Abstract
Here we report a novel Arabidopsis dwarf mutant, fackel-J79, whose adult morphology resembles that of brassinosteroid-deficient mutants but also displays distorted embryos, supernumerary cotyledons, multiple shoot meristems, and stunted roots. We cloned the FACKEL gene and found that it encodes a protein with sequence similarity to both the human sterol reductase family and yeast C-14 sterol reductase and is preferentially expressed in actively growing cells. Biochemical analysis indicates that the fk-J79 mutation results in deficient C-14 sterol reductase activity, abnormal sterol composition, and reduction of brassinosteroids (BRs). Unlike other BR-deficient mutants, the defect of hypocotyl elongation in fk-J79 cannot be corrected by exogenous BRs. The unique phenotypes and sterol composition in fk-J79 indicate crucial roles of sterol regulation and signaling in cell division and cell expansion in embryonic and post-embryonic development in plants.
Keywords: Arabidopsis, brassinosteroids, C-14 sterol reductase, dwarf, embryogenesis, meristem, sterols
Despite the identification of plant steroids more than two decades ago, only recently have the profound effects of brassinosteroids (BRs) on plant growth and development (Clouse and Sasse 1998; Altmann 1999) been fully recognized and detailed biosynthetic pathways established (Fujioka and Sakurai 1997a,b; Yokota 1997). Physiological, biochemical, and molecular genetic studies have demonstrated that BRs are essential for cell expansion, skotomorphogenesis, apical dominance, leaf and chloroplast senescence, male fertility, and gene expression.
Our understanding of the functions of BRs has been facilitated by the isolation and characterization of BR biosynthetic and perception mutants in Arabidopsis thaliana (Altmann 1999). All of the BR-deficient mutants reported thus far are dwarfs, due to a general defect of cell expansion in aerial parts. Molecular characterization has revealed that the de-etiolated2 (det2), constitutive photomorphogenesis and dwarfism (cpd), and dwarf4 (dwf4) mutants of Arabidopsis have specific defects in the BR biosynthetic pathway. DET2 encodes the ortholog of a human steroid 5α-reductase that can perform a similar enzymatic function in a human cell line (Fujioka et al. 1997; Li et al. 1997; Noguchi et al. 1999a). DWF4 encodes a cytochrome P450 that catalyzes the sterol C-22α hydroxylation reaction that is proposed to constitute a rate-limiting step in BR biosynthesis (Choe et al. 1998). CPD is also a cytochrome P450 and catalyzes the sterol C-23α hydroxylation reaction (Szekeres et al. 1996). Defective cell expansion can be corrected by exogenous BRs in these three mutants. Three additional allelic Arabidopsis mutants, bri1 (Clouse et al. 1996), bin (Li and Chory 1997), and cbb2 (Kauschmann et al. 1996), phenotypically resemble BR-deficient mutants, but are insensitive to exogenous BRs. Molecular cloning revealed that BRI encodes a putative leucine-rich repeat receptor kinase involved in BR signal transduction (Li and Chory 1997).
Beside BRs, dozens of other sterols are also found in plants. Major plant sterols such as sitosterol and stigmasterol are similar in structure to the fungal ergosterol and cholesterol in animals. Animal sterols are known to regulate transcriptional and post-transcriptional events, which, in turn, affect lipid synthesis, meiosis, apoptosis, developmental patterning, protein cleavage, and protein degradation (Edwards and Ericsson 1999). In yeast, sterols are found to have specific regulatory and developmental roles in addition to structural roles in determining general membrane properties (Parks et al. 1995). In plants, however, it is presently unclear whether any plant sterols other than BRs play regulatory roles in development. Nor is it clear how sterol biosynthesis in plants is controlled (Yokota 1997; Hartmann 1998). On the basis of studies using Arabidopsis as a model, the sterol biosynthetic pathway in plants can be divided into sterol-specific and BR-specific pathways (Choe et al. 1999b). The phenotypes of several dwarf mutants such as dwf7 and dim1 (cbb1, dwf1), blocked in both BR- and sterol-specific pathways, are similar to those blocked only in the BR-specific pathway (Takahashi et al. 1995; Kauschmann et al. 1996; Klahre et al. 1998; Choe et al. 1999a,b). The defects in dwf7 and dim1 mutants can also be rescued by exogenous BRs, suggesting that no sterols other than BRs are actively involved in the promotion of cell expansion in post-embryonic development.
Here we describe a novel dwarf mutant of Arabidopsis, ell1 (extra-long-lifespan1), that exhibits unique phenotypes caused by abnormal cell division and cell expansion in both embryonic and post-embryonic development. We have found that ell1 (fk-J79) is allelic to a previously described embryonic patterning mutant fk-X224 (Kathrin Schrick, pers. comm.) (Mayer et al. 1991). The deduced FK sequence predicts a protein that is similar to the human lamin B receptor (LBR)/sterol reductase multigene family (Holmer et al. 1998) and various yeast sterol reductases including C-14 sterol reductase (ERG24) (Lorenz and Parks 1992). The C-14 sterol reductase in plants is thought to act early in the biosynthetic pathway, before the divergence of BR- and sterol-specific pathways (Choe et al. 1999b). Both fk-J79 and fk-X224 mutants fail to develop a typical hypocotyl. On the basis of analysis of sterol composition and the failure of exogenous BRs to restore the mutants to wild-type growth and development, we propose that the phenotype of fk-J79 is caused by both the synthesis of abnormal sterols and reduced amounts of BRs and non-BR sterols.
Results
fk-J79 is pleiotropic
Cytokinin is a hormone-controlling cell division, shoot initiation, apical dominance, and senescence in higher plants (D'Agostino and Kieber 1999). We conducted a genetic screen for constitutive cytokinin response mutants on the basis of the observation that cytokinin-treated 12-day-old, dark-grown seedlings of wild-type Arabidopsis showed stunted hypocotyls and roots (Chory et al. 1994) and turn green faster upon illumination. We initially identified the ell1 mutant as a putative constitutive cytokinin response mutant from T-DNA mutagenized Arabidopsis seedlings on MS plates in the absence of exogenous cytokinins. The ell1 mutant displayed pleiotropic phenotypes during both embryonic and postembryonic development. We subsequently found the ell1 mutant to be allelic to fk-X224 (Mayer et al. 1991); ell1 was renamed fk-J79.
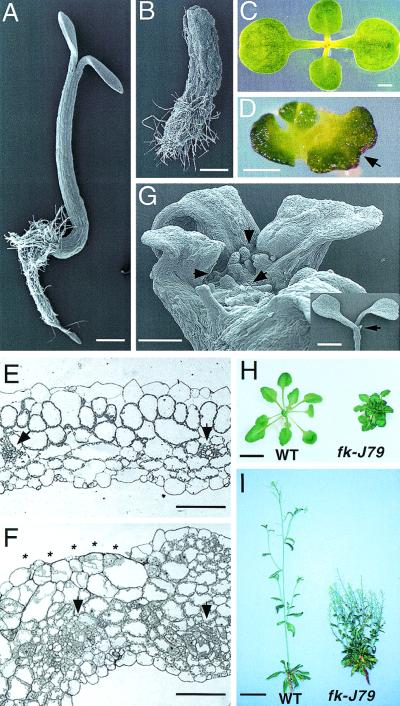
fk-J79 is a near-sterile single nuclear recessive mutant. fk-J79 seedlings were not able to elongate in either light (Fig. 1A,B) or dark (data not shown). In the light, fk-J79 plants developed stunted roots, a short, but thick structure in the region that the hypocotyl normally forms, and thick, irregular cotyledons (Fig. 1C,D). Epidermal cells in the hypocotyl-like region varied greatly in size and shape (Fig. 1B). The thickening of the cotyledons was due to abnormal cell division as evidenced by extra layers of mesophyll cells and a dramatic increase in the number of cytoplasmic-dense phloem cells (Fig. 1E,F). In addition, the continuity of the epidermal cell layer was often disrupted (Fig. 1F). The abundance of chloroplasts and increased cell layers resulted in the dark-green phenotype of fk-J79 (Fig. 1D,F). Cotyledon fusions occurred occasionally as evidenced by the sinuate cotyledons and disrupted phyllotaxis (Fig. 1D), which resembled cup-shaped cotyledon (cuc) mutants (Aida et al. 1997). One unusual feature of fk-J79 is the manifestation of supernumerary cotyledons. fk-J79/+ plants were fully fertile and produced 25% fk-J79 progeny in a self cross, of which ∼65% were dicotyledonary and ∼25% were tricotyledonary (data not shown). Examination of germinating fk-J79 seedlings revealed that individuals with three to four cotyledons often contained two or more adventitious shoot apical meristems (SAM) (Fig. 1G). Dark-grown fk-J79 plants resembled their light-grown counterparts except that they were etiolated and the cotyledons were not expanded. The apical hook was not seen in fk-J79 seedlings due to defective embryogenesis (described below). Notably, fk-J79 plants continued to develop in the dark, producing a number of rosette leaves and a short, branching root system (data not shown).
Figure 1.
Morphological phenotypes of the fk-J79 mutant. (A,B) Scanning electron micrograph (SEM) of a light-grown wild-type (A) or fk-J79 (B) seedling 4 days after germination (DAG). Bar, 0.5 mm. (C,D) Top view of a light-grown wild-type (C) or fk-J79 (D) seedling 7 DAG. Note that leaf axis in the center are perpendicular to the cotyledon in wild type but not in fk-J79. (Arrow) A fused cotyledon. Bar, 1 mm in C and 0.5 mm in D. (E,F) Transverse section of a wild-type (E) or fk-J79 (F) cotyledon. (Arrows) Vascular bundles. (Asterisks) A region missing the epidermal cell layer in fk-J79. Bar, 100 μm. (G) SEM of a light-grown fk-J79 seedling (7 DAG). (Arrows) Three adventitious shoot meristems on a large, flat shoot tip region. Bar, 0.25 mm; a wild-type seedling is shown for comparison (inset). (Arrowhead) The single shoot apical meristem. Bar, 2 mm. (H) Four-week-old greenhouse grown wild-type and fk-J79 plants. Bar, 0.5 cm. (I) Six-week-old wild-type and twelve-week-old fk-J79 plants grown in a greenhouse. Bar, 2.5 cm.
When grown in the greenhouse, the morphology of fk-J79 adult plants was similar to that of BR-deficient mutants, exhibiting small, dark-green rosettes, reduced petioles, loss of apical dominance, and dwarfed growth. However, extra basal rosettes were produced in the fk-J79 mutant due to an abnormal initiation of adventitious SAMs. In contrast, the wild type contained only one rosette (Fig. 1H,I). Compared with the wild type, fk-J79 plants were found to have a prolonged shoot meristematic activity that did not follow a normal senescence program (Fig. 1I). This together with their sterility, may have resulted in the extended lifespan of fk-J79 mutants.
fk-J79 displays an altered pattern of development in the embryonic apical–basal axis
The distorted fk-J79 embryonic organs (Fig. 1B) suggested a defect in embryonic development. Thus, we examined embryogenesis in the fk-J79 mutant. To obtain samples with synchronized development, fk-J79 embryos and neighboring wild-type embryos were taken from fk-J79/+ siliques for comparison. The fk-J79 mutation was found to disrupt normal cell division and expansion during embryogenesis. This caused abnormal morphology of cotyledons, hypocotyl, radicle, and the SAM. In wild-type embryos at the heart stage, cotyledon primordia arose at the apical portion (Fig. 2A). The O′ line, which divides the embryo proper into the apical and central domains, was clearly observed in the wild type because of the regular pattern of cell divisions during earlier stages (West and Harada 1993; Jürgens 1995). At the inner region of the central domain, elongated provascular cells were formed in the wild type (Fig. 2A). In contrast, fk-J79 embryos at the same stage remained globular shaped and failed to develop cotyledon primordia (Fig. 2B). Overall cellular arrangement was irregular and the O′ line was ambiguous, indicating that the mutation disrupted the normal pattern of cell division and expansion. Elongation of the provascular cells in the central domain was incomplete, which might cause the abnormal vasculature observed in seedlings (described below). Some of the cells in the outermost layer divided obliquely, disrupting the continuity of the epidermal layer (arrow in Fig. 2B). The morphology of the suspensor appeared normal.
Figure 2.
fk-J79 mutant is defective in embryogenesis. (A,B) Wild-type (A) and fk-J79 (B) embryos at the early heart stage. Arrow in B denotes oblique cell division in the epidermal layer. (Asterisk) Cotyledon primordium; (arrowhead) O′ line; (pv) provascular cells. (C,D) Wild-type (C) and fk-J79 (D) embryos at the late heart stage. Arrowheads in D denote small protrusions on the embryo surface. (Asterisk) Cotyledon primordium. (E–G) Wild-type (E) and fk-J79 (F,G) embryos at the bending cotyledon stage. (H,J) Magnification of E, F, and G, respectively. (Arrows) The region at the cotyledon base. Bars, 50 μm in A–D; 100 μm in E–J.
When wild-type embryos entered late-heart and early-torpedo stage (Fig. 2C), fk-J79 embryos started to develop cotyledon primordia, whose size was uneven and whose position was usually asymmetric (Fig. 2D). The majority of fk-J79 embryos had two primodia and the remainder had either none or more than two primordia, consistent with the frequency of dicotyledons observed in germinated seedlings. Small protrusions were frequently observed at their surface due to abnormal expansion of cells in the outermost layer (arrowheads in Fig. 2D). Later, at the bending cotyledon stage, wild-type cotyledons elongated and bent over toward the embryo axis (Fig. 2E). The central domain of the wild-type embryos also elongated longitudinally and formed the hypocotyl and radicle. The dome-shaped SAM could be clearly observed at this stage (arrow in Fig. 2H). In contrast, cotyledons of fk-J79 embryos failed to bend (Fig. 2F,G). In addition, the central domain of fk-J79 embryos expanded in the lateral rather than longitudinal direction, so that the length of hypocotyl and radicle was significantly reduced. Overall size varied among the mutant embryos (Fig. 2, cf. F with G). The morphology of the region at the cotyledon base was also variable. Some embryos lacked a typical dome-shaped SAM (arrow in Fig. 2I), whereas the others had large, aberrantly shaped bulges, which might correspond to an enlarged SAM or ectopic leaf primordium (arrow in Fig. 2J). In summary, the fk-J79 mutation disrupts normal cell division and expansion pattern in both apical–basal and lateral axes during embryogenesis, which caused abnormal morphology of cotyledons, hypocotyl, radicle, and the SAM.
FK encodes a homolog of human and yeast sterol reductases
Genetic and molecular analyses revealed fk-J79 as a single, T-DNA-tagged, recessive mutant (Fig. 3A). The inserted locus and its corresponding cDNA and wild-type genomic clones were identified by standard library screening as described in the Materials and Methods. The cDNAs identified predicted a protein with 365 amino acids (Fig. 3B). The predicted protein shared significant identity with the carboxy-terminal, sterol reductase domain (∼350 residues) of the LBR of human (38%), rat (38%), Xenopus (35%), and chicken (34%). In addition, FK also showed identity to various full-length sterol reductase sequences including yeast C-14 sterol reductase (ERG24, 34%), human (SR-2, 34%), Arabidopsis Δ7 sterol C-7 reductase (ST7R, 29%), and yeast C-24 sterol reductase (ERG4, 26%) (Fig. 3C). The predicted secondary structure indicates that FK is a membrane protein with eight transmembrane helices, consistent with the structures of yeast ERG24 and human LBR and sterol reductase (Fig. 3C). The signature motif of sterol reductases (Lecain et al. 1996), LLXSGWWGXXRH, was found on the carboxy-terminal half of the predicted FK sequence (Fig. 3B). Thirteen exons and twelve introns were predicted by comparison between cDNA and genomic sequences (Fig. 3D). The T-DNA was found to be inserted 79 bp upstream of the predicted start codon (ATG) of the deduced FK protein. Another ORF of 1671 bp, designated as D61, was also found at 1713 bp upstream of the T-DNA insertion in the same genomic clone (Fig. 3D).
Figure 3.
Cloning, sequence analysis, and mutant complementation. (A) DNA blot analysis of FK in wild-type, fk-J79/+, and fk-J79 plants. Genomic DNA were digested with BamHI and separated by gel electrophoresis and blotted to a nylon membrane. A 1.1-kb EcoRI–HindIII fragment (λ4-1.2-2) that contained the left border of T-DNA and a flanking Arabidopsis genomic sequence was labeled and used as a probe for the hybridization. (B) Predicted protein sequence of FK. The signature motif of sterol reductase is underlined. (FK sequence has been deposited in GenBank, accession no. AF257178 and AF263244.) (C) FK sequence is similar to human LBR (38%, LBR, GenBank accession no. L25941), yeast C-14 sterol reductase (34%, ERG24, GenBank accession no. P32462), human sterol reductases (39%, SR-1, GenBank accession no. AF096304; 34%, SR-2 GenBank accession no. AF034544 or AF096305), Arabidopsis Δ7 sterol reductase (ST7R, GenBank accession no. U49398) and yeast C-24 sterol reductase (ERG4, GenBank accession no. P25340). Numbers at right represent length of the predicted proteins. Shaded cylinders denote transmembrane helices predicted by the SOSUI Program (http://www.tuat.ac.jp/∼mitaku/adv_sosui). Areas denoted by the arrow bar are the homologous regions aligned on the basis of the result of the BLAST search of the GenBank (Altschul et al. 1997). (D) A T-DNA is inserted 79 bp upstream of the predicted start codon (ATG) of FK in fk-J79 mutant. An ORF (D61) of 1671 bp is transcribed in an opposite direction to FK. The 1786-bp region between FK and D61 genes is a putative shared promoter. A comparison between cDNA and genomic sequences of FK revealed 13 exons (shaded boxes) and 12 introns (lines between hatched boxes). (E) Complementation of fk-J79 mutant. A NotI–EcoO1091 genomic fragment containing a 1786-bp shared promoter and 2734 bp of FK or 1671 bp of D61 was inserted into a binary vector. Each construct was transfected into Agrobacterium tumefaciens and the strain was used to transform fk-J79/+ plants by the vacuum infiltration method. Ratios are the number of fk-J79 to the wild-type plants.
To test whether the fk-J79 mutant could be rescued by FK, a wild-type genomic fragment containing the 1.7-kb promoter and the FK-coding region was used for complementation experiments. As a control, the same promoter region and the upstream gene D61 (1.7 kb) was used (Fig. 3E). No fk-J79-like plants were found among the 63 (T1) and 42 (T2) transgenic lines that contained the introduced wild-type FK gene. This provides clear evidence of complementation by the introduced FK gene, because 25% would be expected to have the fk-J79 mutant phenotype if there were no complementation. In contrast, fk-J79 phenotype was observed in 25% of the transgenic lines that contained the introduced wild-type D61 gene (Fig. 3E). Together these data indicate that FK but not D61 could rescue fk-J79. It was concluded that the T-DNA insertion in the FK sequence was responsible for the fk-J79 mutant phenotype.
fk-J79 is allelic to the apical–basal patterning mutant fk-X224
FK was found to be located in the vicinity of 70.0 cM of Chromosome 3 using segregation analysis of its restriction fragment length polymorphisms in recombinant inbred lines (Lister and Dean 1993; data not shown). Further mapping with IGF BAC filters (Mozo et al. 1998) revealed that FK was located near mi456 (72.8 cM) on chr 3, and hybridized with six BACs (F16E19, F22D13, F1B11, F15H16, F10F15, and F8J2) within this region (data not shown). Because fk-X224 showed striking phenotypic similarity to fk-J79 in embryonic and seedling defects, and because it was reported to be located near 77.0 cM on Chr 3 (Rhee et al. 1998), reciprocal crosses were performed to determine their allelism. The results indicated that fk-J79 and fk-X224 are noncomplementary, and thus allelic (see Materials and Methods). To further confirm that fk-X224 is allelic to fk-J79, the FK cDNA probe was used for RNA blot analysis (Fig. 4A). Whereas the 1.3-kb FK transcript was detected in the wild type, no signal was detected with fk-X224 RNA. The 1.3-kb transcript was much reduced in RNA obtained from fk-J79 plants when compared with the wild-type RNA. An additional transcript of 3.6 kb appeared in the RNA from fk-J79 plants, probably resulting from the T-DNA insertion. The above results suggest that fk-X224, but not fk-J79, is likely to be a null mutant of FK. This is consistent with the result of protein gel blot analysis showing significant (>50%) reduction of the FK in the fk-J79 mutant (Fig. 4B).
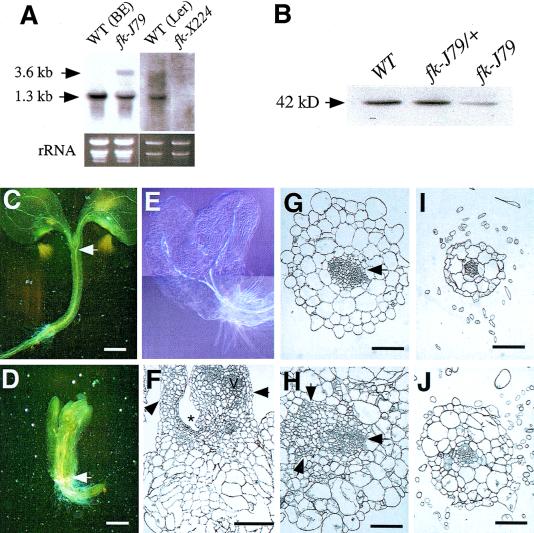
Figure 4.
fk-J79 is allelic to fk-X224 and is defective in cell division, cell expansion, and vascular development. (A) RNA blot analysis of FK in BE (WT), fk-J79, Ler (WT), and fk-X224 plants. RNA samples were prepared from 2-week-old light-grown plants on MS medium. For each sample, 5 μg of total RNA was loaded. Equal loading was determined by the ethidium bromide staining of rRNA bands. A 1.3-kb fragment (Δ13) containing full-length FK cDNA was labeled and used as a probe for the hybridization. (B) Protein gel blot analysis of FK in wild-type, fk-J79/+, and fk-J79 plants. Proteins were prepared from 2-week-old plants, separated on SDS–polyacrylamide gel, blotted to the membrane, and probed with antibody against FK. (C–E) Vasculature pattern in wild-type (C) and fk-J79 (D,E) seedlings. Seedlings were cleared using Hoyer's solution (Berleth and Jürgens 1993). As denoted by arrows, the vascular bundles from cotyledons meet at the top of the hypocotyl in wild type, but at the top of the root in fk-J79. Bars, 0.5 mm in (C–E). (F) Longitudinal section of an fk-J79 seedling (4 DAG). (Arrows) A leaf on the left and a fused organ on the right. (Asterisk) A depressed shoot apical region. Note that the large cortical cells expand in the lateral direction. (V) Vascular bundle; bar, 200 μm. (G–J) Transverse section of the hypocotyl in the wild-type (G) and fk-J79 mutant (H) and root in the wild type (I) and fk-J79 mutant (J). The vascular stele is denoted by a single arrow in the wild type (G). A large and disorganized vascular region is denoted by three arrows in the fk-J79 mutant (H). Bar, 100 μm.
Abnormal cell division, cell expansion, and vascular development in fk-J79 mutant
Because fk-J79 displayed a defect in embryonic apical–basal patterning that was similar to what has been proposed in the fk-X224 mutant (Mayer et al. 1991), we further examined the cellular defects in fk-J79 mutants. In the wild type, the two single vascular bundles in their respective petioles converged at the top of hypocotyl (Fig. 4C; Aida et al. 1999). This pattern was also very obvious in det2 (data not shown), a known BR-deficient dwarf mutant. In contrast, the vascular bundles met at the top of the root system in fk-J79 mutants (Fig. 4D,E), indicating that a typical hypocotyl was missing and the hypocotyl-like structure was fused petioles. In addition, the vasculature in fk-J79 was greatly reduced and discrete in the cotyledons (Fig. 4E). This defect is consistent with an incomplete provascular cell elongation observed during embryogenesis (Fig. 2B).
The dramatic thickening of the hypocotyl-like region in fk-J79 mutants was partly due to an exaggerated cell expansion in the lateral direction (Fig. 4F). Compared with a shallow, dome-shaped SAM in the wild type (Fig. 2H), the SAM region formed a depression in some fk-J79 young seedlings (Fig. 4F). Transverse sections revealed a dramatic size increase of the vascular bundles in fk-J79 that resulted from abnormal division of phloem companion and vascular parenchyma cells and fusion of two or more ectopic vascular bundles (Fig. 4G,H). The number of xylem cells was reduced or they were incompletely differentiated. The pattern of endarch xylem with collateral phloem in vascular bundles was obvious in the wild type but disrupted in fk-J79 (Fig. 4G,H). In addition, cortical cells in the hypocotyl-like region contained more chloroplasts and denser cytoplasm in fk-J79 than the wild type, suggesting this region to be part of the leaf and that incomplete organ differentiation might have occurred. Although the overall radial pattern was preserved, cellular organization was disrupted in fk-J79 as evidenced by the uneven cell size and oblique cell division planes in all cell layers (Fig. 4G,H). The pattern of the vascular stele was similar between fk-J79 and the wild type in the root although abnormal cell division and expansion persisted in cortical cell layers of fk-J79 plants (Fig. 4I,J).
FK is preferentially expressed in actively growing cells
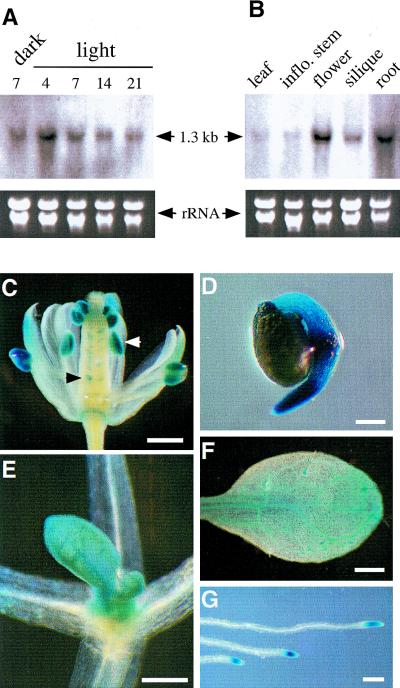
Although the fk-J79 mutant is pleiotropic, its defects appear to be restricted to embryos and meristems. The temporal and spatial expression pattern of FK was examined. By use of the FK cDNA as a probe in RNA blot analyses, a single transcript of 1.3 kb was detected. Its expression level decreased as plants matured. FK expression was not affected by light (Fig. 5A). FK was highest in flowers and roots, moderately abundant in siliques, and lowest in rosette leaves and inflorescence stems (Fig. 5B).
Figure 5.
FK expression. (A) RNA gel blot analysis of FK expression in different stages. RNA samples were prepared from dark-grown 7 DAG, or light-grown 4-, 7-, 14-, or 21-DAG wild type on MS medium. For each sample, 5 μg of total RNA was loaded. Equal loading was determined by the ethidium bromide staining of rRNA bands. A 1.3-kb fragment (Δ13) containing full-length FK cDNA was labeled and used as a probe for the hybridization. (B) RNA blot analysis of FK expression in different tissues. RNA samples were prepared from the wild-type rosette leaves (4-week old), inflorescence stems, flowers, siliques, and root. RNA gel electrophoresis, probe preparation, and blot hybridization are as described in A. (C–G) The expression of GUS (indicated by blue stain) in FK::GUS transgenic plants. (C) A flower showing high levels of GUS expression in anthers (white arrow) and ovules (black arrow); (D) a seedling 24 hr after germination; (E) shoot tip; (F) leaf of a seedling 7 DAG showing blue stain in vascular tissue and trichomes; (G) root-tips. Bars, 500 μm in C and F, 200 μm in D and E, and 300 μm in G.
To gain more insight into FK expression patterns and function, we generated transgenic lines carrying a FK::GUS reporter gene. Consistent with the FK transcript expression patterns determined by RNA blot analysis, high levels of FK::GUS expression were found in the anthers, ovules, germinating seedlings, shoot tips, root tips, and vasculature tissues in which cells would actively divide or expand (Fig. 5C–G). Overall, the expression pattern suggests that FK function is likely important for cell expansion and division.
BR levels are reduced in the fk-J79 mutants
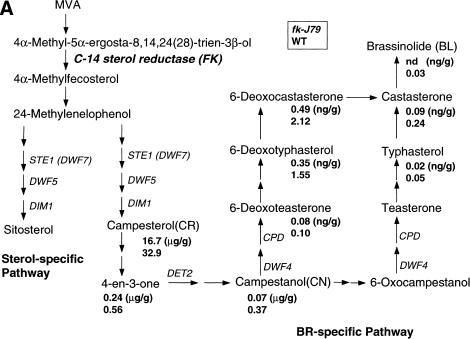
The DNA sequence of FK suggested that the encoded protein might be involved in plant sterol and BR biosynthesis. This hypothesis was supported by the dwarf phenotype of fk-J79, similar to that of the BR-deficient mutants det2, cpd, and dwarfs. To examine the possible biochemical consequences of the fk-J79 mutation, levels of BRs in fk-J79 and wild-type plants were determined by GC–MS analysis using deuterium-labeled internal standards. As summarized in Figure 6A, castasterone, typhasterol, 6-deoxocastasterone, 6-deoxotyphasterol, and 6-deoxoteasterone were detected in fk-J79 mutants, but their levels were significantly reduced compared with those in the wild type. Brassinolide was not present at detectable amounts in the fk-J79 mutants (Fig. 6A).
Figure 6.
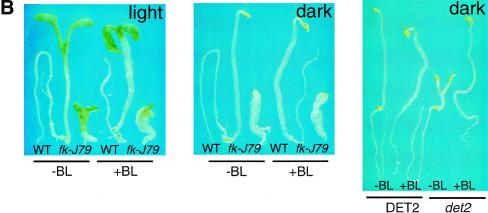
The fk-J79 mutation affects sterol biosynthesis and an application of BRs does not rescue the fk-J79 mutant's defect in hypocotyl elongation. (A) A simplified schematic sterol biosynthetic pathway in Arabidopsis. Values below each compound represent endogenous levels (per gram fresh weight) in fk-J79 (top) and wild type (bottom). (nd) Not detected. (B) The wild-type and fk-J79 or det2 mutant seedlings 7 DAG, treated with (+) or without (−) 1 μm of brassinolide (BL).
Exogenous BRs did not rescue the fk-J79 defect in hypocotyl elongation
Because all of the known BR-deficient mutants can be rescued by exogenous application of BRs, we conducted similar experiments with fk-J79 mutants. When supplied exogenously, neither the end product BLs (0.1–1.0 μm brassinolide or 24-epibrassinolide) nor active intermediates [0.1–1.0 μm campesterol (CR) or castasterone, data not shown] were found to correct the defect of hypocotyl elongation in either fk-J79 or fk-X224 plants (Fig. 6A,B; data not shown for fk-X224). The correction of the det2 mutant phenotype by these compounds in concurrent tests indicated that the experimental conditions were potentially appropriate and effective (Fig. 6B).
Sterols other than BRs might be required for the rescue of fk-J79 because C-14 sterol reductase acts before the divergence point of the sterol-specific and BR-specific pathways (Choe et al. 1999b; Fig. 6A). To test whether other sterols are required for rescue, we performed experiments using various combinations of BRs and sitosterol or stigmasterol, the end product of the sterol-specific pathway. None of the treatments restored hypocotyl elongation in fk-J79 mutants (data not shown). The above results indicate that the fk-J79 mutation has other effects in addition to the reduction of BRs, sitosterol, and stigmasterol. This hypothesis is consistent with the unique fk-J79 phenotypes in embryogenesis, meristem program, and root development not found in other BR-deficient mutants.
The fk-J79 mutant accumulates 8,14-diene sterols
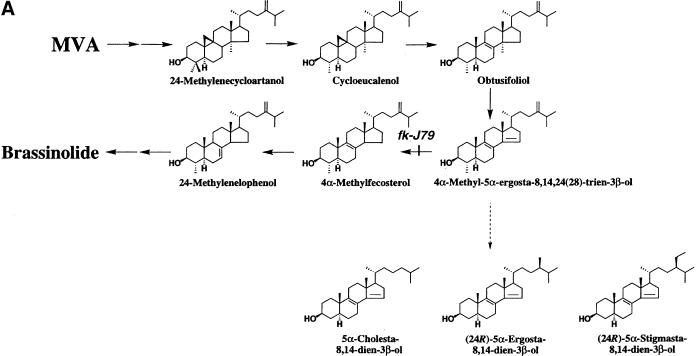
To pinpoint the defective step in the sterol biosynthetic pathway in fk-J79 mutant plants, we have analyzed their sterol composition extensively. Endogenous levels of CR, (24R)-ergost-4-en-3-one (4-en-3-one) and campestanol (CN) in the wild-type and fk-J79 seedlings were determined by GC–MS analysis using deuterium-labeled internal standards. The endogenous levels of CR, 4-en-3-one, and CN in the wild type were 32.9 μg/g fresh weight (fw), 0.56 μg/g fw, 0.37 μg/g fw, respectively (Fig. 6A). However, the levels of these three sterols in fk-J79 plants were diminished to 51%, 43%, and 19% of the wild-type levels, respectively (Fig. 6A). Approximate levels of the other sterols were estimated on the basis of measurement of the total ion currents from the mass spectra data. The levels of sitosterol and sitostanol in fk-J79 mutants were also diminished to ∼50% and 20% of the wild type, respectively. Most significantly, it was found that fk-J79 mutant plants accumulated ∼10 times higher levels of 4α-methyl-5α-ergosta-8,14,24(28)-trien-3β-ol, the substrate of C-14 sterol reductase (Fig. 7A).
Figure 7.
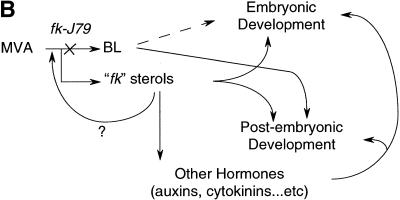
Novel sterols are produced in fk-J79 mutants. (A) fk-J79 mutation causes both the reduction of BRS and the accumulation of three abnormal 8,14-diene sterols. (B) A hypothetical model depicts the effects of fk-J79 mutation on plant development.
In addition, the fk-J79 mutant was found to accumulate high levels of several novel sterols. These fk sterols were not detectable in the wild-type plants. From the mass spectra, the fk sterols appeared to be 8,14-diene-sterols. To elucidate the precise structure of these compounds, we have chemically synthesized several possible 8,14-diene sterols (H. Seto, S. Fujioka, S. Takatsuto, unpubl.). By direct comparison with our synthesized sterols, the fk sterols were found to be (24R)-5α-stigmasta-8,14-dien-3β-ol, (24R)-5α-ergosta-8,14-dien-3β-ol, and 5α-cholesta-8,14-dien-3β-ol) (Fig. 7A). Their mass spectral data were as follows: (24R)-5α-stigmasta-8,14-dien-3β-ol, m/z (relative intensity), 484 [M+] (38), 469 (5), 379 (80), 238 (25), 182 (100); (24R)-5α-ergosta-8,14-dien-3β-ol, m/z 470 [M+] (34), 455(6), 365 (90), 238 (28), 182 (100); 5α-cholesta-8,14-dien-3β-ol, m/z 456 [M+] (28), 441 (7), 351 (83), 238 (28), 182 (100). Their levels were estimated to be ∼60 μg/g fw, 6 μg/g fw, and 0.6 μg/g fw, respectively.
In summary, the C-14 sterol reductase deficiency results in both reduced levels of various BRs and sterols, in accumulation of the substrate of C-14 sterol reductase, and in the accumulation of considerable amounts of novel fk sterols. Because BRs and sitosterol or stigmasterol fail to rescue the mutant phenotype, it is possible that the accumulation of the novel sterols makes a significant contribution to the unique developmental defects in fk-J79 mutants.
Discussion
Analysis of the nonlethal embryonic pattern mutant fk-J79 has provided a unique opportunity to understand the contributions of sterol biosynthesis and regulation to the control of cell division and cell expansion in a multicellular organism. The seedling-lethal phenotype of the null allele fk-X224 indicates that FK is essential for plant growth and development. This is consistent with the fact that we were unable to identify additional viable alleles of fk-J79 in repeated screens. Our molecular and biochemical evidence indicates that FK encodes a C-14 sterol reductase. Features such as dwarfism, loss of apical dominance, compact rosettes, and reduced fertility found in fk-J79 mutants are likely to be due to deficient BRs synthesis. However, the fk-J79 mutant displays unique and strong defects in embryogenesis and patterning that are similar to the null allele fk-X224. These phenotypes appear to be the consequence of changes in specific cellular functions associated with altered sterol regulation and signaling due to abnormal sterol composition.
FK encodes a C-14 sterol reductase
The predicted FK sequence shares significant homology with the carboxy-terminal half of the LBR and various sterol reductases from mammals to yeast. LBR is likely to be a chimeric protein exhibiting C-14 sterol reductase activity (Silve et al. 1998). The similarity between LBR and FK is limited to the sterol reductase domain, not the receptor domain. FK is also similar to the two human sterol reductases, SR-1 and SR-2, localized in the endoplasmic reticulum (Holmer et al. 1998). Whereas SR-1 (TM7SF2) is a putative sterol reductase, SR-2 (DHCR7) has been demonstrated to have Δ7-sterol reductase activity (Moebius et al. 1998). Interestingly, defects in DHCR7 cause the Smith-Lemli-Opitz syndrome (SLOS) that is an inborn disorder of sterol metabolism with characteristic congenital malformations and dysmorphias in humans (Fitzky et al. 1998).
FK is most similar to ERG24, which encodes a C-14 sterol reductase in Saccharomyces cerevisiae. Like FK, the ERG24 sequence is related to human sterol reductases SR-1 and SR-2 (Fig. 3C). The assumed biochemical reaction that FK catalyzes is upstream of 24-methylenelophenol, in which the sterol- and BR-specific biosynthetic pathways diverge (Fig. 6A). To support the notion that FK exhibits C-14 sterol reductase activity, we have detected a 10-fold accumulation of the substrate for C-14 sterol reductase and diminished amounts of various sterols and BRs in fk-J79 mutants.
fk-J79 is distinct from other BR-deficient mutants
Steroid hormones are essential for embryonic and adult development in animals (Evans 1988; Beato et al. 1995; Thummel 1996). Previous studies on BR-deficient or BR-insensitive mutants have revealed that BRs are important for post-embryonic development in plants. Their dwarf phenotype is attributed to reduced cell size, not cell number. However, BRs have been shown to promote cell division in the experiments by use of various cell cultures and protoplasts (Clouse and Sasse 1998). We have shown here that an unbalanced sterol composition and reduced amounts of BRs cause abnormal cell division in the intact plants. It has been shown that extranumerary phloem cell files are produced at the expense of xylem cells in a BR-deficient mutant cpd (Szekeres et al. 1996). This phenomenon is more pronounced in the fk-J79 mutants, presumably due to more drastic changes in sterol composition and regulation.
Compared with BR-deficient mutants, fk-J79 is distinct in its formation of multiple basal rosettes. It is not clear when and how multiple SAMs are formed in the fk-J79 mutant. However, shoot initiation in fk-J79 plants does not seem to cease in post-embryonic development, suggesting that there is a persistent hyperactive shoot meristematic program. Multiple SAMs formation has not been reported from other BR-deficient mutants. Beside a defect in SAM, the activity of the root apical meristem (RAM) in both the fk-J79 and fk-X224 mutants is reduced, as reflected by their highly stunted and branched roots (data not shown). Lateral roots are initiated frequently, suggesting that determinate roots are produced in fk-J79 plants. Thus, the fk-J79 mutant is unique in that both SAM and RAM development are affected. This again is in contrast to the BR-deficient mutants such as dim1, det2, and dwf4, in which no altered root meristem is found. It is known that BRs promote shoot but inhibit root elongation (Clouse and Sasse 1998). Our data suggest that the contrasting root phenotype of fk-J79 mutant is a result of abnormal sterol regulation/signaling, rather than a simple deficiency of BRs. The preferential expression of FK in root tips further implicates its role in root development.
The role of FK in meristem programming and embryonic and vascular patterning
Although a number of genes have been identified as regulators of SAM development in Arabidopsis (for review, see Meyerowitz 1997), the molecular mechanisms that regulate SAM formation and maintenance are not fully understood. There seems to be a correlation between multiple SAMs and supernumerary cotyledons in fk-J79 mutants. Similar to FK, mutations in the Arabidopsis FASS gene cause a strong compression in the apical–basal axis and radial enlargement caused by uncontrolled cell expansion in the hypocotyl region. Supernumerary cotyledon formation also seems to correlate with the width of the hypocotyl in fass mutants (Torres-Ruiz and Jürgens 1994). It has been proposed that FASS gene function is important in organizing cortical microtubules mediated through interactions between microtubules and plasma membrane (McClinton and Sung 1997). This raises the possibility that the fk-J79 mutant has altered membrane functions that are important for the microtubule organization. A detailed immunocytochemical study may reveal the cellular mechanism underlying the aberrant pattern of cell division and expansion in the fk-J79 mutant.
Although the reduced number and disrupted pattern of vascular bundles in the stem has been reported in BR-deficient mutants (Choe et al. 1999b), the causal molecular and cellular mechanisms are unknown. In contrast, emerging studies reveal that the plant hormone auxin tightly controls vascular development. Mutations in the Arabidopsis gene MONOPTEROUS (MP) interfere with the formation of vascular strands and with apical–basal patterning. The MP gene encodes a transcription factor binding to auxin-response elements that is involved in cell axialization and polar auxin transport (Hardtke and Berleth 1998). In mp mutants, cells in the vascular strands are incompletely differentiated. In addition, the discontinuous vascular system is reduced to higher order veins (Przemeck et al. 1996). This has striking similarity to what we have observed in fk-J79 cotyledons (Fig. 4D,E). It is not clear how fk-J79 mutation affects vascular patterning. A future challenge is to determine whether the effects of fk-J79 mutation result from an abnormal sterol regulation/signaling or interactions with other hormones such as auxin (Fig. 7B).
Potential consequences of the block in C-14 sterol reductase
In humans, the severity of the SLOS is correlated with the accumulation of the biosynthetic precursor 7-dehydrocholesterol (Neklason et al. 1999). 7-Dehydrocholesterol has been shown to be a very effective feedback inhibitor of HMG-CoA reductase. This in turn results in a general decrease of plasma total sterols (Honda et al. 1998). It remains to be determined whether there is a similar feedback regulation of sterol biosynthesis in plants. In yeast, mutations and drugs that inhibit steps in ergosterol biosynthesis cause reduction of ergosterol pools and an accumulation of novel sterols. Similar to the fk-J79 mutant, mutations in C-14 sterol reductase (ERG24) resulted in the accumulation of ergosta-8,14-diene-3β-ol (ignosterol) in yeast (Lorenz and Parks 1992). However, the net sterol level is not affected because of a feedback effect of ergosterol on the expression of ergosterol biosynthetic genes (Palermo et al. 1997). Whereas novel sterols can affect bulk functions in membranes, alterations of specific physiological processes are affected by specific mutations in ergosterol biosynthesis. For example, mutations in the ERG3 (C-5 sterol desaturase) gene result in defects in the utilization of non-fermentable carbon sources and resistance to some environmental stresses, whereas mutations in the ERG6 (C-24 sterol methyltransferase) gene cause defects in mating and tryptophan uptake (Parks et al. 1995). Although it remains possible that bulk membrane properities like fluidity and permeability may have been altered in the fk-J79 mutant, it is likely that many of the defects are due to specific changes in cell signaling or response. This notion is supported by the tissue expression pattern of FK in wild-type seedlings that coincides with the locations in which severe defects are found in the fk-J79 mutant. Understanding how plant cells, especially in the embryos and meristems, respond to sterols and change the normal growth and proliferation patterns, remains a future challenge.
Materials and methods
Mutant screen and analysis
A population of 4900 (CS2360) and ∼1200 T-DNA-mutagenized lines derived from Arabidopsis Biological Resource Center (ABRC, Ohio State University, Columbus, OH) and the authors' laboratory, respectively, and 114,000 EMS-mutagenized M2 (represents 14,250 M1, Lehle Seeds, Round Rock, TX) were used for the screen. Mutagenized seeds were germinated and grown on MS medium (GIBCO BRL) in the dark for 12 days. These etiolated plants were then illuminated with white light (75 μEm−2s−1) for 6 hr before putative mutants were selected. The selection was based on reduced elongation of hypocotyl and roots and greening of cotyledons that resemble seedlings treated with high levels of cytokinins. Over 100 putative mutants were obtained, but at least 50% of them were lethal. One of the putative mutants, designated as ell1/fk-J79, survived in the greenhouse conditions for >6 months, and it was used for the study. Because fk-J79 was near sterile, fk-J79/+ was identified from the T2 population to maintain the genetic material. The T-DNA inserted in fk-J79 contained the kanamycin resistance marker and a 35S:AtHXK2 (Jang et al. 1997). However, none of the 35S:AtHXK2 transgenic plants (n = 45) showed a fk-J79 phenotype, indicating that the phenotype was not due to the transgene but rather the T-DNA insertional mutagenesis. fk-X224 seeds (stock no. CS 8149) were obtained from ABRC.
For chemical rescue experiments, seedlings were grown in the light or dark on MS medium containing brassinosteroid compounds (part of the compounds was kindly provided by Steven Clouse, Department of Horticultural Science, North Carolina State University).
Light and scanning electron microscopy
The wild type (Benscheim ecotype) and fk-J79 were germinated and grown on MS medium with a 16/8-hr light/dark period at 25°C. A dissecting (Olympus SZH10) or an inverted (Nikon ECLPSE E800) microscope connected to a CCD camera was used for the observation and documentation of morphology. For scanning electron microscopy, samples were fixed in 3% glutaldehyde and subjected to a sequential dehydration with 50%, 75%, 85%, 95%, and finally 100% ethanol prior to a final critical point drying. Samples were then coated with gold using ion sputter coater before observation. Samples for semi-thin sections were prepared as for SEM except that they were infiltrated and embedded in Spur's low-viscosity resin after dehydration by 100% ethanol.
Genetic analysis
fk-J79/+ plants were crossed to the wild type to generate F1 plants that were 100% wild-type phenotype. The F1 were allowed to self-pollinate and produce F2 seeds. The ratio of wild type and fk-J79 in the resulting F2 population was 3:1, expected for a recessive mutation. The backcross was performed again using fk-J79/+ from the F2 population. Results were consistent with the previous analysis using F1. In the allelism test, pollens from fk-J79/+ were used to pollinate fk-X224/+ and vise versa. Unlike fk-J79, fk-X224 did not carry a kanamycin resistance marker, therefore, the F1 were determined by use of MS plates containing kanamycin. One-half of the F1 were kanamycin resistant, in which approximately one-half of them showed fk-J79 phenotype, indicating the cross was successful. The F1 hybrids (n = 341) between fk-J79/+ and fk-X224/+ plants derived from 18 independent reciprocal crosses contained 16.4%–30.0% of fk-J79 or fk-X224 plants. In contrast, the F1 progeny from crosses between wild type and fk-J79/+ or fk-X224/+ plants yielded no fk-J79 or fk-X224 plants.
Cloning and sequence analysis of FK
The genomic libraries of the wild-type and fk-J79 mutants were constructed using genomic DNA that was partially digested with Sau3AI and by use of a λ FIX II/Gigapack II cloning kit following the manufacturer's recommendations (Stratagene). Library screening was performed using PCR-labeled probe (Jang et al. 1997) and procedures as described (Sheen 1991). Two λ phage genomic clones (λ4-1.1 and λ4-1.2) that contained the T-DNA insertion were identified from a fk-J79 genomic library using a T-DNA probe. A 1.1-kb EcoRI–HindIII fragment (λ4-1.2-2) that contained the left border of T-DNA, and a flanking Arabidopsis genomic sequence was then used to screen a cDNA library (Minét et al. 1992). The cDNA clone identified was then used to screen a wild-type (Benscheim ecotype) genomic library. One genomic clone containing the putative full FK-coding region was used for sequence analyses. Genomic and cDNA fragments were subcloned into pBluescript vector (Stratagene) and were sequenced using an ABI-automated sequencer. Sequence data were analyzed with the GCG program (Genetics Computer Group, Madison, WI) and different programs available from the Internet (http://www.expasy.ch/). Secondary structure prediction was performed with the SOSUI Program (http://www.tuat.ac.jp/∼mitaku/adv_sosui).
Plant transformation
An Agrobacterium-mediated transformation using the vacuum infiltration method (Bechtold and Pelletier 1998) was used for fk-J79 mutant complementation. A wild-type genomic fragment containing the 1.7-kb promoter and the FK-coding region or the same promoter region and the upstream gene D61 (1.7 kb) was cloned into pGTV (provided by L. Sun and H. Goodman, MGH, Boston, MA). The C58 strain of Agrobacterium tumefaciens was transfected with either construct and was used to transform fk-J79/+ plants. pGTV carried the DHFR (encodes dihydrofolate reductase) marker that conferred methotrexate resistance (Becker et al. 1992). The transformed lines were selected on methotrexate and confirmed by PCR. The FK::GUS construct was made by fusing the ∼1.7-kb 5′ region of FK with the β-glucoronidase-coding sequence and cloned it into the pBIN19-based vector. Wild-type Arabidopsis transformed with the FK::GUS were used for the subsequent expression analyses.
Nucleic acid and protein gel blot analysis
DNA and RNA isolation and gel blot analysis were performed as described by Ausubel et al. (1987). For protein gel blot analysis, a polyclonal peptide antibody was custom made using FK peptide sequence 103–116:(C)GRSSSNKGSSLKPH-COOH (Zymed, South San Francisco, CA). Protein extracts were derived from 2-week-old seedlings grown on 1× MS plates with 2% sucrose. The immunoassay was performed using the Phototope-Star immunoblot detection kit (New England Biolabs, Beverly, MA).
Clearing of embryos and seedlings
For vascular visualization, tissues were soaked in a solution of ethanol:acetic acid (6:1) for 1 hr, rinsed with 70% ethanol, and soaked in a solution of chloral hydrate:glycerol:water (8:1:2) for 2–6 hr before observation (Berleth and Jürgens 1993).
Analysis of endogenous sterols and BRs
For sterol analysis, Arabidopsis seedlings (wild-type and fk-J79) were germinated and grown for 21 days on Murashige-Skoog medium containing 0.8% agar and 2% sucrose under a 12-hr light/12-hr dark regime, at 24°C. The same conditions were used for obtaining plant material for BR analysis, except that they were 5 weeks old.
For sterol analysis, plant material (1 gram fw equivalent) from wild-type and fk-J79 mutant was used. Plant material was extracted with 50 mL of MeOH-CHCl3 (4:1) twice, and 30 μg of [2H6]campesterol, 500 ng of [2H6]campestanol, and 500 ng of [2H6](24R)-ergost-4-en-3-one were added to the extract (1 gram fw equivalent) as internal standards. For BR analysis, plant material (50 gram fw equivalent) from wild-type and fk-J79 mutant was used. Plant material was extracted with 500 ml of MeOH-CHCl3 (4:1) twice, and [2H6]brassinolide, [2H6]castasterone, [2H6]typhasterol, [2H6]teasterone, [2H6]6-deoxocastasterone, [2H6]6-deoxotyphasterol, and [2H6]6-deoxoteasterone (100 ng each) were added to the extract as internal standards. Purification and GC–MS analyses were performed according to the method described in Noguchi et al. (1999b).
Acknowledgments
We thank ABRC for providing the fk-X224 seeds; D. Bongard for sequence analysis; S. Clouse for providing BL compounds; H. Goodman and L. Sun for providing the pGTV vector; J.-X. He for assistance in Western analysis; S. Jeong for assistance in GUS assay; M. Minét for pFL61 library; F. Xie for assistance in allelic test; S. Choe, J. Crowley, K. Feldmann, D. Kirsch, D. Meinke, F. Sack, and Z.-B. Yang for stimulating discussion; and D. Bauer, W.-L. Chiu, J. Elhai, B. Seed, and Z.-B. Yang for critical reading of the manuscript. This work was supported by the MLS, OARDC, and PMBB at the Ohio State University to J.-C.J. Salaries and research support provided by state and federal funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University. Manuscript number HCS99-37. A Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Science, Sports and Culture of Japan to S.F. (grant no. 10460050), and Hoechst A. G. to J.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jang.40@osu.edu; FAX (614) 292-7162.
References
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Ishida T, Tasaka M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development. 1999;126:1563–1570. doi: 10.1242/dev.126.8.1563. [DOI] [PubMed] [Google Scholar]
- Altmann T. Molecular physiology of brassinosteroids revealed by the analysis of mutants. Planta. 1999;208:1–11. doi: 10.1007/s004250050528. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: Greene Publishing Associates and Wiley-Interscience; 1987. [Google Scholar]
- Beato M, Herrlich P, Schutz G. Steroid hormone receptors: Many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Berleth T, Jürgens G. The role of the MONOPTEROUS gene in organizing the basal body region of the Arabidopsis embryo. Development. 1993;124:4415–4424. [Google Scholar]
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Gregory BD, Ross AS, Yuan H, Noguchi T, Fujioka S, Takatsuto S, Tanaka A, Yoshida S, et al. The Arabidopsis dwarf1 mutant is defective in the conversion of 24- methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 1999a;119:897–907. doi: 10.1104/pp.119.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Noguchi T, Fujioka S, Takatsuto S, Tissier CP, Gregory BD, Ross AS, Tanaka A, Yoshida S, Tax FE, et al. The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell. 1999b;11:207–221. [PMC free article] [PubMed] [Google Scholar]
- Chory J, Reinecke D, Sim S, Washburn T, Brenner M. A role of cytokinin in de-etiolation in Arabidopsis. Plant Physiol. 1994;104:339–347. doi: 10.1104/pp.104.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. BRASSINOSTEROIDS: Essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino IB, Kieber JJ. Molecular mechanisms of cytokinin action. Curr Opin Plant Biol. 1999;2:359–364. doi: 10.1016/s1369-5266(99)00005-9. [DOI] [PubMed] [Google Scholar]
- Edwards PA, Ericsson J. Sterols and isoprenoids: Signal molecules derived from the cholesterol biosynthetic pathway. Annu Rev Biochem. 1999;68:157–185. doi: 10.1146/annurev.biochem.68.1.157. [DOI] [PubMed] [Google Scholar]
- Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzky BU, Witsch-Baumgartner M, Erdel M, Lee JN, Paik YK, Glossmann H, Utermann G, Moebius FF. Mutations in the Δ7-sterol reductase gene in patients with the Smith-Lemli-Opitz syndrome. Proc Natl Acad Sci. 1998;95:8181–8186. doi: 10.1073/pnas.95.14.8181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Sakurai A. Brassinosteroids. Nat Prod Rep. 1997a;14:1–10. doi: 10.1039/np9971400001. [DOI] [PubMed] [Google Scholar]
- ————— Biosynthesis and metabolism of brassinosteroids. Physiol Plant. 1997b;100:710–715. [Google Scholar]
- Fujioka S, Li J, Choi YH, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J, et al. The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell. 1997;9:1951–1962. doi: 10.1105/tpc.9.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M-A. Plant sterols and the membrane environment. Trends Plant Sci. 1998;3:170–175. [Google Scholar]
- Holmer L, Pezhman A, Worman HJ. The human lamin B receptor/sterol reductase multigene family. Genomics. 1998;54:469–476. doi: 10.1006/geno.1998.5615. [DOI] [PubMed] [Google Scholar]
- Honda M, Tint GS, Honda A, Nguyen LB, Chen TS, Shefer S. 7-dehydrocholesterol down-regulates cholesterol biosynthesis in cultured Smith-Lemli-Opitz syndrome skin fibroblasts. J Lipid Res. 1998;39:647–657. [PubMed] [Google Scholar]
- Jang JC, León P, Zhou L, Sheen J. Hexokinase as a sugar sensor in higher plants. Plant Cell. 1997;9:5–19. doi: 10.1105/tpc.9.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G. Axis formation in plant embryogenesis:cues and clus. Cell. 1995;81:467–470. doi: 10.1016/0092-8674(95)90065-9. [DOI] [PubMed] [Google Scholar]
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996;9:701–713. [Google Scholar]
- Klahre U, Noguchi T, Fujioka S, Takatsuto S, Yokota T, Nomura T, Yoshida S, Chua NH. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell. 1998;10:1677–1690. doi: 10.1105/tpc.10.10.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecain E, Chenivesse X, Spagnoli R, Pompon D. Cloning by metabolic interference in yeast and enzymatic characterization of Arabidopsis thaliana sterol Δ7-reductase. J Biol Chem. 1996;271:10866–10873. doi: 10.1074/jbc.271.18.10866. [DOI] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li J, Biswas MG, Chao A, Russell DW, Chory J. Conservation of function between mammalian and plant steroid 5α-reductases. Proc Natl Acad Sci. 1997;94:3554–3559. doi: 10.1073/pnas.94.8.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Lorenz RT, Parks LW. Cloning, sequencing, and disruption of the gene encoding sterol C-14 reductase in Saccharomyces cerevisiae. DNA Cell Biol. 1992;11:685–692. doi: 10.1089/dna.1992.11.685. [DOI] [PubMed] [Google Scholar]
- Mayer U, Torres-Ruiz RA, Berleth T, Miséra S, Jürgens G. Mutations affecting body organization in the Arabidopsis embryo. Nature. 1991;353:402–407. [Google Scholar]
- McClinton RS, Sung ZR. Organization of cortical microtubules at the plasma membrane in Arabidopsis. Planta. 1997;201:252–260. doi: 10.1007/s004250050064. [DOI] [PubMed] [Google Scholar]
- Meyerowitz EM. Genetic control of cell division patterns in developing plants. Cell. 1997;88:299–308. doi: 10.1016/s0092-8674(00)81868-1. [DOI] [PubMed] [Google Scholar]
- Minét M, Dufour ME, Lacroute F. Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 1992;2:417–422. doi: 10.1111/j.1365-313x.1992.00417.x. [DOI] [PubMed] [Google Scholar]
- Moebius FF, Fitzky BU, Lee JN, Paik YK, Glossmann H. Molecular cloning and expression of the human Δ7-sterol reductase. Proc Natl Acad Sci. 1998;95:1899–1902. doi: 10.1073/pnas.95.4.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozo T, Fischer S, Meier-Ewert S, Lehrach H, Altmann T. Use of the IGF BAC library for physical mapping of the Arabidopsis thaliana genome. Plant J. 1998;16:377–384. doi: 10.1046/j.1365-313x.1998.00299.x. [DOI] [PubMed] [Google Scholar]
- Neklason DW, Andrews KM, Kelley RI, Metherall JE. Biochemical variants of Smith-Lemli-Opitz syndrome. Am J Med Genet. 1999;85:517–523. doi: 10.1002/(sici)1096-8628(19990827)85:5<517::aid-ajmg18>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Takatsuto S, Sakurai A, Yoshida S, Li J, Chory J. Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-en-3-one to (24R)-24-methyl-5α-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 1999a;120:833–839. doi: 10.1104/pp.120.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999b;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo LM, Leak FW, Tove S, Parks LW. Assessment of the essentiality of ERG genes late in ergosterol biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1997;32:93–99. doi: 10.1007/s002940050252. [DOI] [PubMed] [Google Scholar]
- Parks LW, Smith SJ, Crowley JH. Biochemical and physiological effects of sterol alterations in yeast-a review. Lipids. 1995;30:227–230. doi: 10.1007/BF02537825. [DOI] [PubMed] [Google Scholar]
- Przemeck GKH, Mattsson J, Hardtke CS, Sung ZR, Berleth T. Studies on the role of Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta. 1996;200:229–237. doi: 10.1007/BF00208313. [DOI] [PubMed] [Google Scholar]
- Rhee SY, Weng S, Flanders D, Cherry JM, Dean C, Lister C, Anderson M, Koornneef M, Meinke DW, Nickle T, Smith K, Rounsley SD. Genome maps 9. Arabidopsis thaliana. Wall chart. Science. 1998;282:663–667. doi: 10.1126/science.282.5389.663. [DOI] [PubMed] [Google Scholar]
- Sheen J. Molecular mechanisms underlying the differential expression of maize pyruvate, orthophosphate dikinase genes. Plant Cell. 1991;3:225–245. doi: 10.1105/tpc.3.3.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silve S, Dupuy PH, Ferrara P, Loison G. Human lamin B receptor exhibits sterol C14-reductase activity in Saccharomyces cerevisiae. Biochim Biophys Acta. 1998;1392:233–244. doi: 10.1016/s0005-2760(98)00041-1. [DOI] [PubMed] [Google Scholar]
- Szekeres M, Nèmeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Rèdei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Gasch A, Nishizawa N, Chua NH. The DIMINUTO gene of Arabidopsis is involved in regulating cell elongation. Genes & Dev. 1995;9:97–107. doi: 10.1101/gad.9.1.97. [DOI] [PubMed] [Google Scholar]
- Thummel CS. Flies on steroid-Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet. 1996;12:306–310. doi: 10.1016/0168-9525(96)10032-9. [DOI] [PubMed] [Google Scholar]
- Torres-Ruiz RA, Jürgens G. Mutations in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development. 1994;120:2967–2978. doi: 10.1242/dev.120.10.2967. [DOI] [PubMed] [Google Scholar]
- West MAL, Harada JJ. Embryogenesis in higher plants: An overview. Plant Cell. 1993;5:1361–1369. doi: 10.1105/tpc.5.10.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T. The structure, biosynthesis and function of brassinosteroids. Trends Plant Sci. 1997;2:137–143. [Google Scholar]