Abstract
An interaction between the origin recognition complex (ORC) and Cdc6p is the first and a key step in the initiation of chromosomal DNA replication. We describe the assembly of an origin-dependent complex containing ORC and Cdc6p from Saccharomyces cerevisiae. Cdc6p increases the DNA binding specificity of ORC by inhibiting non-specific DNA binding of ORC. Cdc6p induces a concomitant change in the conformation of ORC and mutations in the Cdc6p Walker A and Walker B motifs, or ATP-γ-S inhibited these activities of Cdc6p. These data suggest that Cdc6p modifies ORC function at DNA replication origins. On the basis of these results in yeast, we propose that Cdc6p may be an essential determinant of origin specificity in metazoan species.
Keywords: ORC, Cdc6p, ATPase, sequence specificity, DNA replication, S. cerevisiae
Initiation of chromosomal DNA replication in both eukaryotic and prokaryotic cells is achieved by the stepwise assembly of cell-cycle-regulated protein complexes at origins of DNA replication (Kornberg and Baker 1992; Diffley 1996; Stillman 1996; Leatherwood 1998; Donaldson and Blow 1999). The mechanism and regulation of the assembly are well known for bacteria but not for eukaryotic cells. This situation is in part due to lack of an origin-dependent system for eukaryotic chromosomal DNA replication.
One of the central proteins involved in the process of initiation of DNA replication is the Origin Recognition Complex (ORC), originally identified as a six-protein complex that bound specifically to Saccharomyces cerevisiae origins of DNA replication (Bell and Stillman 1992). ORC is bound to chromatin at origins of chromosomal DNA replication throughout the cell cycle and is thought to function as a landing pad for the assembly of a series of cell-cycle-regulated protein complexes (Diffley et al. 1994; Cocker et al. 1996; Aparicio et al. 1997; Donovan et al. 1997; Liang and Stillman 1997; Tanaka et al. 1997; Wang et al. 1999). On the basis of studies using yeast and Xenopus cells, Cdc6p most likely binds to ORC and then the ORC–Cdc6p complex (and perhaps other proteins) recruits the six minichromosome maintenance (MCM) proteins to form a pre-replication complex (pre-RC) at each origin (Cocker et al. 1996; Santocanale and Diffley 1996; Aparicio et al. 1997; Donovan et al. 1997; Liang and Stillman 1997; Tanaka et al. 1997; Perkins and Diffley 1998; Weinreich et al. 1999).
MCM loading on to chromatin and pre-RC formation is dependent on Cdc6p in S. cerevisiae (Santocanale and Diffley 1996; Donovan et al. 1997; Tanaka et al. 1997) as well as in Xenopus and mammalian cells (Coleman et al. 1996; Williams et al. 1997; Stoeber et al. 1998; Yan et al. 1998). In the fission yeast Schizosaccharomyces pombe, the Cdc18 protein, the homolog of S. cerevisiae Cdc6p, also functions to control initiation of DNA replication (Kelly et al. 1993; Nishitani and Nurse 1995; Jallepalli and Kelly 1996; Muzi-Falconi et al. 1996; Leatherwood 1998; Lopez-Girona et al. 1998).
As part of the mechanism to ensure that initiation occurs once per cell cycle, Cdc6p (or Cdc18) functions in establishing the pre-RC only during a window that lasts from exit from mitosis to late G1, a period when cyclin-dependent protein kinases (CDKs) are inactive during the cell cycle (Kelly et al. 1993; Dahmann et al. 1995; Muzi-Falconi et al. 1996; Piatti et al. 1996; Santocanale and Diffley 1996; Brown et al. 1997; Detweiler and Li 1997, 1998; Liang and Stillman 1997). At other times during the cell cycle, formation of the pre-RC is negatively controlled by cyclin-dependent protein kinases (Dahmann et al. 1995; Piatti et al. 1996; Brown et al. 1997; Lopez-Girona et al. 1998). Cdc6p can also regulate the entry into mitosis (Kelly et al. 1993; Piatti et al. 1995; Weinreich et al. 1999). Therefore, Cdc6p is a key regulatory factor for pre-RC formation and thus, for initiation of DNA replication in yeast, Xenopus, and mammalian cells.
One of the first events, and certainly a key event, in pre-RC formation is believed to be Cdc6p binding to origin-bound ORC, but even for this first step of chromosomal DNA replication, an in vitro assay system has not been established. We suggest that such a biochemical system will be important, not only for understanding ORC–Cdc6p interaction, but also for reconstitution of the pre-RC and eventual reconstitution of eukaryotic chromosomal DNA replication in vitro, a precursor to fully appreciating the complexities of this process and how it is regulated. In this study, we report the development of an in vitro assay system for the origin-dependent interaction between ORC and Cdc6p. Cdc6p increased the sequence specificity of ORC by restricting ORC binding to functional origin sequences and induced a conformational change in ORC in vitro. Experiments with ATP-γ-S and altered Cdc6p proteins suggested that the ATPase activity of Cdc6p is involved in both of these activities.
Results
Development of an origin-dependent assay system for the interaction between ORC and Cdc6p
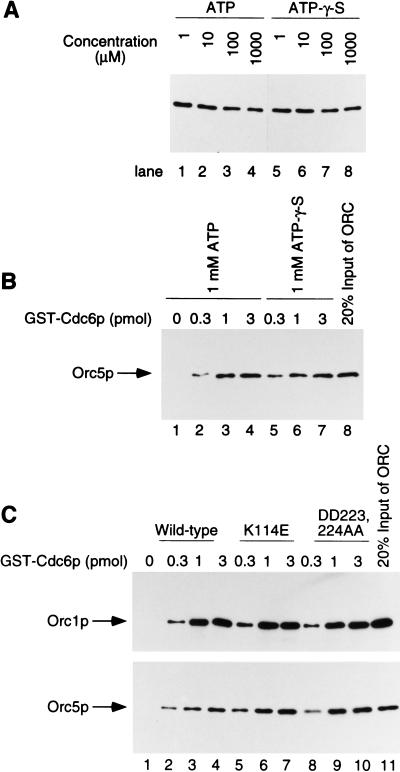
Cdc6p precipitation experiments were used to examine the binding of S. cerevisiae ORC to Cdc6p by use of ORC purified from recombinant baculovirus infected insect cells (Bell et al. 1995) and glutathione-S-transferase (GST)-fused Cdc6p purified from Escherichia coli cells. Near stoichiometric amounts of GST–Cdc6p, ORC, and origin DNA were incubated under various conditions, and the GST–Cdc6p was precipitated with glutathione–agarose beads (pulldown experiments). The efficiency of the interaction was determined by immunoblotting of one or more of the ORC subunits, (Orc5p in most figures, or Orc5p and Orc1p in Fig. 5), or by measurement of the amount of 32P-labeled origin DNA in the precipitated fraction.
Figure 5.
ATPase activity of Cdc6p is not required for its interaction with ORC. (A–C) Incubation was performed with 1 pmole of wild-type ARS1 DNA fragment, ORC (1 pmole), and GST–Cdc6p (3 pmole) at 30°C for 10 min unless noted. ORC pulldown with glutathione–agarose beads was visualized by immunoblotting of the Orc5p subunit or the Orc1p subunit. (A) ATP-γ-S did not inhibit the interaction between ORC and Cdc6p. ORC and GST–Cdc6p were incubated in the presence of the indicated concentrations of ATP or ATP-γ-S prior to the GST–Cdc6p precipitation. (B) ATP-γ-S did not effect the efficiency of the interaction between ORC and Cdc6p. Indicated amounts of GST–Cdc6p and 1 pmole of ORC were incubated in the presence of 1 mm ATP or ATP-γ-S prior to the GST–Cdc6p precipitation. (C) Effect of mutations in Cdc6p on the interaction between ORC and Cdc6p. ORC was incubated with the indicated concentrations of wild-type and altered GST–Cdc6p in the presence of 1 mm ATP.
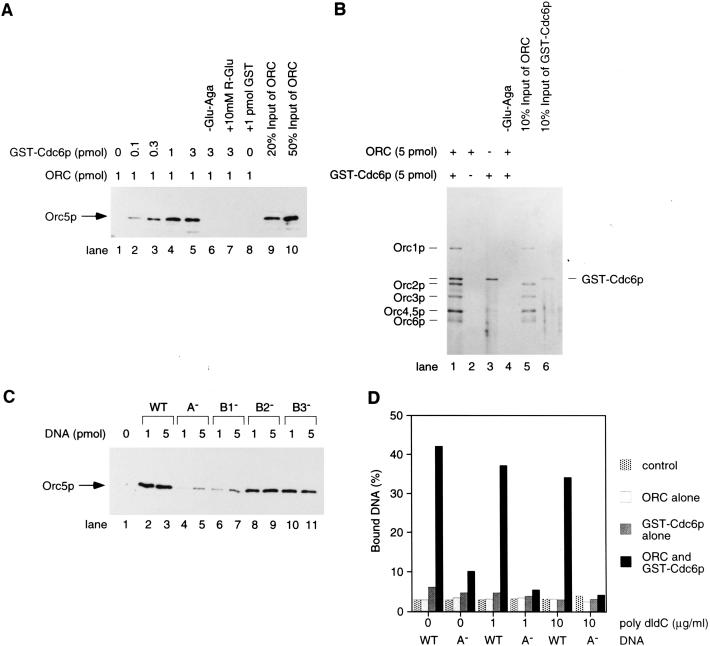
In the presence of ARS1 DNA (Marahrens and Stillman 1992), ∼20% of the added Orc5p was precipitated with glutathione–agarose in a manner dependent on GST–Cdc6p (Fig. 1A, lanes 1–5). Maximal binding occurred at equimolar concentrations of ORC, Cdc6p, and origin DNA. The Orc5p precipitation was dependent on glutathione–agarose beads (Fig. 1A, lane 6) and inhibited by the reduced form of glutathione (a competitor of GST binding to glutathione–agarose beads; Fig. 1A, lane 7). GST alone did not pull down ORC (Fig. 1A, lane 8). Using direct staining of the proteins, all the subunits of ORC were shown to be precipitated equally in the presence of GST–Cdc6p (Fig. 1B, lane 1) but not in its absence (Fig. 1B, lane 2), indicating that ORC could directly bind to GST–Cdc6p.
Figure 1.
ARS-dependent interaction between ORC and Cdc6p. (A–D) Glutathione bead precipitation (pulldown) experiments were done in the presence of 1 pmole of ORC, 3 pmole of GST–Cdc6p, 1 mm ATP, and ARS1 DNA at an equimolar concentration with ORC (unless otherwise noted). Wild-type and mutant ARS1 DNA fragments (290 bp) were prepared as described (Rao and Stillman 1995). Radiolabeled DNA fragments were prepared with radiolabeled primers as described (Rao and Stillman 1995). (A) GST–Cdc6p-dependent precipitation of Orc5p with glutathione–agarose beads. The GST–Cdc6p precipitated with glutathione–agarose beads was visualized by immunoblotting with anti-Orc5p antibodies. (B) All the subunits of ORC precipitate with glutathione–agarose beads in a manner dependent on GST–Cdc6p. ORC and GST–Cdc6p pulldown with glutathione–agarose beads (lanes 1–4) were visualized by silver stain. (Lanes 5,6) Input amounts of ORC and GST–Cdc6p, respectively. (C) Requirement for a functional ARS. Wild-type and mutant derivatives of ARS1 DNA were as described (Marahrens and Stillman 1992; Rao and Stillman 1995). GST–Cdc6p pulldown with glutathione–agarose beads was visualized by immunoblotting the Orc5p subunit. (D) GST–Cdc6p can interact with ARS DNA only though ORC. ORC and GST–Cdc6p were incubated with 1 pmole of either wild-type or A− ARS1 radiolabeled DNA fragments in the presence of the indicated concentrations of poly[d(I–C)]. The amount of DNA fragment pulled down with glutathione–agarose beads was determined by measurement of radioactivity in a liquid scintillation counter. DNA was extracted from glutathione–agarose precipitates with 2% SDS solution.
The interaction between ORC and GST–Cdc6p required DNA containing a functional ARS (Fig. 1C). Both the A and B1 regions of ARS1 were important to support an efficient interaction between ORC and GST–Cdc6p (Fig. 1C, lanes 4–7), as they are for initiation of DNA replication in vivo (Marahrens and Stillman 1992). Mutations in the B2 and B3 elements did not affect the interaction (Fig. 1C, lanes 8–11). Because the A and B1 elements were required for ORC to bind to origins in a sequence-specific manner (Bell and Stillman 1992; Rao and Stillman 1995; Rowley et al. 1995), the data suggest that the binding of GST–Cdc6p to ORC required prior ORC binding to the ARS. In the presence of ATP-γ-S, we also observed a DNA-dependent interaction (data not shown), suggesting that ATP hydrolysis was not required for the ORC–Cdc6p interaction on DNA. Vigorous washing of the glutathione bead precipitate was essential to observe the origin-dependent interaction between ORC and Cdc6p. Under conditions without the washing procedure, ORC precipitated with Cdc6p even in the absence of origin DNA (data not shown). The observation that a functional origin was required to maintain the ORC–Cdc6p interaction under vigorous washing conditions suggests that origin DNA may stabilize the ORC–Cdc6p complex. This stabilization might explain why the dependency was not observed in previous reports (Liang et al. 1995; Wang et al. 1999)
Next, we tested whether GST–Cdc6p could interact stably with DNA in the absence of ORC. Glutathione bead precipitation experiments using radiolabeled ARS1 DNA fragments showed not only that ORC, GST–Cdc6p, and ARS1 DNA formed a complex, but also that GST–Cdc6p could interact with ARS1 DNA only through binding to ORC (Fig. 1D). GST–Cdc6p alone did not interact with DNA, and the precipitation of DNA with the glutathione beads in the presence of ORC and GST–Cdc6p required functional origin sequences. It is possible, however, that Cdc6p may contact DNA directly either on its own or with ORC when present in the complex. Nevertheless, by use of these assay conditions, GST–Cdc6p appears not to bind to ARS DNA directly, a conclusion that is supported by a gel electrophoretic mobility shift assays (GEMSAs) and a DNase I protection assays (Fig. 2A and 7, below).
Figure 2.
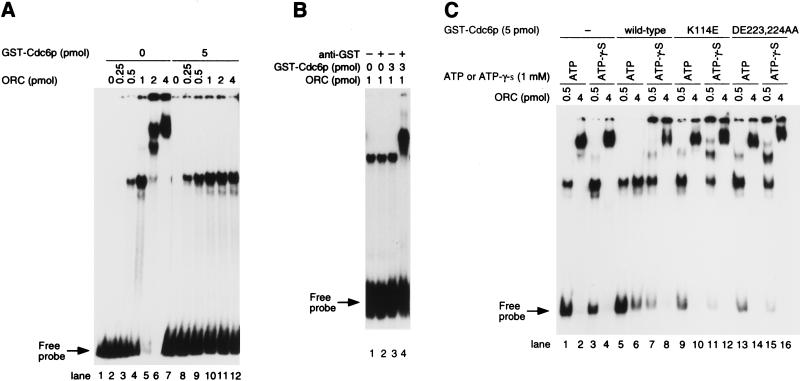
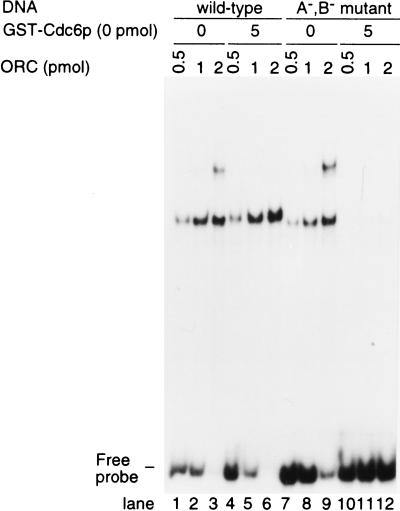
Effect of Cdc6p on the manner of ORC binding to ARS1 DNA. (A) Effect of Cdc6p on ORC–DNA multimerization. Increasing amounts of ORC were added to origin DNA (1 pmole) in the presence (lanes 1–6) or absence (lanes 7–12) of GST–Cdc6p. (B) ORC with or without GST–Cdc6p was incubated for 10-min with DNA probe and then antibody against GST was added as indicated for 30 min at 4°C. (C) Requirement for the Walker A and B motifs in Cdc6p for the inhibition of ORC–DNA multimerization by Cdc6p. Effect of wild-type GST–Cdc6p, GST–Cdc6pK114E, and GST–Cdc6pDE223, 224AA on the multimerization of the ORC–DNA complex were examined in the presence of 1 mm of ATP or ATP-γ-S. (A–C) GEMSA was done in the presence of 10 μg/ml poly[I–C].
Figure 7.
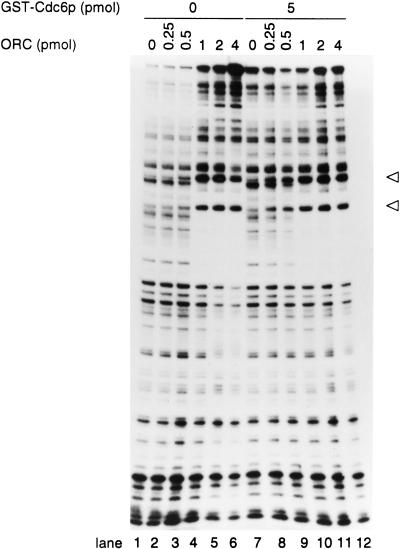
DNase I protection assay in the presence of increasing amounts of ORC, with and without GST–Cdc6p. (Open arrowheads) ORC-induced DNase I hypersensitive sites.
Cdc6p inhibits the multimerization of ORC on DNA by increasing the sequence-specific binding of ORC to origin DNA
To examine whether the binding of Cdc6p to ORC affects the way in which ORC is bound to ARS DNA, GEMSAs were employed. Increasing amounts of ORC were mixed with 1 pmole of labeled origin DNA, and when the amount of ORC exceeded the amount of DNA, slower migrating bands were visible (Fig. 2A, lanes 4–6). As shown below (Fig. 3), the additional bands seemed to be due to ORC interacting with non-origin sequences in addition to specific origin binding. In contrast, in the presence of GST–Cdc6p, the supershifted bands were not present (Fig. 2A) and only a single complex was observed, even at higher concentrations of ORC (Fig. 2A, lanes 10–12). Even though the migration of the complex with 1 pmole of ORC was indistinguishable in the presence or absence of GST–Cdc6p, GST–Cdc6p was present in the former complex because antibody against GST caused a supershift of the band only in the presence of GST–Cdc6p (Fig. 2B).
Figure 3.
Cdc6p increases the sequence-specific binding of ORC to origin DNA. Wild-type ARS1 DNA or ARS1 DNA lacking functional A and B1 elements were used to examine the effect of Cdc6p on the specificity of ORC–DNA binding. The wild-type DNA (290 bp) contained the ARS1 A, B1, B2, and B3 elements, whereas the A, B1 deleted mutant DNA was the same length, except that it contained the B2 and B3 elements plus flanking vector DNA. The GEMSA was done in the presence of 3 μg/ml poly[d(I–C)].
Next, we examined the effect of altered forms of Cdc6p on their ability to inhibit the multimerization of ORC on DNA. GST–Cdc6p with either a defective Walker A or a defective walker B motif (Cdc6p K114E and DE223, 224AA, respectively; see Weinreich et al. 1999) was purified and shown not to inhibit ORC–DNA multimerization (Fig. 2B, lanes 5-6 compared to lanes 9-10 and 13-14), demonstrating the importance of the putative nucleotide-binding motifs of Cdc6p for its inhibitory effect on the multimerization of ORC. Because alteration of the Walker A and B motifs should block the ATPase activity of GST–Cdc6p, we tested whether GST–Cdc6p modulation of ORC DNA binding could occur in the presence of ATP-γ-S, an analog of ATP that is difficult to hydrolyze. In the presence of ATP-γ-S, the wild-type Cdc6p could not inhibit the ORC multimerization on DNA (Fig. 2B, lanes 7,8). Altered GST–Cdc6p did not inhibit the ORC multimerization on DNA in the presence of ATP-γ-S (Fig. 2B, lanes 11,12, 15, 16). Because ATP-γ-S did not affect the ORC multimerization on DNA without Cdc6p (Fig. 2B, lanes 3,4), the inhibition of the ORC multimerization by ATP-γ-S may be due to its effect on Cdc6p, but not on ORC. It is still possible that the ATPase activity of ORC is activated by Cdc6p and this activation is involved in the inhibition of ORC multimerization. However, combined with data using altered GST–Cdc6p, these data suggest that the ATPase activity of Cdc6p is involved in a function of Cdc6p that inhibits or reverses the ORC multimerization on DNA.
Under the conditions employed in these experiments, supershifted bands with near equimolar amounts of ORC and DNA were also observed with a mutant DNA probe that lacked the A and B1 elements of ARS1 (Fig. 3, lanes 7–9). In the absence of Cdc6p, all bands with mutant ARS1 DNA, but not the wild-type DNA, disappeared by increases in the poly[d(I–C)] competitor (data not shown, also see Rao and Stillman 1995). The addition of GST–Cdc6p increased the specific binding of ORC to DNA because ORC still bound to the wild-type ARS1 DNA, but it no longer bound to the mutant DNA probe (Fig. 3, cf. lanes 4–6 and 10–12). Similar results were obtained with ARS1 DNA containing only an A− mutation and with completely nonspecific DNA (a DNA fragment from pUC19; data not shown). Thus, under these conditions with stoichiometric ORC, Cdc6p and DNA, ORC bound only to origin DNA with high affinity.
Because GST–Cdc6p blocked binding of ORC to nonorigin sequences, and it also blocked the multimeric forms of ORC–DNA complexes, we suggest that the supershifted bands observed in Figures 2 and 3 were due to ORC binding not only ARS1 sequences, but also to adjacent, nonspecific sites on the DNA. Thus, Cdc6p appears to block multimerization by restricting ORC binding only to the functional origin by inhibiting binding to nonspecific DNA sequences. Furthermore, altered forms of Cdc6p with defective nucleotide binding motifs were unable to prevent nonspecific binding of ORC to DNA and wild-type GST–Cdc6p could not block ORC binding to nonspecific DNA in the presence of ATP-γ-S (data not shown). Therefore, we suggest that Cdc6p increased the DNA sequence specificity of ORC binding to ARS1 DNA by binding ATP and possibily hydrolyzing ATP.
Mechanism of modulation of the DNA binding activity of ORC by Cdc6p
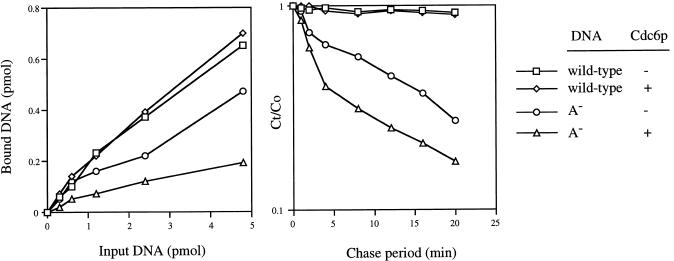
Next, we performed a series of experiments to explore how Cdc6p increased the sequence specificity of ORC DNA binding. First, a filter binding assay was used to determine the effect of Cdc6p on the association or dissociation of either specific wild-type ARS1 or nonspecific (A− mutant ARS1) DNAs to or from ORC, respectively. GST–Cdc6p inhibited the association of ORC to the nonspecific DNA, but did not affect binding to the specific DNA (Fig. 4, left), confirming the results obtained by use of GEMSA (Fig. 3). In a second approach, chase experiments with excess DNA added to the reaction after formation of the ORC–DNA complexes revealed that Cdc6p stimulated the rate of dissociation of ORC from the nonspecific DNA, but did not affect the rate of dissociation of ORC from the specific DNA (Fig. 4, right). These results suggested that Cdc6p increased the sequence specificity of ORC–DNA interaction by inhibiting stable binding of ORC to nonspecific DNAs rather than by stimulating an ORC–origin DNA interaction.
Figure 4.
Effect of Cdc6p on the association of ORC with DNA or on the rate of dissociation of DNA from ORC (filter binding assay). (Left) Indicated amounts of radiolabeled DNA were incubated for 10 min at 30°C with ORC (5 pmole) in the presence and absence of Cdc6p (15 pmole). (Right) Radiolabeled DNA (2.5 pmole) was incubated for 10 min at 30°C with ORC (2.5 pmole). Then, 50 pmole of each unlabeled DNA was added with and without Cdc6p (7.5 pmole) and the reactions incubated further at 30°C for indicated periods. (Ct and Co) Concentrations of DNA bound to ORC retained and the initial concentrations, respectively. (□) Wild-type ARS1, minus Cdc6p; (⋄) wild-type ARS1 plus Cdc6p; (○) A− ARS1 minus Cdc6p; (▵) A− ARS1 plus Cdc6p.
Glutathione-bead pulldown experiments in the presence of wild-type ARS1 origin DNA showed that ATP-γ-S did not inhibit the binding between ORC and GST–Cdc6p (Fig. 5A,B). Furthermore, GST–Cdc6p with defective Walker A and B motifs (Cdc6p K114E and DE223, 224AA, respectively) bound to ORC with an efficiency similar to that of the wild-type protein (Fig. 5C). These results are consistent with the observation that Cdc6pK114E and Cdc6pDE223, 224AA proteins could bind chromatin in vivo (Perkins and Diffley 1998; Weinreich et al. 1999). The results also imply that the inability of these altered Cdc6p proteins to increase the DNA sequence specificity of ORC, or the ability of ATP-γ-S to prevent wild-type Cdc6p from increasing the sequence specificity of ORC to origin DNA cannot be explained by lack of the binding between ORC and Cdc6p. After binding to ORC, GST–Cdc6p may affect the conformation of ORC and/or bind to some regions of the origin DNA to increase the sequence specificity of the ORC–DNA interaction.
Cdc6p alters the structure of ORC
The observation that Cdc6p and ORC interact with each other suggested that Cdc6p may alter the structure of ORC, which, in turn, causes a change in the manner in which ORC binds to DNA. To test for this possibility, a protease partial digestion assay was used to detect any conformational change in ORC upon its interaction with Cdc6p. A similar assay was employed to show that human Cdc6p becomes resistant to trypsin in the presence of ADP (Herbig et al. 1999). In our assay, it was necessary to titrate the amounts of the protease for each subunit of ORC that we tested as they had differential trypsin sensitivities.
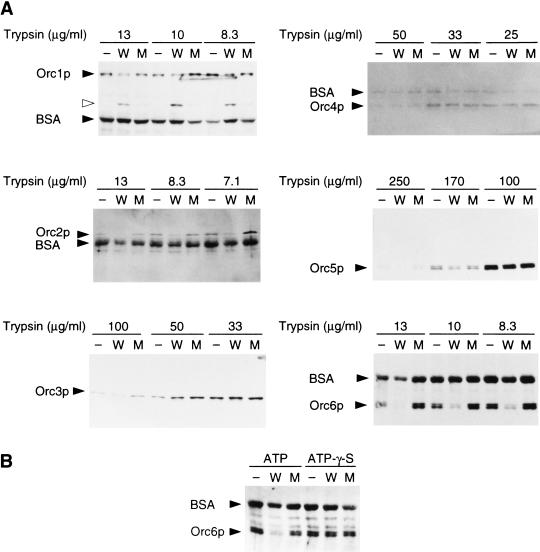
The wild-type GST–Cdc6p, but not GST–Cdc6pK114E stimulated the ability of trypsin to digest some subunits of ORC (Fig. 6A). Increasing amounts of trypsin were added in the absence of GST–Cdc6p, or in the presence of either wild-type or a mutant version of GST–Cdc6p. Following digestion, proteins were subjected to gel electrophoresis and blots were probed with antibodies directed against all six subunits of ORC. Because large amounts of BSA were included in these experiments to buffer the protease, it cross-reacted during immuno-staining.
Figure 6.
Trypsin digestion assay for ORC–Cdc6p interaction. (A–B) ORC (0.3 pmole) and 0.3 pmole of wild-type ARS1 DNA fragment were incubated in the absence (−) or presence of 1 pmole of wild-type (W) or K114E altered (M) GST–Cdc6p in 10 μl of the same reaction cocktail as used for the pulldown experiments for 10 min at 30°C. Then, the indicated final concentration of trypsin was added and the reactions incubated further for 10 min at 30°C. (A) Effect of Cdc6p on digestion of each subunit of ORC by trypsin. Incubation was done in the presence of 1 mm ATP. (Open arrowhead) Degraded product of Orc1p, which is increased by Cdc6p. (B) Effect of ATP-γ-S on the stimulation of trypsin digestion of Orc6p by Cdc6p. Incubation was performed with 1 mm ATP or ATP-γ-S. The trypsin concentration was 10 μg/ml.
The Orc6p subunit was rendered more protease sensitive in the presence of wild- type GST–Cdc6p compared with either no GST–Cdc6p or a mutant version of GST–Cdc6p (Fig. 6A, lower right panel). A reproducible effect was also observed with the Orc2p subunit, although, in this case, the antibody used to detect Orc2p in immunoblots was weaker than the anti-Orc6p antibody (Fig. 6A, middle left panel). The pattern of digestion of Orc1p by trypsin was also affected by the wild-type GST–Cdc6p, but not by Cdc6pK114E (Fig. 6A, top left panel). In this case, however, the effect was observed only by the appearance of a smaller Orc1p fragment (Fig. 6A, open arrowhead). In contrast, we did not see any enhanced protease sensitivity of the Orc3p, Orc4p, or Orc5p subunits in the presence of GST–Cdc6p (Fig. 6A).
Because the Walker A box mutant of Cdc6p was inactive in this assay, we tested if the increased protease sensitivity of Orc6p in the presence of wild type GST–Cdc6p was inhibited by ATP-γ-S. As shown in Figure 6B, the Orc6p subunit was only more sensitive to trypsin in the presence of ATP and wild-type Cdc6p, and ATP-γ-S blocked the effect of Cdc6p. On the basis of all of these observations, we consider that GST–Cdc6p changes the conformation of ORC by using its ATPase activity and as a result, increases the binding specificity of ORC binding to DNA. Alternatively, the Cdc6p–ATP complex might induce ATPase activity of ORC.
DNaseI footprinting
Next, we tested whether the addition of GST–Cdc6p to ORC altered the pattern of the DNase I footprint of ORC on DNA. However, GST–Cdc6p did not significantly affect the pattern of DNase I protection at the ARS1 origin (Fig. 7), suggesting that Cdc6p did not increase the sequence-specific binding of ORC to origin DNA by direct binding to some region of ARS1 DNA. The results from the DNase I protection assay also suggest that the interaction between ORC and Cdc6p at an origin does not mimic formation of the a pre-RC, which was defined as a change in the pattern of DNase I protection in vivo (Diffley et al. 1994). This result was expected as it is known that the MCM proteins are required for this effect. Because little change in the DNase I footprint was observed (Fig. 7) under the conditions where we see ORC multimerization (i.e., without GST–Cdc6p; Fig. 2A), ORC binding to nonspecific DNA must be randomly distributed along the probe, except at the functional origin where a clear DNase I footprint was observed.
Discussion
In this study, we have reconstituted with purified proteins the formation of a complex between Cdc6p, ORC, and origin DNA, a step that is essential for establishing a competent state of chromosomes prior to initiation of DNA replication in eukaryotic cells. Because near stoichiometric amounts of ORC, Cdc6p, and DNA were added in these assays, the formation of this complex is very efficient. In addition to assembly of this purified complex, we have preliminary results showing that the MCM proteins present in an extract from G1 phase cells, but not from G2 phase cells, can bind to the complex containing Cdc6p, ORC, and origin DNA. The establishment of the in vitro system described herein will allow us to build on this complex with other purified replication proteins, leading to further understanding of the mechanism and regulation of initiation of DNA replication in eukaryotes.
Cdc6p is a key factor for the regulation of initiation of DNA replication, but the biochemical function of this protein has been unclear. Studies in vivo suggest that Cdc6p participates in the recruitment of the MCM protein to the pre-RC (Cocker et al. 1996; Coleman et al. 1996; Santocanale and Diffley 1996; Aparicio et al. 1997; Detweiler and Li 1997; Donovan et al. 1997; Liang and Stillman 1997; Tanaka et al. 1997; Perkins and Diffley 1998; Stoeber et al. 1998; Weinreich et al. 1999). On the basis of these observations and our demonstration of direct binding of Cdc6p to ORC on origin DNA, we suggest that it is in this complex that Cdc6p recruits the MCM proteins. Human Cdc6p was shown to contain an intrinsic ATPase activity (Herbig et al. 1999). In this report, we uncovered three biochemical activities of Cdc6p that may contribute to its role in initiation of DNA replication. The first of these demonstrates that ORC and Cdc6p interact directly with each other.
Second, Cdc6p modulates the DNA binding activity of ORC by restricting DNA binding to functional origin sequences. The increased DNA binding specificity of ORC appears to be accomplished by Cdc6p increasing the rate of dissociation of ORC from nonorigin DNA sequences and by decreasing the rate of association of ORC to nonorigin DNA. Both ATP and a functional nucleotide-binding motif within Cdc6p are required for Cdc6p blocking nonspecific DNA binding of ORC to DNA, suggesting that Cdc6p ATPase activity may be required for this activity. At present we do not know whether Cdc6p is involved in ORC loading on origin DNA in vivo. In vivo footprinting and CHIP assays suggest that ORC is localized to origins throughout the cell division cycle. But it is not known whether both sister chromatids bind ORC at origins immediately after DNA replication, when Cdc6p is not present.
It is not known when newly synthesized ORC complexes bind to DNA during the cell division cycle in vivo and, therefore, it is not clear when the cooperativity between ORC and Cdc6p results in increased DNA binding specificity. In S. cerevisiae, all six ORC subunits are present at constant levels throughout the entire cell cycle (Liang and Stillman 1997; Weinreich et al. 1999). On the other hand, Cdc6p is an unstable protein that varies in amount during the cell cycle, with the protein made just prior to entry into mitosis or in late G1 phase of the cell cycle just before S phase (Zhou and Jong 1990; Zwerschke et al. 1994; Piatti et al. 1995; Elsasser et al. 1999; Drury et al. 2000). In rapidly proliferating cells, Cdc6p first binds to chromatin as cells exit mitosis, but in cells that come out of stationary phase Cdc6p is loaded in late G1, prior to activation of the cyclin-dependent protein kinases (Piatti et al. 1995; Detweiler and Li 1997; Donovan et al. 1997; Liang and Stillman 1997; Weinreich et al. 1999). Thus it is possible that ORC and Cdc6p interact and bind to vacant origins late in the cell division cycle. The fact that Cdc6p hinders ORC binding to nonorigin sequences in vitro suggests that one function of Cdc6p is to restrict replication only to functional origins. This activity of Cdc6p would increase the probability that all origins have the potential for forming pre-RCs after anaphase of mitosis.
In Xenopus egg extracts, ORC can bind to DNA in the absence of Cdc6p (Coleman et al. 1996), but it is well known that under these conditions initiation of DNA replication occurs in a sequence independent manner (Mahbubani et al. 1992).
It has been reported that the Cdc6p nucleotide-binding motif is essential for interaction with the Orc1p subunit of ORC (Wang et al. 1999), but it is not clear how this interaction relates to the observations described herein. Both Orc5p and Orc1p were precipitated with GST–Cdc6p that contained altered Walker A or Walker B motifs (Fig. 5C). We do not observe an effect of the Cdc6p Walker A and B motifs or ATP-γ-S on the interaction between Cdc6p and ORC in the absence of origin DNA, and it is known that the altered Cdc6p proteins can bind to chromatin in vivo (Perkins and Diffley 1998; Weinreich et al. 1999). It is possible that the Orc1p–Cdc6p interaction controls a later stage of DNA replication, such as MCM protein loading (see below).
Cdc6p remains bound to chromatin, most probably via its interaction with ORC, throughout the G1 phase of the cell cycle (Weinreich et al. 1999). During this time, a window exists that allows the MCM complex to bind to origins and form the pre-RC (Piatti et al. 1996). We have shown that a third biochemical function intrinsic to Cdc6p is its ability to remodel the ORC in vitro so that certain ORC subunits become hypersensitive to protease digestion. The ORC remodeling activity of Cdc6p also seems to require ATP and a functional nucleotide-binding motif in Cdc6p, suggesting that an ATPase of Cdc6p mediates the ORC remodeling.
It is possible that the conformational change in ORC is involved in recruitment of the MCM proteins and thus, formation of the pre-RC. In support of this suggestion, genetic analyses suggest that ATP binding and possibly ATP hydrolysis activities of Cdc6p are required for the MCM loading on chromatin (Perkins and Diffley 1998; Weinreich et al. 1999). Thus it is possible that the remodeled form of ORC is essential for MCM loading, and the role of Cdc6p in MCM loading is indirect via its interaction with ORC. Alternatively, both proteins might cooperate to recruit the MCM complex.
The identity of the ORC subunits that are remodeled is of considerable interest. On the basis of DNA cross-linking measurements, Orc2p and Orc6p appear to be located on the B2 element proximal side (relative to the A domain) of ARS1 (Lee and Bell 1997), adjacent to the site of initiation of DNA replication (Bielinsky and Gerbi 1998). This region of the origin is most likely the site of initial unwinding of the double helix. Although it is not known where the MCM proteins interact with the origin DNA, it is likely that it is near the B2 element for a number of reasons. First, the MCM proteins have intrinsic DNA helicase activity (Ishimi 1997; Kelman et al. 1999; Chong et al. 2000). Second, the B2 element region of ARS1 has an intrinsic ability to unwind in supercoiled plasmid DNA (Lin and Kowalski 1997). Thus it is possible that the MCM protein complex is loaded on to the origin DNA near the B2 element by interacting with ORC that has been remodeled by the ATPase activity of Cdc6p. Furthermore, because Orc1p, Orc2p, and Orc6p, but not other subunits of ORC are phosphorylated in vitro by the S phase cyclin–CDK Cdc28/Clb5 (M. Weinreich and B. Stillman, unpubl.), phosphorylation of these subunits after START may affect the ORC–Cdc6p–ARS DNA interaction and/or the pre-RC.
Cdc6p, also a target of the cyclin–CDKs, is released from the ORC–origin complex and degraded after cells commit to a new round to DNA replication after passage through START (Piatti et al. 1996; Detweiler and Li 1997; Liang and Stillman 1997; Elsasser et al. 1999; Drury et al. 2000). Therefore, another possibility is that release of Cdc6p from the pre-RC triggers a conformational change in ORC that allows subsequent events during initiation.
These in vitro activities of Cdc6p might explain previous genetic results in which overproduction of Cdc6p suppresses the temperature sensitivity of an orc5-1 mutant (Liang et al. 1995). In the orc5-1 mutant, the frequency of initiation of DNA replication at each origin was sufficiently low that S phase was compromised, but the frequency increased when higher levels of Cdc6p were present. We suggest that Cdc6p overexpression helped the altered ORC to bind better to origin DNA or attract other replication proteins, increasing the probability of origin firing in this mutant. Thus, we conclude that Cdc6p and ORC cooperatively recognize DNA replication origins and thereby determine the frequency of initiation of DNA replication in cells (Liang et al. 1995).
The biochemical activities of Cdc6p in modulating ORC function suggest a way to alter the frequency of origins along chromosomes that has been observed during development in Drosophila and Xenopus (Spradling and Orr-Weaver 1987). At early stages of development, where origins are close together and the ORC to DNA ratio is relatively high, Cdc6p may not be as important for determining origin specificity. At later stages of development when origins are less frequent along chromosomes, Cdc6p could be an essential component in origin determination. Initial studies with purified ORC from both Drosophila and mammalian ORC suggests that they have relatively weak DNA intrinsic binding specificity (Chesnokov et al. 1999; X.H. Yang and B. Stillman, unpubl.). On the basis of our results with yeast ORC, we suggest that in other eukaryotic species Cdc6p functions with ORC to determine the location of DNA replication origins in chromosomes.
Materials and methods
Protein preparation
Wild-type ORC was expressed in Sf9 cells infected with recombinant baculovirus expressing these proteins as described (Bell et al. 1995). The method of ORC purification was as described previously (Klemm et al. 1997). Wild-type and altered GST–Cdc6p were purified from E. coli cells (DH5α) transformed with their overproducing plasmids (M. Weinreich and B. Stillman, unpubl.) by use of glutathione–agarose column chromatography. About 10% of total protein in the crude extract was GST–Cdc6p. The yield was about 30%. Briefly, cells were lysed by digestion with lysozyme and centrifugation, and the supernatant was subjected to glutathione–agarose chromatography as specified by the Sigma Corporation. The GST–Cdc6p was eluted with 10 mm reduced form of glutathione and the protein was dialyzed to remove glutathione against buffer A (20 mm HEPES-KOH at pH 7.6, 0.1 m KCl, 1 mm EDTA, 1 mm DTT, and 20% glycerol].
Glutathione bead precipitation assay (pulldown assay)
ORC, GST–Cdc6p, and DNA were incubated at 30°C for 10 min in 25 μl of reaction cocktail containing 50 mm HEPES-KOH (pH 7.5), 0.2 m KCl, 1 mm EDTA, 1 mm EGTA, 5 mm MgOAc, 0.02% NP-40, 10% glycerol, and 2 mg/ml BSA. Adenine nucleotides and ARS1 DNA were added as indicated. The ARS1 DNA was a 290-bp fragment generated by PCR from either a plasmid pARS1WT, or in Figure 1, its mutant derivatives containing A− (858–865), B1− (835–842), B2− (802–808), and B3− (756 plus 758) mutations (Marahrens and Stillman 1992; Rao and Stillman 1995). The mixture was diluted with reaction buffer without BSA and then, 40 μl of glutathione–agarose beads (50% slurry; Sigma) was added. After a 2-hr rotation at 4°C and three vigorous washes (e.g. 3 × 30 sec on a vortex at maximum strength) with the same buffer as used for the dilution (see above; 1.5 ml), the precipitates were suspended with SDS sample buffer and run on a 7.5% SDS–polyacrylamide gel followed by immunoblotting with anti-Orc5p monoclonal antibody SB5 (S.P. Bell and B. Stillman, unpubl.). The buffers used for dilution and washing contained the same concentrations of adenine nucleotides as the reaction cocktail.
GEMSA and a DNase I footprint assay
GEMSA and DNase I footprinting were done as described with some modifications (Rao and Stillman 1995; Bell and Stillman 1992, respectively). ORC and GST–Cdc6p were incubated with radiolabeled DNA fragments (1 pmole) for 10 min at 30°C in 25 μl of the same reaction cocktail as used for the pulldown experiments, except for the presence of competitor. The reaction sample was loaded onto a 3.5% polyacrylamide gel containing 0.5× TBE (0.045 m Tris-borate at pH 8.3, 0.1 mm EDTA) and the gel was run for 12 hr at a constant 80 volts at 4°C and dried and autoradiographed (GEMSA). After DNase I digestion in the presence of 10 μg/ml poly[I–C] competitor, DNA fragments were precipitated with ethanol and run on a 6% polyacrylamide, 8M urea gel followed by autoradiography (a DNase I footprint assay).
Filter binding assay
Radiolabeled DNA (wild-type or A− mutant ARS1) fragments (290 bp) were incubated with ORC in 25 μl of the same reaction cocktail as used for the pulldown experiments, except for the presence of 3 μg/ml poly[d(I–C)] competitor. Samples were passed through nitrocellulose membranes (Millipore HA, 0.45 μm) and washed in the same buffer as reaction cocktails (ice-cold). The radioactivity remaining on the filters was counted in a liquid scintillation counter.
Trypsin digestion assay
After incubation of ORC, Cdc6p, and DNA for 10 min at 30°C, various concentrations of trypsin were added and the reactions incubated further for 10 min at 30°C. Samples were suspended with SDS sample buffer and run on a 7.5% SDS–polyacrylamide gel followed by immunoblotting with anti-Orc1p, Orc2p, Orc3p, Orc4p, Orc5p, or Orc6p monoclonal antibodies SB 16 and 35, 46, 3, 12, 5, or 49, respectively (S.P. Bell and B. Stillman, unpubl.).
Acknowledgments
We thank Michael Weinreich for providing unpublished plasmids, Lee Zou for helpful suggestions and Shinji Tsutsumi for technical advice. This research was supported by a grant from the National Institute of General Medical Sciences (GM45436). T.M. is a research Fellow of the Japan Society for the Promotion of Science (JSPS).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL stillman@cshl.org; FAX (516) 367-8879.
References
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Bell SP, Mitchell J, Leber J, Kobayashi R, Stillman B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP dependent recognition of eukaryotic origins of DNA replication by a multi-protein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Bielinsky A-K, Gerbi SA. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science. 1998;279:95–98. doi: 10.1126/science.279.5347.95. [DOI] [PubMed] [Google Scholar]
- Brown GW, Jallepalli PV, Huneycutt BJ, Kelly TJ. Interactions of the S phase regulator Cdc18 with cyclin-dependent kinase in fission yeast. Proc Natl Acad Sci. 1997;94:6142–6147. doi: 10.1073/pnas.94.12.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov I, Gossen M, Remus D, Botchan M. Assembly of functionally active Drosophila origin recognition complex from recombinant proteins. Genes & Dev. 1999;13:1289–1296. doi: 10.1101/gad.13.10.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JP, Hayashi MK, Simon MN, Xu RM, Stillman B. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc Natl Acad Sci. 2000;97:1530–1535. doi: 10.1073/pnas.030539597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JF. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Diffley JFX, Nasmyth KA. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- Detweiler CS, Li JJ. Cdc6p establishes and maintains a state of replication competence during G1 phase. J Cell Sci. 1997;110:753–763. doi: 10.1242/jcs.110.6.753. [DOI] [PubMed] [Google Scholar]
- ————— Ectopic induction of Clb2 in early G1 phase is sufficient to block prereplicative complex formation in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1998;95:2384–2389. doi: 10.1073/pnas.95.5.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF. Once and only once upon a time: Specifying and regulating origins of DNA replication in eukaryotic cells. Genes & Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Donaldson AD, Blow JJ. The regulation of replication origin activation. Curr Opin Genet Dev. 1999;9:62–68. doi: 10.1016/s0959-437x(99)80009-4. [DOI] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JFX. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr Biol. 2000;10:231–240. doi: 10.1016/s0960-9822(00)00355-9. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Chi Y, Yang P, Campbell JL. Phosphorylation controls timing of Cdc6p destruction: A biochemical analysis. Mol Biol Cell. 1999;10:3263–3277. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig U, Marlar CA, Fanning E. The Cdc6 nucleotide-binding site regulates its activity in DNA replication in human cells. Mol Biol Cell. 1999;10:2631–2645. doi: 10.1091/mbc.10.8.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- Jallepalli P, Kelly T. RumI and Cdc18 link inhibition of cyclin-dependent kinases to the initiation of DNA replication in S. pombe. Genes & Dev. 1996;10:541–552. doi: 10.1101/gad.10.5.541. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Kelman Z, Lee JK, Hurwitz J. The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum DeltaH contains DNA helicase activity. Proc Natl Acad Sci. 1999;96:14783–14788. doi: 10.1073/pnas.96.26.14783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm RD, Austin RJ, Bell SP. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- Kornberg A, Baker TA. DNA replication. New York, NY: W. H. Freeman and Company; 1992. [Google Scholar]
- Leatherwood J. Emerging mechanisms of eukaryotic DNA replication initiation. Curr Opin Cell Biol. 1998;10:742–748. doi: 10.1016/s0955-0674(98)80117-8. [DOI] [PubMed] [Google Scholar]
- Lee DG, Bell SP. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes & Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Weinreich M, Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–776. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- Lin S, Kowalski D. Functional equivalency and diversity of cis-acting elements among yeast replication origins. Mol Cell Biol. 1997;17:5473–5484. doi: 10.1128/mcb.17.9.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Girona A, Mondesert O, Leatherwood J, Russell P. Negative regulation of Cdc18 DNA replication protein by Cdc2. Mol Biol Cell. 1998;9:63–73. doi: 10.1091/mbc.9.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahbubani, H.M., Paull, T., Elder, J.K., and Blow, J.J. DNA replication initiates at multiple sites on plasmid DNA in Xenopus egg extracts. Nucleic Acids Res. 20: 1457–1462 [DOI] [PMC free article] [PubMed]
- Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- Muzi-Falconi M, Brown GW, Kelly TJ. cdc18+ regulates initiation of DNA replication in Schizosaccharomyces pombe. Proc Natl Acad Sci. 1996;93:1566–1570. doi: 10.1073/pnas.93.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Nurse P. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- Perkins G, Diffley JF. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a “reductional” anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Bohm T, Cocker JH, Diffley JFX, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes & Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- Rao H, Stillman B. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc Natl Acad Sci. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley A, Cocker JH, Harwood J, Diffley JF. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J. 1995;14:2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J. 1996;15:6671–6679. [PMC free article] [PubMed] [Google Scholar]
- Spradling A, Orr-Weaver T. Regulation of DNA replication during Drosophila development. Annu Rev Genet. 1987;21:373–403. doi: 10.1146/annurev.ge.21.120187.002105. [DOI] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Stoeber K, Mills AD, Kubota Y, Krude T, Romanowski P, Marheineke K, Laskey RA, Williams GH. Cdc6 protein causes premature entry into S phase in a mammalian cell- free system. EMBO J. 1998;17:7219–7229. doi: 10.1093/emboj/17.24.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Wang B, Feng L, Hu Y, Huang SH, Reynolds CP, Wu L, Jong AY. The essential role of Saccharomyces cerevisiae CDC6: Nucleotide-binding site in cell growth, DNA synthesis and Orc1 association. J Biol Chem. 1999;274:8291–8298. doi: 10.1074/jbc.274.12.8291. [DOI] [PubMed] [Google Scholar]
- Weinreich M, Liang C, Stillman B. The Cdc6p nucleotide-binding motif is required for loading mcm proteins onto chromatin. Proc Natl Acad Sci. 1999;96:441–446. doi: 10.1073/pnas.96.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Shohet RV, Stillman B. A human protein related to yeast Cdc6p. Proc Natl Acad Sci. 1997;94:142–147. doi: 10.1073/pnas.94.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, DeGregori J, Shohet R, Leone G, Stillman B, Nevins JR, Williams S. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc Natl Acad Sci. 1998;95:3603–3608. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Jong A. CDC6 mRNA fluctuates periodically during the yeast cell cycle. J Biol Chem. 1990;265:19904–19909. [PubMed] [Google Scholar]
- Zwerschke W, Rottjakob H-W, Kuntzel H. The Saccharomyces cerevisiae CDC6 gene is transcriped at late mitosis and encodes an ATP/GTPase controlling S phase initiation. J Biol Chem. 1994;269:23352–23356. [PubMed] [Google Scholar]