Abstract
RecQ helicase is important to homologous recombination and DNA repair in Escherichia coli. We demonstrate that RecQ helicase, in conjunction with RecA and SSB proteins, can initiate recombination events in vitro. In addition, RecQ protein is capable of unwinding a wide variety of DNA substrates, including joint molecules formed by RecA protein. These data are consistent with RecQ helicase assuming two roles in the cell; it can be (1) an initiator of homologous recombination, or (2) a disrupter of joint molecules formed by aberrant recombination. These findings also shed light on the function of the eukaryotic homologs of RecQ helicase, the Sgs1, Blm, and Wrn helicases.
Keywords: Recombination, DNA repair, RecQ, RecA, helicase
Cells possess the capacity to repair DNA lesions brought on by exposure to DNA-damaging agents (Friedberg et al. 1995). Frequently, DNA repair is achieved by homologous recombination, particularly when the damage results in a potentially lethal double-stranded DNA (dsDNA) break. In general, repair mediated by homologous recombination requires that the dsDNA suffering the lesion be separated into its component DNA strands. The single-stranded DNA (ssDNA) formed by this processing step can then be used by homologous pairing proteins such as the prokaryotic RecA protein and the eukaryotic Rad51 protein to promote homologous pairing and DNA strand invasion (Sung 1994; Maeshima et al. 1995; Baumann et al. 1996; Gupta et al. 1997; for review, see Kowalczykowski and Eggleston 1994; Camerini-Otero and Hsieh 1995).
The processing step of recombination can be mediated by a DNA helicase, a dsDNA nuclease, or a combination of these two activities. In wild-type Escherichia coli, RecBCD enzyme is the helicase/nuclease responsible for initiating the majority of recombination events (for review, see Smith 1988; Kowalczykowski et al. 1994). Mutations at the recB or recC loci result in a 100- to1000-fold decrease in recombination frequency, in addition to a dramatic increase in sensitivity to ionizing radiation (Howard-Flanders and Theriot 1966; Emmerson 1968). However, both recombination proficiency and resistance to ionizing radiation are restored to wild-type levels in a recBC background by extragenic sbcBC (suppressor of recBC) mutations that allow recombination to proceed by an alternate pathway referred to as the RecF pathway (Kushner et al. 1971; Lloyd and Buckman 1985). The RecF pathway defines a set of gene products that are required for homologous recombination and recombinational repair in a recBCsbcBC background (for review, see Clark and Low 1988; Mahajan 1988; Kowalczykowski et al. 1994). Recombination promoted by these proteins is not confined solely to this specific genetic background, but is also important for a number of recombination processes in wild-type cells: Crossovers occurring at internal regions of duplex DNA, such as at ssDNA gaps, are believed to be mediated by RecF pathway gene products (Lanzov et al. 1991; Lloyd and Buckman 1995). In addition, these gene products play an important role in repair of broken replication forks in wild-type cells (Courcelle et al. 1997). Thus, rather than simply representing an alternate pathway, recombination directed by these proteins is a normal and important means of genomic maintenance.
Nakayama et al. (1984) identified a component of this repair pathway that is designated recQ. A null mutation at this locus conferred both a significant reduction in recombination frequency (∼100-fold) and an increase in UV sensitivity (∼20-fold) in a recBCsbcBC background (Nakayama et al. 1984, 1985). In addition, recQrecBCsbcBC cells are sensitive to the DNA damaging agents methyl-methane sulfonate (Mendonca et al. 1995) and hydrogen peroxide (Kusano et al. 1994).
The RecQ protein is a 3′ → 5′ DNA helicase that acts on both partially dsDNA and fully duplex DNA (Umezu et al. 1990). The ability to act at a flush dsDNA end is shared among a small number of other helicases, including the major initiator of homologous recombination, the RecBCD enzyme (Taylor and Smith 1985). Recombination initiated by the RecBCD enzyme begins from such a blunt dsDNA end present at a dsDNA break, produced as the result of DNA damage, a broken replication fork, or conjugal DNA transfer. After binding to this end, the helicase and nuclease activities of RecBCD enzyme convert the dsDNA to a 3′-ssDNA overhang, a product that is the preferred substrate for RecA protein-mediated strand invasion (Konforti and Davis 1987; Anderson and Kowalczykowski 1997a,b; for review, see Kowalczykowski and Eggleston 1994). Because RecQ helicase also acts on blunt dsDNA ends and is needed for recombination in a recBCsbcBC background, it was proposed to act as a functional analog of RecBCD enzyme during this processing step (Clark and Sandler 1994; Kowalczykowski et al. 1994; Mendonca and Matson 1995). Thus, the genetic data argue that, with the exception of a minor contribution from both helicases II and IV, RecQ protein is the only helicase of the 12 helicases in E. coli that can substitute for RecBCD enzyme function in genetic recombination.
Recently, homologs of the RecQ helicase were identified in a wide variety of organisms including the budding yeast Saccharomyces cerevisiae (Gangloff et al. 1994; Watt et al. 1995), the fission yeast Schizosaccharomyces pombe (Stewart et al. 1997), Caenorhabditis elegans, and Homo sapiens (Puranam and Blackshear 1994; Seki et al. 1994; Ellis et al. 1995; Yu et al. 1996). The yeast homolog, Sgs1 protein, plays a key role in recombination, chromosome partitioning, and genome stability in S. cerevisiae. Three proteins from human, encoded by the RECQL, BLM, and WRN genes, are also highly homologous (∼40% identical among the helicase domains) to E. coli RecQ helicase and Sgs1 protein (Puranam and Blackshear 1994; Seki et al. 1994; Ellis et al. 1995; Yu et al. 1996). Mutations in the RECQL gene do not result in a discernible phenotype, but mutations at the BLM and WRN loci are linked to Bloom’s syndrome and Werner’s syndrome, respectively. Both of these disorders are rare, inherited diseases that share similar features including DNA replication abnormalities and pronounced genomic instability (Ellis et al. 1995; Yu et al. 1996). The phenotypic similarity of blm and wrn mutants and yeast sgs1 mutants suggests that all these helicases play similar roles in DNA metabolism; the structural similarity of these three eukaryotic proteins and the prokaryotic RecQ helicase suggests that each may share a common biochemical function. The potential for functional similarity among this group of proteins is further supported by the recent identification of the Sgs1, Wrn, and Blm proteins as 3′ → 5′ DNA helicases (Lu et al. 1996; Gray et al. 1997; Karow et al. 1997; Suzuki et al. 1997).
For these reasons, in vitro analysis of purified RecQ helicase was carried out to determine whether this protein can mediate steps essential to homologous recombination. The results presented here demonstrate that the RecQ helicase is a multifunctional helicase that is capable of both initiating homologous recombination and disrupting nascent joint molecules. These findings also provide insight into the potential roles of the eukaryotic homologs of RecQ helicase in DNA metabolism.
Results
RecQ helicase initiates homologous pairing in vitro
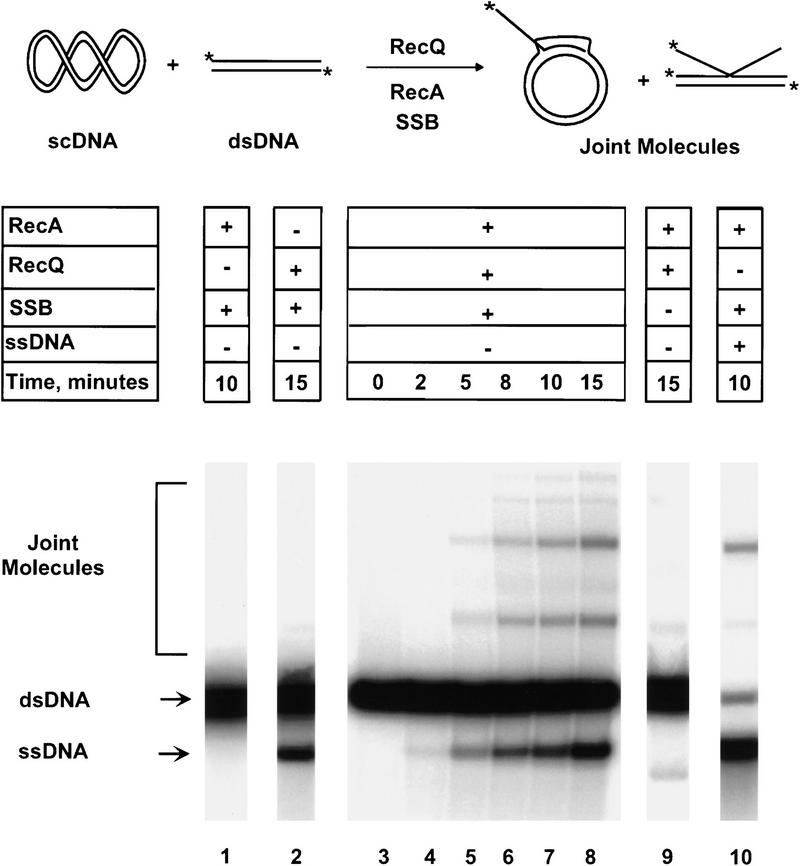
A DNA helicase can act at either the initiation or DNA heteroduplex extension phase of homologous recombination. To address the possibility that RecQ helicase initiates homologous recombination, we tested its ability to produce a DNA substrate for RecA protein-promoted joint molecule formation. In our assay system, used previously to define an initiation function for the RecBCD enzyme (Roman et al. 1991), RecA protein is provided with a pair of homologous DNA substrates: Supercoiled DNA (scDNA) and 5′–end labeled linear dsDNA (Fig. 1). However, RecA protein is unable to pair these substrates because neither contains ssDNA; thus, processing of the linear dsDNA to its component ssDNA strands is required. If RecQ helicase can make ssDNA available to RecA protein, then homologous pairing products, joint molecules, will be observed by agarose gel electrophoresis as species that migrate more slowly than either of the dsDNA substrates. The expectation was that joint molecules will form only when RecQ helicase, RecA protein, and single-stranded DNA binding (SSB) protein are present together in a reaction, hereafter referred to as the coupled pairing reaction.
Figure 1.
RecQ helicase, in conjunction with RecA and SSB proteins, initiates homologous pairing. Labeled, linear pUC19 DNA and pUC1950 scDNA were incubated with: (Lane 1) RecA and SSB proteins; (lane 2) RecQ helicase and SSB protein; (lanes 3–8) all three proteins; (lane 9) RecQ helicase and RecA protein alone. Slower migrating species that represent joint molecules, labeled Joint Molecules, are observed only in the presence of all three proteins. In a control reaction, RecA and SSB proteins were incubated with labeled linear pUC19 ssDNA, cold linear pUC19 dsDNA, and pUC1950 scDNA to produce bona fide joint molecules (lane 10). All reactions were carried out as in Materials and Methods. Bands corresponding to linear ssDNA and dsDNA are also indicated to the left.
As expected, the coupled pairing reaction produced several strong joint molecule product bands with a mobility retarded relative to the linear dsDNA substrate (Fig. 1, lanes 3–8). RecA protein alone failed to pair the two dsDNAs, defining RecQ helicase as an essential component (Fig. 1, lane 1). In the absence of RecA protein, RecQ helicase produced only unit-length ssDNA (Fig. 1, lane 2): Demonstrating both the need for RecA protein and that the processivity of RecQ helicase is sufficient for it to unwind plasmid-length DNA molecules (F.G. Harmon and S.C. Kowalczykowski, unpubl.). The assignment of these slower-migrating bands as bona fide pairing intermediates was confirmed by comparison with authentic joint molecules created by the pairing of heat-denatured 5′-end labeled pUC19 DNA with both linear pUC19 dsDNA and supercoiled pUC1950 DNA by use of RecA and SSB proteins (Fig. 1, cf. lanes 7 and 8 with 10). Finally, SSB protein is also an important component of the pairing reaction, because very few joint molecules were observed in its absence (Fig. 1, lane 9). This poor yield is a consequence of the low amounts of ssDNA that are liberated by RecQ helicase when the SSB protein is absent; SSB protein is needed to both trap the individual DNA strands and prevent their reannealing (Umezu and Nakayama 1993). The same dependence on SSB protein was observed previously in the coupled RecABCD pairing reactions (Roman et al. 1991). Therefore, joint molecule formation requires the combined actions of RecA, RecQ, and SSB proteins. Furthermore, these data clearly show that the RecQ helicase is capable of initiating joint molecule formation.
The coupled pairing reaction produces an unexpected variety of joint molecule products
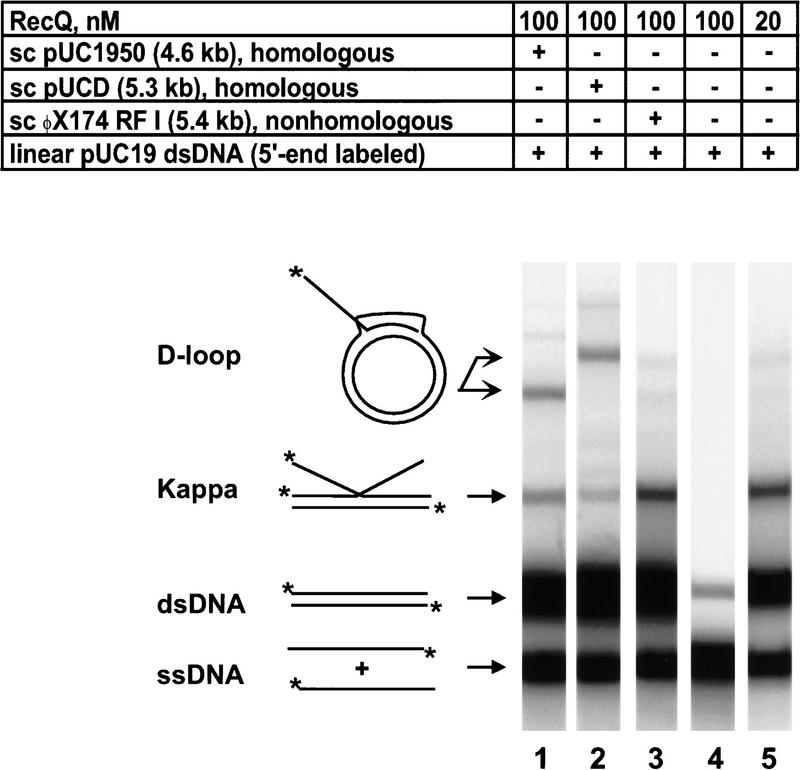
Surprisingly, the combined actions of RecQ helicase, RecA protein, and SSB protein produced four species of joint molecules (Fig. 1). On the basis of previous results with the coupled RecABCD-pairing reactions (in the presence of SSB protein), invasion of the scDNA by the linear ssDNA to form displacement-loops or D-loop joint molecules was anticipated (Dixon and Kowalczykowski 1991; Roman et al. 1991); simple D-loop products could account for one or two, but not all, of the observed joint molecules. To determine which species in the coupled pairing reaction arose from invasion of the scDNA, the scDNA recipient was changed to pUCD, a homologous DNA that is 700 bp larger than pUC1950 (Fig. 2). With this larger DNA, the migration of scDNA-dependent D-loop products will decrease relative to those formed in the standard coupled reaction. For comparison, the products of a standard coupled reaction are shown in lane 1 of Figure 2, with the deduced D-loop band indicated on the left. In the coupled reaction in which pUCD is present, the migration of the assigned D-loop band is reduced (Fig. 2, lane 2). To confirm that homologous pairing is required to produce this species, coupled reactions were repeated with nonhomologous φX174 RF I as the scDNA recipient. Because φX174 RF I is approximately the same size as pUCD, a φX174 RF I-dependent D-loop should comigrate with the pUCD D-loop. As expected, no D-loop species is formed in this case (Fig. 2, lane 3); therefore, homology between the linear dsDNA and scDNA recipient is required.
Figure 2.
The RecAQ-coupled pairing reaction produces two classes of joint molecules. Each lane represents a 10 min time point taken from the indicated reaction. (Lanes 1–3) scDNA recipient in each reaction was different to determine the position of the band corresponding to scDNA-dependent D-loops. (Lane 1) Taken from a standard coupled reaction, with the deduced migration positions of each joint molecule indicated to the left. The products shown in lanes 2 and 3 are from reactions with larger, homologous pUCD scDNA and nonhomologous φX174 RF I, respectively. The migration of the D-loop band is slower for the larger pUCD but the same band is completely absent when φX174 RF I is used in its place. (Lanes 4,5) scDNA was omitted from reactions to identify scDNA-independent Kappa intermediates. (Lane 4) Band corresponding to the Kappa intermediate is absent at the prevailing RecQ helicase concentration (100 nm); however, the band is present with fivefold less RecQ helicase (lane 5), confirming that it is scDNA-independent.
Unexpectedly, the migration and presence of the second major joint molecule species was independent of the scDNA added to the reaction, indicating that its formation did not involve invasion of the scDNA (Fig. 2; cf. lanes 1–3, species labeled Kappa). The properties of this joint molecule suggested that it could be composed of a linear dsDNA molecule that was invaded by homologous ssDNA; the resultant joint molecule would resemble the letter K and, hence, is referred to as a Kappa intermediate. When scDNA was omitted from a coupled pairing reaction, no joint molecules were present; however, a greater proportion of the dsDNA was unwound by RecQ helicase (Fig. 2, lane 4). The increased DNA unwinding by RecQ helicase in this reaction is consistent with our findings that RecQ helicase binds to, and is sequestered by, scDNA (F.G. Harmon and S.C. Kowalczykowski, in prep.). Therefore, it suggested to us that the Kappa intermediate was being produced, but was also a substrate for RecQ protein-mediated unwinding and, therefore, was being quickly destroyed when more free helicase was present. To investigate this possibility, coupled pairing reactions in which the scDNA was omitted were repeated by use of fivefold less (20 nm) RecQ helicase (Fig. 2, lane 5). At this RecQ helicase concentration, the Kappa intermediate was formed to nearly the same extent as in the standard coupled reaction. Addition of larger, homologous linear dsDNA to similar reactions yields a slower migrating Kappa species that is absent when a similar-sized, but non-homologous, linear DNA is substituted, confirming the need for homology in the recipient linear dsDNA (data not shown). Therefore, a Kappa intermediate, derived from the invasion of linear dsDNA by homologous ssDNA is also a major product of the coupled RecAQ pairing reaction. The nature of the more complex joint molecules formed in these reactions is currently being investigated (F.G. Harmon and S.C. Kowalczykowski, unpubl.). Taken together, the formation of these two classes of intermediates in the coupled reaction demonstrates that the RecQ helicase, in a manner similar to RecBCD enzyme, provides RecA protein with a ssDNA substrate that it can use to invade a homologous counterpart. These findings provide strong support for the proposal that RecQ helicase can serve as an initiator of homologous recombination.
RecQ helicase can disrupt joint molecules
The ability to unwind joint molecules is a hallmark of branch migration proteins such as the RuvAB and RecG helicases (Tsaneva et al. 1992; Whitby et al. 1993). In the above experiments, RecQ helicase appeared to dissociate Kappa intermediates, suggesting that it also has branch migration activity. If RecQ helicase possesses such an activity, then the average lifetime and steady-state yield of joint molecules produced in the coupled pairing reaction should be reduced at a higher concentration of RecQ helicase. In agreement, the Kappa intermediates and D-loops formed in the presence of fivefold more RecQ helicase (500 nm) are more transient, and they form to maximal extents that are significantly lower than in the standard coupled pairing reaction (cf. Fig. 3A, lanes 1–5 with Fig. 1). The band corresponding to Kappa intermediates also shifts to a faster migrating species over the course of the reaction (Fig. 3A, lanes 3–5). The alteration in migration observed for this intermediate likely reflects RecQ helicase-induced changes in the position of the branch point in this molecule. In separate experiments, RecQ helicase was found to extend the initial region of pairing in Kappa intermediates to generate intact, heteroduplex, linear dsDNA (F.G. Harmon and S.C. Kowalczykowski, unpubl.).
Figure 3.
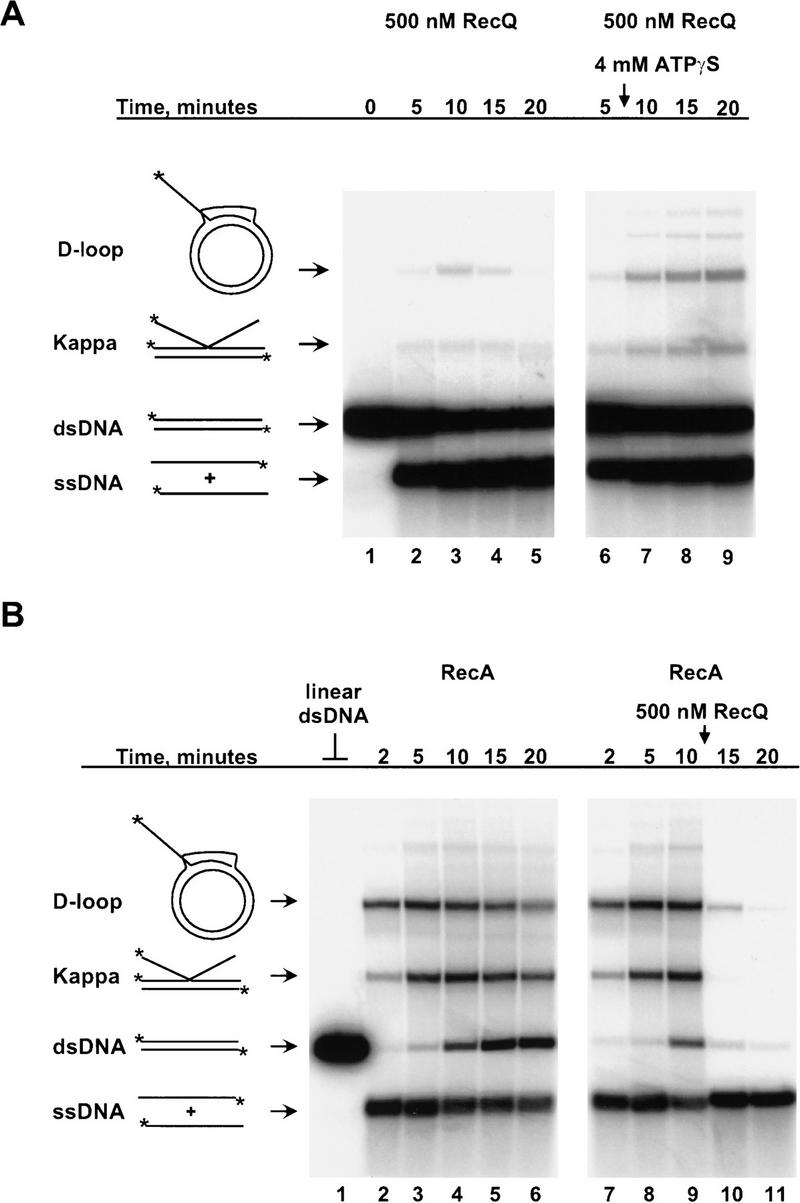
RecQ helicase dissociates joint molecules. (A) RecQ helicase unwinds joint molecules during the course of the coupled pairing reaction. The addition of fivefold more RecQ protein (500 nm) to the coupled pairing reaction results in transient intermediates and a lower steady-state yield of these species (lanes 2–5). The helicase activity of RecQ helicase is required to observe this phenomenon because addition of ATPγS, a nonhydrolyzable analog of ATP, affords protection from disruption to the pairing intermediates formed in the reaction (lanes 6–9). The time at which 4 mm ATPγS was added (8 min) is indicated by the vertical arrow. (B) RecQ helicase binds to and unwinds joint molecules formed in its absence. (Lane 1) Linear pUC19 dsDNA marker. (Lanes 2–6) RecA and SSB proteins were incubated with labeled linear pUC19 ssDNA, unlabeled linear pUC19 dsDNA, and pUC1950 scDNA to produce homologous pairing intermediates. RecQ helicase added to similar reactions dissociated the joint molecules formed by RecA protein (lanes 7–11). RecQ helicase (500 nm) was added after taking the 10 min time point in lane 9.
To directly demonstrate that RecQ helicase was actively dissociating these joint molecules, the stability of D-loops and Kappa intermediates was determined following addition of ATPγS (Fig. 3A). ATPγS, a nonhydrolyzable ATP analog, inhibits the helicase activity of RecQ protein (F.G. Harmon and S.C. Kowalczykowski, unpubl.) and, therefore, should protect joint molecules from dissociation. Addition of ATPγS to an ongoing coupled pairing reaction results in a greater yield and increased lifetime for both species of pairing intermediates (Fig. 3A, cf. lanes 3–5 with 7–9). ATPγS also clearly inhibited the helicase activity of RecQ helicase, because there was no further accumulation of free ssDNA following addition of inhibitor (Fig. 3A, lanes 3–5). These findings were consistent with RecQ helicase actively unwinding these pairing products.
To determine if RecQ helicase can recognize and initiate unwinding of joint molecules formed in its absence, RecQ helicase was added to ongoing RecA protein-mediated homologous pairing reactions (containing heat-denatured DNA) (Fig. 3B). In the absence of RecQ helicase, both D-loops and Kappa intermediates were rapidly formed and then persisted over a 20 min time course (Fig. 3B, lanes 1–6). In contrast, when 500 nm RecQ helicase was added 10 min after starting the reaction, the majority of joint molecules were rapidly unwound in <5 min (Fig. 3B; lanes 7–11). Thus, RecQ helicase binds to and unwinds joint molecules, suggesting it has dual capabilities: Initiation of RecA-promoted homologous pairing and disruption of joint molecules by branch migration.
RecQ protein is a promiscuous DNA helicase
The branch migration proteins, RuvAB and RecG helicases, act on a limited subset of DNA substrates that contain a branch point, such as the four-way junction DNA substrate that mimics a Holliday junction (Iwasaki et al. 1992; Parsons et al. 1992; Lloyd and Sharples 1993b; Whitby et al. 1994). If RecQ protein acted primarily during branch migration, then it was reasonable to expect that it would display specificity toward similar DNA substrates. To test the substrate specificity of RecQ helicase, the rate of unwinding was determined for a series of oligonucleotide-derived DNA substrates that resemble the structures encountered during homologous recombination (Table 1).
Table 1.
The RecQ helicase can bind to and unwind a wide variety of DNA substrates
| DNA substrate
|
Structure (annealed oligonucleotides)a
|
Unwinding rateb (nm (bp)/nm RecQ/sec)
|
Relative binding affinityc
|
|---|---|---|---|
| 3-way junction | 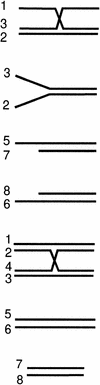 |
2.9 | 7 |
| Flayed duplex | 2.2 | 8 | |
| 5′-ssDNA overhang | 1.2 | 2 | |
| 3′-ssDNA overhang | 1.1 | 12 | |
| 4-way junction | 1.0 | 1 | |
| 63-bp blunt duplex | 0.8 | N/D | |
| 48-bp blunt duplex | 0.2 | 1 |
Reactions were performed and analyzed as described in Materials and Methods.
Numbers correspond to the oligonucleotides in Table 2 annealed as described in Materials and Methods to form the indicated DNA substrate.
Calculated from the linear slope of a plot of unwinding rate [nm (bp)/sec] vs. RecQ helicase concentration (nm). The relative error for each rate is approximately ±5%.
Determined as in Materials and Methods and normalized to the binding value of 5 × 106/m obtained for the 48-bp blunt duplex substrate. The relative error for each value is approximately ±5%.
The helicase assays were performed over a range of RecQ helicase concentrations, and the results are shown in Table 1. In general, RecQ helicase had little difficulty unwinding any of the DNA substrates, with the exception of the 48-bp blunt duplex, which was unwound at a fivefold slower rate (Table 1). This difference is probably the result of the short length of this DNA substrate because RecQ helicase is capable of unwinding a 63-bp blunt molecule (Table 1), as well as plasmid-length linear dsDNA (see above). The unwinding rates obtained for the other DNA substrates were very similar to each other, indicating that RecQ helicase has no obvious preference for a particular type of DNA structure (Table 1). Binding studies with the same oligonucleotide DNA substrates indicate that RecQ helicase also has a broad DNA binding specificity (Table 1). The lack of a strong DNA substrate specificity by RecQ helicase is in contrast to that of RecG helicase, which is unable to unwind blunt duplex DNA at protein concentrations that result in complete dissociation of an equal amount of four-way junction DNA (Lloyd and Sharples 1993a). In addition, RuvAB helicase displays only limited activity on dsDNA substrates other than four-way junctions (Iwasaki et al. 1992; Lloyd and Sharples 1993a). From this standpoint, it is clear that RecQ helicase is not specific for branched molecules. However, these findings also imply that RecQ helicase has the potential to initiate recombination from almost any type of DNA substrate presented to it, unlike the RecBCD enzyme that must initiate from a nearly blunt dsDNA end. Further, because RecQ helicase has the capacity to unwind a broad spectrum of DNA substrates, its role in recombination is not limited to the early initiation steps, but could also include the dissociation of aberrant joint molecules.
Discussion
Using a coupled in vitro system consisting of RecQ, RecA, and SSB proteins, we have demonstrated the formation of homologously paired joint molecules. The RecQ helicase is capable of initiating homologous pairing by processing a linear dsDNA substrate to provide a ssDNA substrate for RecA protein (Figs. 1 and 2). Thus, RecQ helicase is likely to act as an initiator during the early steps of homologous recombination. Because RecQ helicase can also disrupt the homologous pairing products formed in these reactions (Fig. 3), it may have additional roles in DNA metabolism. However, RecQ protein is not a branch migration-specific helicase because, in contrast to the RuvAB and RecG helicases, it displays no greater helicase activity on a four-way junction DNA substrate than it does on other DNA substrates (Table 1). In addition, RecQ helicase displayed a broad DNA binding specificity (Table 1). Overall, the data demonstrate that RecQ helicase is capable of acting on a wide variety of DNA substrates and, thus, has the potential to play multiple roles in recombination.
The model in Fig. 4 outlines the series of molecular events that comprise the coupled homologous pairing reaction involving RecA, RecQ, and SSB proteins. The linear dsDNA substrate was first processed by RecQ helicase to produce linear ssDNA (Fig. 4A). This DNA is expected to be complexed with the SSB protein because it plays an integral part in the RecQ helicase unwinding reaction (Umezu and Nakayama 1993). In this capacity, SSB protein acts to prevent reannealing of the strands behind the helicase (Roman et al. 1991), but an SSB protein-coated ssDNA is a poor binding substrate for RecA protein (Kowalczykowski et al. 1987). Therefore, RecA protein must displace SSB protein from the ssDNA to form a nucleoprotein filament that is capable of initiating joint molecule formation. Under the conditions used, RecA protein can displace the SSB protein, assemble a filament of the ssDNA, and form two major classes of joint molecules: scDNA-dependent D-loops and linear Kappa intermediates (Fig. 4B). D-loops are also the major product of the in vitro coupled RecABCD-pairing reactions by use of similar DNA substrates (Dixon and Kowalczykowski 1991; Roman et al. 1991). Kappa intermediates, however, are specific to the RecAQ-coupled pairing reactions. These joint molecules are probably formed more readily in the presence of PEG, the volume excluding agent used here, because of the enhanced ability of RecA protein to assemble on SSB-protein complexed ssDNA under these conditions (Lavery and Kowalczykowski 1992). These findings clearly indicate that RecQ helicase, like the RecBCD enzyme, is capable of processing dsDNA to ssDNA as required for the early steps of homologous recombination. The idea that RecQ helicase initiates homologous recombination is also consistent with the genetic data that show a requirement for recQ function for recombination and DNA repair in the absence of RecBCD enzyme function (Nakayama et al. 1984, 1985; Kusano et al. 1994; Mendonca and Matson 1995; Mendonca et al. 1995).
Figure 4.
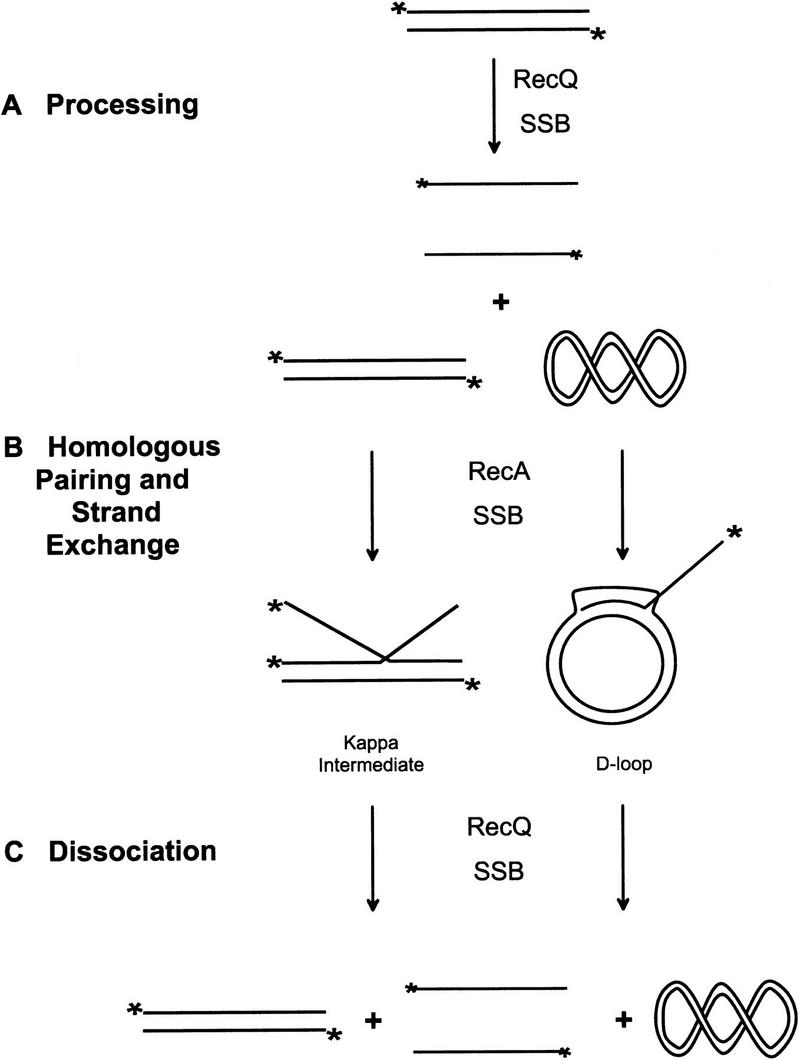
Model for RecQ helicase action in homologous recombination. (A) RecQ helicase initially bound to the 5′-end labeled linear dsDNA substrate unwinds these molecules to produce linear ssDNA. During the unwinding process, SSB protein binds to the liberated ssDNA, trapping it and, thereby, preventing the strands from reannealing. (B) This SSB protein-coated ssDNA is then bound by RecA protein to form a nucleoprotein filament competent for homologous pairing. The RecA nucleoprotein filament initiates homologous pairing and strand invasion to form joint molecules. The major species of joint molecules formed in these reactions are scDNA-dependent D-loops and linear scDNA-independent Kappa intermediates. (C) After the initial unwinding of linear dsDNA, RecQ helicase is available to bind to and dissociate the accumulated joint molecules.
RecQ protein, however, is not simply a backup for RecBCD enzyme, because the former acts on a wider variety of dsDNA substrates than does the latter. Thus, RecQ helicase has the potential to initiate recombination events in wild-type cells from DNA substrates that are not suitable for RecBCD enzyme; for example, many of the recombination events promoted by the RecF pathway are thought to begin from nicks or gaps present in the donor DNA (Lloyd and Thomas 1984; Lanzov et al. 1991). Conceivably, RecQ protein could act at these internal sites to initiate recombination much as it does from flush dsDNA ends. For recombination following bacterial conjugation, a consequence of such internally initiated crossover events is a shorter distance between adjacent crossovers and, therefore, a higher frequency of recombination exchanges per unit length of DNA than recombination promoted solely from the ends of the donor DNA (Lanzov et al. 1991). In complete agreement with the idea of RecQ helicase initiating recombination from internal sites in DNA is the finding that otherwise wild-type recQ mutants show a decreased number of exchanges per unit length of DNA (Lanzov et al. 1991; Lloyd and Buckman 1995). Additionally, recFrecQ double mutants produce recombination products with crossovers confined solely to the ends of the DNA, which is consistent with initiation occurring only at the dsDNA ends by RecBCD enzyme (Lanzov et al. 1991). These findings support the view that RecQ helicase initiates recombination crossover events from sites internal to the donor DNA.
Further support for the proposal that RecQ helicase acts to initiate recombination comes from an investigation of conjugal recombination in resolvase-deficient ruv recG strains (Ryder et al. 1994). In a ruvArecG background, recombination frequency is reduced to 0.1% of wild type and cell viability is much lower than that of wild-type E. coli. Because both the RuvAB and RecG proteins are Holliday junction-specific binding proteins, the observed deficiency in recombination and reduced viability of these cells is likely the result of the accumulation of stalled recombination intermediates that arise from a block in branch migration. Inclusion of a recQ mutation in the ruvArecG background results in a 25-fold increase in recombinational frequency and a concomitant increase in cell viability. A similar suppressor effect was also observed with the disruption of the recF, recJ, recO, or recR genes. Ryder et al. explained the suppressor effect of the mutations with a model in which the inactivation of proteins needed for initiation of RecF pathway-dependent recombination (e.g., RecQ helicase) redirects some of the potential recombination substrates toward RecBCD enzyme-dependent initiation, thereby avoiding the formation of crossovers that would require resolution by either RuvAB or RecG helicases. In this model, the intermediates by RecBCD enzyme-dependent initiation must be resolved by a separate, but unknown mechanism. The different requirements for resolution between the RecQ helicase- and RecBCD enzyme-dependent initiation observed by this group may represent the differences in the intermediates formed by recombination initiated from internal regions in the donor DNA and those begun from its ends (see above). These genetic data clearly indicate a key role for RecQ helicase in the initiation of recombination; however, the biochemical attributes that distinguish RecQ protein from other helicases in this capacity remain unclear. Two important properties are likely to include the ability to effectively act on recombinogenic lesions (e.g., gapped and blunt-ended dsDNA), as well as to allow access of RecA protein to the ssDNA produced by unwinding. Regardless, these genetic findings and our biochemical data support the idea that RecQ helicase initiates homologous recombination.
RecQ helicase action, however, was not solely confined to an initiation role in the coupled pairing reactions; it also displayed a late role with its propensity to unwind the D-loops and Kappa intermediates formed by RecA protein. In our view, RecQ helicase is free to act on any DNA substrate present in the reaction following the first round of linear dsDNA unwinding, including the homologously paired joint molecules (Fig. 4C). This nonspecific facet of RecQ helicase activity is a direct result of its relaxed DNA substrate specificity. Thus, RecQ helicase can initiate recombination as well as act later to either promote DNA heteroduplex extension or impede homologous recombination by disruption of short DNA heteroduplex tracts, depending on the DNA substrates it encounters.
In support of the multifunctional capacity of RecQ helicase, Hanada et al. (1997) found that this protein also acts as a suppressor of illegitimate recombination in E. coli. In this study recombination between λ spi phage and the E. coli chromosome took place with as little as 8 bp of contiguous, shared homology. In the absence of a functional recQ gene, this type of aberrant recombination was elevated by 30- to 300-fold. Interestingly, enhanced illegitimate recombination in the absence of RecQ helicase was also observed in both recA and recBCsbcBC backgrounds. This suggests the suppressor activity of RecQ helicase is independent of its initiation function in recombination. It was proposed that RecQ helicase suppresses illegitimate recombination by dissociating nascent joint molecules formed between the phage and chromosomal DNA (Hanada et al. 1997). This idea is consistent with the data presented here, which demonstrate that RecQ helicase is capable of both disrupting joint molecules and unwinding a variety of partially duplex DNA substrates.
The recent identification of the RecQ helicase-like proteins in eukaryotic organisms, including the Sgs1 (budding yeast), Blm (human), and Wrn (human) helicases, raises the possibility that RecQ helicase function is conserved in higher organisms. Mutations at the BLM and WRN loci result in the inherited genetic diseases known as Bloom’s syndrome and Werner’s syndrome, respectively (Ellis et al. 1995; Yu et al. 1996). The sgs1, blm, and wrn mutants all display similar phenotypes: Chromosomal aberrations, chromosomal nondisjunction, hyper-recombination, and alterations in DNA replication (Gangloff et al. 1994; Ellis et al. 1995; Watt et al. 1995, 1996; Yu et al. 1996). If these eukaryotic helicases are functional, as well as structural, homologs of E. coli RecQ helicase, then it is possible that they are involved in initiation of homologous recombination or act as antagonists to illegitimate recombination. The latter possibility is supported by the fact that aberrant recombination events are elevated in cells from both Bloom’s syndrome and Werner’s syndrome patients (Ellis et al. 1995; Yu et al. 1996). In addition, Δsgs1 cells display a hyper-recombination phenotype (Gangloff et al. 1994). Therefore, it is reasonable to propose that these proteins, like RecQ helicase, are responsible for removing aberrant crossovers that might jeopardize key steps in DNA replication and chromosome partitioning during mitosis and meiosis. In support of this idea, the chromosomal breakage, nondisjunction, and other chromosomal abnormalities common to Bloom’s and Werner’s syndrome are readily explained by the presence of recombination intermediates during segregation of chromosomes in mitosis and meiosis I (Ellis et al. 1995; Yu et al. 1996). In addition, Δsgs S. cerevisiae cells have a marked increase in chromosomal nondisjunction during both mitosis and meiosis I (Watt et al. 1995). Therefore, the Sgs1, Blm, and Wrn proteins, like E. coli RecQ helicase, may be required to disrupt intermediates formed by nonhomologous and aberrant recombination events; However, our findings do not preclude that future biochemical and genetic studies will demonstrate an initiation function for the eukaryotic homologs.
Materials and methods
Reagents
Chemicals were reagent grade and all solutions were prepared by use of Barnstead Nanopure water. PEP and ATP were purchased from Sigma. ATPγS was purchased from Boehringer Mannheim. ATPγS and ATP were dissolved as concentrated stock solutions at pH 7.5. Nucleotide concentrations were determined spectrophotometrically by use of an extinction coefficient of 1.54 × 104/m per cm at 260 nm. Polyethylene glycol (8000 molecular weight; PEG) was purchased from Sigma and dissolved in water as a 40% (wt/vol) stock.
RecQ helicase
Running buffer for all columns, except hydroxylapatite, was buffer A [20 mm Tris-HCl (pH 8.0), 1 mm EDTA, 1 mm dithiothreitol (DTT)] with 100 mm NaCl. Hydroxylapatite running buffer consisted of buffer B [50 mm KPi (pH 6.8) and 1 mm DTT)]. RecQ helicase expression vector pSQ211 was a kind gift of Dr. Hiroshi Iwasaki (Osaka University, Japan). pSQ211 is a derivative of pET8C with the recQ gene downstream of the T7 polymerase φ10 promoter. Strain SCK225 was constructed by transforming BL21(DE3)recA with pSQ211. SCK225 was grown at 37°C in terrific broth (Sambrook et al. 1989) supplemented with 150 μg ampicillin/ml to mid-log phase (A600 = 1.0) when the recQ gene was induced with 1 mm IPTG for 2 hr. Following harvest, cells were resuspended in 5.5 vol of buffer A, lysed with one passage through a French pressure cell and the lysate cleared by centrifugation at 77,000g. DNA was removed by centrifugation at 9000g following drop-wise addition of 0.31 volumes of 10% streptomycin sulfate. This supernatant was loaded onto Q-Sepharose (330 ml; flow rate of 5.5 ml/min). RecQ helicase, identified by ssDNA-dependent ATP hydrolysis activity and a 65-kD band on SDS-PAGE, eluted at ∼300 mm NaCl in a 1500 ml linear gradient (100–500 mm NaCl). The pooled peak fractions from Q-Sepharose were loaded onto ssDNA-cellulose (225 ml; flow rate of 6.4 ml/min) and RecQ helicase was batch eluted with 1000 mm NaCl. The ssDNA cellulose pool was loaded onto hydroxylapatite (Bio-Gel HTP, BioRad; 40 ml; flow rate of 1.4 ml/min) and the column resolved with a 400 ml linear gradient from 50 to 400 mm KPi. RecQ helicase eluted from this column at ∼125 mm KPi. The pooled peak fractions from hydroxylapatite were loaded onto MonoQ HR10/10 (Pharmacia; flow rate of 2 ml min). RecQ helicase eluted at 200 mm NaCl in a 160 ml linear gradient (150 to 400 mm NaCl). The pool from MonoQ was concentrated by dialysis into storage buffer [20 mm Tris-HCl (pH 8.0), 1 mm EDTA, 0.1 mm DTT, 100 mm NaCl, 50% (vol/vol) glycerol]. Protein concentration was determined spectrophotometrically with an extinction coefficient for RecQ helicase of 1.48 × 104/m per cm at 260 nm, calculated from its primary amino acid sequence. The resulting protein was >95% RecQ helicase by Coomassie staining. The protein is free of exonuclease activity as judged by the release of <5% TCA soluble counts from 10 μm uniformly 3H-labeled ds- or ssDNA in 1 hr at 37°C in the presence of 1 μm RecQ protein.
Other proteins
RecA protein was purified from E. coli strain GE1710, obtained from Dr. George A. Weinstock (University of Texas Medical School, Houston), by use of a preparative protocol on the basis of spermidine acetate precipitation (Griffith and Shores 1985). RecA protein concentration was determined spectrophotometrically by use of an extinction coefficient of 2.7 × 104/m per cm at 280 nm. SSB protein was purified from E. coli strain RLM727 as described (LeBowitz 1985) and its concentration was determined spectrophotometrically by use of an extinction coefficient at 280 nm of 3.0 × 104/m per cm.
DNA
Covalently closed circular pUC19, pUC1950, and pUCD DNA were purified with alkaline lysis followed by CsCl-ethidium bromide equilibrium centrifugation (Sambrook et al. 1989). pUC1950 is a derivative of pUC19 with a nonhomologous 1950-bp insert between the HindIII and EcoRI sites in pUC19. pUCD is a stable dimer of pUC19. The molar nucleotide concentration of each was determined by use of the extinction coefficient of 6500/m per cm at 260 nm. DNA was linearized by digestion with HindIII restriction endonuclease (New England Biolabs), then gel purified by use of the EluTrap (Schleicher & Schuell). Linear dsDNA was 5′-end labeled with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (NEN). φX174 RF I was purchased from New England Biolabs.
Oligonucleotides
The DNA sequences of the oligonucleotides are shown in Table 2. Oligonucleotides were synthesized on a Milligen/ Biosearch Cyclone Plus DNA synthesizer and purified by use of denaturing polyacrylamide gel electrophoresis. Briefly, oligonucleotide was separated from aborted products by electrophoresis through an 8% polyacrylamide (20:1 acrylamide/bisacrylamide)/50% urea gel in TBE (89 mm Tris, 89 mm sodium borate, 1 mm EDTA). The DNA was recovered by soaking the excised band in 0.3 mm sodium acetate overnight at 25°C followed by ethanol precipitation. The recovered DNA pellet was dissolved in TE [10 mm Tris-HCl (pH 8.0) and 1 mm EDTA]. The concentration of the purified oligonucleotides were determined spectrophotometrically by use of a molar extinction coefficient at 260 nm calculated by the formula ε = [(no. of G × 11.7) + (no. of C × 7.3) + (no. of A × 15.4) + (no. of T × 8.8)].
Table 2.
Oligonucleotides used to construct DNA substrates
| 1a | 5′- | G | A | C | G | C | T | G | C | C | G | A | A | T | C | C | T | A | C | C | A | G | T | G | C | C | T | T | G | C | T | A | G | G | A | C | A | T | C | T | T | T | G | C | C | C | A | C | C | T | G | C | A | G | G | T | T | C | A | C | C | C | -3′ | |||
| 2a | 5′- | T | G | G | G | T | G | A | A | C | C | T | G | C | A | G | G | T | G | G | G | C | A | A | A | G | A | T | G | T | C | C | T | A | G | C | A | A | T | G | T | A | A | T | C | G | T | C | A | A | G | C | T | T | T | A | T | G | C | C | G | T | T | -3′ | ||
| 3a | 5′- | C | A | A | C | G | G | C | A | T | A | A | A | G | C | T | T | G | A | C | G | A | T | T | A | C | A | T | T | G | C | T | A | G | G | A | C | A | T | G | C | T | G | T | C | T | A | G | A | G | G | A | T | C | C | G | A | C | T | A | T | C | G | A | -3′ | |
| 4a | 5′- | A | T | C | G | A | T | A | G | T | C | G | G | A | T | C | C | T | C | T | A | G | A | C | A | G | C | A | T | G | T | C | C | T | A | G | C | A | A | G | G | C | A | C | T | G | G | T | A | G | A | A | T | T | C | G | G | C | A | G | C | G | T | -3′ | ||
| 5 | 5′- | A | C | A | G | C | A | C | C | A | G | A | T | T | C | A | G | C | A | A | T | T | A | A | G | C | T | C | T | A | A | G | C | C | A | T | C | C | G | C | A | A | A | A | A | T | G | A | C | C | T | C | T | T | A | T | C | A | A | A | A | G | G | A | -3′ | |
| 6 | 5′- | T | C | C | T | T | T | T | G | A | T | A | A | G | A | G | G | T | C | A | T | T | T | T | T | G | C | G | G | A | T | G | G | C | T | T | A | G | A | G | C | T | T | A | A | T | T | G | C | T | G | A | A | T | C | T | G | G | T | G | G | C | T | G | T | -3′ |
| 7 | 5′- | T | C | C | T | T | T | T | G | A | T | A | A | G | A | G | G | T | C | A | T | T | T | T | T | G | C | G | G | A | T | G | G | C | T | T | A | G | A | G | C | T | T | A | A | T | T | G | C | -3′ | ||||||||||||||||
| 8 | 5′- | G | C | A | A | T | T | A | A | G | C | T | C | T | A | A | G | C | C | A | T | C | C | G | C | A | A | A | A | A | T | G | A | C | C | T | C | T | T | A | T | C | A | A | A | A | G | G | A | -3′ |
These sequences are the same as those for oligonucleotides 1–4 described in Benson and West (1994).
Construction of oligonucleotide substrates
Synthetic DNA substrates were prepared by annealing the combinations of oligonucleotides indicated in Table 1. Equimolar concentrations of oligonucleotides were mixed in annealing buffer [20 mm Tris-acetate (pH 7.9), 10 mm magnesium acetate, 50 mm potassium acetate and 1 mm DTT], heated to 100°C for 7 min, then allowed to cool at room temperature to 25°C over a 5 hr period. Because this procedure resulted in >95% annealing of the oligonucleotides, the DNA substrates were not further purified and were used directly. For DNA binding, oligonucleotides were 5′-32P-end labeled with T4 polynucleotide kinase and [γ-32P]ATP. Unincorporated [γ-32P]ATP was removed using MicroSpin G-25 columns (Pharmacia Biotech).
Pairing reactions
Unless otherwise indicated, all reactions were carried out at 37°C in a buffer consisting of 25 mm Tris-acetate (pH 7.5), 1 mm magnesium acetate, 0.1 mm DTT, 1 mm ATP, 1 mm PEP, and 10% (wt/vol) PEG. Pyruvate kinase (Sigma) was added to a final concentration of 20 U/ml. Coupled reactions consisted of 10 μm 5′-end labeled HindIII-cut pUC19 DNA, 20 μm supercoiled DNA (when present), 2 μm SSB protein, 5 μm RecA protein, and RecQ helicase at the indicated concentration. All reaction components were preincubated for 3 min at 37°C before initiating the reaction by addition of ATP. Pairing reactions having only RecA protein were carried out in the same buffer as above and consisted of 5 μm heat-denatured 5′-end labeled linear pUC19, 5 μm unlabeled HindIII-cut pUC19, 20 μm supercoiled pUC1950, 2 μm SSB protein, and 5 μm RecA protein. Reactions were started by the addition of ATP. Aliquots were taken from the reactions at the indicated times, stopped with 40 mm EDTA/0.8% SDS and stored on ice. The samples were deproteinized with 1.5 μg/μl proteinase K (Boehringer Mannheim) at room temperature for 10 min prior to loading on a 0.8% agarose 1× TAE gel and run for 600 V ⋅ hr. Gels were dried and visualized with a Molecular Dynamics Storm 840 PhosphorImager. The quantity of DNA in each band was determined by use of ImageQuant software.
Gel band-shift assays
Binding of RecQ helicase to the synthetic DNA substrates was analyzed by use of nondenaturing PAGE band-shift assays. Reactions containing 20 nm DNA molecules and the indicated concentration of RecQ helicase were incubated at 37°C for 4 min in binding buffer [25 mm Tris-acetate (pH 7.5), 1.2 mm magnesium acetate, 28 mm NaCl, 0.1 mm DTT, and 14% glycerol] then immediately loaded onto a 6% polyacrylamide gel (30:1 acrylamide/bisacrylamide). Electrophoresis was carried out at 380 V for 1 to 1.5 hr in a Tris-glycine buffer (25 mm Tris, 190 mm glycine, 0.1 mm EDTA). Gels were dried onto DEAE paper (Whatman) and visualized by use of a Molecular Dynamics Storm PhosphorImager. The concentration of DNA in bands corresponding to free DNA substrate was determined by use of ImageQuant software. Binding affinity (per molar) was calculated as the reciprocal of the RecQ helicase concentration at which half the available DNA was free.
Helicase assays
The rate of RecQ helicase-mediated unwinding of the synthetic DNA substrates was determined by use of a spectrofluorimetric dye-displacement assay as described previously (Eggleston et al. 1996). Reactions were carried out at 37°C in a buffer consisting of 25 mm Tris-acetate (pH 7.5), 1.2 mm magnesium acetate, 1 mm ATP, 1 mm PEP, and 20 U/ml pyruvate kinase. DNA substrate [480 nm (bp)] 70 nm Hoechst 33582 dye, and RecQ helicase at 5–25 nm were preincubated for 2 min before starting the reaction by the addition of ATP. SSB protein (in monomers) was also present at 20% the concentration of the total nucleotides present in the reaction. The unwinding rate in nm (bp)/sec at each RecQ helicase concentration was calculated from the initial slope of the unwinding curve as described previously (Eggleston et al. 1996). For each DNA substrate, the unwinding rate per RecQ helicase monomer was calculated as the slope of straight line fit to a plot of rate versus RecQ helicase concentration.
Acknowledgments
We are grateful to Dr. Hiroshi Iwasaki for providing us with plasmid pSQ211 and the following members of this laboratory for their insightful comments and critical reading of this manuscript: Dan Anderson, Deana Arnold, Piero Bianco, Joel Brockman, Frederic Chedin, Jason Churchill, Noriko Kantake, Alex Mazin, Jim New, Erica Seitz, Tomohiko Sugiyama, and Bob Tracy. This work is supported by funds from the National Institutes of Health grant (GM-41347).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sckowalczykowski@ucdavis.edu; FAX (916) 752-5939.
References
- Anderson DG, Kowalczykowski SC. The recombination hot spot χ is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes & Dev. 1997a;11:571–581. doi: 10.1101/gad.11.5.571. [DOI] [PubMed] [Google Scholar]
- ————— The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell. 1997b;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- Benson FE, West SC. Substrate specificity of the Escherichia coli RuvC protein. Resolution of three- and four-stranded recombination intermediates. J Biol Chem. 1994;269:5195–5201. [PubMed] [Google Scholar]
- Camerini-Otero RD, Hsieh P. Homologous recombination proteins in prokaryotes and eukaryotes. Annu Rev Genet. 1995;29:509–552. doi: 10.1146/annurev.ge.29.120195.002453. [DOI] [PubMed] [Google Scholar]
- Clark AJ, Low KB. Pathways and systems of homologous recombination in Escherichia coli. In: Low KB, editor. The recombination of genetic material. San Diego, CA: Academic Press; 1988. pp. 155–215. [Google Scholar]
- Clark AJ, Sandler SJ. Homologous genetic recombination: The pieces begin to fall into place. Crit Rev Microbiol. 1994;20:125–142. doi: 10.3109/10408419409113552. [DOI] [PubMed] [Google Scholar]
- Courcelle J, Carswell-Crumpton C, Hannawalt PC. recF and recR are required for resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci. 1997;94:3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DA, Kowalczykowski SC. Homologous pairing in vitro stimulated by the recombination hotspot, Chi. Cell. 1991;66:361–371. doi: 10.1016/0092-8674(91)90625-9. [DOI] [PubMed] [Google Scholar]
- Eggleston AK, Rahim NA, Kowalczykowski SC. A helicase assay based on the displacement of fluorescent, nucleic acid-binding ligands. Nucleic Acids Res. 1996;24:1179–1186. doi: 10.1093/nar/24.7.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- Emmerson PT. Recombination deficient mutants of Escherichia coli K12 that map between thyA and argA. Genetics. 1968;60:19–30. doi: 10.1093/genetics/60.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MD, Shen JC, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA. The Werner syndrome protein is a DNA helicase. Nature Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- Griffith J, Shores CG. RecA protein rapidly crystallizes in the presence of spermidine: A valuable step in its purification and physical characterization. Biochemistry. 1985;24:158–162. doi: 10.1021/bi00322a022. [DOI] [PubMed] [Google Scholar]
- Gupta RC, Bazemore LR, Golub EI, Radding CM. Activities of human recombination protein Rad51. Proc Natl Acad Sci. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Toshiyuki U, Kohno Y, Saito K, Kato J, Ikeda H. RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc Natl Acad Sci. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Flanders P, Theriot L. Mutants of Escherichia coli K-12 defective in DNA repair and in genetic recombination. Genetics. 1966;53:1137–1150. doi: 10.1093/genetics/53.6.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Takahagi M, Nakata A, Shinagawa H. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes & Dev. 1992;6:2214–2220. doi: 10.1101/gad.6.11.2214. [DOI] [PubMed] [Google Scholar]
- Karow JK, Chakravarty RK, Hickson ID. The Bloom’s syndrome gene protein product is a 3′–5′ DNA helicase. J Biol Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- Konforti BB, Davis RW. 3′ homologous free ends are required for stable joint molecule formation by the RecA and single-stranded binding proteins of Escherichia coli. Proc Natl Acad Sci. 1987;84:690–694. doi: 10.1073/pnas.84.3.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Eggleston AK. Homologous pairing and DNA strand-exchange proteins. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Clow JC, Somani R, Varghese A. Effects of the Escherichia coli SSB protein on the binding of Escherichia coli RecA protein to single-stranded DNA: Demonstration of competitive binding and the lack of a specific protein-protein interaction. J Mol Biol. 1987;193:81–95. doi: 10.1016/0022-2836(87)90629-2. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano K, Sunohara Y, Takahashi N, Yoshikura H, Kobayashi I. DNA double-strand break repair: Genetic determinants of flanking crossing-over. Proc Natl Acad Sci. 1994;91:1173–1177. doi: 10.1073/pnas.91.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner SR, Nagaishi H, Templin A, Clark AJ. Genetic recombination in Escherichia coli: The role of exonuclease I. Proc Natl Acad Sci. 1971;68:824–827. doi: 10.1073/pnas.68.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzov V, Stepanova I, Vinogradskaja G. Genetic control of recombination exchange frequency in Escherichia coli K-12. Biochimie. 1991;73:305–312. doi: 10.1016/0300-9084(91)90217-o. [DOI] [PubMed] [Google Scholar]
- Lavery PE, Kowalczykowski SC. Enhancement of recA protein-promoted DNA strand exchange activity by volume-occupying agents. J Biol Chem. 1992;267:9307–9314. [PubMed] [Google Scholar]
- LeBowitz J. “Biochemical mechanism of strand initiation in bacteriophage lambda DNA replication.” Ph.D. thesis. Baltimore, MD: Johns Hopkins University; 1985. [Google Scholar]
- Lloyd RG, Buckman C. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J Bacteriol. 1985;164:836–844. doi: 10.1128/jb.164.2.836-844.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Conjugational recombination in Escherichia coli: Genetic analysis of recombinant formation in Hfr x F- crosses. Genetics. 1995;139:1123–1148. doi: 10.1093/genetics/139.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd RG, Sharples GJ. Dissociation of synthetic Holliday junctions by E. coli RecG protein. EMBO J. 1993a;12:17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Processing of recombination intermediates by the RecG and RuvAB proteins of Escherichia coli. Nucleic Acids Res. 1993b;21:1719–1725. doi: 10.1093/nar/21.8.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd RG, Thomas A. A molecular model for conjugational recombination in E. coli K12. Mol & Gen Genet. 1984;197:328–336. doi: 10.1007/BF00330981. [DOI] [PubMed] [Google Scholar]
- Lu J, Mullen JR, Brill SJ, Kleff K, Romeo AM, Strenglantz R. Human homologues of yeast helicase. Nature. 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- Maeshima K, Morimatsu K, Shinohara A, Horii T. RAD51 homologues in Xenopus laevis: Two distinct genes are highly expressed in ovary and testis. Gene. 1995;160:195–200. doi: 10.1016/0378-1119(95)00148-y. [DOI] [PubMed] [Google Scholar]
- Mahajan SK. Pathways of homologous recombination in Escherichia coli. In: Kucherlapati R, Smith GR, editors. Genetic recombination. Washington, D.C.: American Society for Microbiology; 1988. pp. 87–140. [Google Scholar]
- Mendonca VM, Matson SW. Genetic analysis of ΔhelD and ΔuvrD mutations in combination with other genes in the RecF recombination pathway in Escherichia coli: Suppression of a ruvB mutation by a uvrD deletion. Genetics. 1995;141:443–452. doi: 10.1093/genetics/141.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonca VM, Klepin HD, Matson SW. DNA helicases in recombination and repair: Construction of a ΔuvrD ΔhelD ΔrecQ mutant deficient in recombination and repair. J Bacteriol. 1995;177:1326–1335. doi: 10.1128/jb.177.5.1326-1335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H, Nakayama K, Nakayama R, Irino N, Nakayama Y, Hanawalt PC. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K-12: Identification of a new mutation (recQ1) that blocks the recF recombination pathway. Mol & Gen Genet. 1984;195:474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Irino N, Nakayama H. The recQ gene of Escherichia coli K12: Molecular cloning and isolation of insertion mutants. Mol & Gen Genet. 1985;200:266–271. doi: 10.1007/BF00425434. [DOI] [PubMed] [Google Scholar]
- Parsons CA, Tsaneva I, Lloyd RG, West SC. Interaction of Escherichia coli RuvA and RuvB proteins with synthetic Holliday junctions. Proc Natl Acad Sci. 1992;89:5452–5456. doi: 10.1073/pnas.89.12.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranam KL, Blackshear PJ. Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J Biol Chem. 1994;269:29838–29845. [PubMed] [Google Scholar]
- Roman LJ, Dixon DA, Kowalczykowski SC. RecBCD-dependent joint molecule formation promoted by the Escherichia coli RecA and SSB proteins. Proc Natl Acad Sci. 1991;88:3367–3371. doi: 10.1073/pnas.88.8.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder L, Whitby MC, Lloyd RG. Mutation of recF, recJ, recO, recQ, or recR improves Hfr recombination in resolvase-deficient ruv recG strains of Escherichia coli. J Bacteriol. 1994;176:1570–1577. doi: 10.1128/jb.176.6.1570-1577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Seki M, Miyazawa H, Tada S, Yanagisawa J, Yamaoka T, Hoshino S, Ozawa K, Eki T, Nogami M, Okumura K, et al. Molecular cloning of cDNA encoding human DNA helicase Q1 which has homology to Escherichia coli Rec Q helicase and localization of the gene at chromosome 12p12. Nucleic Acids Res. 1994;22:4566–4573. doi: 10.1093/nar/22.22.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GR. Homologous recombination in procaryotes. Microbiol Rev. 1988;52:1–28. doi: 10.1128/mr.52.1.1-28.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E, Chapman CR, Al-Khodairy F, Carr AM, Enoch T. rqh1+, a fission yeast gene related to the Bloom’s and Werner’s syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Shimamoto A, Imamura O, Kuromitsu J, Kitao S, Goto M, Furuichi Y. DNA helicase activity in Werner’s syndrome gene product synthesized in a baculovirus system. Nucleic Acids Res. 1997;25:2973–2978. doi: 10.1093/nar/25.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AF, Smith GR. Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J Mol Biol. 1985;185:431–443. doi: 10.1016/0022-2836(85)90414-0. [DOI] [PubMed] [Google Scholar]
- Tsaneva IR, Muller B, West SC. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell. 1992;69:1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- Umezu K, Nakayama H. RecQ DNA helicase of Escherichia coli. Characterization of the helix-unwinding activity with emphasis on the effect of single-stranded DNA-binding protein. J Mol Biol. 1993;230:1145–1150. doi: 10.1006/jmbi.1993.1231. [DOI] [PubMed] [Google Scholar]
- Umezu K, Nakayama K, Nakayama H. Escherichia coli RecQ protein is a DNA helicase. Proc Natl Acad Sci. 1990;87:5363–5367. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt PM, Louis EJ, Borts RH, Hickson ID. Sgs1: A eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- Watt PM, Hickson ID, Borts RH, Louis EJ. SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby MC, Ryder L, Lloyd RG. Reverse branch migration of Holliday junctions by RecG protein: A new mechanism for resolution of intermediates in recombination and DNA repair. Cell. 1993;75:341–350. doi: 10.1016/0092-8674(93)80075-p. [DOI] [PubMed] [Google Scholar]
- Whitby MC, Vincent SD, Lloyd RG. Branch migration of Holliday junctions: Identification of RecG protein as a junction specific DNA helicase. EMBO J. 1994;13:5220–5228. doi: 10.1002/j.1460-2075.1994.tb06853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner’s syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]