Abstract
Anticipatory ligand binding through massive protein sequence variation is rare in biological systems, having been observed only in the vertebrate adaptive immune response and in a phage diversity-generating retroelement (DGR). Earlier work has demonstrated that the prototypical DGR variable protein, major tropism determinant (Mtd), meets the demands of anticipatory ligand binding by novel means through the C-type lectin (CLec) fold. However, because of the low sequence identity among DGR variable proteins, it has remained unclear whether the CLec fold is a general solution for DGRs. We have addressed this problem by determining the structure of a second DGR variable protein, TvpA, from the pathogenic oral spirochete Treponema denticola. Despite its weak sequence identity to Mtd (∼16%), TvpA was found to also have a CLec fold, with predicted variable residues exposed in a ligand-binding site. However, this site in TvpA was markedly more variable than the one in Mtd, reflecting the unprecedented approximate 1020 potential variability of TvpA. In addition, similarity between TvpA and Mtd with formylglycine-generating enzymes was detected. These results provide strong evidence for the conservation of the formylglycine-generating enzyme-type CLec fold among DGRs as a means of accommodating massive sequence variation.
Keywords: x-ray crystallography, lipoprotein, periodontal disease
Massive protein sequence variation is rare in biological systems. It has been observed only in the adaptive immune system of vertebrates and in a diversity-generating retroelement (DGR) of Bordetella bacteriophage. In the case of the adaptive immune system, as many as approximately 1014–16 sequences are accommodated by variable proteins (e.g., antibodies and T-cell receptors) of the Ig and leucine-rich repeat families (1, 2). This massive scale of variation is required for anticipatory binding of novel ligands (3). The same holds true for the Bordetella bacteriophage DGR (4–6). Its variable protein, major tropism determinant (Mtd), accommodates approximately 1013 sequences by using a C-type lectin (CLec) fold (5). Mtd serves as the receptor-binding protein of the phage, and its variability enables the phage to keep pace with environmentally programmed changes in Bordetella (7).
Approximately 100 DGRs similar to the prototypical Bordetella bacteriophage DGR have been identified to date in bacterial and phage genomes (7, 8). These DGRs have in common a distinctive reverse transcriptase and two nearly identical repeat regions, the template region (TR) and variable region (VR) (Fig. S1). The Bordetella bacteriophage DGR reverse transcriptase has been shown to mediate the diversification of the protein-coding VR in Mtd through the transfer of sequence information from the invariant TR (4, 8, 9). Adenines are transmitted from the TR to the VR with particularly poor fidelity, resulting in random sequence variation of adenine-containing codons. This adenine-directed mechanism yields 12 variable amino acids in Mtd, which, despite being interspersed with invariant ones in the primary sequence, are organized by the CLec fold of Mtd into a continuous, solvent-exposed ligand-binding site (6).
The VRs and TRs of other DGRs also differ mainly at adenines, suggesting that the adenine-directed mechanism of variation is conserved (7, 8). Despite these similarities, it has remained uncertain whether the CLec fold is conserved among DGRs as a means to accommodate massive sequence variation. DGR variable proteins are surprisingly divergent (∼17% sequence identity) and have only two features in common. The first is a “GXXW” motif in the VR (which lacks a clear structural or functional role), and the second is a C-terminal location of the VR (5). These patterns are suggestive of, but not definitive evidence for, conservation of the CLec fold in DGR variable proteins.
We have addressed this issue and report here the structure of a DGR variable protein from Treponema denticola, TvpA (Treponema variable protein A). TvpA shares only approximately 16% sequence identity with Mtd, and its structure is only the second to be determined for a DGR variable protein. T. denticola is an anaerobic Gram-negative spirochete associated with periodontal disease (10), and the sequenced strain has a DGR that potentially diversifies seven variable proteins, including TvpA. These seven variable proteins are related in sequence (25–67% identity), and several, including TvpA, have predicted lipoprotein signal sequences that likely target these proteins to the outer surface of the spirochete (11). The function of TvpA and the other T. denticola DGR variable proteins is unknown, but their potential surface localization suggests a role in mediating interactions with other organisms.
Results
Overall Structure of TvpA.
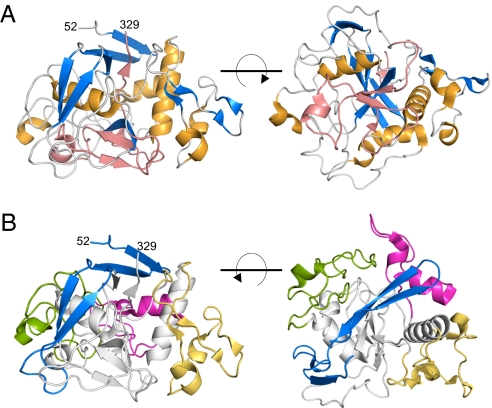
TvpA was expressed in Escherichia coli, purified to homogeneity, and crystallized. The structure of TvpA was determined by single-wavelength anomalous dispersion (SAD) from a mercury derivative (Table S1) and refined to 1.4 Å resolution limit. The electron density calculated from the SAD phases was unambiguous and permitted the chain to be traced from residue 52 to the C-terminal residue, 329. No electron density was evident for residues 39 to 51, presumably because of the inherent flexibility of this region, and residues 1 to 38 were excluded from the expression construct because they likely constitute the cleaved lipoprotein signal sequence.
TvpA, at first glance, is strikingly different from Mtd. TvpA consists of a single globular domain (Fig. 1), whereas Mtd, in contrast, consists of three domains (β-prism, β-sandwich, and CLec) (5). In addition, TvpA exists as a monomer in the crystal structure, whereas Mtd forms a highly intertwined, constitutive trimer (5). We note that a dimeric form of TvpA was evident along with the monomeric form during purification (Fig. S2), raising the possibility that TvpA exists as a dimer when attached to the T. denticola membrane. However, unlike Mtd, TvpA is not an obligatory oligomer and is able to exist stably as a monomer.
Fig. 1.
Structure of TvpA. (A) Two views of TvpA in ribbon representation (α-helices are gold, β-sheets blue, and loops are gray, except for the VR, which is entirely pink). (B) Two orientations of TvpA in ribbon representation with the β-flap (residues 52–94) in blue, insert 1 (residues 103–136) in magenta; insert 1′ (residues 153–195) in light orange; and insert 2 (residues 212–258) in green. Other portions of TvpA are in gray.
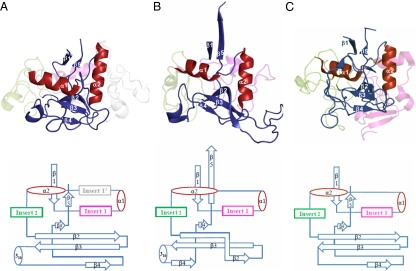
Despite these global differences, close inspection indicates a striking relationship between TvpA and Mtd. The single domain of TvpA has the same fold as the CLec domain of Mtd (rmsd, 2.6 Å; 145 Cα; Z = 11.8; Fig. 2) (12), indicating that TvpA accommodates massive sequence variation by using the CLec fold as well. This result provides strong evidence for the conservation of the CLec fold among DGR-variable proteins. TvpA, however, is most structurally similar to a family of formylglycine-generating enzymes (FGEs; rmsd, 1.9 Å with human FGE or hFGE; 212 Cα; Z = 26.2; 22% sequence identity; Fig. 2). FGEs are responsible for carrying out the conversion of a cysteine to a formylglycine in sulfatases (13, 14). We note that the FGE fold is a subtype of the CLec fold (15). Mtd also has a stronger structural resemblance to hFGE (rmsd, 2.5 Å; 148 Cα; Z = 13.1; 19% sequence identity) than to the archetypal CLec domain in macrophage mannose receptor (rmsd, 3.0 Å; 106 Cα; Z = 5.9; 8% sequence identity).
Fig. 2.
The FGE-type CLec fold of TvpA. Ribbon representation (Top; β-strands in blue and α-helices in red) and topology diagram (Bottom) of the CLec fold in (A) TvpA (2Y3C), (B) Mtd (1YU0), and (C) hFGE (2HI8). Disulfide bonds in hFGE are displayed as sticks. For clarity, inserts are ghosted, the β-flap of TvpA is not shown, and only the C-terminal domain of Mtd is displayed.
Protein Fold of TvpA.
The FGE-type CLec domain of TvpA begins at residue 95 and continues through to the C terminus (Fig. 2). As with all CLec domains, the N- and C-termini of the domain form β-strands (β1 and β5) that pair with one another. Preceding β1 in TvpA is a “β-flap” (residues 52–94), which is composed of short β-strands that wrap around the CLec domain (Fig. 1B). The β-flap serves to seal off exposed hydrophobic regions of the CLec domain, which in Mtd is accomplished by interdomain contacts (i.e., with the β-sandwich domain). As is characteristic of the CLec fold, between β1 and β5 are two α-helices, α1 and α2, which are roughly perpendicular to one another, followed by a three-stranded, antiparallel β-sheet (β2, β3, β4; Fig. 2). This β-sheet forms a ligand-binding site in Mtd and other CLec-fold proteins (5, 6). A short β4′-strand as well as a short 310-helix further extend the antiparallel β-sheet (Fig. 2); equivalents of these are found in Mtd. TvpA has three short regions inserted between secondary structure elements of the CLec fold, somewhat like Mtd (as detailed later).
VR.
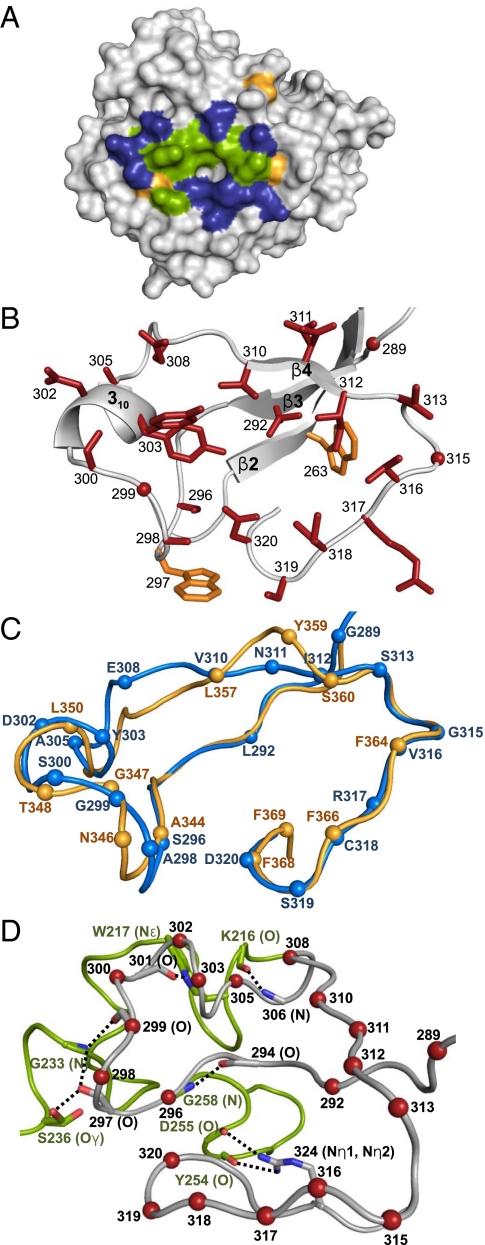
As with all DGR-variable proteins, the VR of TvpA is located at its C terminus (residues 284–329) (8). The TvpA VR is 46 residues long, which is comparable to the 45-residue VR of Mtd. The variable residues of both TvpA and Mtd are located within the CLec ligand-binding site, i.e., between β3 and β4′ on the external face of the β2β3β4β4′ sheet. As in Mtd, the variable residues in TvpA are solvent exposed and form a shallow binding site, with variable hydrophobic residues in the middle surrounded by variable hydrophilic residues (Fig. 3 A and B). Further like Mtd, TvpA has two nonvariable aromatic residues within the site that may contribute to binding (W263 and W297; Fig. 3B).
Fig. 3.
(A) Surface representation of TvpA, with the VR facing the viewer. Variable hydrophobic residues (A, V, L, I, Y) are green, variable hydrophilic residues (S, N, D, E, R, C) blue, and variable glycines pale orange. (B) VR of TvpA in ribbon representation. The main chain is in gray, side chains of variable residues are in red (red dots are variable glycines), and nonvariable aromatic residues are in orange. (C) Superposition of the VR of TvpA (blue) and Mtd-P1 (light orange) in Cα representation. The spheres represent the position of variable residues in each protein. (D) Stabilization of the VR (gray, variable residues indicated by red spheres) by insert 2 (green) in Cα representation. Dashed line indicates hydrogen bonds.
Despite the low sequence identity between the VRs of TvpA and Mtd (18%), the structures of these regions have remarkably similar conformations (rmsd, 1.2 Å; 45 Cα; Fig. 3C). Whereas Mtd has 12 potentially variable residues, TvpA has 20, which provides TvpA with a potential diversity of 6 × 1020, considerably much larger than that of Mtd (4) and indeed greater than that of antibodies, T-cell receptors, and variable lymphocyte receptors of agnathous fish. Eleven of the 20 variable residues of TvpA have structural equivalents in Mtd (Fig. 3C). One variable residue in Mtd (369 on β4′) has no structural counterpart in TvpA, increasing to at least 21 the number of potentially variable residues accommodated between β3 and β4′ in the FGE-type CLec-fold. Ten of the 11 residues in TvpA with equivalents in Mtd are encoded by AAC or AAT codons in the TR (A298, G299, S300, D302, V310, N311, I312, V316, C318, and D320). As previously noted, adenine-directed variation of such codons captures the gamut of chemical character and precludes nonsense codons (5). The 11th residue in common with Mtd, S296, is encoded by AGC, which permits substitution by three other amino acids.
The nine variable residues of TvpA without equivalents in Mtd are dispersed throughout the VR, highlighting the extraordinary accommodation of variation by this portion of the CLec fold. Only three of these nine variable residues are encoded by AAC in the TR (G289, Y303, S319), and two others are encoded by AAA (L292 and E308), which permits substitution by any of the other 19 residues but also permits nonsense codons. The remaining four are encoded by codons that permit substitution by three other amino acids (A305, S313, G315, and R317). Overall, a staggering 72% of the region between β3 and β4′ is variable in TvpA, compared with 46% in Mtd.
The last nine amino acids (321–329) of the TvpA VR are located along the buried β5-strand (Fig. 2), and do not contain any adenine-encoded residues and are thus invariant. The nucleic acid sequence corresponding to these residues likely functions as the initiation of mutagenic homing (IMH) element, which has been shown to set the directionality of information transfer in the Bordetella bacteriophage DGR (8).
Inserts.
The inserts of TvpA (Fig. 1B), as in Mtd, surround and brace the VR with hydrogen bonds. Whereas Mtd has two inserts, TvpA has three. Two of these three occur in the same topological location as in Mtd (Fig. 2) and are of comparable lengths: insert 1 (residues 103–136) is located between β1 and α1, and insert 2 (residues 212–258) is between α2 and β2. FGEs also have inserts at these same topological locations, although insert 1 is twice as long in hFGE as in TvpA and Mtd (Fig. 2). Although insert 1 has no statistically significant sequence or structural homology among these proteins, insert 2 of TvpA is quite similar to the second inserts of hFGE (rmsd, 1.71 Å; 46 Cα; 31% sequence identity) and Mtd (rmsd, 2.57 Å; 31 Cα; 16% sequence identity). Consistent with this conservation, insert 2 makes the most contacts to the VRs in TvpA (Fig. 3D) and Mtd (5).
A third insert, insert 1′ (residues 153–195), is unique to TvpA and is located between α1 and α2 (Fig. 2). Unlike inserts 1 and 2, whose sequences are identifiable in a number of CLec-fold proteins of the FGE-type in T. denticola and other organisms, insert 1′ is rare and identifiable in only one other protein, a T. denticola FGE-type CLec-fold protein (TDE0544). The role of this additional insert appears to be structural. Insert 1′ in TvpA wraps around α1 and appears, in conjunction with insert 1, to provide stabilization to α1. In Mtd, the role of insert 1′ is fulfilled by interprotomer contacts within the Mtd trimer, and in hFGE by a disulfide bond (Fig. S3).
Relationship to FGE.
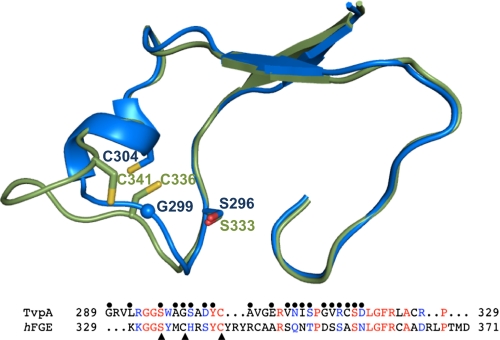
TvpA and hFGE have the greatest sequence and structural homology in the portion that is the VR of TvpA and the catalytic site of hFGE (rmsd, 0.98 Å; 46 Cα; 26% identity; Fig. 4). By comparison, the VR of Mtd is less structurally similar to the catalytic site of hFGE (rmsd, 1.61 Å; 41 Cα; 22% identity; Fig. S4). FGEs carry out a multistep redox reaction involving molecular oxygen and two catalytic cysteines and a serine, which, in hFGE, are C336, C341, and S333 (16). Two of these three catalytic residues are present in TvpA, and the third can be generated by DGR variation. The catalytic Cys336 of hFGE is equivalent to residue 299 of TvpA, which is a glycine but can be replaced by a cysteine through adenine-directed variation. The catalytic Cys341 of hFGE is equivalent to the invariant TvpA residue Cys304, although the position of the two residues is slightly different because, in TvpA, the Cys is on a 310-helix as opposed to a loop, as in hFGE (Fig. 4). The catalytic Ser333 of hFGE is equivalent to the TvpA variable residue Ser296; these two residues superpose precisely. It is unlikely, however, because of the anaerobic nature of T. denticola, that TvpA or any of its variants could carry out the oxygen-dependent formylglycyl reaction. Rather, the similarities between the VR of TvpA and the catalytic site of hFGE more likely reflect the common ancestry of a functionally important site.
Fig. 4.
Relationship to FGE. Superposition of the VR of TvpA (blue) and the catalytic site of hFGE (green) in Cα representation. Bottom: Sequence alignment of the regions shown above. TvpA variable residues denoted by spheres and hFGE catalytic residues by arrows. Identical residues are in red, and chemically similar ones in blue.
Discussion
The vertebrate adaptive immune response has made wide use of the Ig fold to accommodate massive sequence variation, and, to a more restricted extent, the leucine-rich repeat fold (3). The CLec fold of Mtd has added a third fold to this short list. Although DGRs are numerous and widespread in bacterial and phage genomes (7, 8), it has remained unclear whether the CLec fold is the general means by which DGRs accommodate massive sequence variation (5). This is because DGR-variable proteins are sequence-divergent and have limited similarities. The first of two similarities is the location of the VR, which includes the invariant IMH, to the approximately 45 C-terminal residues of variable proteins. In the CLec fold, this location enables surface presentation of variable residues for engagement with ligands, and the residues corresponding to the invariant IMH are buried on the β5-strand and serve a structural rather than functional role. The second similarity is a GXXW motif in the VR. The GXXW motif in Mtd is at a turn region at the edge of the ligand-binding site, but in the structure of Mtd-P1 bound to its receptor pertactin, this motif makes no contacts with pertactin (6). The reason for the conservation of the GXXW motif is not apparent, and continues to remain obscure with the structure of TvpA. These conserved patterns are consistent with the CLec fold but could conceivably be fulfilled by other folds. To address this issue, we pursued structure determination of a second DGR-variable protein.
We chose TvpA because of its low sequence identity with Mtd, which is typical of DGR variable proteins in general. Four major conclusions follow from the structure of TvpA. First, TvpA is a CLec fold protein, providing direct evidence for the conservation of the CLec-fold in T. denticola DGR-variable proteins and suggesting strongly that the CLec fold is conserved in DGRs as a general means for accommodating massive sequence variation. Second, the potential variability of TvpA (∼6 × 1020) is immense, much larger than that of variable proteins of the adaptive immune response. The structure of TvpA reveals that a remarkable three fourths of the residues between the β3- and β4′-strands, which form a ligand-binding site, are potentially variable. To our knowledge, TvpA is the most diverse naturally occurring protein described to date. Third, TvpA and Mtd belong to the FGE type of the CLec fold. Not only are the essential elements of the CLec fold conserved, but so too are inserts 1 and 2, suggesting a common ancestor for these proteins. Furthermore, the resemblance to FGEs raises the possibility that some DGR-variable proteins may confer enzymatic activity, just as antibodies have been shown by engineering to be capable of catalysis (17). Finally, TvpA exists stably in monomeric form, indicating that oligomerization in DGR-variable proteins is not obligatory.
The function of TvpA and the six other related T. denticola DGR-variable proteins is unknown, but the presence of lipopeptide signal sequences is suggestive. In spirochetes, the lipoprotein signal sequence targets proteins by default to the outer leaflet of the outer membrane (11), where such proteins are in position to mediate interactions with other bacteria or with host cells. T. denticola associates with and multiplies in the subgingival plaque, a biofilm composed of hundreds of bacterial species (18, 19). Bacteria add to the biofilm in sequential fashion through receptor–ligand interactions that promote interspecies coaggregation (19, 20). Primary (i.e., early) colonizers adhere to a pellicle formed on the tooth surface and to each other, followed by secondary (i.e., late) colonizers that attach to primary colonizers, extracellular polymeric substances within the biofilm produced by primary colonizers, or other bacteria that bridge the primary and secondary colonizers. T. denticola is a late colonizer, being found on the surface layers of the biofilm located in moderately deep subgingival pockets (18, 19). The incorporation of T. denticola into the subgingival plaque is likely dependent on various factors, most prominently the specific multispecies composition of the plaque, which notably varies from individual to individual (19). Massive sequence variability of surface-exposed TvpA and related T. denticola DGR-variable proteins would enable anticipatory binding to novel biofilm ligands or other bacteria to promote incorporation of T. denticola into the biofilm. As described for Mtd, tethering of variable proteins to the T. denticola membrane would also ensure that these interactions are driven by avidity, a conserved feature of anticipatory binding (6). As in the adaptive immune response and as shown for the Bordetella bacteriophage, massive sequence variability is an excellent strategy for surviving in a varying and unpredictable world.
Materials and Methods
Cloning, Expression, and Purification.
The coding sequence of TvpA (TDE2269, residues 39–329) was amplified from genomic DNA by standard PCR methods and inserted between the NdeI and XhoI restriction sites of pET-28b (Novagen), resulting in the addition of an N-terminal His-tag to TvpA. The plasmid was transformed into E. coli BL21(DE3), and TvpA expression was induced at 18 °C at an OD600 of 0.6 to 0.8 with 1 mM isopropyl β-d-1-thiogalactopyranoside. Induced bacteria were then grown overnight, after which time they were pelleted by centrifugation (20 min at 5,500 × g; 4 °C), resuspended in 10 mL per gram of cell pellet of 500 mM NaCl, 50 mM sodium phosphate buffer, pH 8, 20 mM imidazole (buffer A) with 1 mM PMSF, and lysed by sonication. Lysed bacteria were centrifuged (15 min at 10,000 × g; 4 °C), and the supernatant was applied to a Ni2+-charged POROS MC 20-μm column. The column was washed with buffer A, and TvpA was eluted from the column with a 20- to 500-mM imidazole gradient. The N-terminal His-tag was cleaved from TvpA by thrombin (in 50 mM Tris, pH 8, 150 mM NaCl, 1 mM β-mercaptoethanol at a 1:5 thrombin:TvpA mass ratio) overnight at room temperature. The digested sample was reapplied to the Ni2+-column, and the flow-through containing thrombin and digested TvpA was applied to a benzamidine column following the manufacturer's protocol (GE Healthcare) to capture thrombin. Finally, TvpA was purified by size-exclusion chromatography (Superdex 75 in 150 mM NaCl, 50 mM Tris, pH 8). Purified TvpA was concentrated to 165 mg/mL in 20 mM NaCl, 10 mM Tris, pH 8. A calculated extinction coefficient of 82,765 M−1cm−1 at 280 nm was used for the determination of TvpA concentration.
Crystallization and Data Collection.
Crystals of TvpA were grown at 20 °C by mixing 1.6 μL of TvpA (165 mg/mL) with 0.4 μL of 100 mM Bicine, pH 7, 2.7 to 3 M NaCH3COO, pH 7, by using the sitting drop, vapor diffusion method. Crystals were cryoprotected by soaking in 4 M NaCH3COO, pH 7, for 10 s. Crystals were derivatized by soaking in 25 mM HgCl2, 100 mM Bicine, pH 7, 3 M NaCH3COO, pH 7, for 2 h at 20 °C.
Diffraction data for native and Hg-derivatived TvpA crystals were collected at beamlines 23 ID-B and -D at the Advanced Photon Source (Argonne, IL), respectively, and processed with MOSFLM (21) and Scala (22).
Structure Determination, Refinement, and Analysis.
Phase information was obtained by SAD from the Hg derivative of TvpA (Table S1). Phases were calculated and refined by using Phenix (23), which was also used for automated model building. Residues 53 to 326 were built automatically, and residues 52 and 327 to 329 were manually added by using COOT (24). Several cycles of maximum likelihood restrained refinement against data from the Hg derivative were carried out with REFMAC5 (25), and the model was then refined against the high-resolution, native data set. The molecular replacement protocol in Phaser (26) was used to provide an initial set of phases for the native data set. This yielded a better Rfree than a simple transfer of the model to the native data set followed by rigid body and restrained refinement. Simulated annealing refinement (starting temperature, 5,000 K; final temperature, 100 K; cooling rate, 50 K per cycle of dynamics) was then carried out by using CNS (27, 28). After this point, 10 cycles of maximum likelihood restrained refinement by using REFMAC5, followed by manual rebuilding into σA-weighted 2mFo-DFc and mFo-DFc maps by using COOT, was carried out. Waters and an acetate were added in the later stages of refinement into at least 3σ mFo-DFc electron density. A molecule of β-mercaptoethanol was modeled as covalently bound to C318 by using the Dundee PRODRG2 Server (29). The presence of β-mercaptoethanol was confirmed by MS.
Structure validation was performed using Procheck and MolProbity. In the final TvpA model, which spans residues 52 to 329, 98.7% and 99.7% of residues were in allowed and generously allowed Ramachandran regions, respectively. The final map had correlation coefficients of 0.932 and 0.914 for the main chain and side chains, respectively, as calculated with OVERLAPMAP. The MolProbity clash score was 0.91 (99th percentile), and overall score was 1.10 (99th percentile).
Molecular graphics were made using PyMol (http://www.pymol.org/). The crystal structure and structure factors have been deposited to the Protein Data Bank (accession no. 2Y3C).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R01 AI072504 (to P.G.). Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under Contract DE-AC02-06CH11357. The National Institute of General Medical Sciences and National Cancer Institute Collaborative Access Team (GM/CA-CAT) has been funded in whole or in part with federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104).
Footnotes
The authors declare no conflict of interest.
Data deposition: The crystallographic data reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2Y3C).
This article is a PNAS Direct Submission. F.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105613108/-/DCSupplemental.
References
- 1.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 2.Alder MN, et al. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005;310:1970–1973. doi: 10.1126/science.1119420. [DOI] [PubMed] [Google Scholar]
- 3.Pancer Z, Cooper MD. The evolution of adaptive immunity. Annu Rev Immunol. 2006;24:497–518. doi: 10.1146/annurev.immunol.24.021605.090542. [DOI] [PubMed] [Google Scholar]
- 4.Liu M, et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science. 2002;295:2091–2094. doi: 10.1126/science.1067467. [DOI] [PubMed] [Google Scholar]
- 5.McMahon SA, et al. The C-type lectin fold as an evolutionary solution for massive sequence variation. Nat Struct Mol Biol. 2005;12:886–892. doi: 10.1038/nsmb992. [DOI] [PubMed] [Google Scholar]
- 6.Miller JL, et al. Selective ligand recognition by a diversity-generating retroelement variable protein. PLoS Biol. 2008;6:e131. doi: 10.1371/journal.pbio.0060131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medhekar B, Miller JF. Diversity-generating retroelements. Curr Opin Microbiol. 2007;10:388–395. doi: 10.1016/j.mib.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doulatov S, et al. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature. 2004;431:476–481. doi: 10.1038/nature02833. [DOI] [PubMed] [Google Scholar]
- 9.Guo H, et al. Diversity-generating retroelement homing regenerates target sequences for repeated rounds of codon rewriting and protein diversification. Mol Cell. 2008;31:813–823. doi: 10.1016/j.molcel.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loesche WJ, Grossman NS. Periodontal disease as a specific, albeit chronic, infection: Diagnosis and treatment. Clin Microbiol Rev. 2001;14:727–752. doi: 10.1128/CMR.14.4.727-752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schulze RJ, Zückert WR. Borrelia burgdorferi lipoproteins are secreted to the outer surface by default. Mol Microbiol. 2006;59:1473–1484. doi: 10.1111/j.1365-2958.2006.05039.x. [DOI] [PubMed] [Google Scholar]
- 12.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(web server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dierks T, et al. Molecular basis for multiple sulfatase deficiency and mechanism for formylglycine generation of the human formylglycine-generating enzyme. Cell. 2005;121:541–552. doi: 10.1016/j.cell.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Bojarová P, Williams SJ. Sulfotransferases, sulfatases and formylglycine-generating enzymes: A sulfation fascination. Curr Opin Chem Biol. 2008;12:573–581. doi: 10.1016/j.cbpa.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 15.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 16.Roeser D, et al. A general binding mechanism for all human sulfatases by the formylglycine-generating enzyme. Proc Natl Acad Sci USA. 2006;103:81–86. doi: 10.1073/pnas.0507592102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tramontano A, Janda KD, Lerner RA. Catalytic antibodies. Science. 1986;234:1566–1570. doi: 10.1126/science.3787261. [DOI] [PubMed] [Google Scholar]
- 18.Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev. 2007;71:653–670. doi: 10.1128/MMBR.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolenbrander PE, et al. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 20.Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. Bacterial coaggregation: An integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11:94–100. doi: 10.1016/s0966-842x(02)00034-3. [DOI] [PubMed] [Google Scholar]
- 21.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;Vol 26 [Google Scholar]
- 22.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 23.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 28.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Schüttelkopf AW, van Aalten DM. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.