult in the presence of millions of domestic ducks in rural landscapes. Domestic ducks show fewer clinical signs of disease than chickens, and can shed virus up to 17 days (Hulse-Post et al., 2005). Vaccination-based control strategies are difficult to implement in ducks because they have longer production cycles and lower vaccine efficiency (compared to chickens), requiring repeated injections throughout the animal’s life (Tian et al., 2005). Second, poor veterinary services and low bio-security measures in farms, wet markets, and along market chains are other important factors contributing to HPAI H5N1 virus persistence in South Asia (Sims, 2007). Third, even in the absence of those conditions, new introduction from endemically infected countries through regular poultry trade (Gauthier-Clerc et al., 2007), smuggling (Van Borm et al., 2005), contaminated fomites (Yamamoto et al., 2008), or migratory birds contributes to HPAI H5N1 reintroduction and persistence (Ducatez et al., 2006; Starick et al., 2008; Globig et al., 2009).
Another unique feature of HPAI H5N1 virus is the spill-over from domestic poultry to wild birds. HPAI H5N1 can be highly pathogenic to many wild bird species, for example, wild bird outbreaks in Qinghai Lake, China in May 2005 (Chen et al., 2005; Liu et al., 2005), and it can also cause subclinical infections in others (Keawcharoen et al., 2008). Several field studies have looked at detailed migration patterns (Gaidet et al., 2008; Yamaguchi et al., 2008; Prosser et al., 2009; Takekawa et al., 2010), and experimental infection studies have examined the clinical impact and asymptomatic shedding of HPAI H5N1 for a range of species (Brown et al., 2008; Keawcharoen et al., 2008; Kwon and Swayne, 2008), which supported the hypothesis that wild waterfowl may be involved in long-distance transmission of the virus (Kalthoff et al., 2008).
A hypothesis is that the global persistence of HPAI H5N1 results from the interplay between the high capacity to persist in domestic poultry in localized areas and sporadic long-distance introduction events through migratory birds. To date, this hypothesis has never been presented with supporting and integrated empirical data. Here, we studied those two processes, i.e., persistence and long-distance spread, using two approaches: (1) quantifying and mapping HPAI H5N1 risks in South Asia (India, Bangladesh, and Myanmar) and Southeast Asia (Thailand, Cambodia, Laos, and Vietnam); and (2) tracking wild bird migration from regions identified at risk in South Asia through western China to Mongolia and Russia (Central Asia flyway) by satellite telemetry.
Materials and Methods
Ethics Statement
Bird capture and marking was conducted under the U.S. Geological Survey (USGS) Western Ecological and Patuxent Wildlife Research Center’s Animal Care and Use Committee Review.
Epidemiological Data
All data on HPAI H5N1 outbreaks were extracted from the EMPRES-I database maintained by the Food and Agriculture Organization of the United Nations, except data on HPAI H5N1 outbreaks prior to December 2005 in Thailand and Vietnam that originated from the Thailand Department of Livestock Development (DLD, Bangkok, Thailand) and the Vietnam Department of Animal Health (DAH, Hanoi, Vietnam), respectively. Table S1 presents the sources of poultry data used in the region to derive the layers of chicken and duck density. Human population data were extracted from the GRUMP database (CIESIN et al., 2005). Data on average cropping intensity were derived from annual maps of cropping intensity and paddy rice agriculture, at 500-m spatial resolution, in 2005 using satellite images from the Moderate Resolution Imaging Spectroradiometer (MODIS) onboard the NASA Terra platform (Biradar and Xiao, 2010). Elevation data was obtained from the GTOPO30 database (LDAAC, 2004). All data were resampled or aggregated at a spatial resolution of 0.04166 decimal degrees (0.041666 dd is approximately 5 km at equator). HPAI H5N1 outbreak data were converted into presence or absence in each pixel, animal and human density data were converted into a number per pixel, and the original 500-m resolution cropping intensity data were aggregated by computing the mean number of cropping season in each 0.04166 dd pixel. Outbreak data from Thailand and Vietnam were all geolocated at the subdistrict and commune level, respectively (median size 16 km2). In the other countries, a proportion of the outbreaks were located in EMPRES-I at the village location level, and a fraction was geolocated at the administrative level 3 level. The fraction of allocation to an exact location represented 44%, 100%, 87%, 77%, and 100% for Bangladesh, Cambodia, India, Laos, and Myanmar, respectively.
Wildbird Capture and Satellite Telemetry
Captures of wild waterfowl were made with monofilament nooses, or by herding birds into corrals during wing molt, when many waterfowl are flightless. We fit birds with solar-powered Platform Terminal Transmitters (PTTs; Microwave Telemetry Inc., Columbia, MD). The transmitters weighed 9.5–45 g, and some were equipped with internal GPS receivers, solar panels, temperature and voltage sensors, and external antennas. Transmitters were attached dorsally with Teflon ribbon (Bally Ribbon Mills, Bally, PA) tied with knots secured by super glue (Henkel Loctite Corp., Rocky Hill, CT).
In Qinghai Lake, China, we marked 14 pre-breeding Bar-headed Geese (Anser indicus) in March–April 2007, and 12 postbreeding Ruddy Shelducks (Tadorna ferruginea) in September 2007 (before fall migration starts). We also marked 15 Bar-headed Geese and 4 Ruddy Shelducks in spring 2008 and 10 Ruddy Shelducks in fall 2008. We used 30 g and 45 g backpack GPS Argos solar transmitters (Microwave Telemetry, Inc.). Bar-headed Geese and Ruddy Shelducks were selected for marking, because these species comprised the largest numbers of waterfowl that died in the 2005 outbreak at Qinghai Lake. Tracheal and cloacal swab samples were taken for each bird and stored at 4°C before being frozen in liquid nitrogen (maximum 3 hours postcollection) to test for influenza A. All the 55 birds appeared healthy, showing no symptoms of influenza. Twenty-two of 29 marked Bar-headed Geese and 24 of 26 marked Ruddy Shelduck were sampled for avian influenza virus (cloacal and tracheal swabs, blood serology) following standard procedures (FAO, 2007). Virology analyses, conducted by the Chinese Academy of Sciences, Wuhan Institute of Virology, included: (1) type A influenza with an ELISA test (Optical Density 630 above 0.23 as positive); (2) H5 subtype with RT-PCR (Fouchier et al., 2000); and (3) H5, H7, H9, and H10 antibodies with hemagglutinin inhibition (OIE, 2004). One of the samples was positive for influenza A virus through ELISA, and none were positives by RT-PCR.
In India, 15 Bar-headed Geese, 4 Ruddy Shelducks, and 41 individuals of other 8 duck species were marked at Chilika Lagoon and Koonthankulum Reserves during December 2008 and January 2009. All the 61 birds marked with satellite transmitters in India have tested negative through RT-PCR for the H5N1 virus, using a similar sampling procedure as the birds marked in China.
Transmitters were programmed to collect GPS locations at 2-hour intervals throughout the 24-hour day, and transmit signals to Argos satellites every 65 seconds for an 8-hour on-cycle, followed by a 48-hour off-cycle to facilitate solar recharging. Data were recovered from the Argos Data Collection and Location System (CLS America Inc., Largo, MD) via receivers aboard polar-orbiting weather satellites. CLS calculated PTT location estimates that were derived from the perceived Doppler-effect shifts in transmission frequency during a satellite overpass. The accuracy of each location was rated by class. Class G locations indicated that the position was a GPS location with mean accuracy ± 18.5 m. Conventional Argos location classes 0, 1, 2, and 3 indicated the location was derived from ≥ 4 transmissions, with 1-sigma accuracy >1000 m, 350–1000 m, 150–350 m, and ≤ 150 m, respectively. Location classes A (3 transmissions) and B (2 transmissions) are not assigned accuracy estimates by CLS, and location class Z indicates that no locations were obtained.
We compiled and validated our Doppler-derived location data using the Argos Filter Algorithm (D. Douglas, Version 7.03, http://alaska.usgs.gov/science/biology/spatial/). The filtering algorithm flags improbable locations based on user-defined distance and velocity thresholds. We used the algorithm to compile two datasets for analysis. The first dataset included one location per duty cycle, based on the highest-accuracy location class. Quality of signals was judged first on the basis of location class (G > 3 > 2 > 1 > 0 > A > B), and then by indices of residual frequency error. Our primary interest for compiling a dataset with one location per duty cycle was to evaluate broad-scale migration routes and movement chronology. For finer scale spatial analyses, we constructed a comprehensive dataset that included all GPS locations and Argos readings of class 1 or better.
Analysis
We used an autologistic multiple regression model to analyze the relationship between HPAI H5N1 presence and the sets of predictors (Gilbert et al., 2008). This method was chosen so that our results could be compared with previous work carried out in Southeast Asia (Gilbert et al., 2008). The autologistic approach adds an autoregressive term to the standard multiple logistic regression to account for spatial autocorrelation in the response variable (Augustin et al., 1996). In addition, low prevalence values for the response variable (<10%) tend to bias multiple logistic regression performance metrics (McPherson et al., 2004). For each model, we selected all HPAI H5N1 outbreak present pixels in addition to nine randomly selected negative pixels, and bootstrapped this operation 250 times so that the obtained model parameters and performance metrics would not depend on a particular set of negatives. For each model, and each set of negatives, we estimated the coefficient of each variable and their corresponding odd-ratio, the χ2 resulting from log-likelihood ratio tests between the full model and a model with each variable removed. As indicators of goodness-of-fit, we estimated the area under the curve (AUC) of the receiver-operating characteristic (ROC) plots, Cohen’s Kappa index (Cohen, 1960) with a threshold probability of 0.9 to be classified as positive, and the correlation coefficient between observed presence/absence and fitted values (Elith et al., 2006). Although the use of AUC as a single measure of goodness-of-fit in distribution modeling has been recently questioned (Lobo et al., 2008), it was used in this study because we needed to be able to compare the accuracy of models based on very different prevalence in the training set. It should, however, be interpreted cautiously, as we compare it for analyses over different extents. We then averaged these estimates over the 250 bootstraps. The autologistic approach allows obtaining unbiased estimates of each model variable coefficient and significance, but it tends to artificially inflate the autologistic model performance metrics index because it incorporates information on the neighboring sites in the same data set. Therefore, the individual variable coefficient and log-likelihood ratio tests were estimated based on an autologistic model, while the AUC, Kappa, and correlation coefficient were estimated based on the same model without the autoregressive term.
Results
The first occurrence of HPAI H5N1 outbreak in South Asia was reported in India in early 2006 (Dubey et al., 2009) in the western part of the country and was quickly contained (Fig. 1a). In 2007, H5N1 outbreaks were reported in Bangladesh (n outbreaks = 69) and in surrounding India states (n = 4), and the virus reoccurred in those two regions in 2008 (n = 288) and in May 2009 (n = 38). Our previous studies of HPAI H5N1 risk in Southeast Asia showed that domestic duck density, cropping intensity, human population, and chicken density were important risk factors (Gilbert et al., 2008). When applied to South Asia, the previous risk model correctly predicted high risk in Bangladesh but tended to over-predict areas at risk in India (see Fig. S1B). In Southeast Asia, cropping intensity was heavily weighted in the model because it is a good indicator of domestic duck distribution. A strong association between duck, paddy rice, and double cropping occurs in Southeast Asia (Gilbert et al., 2007), and an identical association is apparent in Bangladesh and surrounding regions of northeastern India (Hossain et al., 2005). However, cropping intensity does not closely relate to duck distribution in other parts of India where vegetarian diets dominate (see Fig. S1), and hence, cropping intensity has a comparatively lower importance as a risk indicator. What remained was that the distribution of domestic ducks, one of the most important risk factors in the Southeast Asia Model, appeared to match the distribution of HPAI H5N1 outbreak cases in Bangladesh and India (Fig. 1a,b). Exploratory analysis showed that incremental increases in duck density were consistently associated with incremental HPAI H5N1 risk in Southeast Asia, but HPAI H5N1 risk quickly reaches a plateau as duck density increases in South Asia (Fig. S2). This prompted the development of a new model where the effect of risk factors was considered separately for regions where duck farming was present at relatively high densities (>20 ducks/km2), and for regions where it was considered low or absent (≤ 20 ducks/km2). Two spatial epidemiological models were developed: one with combined H5N1 outbreak data from India, Bangladesh, Myanmar, Thailand, Cambodia, Laos, and Vietnam (South and Southeast Asia Model), and a second one using only data from India, Bangladesh, and Myanmar (South Asia Model).
Fig. 1.
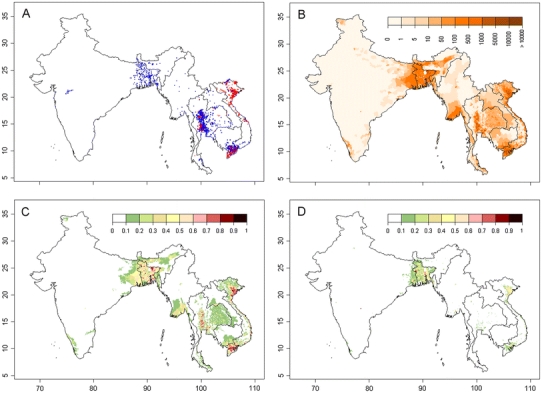
Distribution of outbreaks, duck density, and HPAI H5N1 presence probability predicted by two risk models. a: Distribution of HPAI H5N1 virus outbreaks in Bangladesh, India, Myanmar, Thailand, Cambodia, Laos, and Vietnam (data for Thailand and Vietnam differentiate records from the second wave [in blue] from records that took place afterward [in red]). b: Distribution of domestic duck density (heads/km2). c, d: Predicted probability of HPAI H5N1 virus presence by the South and Southeast Asia Model, and the South Asia Model, respectively.
The South and Southeast Asia Model showed a very good capacity to discriminate HPAI H5N1 presence and absence over all seven countries (Table 1), and highly significant risk factors included chicken density, duck density, human population density, and high versus low duck farming areas (Table 2). Four distinct subregions at risk of HPAI H5N1 were identified (Fig. 1c): Bangladesh and northeastern India, southern Myanmar, the central plain of Thailand, and the Mekong and Red River Deltas in Vietnam. The South Asia Model compares well to the combined model except that the continuous variable of duck density was no longer identified as a significant risk factor. However, duck versus no-duck farming remained a highly significant factor (Table 2), and details of model parameters (see Table S2) showed that chicken density in South Asia was a significant risk factor only when duck farming was present at relatively high densities (>20 ducks /km2; significant interaction term). The distribution of HPAI H5N1 predicted by the second model is concentrated in Bangladesh and northeastern India, the Mekong and Red River Deltas in Vietnam, and to a lesser extent in the central plain of Thailand and in Myanmar.
Table 1.
Mean Area Under Curve (AUC), Cohen’s Kappa (κ), and Correlation Coefficient (r) of 250 Bootstrapped Autologistic Regression Models (the South and Southeast Asia Model, and the South Asia Model), when applied to all countries, India and Bangladesh, Thailand and Vietnama
| Model | All countries | India and Bangladesh | Thailand and Vietnamb | Thailand and Vietnamc | |
|---|---|---|---|---|---|
| South and Southeast Asia | AUC | 0.924 ± 0.003 | 0.916 ± 0.011 | 0.837 ± 0.006 | 0.861 ± 0.010 |
| κ | 0.413 ± 0.005 | 0.223 ± 0.012 | 0.358 ± 0.014 | 0.341 ± 0.023 | |
| r | 0.620 ± 0.006 | 0.398 ± 0.018 | 0.578 ± 0.011 | 0.608 ± 0.016 | |
| South Asia | AUC | 0.872 ± 0.012 | 0.935 ± 0.009 | 0.804 ± 0.012 | 0.858 ± 0.011 |
| κ | 0.387 ± 0.017 | 0.382 ± 0.016 | 0.305 ± 0.022 | 0.536 ± 0.027 | |
| r | 0.423 ± 0.012 | 0.504 ± 0.022 | 0.375 ± 0.012 | 0.564 ± 0.023 |
aModels average coefficients are presented in Table S1.
bEstimated using HPAI H5N1 records from July 2004–2005.
cEstimated using HPAI H5N1 records from 2006–present.
Table 2.
Mean χ2 resulting from log-likelihood ratio tests applied to 250 bootstrapped autologistic regression models upon removal of each variable, presented for the South and Southeast Asia Model and for the South Asia Model, respectivelya
| Term dropped | χ2 | df | P | Significanceb |
|---|---|---|---|---|
| South and Southeast Asia Model | ||||
| Chicken density (Log10) | 35.706 ± 9.414 | 2 | <0.001 | *** |
| Duck density (Log10) | 50.756 ± 12.851 | 2 | <0.001 | *** |
| Human population density (Log10) | 15.245 ± 4.252 | 2 | <0.001 | *** |
| No. of crop cycles | 6.760 ± 4.018 | 2 | 0.0341 | * |
| Presence of duck farming (>20 ducks/km2) | 224.336 ± 33.287 | 6 | <0.001 | *** |
| South Asia Model | ||||
| Chicken density (Log10) | 18.707 ± 8.576 | 2 | <0.001 | *** |
| Duck density (Log10) | 2.836 ± 3.025 | 2 | 0.2422 | ns |
| Human population density (Log10) | 56.829 ± 11.340 | 2 | <0.001 | *** |
| No. of crop cycles | 6.464 ± 4.331 | 2 | 0.0395 | * |
| Presence of duck farming (>20 ducks/km2) | 56.280 ± 15.023 | 6 | <0.001 | *** |
aModels average coefficients are presented in Table S2.
b* significant, *** very highly significant, ns nonsignificant.
The wild waterfowl marked with satellite transmitters along the Central Asia flyway began their spring migration in March, and converged at the Qinghai Lake region in April–May with several birds continuing north to Mongolia (Fig. 2a). Among the birds that had been marked, two Bar-headed Geese and one Ruddy Shelduck spent time in Bangladesh in the “hot-spot” (high risk) areas identified by our risk model during the timeframe when several HPAI H5N1 outbreaks in poultry were reported (Fig. 2b,c).
Fig. 2.
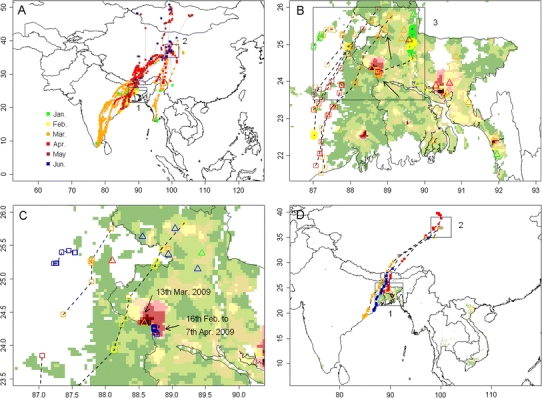
Movements of satellite-tracked wild waterfowl along the Central Asia flyway, and in India and Bangladesh. a: Distribution of all satellite-tracked wild waterfowl along the Central Asia flyways in 2009 color-coded by month (1 marks the region displayed in panel b, and 2 marks the Qinghai Lake region, China; credit to FAO–USGS Avian Influenza Program). b: Distribution of satellite-tracked wild waterfowl (open squares) and HPAI H5N1 virus outbreaks (open triangles) in northeastern India and Bangladesh in 2009, color-coded by month. Two points of coexistence of wild waterfowl and HPAI H5N1 outbreaks are indicated by the arrows, the black rectangle (labeled as 3) marks the region displayed in panel c. c: Same as panel b, with different color coding (green, yellow, orange, red, brown, blue for March 1–4, 5–9, 10–14, 15–19, 20–24, 25–31, respectively). d: Movement recorded for a Ruddy Shelduck and two Bar-headed Geese from March to June 2009 (1 marks the region displayed in panel b, and 2 marks the Qinghai Lake region, China). Background color in panels b and c presents the distribution of HPAI H5N1 presence probability predicted by the South Asia Model, with the same legend as in Figure 1c and d.
The Ruddy Shelduck spent more than 7 weeks, between February 16 and April 7, in the area predicted as highest risk for HPAI H5N1 presence by our risk model (Fig. 2c), at a distance <20 km from an HPAI H5N1 poultry outbreak reported on March 13, 2009. The migration route of two Bar-headed Geese passed near the poultry outbreak locations, but in this case, the HPAI H5N1 poultry outbreaks were reported 9 and 16 days after the birds’ presence. These three birds then undertook their northward migration toward Qinghai Lake (Fig. 2d). The Ruddy Shelduck arrived after 3 days at the Qinghai Lake region of China on April 9, 2009 (Fig. S3). The short duration of its migration in comparison to the duration of excretion reported in experimentally infected ducks (Brown et al., 2008; Latorre-Margalef et al., 2009) would allow for introduction of the virus from Bangladesh to China. A month later in May 2009, a new outbreak of HPAI H5N1 in wild birds was reported in the Qinghai Lake area of China. In comparison with the Ruddy Shelduck, the two Bar-headed Geese took a much longer period (39–50 days) to migrate from Bangladesh, and stayed only briefly at the Qinghai Lake area of China.
We summarized wild bird outbreaks reported in the OIE database from 2005–2009 in the region (Table S3; http://www.oie.int/downld/AVIAN%20INFLUENZA/A2009_AI.php). The pattern of outbreaks was very similar in 2005 and 2009, spreading from southern poultry farming areas in South Asia to areas with little poultry in the northern Central Asia flyway in China, Mongolia, and then Siberia. On July 27, 2009, an HPAI H5N1 wild bird outbreak was reported at Doroo Lake (Doroo nuur), a freshwater lake in central Mongolia. Thirty birds including Bar-headed Geese, Ruddy Shelduck, Whooper Swan (Cynus cygnus), Common Goldeneye (Bucephala clangula), and Mongolian Gulls (Larus vegae mongolicus) were found sick or dead. Two of the mortalities included Bar-headed Geese captured and banded by our research team in July 2008 at Terkhiin Tsagaan Lake, located 150 km to the southwest. Samples were taken, analyzed, and compared to previous ones circulating in Asia (Kang et al., 2010; Sakoda et al., 2010), but unfortunately the comparison did not include any isolates from South Asia.
Discussion
Our results reveal a direct spatio-temporal link between areas identified at highest HPAI H5N1 risk in India and Bangladesh, in March 2009, and the wild bird outbreaks, in May through July 2009, in China (Qinghai Lake region), Mongolia, and Russia.
The spatial prediction of HPAI H5N1 risk was comparatively less strongly related to duck density in South Asia than it was in Southeast Asia. A strong association between duck and rice farming is observed in Southeast Asia (Gilbert et al., 2007), and an identical association is apparently found in Bangladesh and surrounding regions of northeastern India (Hossain et al., 2005). However, the production systems may differ. Duck and poultry production in Bangladesh is ubiquitous, and most small farmers keep ducks under a family subsistence level of management (Das et al., 2008). In contrast, duck production is far more intensified in terms of flock size, movements, and trade in Thailand and Vietnam, with some flocks being moved over 100-km by trucks (Songserm et al., 2006). Duck movement and active trade of products may have contributed to regional HPAI H5N1 spread in Southeast Asia (Thailand and Vietnam) during the first two waves of epidemics, which would explain the higher statistical relationship between duck density and HPAI H5N1 presence as compared to South Asia. The South Asia Model provided highest predictive capacity for India and Bangladesh (Table 1 and Fig. 1d), but also performed well on the Thailand and Vietnam 2006 outbreaks (Table 1 and Fig. 1; compare red dots in Fig. 1a to predicted risk in Fig. 1d), a period when Thailand focused on active surveillance, pre-movement testing, and restricted movements of free-grazing ducks, while Vietnam implemented massive vaccination in chicken and duck populations. We propose that domestic ducks merely act as a reservoir of HPAI H5N1 in South Asia and are less implicated in spread, compared to in Southeast Asia prior to control targeted on ducks. This may explain why a minimal density of domestic ducks is required for persistence, but that their abundance does not proportionally relate to HPAI H5N1 risk.
In this study, we used an autologistic regression approach to compare the analysis of Bangladesh and India data to previously published similar models based on Thailand and Vietnam data. Alternative approaches have been used to model the distribution of HPAI H5N1 in other regions (Williams et al., 2008), and distribution modeling can be achieved through many different statistical techniques (Elith et al., 2006). In particular, the logistic regression framework has its limitations for modeling nonlinearity of effect of the continuous-scale risk factors on the logit form of the outcome variable. Alternative methods such as boosted regression trees (Elith et al., 2008) of maximum entropy models should be considered in future research.
Three of the GPS-marked birds stayed for extended periods of time in, or close to, areas of high HPAI H5N1 risk in South Asia, one of them staying at a distance <20 km from a documented concurrent HPAI H5N1 outbreak, prior to undertaking their northward migration, which included stopping over in Qinghai Lake, China. We could not document that these three birds spread HPAI H5N1 from Bangladesh to Qinghai Lake area. In addition, one should note that the bird that stayed closest to a documented HPAI H5N1 outbreak was a Ruddy Shelduck. Recent inoculation experiments carried out on that species resulted in 100% mortality, with birds surviving in rather poor physical conditions, though shedding virus for several days (Kwon et al., 2010). In addition, Kwon et al. (2010) failed to detect H5N1 in a noninoculated Ruddy Shelduck that was in contact with other ones inoculated with HPAI H5N1. So, the available evidence suggests that it is quite unlikely that an HPAI H5N1-infected Ruddy Shelduck could have migrated over such long distances and transmitted the disease to conspecifics. However, daily regional movements for these species may be >100 km per day. GPS-equipped birds represent a tiny fraction of the waterfowl population; we postulate that these birds are indicative of hundreds to thousands of migratory birds that could have been in contact with outbreak sites when poultry outbreaks were taking place in Bangladesh, and could have spread the virus along their northward migration to Qinghai Lake of China and into Mongolia and Russia. On July 27, 2009, an outbreak documented in wild birds at Doroo Lake in central Mongolia, where there are no poultry, was attributed to H5N1, including two Bar-headed Geese marked by our team during the previous year. Furthermore, other waterfowl, including several duck species not studied here, migrate through these regions and may have distinct movement patterns and chronology. Species with different routes and timing may provide more opportunity than we have indicated for exchange of virus among birds or by fecal–oral transmission in wetlands used during migration.
Three conditions would have been necessary to allow this to happen. First, birds would need to be infected shortly before their departure so to arrive at a time when they were still shedding virus; second, exposed and infected birds must have maintained their ability to migrate. Third, other susceptible birds must be present when infected birds arrive at the Qinghai region or Mongolia. The first and third conditions are both plausible: Our results indicate that many birds could have departed from a high HPAI H5N1 risk area in Bangladesh or India at the time of concurrent outbreaks, some of which may have been infected. Upon arrival in Qinghai Lake or Mongolia, transmission to other birds would have been facilitated by the high densities and aggregation of birds, and the relatively cold temperatures that favor persistence of the virus in the environment (Brown et al., 2007; Rohani et al., 2009). With regard to migration capacity, recent inoculation experiments of wild waterfowls with HPAI H5N1 virus indicate a high variability in the pathobiology of infections (Kwon et al., 2010), depending on species. Only a limited number of species have been tested so far, and more detailed studies on the pathobiology of HPAI H5N1 and their capacity to transmit the virus to conspecifics of other species would shed more light on their capacity to transmit the virus over long-distances. Despite these uncertainties, given that some species of experimentally infected wild waterfowls do not become symptomatic, or are asymptomatic for days (3–7 days) before becoming symptomatic, one cannot exclude that exposed wild birds of less susceptible species could make long migratory movements of hundreds to thousands of kilometers (Gaidet et al., 2010). Furthermore, the individual ability to perform long-distance migration may be less critical to virus spread when large numbers of waterfowl are migrating along the same route and may infect each other in a series of infection cycles. Finally, recent information suggests that exposure to low pathogenic avian influenza viruses provides possible resistance to HPAI H5N1, making migration more easily feasible for infected birds (Kalthoff et al., 2008). We conclude that all three conditions could have been met in spring 2009 to support introduction of HPAI H5N1 to the Qinghai Lake region by migratory waterfowl from a region of high HPAI H5N1 risk in domestic poultry in South Asia. This link between HPAI H5N1 viruses circulating in South Asia, Qinghai Lake area of China, Mongolia, and Russia, (Fig. 2a, Table S3) is consistent with phylogenetic studies that report a high homology between HPAI H5N1 isolates found in northeast India in 2007 and viruses isolated from wild waterfowl in Russia, China, and Mongolia (Wallace et al., 2007; Mishra et al., 2009).
Our study highlights the potential for integrating HPAI H5N1 risk modeling with wild waterfowl migration ecology data to map and track (and potentially forecast) hot-spots, introduction, persistence, and spread of HPAI H5N1 along the Asian flyways. In this study, we failed to integrate the temporal dimension in the HPAI H5N1 risk modeling and this limits the applicability of this type of model for establishing areas and time at risk of HPAI H5N1 infection. For example, we found that a Ruddy Shelduck spent several weeks in an area at risk, but in the absence of an explicit reference to time in the model, it is only because it stayed at <20 km from a documented concurrent HPAI H5N1 outbreak that we could consider that it was at high risk of infection. Future research would hence gain from the development of space–time statistical models that could generate not only “hot-spots” but also “hot-times” predictions to be matched against wild bird space–time data.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This study was supported by grants from the U.S. National Institutes of Health (R01-TW007869), the U.S. National Aeronautics and Space Administration (NNG05GH80G), and the Food and Agriculture Organization of the United Nations (FAO/UN) AGAH-EMPRES Wildlife Unit; donations from the governments of Sweden, Canada, and the United Kingdom; the U.S. Geological Survey Avian Influenza Program, and National Science Foundation Small Grants for Exploratory Research (0713027). Additional support was also provided by the Max Planck Institute for Ornithology, Germany, and the Biotechnology and Biological Sciences Research Council, UK (BB/F015615/1). We thank J. Domenech, J. Lubroth (FAO Animal Health Service and EMPRESS Program), S. Haseltine, R. Kearney, P. Bright, S. Schwarzbach, and J. Howell of USGS for supporting this project. We thank Martin Wikelski (Max Planck Institute for Ornithology), Charles Bishop (University of Wales, Bangor), and Patrick Butler (University of Birmingham, U.K) for the contribution of transmitters and migration pathways of geese from India. Bird capture and marking was conducted under the USGS Western Ecological and Patuxent Wildlife Research Center’s Animal Care and Use Committee Review. For field assistance with wild bird studies in India, we thank S. Heath (USGS), L. Hawkes (UWales), T. McCracken (UNFAO), P. Sathiyaselvam (BNHS), Gangaiamaran (BNHS), A. George (BNHS); and in China, we thank S. Li, D. Zhang, T. Li, Y. Li, F. Lei, N. Kong, and D. Zhao. GIS-based wild bird database was managed by D.C. Douglas and W.M. Perry (USGS), and the HPAI H5N1 outbreaks and EMPRES database support were provided by C. Hamilton and J. Siembieda (FAO). The authors thank two anonymous reviewers for their insightful reviews on a previous version of the manuscript. Any use of trade, product, or firm names does not imply endorsement by the U.S. Government.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Alexander D. Summary of avian influenza activity in Europe, Asia, Africa, and Australasia, 2002–2006. Avian Diseases. 2007;51:161–166. doi: 10.1637/7602-041306R.1. [DOI] [PubMed] [Google Scholar]
- Augustin NH, Mugglestone MA, Buckland ST (1996) An autologistic model for the spatial distribution of wildlife. Journal of Applied Ecology 33:339–347
- Biradar C, Xiao X (2010) Quantifying the area and spatial distribution of double and triple-cropping croplands in India with multi-temporal MODIS data in 2005. International Journal of Remote Sensing (in press)
- Brown J, Stallknecht DE, Swayne DE. Experimental infection of swans and geese with highly pathogenic avian influenza virus (H5N1) of Asian lineage. Emerging Infectious Diseases. 2008;14:136–142. doi: 10.3201/eid1401.070740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J, Swayne D, Cooper R, Burns R, Stallknecht DE. Persistence of H5 and H7 avian influenza viruses in water. Avian Diseases. 2007;51:285–289. doi: 10.1637/7636-042806R.1. [DOI] [PubMed] [Google Scholar]
- Chen H, Smith GJD, Zhang SY, Qin K, Wang J, Li KS, et al. H5N1 virus outbreak in migratory waterfowl. Nature. 2005;436:191–192. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- CIESIN, IPFRI, CIAT (2005) Global Rural-Urban Mapping Project (GRUMP), Alpha Version. Center for International Earth Science Information Network (CIESIN), Columbia University; International Food Policy Research Institute (IPFRI); The World Bank; Centro Internacional de Agricultura Tropical (CIAT). http://sedac.ciesin.columbia.edu/gpw/global.jsp. Accessed September 2010
- Cohen J. A coefficient of agreement for nominal scales. Educational and Psychological Measurement. 1960;20:37–46. doi: 10.1177/001316446002000104. [DOI] [Google Scholar]
- Das S, Chowdhury S, Khatun M, Nishibori M, Isobe N, Yoshimura Y. Poultry production profile and expected future projection in Bangladesh. World’s Poultry Science Journal. 2008;64:99–118. doi: 10.1017/S0043933907001754. [DOI] [Google Scholar]
- Dubey S, Nagarajan S, Tosh C, Bhatia S, Krishna L. Avian influenza: a long-known disease and its current threat. Indian Journal of Animal Sciences. 2009;79:113–140. [Google Scholar]
- Ducatez M, Olinger C, Owoade A, De Landtsheer S, Ammerlaan W, Niesters H, et al. (2006) Multiple introductions of H5N1 in Nigeria—phylogenetic analysis reveals that this deadly virus first arrived in Africa from different sources. Nature 442:37 [DOI] [PubMed]
- Elith J, Graham CH, Anderson RP, Dudík M, Ferrier S, Guisan A, et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography. 2006;29:129–151. doi: 10.1111/j.2006.0906-7590.04596.x. [DOI] [Google Scholar]
- Elith J, Leathwick JR, Hastie T. A working guide to boosted regression trees. Journal of Animal Ecology. 2008;77:802–813. doi: 10.1111/j.1365-2656.2008.01390.x. [DOI] [PubMed] [Google Scholar]
- FAO (2007) Wild birds and avian influenza: an introduction to applied field research and disease sampling techniques. Food and Agriculture Organization of the UN (FAO). Rome, Italy
- Fouchier RA, Bestebroer TM, Herfst S, Van Der Kemp L, Rimmelzwaan GF, Osterhaus AD (2000) Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. Journal of Clinical Microbiology 38:4096–4101 [DOI] [PMC free article] [PubMed]
- Gaidet N, Cappelle J, Takekawa JY, Prosser DJ, Iverson SA, Douglas DC, et al. Potential spread of highly pathogenic avian influenza H5N1 by wildfowl: dispersal ranges and rates determined from large-scale satellite telemetry. Journal of Applied Ecology. 2010;47:1147–1157. doi: 10.1111/j.1365-2664.2010.01845.x. [DOI] [Google Scholar]
- Gaidet N, Newman S, Hagemeijer W, Dodman T, Cappelle J, Hammoumi S, et al. Duck migration and past influenza A (H5N1) outbreak areas. Emerging Infectious Diseases. 2008;14:1164–1166. doi: 10.3201/eid1407.071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier-Clerc M, Lebarbenchon C, Thomas F. Recent expansion of highly pathogenic avian influenza H5N1: a critical review. Ibis. 2007;149:202–214. doi: 10.1111/j.1474-919X.2007.00699.x. [DOI] [Google Scholar]
- Gilbert M, Xiao X, Chaitaweesub P, Kalpravidh W, Premashthira S, Boles S, et al. Avian influenza, domestic ducks and rice agriculture in Thailand. Agriculture Ecosystems & Environment. 2007;119:409–415. doi: 10.1016/j.agee.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M, Xiao X, Pfeiffer D, Epprecht M, Boles S, Czarnecki C, et al. (2008) Mapping H5N1 highly pathogenic avian influenza risk in Southeast Asia. Proceedings of the National Academy of Sciences of the United States of America 105:4769–4774 [DOI] [PMC free article] [PubMed]
- Globig A, Staubach C, Beer M, Koppen U, Fiedler W, Nieburg M, et al. Epidemiological and ornithological aspects of outbreaks of highly pathogenic avian influenza virus H5N1 of Asian lineage in wild birds in Germany, 2006 and 2007. Transboundary and Emerging Diseases. 2009;56:57–72. doi: 10.1111/j.1865-1682.2008.01061.x. [DOI] [PubMed] [Google Scholar]
- Hossain S, Sugimoto H, Ahmed G, Islam M. Effect of integrated rice-duck farming on rice yield, farm productivity, and rice-provisioning ability of farmers. Asian Journal of Agriculture and Development. 2005;2:79–86. [Google Scholar]
- Hulse-Post D, Sturm-Ramirez K, Humberd J, Seiler P, Govorkova E, Krauss S, et al. Role of domestic ducks in the propagation and biological evolution of highly pathogenic H5N1 influenza viruses in Asia. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10682–10687. doi: 10.1073/pnas.0504662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalthoff D, Breithaupt A, Teifke J, Globig A, Harder T, Mettenleiter T, et al. Highly pathogenic avian influenza virus (H5N1) in experimentally infected adult mute swans. Emerging Infectious Diseases. 2008;14:1267–1270. doi: 10.3201/eid1407.071468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Batchuluun D, Kim MC, Choi JG, Erdene-Ochir TO, Aek MR, et al. (2010) Genetic analyses of H5N1 avian influenza virus in Mongolia, 2009 and its relationship with those of eastern Asia. Veterinary Microbiology. 147(1–2):170–175 [DOI] [PubMed]
- Keawcharoen J, van Riel D, van Amerongen G, Bestebroer T, Beyer W, van Lavieren R, et al. Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5NI) Emerging Infectious Diseases. 2008;14:600–607. doi: 10.3201/eid1404.071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y, Swayne D. Susceptibility of five migratory aquatic birds to H5N1 highly pathogenic avian influenza virus (A/Chicken/Korea/IS/06) Poultry Science. 2008;87:164. [Google Scholar]
- Kwon YK, Thomas C, Swayne DE. Variability in pathobiology of South Korean H5N1 high-pathogenicity avian influenza virus infection for 5 species of migratory waterfowl. Veterinary Pathology. 2010;47:495–506. doi: 10.1177/0300985809359602. [DOI] [PubMed] [Google Scholar]
- Latorre-Margalef N, Gunnarsson G, Munster V, Fouchier R, Osterhaus A, Elmberg J, et al. (2009) Effects of influenza A virus infection on migrating mallard ducks. Proceedings of the Royal Society of London. Series B: Biological Sciences (London) 276:1029–1036 [DOI] [PMC free article] [PubMed]
- LDAAC (2004) Global 30 Arc—Second Elevation Data Set GTOPO30. Land Process Distributed Active Archive Center. http://eros.usgs.gov/#/Find_Data/Products_and_Data_Available/gtopo30_info. Accessed September 2010
- Li K, Guan Y, Wang J, Smith G, Xu K, Duan L, et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- Liu J, Xiao J, Lei F, Zhu Q, Qin K, Zhang X, et al. (2005) Highly pathogenic H5N1 influenza virus infection in migratory birds. Science 309:1206 [DOI] [PubMed]
- Lobo JM, Jiménez-Valverde A, Real R. AUC: a misleading measure of the performance of predictive distribution models. Global Ecology and Biogeography. 2008;17:145–151. doi: 10.1111/j.1466-8238.2007.00358.x. [DOI] [Google Scholar]
- McPherson JM, Jetz W, Rogers DJ. The effects of species’ range sizes on the accuracy of distribution models: ecological phenomenon or statistical artefact? Journal of Applied Ecology. 2004;41:811–823. doi: 10.1111/j.0021-8901.2004.00943.x. [DOI] [Google Scholar]
- Mishra A, Cherian S, Chakrabarti A, Pawar S, Jadhav S, Pal B, et al. (2009) A unique influenza A (H5N1) virus causing a focal poultry outbreak in 2007 in Manipur, India. Virology Journal 6:26 [DOI] [PMC free article] [PubMed]
- OIE . Manual of diagnostic tests and vaccines for terrestrial animals (mammals, birds and bees) Paris: Office International Des Epizooties; 2004. [PubMed] [Google Scholar]
- Prosser D, Takekawa J, Newman S, Yan B, Douglas D, Hou Y, et al. Satellite-marked waterfowl reveal migratory connection between H5N1 outbreak areas in China and Mongolia. Ibis. 2009;151:568–576. doi: 10.1111/j.1474-919X.2009.00932.x. [DOI] [Google Scholar]
- Rohani P, Breban R, Stallknecht D, Drake J (2009) Environmental transmission of low pathogenicity avian influenza viruses and its implications for pathogen invasion. Proceedings of the National Academy of Sciences of the United States of America 106:10365–10369 [DOI] [PMC free article] [PubMed]
- Sakoda Y, Sugar S, Batchluun D, Erdene-Ochir T, Okamatsu M, Isoda N, et al. Characterization of H5N1 highly pathogenic avian influenza virus strains isolated from migratory waterfowl in Mongolia on the way back from southern Asia to their northern territory. Virology. 2010;406:88–94. doi: 10.1016/j.virol.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Sims L. Lessons learned from Asian H5N1 outbreak control. Avian Diseases. 2007;51:174–181. doi: 10.1637/7637-042806R.1. [DOI] [PubMed] [Google Scholar]
- Songserm T, Jam-on R, Sae-Heng N, Meemak N, Hulse-Post D, Sturm-Ramirez K, et al. Domestic ducks and H5N1 influenza epidemic, Thailand. Emerging Infectious Diseases. 2006;12:575–581. doi: 10.3201/eid1204.051614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starick E, Beer M, Hoffmann B, Staubach C, Werner O, Globig A, et al. Phylogenetic analyses of highly pathogenic avian influenza virus isolates from Germany in 2006 and 2007 suggest at least three separate introductions of H5N1 virus. Veterinary Microbiology. 2008;128:243–252. doi: 10.1016/j.vetmic.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Takekawa JY, Newman SH, Xiao X, Prosser DJ, Spragens KA, Palm EC, et al. Migration of waterfowl in the east Asian flyway and spatial relationship to HPAI H5N1 outbreaks. Avian Diseases. 2010;54:466–476. doi: 10.1637/8914-043009-Reg.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Zhang S, Li Y, Bu Z, Liu P, Zhou J, et al. Protective efficacy in chickens, geese and ducks of an H5N1-inactivated vaccine developed by reverse genetics. Virology. 2005;341:153–162. doi: 10.1016/j.virol.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Van Borm S, Thomas I, Hanquet G, Lambrecht B, Boschmans M, Dupont G, et al. Highly pathogenic H5N1 influenza virus in smuggled Thai eagles, Belgium. Emerging Infectious Diseases. 2005;11:702–705. doi: 10.3201/eid1105.050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RG, HoDac HM, Lathrop RH, Fitch WM (2007) A statistical phylogeography of influenza A H5N1. Proceedings of the National Academy of Sciences of the United States of America 104:4473–4478 [DOI] [PMC free article] [PubMed]
- Williams RAJ, Fasina FO, Peterson AT. Predictable ecology and geography of avian influenza (H5N1) transmission in Nigeria and West Africa. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2008;102:471–479. doi: 10.1016/j.trstmh.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Hiraoka E, Fujita M, Hijikata N, Ueta M, Takagi K, et al. Spring migration routes of mallards (Anas platyrhynchos) that winter in Japan, determined from satellite telemetry. Zoological Science. 2008;25:875–881. doi: 10.2108/zsj.25.875. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Nakamura K, Okamatsu M, Yamada M, Mase M. Avian influenza virus (H5N1) replication in feathers of domestic waterfowl. Emerging Infectious Diseases. 2008;14:149–151. doi: 10.3201/eid1401.071036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


