Abstract
In eukaryotes the sites for the initiation of chromosomal DNA replication are believed to be determined in part by the binding of a heteromeric origin recognition complex (ORC) to DNA. We have cloned the genes encoding the subunits of the Drosophila ORC. Each of the genes is unique and can be mapped to discrete chromosomal locations implying that the pattern and developmental regulation of origin usage in Drosophila is not regulated solely by a large family of different ORC proteins. The six-subunit ORC can be reconstituted with recombinant proteins into a complex that restores DNA replication in ORC-depleted Drosophila or Xenopus egg extracts.
Keywords: DNA replication, ORC, Drosophila, gene mapping
Each chromosome in a dividing eukaryotic cell has many DNA sites that serve to define an origin of DNA replication (ori). These ori sites interact with a variety of proteins that must orchestrate both the mechanisms of DNA synthesis and the regulation of the process throughout the cell cycle (Diffley 1996; Stillman 1996; Dutta and Bell 1997). For multicellular eukaryotes the utilization of these sites changes during development, and though the program of such spatial and temporal activation is poorly understood the process is known to affect both gene expression programs and chromosome folding. Thus, the nature and complexity of these cis-acting replicator elements in metazoans is of considerable interest. It is also of practical significance to define these sequences for use in detecting the local changes in DNA structure that prepare the template for synthesis.
Ori sites in metazoans are poorly defined mainly because of a lack of a simple and decisive biochemical or genetic assay (Aladjem et al. 1998; Gilbert 1998). The discovery of the heteromeric origin recognition complex (ORC) in Saccharomyces cerevisiae (Bell and Stillman 1992) and homologous proteins in metazoans (Gavin et al. 1995; Gossen et al. 1995) provides another approach to the definition of such replicator sequences. It is conceivable that ORC-like proteins bind to specific DNA elements in the regions known to contain chromosomal ori elements. In S. cerevisiae ORC is a six-subunit site-specific DNA-binding protein that forms a platform on which other cellular proteins create prereplication (Diffley et al. 1994; Aparicio et al. 1997) and preinitiation complexes (Zou and Stillman 1998). Extensive genetic dissection of ori sequences in budding yeast distinguishes ORC binding sites as the defining core element of a replicator (Bell and Stillman 1992; Newlon 1997). Activity at the locus is highly regulated. For example, the yeast ORC complex requires ATP as an allosteric effector to bind to specific DNA, but hydrolysis is limited by ori DNA binding; presumably other proteins are required to drive transitions to a subsequent functional state (Klemm et al. 1997). Although ORC1 contains the subunit critical for this ATP binding function, other subunits are also required for site-specific DNA binding (Lee and Bell 1997).
To date, a wide variety of eukaryotic organisms have been shown to contain orthologs of the individual ORC1, ORC2, ORC4, and ORC5 subunits. These include Schizosaccharomyces pombe, Kluyveromyces lactis, Arabidopsis thaliana, Xenopus laevis, and Homo sapiens (Muzi-Falconi and Kelly 1995; Grallert and Nurse 1996; Leatherwood et al. 1996; Ohtani et al. 1996; Ishiai et al. 1997; Quintana et al. 1998). The subunit composition of the metazoan complexes and associated activities remains largely unresolved and only in Drosophila and Xenopus have studies directly addressed the protein complexity of ORC. Immunoprecipitation and immunoaffinity purification of ORC from Xenopus egg extracts have shown that ORC1 and ORC2 are associated with at least four additional polypeptides (Romanowski et al. 1996; Carpenter and Dunphy 1998; Tugal et al. 1998). In general, resolution of this sort of biochemical question requires extensive in vitro functional or genetic dissection. Significantly, antibodies to ORC2 or ORC1 proteins have been used to immunodeplete Xenopus egg extract of cross-reacting material, and this depletion destroys DNA replication activity in this cell-free system (Carpenter et al. 1996; Romanowski et al. 1996; Rowles et al. 1996; Hua and Newport 1998). However, it is not known whether additional proteins were depleted, which could contribute to the loss of DNA replication activity, along with the immunologically targeted polypeptides in these experiments.
In Drosophila we used a polyclonal antiserum directed against ORC2 to follow and purify the protein through a multistep biochemical protocol. We were able to show that the protein cosedimented in a glycerol gradient with five other polypeptides. From that complex, protein sequence information allowed us to identify the Drosophila orthologs of ORC5 (Gossen et al. 1995) and ORC1 (Pak et al. 1997). Genetic studies are consistent with the hypothesis that this complex participates in DNA replication as does the yeast complex. Mutants containing lethal or hypomorphic alleles in either ORC2 or ORC5 show defects in DNA replication patterns in larvae or in the amplification of the chorion genes in ovarian follicle cells (Landis et al. 1997; M. Pflumm and M.R. Botchan, unpubl.). To investigate the genomic complexity of the functions of ORC in Drosophila DNA replication and to reconstitute a functional complex from recombinant proteins, we have endeavored to identify all of the genes encoding the proteins of this complex.
Results and Discussion
Cloning of ORC subunits
To isolate the genes encoding the unknown subunits of Drosophila ORC we purified ORC from Drosophila embryos (0–12 hr of development) through the several steps of conventional chromatography described previously (Gossen et al. 1995). Proteins corresponding to Drosophila DmORC3 (79 kD), DmORC4 (42 kD), and DmORC6 (30 kD) were isolated and tryptic peptides sequenced. On the basis of sequence information, degenerate primers were designed and used to amplify the genomic DNA that encoded the peptide. These DNAs were used to probe a Drosophila melanogaster cDNA library (Brown and Kafatos 1988). Two different peptides from each subunit band were used to make such genomic probes. For each set, a single ORF was identified that encoded all of the peptides derived from the appropriate subunit of ORC (Materials and Methods). An intact cDNA for each subunit included a putative initiator ATG preceded by stop codons in all three reading frames. In combination with previously described DmORC1, DmORC2, and DmORC5 genes, the isolation of the Drosophila cDNAs for ORC3, ORC4, and ORC6 completes the identification of the genes encoding for this complex.
Translation of full-length cDNA clones for each subunit predicts a range of amino acid identities with yeast ORC components (24% for ORC4, 21% for ORC3, and 19% for ORC6; Fig. 1). The alignments of amino acid identities between Drosophila ORC3 and ORC6 components with counterparts in the budding yeast complex are not compelling and one must hold open the possibility that selection might have substantially allowed for divergence of certain functions. Alternatively, the subunits for ORC3 and ORC6 described for Drosophila might not be orthologs with the S. cerevisiae genes at all and may have derived from another evolutionary branch. This is more likely to be the case for ORC6, where the yeast and Drosophila proteins have very different sizes and show no patches of statistically significant identity or homology (Fig. 1C). It will be interesting to learn if a similar gene to Drosophila ORC6 is found in other organisms. The ORC3 alignments in Figure 1A do show that both humans and Xenopus have orthologs to the Drosophila ORC3. The Xenopus p81 protein was initially identified as an ORC2-associated protein (Carpenter and Dunphy 1998), and recent work confirms that this protein is part of a larger ORC complex in Xenopus (Tugal et al. 1998).
Figure 1.
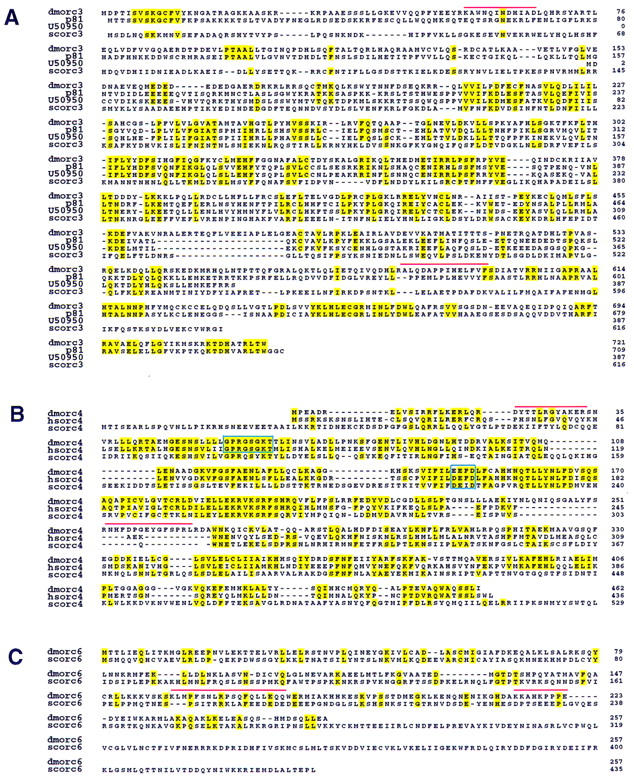
Drosophila homologs of S. cerevisiae, X. laevis, and H. sapiens ORC proteins. (A) The deduced amino acid sequence encoded by the DmORC3 is shown in alignment with S. cerevisiae ORC3, X. laevis p81, and an expressed sequence tag obtained from GenBank (accession no. U50950). Identical residues are highlighted. Red bars indicate tryptic polypeptides that were used for creation of the degenerate PCR primers used for cloning as described. (B) The deduced amino acid sequence encoded by DmORC4 is shown in alignment with S. cerevisiae ORC4 and H. sapiens ORC4. The putative nucleotide binding site and hydrolysis motifs are boxed. (C) The deduced amino acid sequence encoded by DmORC6 is shown in alignment with S. cerevisiae ORC6.
The metazoan ORC4 genes show several regions of peptide identity to each other and to the homologous protein in yeast. In particular the ORC4 proteins of human (Quintana et al. 1997) and Drosophila, and a recently characterized Xenopus homolog (Tugal et al. 1998) conserve ATP-binding and hydrolysis motifs constituting the Walker A and B boxes and other extensive homologies in this central domain, as indicated in Figure 1B. The S. cerevisiae ORC4 protein maintains good homology around the Walker B motif, whereas more divergence is apparent at the A box. It seems possible that an unknown ORC4 ATP binding function was shared in a common ancestor and perhaps preserved in the metazoans. This activity might have been lost in the budding yeast as mutation of these more divergent motifs in S. cerevisiae seems to be of no functional consequence (Klemm et al. 1997).
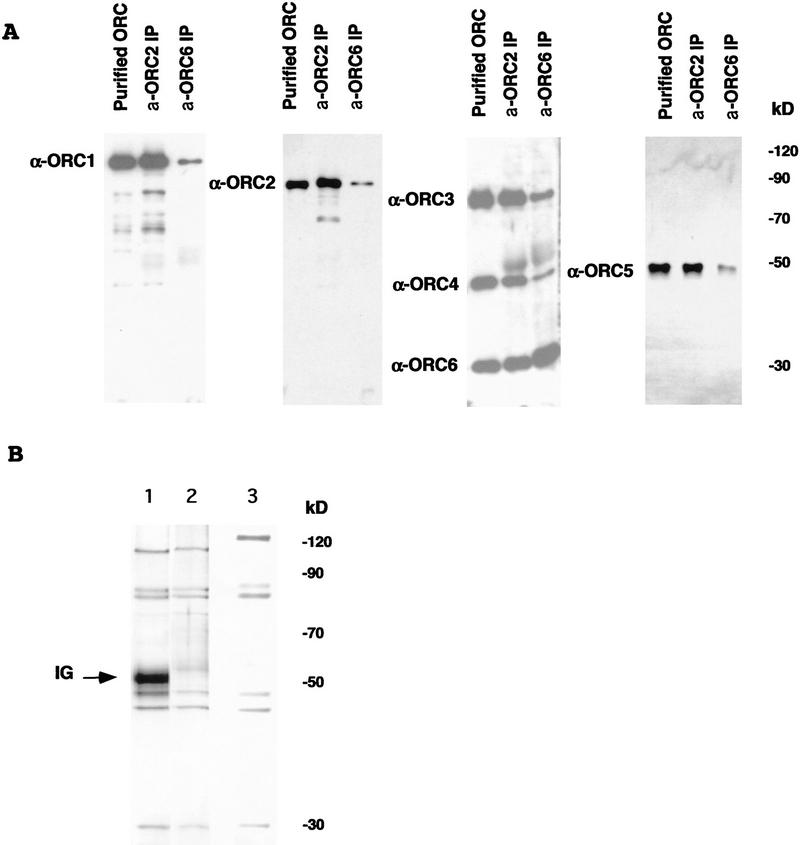
To confirm that the genes identified are those encoding the proteins copurifying in the complex, we raised polyclonal antisera specific for each of the individual full-length proteins. Purified ORC was subjected to SDS-PAGE analysis together with embryo extracts immunoprecipitated with either ORC2 or ORC6 antibodies. The proteins were prepared for immunoblot analysis using individual anti-ORC subunit antibodies. Figure 2A shows that each of the reagents recognized the expected proteins specifically. An interesting point is that in comparison to other ORC subunits there seems to be a free pool of ORC6 unassociated with the other components. This is indicated by the overabundance of ORC6 in the lane containing the ORC6 immunoprecipitated (IP) material. Biochemical fractionation indicated that ORC6 was the only subunit maintained in the extracts in a low molecular weight form unassociated with other DmORC components (data not shown).
Figure 2.
(A) Immunoblot analysis of six-subunit Drosophila ORC using antibodies raised against individual ORC subunits. Protein samples (∼20 ng of total ORC estimated by silver-stained material in the samples) corresponding to purified Drosophila embryonic ORC and immunoprecipitated by α-ORC2 or α-ORC6 antibodies, material was separated using SDS-PAGE and transferred to a PVDF membrane. Membrane strips were incubated with antibodies raised against individual Drosophila ORC subunits as indicated by the side labels. Signals with anti-ORC3 and anti-ORC6 sera were obtained first and the same filter was subsequently probed with the ORC4 reagent. Low levels of specific ORC1 and ORC2 proteolytic fragments are also visible. Protein bands were visualized by subsequent ECL assay. (B) Silver-stained gel of Drosophila ORC immunoprecipitated by α-ORC2 (lane 1) and recombinant baculovirus-expressed Drosophila ORC as eluted from a Ni column (lane 2) and subsequently purified on a glycerol gradient (lane 3). Lanes 1 and 2 are taken from the same gel. DmORC1 has an altered electophoretic mobility due to addition of a His tag.
DmORC genes are unique
In principle, the genetic complexity of ori usage in Drosophila might be explained by the existence of a large family of ORC genes, each with a distinct pattern of temporal or tissue expression and the ability of different ORC complexes to recognize different DNA elements. No data consistent with this thought were obtained. Genomic blot analysis employing restriction enzymes that did not recognize the respective cDNA probes revealed single bands for each of the ORC genes (Fig. 3B). This apparent uniqueness allowed us to unambiguously identify the locations of each of the ORC genes in the polytene chromosome cytological map (Fig. 3A) (Materials and Methods). As anticipated, large stores of mRNA for each of the ORC genes are maternally deposited and the level of such mRNA decreases through development (Fig. 3C). Undoubtedly, zygotic induction of ORC genes is highly regulated in a tissue-specific manner and our inability to detect mRNA in the latter stages is probably due to the insensitivity of the Northern blotting procedure. A hint of such differential and complex regulation is indicated by an increase in abundance for ORC2 and ORC4 mRNAs at ∼6 hr of development, relative to that detected at 4–6 hr. Also a second transcript for both ORC4 and ORC6 becomes more apparent at these times. The biological significance of these second transcripts requires further study. We suspect that they represent alternate start sites or 3′ processing of the mRNAs encompassing the single ORFs, because protein patterns from the IPs of the ORC material were identical throughout the early staged times (data not shown), and our attempts to clone cDNAs from other staged libraries yielded the same ORFs.
Figure 3.
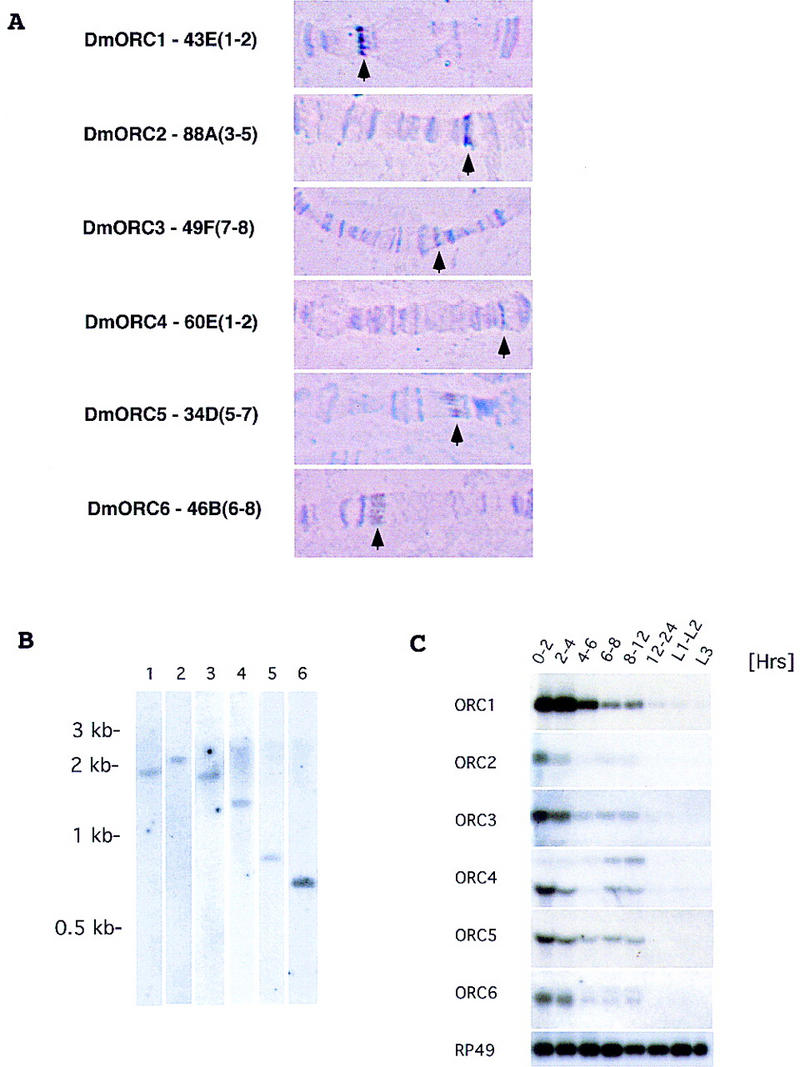
(A) Cytogenetic localization of Drosophila ORC genes based on results of fluorescent in situ hybridization. Chromosomal assignments for each gene are provided along with a representative picture of the polytene bands in the region of assignment (arrows point to specific bands). (B) Genomic blot using specific cDNA probes for Drosophila ORC genes. Genomic DNA (10 μg) was digested with HindIII and KpnI, separated on 0.8% agarose gel, and transferred to Zeta-probe blotting membrane (Bio-Rad). Membranes were hybridized with specific cDNA probes for each of the Drosophila ORC genes: (Lane 1) ORC1; (lane 2) ORC2; (lane 3) ORC3; (lane 4) ORC4; (lane 5) ORC5; (lane 6) ORC6. (C) Expression of Drosophila ORC genes during development. Northern blot analysis of total RNA isolated from Drosophila embryos and larvae extracted at different times of development was performed using radiolabeled probes corresponding to the individual ORC genes. RP49 (ribosomal protein 49) serves as a loading control throughout. Sizes of mRNAs for individual ORC genes are as follows—ORC1, 2.7 kb; ORC2, 2.2 kb; ORC3, 2.3 kb; ORC4, 2.3 and 1.5 kb; ORC5, 1.5 kb; ORC6, 1.2 and 1.1 kb—and were estimated by comparison to a standard set of markers (GIBCO BRL RNA ladder).
Reconstitution of recombinant ORC
With complete cDNAs for each of the Drosophila ORC subunits available we wanted to determine if coexpression of the genes from baculovirus vectors would be sufficient for complex formation. We expressed each of the genes individually and found only ORC2 and ORC6 to be readily soluble proteins (Materials and Methods). However, upon coinfection of all six viral vectors, each of which carried a unique ORC subunit gene, all other proteins (i.e., ORC1, ORC3–ORC5) remained soluble and readily formed a complex. We used a His-tagged version of ORC1 to simplify purification, and a silver-stained SDS-PAGE analysis of the material eluted from the affinity resin is shown in Figure 2B (lane 2). This material was purified further by sedimentation through a glycerol gradient, as described previously for the embryonic ORC. We found that the six subunits cosedimented as did the native material, and the protein pattern observed in the peak fractions is shown in Figure 2B (lane 3).
Biochemical assays for ORC function
To date, the best understood biochemical activity of S. cerevisiae ORC is its ATP-dependent sequence-specific DNA binding. Employing an immunoprecipitation method and DNA restriction fragments that span the ACE3 and oriβ elements of the chorion gene cluster of Drosophila (Heck and Spradling 1990) we have not been able to obtain any evidence for site-specific DNA-binding activity for the recombinant or embryonic DmORC (M. Gossen and M. Botchan, unpubl.). These results may be as anticipated from the known rapid and permissive replication ori usage in early embryogenesis.
A soluble DNA replication system that is dependent on the Drosophila ORC protein would be the most direct assay for the functional integrity of the reconstituted complex. X. laevis soluble egg extracts have provided a powerful tool to study cell-cycle and DNA replication proteins; therefore, we sought to mimic such protocols with early embryonic extracts of Drosophila (Materials and Methods). We employed Xenopus demembraned sperm DNA as a template for replication to be mediated by Drosophila 0- to 2-hr embryonic extracts. We found, as have others (Crevel and Cotterill 1991), that DNA synthesis in these extracts is at least 5–10 times less efficient than that by the synchronized Xenopus egg extract in side-by-side reactions (data not shown). The formation of nuclei around the sperm chromatin in Drosophila extracts was low compared to that observed in Xenopus extracts. Accordingly, we observed a severalfold enhancement of DNA replication when we added a Xenopus membrane fraction (Blow and Laskey 1988; Sheehan et al. 1988) to the soluble Drosophila extracts. DNA replication in these extracts is ORC dependent (Fig. 4).
Figure 4.
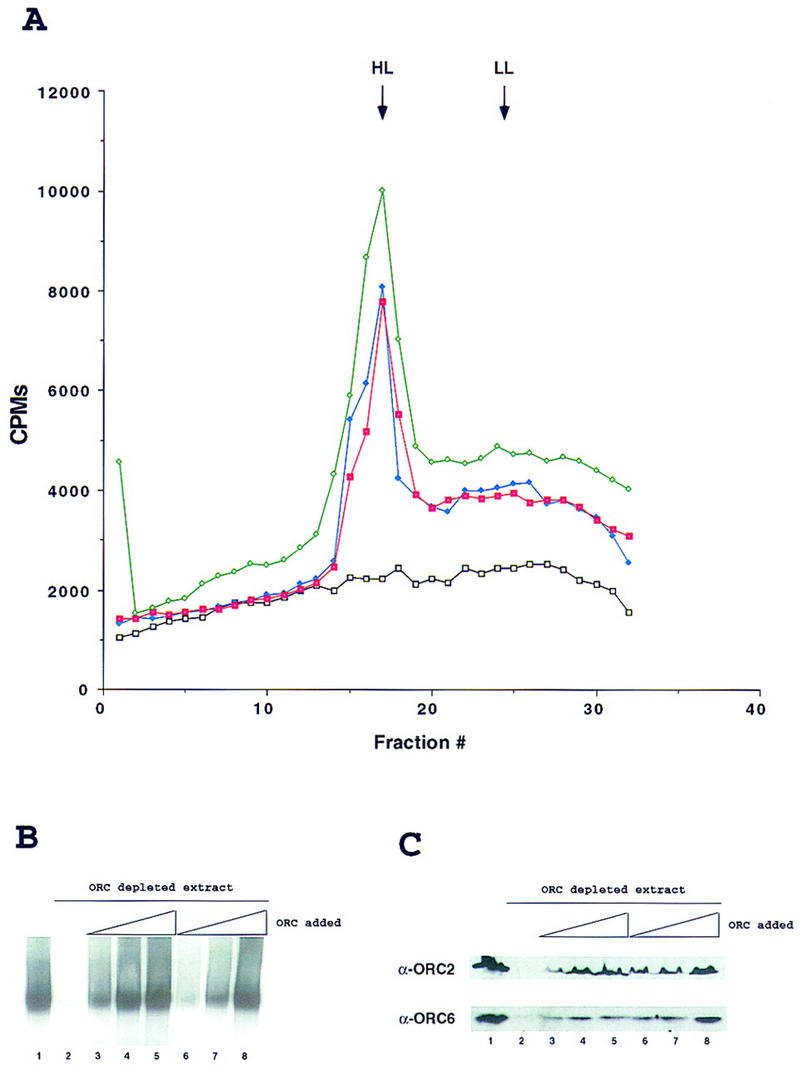
In vitro replication in Drosophila egg extracts is ORC dependent. (A) Density substitution analysis of replicated DNA. Demembraned Xenopus sperm DNA was incubated for 1 hr in Drosophila egg extract at a concentration of 10 ng/μl in the presence of BrdUTP and [α-32P]dCTP. DNA was extracted and subjected to centrifugation through a gradient of CsCl. Xenopus sperm DNA in Drosophila egg extract (green), in ORC-depleted Drosophila extract (black) and after addition to ORC-depleted extract of 100 ng of purified DmORC (blue) or baculovirus-expressed recombinant DmORC (red) are presented on the density profiles. (B) DNA replication in Drosophila extracts. Xenopus sperm DNA was incubated for 1 hr in Drosophila extract at a concentration of 2–5 ng/μl in the presence of [α-32P]dCTP. Where indicated, extracts were depleted for ORC using antibodies raised against DmORC2 and DmORC6. The reconstitution experiment was performed by addition of increasing amounts of purified DmORC [(lane 3) 25 ng; (lane 4) 50 ng; (lane 5) 100 ng] or recombinant baculovirus-expressed DmORC [(lane 6) 25 ng; (lane 7) 50 ng; (lane 8) 100 ng]. (C) Control for depletion and reconstitution experiments. Extracts used for in vitro replication in A and B were subjected to immunoblot analysis using antibodies raised against ORC2 and ORC6 (same order of lanes as in B).
The polyclonal antisera directed against ORC2 and ORC6 were affinity purified and coupled separately to protein A beads. The Drosophila extracts were subjected to one round of immunodepletion with each reagent, and immunoblot analysis showed that this process depletes the extracts completely of ORC2 and almost entirely of ORC6 (Fig. 4C, lanes 1,2). The subsequent replication reactions for depleted or mock-depleted reactions were analyzed by CsCl density gradient fractionation of BrdU and [α-32P]dCTP-labeled DNA (Fig. 4A) and by gel electrophoresis of the sperm DNA extracted from the extracts (Fig. 4B). The average shear size of the DNA was >30 kb, which places all of the labeled DNA to one zone in the electrophoresis (Fig. 4B) and allows for a relatively discrete separation of semiconservatively replicated DNA at a heavy/light (HL) density from the DNA centered at the unsubstituted light/light (LL) position (Fig. 4A), which may arise from incomplete duplex or repair synthesis. DmORC-depleted extracts quantitatively maintained the ability to support complementary strand synthesis of single-stranded M13 DNA (data not shown), suggesting that the ORC is specifically required at the initiation stage of synthesis for duplex DNA and that the immunodepletion did not remove DNA polymerase activity from the extracts. The ability of the ORC-depleted extracts to replicate the sperm chromatin could be complemented by addition of increasing amounts of purified embryonic DmORC (Fig. 4B, lanes 3–5) or the purified recombinant DmORC (Fig. 4B, lanes 6–8). In various experiments activity was restored to 80%–100%. Comparing the specific activity of different preparations of the recombinant complex to that of the endogenous activity revealed no significant differences.
The incorporation of biotin-16–UTP into DNA during the replication reactions can be visualized in these reconstituted extracts by confocal microscopy (Fig. 5). Nuclear formation and replication in mock-depleted Drosophila extracts parallels the same events detected in Xenopus extracts (Fig. 5A). Approximately 10%–20% of Xenopus sperm DNA (depending on batch of extract used) proceeded to form nuclei surrounded by nuclear membrane. All of these nuclei (100%) were positive for DNA replication. The remaining sperm chromatin showed various degrees of decondensation, and the biotin–16-UTP incorporation was significantly lower for these structures. Depletion of the initial Drosophila extracts with DmORC2 and DmORC6 reagents or Xenopus extracts with XlORC1 and XlORC2 reagents blocks DNA replication without affecting the capacity to form nuclei (Fig. 5B). Judging from the fluorescence signal, purified DmORC was able to reconstitute DNA replication in 100% of the Drosophila nuclei to approximately wild-type levels. Interestingly we observed discrete replication foci throughout the Xenopus nuclei when we added the DmORC to the depleted frog extracts (Fig. 5C). Quantitation of DNA incorporation in such heterologous complementation experiments showed that the replication levels were at best 15%–20% of wild type. This type of punctate pattern can be detected upon input of limiting amounts of DmORC in the Drosophila reactions (data not shown) and thus may represent a reduced level of origin starts in the heterologous experiment, even at highest ORC input. Other more complex explanations may account for the inability of the Drosophila ORC to fully complement the replication in the Xenopus extracts. In any case, these experiments show that the ORC can be reconstituted to maintain its activity in DNA replication. This is a prerequisite for further studies needed to probe directly the activities of each of the subunits of this pivotal regulatory complex.
Figure 5.
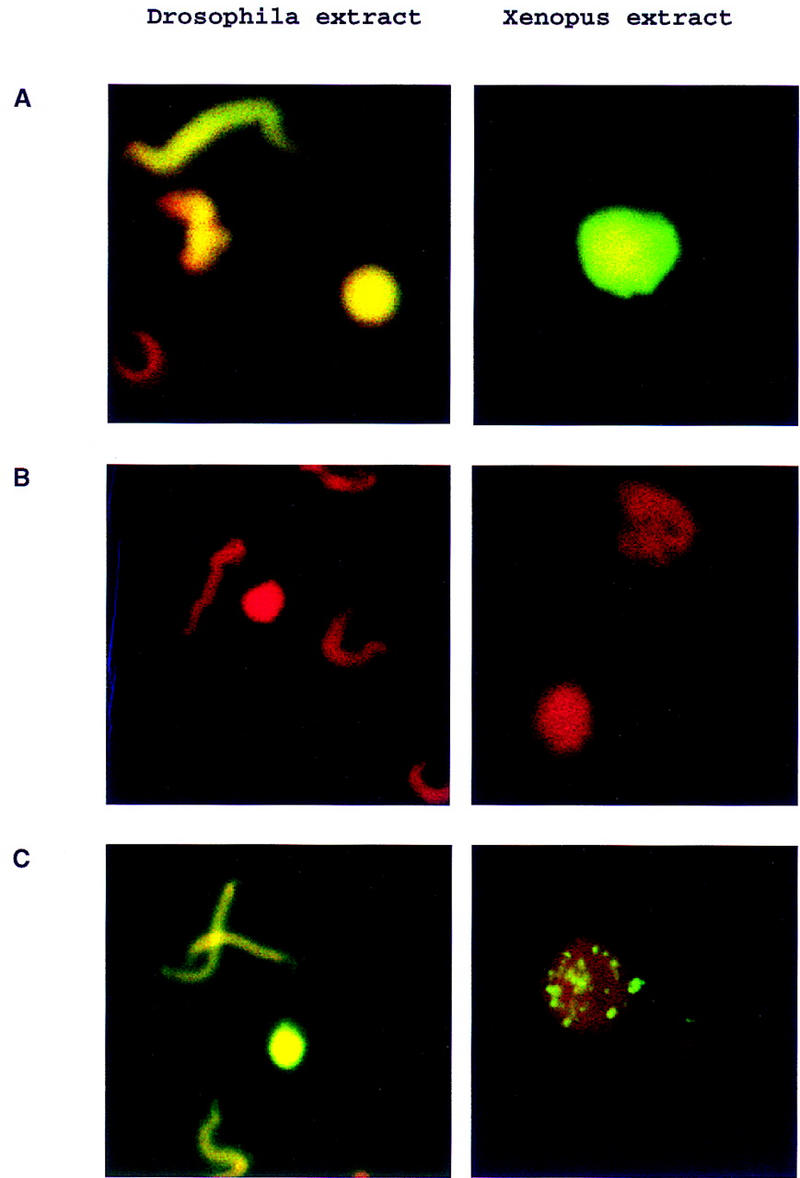
Visualization of ORC-dependent DNA replication in Drosophila and Xenopus extracts. Xenopus sperm DNA (10 ng/μl) was incubated for 2 hr in Drosophila or Xenopus egg extracts in the presence of biotin–16-UTP. In Drosophila, the extent of the sperm decondensation was variable and many of the replicating chromatin structures were elongated in shape but were clearly decondensed relative to sperm. The reactions were fixed and stained for detection of the incorporated UTP analog with fluorescein-conjugated streptavidin. DNA was counterstained with propidium iodide. Merged confocal images are presented. (A) DNA replication in mock-depleted Drosophila or Xenopus egg extracts; (B) DNA replication in ORC-depleted Drosophila or Xenopus egg extracts; (C) DNA replication in ORC-depleted Drosophila or Xenopus egg extract after readdition of purified DmORC (100 ng).
Materials and methods
Purification of Drosophila ORC was performed as described previously (Gossen et al. 1995). Purified ORC was separated by SDS-PAGE and protein bands corresponding to subunits 3, 4, and 6 were excised, cleaved with trypsin, and sequenced. We obtained six peptides from the ORC3 subunit, six peptides from the ORC6 subunit, and four peptides from the ORC4 subunit. Using peptide sequences as a guide, we synthesized degenerate oligonucleotides to amplify the genomic DNAs that encoded peptides shown in Figure 1. The resulting PCR fragments were cloned, sequenced, and used to probe a Drosophila cDNA library (Brown and Kafatos 1988). Positive clones were sequenced. Additional peptide sequences were used to confirm that the correct ORF was identified. Alignments were performed with the ClustalW program (Thompson et al. 1994). In situ hybridization on polytene chromosomes was performed as described by Todd Laverty (http://fruitfly.berkeley.edu/methods/cytogenetics.html).
For recombinant ORC reconstitution, individual ORC genes were initially subcloned in pFastBac plasmid (GIBCO BRL). His tag was added to the amino terminus of DmORC1 during subcloning. Obtained clones were subsequently expressed simultaneously in either Sf9 or High5 cells (Invitrogen) according to manufacturers’ recommendations. The resulting complex was purified on a Ni column (Qiagen). Xenopus egg extracts and demembraned sperm chromatin were prepared essentially as described (Blow and Laskey 1988). The membrane fraction was prepared as described (Sheehan et al. 1988). The preparation of Drosophila egg extracts was based on a procedure described previously (Crevel and Cotterill 1991). In brief, embryos (0–2 hr) were washed with extraction buffer, cold treated, and homogenized. The homogenate was centrifuged for 20 min at 20,000 rpm in a TLA100 Beckmann rotor. The middle layer was collected and recentrifuged. The supernatant was collected and made 5% with respect to glycerol and 1 mm with respect to ATP. The extract was frozen in 20-μl beads in liquid nitrogen.
Extract beads were thawed and supplemented with an ATP-regenerating system (60 mm PC and 150 μg/ml CPK) and 1 μl of Xenopus membrane fraction. Reactions were carried out in 20 μl at room temperature. DNA template was added to a final concentration of 1–10 ng/μl. Immunodepletions of Drosophila and Xenopus extracts were performed using α-DmORC2 and α-DmORC6 (Drosophila) or α-XlORC1 and α-XlORC2 (Xenopus) antibodies (Romanowski et al. 1996) coupled with protein A beads. Depletions were performed in a cold room with rotation for 1 hr two times. Mock depletion was performed with BSA-blocked protein A beads. Completeness of immunodepletions was monitored by Western blotting. For DNA synthesis experiment 5 μCi of labeled dCTP was added. For density substitution experiments (Blow and Laskey 1986), in addition to labeled dCTP, BrdUTP was added to a concentration of 1 mm. Positions of LL and HL peaks were determined in parallel experiments using single-stranded M13 DNA as a template.
Microscopy and indirect immunofluorescence experiments were performed as described previously (Romanowski et al. 1996).
Acknowledgments
We thank Sharleen Zhou for her work in providing peptide sequences for ORC proteins, Todd Laverty for help in the identification of polytene loci, and Aria Adeli and Siavash Karimzadegan for DNA sequencing. Ron Laskey and members of his laboratory, especially Kai Stoeber for discussions and advice in establishing the in vitro replications, are gratefully acknowledged. Support for this research was provided by funds from National Institutes of Health grant CA30490 (to M.R.B.) and a training grant (CA09041) to the Cancer Research laboratory of UC Berkeley. M.G. is supported by a Senior Postdoctoral Fellowship of the American Cancer Society, California Division.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Note
The cDNA sequences of Drosophila ORC3, ORC4, and ORC6 genes have been submitted to GenBank under accession nos. AF139062, AF139063, and AF139065, respectively.
Footnotes
E-MAIL mbotchan@uclink4.berkeley.edu; FAX (510) 643-6334.
References
- Aladjem MI, Rodewald LW, Kolman JL, Wahl GM. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- ————— A role for the nuclear envelope in controling DNA replication within the cell cycle. Nature. 1988;332:546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- Brown NH, Kafatos FC. Functional cDNA libraries from Drosophila embryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- Carpenter PB, Dunphy WG. Identification of a novel 81-kDa component of the Xenopus origin recognition complex. J Biol Chem. 1998;273:24891–24897. doi: 10.1074/jbc.273.38.24891. [DOI] [PubMed] [Google Scholar]
- Carpenter PB, Mueller PR, Dunphy WG. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature. 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- Crevel G, Cotterill S. DNA replication in cell-free extracts from Drosophila melanogaster. EMBO J. 1991;10:4361–4369. doi: 10.1002/j.1460-2075.1991.tb05014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF. Once and only once upon a time: Specifying and regulating origins of DNA replication in eukaryotic cells. Genes & Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Dutta A, Bell SP. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- Gavin KA, Hidaka M, Stillman B. Conserved initiator proteins in eukaryotes. Science. 1995;270:1667–1671. doi: 10.1126/science.270.5242.1667. [DOI] [PubMed] [Google Scholar]
- Gilbert DM. Replication origins in yeast versus metazoa: Separation of the haves and the have nots. Curr Opin Genet Dev. 1998;8:194–199. doi: 10.1016/s0959-437x(98)80141-x. [DOI] [PubMed] [Google Scholar]
- Gossen M, Pak DT, Hansen SK, Acharya JK, Botchan MR. A Drosophila homolog of the yeast origin recognition complex. Science. 1995;270:1674–1677. doi: 10.1126/science.270.5242.1674. [DOI] [PubMed] [Google Scholar]
- Grallert B, Nurse P. The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes & Dev. 1996;10:2644–2654. doi: 10.1101/gad.10.20.2644. [DOI] [PubMed] [Google Scholar]
- Heck MM, Spradling AC. Multiple replication origins are used during Drosophila chorion gene amplification. J Cell Biol. 1990;110:903–914. doi: 10.1083/jcb.110.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua XH, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J Cell Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiai M, Dean FB, Okumura K, Abe M, Moon KY, Amin AA, Kagotani K, Taguchi H, Murakami Y, Hanaoka F, O’Donnell M, Hurwitz J, Eki T. Isolation of human and fission yeast homologues of the budding yeast origin recognition complex subunit ORC5: Human homologue (ORC5L) maps to 7q22. Genomics. 1997;46:294–298. doi: 10.1006/geno.1997.5003. [DOI] [PubMed] [Google Scholar]
- Klemm RD, Austin RJ, Bell SP. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- Landis G, Kelley R, Spradling AC, Tower J. The k43 gene, required for chorion gene amplification and diploid cell chromosome replication, encodes the Drosophila homolog of yeast origin recognition complex subunit 2. Proc Natl Acad Sci. 1997;94:3888–3892. doi: 10.1073/pnas.94.8.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherwood J, Lopez-Girona A, Russell P. Interaction of Cdc2 and Cdc18 with a fission yeast ORC2-like protein. Nature. 1996;379:360–363. doi: 10.1038/379360a0. [DOI] [PubMed] [Google Scholar]
- Lee DG, Bell SP. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzi-Falconi M, Kelly TJ. Orp1, a member of the Cdc18/Cdc6 family of S-phase regulators, is homologous to a component of the origin recognition complex. Proc Natl Acad Sci. 1995;92:12475–12479. doi: 10.1073/pnas.92.26.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon CS. Putting it all together: Building a prereplicative complex. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- Ohtani K, DeGregori J, Leone G, Herendeen DR, Kelly TJ, Nevins JR. Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol Cell Biol. 1996;16:6977–6984. doi: 10.1128/mcb.16.12.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- Rowles A, Chong JP, Brown L, Howell M, Evan GI, Blow JJ. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- Sheehan MA, Mills AD, Sleeman AM, Laskey RA, Blow JJ. Steps in the assembly of replication-competent nuclei in a cell-free system from Xenopus eggs. J Cell Biol. 1988;106:1–12. doi: 10.1083/jcb.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugal T, Zou-Yang XH, Gavin K, Pappin D, Canas B, Kobayashi R, Hunt T, Stillman B. The orc4p and orc5p subunits of the xenopus and human origin recognition complex are related to orc1p and cdc6p. J Biol Chem. 1998;273:32421–32429. doi: 10.1074/jbc.273.49.32421. [DOI] [PubMed] [Google Scholar]
- Quintana DG, Hou Z-H, Thome KC, Hendricks M, Saha P, Dutta A. Identification of Hs ORC4, a member of the human origin of replication recognition complex. J Biol Chem. 1997;272:28247–28251. doi: 10.1074/jbc.272.45.28247. [DOI] [PubMed] [Google Scholar]
- Quintana DG, Thome KC, Hou Z-H, Ligon AH, Morton CC, Dutta A. ORC52, a new member of the human origin recognition complex, is deleted in uterine leiomyomas and malignant myeloid diseases. J Biol Chem. 1998;273:27137–27145. doi: 10.1074/jbc.273.42.27137. [DOI] [PubMed] [Google Scholar]
- Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]