Abstract
Neurons establish specific synaptic connections with their targets, a process that is highly regulated. Numerous cell adhesion molecules have been implicated in target recognition, but how these proteins are precisely trafficked and targeted is poorly understood. To identify novel components that affect synaptic specificity we carried out a forward genetic screen in the Drosophila eye. We identified a novel gene, named ric1 homologue (rich), whose loss leads to synaptic specificity defects. Loss of rich leads to reduction of N-Cadherin in the photoreceptor cell synapses but not of other proteins implicated in target recognition, including Sec15, DLAR, Jelly belly, and PTP69D. The Rich protein binds to Rab6 and Rab6 mutants display very similar phenotypes as the rich mutants. The active form of Rab6 strongly suppresses the rich synaptic specificity defect, indicating that Rab6 is regulated by Rich. We propose that Rich activates Rab6 to regulate N-Cadherin trafficking and affects synaptic specificity.
Keywords: photoreceptor R7, target specificity, GEF, Rab6, N-Cadherin
Introduction
Neurons establish precise synaptic connections with their targets, which is crucial for the assembly of functional neural circuits. To achieve this, neurons integrate numerous signals that allow them to decide when to extend their growth cones, to follow a specific route, to determine when to fasciculate or defasciculate, and when to stop and form synaptic connections. Improper synapse formation may lead to cognitive diseases and mental retardation such as autism spectrum disorders (McAllister, 2007; Sudhof, 2008) or neurodegenerative disorders (Haass and Selkoe, 2007). Hence, providing a molecular framework to understand how neurons form proper synapses remains an important endeavor.
The Drosophila visual system is an excellent model to untangle this type of question because of its stereotyped structure, well documented cellular behavior, accessibility to genetic manipulation, and because the homologues of numerous fly proteins play similar roles in vertebrates (Kunes and Steller, 1993; Meinertzhagen, 1993; Sanes and Zipursky, 2010). The adult Drosophila compound eye contains ~ 800 small units called ommatidia, each of which comprises eight photoreceptor (PR) cells, R1–R8. R1–R6 cells are outer PR cells, that synapse in the first optic ganglion, the lamina, to form a primary visual map. In the lamina, terminals of PR cells and post-synaptic neurons form repeated modules called cartridges. Each cartridge contains 6 PR terminals that originate from 6 different ommatidia. Hence, each cartridge receives input from a single point in space. Improper organization of the cartridges often leads to visual map disruption and abnormal optomotor behavior (Clandinin and Zipursky, 2000). The inner PR cells, R7 and R8, project their axons through the lamina and stop in two distinct layers, M6 and M3, in the medulla where they make precise synaptic connections with the post-synaptic cells (Kunes and Steller, 1993; Meinertzhagen, 1993; Sanes and Zipursky, 2010; Ting and Lee, 2007; Tomasi et al., 2008). The formation of specific synaptic connections between R cells and post-synaptic cells relies upon a complex bidirectional interaction between R cells and their targets. To date, many molecules have been identified that play pivotal roles in this targeting process (Giagtzoglou Nikolaos, 2009), including cell adhesion molecules (Lee et al., 2001; Lee et al., 2003; Senti et al., 2003), signaling molecules (Bazigou et al., 2007; Clandinin et al., 2001; Garrity et al., 1999; Hofmeyer et al., 2006; Newsome et al., 2000; Ruan et al., 1999), transcriptional factors (Morey et al., 2008; Petrovic and Hummel, 2008; Rao et al., 2000; Senti et al., 2000), and molecules that affect protein trafficking (Mehta et al., 2005).
N-Cadherin (CadN) is a Ca2+ dependent cell adhesion molecule (Shapiro et al., 2007) that plays an important role in synapse formation in the developing nervous system (Clandinin et al., 2001; Lee et al., 2001; Nern et al., 2005; Prakash et al., 2005; Ting et al., 2005). In Drosophila eyes, loss of CadN leads to targeting defects of the photoreceptors in lamina and medulla: R1–R6 growth cones fail to extend from the ommatidial bundle and are not able to select appropriate synaptic partners (Prakash et al., 2005); moreover, R7 cells often terminate in an improper medulla layer. In addition, the lamina neurons require CadN for layer specific termination of their axons (Nern et al., 2008). CadN mediated homophilic interactions between pre- and post synaptic cells are thought to be key for synaptic partner selection. However, it is still unclear how the cell type specific function of CadN is achieved since CadN is widely expressed in the brain (Lee et al., 2001). In cultured mouse hippocampal neurons, CadN undergoes both constitutive and activity dependent endocytosis and recycling, which is critical for synaptic plasticity (Tai et al., 2007). It has been reported that the CadN expression pattern is highly dynamic during fly eye development but how it is regulated is unknown (Matthews et al., 2008; Nern et al., 2008). It is therefore interesting to establish how trafficking of CadN contributes to the dynamic changes of its expression pattern and determines PR cell targeting specificity. Here, we present evidence that Rab6, and its potential GEF protein Rich, control PR cell target selection by regulating CadN.
Rab6 is a small GTPase that is present in the Golgi apparatus as well as cytoplasmic vesicles (Del Nery et al., 2006; Martinez et al., 1994; Opdam et al., 2000; Utskarpen et al., 2006). Rab6 recruits the dynein motor complex and plays an important role in microtubule dependent retrograde transport from the endosome to the Golgi and from the Golgi to the ER (Girod et al., 1999; Martinez et al., 1997; Matanis et al., 2002; Short et al., 2002; White et al., 1999). In addition, Rab6 also directs targeting of secretory vesicles to the plasma membrane (Coutelis and Ephrussi, 2007; Grigoriev et al., 2007; Januschke et al., 2007). Like other GTPases, Rab6 alternates between a GTP-bound active form and a GDP-bound inactive form. The GEF proteins unload GDP from Rab GTPases to generate active GTP loaded Rabs (Stenmark, 2009). To date, the only Rab6 GEF that has been characterized is a yeast protein complex comprised of two proteins Ric1p and Rgp1p. The Ric1p-Rgp1p complex binds to Ypt6p, the yeast Rab6 homolog, in a nucleotide-dependent manner.
The purified complex stimulates guanine nucleotide exchange on Ypt6p (Siniossoglou et al., 2000). Rab6 is highly conserved in eukaryotes. However, both Ric1 and Rgp1 are only present in budding yeast, although related proteins containing RIC1 domains as well as other domains or Rgp1 domains can be identified in higher eukaryotes.
In a forward genetic screen, we isolated a complementation group in which the PR cells display defects in synaptic specificity. We mapped the lesions to a novel gene CG9063 that encodes a protein containing a RIC1 domain as well as other domains. Our data indicate that this protein is required for proper synaptic specificity in the eye and antenna lobe (AL) by regulating CadN trafficking via Rab6.
Results
Identification of a new complementation group that affects photoreceptor cell specificity
To isolate new genes involved in synapse development and function, we carried out an F1 genetic screen in the Drosophila visual system (Ohyama et al., 2007; Verstreken et al., 2003). By creating clones of randomly induced mutations with the eyFLP system we identify mutations that fail to evoke a post synaptic response in electroretinograms (ERGs). Lack of an “on” and “off” response indicates aberrant communication between the presynaptic photoreceptors (R1–R6) and the post-synaptic lamina neurons (L1-3). These defects can be due to defects in synaptic transmission or a failure in synapse formation or synaptic partner selection (Mehta et al., 2005). From a screen of about 50,000 mutagenized chromosomes on arm 3L, we isolated several essential complementation groups, including one which consists of two homozygous lethal alleles: 3L61 and 3L62. The eyFLP mosaic mutant animals display small “on” and “off” transients in ERGs (Figure 1A), indicating that the mutant photoreceptors fail to transmit information to their targets. Note that the amplitude of depolarizations in these mutant are fairly normal, indicating that the mutant photoreceptors can capture photons and produce graded potentials, suggesting that the phototransduction pathway is essentially normal.
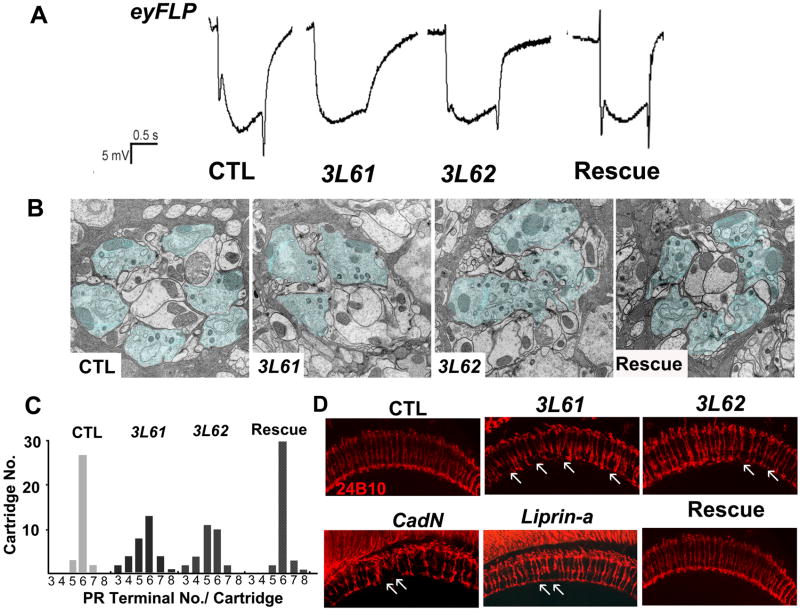
Figure 1. Isolation of 3L6 mutants in an eyFLP screen.
(A) Electroretinogram traces of control flies (CTL), mutant flies (3L61, 3L62), and mutant flies carrying a genomic rescue fragment (Rescue). The ERG profiles of the mutant flies show diminished “on and off” transients but normal depolarization in response to light. (B) TEM of cartridges from a control, 3L61, 3L62, and a rescued fly. The photoreceptor terminals are marked in blue. (C) Distribution of photoreceptor terminals per-cartridge for wild-type (n=31 cartridges from 3 animals), mutant (3L61: n=52 from 3 animals; 3L62: n=32 from 3 animals) and rescued flies (n=43 cartridges from 3 animals). (D) The R7 and R8 projection patterns (24B10 staining) of a control animal, a 3L61 mutant, a 3L62 mutant, a CadNΔ14 mutant, a Liprin αe mutant, and a 3L61 rescued fly. The arrows indicate the mistargeted terminals. The genotypes of flies used in this figure: control flies (y w eyFLP GMR-lacZ; FRT80B iso/FRT80B cl, ubiGFP); 3L61 (y w eyFLP GMRLacZ; FRT80B 3L61/FRT80B cl, ubiGFP); 3L62 (y w eyFLP GMRLacZ; FRT80B 3L61 or 3L62/FRT80B cl, ubiGFP); rescued flies: (y w; rich-gRE::VK37/CyO; FRT80B 3L61); CadN (y w eyFLP GMRlacZ; FRT40A CadNΔ14/FRT40A ubiGFP) and Liprin α (y w eyFLP GMRlacZ; FRT40A Liprin αe/FRT40A cl). (See also Figure S1)
To determine whether the failure to evoke a postsynaptic response is due to defects in R cell connectivity, we investigated the morphology of the terminals of the outer PR cells, R1–R6, in the lamina. We labeled the R2–R5 with Ro-tau-lacZ (Garrity et al., 1999) (Figures S1A and S1B) in the 3rd instar larval brains and the R1–R6 with Rh1 GFP (Ratnakumar and Desplan, 2004b) (Figures S1C and S1D) in the adult brains. In the eyFLP; 3L61 mutant animals, the outer PR axons correctly target to the lamina layer at both stages, indicating that 3L61 is not required for the lamina layer targeting of the outer PR cells.
To analyze cartridge organization and ultrastructural features of the R1–R6 axon termini, we performed transmission electron microscopy (TEM) of the lamina. Photoreceptor terminals were identified based on the presence of glial invaginations (capitate projections) (Meinertzhagen, 1993). Although the mutant epithelial glial cells are thinner than in wildtype, the cartridges are readily identifiable in 3L61 and 3L62 mutants. The mutant cartridges contain a highly variable number of PR cell terminals ranging from 3 to 8 (Figures 1B and 1C). However, in these mis-sorted cartridges, active zone integrity, vesicle density, and capitate projections are normal (Figure 1B) as is often observed in targeting mutants (Hiesinger et al., 2006). In summary, the axons of the outer PR target to the correct layer but proper cartridge formation is impaired.
R7 cells fail to target to the M6 layers in 3L6 mutants
To determine if the terminals of R7 and R8 are layered correctly in the M6 and M3 layers of the lamina, we revealed them with Chaoptin (mAb24B10) (Fujita et al., 1982). Staining of the eyFLP; 3L6 mutant medulla revealed a few “gaps” as if some R7 terminals are “missing” (Figure 1D). This pattern is similar to that observed in CadN (Lee et al., 2001) and Liprin α mutants (Choe et al., 2006; Hofmeyer et al., 2006) in which R7 axons fail to terminate at the M6 layer, but rather target the M3 layer. However, the observed “gaps” may also be caused by a loss of R7 cells. We therefore stained 3rd instar larval eye discs with anti-Elav (Figures S2A and S2B) and 24B10 antibodies (Figures S2C and S2D) to reveal the differentiation of neurons and R cells. To determine if specific PR cells were properly identified, we also labeled R4 cells with mΔGFP (Cooper and Bray, 1999) (Figures S2E and S2F), R7 cells with 181Gal4 GFP (Lee et al., 2001) (Figures S2C and S2D), and R8 cells with anti-Senseless (Nolo et al., 2000) (Figures S2A and S2B). We observed no difference in staining pattern for any of the markers between 3L6 mutant and wild type cells, indicating that the differentiation of PR cells is not affected in the mutants. We then analyzed retinal thick sections of adult flies and did not observe loss of R7 cells in the 3L6 mutants (Figures S2G and S2H), although a rare ommatidium has an abnormal morphology. In contrast, labeling of the R7 terminals with UAS-Synaptotagmin GFP (SytGFP) drived by Pan-R7-Gal4 (Ratnakumar and Desplan, 2004a), showed that about 20% (19.7 ± 3%, n=268) of all R7 cell terminals fail to reach their target layer M6 but target the M3 layer (Figures 2A, 2B, 2A′ and 2B′) in the adult brains of eyFLP; 3L61 mutants. Note that the targeting defect is an underestimate (see Figure 5B) since we did not label the mutant clones and the 3L61 mutant clones are small because of a growth disadvantage in respect to heterozygous cells. Finally, we assessed the projection pattern of R8 cells by labeling them with Rh6-GFP (Ratnakumar and Desplan, 2003). 3L6 mutant animals do not exhibit any obvious R8 targeting defects (Figures 2C, 2D, 2C′and 2D′).
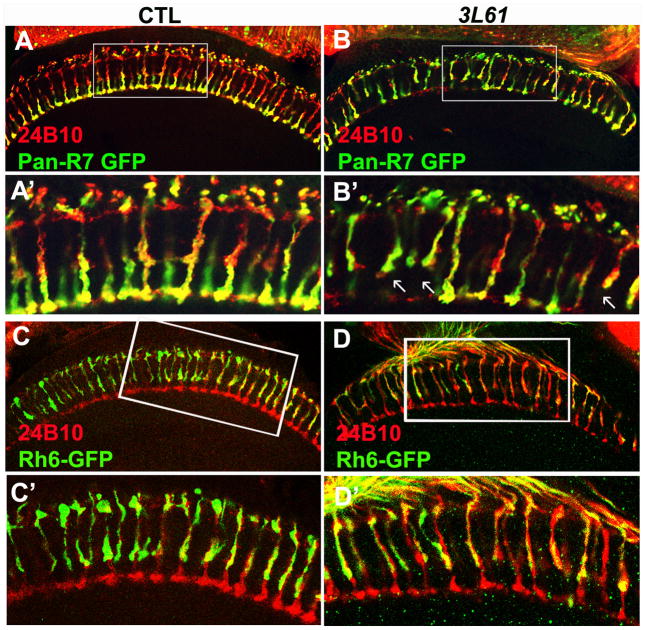
Figure 2. The R7 cells exhibit targeting defects in the 3L6 mutant animals.
(A, B, A′, B′) A portion (19.7±3%, n=268) of the R7 cells in the 3L61 mutant animals fail to target to the M6 layers but instead terminate at the M3 layers. The R7 cells were labeled with pan-R7 Gal4 that drives UAS-SytGFP (green). The arrows indicate the mistargeted R7s. (C, D, C′, D′) The R8 cells have intact target patterns. The R8 cells were labeled with Rh6-GFP (green), the photoreceptor cells were labeled with 24B10 staining (red). Panels (A′ –D′) are the enlarged views of the boxed regions in panel (A –D). The genotypes of flies used in this figure: Control (A, A′) (y w eyFLP GMRlacZ; PanR7 Gal4/UAS-SytGFP; FRT80B iso/FRT80B cl); 3L61 (B, B′) (y w eyFLP GMRlacZ; PanR7 Gal4/UAS-SytGFP; FRT80B 3L61/FRT80B cl); Control (C, C′) (y w eyFLP GMRlacZ; Rh6GFP/CyO; FRT80B iso/FRT80B cl). 3L61 (D, D′) (y w eyFLP GMRlacZ; Rh6GFP/CyO; FRT80B 3L61/FRT80B cl). (See also Figure S2)
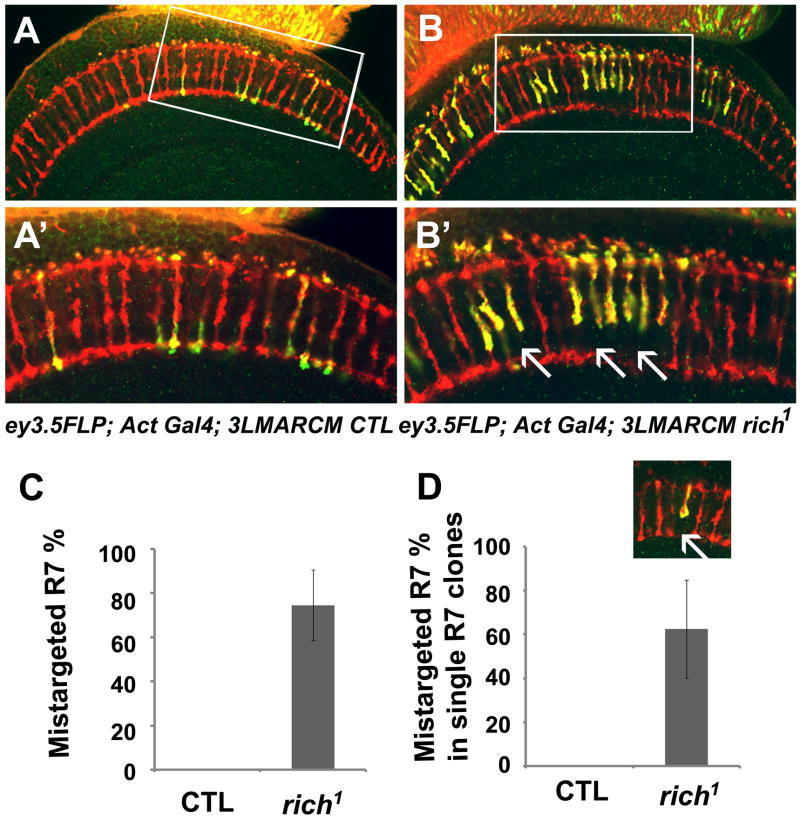
Figure 5. rich is required for synaptic specificity of R7.
Single optic section taken through a medulla of either (A, A′) control (ey3.5FLP; Act>Gal4 UAS-SytGFP/UAS-SytGFP; FRT80B M (3), tub-Gal80/FRT80B iso) or (B, B′) rich mutant (ey3.5FLP; Act>Gal4 UAS-SytGFP/UAS-SytGFP; FRT80B M (3), tub-Gal80/FRT80B rich1) animals. (A′, B′) are the enlarged images of the boxed region in (A, B). Many of the mutant R7 cells fail to target to the M6 layers (arrows). (C) Quantification of the R7 targeting defects in the ey3.5 animals (0 in control n=159;74±16% in rich n=175) illustrated in A and B. (D) 62.5 ±22% (n=37) R7 cells fail to target to the correct layer even when they are surrounded by the wild-type R7 and R8 neighbors. The small inset illustrates the single R7 mutant clone (see also Figure S4).
3L6 mutants corresponds to CG9063
Since the phenotypes associated with loss of 3L6 are specific and interesting, we performed meiotic recombination mapping using P-elements (Zhai et al., 2003). Rough mapping placed 3L6 in the 77A4-79F4 cytological interval. Deficiency mapping mapped 3L6 to 79C2-80A4. As recombination frequencies are extremely low in this interval, we generated 4 small overlapping deficiency using FRT bearing P-elements and PiggyBac insertions (Parks et al., 2004; Thibault et al., 2004) (Figure 3A). Complementation tests narrowed the putative gene down to 6 genes and sequencing revealed two premature stop codons in CG9063 at amino acid (aa) 380 (3L61) and 1196 (3L62) (Figure 3A). Since 3L61 has an early stop codon, it is probably a null or a strong hypomorphic allele whereas the 3L62 allele contains a late stop codon, indicating that it may be a hypomorphic allele of CG9063. These data are in agreement with all the phenotypic data. Note that the 3L62 allele causes a weaker ERG and R7 targeting defect than the 3L61 allele (Figures 1A, 1D, 7E, 7F, and 7M).
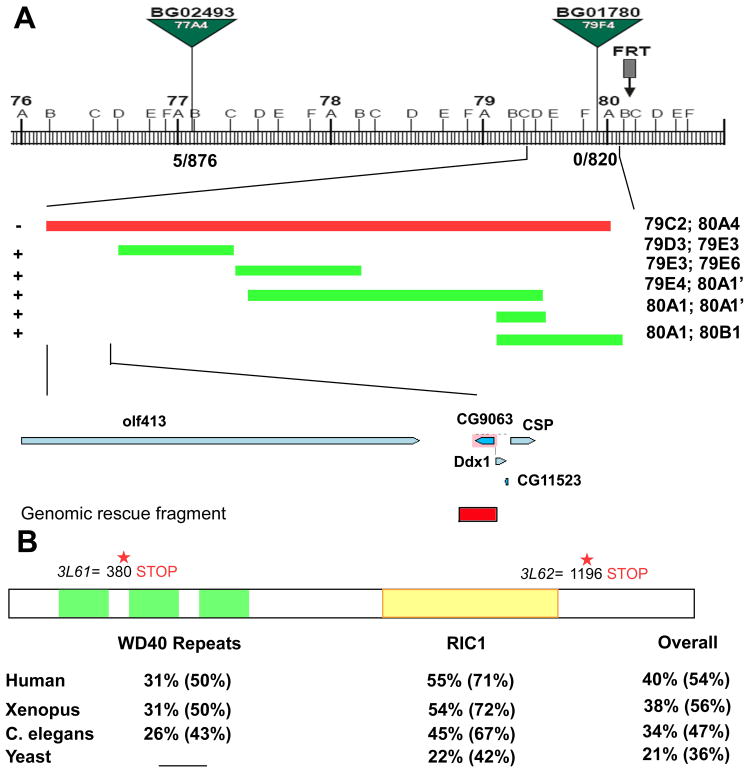
Figure 3. 3L6 encodes an evolutionarily conserved gene CG9063 (rich).
(A) Schematic showing the cytological interval and the position of the P-elements and the overlapping deficiencies used to identify the genomic region of the 3L6. The recombination rate is 5/876 for BG02493 and 0/820 for BG01780. “−” indicates “fail to complement” and “+” indicates “complements” 3L61 and 2. The lower panel shows genes in the region identified by deficiency mapping and the genomic rescue fragment of rich (Red box). (B) Schematic of the Rich protein showing the predicted domains and the locations of the stop codons in 3L61 and 3L62. The bottom panel shows the homology (identity %; similarity %) of WD40 repeats domain, RIC1 domain, and overall protein sequence between Rich and its homologs in humans, frogs, worms, and yeast.
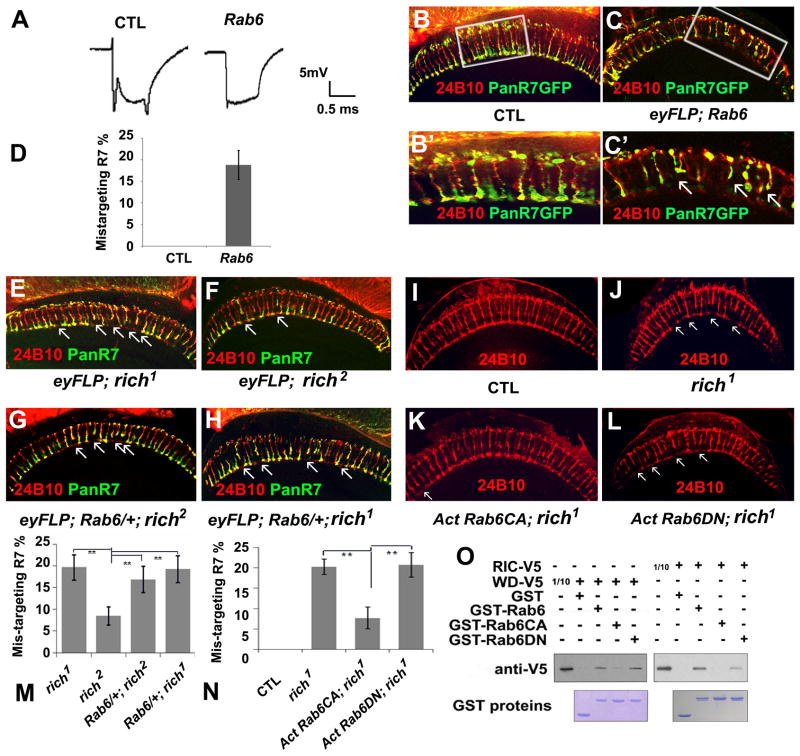
Figure 7. Rich interacts with Rab6 and positively regulates Rab6 activity in fly eyes.
(A) The ERG profiles of control animals (CTL) (y w eyFLP GMRlacZ; FRT40A iso/FRT40A cl) and Rab6 mutant animals (y w eyFLP GMRlacZ; FRT40A Rab6D23D/FRT40A cl). Rab6 mutants have no “on” and “off” transients but exhibit a normal depolarization. (B, B′, C, C′) A portion (18.8±3%, n=265) of the R7 cells in the Rab6 mutant animals (y w eyFLP GMRlacZ; FRT40A Rab6D23D/FRT40A cl; PanR7 Gal4/UAS-SytGFP) fail to target to the M6 layers as the controls (y w eyFLP GMRlacZ; FRT40A iso/FRT40A cl; PanR7 Gal4/UAS-SytGFP), but instead terminate at the M3 layers. The R7 cells were labeled with pan-R7 GFP (Green). The arrows indicate mistargeted R7s. (B′, C′) are the enlarged images of the boxed regions in image (B, C). (D) Quantification of the mistargeting R7s in Rab6 mutants. (E–H) Loss of one copy of Rab6 enhanced the phenotype of the hypomorphic allele rich2 but not the phenotype of the null allele rich1, indicating that Rich and Rab6 function in the same pathway. Single optic sections of medulla in rich1 (E) (y w eyFLP GMRlacZ; PanR7 Gal4/UAS-SytGFP; FRT80B rich1/FRT80B cl); rich2 (F) (y w eyFLP GMRlacZ; PanR7 Gal4/UAS-SytGFP; FRT80B rich2/FRT80B cl); rich2 with one copy of Rab6 (G) (y w eyFLP GMRlacZ/UAS-SytGFP; PanR7 Gal4/FRT40A Rab6D23D; FRT80B rich2/FRT80B cl), and rich1 with one copy of Rab6 (H) (y w eyFLP GMRlacZ/UAS-SytGFP; PanR7 Gal4/FRT40A Rab6D23D; FRT80B rich1/FRT80B cl). The R7 cells were labeled with pan-R7 GFP (green) and 24B10 staining indicates the projection patterns of the photoreceptor cells. (I–J) Expression of Rab6CA but not Rab6DN suppresses the targeting phenotypes of rich1 mutant, indicating Rich positively regulate Rab6. Single optic sections of medulla in control animals (I) (y w eyFLP; Act>Gal4 UAS-SytGFP/UAS-SytGFP; FRT80B M (3), tub-Gal80/FRT 80B iso), rich1 mutants (J) (y w eyFLP; Act>Gal4 UAS-SytGFP/UAS-SytGFP; FRT80b M (3), tub-Gal80/FRT 80b rich1), rich1 mutants in which the constitutively active form of Rab6 (Rab6CA) is expressed in the mutant cells (y w eyFLP/UAS-YFPRab6 Q71L; Act>Gal4 UAS-SytGFP/CyO; FRT80B M (3), tub-Gal80/FRT 80B rich1), and rich1 in which the dominant negative form of Rab6 (Rab6DN) is expressed in the mutant cells (eyFLP; Act>Gal4 UAS-SytGFP/UAS-YFPRab6 T26N; FRT80B M (3), tub-Gal80/FRT 80B rich1). Projection patterns are shown with 24B10 staining. The arrows indicate the mistargetted R cells. (M) Quantification of the R7 targeting phenotypes in the mutants illustrated in (E–H). 19.7±3% (n=268) R7 cells in rich1 mutant (E); 8.5±2% (n=329) R7 cells in rich2 mutants (F) fail to target to the correct layer. Upon removing one copy of Rab6, 19.0±3% (n=308) R7 cells in rich1 (H) and 17.0±3% (n=483) R 7 cells in rich2 (G) fail to target to the correct layers. (N) Quantification of the R7 targeting phenotypes in the flies illustrated in (I–L). In rich1 mutants (J), 20.24±2% (n=252) R7 cells fail to target to the corresponding layer. In the rich1 mutant with Rab6CA expression animals (K), 7.69±3% (n=296) R7 cells fail to target to the correct layer. In the rich1 mutant with Rab6DN expression animals (L), 20.8±4% (n=233) R7 cells fail to target to the correct layer. (O) GST pull-down experiments show that both the RIC1 domain and the WD40 domain interact with Rab6 physically. The RIC1 and WD40 domain have high affinity for the wild-type Rab6 and Rab6DN but have low affinity for the Rab6CA. 1/10 input of the RIC1 domain and WD40 domain were loaded as control (see also Figure S5).
CG9063 is evolutionarily conserved over the full length of the protein from worm to human and exhibits 40% identity and 54% aa similarity between the fly and human homologue (Figure 3B). However, their functions have not been studied in these species. The protein encoded by CG9063 is predicted to have WD40 domains as well as a RIC1 domain. The RIC1 domain, but not the WD40 domain, is present in a yeast protein RIC1p that interacts with Rgp1p. Together they form a GEF complex for Ypt6p, a homolog of Rab6 in yeast (Siniossoglou et al., 2000). Since the RIC1 domain is highly conserved across species and the protein encoded by CG9063 is the only protein containing a RIC1 domain in the fly, we named CG9063 rich, for ric1 homolog. We renamed 3L61 as rich1 and 3L62 as rich2. A genomic fragment that only contains rich rescues both the lethality and the targeting phenotypes (Figure 1) of both alleles, indicating that rich is the gene responsible for the mutant phenotypes.
Expression of Rich in the developing visual system
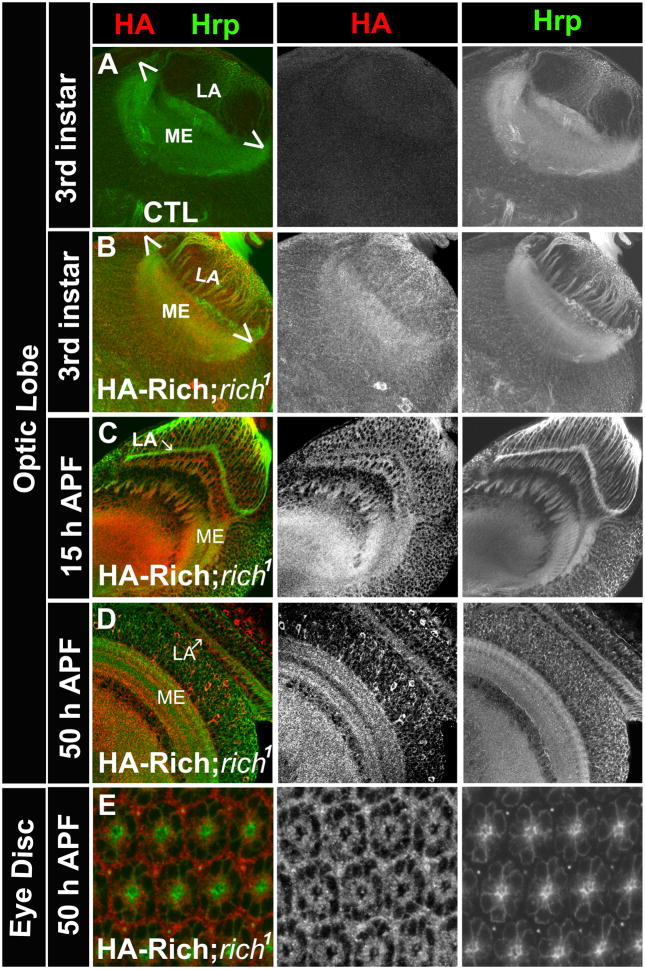
To assess the expression pattern of Rich, we generated antibodies against the N- and C- termini of the protein. As all antibodies performed poorly in all assays, we generated a genomic rescue construct with a triple HA tag inserted at the N-terminus and introduced it into the rich1 mutant background. This construct rescues both the lethality and targeting phenotypes of rich1 mutants, indicating that the tagged protein is functional. At the 3rd instar larval stage (Figures 4A and 4B), Rich is detected in most cells of the optic lobes. The enrichment in the lamina and medulla neuropil is consistent with its role in PR cell axon targeting. At the pupal stage, Rich is expressed in PR cells, as well as the post-synaptic cells that form the lamina plexus and medulla (Figures 4C and 4D). The expression pattern of Rich substantially overlaps with that of CadN in the optic lobe (Figures S3A and S3B). In summary, Rich is present at the proper time and in the proper cells to account for the observed targeting defects.
Figure 4. Rich is expressed in the developing eye.
(A) Hrp (green) and HA (red) staining of an optic-lobe of a control animal (y w) at the 3rd instar larval stage. The HA antibody does not detect non-specific staining in the brain. (B) The optic lobe of a rich mutant animal carrying a HA-Rich-genomic rescue construct (rescue animal: HA-Rich-gRE::VK37; FRT80B rich1) at the 3rd instar larval stage. At this stage, Rich is enriched in the developing lamina (LA) and medulla (ME). (C, D) The optic lobe of rescued animals at 15 hrs and 50 hrs after pupa formation (APF). Rich is enriched in the lamina and medulla neuropil. (E) The eye disc of the rescued animal at 50 hrs APF. Rich is expressed in the PR cells. (See also Figure S3).
Rich is required in PR cells for targeting specificity
Since Rich is present in the PR cells as well as the post-synaptic cell types and the eyFLP system generates mutant clones in both cell types, it is not obvious whether Rich is required cell autonomously in the PR cells. To determine if Rich is required in PR cells, we used ey3.5FLP (Chotard et al., 2005) in combination with MARCM (Lee and Luo, 1999) to generate clones of GFP-labeled rich1 homozygous mutant PR cells. In the medulla, 74% of the mutant R7 cells (n=175) fail to target to the M6 layers showing that Rich is required in R cells for targeting specificity (Figures 5A, 5B, 5A′, 5B′ and 5C). We also generated single R7 cell mutant clones using the GMR FLP system (Lee et al., 2001). These clones are induced late in PR development and only affect 15% of the R7 cells. Interestingly, we did not observe any mistargeting phenotype, possibly because of perdurance (Figure S4). We therefore assessed the phenotype of single mutant R7 cells surrounded by wild type R7s and R8s in the ey3.5 FLP mutant animals. As shown in Figure 5D, 62.5% of the mutant R7 cells (n=37) fail to target to the M6 layer suggesting that Rich functions cell autonomously in R7 cells. Similar to eyFLP, rich1 animals, ey3.5 FLP, rich1 animals also display improper cartridge organization in the lamina (data not shown).
Rich is enriched in the Golgi and co-localizes with Rab6
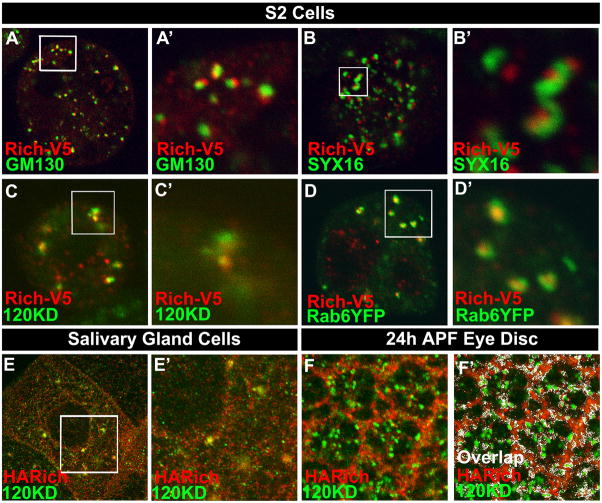
In yeast, the RIC1p is part of a Rab6 GEF (Siniossoglou et al., 2000) and Rab6 is localized to the Golgi as well as cytoplasmic vesicles (Januschke et al., 2007; Martinez et al., 1997; Martinez et al., 1994). Hence, Rich may function as a Golgi-associated Rab6 GEF component and interact with Rab6. To localize Rich, we expressed V5-tagged protein in Drosophila S2 cells and stained with anti-V5 antibody and different Golgi markers, including the cis-cisternal marker dGM130 (Kondylis et al., 2001), the trans-cisternal marker dSyntaxin16 (dSyx16) (Xu et al., 2002), and the medial-cisternal marker 120KD protein (Stanley et al., 1997), as well as the ER marker Boca (Culi and Mann, 2003). In Drosophila, Golgi cisternae are stacked but not connected to form a ribbon like structure. The different Golgi markers are closely apposed but do not fully overlap (Yano et al., 2005). As shown in Figure 6A–C, A′–C′, Rich localizes next to the Golgi marker GM130 and Syx16 and partially colocalizes with the 120KD protein, indicating it is in the Golgi apparatus. In contrast, there is no overlap between Rich and Boca (data not shown). As overexpression of Rich may affect its subcellular expression pattern, we also determined the localizing of the genomic-tagged Rich protein in vivo in salivary gland cells and pupal eye imaginal discs (Figures 6E, 6F, 6E′ and 6F′). Consistent with the S2 cell data, Rich partially colocalizes with the 120KD protein. To test whether Rab6 and Rich are colocalized, we expressed YFP-tagged Rab6 and V5-tagged Rich in S2 cells and stained the cells with anti-GFP and anti-V5 antibody. The signals of Rab6 and Rich overlap extensively, suggesting a possible association of Rab6 and Rich (Figures 6D and 6D′). Note that Rich is present in the neuropil of the optic lobes. Since the Golgi apparatus is barely detectable, in the neuropil, Rich may also be present in vesicles in axons and synapses (Figure 4). Interestingly, Rab6 is also localized in the neuropil of optic lobes when we express Rab6YFP in the brain, consistent with the presence of Rab6 in cytoplasmic vesicles in addition to Golgi (Figures S3C, S3D, and S3E).
Figure 6. Rich is enriched in Golgi apparatus and co-localizes with Rab6.
(A–C; A′–C′) S2 cells expressing V5-tagged Rich were stained with anti-V5 (red) and different Golgi markers (green): GM130 (A, A′), SYX16 (B, B′), and 120KD protein (C, C′). (A′–C′) are the enlarged images of the boxed regions in image (A–C). Noticed that Rich is expressed in punctae that are co-localized or juxtaposed to the Golgi markers, indicating that Rich is enriched in Golgi apparatus. (D, D′) S2 cells expressing V5-tagged Rich and YFP tagged Rab6 were stained with anti-V5 (red) and anti-GFP (green) antibody. Rich and Rab6 are substantially co-localized in S2 cells. (D′) is the enlarged image of the boxed region in (D). (E, E′) Staining of salivary gland cells from a rescued animal (HA-Rich-gRE::VK37; FRT80B rich1) with anti-HA (red: shows the pattern of Rich) and anti-120KD protein (green). Rich partially colocalizes with the 120KD protein, indicating Rich is enriched in the Golgi when it is expressed at the endogenous level. (E′) is the enlarged image of the boxed region in (E). (F, F′) Rich is enriched in the Golgi of photoreceptor cells. Eye disc of a rescued animal (HA-Rich-gRE::VK37; FRT80B rich1) at 24 hrs after pupal formation was stained with anti-HA (red), anti-120KD (green). (F′) The colocalized signals of HA and 120KD protein shown in (F) were analyzed with the ImageJ and indicated with a pseudo-color (white).
rich and Rab6 mutants display similar phenotypes
GTPases interconvert between GTP-bound and GDP-bound forms. The GTP bound form activates downstream effectors. The GDP bound form is an “inactive” form that has a high affinity for the GEF protein. Once the GEF protein unloads the GDP, GTP will bind the small GTPases and activate it (Stenmark, 2009). If Rich is a Rab6 GEF, we expect that Rab6 cannot be activated in rich mutants and that they may display similar phenotypes. Indeed, eyFLP; Rab6 mutants display no “on” and “off” transients in the ERG recording profile (Figure 7A), very similar to the ERG profiles of the rich mutants (Figure 1A). Before assessing whether Rab6 mutants have PR targeting phenotypes, we first assessed whether Rab6 mutant eyes have differentiation defects. We stained the eye discs of the third instar eyFLP;Rab6 mutant larvae with anti-24B10, anti-Elav, anti-Senseless (labels R8), and anti-Prospero (labels R7). The staining patterns all appear normal, indicating that there are no differentiation defects in Rab6 mutant eyes (data not shown). We then labeled R7 cells with Pan-R7 Gal4 and assessed the R7 targeting phenotypes of Rab6 mutants. 18.8 ± 3% (n=265) of the R7 cells in Rab6 mutants (versus 19.7 ± 3 % (n=268) for rich1 mutants) fail to target to the correct layers in the medulla (Figures. 7C, 7C′, and 7D).
The similarity in phenotype between Rab6 and rich mutant PR suggests that they function in the same pathway. To test this hypothesis, we removed one copy of Rab6 in the eyFLP; rich1 or eyFLP; rich2 mutant animals and quantified the R7 targeting defects in the adult medulla. Rab6 heterozygous animals do not have R7 targeting defects, while the hypomorphic allele rich2 only has weak R7 targeting defects (8.5 ± 2%, n=329) (Figures. 7F and 7M). We reasoned that loss of one copy of Rab6 in the partial loss of function mutant rich2 would enhance the phenotype. Indeed, loss of a copy of Rab6 enhanced the R7 targeting defects in eyFLP; rich2 animals (17.0 ± 3%, n=483) similar to the rich1 mutant phenotype (Compare Figures 7E and 7F to 7G; Figure 7M). However, loss of a copy of Rab6 does not modify the R7 targeting defects of the null allele rich1 (19.0 ± 3%; n=308) (compare Figures 7E to 7H; Figure 7M). Hence, the phenotypic and genetic data strongly argue that rich and Rab6 indeed function in a common pathway.
To provide additional evidence that Rich and Rab6 function together, we overexpressed a constitutively active form of Rab6 (Rab6CA) in rich1 mutant cells using the MARCM system. This greatly suppressed the R7 targeting defects caused by loss of rich. Moreover, overexpression of a dominant negative form of Rab6 (Rab6DN) did not suppress or enhance the rich1 phenotype, indicating that Rich positively regulates Rab6 activity, possibly as a GEF (Figures 7I–L, and 7N). To determine if Rab6 physically interacts with Rich, we overexpressed the WD40 domain (AA8-AA521) and RIC1 domain (AA711-AA995) of Rich in S2 cells and performed GST pull-down experiments using GST fusion proteins containing either wildtype Rab6 (Rab6WT), Rab6CA or Rab6DN. A typical GEF protein tends to bind to the nucleotide-free form (e.g., Rab6WT) or GDP-bound form (e.g., Rab6DN). Consistent with this, both the WD40 domain and the RIC1 domain have a strong affinity for Rab6WT and Rab6DN but not Rab6CA, suggesting that Rich directly regulates Rab6 (Figure 7O).
To directly assess the GEF activity of Rich, we overexpressed Rich in S2 cells and performed GTP exchange assay with the cell lysate. In this experimental setting, a positive control, Tiam1, showed proper GTP exchange activity for Rac1. However, we did not observe an obvious GEF activity for Rich (Figure S5A). A possible explanation is that similar to yeast, one or more other proteins are required for Rich to control its GEF activity: e.g., Ric1p in yeast has to interact with Rgp1p to stimulate GTP exchange of Ypt6p. In flies and vertebrates, there are Rgp1-related genes that encode proteins containing a Rgp1 domain. We cloned the sole Drosophila homologue of Rgp1, CG1116, and expressed the CG1116-PB in S2 cells alone or together with Rich. We performed the GEF assay with the cell lysates and did not observe any GEF activity. Similar results were obtained when we coexpressed yeast Rgp1 together with Drosophila Rich. In yeast, Rgp1p and Ric1p tightly interact with each other, but neither CG1115-PB or Rgp1p bind to Rich in IP experiments (Figure S5B), suggesting that Rich uses a different interactor to regulate Rab6 activity.
rich and CadN function in a common pathway
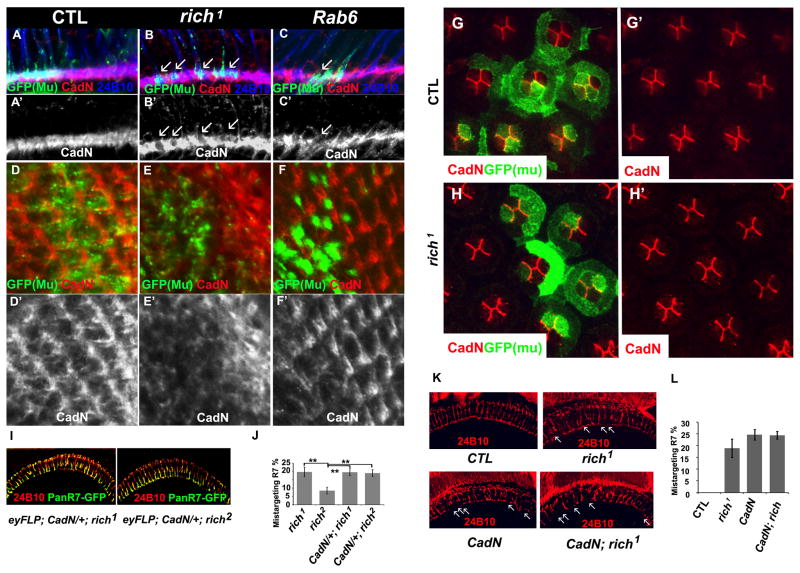
Since Rab6 affects protein trafficking, we wondered whether the targeting defects in rich and Rab6 mutants are due to mistrafficking of proteins that are essential for PR cell targeting. We generated rich and Rab6 mutant eyes and marked the mutant cells with SytGFP using MARCM. We stained the lamina at 24h after pupa formation (APF) for proteins that have been implicated in PR targeting, including CadN (Lee et al., 2001), Sec15 (Metha et al., 2005), DLAR (Clandinin et al., 2001; Maurel-Zaffran et al., 2001), PTP69D (Garrity et al., 1999; Newsome et al., 2000), and Jelly belly (Jeb) (Bazigou et al., 2007). Only CadN was found to be reduced inside the mutant terminals (Figures 8 and S6A), while the distribution of the other tested proteins are normal. We also performed real-time PCR of CadN in rich mutants heads and did not observe an obvious change in RNA levels of CadN, indicating that the reduction of CadN in the mutant terminals is not due to decreased transcription (Figure S7B). Similarly, overexpression of CadN in rich mutant clones does not rescue the targeting phenotypes (Figure S8). We also did not observe any obvious accumulation of CadN in PR axons or PR cell bodies (Figures 8A–C and 8A′–C′, data not shown), suggesting that mis-trafficked CadN is degraded. The selective disruption of CadN among the tested proteins suggests that the different proteins required for targeting use different trafficking routes.
Figure 8. Rich, Rab6, and CadN function in a common pathway.
(A–F′) N-Cadherin is reduced in the lamina of rich and Rab6 mutants. Single optic slides of lamina of the fly eye at 24h APF stained with anti-CadN (red), 24B10 (blue), and GFP (green, mark the mutant cells). (A–C, A′–C′) show a side view of the lamina. (D–F, D′–F′) show a frontal view of the lamina. (A, A′, D, D′) control (y w eyFLP; Act>Gal4 UAS-SytGFP/UAS-SytGFP; FRT80B M (3), tub-Gal80/FRT 80B iso). (B, B′, E, E′) rich1 mutant (y w eyFLP; Act>Gal4 UAS-SytGFP/UAS-SytGFP; FRT80b M (3), tub-Gal80/FRT 80B rich1). (C, C′, F, F′) Rab6 mutant (y w eyFLP; FRT40A tub-Gal80/FRT40A Rab6D23D; tubGal4/UAS-SytGFP). (A′–F′) are red channels (CadN) of (A–F). (G–H′) CadN distribution is not affected in rich1 mutant cone cells. The pupal eye discs at 36APF were stained with anti-GFP (green) and anti-CadN (red) antibody. The mutant cells were marked with CD8GFP. The animal genotypes are: CTL (y w eyFLP; Act>UAS-SytGFP/UAS-CD8GFP; FRT80B M (3), tub-Gal80/FRT80B iso). rich1 (y w eyFLP; Act>UAS-SytGFP/UAS-CD8GFP; FRT80B M (3), tub-Gal80/FRT 80B rich1). (I) The R7 cell targeting patterns of rich1 and rich2 upon removal of one copy of CadN (y w UAS-SytGFP; FRT40A CadN405/CyO; FRT80B rich1/TM6b; and y w UAS-SytGFP; FRT40A CadN405/CyO; FRT80B rich2/TM6B). Compared with Figure 7(E) and (F), removal of one copy of CadN enhances the R7 targeting phenotypes of the hypomorphic allele rich2 but not the phenotypes of the null allele rich1, indicating Rich and CadN function in a same pathway to regulate R7 targeting. (J) The quantification of R7 targeting phenotypes of rich1 mutants (Figure 7E), rich2 mutants (Figure 7F) and mutants of rich1 and rich2 with one copy of CadN. Upon removal of one copy of CadN, 19.6 ± 2% (n=384) of R7 cells in rich1 and 19.0 ± 2 % (n=493) of R7 cells in rich2 mutants fail to target to the correct layer. (K) The CadN, rich1 double mutants have similar eye targeting phenotypes as CadN single mutants. The genotypes are: (CTL) y w eyFLP;; FRT80B cl ubi-GFP/FRT80B iso. (rich1): y w eyFLP;; FRT80B cl ubi-GFP/FRT80B rich1. (CadN): y w eyFLP; FRT40A CadNΔ14/FRT40A ubi-GFP. (CadN; rich1): y w eyFLP; FRT40A rich-gRE::VK37/FRT40A CadNΔ14; FRT80B rich1/3L6 deletion. (L) Quantification of mistargeted R7 of control flies (CTL): 0%, n=458; rich1 mutants, 18.9± 3.9%, n=450; CadN mutants, 24.6±2.2%, n= 244; CadN; rich1 mutants: 24.4±1.6%, n=237. (See also Figure S6–S8)
To determine whether the subcellular localization of CadN is also regulated by Rich in other cells than PRs, we examined rich mutant phenotypes in the developing eye. Each ommatidium consists of twenty cells, including four cone cells. The cone cells form a plate on the top of the photoreceptors, and previous studies have shown that CadN is localized at the adherens junction of cone cell interfaces and plays a role in regulating cone cell patterns (Hayashi and Carthew, 2004). We therefore created mutant clones of rich in cone cells and stained for CadN in developing eyes 36 hrs after puparium formation (Figure 8G–H′). We did not observe any obvious changes of CadN distribution between cone cells in wild type and mutant cells. These data indicate that Rich regulates CadN in a cell type specific manner.
Loss of CadN leads to defects in cartridge formation in lamina and mistargeting of R7 cells (Lee et al., 2001), similar to the defects we observed in rich and Rab6 mutants, suggesting that Rich and Rab6 function in a common pathway to regulate CadN trafficking. To test this, we removed one copy of CadN from eyFLP; rich1 or eyFLp; rich2 mutant animals. Loss of one copy of CadN greatly enhanced the targeting phenotype of the hypomorphic allele rich2 but not the of the null allele rich1, providing further evidence that rich and CadN function in a common pathway (Figures 8I–L). Moreover, homozygous double mutants for rich and CadN exhibit very similar phenotypes to CadN mutants. The data therefore indicate that Rich and Rab6 regulate CadN trafficking to affect axon target selection in the eye. To assess the specificity of this genetic interaction we also tested weather rich interacted genetically with other genes, including DLAR, liprin α, or Jeb. We did not observe any interactions between rich and DLAR, liprin α, or Jeb (Figure S6B), consistent with our previous data.
CadN is broadly expressed in the fly CNS and also plays important roles in determining synaptic specificity of olfactory receptor neurons (ORNs) (Hummel and Zipursky, 2004; Zhu and Luo, 2004). To test whether rich and Rab6 mutants exhibit similar phenotypes in other neurons we focused on the ORNs. In Drosophila, around 1500 ORNs are present in the antenna and the maxillary palps. The ORNs send their axons into the antenna lobe (AL), where they form around 50 highly organized neuropilar structures, the glomeruli (Laissue et al., 1999). The axons of ORNs in CadN mutant typically target the appropriate region of the antennal lobe but fail to converge on a single glomerulus and instead spread out on the surface of different glomeruli (Hummel and Zipursky, 2004; Zhu and Luo, 2004). We generated mosaic flies with rich or Rab6 mutant ORN and wild type AL targets using the MARCM system. eyFLP was used to induce mitotic recombination in the ORN progenitor cells but not their targets (Hummel and Zipursky, 2004). To distinguish different subclasses of ORNs, different olfactory receptor Gal4s were used to label the mutant ORNs. We tested 3 different subclasses of ORNs, including two from the antenna (OR22a, Or47b) and one from maxillary palps (Or46a). These three ORNs were previously shown to require CadN to establish proper connections with their targets (Hummel and Zipursky, 2004). In both rich and Rab6 mutants, the Or47b and Or46a neurons fail to converge their axons into a single glomerulus, very similar to CadN mutants, indicating that rich, Rab6, and CadN regulate a common process in the antenna (Figure S8C). However, the Or22a neurons do not have any obvious defects when rich or Rab6 is lost (Figure S8C), in contrast to CadN mutants. Hence, the ORNs seem to have different sensitivities to a reduction in CadN. The difference between the Or47b, the Or46a and the Or22a neurons in rich and Rab6 mutants may reflect this difference.
Disscussion
Here we report the identification of mutations in rich, a gene that is evolutionarily conserved from worms to human. rich is required for synaptic specificity in Drosophila eyes and olfactory receptor neurons and acts together with Rab6. Our data define a novel role for Rich and Rab6 in regulating axon targeting in the eye by regulating CadN trafficking in a subset of neurons to control target specificity.
How does Rich Regulate Rab6
Rab6 has been implicated in multiple membrane trafficking pathway and numerous “downstream” effectors have been identified (Valente et al., 2010). However, the “upstream” regulators have not yet been identified in higher eukaryotes. The Ric1p forms a complex with Rgp1p in yeast and promotes GTP exchange for the yeast Rab6 homologue Ypt6 (Siniossoglou et al., 2000). Surprisingly, although Rab6 family proteins are highly conserved (similarity between Rab6 and Ypt6 is 84%), the Ric1p and Rgp1p only exhibit limited similarity with the fly and vertebrate homologues (Fig. 3B). In addition, fly Rich contains several WD40 domains not found in the yeast protein, yet both the RIC1 and WD40 domains bind to Rab6. Importantly, rich and Rab6 show obvious genetic interactions and similarities in phenotypes in flies. However, we were not able to detect GTP exchange activity of Rich, nor were we able to find an interaction between Rich and the Drosophila Rgp1p like protein. Therefore, it is likely that Rich is using other interactors to regulate Rab6 activity. Moreover, we found that Rich/Rab6 regulates CadN trafficking in a cell type specific manner, yet both Rab6 and Rich are broadly expressed in brains. Hence, the other Rich interactors might be key to modulate Rab6 activity differentially in various cell types.
Rab6 and CadN Trafficking
In the medulla, several cell surface proteins have been identified that regulate R7 or R8 targeting in a cell type specific manner. For example, CadN (Lee et al., 2001), as well as DLAR (Clandinin et al., 2001) and PTP69D (Newsome et al., 2000), mainly regulate R7 but not R8 synaptic specificity. On the other hand, Jeb (Bazigou et al., 2007), together with Flamingo (fmi) (Senti et al., 2003) and Golden goal (gogo) (Tomasi et al., 2008) direct R8 but not R7 targeting. The expression patterns of these cell surface molecules are broad, whereas their cell type specific functions are quite defined. It is therefore likely that these proteins depend on a regulated set of trafficking rules to achieve synaptic specificity. However, so far, only Sec15 has been shown to affect synaptic specificity, and in sec15 mutants, the synaptic specificity of both R7 and R8 are affected (Mehta et al., 2005). Here, we established a role for two proteins that have not yet been implicated in trafficking of important cell surface proteins in the CNS like CadN. The role of CadN at numerous synapses has been extensively studied (Dalva et al., 2007; Ting and Lee, 2007) but the molecular pathways by which CadN is trafficked have not yet been identified. Our data indicate that both Rich and Rab6 regulate in a specific manner the function of CadN to mediate R7 but not R8 target specificity. Similarly, synapse specificity of ORNs is selectively affected in a subpopulation of ORNs. Moreover, in the visual system, none of the other tested proteins (DLAR, PTP69D, and Jeb) are mislocalized in rich and Rab6 mutants. We find this specificity surprising as Rab6 is known to play multiple roles in membrane trafficking. It controls not only retrograde transport from the endosome to the Golgi or from the Golgi to the ER (Del Nery et al., 2006; Girod et al., 1999; White et al., 1999; Yano et al., 2005) but also targeting of secretory vesicles to the plasma membrane (Grigoriev et al., 2007) and affects localization of Yolkless and Gurken to the oocyte membrane in flies (Coutelis and Ephrussi, 2007). Hence, it is possible that many different proteins that are required for synaptic specificity use unique trafficking routes. It has been previously shown that overexpression of a dominant negative form of Rab6 affects Rhodopsin transport and that this overexpression induces an age-dependent retinal degeneration (Shetty et al., 1998). However, we did not observe a retinal degeneration in both rich and Rab6 mutant eye clones based on ERGs and TEM in 1 day old and 3 week old flies.
Since Rab6 is not only involved in retrograde trafficking (Del Nery et al., 2006; Girod et al., 1999; White et al., 1999; Yano et al., 2005), but also regulates exocytosis (Grigoriev et al., 2007), it remains to be determined how CadN is mistrafficked. It has been reported that CadN undergoes constitutive endocytosis in hippocampal neurons which maybe affected by Rich/Rab6. Alternatively, the cell surface presentation of newly synthesized CadN may require Rab6 dependent exocytosis. Upon activation of NMDA receptors, the rate of CadN endocytosis is significantly reduced in hippocampus neurons, resulting in the accumulation of CadN at the cell surface (Tai et al., 2007). It is possible that Rich/Rab6 is required for this activity as well.
CadN Trafficking and Synaptic Specificity
Neural circuit assembly is a complex process. The conventional model of synaptic specificity is that axons and their target bear labels that act as “locks and keys” to match pre- and post-synaptic partners. This model assumes that each neuron has its unique identifier that is critical to specify its synapses Recent studies suggest that alternative splicing of Dscam and protocadherins (Pcdhs), provide unique identifiers to recognize self versus non-self, but they do not seem to play a critical role in defining synaptic specificity (Sanes and Zipursky, 2010). In many cases, the specificity seems to be mediated by repeated use of a few key molecules like CadN. Hence, local quantitative differences of these key adhesion molecule affect synaptic specificity. In flies, the CadN expression pattern is very dynamic in the developing medulla (Nern et al., 2008). CadN is required in each lamina neuron for specific aspects of targeting. The subtle difference in cell surface levels of CadN in different cell types enables this widely expressed protein to mediate specific local interactions during circuit assembly (Nern et al., 2008). It remains an enigma how the quantitative difference of CadN on the cell surface is achieved. Our data suggest that CadN trafficking via Rich/Rab6 modulates synaptic specificity in a subset of neurons, thereby providing a remarkable specificity for a Rab when compared to other Rabs like Rab5 and Rab11 that affect numerous processes.
Experimental procedures
Molecular biology
We generated a genomic rescue construct including a 7.5 kb fragment retrieved from BACR48E10 (BACPAC resources) in attB-P[acman]-ApR using recombineering as described (Venken et al., 2006). The 3XHA-tagged genomic rescue construct was generated by PCR amplification of the genomic fragment and introduction of an AgeI site before the ATG start codon of CG9063. Transgenes were obtained by germline transformation using phiC31 mediated transgenesis in the VK37 docking site on the second chromosome. To generate the V5 tagged expression vector of full length, WD40, or RIC1 domain of Rich protein, we amplified the cDNA fragments from the cDNA clone (GH03694) using PCR and inserted these fragments into the pAc-V5His (Invitrogen) vector. We also constructed expression vectors for GST fusion proteins containing Rab6WT, Rab6CA, and Rab6DN by PCR amplifying Rab6 fragments from pUASYFPRab6, Rab6CA and RabDN (Zhang et al., 2007) respectively, and inserting the PCR fragments into the pGEX4T-1 vector.
Genetics
rich mutants were isolated from an ey-FLP ethane methylsulfonate (EMS) screen as described previously (Ohyama et al., 2007). The rough mapping using P-element meiotic recombination was performed as described (Bellen et al., 2004; Zhai et al., 2003). The genotypes of the fly strains generated in the paper are described in supplemental data.
Immunohistochemistry
Eye discs, salivary gland, and larval, pupal, and adult brains were fixed in PBS with 3.7% formaldehyde for 20 mins, followed by washing with PBX (PBS + 0.4% Triton X-100) 3 times. The tissues were incubated with primary antibody overnight in 4°C followed by extensive washing and incubated with 2nd antibody for 2 hrs. at RT. For pupal and adult brains, we also incubated with 2nd antibody overnight at 4°C. After extensive washing the samples were mounted in vectorshield followed by microscopy. S2 cells were fixed in 3.7% formaldehyde for 10 mins, and both the primary and secondary antibody incubation was 1hr. at RT.
S2 cell transfection
S2 cells were cultured under standard conditions. The transfections were performed using Lipofectamin LTX (Invitrogen). Two days after transfection, the cells were harvested and used for staining, GST pull down, GEF activity, or immunoprecipitation assay.
GST pull down and western blot
S2 cells were transfected with either pAcRIC1 or pAcWD40. The cell lysate was made by incubating cells in lysis buffer (50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 10 mM NaF, 1 mM Na3VO4,1% NP40, 10% glycerol, protease inhibitor tablets (Roche)) for 30 min followed by centrifuging at 14,000rpm for 10 min at 4°C. GST fusion proteins were made as described in the manufacturer’s manual. The sepharose beads with GST or GST fusion Rab6, Rab6CA, or Rab6 DN were incubated with the cell lysate for 1 h at RT and followed by washing 3 times with lysis buffer. The resulting beads-protein complexes were resolved with SDS-PAGE. The western blot was performed as described before. The anti-V5 antibody (Invitrogen) dilution is 1:5000.
Immunoprecipitation (IP)
The IP experiments were performed as described before (Tong and Jiang, 2007). The anti-V5 antibody (Invitrogen) (dilution: 1:250) was used to pull down the yRic1p or Rich protein.
ERG recording
ERG recordings were performed as described before (Verstreken et al., 2003).
Transmission Electron Microscopy
TEM was performed as described previously (Verstreken et al., 2003). For PR terminal distribution, PR terminal was determined by presence of capitate projections.
Supplementary Material
Acknowledgments
We are grateful to S. L. Zipursky, CH. Lee, T. R. Clandinin, I. Salecker, P. R. Hiesinger, A. Ephrussi, R. H. Palmer, T. Hummel, the Bloomington Drosophila Stock Center and the Developmental Studies Hybridoma Bank for providing reagents. We thank Yuchun He, Hong-Lin Pan for injections to generate transgenic flies. We thank P. R. Hiesinger, N. Giagtzoglou, V. Bayat, for comments. H. J. Bellen is an investigator of the Howard Hughes Medical Institute. C. Tong is supported by a T32 from the National Institute of Neurological Disorders. Confocal microscopy was supported by the Mental Retardation and Developmental Disabilities Research Center at Baylor College of Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bazigou E, Apitz H, Johansson J, Loren CE, Hirst EM, Chen PL, Palmer RH, Salecker I. Anterograde Jelly belly and Alk receptor tyrosine kinase signaling mediates retinal axon targeting in Drosophila. Cell. 2007;128:961–975. doi: 10.1016/j.cell.2007.02.024. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KM, Prakash S, Bright A, Clandinin TR. Liprin-alpha is required for photoreceptor target selection in Drosophila. Proc Natl Acad Sci U S A. 2006;103:11601–11606. doi: 10.1073/pnas.0601185103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotard C, Leung W, Salecker I. glial cells missing and gcm2 cell autonomously regulate both glial and neuronal development in the visual system of Drosophila. Neuron. 2005;48:237–251. doi: 10.1016/j.neuron.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, Lee CH, Herman T, Lee RC, Yang AY, Ovasapyan S, Zipursky SL. Drosophila LAR regulates R1–R6 and R7 target specificity in the visual system. Neuron. 2001;32:237–248. doi: 10.1016/s0896-6273(01)00474-3. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, Zipursky SL. Afferent growth cone interactions control synaptic specificity in the Drosophila visual system. Neuron. 2000;28:427–436. doi: 10.1016/s0896-6273(00)00122-7. [DOI] [PubMed] [Google Scholar]
- Cooper MT, Bray SJ. Frizzled regulation of Notch signalling polarizes cell fate in the Drosophila eye. Nature. 1999;397:526–530. doi: 10.1038/17395. [DOI] [PubMed] [Google Scholar]
- Coutelis JB, Ephrussi A. Rab6 mediates membrane organization and determinant localization during Drosophila oogenesis. Development. 2007;134:1419–1430. doi: 10.1242/dev.02821. [DOI] [PubMed] [Google Scholar]
- Culi J, Mann RS. Boca, an endoplasmic reticulum protein required for wingless signaling and trafficking of LDL receptor family members in Drosophila. Cell. 2003;112:343–354. doi: 10.1016/s0092-8674(02)01279-5. [DOI] [PubMed] [Google Scholar]
- Dalva MB, McClelland AC, Kayser MS. Cell adhesion molecules: signalling functions at the synapse. Nat Rev Neurosci. 2007;8:206–220. doi: 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Nery E, Miserey-Lenkei S, Falguieres T, Nizak C, Johannes L, Perez F, Goud B. Rab6A and Rab6A′ GTPases play non-overlapping roles in membrane trafficking. Traffic. 2006;7:394–407. doi: 10.1111/j.1600-0854.2006.00395.x. [DOI] [PubMed] [Google Scholar]
- Garrity PA, Lee CH, Salecker I, Robertson HC, Desai CJ, Zinn K, Zipursky SL. Retinal axon target selection in Drosophila is regulated by a receptor protein tyrosine phosphatase. Neuron. 1999;22:707–717. doi: 10.1016/s0896-6273(00)80730-8. [DOI] [PubMed] [Google Scholar]
- Giagtzoglou Nikolaos CVL, Bellen Hugo J. Cell Adhesion, the backbone of synapse: “vertebrate” and “invertebrate” perspectives. Cold Spring Harbor Perspectives in Biology. 2009 doi: 10.1101/cshperspect.a003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod A, Storrie B, Simpson JC, Johannes L, Goud B, Roberts LM, Lord JM, Nilsson T, Pepperkok R. Evidence for a COP-I-independent transport route from the Golgi complex to the endoplasmic reticulum. Nat Cell Biol. 1999;1:423–430. doi: 10.1038/15658. [DOI] [PubMed] [Google Scholar]
- Grigoriev I, Splinter D, Keijzer N, Wulf PS, Demmers J, Ohtsuka T, Modesti M, Maly IV, Grosveld F, Hoogenraad CC, et al. Rab6 regulates transport and targeting of exocytotic carriers. Dev Cell. 2007;13:305–314. doi: 10.1016/j.devcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Hiesinger PR, Zhai RG, Zhou Y, Koh TW, Mehta SQ, Schulze KL, Cao Y, Verstreken P, Clandinin TR, Fischbach KF, et al. Activity-independent prespecification of synaptic partners in the visual map of Drosophila. Curr Biol. 2006;16:1835–1843. doi: 10.1016/j.cub.2006.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeyer K, Maurel-Zaffran C, Sink H, Treisman JE. Liprin-alpha has LAR-independent functions in R7 photoreceptor axon targeting. Proc Natl Acad Sci U S A. 2006;103:11595–11600. doi: 10.1073/pnas.0604766103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T, Zipursky SL. Afferent induction of olfactory glomeruli requires N-cadherin. Neuron. 2004;42:77–88. doi: 10.1016/s0896-6273(04)00158-8. [DOI] [PubMed] [Google Scholar]
- Januschke J, Nicolas E, Compagnon J, Formstecher E, Goud B, Guichet A. Rab6 and the secretory pathway affect oocyte polarity in Drosophila. Development. 2007;134:3419–3425. doi: 10.1242/dev.008078. [DOI] [PubMed] [Google Scholar]
- Kondylis V, Goulding SE, Dunne JC, Rabouille C. Biogenesis of Golgi stacks in imaginal discs of Drosophila melanogaster. Mol Biol Cell. 2001;12:2308–2327. doi: 10.1091/mbc.12.8.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunes S, Steller H. Topography in the Drosophila visual system. Curr Opin Neurobiol. 1993;3:53–59. doi: 10.1016/0959-4388(93)90035-w. [DOI] [PubMed] [Google Scholar]
- Laissue PP, Reiter C, Hiesinger PR, Halter S, Fischbach KF, Stocker RF. Three-dimensional reconstruction of the antennal lobe in Drosophila melanogaster. J Comp Neurol. 1999;405:543–552. [PubMed] [Google Scholar]
- Lee CH, Herman T, Clandinin TR, Lee R, Zipursky SL. N-cadherin regulates target specificity in the Drosophila visual system. Neuron. 2001;30:437–450. doi: 10.1016/s0896-6273(01)00291-4. [DOI] [PubMed] [Google Scholar]
- Lee RC, Clandinin TR, Lee CH, Chen PL, Meinertzhagen IA, Zipursky SL. The protocadherin Flamingo is required for axon target selection in the Drosophila visual system. Nat Neurosci. 2003;6:557–563. doi: 10.1038/nn1063. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Martinez O, Antony C, Pehau-Arnaudet G, Berger EG, Salamero J, Goud B. GTP-bound forms of rab6 induce the redistribution of Golgi proteins into the endoplasmic reticulum. Proc Natl Acad Sci U S A. 1997;94:1828–1833. doi: 10.1073/pnas.94.5.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez O, Schmidt A, Salamero J, Hoflack B, Roa M, Goud B. The small GTP-binding protein rab6 functions in intra-Golgi transport. J Cell Biol. 1994;127:1575–1588. doi: 10.1083/jcb.127.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matanis T, Akhmanova A, Wulf P, Del Nery E, Weide T, Stepanova T, Galjart N, Grosveld F, Goud B, De Zeeuw CI, et al. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat Cell Biol. 2002;4:986–992. doi: 10.1038/ncb891. [DOI] [PubMed] [Google Scholar]
- Matthews BJ, Corty MM, Grueber WB. Of cartridges and columns: new roles for cadherins in visual system development. Neuron. 2008;58:1–3. doi: 10.1016/j.neuron.2008.03.024. [DOI] [PubMed] [Google Scholar]
- Maurel-Zaffran C, Suzuki T, Gahmon G, Treisman JE, Dickson BJ. Cell-autonomous and -nonautonomous functions of LAR in R7 photoreceptor axon targeting. Neuron. 2001;32:225–235. doi: 10.1016/s0896-6273(01)00471-8. [DOI] [PubMed] [Google Scholar]
- McAllister AK. Dynamic aspects of CNS synapse formation. Annu Rev Neurosci. 2007;30:425–450. doi: 10.1146/annurev.neuro.29.051605.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta SQ, Hiesinger PR, Beronja S, Zhai RG, Schulze KL, Verstreken P, Cao Y, Zhou Y, Tepass U, Crair MC, et al. Mutations in Drosophila sec15 reveal a function in neuronal targeting for a subset of exocyst components. Neuron. 2005;46:219–232. doi: 10.1016/j.neuron.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Hanson TE. The development of the optic lobe. In: Martinez-Arias MBaA., editor. The Development of Drosophila Melanogaster. Cold Sprin Harbor, NY: Cold Spring Harbor Press; 1993. pp. 1363–1491. [Google Scholar]
- Morey M, Yee SK, Herman T, Nern A, Blanco E, Zipursky SL. Coordinate control of synaptic-layer specificity and rhodopsins in photoreceptor neurons. Nature. 2008;456:795–799. doi: 10.1038/nature07419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Nguyen LV, Herman T, Prakash S, Clandinin TR, Zipursky SL. An isoform-specific allele of Drosophila N-cadherin disrupts a late step of R7 targeting. Proc Natl Acad Sci U S A. 2005;102:12944–12949. doi: 10.1073/pnas.0502888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Zhu Y, Zipursky SL. Local N-cadherin interactions mediate distinct steps in the targeting of lamina neurons. Neuron. 2008;58:34–41. doi: 10.1016/j.neuron.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–362. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Verstreken P, Ly CV, Rosenmund T, Rajan A, Tien AC, Haueter C, Schulze KL, Bellen HJ. Huntingtin-interacting protein 14, a palmitoyl transferase required for exocytosis and targeting of CSP to synaptic vesicles. J Cell Biol. 2007;179:1481–1496. doi: 10.1083/jcb.200710061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdam FJ, Echard A, Croes HJ, van den Hurk JA, van de Vorstenbosch RA, Ginsel LA, Goud B, Fransen JA. The small GTPase Rab6B, a novel Rab6 subfamily member, is cell-type specifically expressed and localised to the Golgi apparatus. J Cell Sci. 2000;113(Pt 15):2725–2735. doi: 10.1242/jcs.113.15.2725. [DOI] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Petrovic M, Hummel T. Temporal identity in axonal target layer recognition. Nature. 2008;456:800–803. doi: 10.1038/nature07407. [DOI] [PubMed] [Google Scholar]
- Prakash S, Caldwell JC, Eberl DF, Clandinin TR. Drosophila N-cadherin mediates an attractive interaction between photoreceptor axons and their targets. Nat Neurosci. 2005;8:443–450. doi: 10.1038/nn1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Y, Pang P, Ruan W, Gunning D, Zipursky SL. brakeless is required for photoreceptor growth-cone targeting in Drosophila. Proc Natl Acad Sci U S A. 2000;97:5966–5971. doi: 10.1073/pnas.110135297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnakumar K, Desplan C. Rh-GAL4 and Rh-GFP insertions 2003 [Google Scholar]
- Ratnakumar K, Desplan C. P{ninaE-EGFP.P} and P{Pan-R7-GAL4} insertions 2004a [Google Scholar]
- Ratnakumar K, Desplan C. P{rh1-GAL4} and P{Rh4-GAL4} insertions 2004b [Google Scholar]
- Ruan W, Pang P, Rao Y. The SH2/SH3 adaptor protein dock interacts with the Ste20-like kinase misshapen in controlling growth cone motility. Neuron. 1999;24:595–605. doi: 10.1016/s0896-6273(00)81115-0. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Zipursky SL. Design principles of insect and vertebrate visual systems. Neuron. 2010;66:15–36. doi: 10.1016/j.neuron.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti K, Keleman K, Eisenhaber F, Dickson BJ. brakeless is required for lamina targeting of R1–R6 axons in the Drosophila visual system. Development. 2000;127:2291–2301. doi: 10.1242/dev.127.11.2291. [DOI] [PubMed] [Google Scholar]
- Senti KA, Usui T, Boucke K, Greber U, Uemura T, Dickson BJ. Flamingo regulates R8 axon-axon and axon-target interactions in the Drosophila visual system. Curr Biol. 2003;13:828–832. doi: 10.1016/s0960-9822(03)00291-4. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Love J, Colman DR. Adhesion molecules in the nervous system: structural insights into function and diversity. Annu Rev Neurosci. 2007;30:451–474. doi: 10.1146/annurev.neuro.29.051605.113034. [DOI] [PubMed] [Google Scholar]
- Short B, Preisinger C, Schaletzky J, Kopajtich R, Barr FA. The Rab6 GTPase regulates recruitment of the dynactin complex to Golgi membranes. Curr Biol. 2002;12:1792–1795. doi: 10.1016/s0960-9822(02)01221-6. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Peak-Chew SY, Pelham HR. Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. Embo J. 2000;19:4885–4894. doi: 10.1093/emboj/19.18.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley H, Botas J, Malhotra V. The mechanism of Golgi segregation during mitosis is cell type-specific. Proc Natl Acad Sci U S A. 1997;94:14467–14470. doi: 10.1073/pnas.94.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai CY, Mysore SP, Chiu C, Schuman EM. Activity-regulated N-cadherin endocytosis. Neuron. 2007;54:771–785. doi: 10.1016/j.neuron.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- Ting CY, Lee CH. Visual circuit development in Drosophila. Curr Opin Neurobiol. 2007;17:65–72. doi: 10.1016/j.conb.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Ting CY, Yonekura S, Chung P, Hsu SN, Robertson HM, Chiba A, Lee CH. Drosophila N-cadherin functions in the first stage of the two-stage layer-selection process of R7 photoreceptor afferents. Development. 2005;132:953–963. doi: 10.1242/dev.01661. [DOI] [PubMed] [Google Scholar]
- Tomasi T, Hakeda-Suzuki S, Ohler S, Schleiffer A, Suzuki T. The transmembrane protein Golden goal regulates R8 photoreceptor axon-axon and axon-target interactions. Neuron. 2008;57:691–704. doi: 10.1016/j.neuron.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Tong C, Jiang J. Using immunoprecipitation to study protein-protein interactions in the Hedgehog-signaling pathway. Methods Mol Biol. 2007;397:215–229. doi: 10.1007/978-1-59745-516-9_15. [DOI] [PubMed] [Google Scholar]
- Utskarpen A, Slagsvold HH, Iversen TG, Walchli S, Sandvig K. Transport of ricin from endosomes to the Golgi apparatus is regulated by Rab6A and Rab6A′. Traffic. 2006;7:663–672. doi: 10.1111/j.1600-0854.2006.00418.x. [DOI] [PubMed] [Google Scholar]
- Valente C, Polishchuk R, De Matteis MA. Rab6 and myosin II at the cutting edge of membrane fission. Nat Cell Biol. 2010;12:635–638. doi: 10.1038/ncb0710-635. [DOI] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Verstreken P, Koh TW, Schulze KL, Zhai RG, Hiesinger PR, Zhou Y, Mehta SQ, Cao Y, Roos J, Bellen HJ. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 2003;40:733–748. doi: 10.1016/s0896-6273(03)00644-5. [DOI] [PubMed] [Google Scholar]
- White J, Johannes L, Mallard F, Girod A, Grill S, Reinsch S, Keller P, Tzschaschel B, Echard A, Goud B, et al. Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J Cell Biol. 1999;147:743–760. doi: 10.1083/jcb.147.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Boulianne GL, Trimble WS. Drosophila syntaxin 16 is a Q-SNARE implicated in Golgi dynamics. J Cell Sci. 2002;115:4447–4455. doi: 10.1242/jcs.00139. [DOI] [PubMed] [Google Scholar]
- Yano H, Yamamoto-Hino M, Abe M, Kuwahara R, Haraguchi S, Kusaka I, Awano W, Kinoshita-Toyoda A, Toyoda H, Goto S. Distinct functional units of the Golgi complex in Drosophila cells. Proc Natl Acad Sci U S A. 2005;102:13467–13472. doi: 10.1073/pnas.0506681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai RG, Hiesinger PR, Koh TW, Verstreken P, Schulze KL, Cao Y, Jafar-Nejad H, Norga KK, Pan H, Bayat V, et al. Mapping Drosophila mutations with molecularly defined P element insertions. Proc Natl Acad Sci U S A. 2003;100:10860–10865. doi: 10.1073/pnas.1832753100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Schulze KL, Hiesinger PR, Suyama K, Wang S, Fish M, Acar M, Hoskins RA, Bellen HJ, Scott MP. Thirty-one flavors of Drosophila rab proteins. Genetics. 2007;176:1307–1322. doi: 10.1534/genetics.106.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Luo L. Diverse functions of N-cadherin in dendritic and axonal terminal arborization of olfactory projection neurons. Neuron. 2004;42:63–75. doi: 10.1016/s0896-6273(04)00142-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.