Abstract
Saccharomyces telomeres consist of ∼350 bp of C1-3A/TG1-3 DNA. Most of this ∼350 bp is replicated by standard, semiconservative DNA replication. After conventional replication, the C1-3A strand is degraded to generate a long single strand TG1-3 tail that can serve as a substrate for telomerase. Cdc13p is a single strand TG1-3 DNA-binding protein that localizes to telomeres in vivo. Genetic data suggest that the Cdc13p has multiple roles in telomere replication. We used two hybrid analysis to demonstrate that Cdc13p interacted with both the catalytic subunit of DNA polymerase α, Pol1p, and the telomerase RNA-associated protein, Est1p. The association of these proteins was confirmed by biochemical analysis using full-length or nearly full-length proteins. Point mutations in either CDC13 or POL1 that reduced the Cdc13p–Pol1p interaction resulted in telomerase mediated telomere lengthening. Over–expression of the carboxyl terminus of Est1p partially suppressed the temperature sensitive lethality of a cdc13-1 strain. We propose that Cdc13p's interaction with Est1p promotes TG1-3 strand lengthening by telomerase and its interaction with Pol1p promotes C1-3A strand resynthesis by DNA polymerase α.
Keywords: Telomere replication, telomerase, DNA polymerase, CDC13, POL1, EST1
In most eukaryotes, telomeres, the physical ends of chromosomes, consist of simple repeated DNA sequences and their associated proteins. Chromosomes in Saccharomyces cerevisiae end with ∼350 bp of duplex C1-3A/TG1-3 DNA. In organisms where the structure at the very end of the chromosome has been examined, the G-rich strand is extended to form a single-strand tail. Telomere-associated proteins include proteins that directly bind to either duplex or single-stranded telomeric DNA as well as those that are brought to the telomere by protein–protein interactions. The major Saccharomyces telomere protein is the duplex C1-3A/TG1-3 binding protein Rap1p (Conrad et al. 1990; Wright et al. 1992; Wright and Zakian 1995). The Cdc13p protein, which binds ss TG1-3 DNA in vitro (Lin and Zakian 1996; Nugent et al. 1996), is also localized to telomeres in vivo (Bourns et al. 1998). Unlike Rap1p, Cdc13p does not bind to internal tracts of C1-3A/TG1-3 DNA (Bourns et al. 1998), suggesting that its association with the telomere is due to its ability to bind ss TG1-3 DNA.
Telomeres are required for the complete replication of linear chromosomes. After replication by a conventional DNA polymerase, removal of the terminal RNA primer leaves an 8–12 base gap at the 5′ end of newly replicated DNA molecules, a gap that can not be repaired by a conventional DNA polymerase. In most eukaryotes, including yeast, the end replication problem is solved by telomerase, a reverse transcriptase that is able to extend the short G-tail left after RNA primer removal, using an integral RNA as its template. There are at least five Saccharomyces genes that are required for the telomerase pathway in vivo, EST1, EST2, EST3, TLC1, and CDC13 (for review, see Nugent and Lundblad 1998). Mutation of any one of these genes causes a gradual loss of telomeric DNA or est (ever shorter telomere) phenotype, consistent with incomplete replication of chromosome ends. However, the products of only two of these genes, TLC1, which encodes the RNA component of telomerase, and EST2, the gene for its catalytic component, are essential for telomerase activity in vitro (for review, see Nugent and Lundblad 1998). Est1p is associated with TLC1 RNA (Lin and Zakian 1995; Steiner et al. 1996) and also binds ss TG1-3 DNA in vitro (Virta-Pearlman et al. 1996; Zhou et al. 2000). EST1 mutations that reduce Est1p binding to telomerase RNA have an est phenotype (Zhou et al. 2000). Given that Cdc13p, an in vivo telomere-binding protein (Bourns et al. 1998), is not required for telomerase activity in vitro (Lingner et al. 1997a), an appealing model is that Cdc13p recruits telomerase to the telomere. This model is supported by the demonstration that a Cdc13p–Est2p fusion protein bypasses the requirement for both Est1p and Est2p (Evans and Lundblad 1999).
Analysis of telomere replication intermediates revealed an unexpected step in telomere replication. Conventional semiconservative replication of the telomeric C1-3A/TG1-3 repeats occurs late in S phase (Wellinger et al. 1993a). After conventional replication is complete, both ends of individual chromosomes acquire 50–100 base ss TG1-3 tails (Wellinger et al. 1993a, b). Because these G-tails are detected in cells lacking telomerase (Dionne and Wellinger 1996; Wellinger et al. 1996), they must be generated by C-strand degradation. As Cdc13p binds efficiently to both 43 and 270 base TG1-3 tails in vitro (Lin and Zakian 1996), these long G-tails are suitable substrates for Cdc13p; and indeed, genetic data reveal that Cdc13p regulates C-strand degradation. When cdc13-1 cells are grown at restrictive temperatures, they accumulate ssDNA that extends from the telomere many kilobases towards the center of the chromosome with the C-strand being preferentially degraded (Garvik et al. 1995). C-strand degradation was unanticipated as it exacerbates the end problem of replication by creating a large gap at both ends of individual chromosomes.
It is unclear when or how often telomerase acts on a given chromosome end. As the rate of telomerase lengthening is inversely proportional to telomere length (Marcand et al. 1999), it is possible that only short telomeres are substrates for telomerase. Although telomerase activity can be detected in extracts prepared from cells throughout the cell cycle, formation of new telomeres is restricted to late G2/M phase (Diede and Gottschling 1999). Regardless of whether or not a given telomere is acted upon by telomerase, its C-strand must be resynthesized as long ss TG1-3 tails are not detected in post S/G2 phase cells (Wellinger et al. 1993a,b). Presumably, C-strand resynthesis is accomplished by a conventional DNA polymerase, although the polymerase that carries out this step has not been identified, nor is it known how DNA polymerase is recruited to the transient G-tail. After the 8–12 base RNA that is expected to prime C-strand resynthesis is removed, newly replicated telomeres should bear an 8–12 base ss TG1-3 tail. As duplex telomeric DNA bearing a five or 10 base TG1-3 tail is bound by Cdc13p in vitro (Lin and Zakian 1996), newly replicated telomeres are suitable substrates for Cdc13p.
Cdc13p appears to be a key regulatory protein in telomere replication as it functions at two critical steps in yeast telomere metabolism, C-strand degradation and G-strand elongation. To gain insight into how Cdc13p regulates these steps, we identified proteins that interact with Cdc13p in vivo. We found that Cdc13p interacts with the catalytic subunit of DNA polymerase α, Pol1p, and Est1p, by both two hybrid and biochemical criteria. Mutations in either CDC13 or POL1 that disrupt the Cdc13p–Pol1p interaction affect telomere length. Overexpression of the portion of Est1p that interacted with Cdc13p by two hybrid criteria partially suppressed the temperature sensitive lethality of a cdc13-1 strain. The interaction of Cdc13p with Est1p provides additional support for a model in which Cdc13p promotes the telomerase pathway by recruiting Est1p to the telomere. We propose that Cdc13p's interaction with Pol1p promotes C-strand resynthesis.
Results
Cdc13p interacts with Pol1p by two hybrid criteria
A two hybrid screen (Fields and Song 1989; Gyuris et al. 1993) was used to identify proteins that interact with Cdc13p in vivo. The first 600 amino acids of the 924 amino acid Cdc13p were expressed as a fusion to the LexA DNA binding domain and used as bait in the yeast strain YEM1α, which carries two reporter genes, LEU2 and LacZ (Fig. 1; the fusion polypeptide encoded by this plasmid is called Cdc13Np). The amino acid changes that confer the cdc13-1 and cdc13-2 phenotypes are both contained within this amino-terminal segment of Cdc13p (Lin and Zakian 1996; Nugent et al. 1996), and this region is sufficient to target Cdc13p to telomeres in vivo (Bourns et al. 1998). A library that produces galactose-inducible proteins as fusions of polypeptides encoded by segments of yeast chromosomal DNA and a transcriptional activation domain from Escherichia coli (Watt et al. 1995) was introduced into YEM1α cells carrying the bait plasmid. If a library plasmid produces a fusion protein that interacts with Cdc13Np, it will activate the LEU2 gene, allowing growth on media lacking leucine. A fusion protein that interacts strongly with Cdc13Np will also activate the lacZ gene, generating blue color when cells are assayed for β-galactosidase activity. Colonies expressing interacting fusion proteins were selected by their ability to grow on galactose media lacking leucine (Fig. 2A) and then screened for production of blue color by a filter LacZ assay (Breeden and Nasmyth 1985).
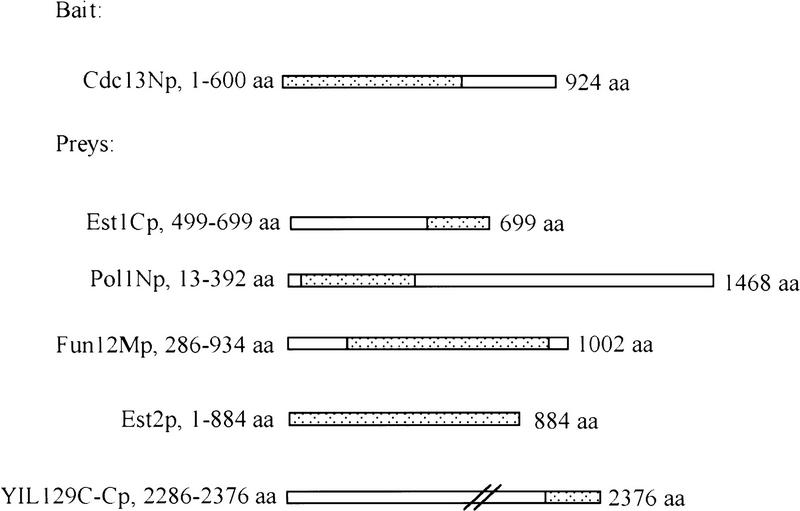
Figure 1.
Polypeptides used in the two-hybrid assay. Dotted regions indicated the portion of Cdc13p expressed from the bait plasmid pEG202 and the portions of proteins expressed from the prey vector pJG4-5 (not to scale). The fusion of the amino-terminal region of Cdc13p with the LexA DNA binding domain is referred to as Cdc13Np in the text. Prey proteins are similarly named; e.g., Pol1Np refers to the fusion of 379 amino acids from near the amino-terminus of Pol1p to the activation domain peptide. The amino acid numbers after the name of each protein indicates the specific amino acids expressed from the two-hybrid vector. The numbers to the right of the schematic for each protein indicate the number of amino acids in the full-length protein.
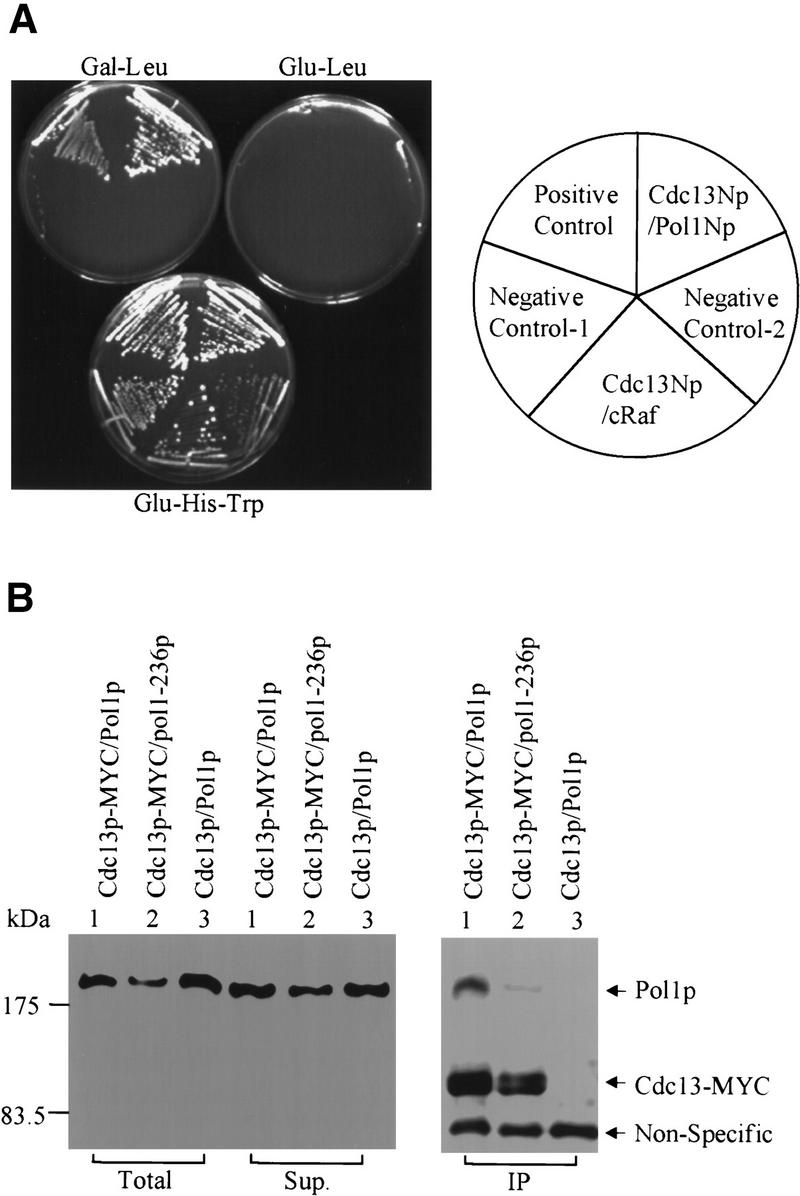
Figure 2.
Cdc13p interacted with Pol1p, the catalytic subunit of DNA polymerase α. (A) Cells expressing prey and/or bait proteins were streaked on galactose plates lacking leucine (Gal–Leu). Galactose induces expression of prey proteins and growth in the absence of leucine requires interaction of bait and prey polypeptides. Alternatively, cells were streaked on glucose plates lacking leucine (Glu-Leu) where prey proteins are not expressed or glucose plates lacking histidine and tryptophan (Glu-His-Trp), which selects for maintenance of both the prey and bait plasmids but not for interaction of their products. The positive control was cells expressing two subunits of RNA polymerase II, Rpb4p and Rpb7p. The negative controls were cells expressing Cdc13Np and carrying the empty prey vector pJG4-5 (Negative Control–1), cells expressing Pol1Np and carrying the empty bait vector pEG202 (Negative Control-2), and cells expressing Cdc13Np and a LexA::cRafp fusion protein (gift from E. Golemis) (Cdc13Np/cRaf). (B) Extracts from cells carrying an MYC3-tagged Cdc13p were immunoprecipitated by monoclonal anti-MYC antibody immobilized on protein-A and protein-G beads. The extract prior to immunoprecipitation (Total), the supernatant from the immunoprecipitate (Sup.), and the immunoprecipitate (IP) were analyzed by Western blotting using both anti-MYC and antiPol1p monoclonal antibodies. Although Cdc13p was not visible in the total cell extract in this gel, it was detectable when more protein was loaded. Extracts were prepared from cells containing 3XMYC (lanes 1,2) or untagged (lane 3) Cdc13p. Cells had either the wild-type POL1 (lanes 1,3) or the pol1-236 allele (lane 2). The nonspecific band was detected by the anti-MYC serum.
From a screening of more than 5 × 106 colonies, we obtained three positive clones. DNA sequence analysis revealed that the plasmids contained amino acids 13–392 from the 1468 amino acid Pol1p, amino acids 286–934 from the 1002 amino acid Fun12p, and amino acids 2286–2376 from the 2376 amino acid YIL129C (Fig. 1; the fusion polypeptides encoded by these prey plasmids were named Pol1Np, Fun12Mp, and YIL129C-Cp). The essential gene FUN12 encodes a protein that is 27% identical to the E. coli IF2 protein and, like its bacterial counterpart, is involved in the initiation of protein synthesis (Choi et al. 1998). Virtually nothing is known about the function of YIL129C. Pol1p is the catalytic subunit of DNA polymerase α, one of three yeast polymerases required for chromosomal DNA replication (Waga and Stillman 1998). Given its central role in DNA replication and the fact that certain POL1 mutations cause telomere lengthening (Carson and Hartwell 1985; Adams and Holm 1996), we decided to focus on the interaction between Pol1p and Cdc13p.
By the criteria of a two hybrid assay, the interaction of Pol1Np with Cdc13Np was both specific and strong (Fig. 2A). Activation was seen only on galactose medium, not for glucose-grown cells, and required the presence of both the bait and prey plasmids (negative controls 1 and 2; Fig. 2A). The POL1 polypeptide did not activate in cells expressing other baits, such as LexA::cRafp (a gift from E. Golemis) (Fig. 2A, cRaf). Cells expressing the interacting Pol1Np and Cdc13Np polypeptides grew as well on plates lacking leucine as the strongly interacting proteins Rpb4p and Rpb7p, two subunits of RNA polymerase II (Khazak et al. 1995) (Fig. 2A, positive control), and produced a similar amount of β-galactosidase by a LacZ filter assay (data not shown).
Full-length Cdc13p and Pol1p interact by biochemical criteria
Immunoprecipitation (IP) was used to determine if the interaction between Cdc13Np and Pol1Np detected by two-hybrid analysis reflected an in vivo association of full-length proteins (Fig. 2B). Extracts were prepared from cells carrying the endogenous copies of both genes expressed from their own promoters with the only modification being the introduction of three MYC epitopes at the Cdc13p carboxyl terminus. Cells with the MYC3-tagged CDC13 allele had a normal growth rate and no detectable change in telomeric DNA (data not shown). Monoclonal anti-MYC antibody was used to immunoprecipitate MYC3-tagged Cdc13p. The precipitate was analyzed by Western blotting using both a monoclonal anti-Pol1p (a generous gift from Dr. P. Plevani) and anti-MYC antibodies. MYC3–Cdc13p was readily detected in the anti-MYC immunoprecipitate (Fig. 2B). Cdc13p appeared as a doublet, because of its existing in different phosphorylated states in vivo (A. Taggart and V.A. Zakian, unpubl.) (Fig. 2B, panel IP). Pol1p was detectable in the anti-MYC immunoprecipitate (Fig. 2B, lane 1 IP) but only if the extract was prepared from a strain with an MYC3-tagged Cdc13p (Fig. 2B, cf. lanes 1 and 3, IP). We conclude that full-length Pol1p and Cdc13p interact in vivo.
Identification of Pol1Np mutations that reduce its ability to interact with Cdc13Np
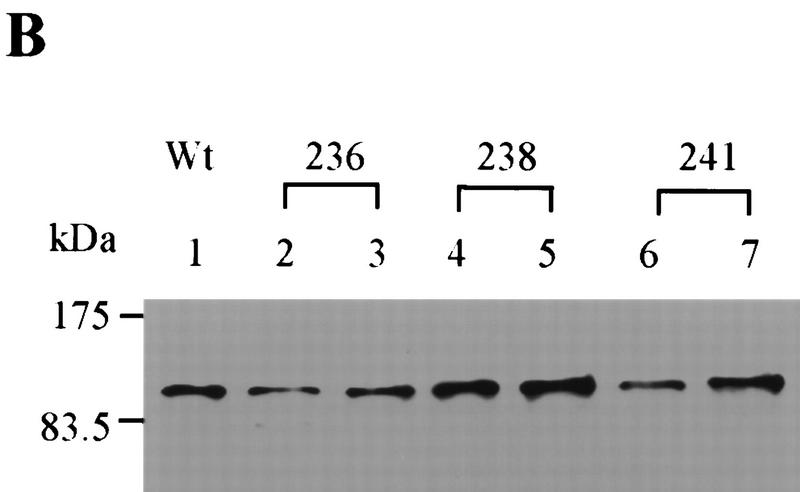
To identify point mutations in Pol1Np that disrupted the Cdc13p–Pol1p interaction, we used PCR to mutagenize the 379 amino acid fragment of Pol1p identified in the two-hybrid screen (Leung et al. 1989; Zhou et al. 1991). The mutagenized fragments were then assessed for their ability to interact with Cdc13Np in the two-hybrid assay. Transformants that produced white colonies in the lacZ filter assay were chosen for further analysis. Out of ∼106 colonies, three mutants gave white or pale blue colored colonies in the LacZ filter assay and produced full-length polypeptides by Western analysis (Fig. 3B). As each of these mutants produced normal amounts of Pol1Np, their failure to interact in the two hybrid assay was not attributable to failure in protein expression. DNA sequencing showed that each of the three had a single amino acid substitution: aspartic acid to asparagine at residue 236 (D236N), glutamic acid to lysine at residue 238 (E238K), and proline to threonine at residue 241 (P241T), respectively (Fig. 3A). Mutations were named pol1-236, 238, and 241 according to the amino acid residue that was mutated.
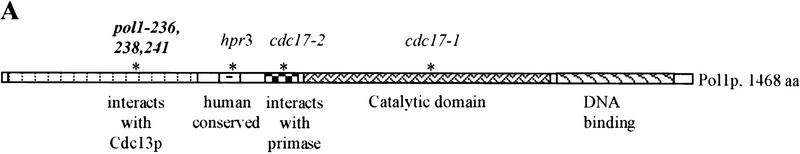
Figure 3.
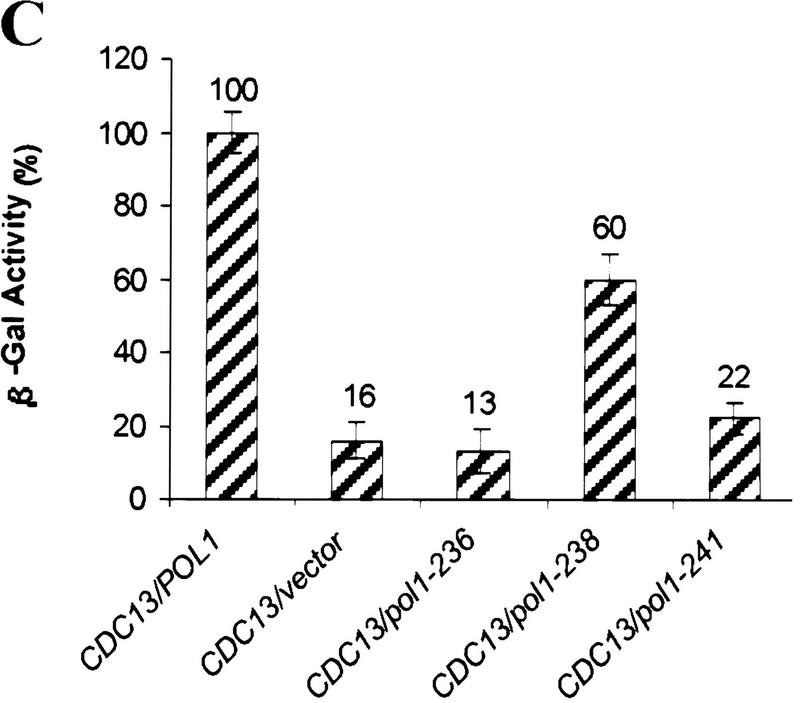
Analysis of pol1 alleles that disrupt interaction of Pol1p with Cdc13p. (A) Positions of functional domains and locations of various mutations within Pol1p are indicated. Mutations in bold disrupted Pol1p interaction with Cdc13p. Positions or regions containing mutations characterized in other labs are also shown: hpr3 (G439E), cdc17-2 (G637D), cdc17-1 (G904D) (Pizzagalli et al. 1988; Lucchini et al. 1990). (B) The wild-type (lane 1) or mutant (lanes 2–7) proteins were expressed from the pJG4-5 vector under control of a GAL1 promoter. The allele number of the expressed protein is indicated above the lanes. Proteins expressed from pJG4-5 are fused to a transcriptional activation domain and an HA epitope. Cell extracts were analyzed from two different isolates for each mutation using Western blotting with an anti-HA antibody. (C) Extracts were produced from strains expressing both Cdc13Np and either the pJG4-5 vector alone or pJG4-5 with the wild-type POL1 segment or one of the point mutants, as indicated below. Data are the average of three independent β-galactosidase measurements. Error bars, s.d.
To estimate the extent of loss of the Pol1Np–Cdc13Np two-hybrid interaction in each of the pol1 mutants, a quantitative liquid lacZ assay was performed (Guarente 1983). Using this assay, interaction with Cdc13Np was essentially abolished in pol1-236 (D236N), reduced ∼50% in pol1-238 (E238K) and reduced ∼90% in pol1-241 (P241T) (Fig. 3C).
Mutations in Pol1p that disrupt the interaction with Cdc13p result in longer telomeres
To determine the importance of the Cdc13p–Pol1p interaction, each of the three POL1 mutations that reduced or eliminated this interaction by two-hybrid criteria was introduced into the chromosomal copy of POL1, using the integration plasmid pCM1 (Lucchini et al. 1988). In these pol1 haploid isolates, the mutant allele was the only copy of POL1, and Pol1p was expressed from its own promoter. Strains carrying the pol1-236, 238, or 241 alleles grew as well as wild-type cells at 23°C, 30°C and 37°C (data not shown). Thus, none of the mutant alleles eliminated an essential function of Pol1p.
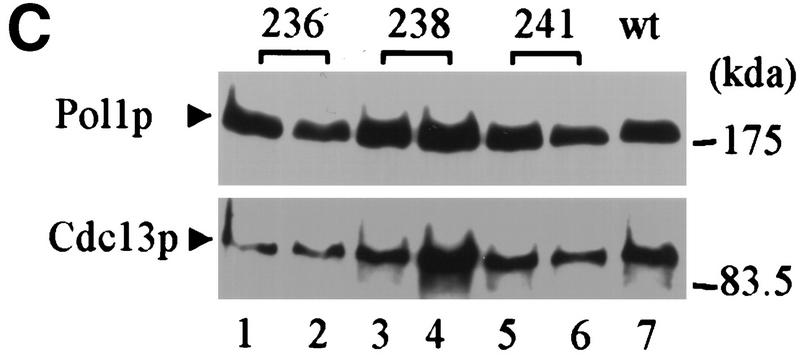
To determine if telomere length is altered in the pol1 mutant strains, DNA was prepared from wild-type and mutant cells, digested with XhoI, and analyzed by Southern blotting using a telomere probe (Fig. 4A). XhoI digestion generates a ∼1.3-kb terminal fragment from Y′-bearing telomeres, which make up about two-thirds of the telomeres in yeast, and multiple larger sized bands from X-bearing telomeres. Each of the pol1 mutants caused an increase in the length of X′ and Y′ telomeres (Fig. 4A). Cells with the pol1-236 allele, the allele that essentially eliminated the Pol1Np–Cdc13Np interaction by two-hybrid criteria (Fig. 3C), had telomeres that averaged 150-bp longer than wild type (Fig. 4A, lanes 2,3). The pol1-238 cells had telomeres that were ∼40 bp longer than wild type (Fig. 4A, lanes 5,6) and an ∼50% reduction in the Pol1Np–Cdc13Np interaction (Fig. 3C). The pol1-241 cells had telomeres that were ∼90 bp longer than wild type (Fig. 4A, lanes 8,9) and an ∼90% reduction in the Pol1Np–Cdc13Np interaction (Fig. 3C). The increase in telomere length seen in these mutants was not progressive, as telomeres were equally long at ∼55 and ∼130 divisions after sporulation (Fig. 4B). This telomere lengthening was telomerase-mediated as lengthening was not seen in a tlc1 pol1-236 double mutant strain (data not shown). As Western analysis showed that each of the mutant alleles produced essentially wild-type levels of both Pol1p and Cdc13p (Fig. 4C), the differences in telomere lengths are unlikely to be attributable differences in the abundance of either protein.
Figure 4.
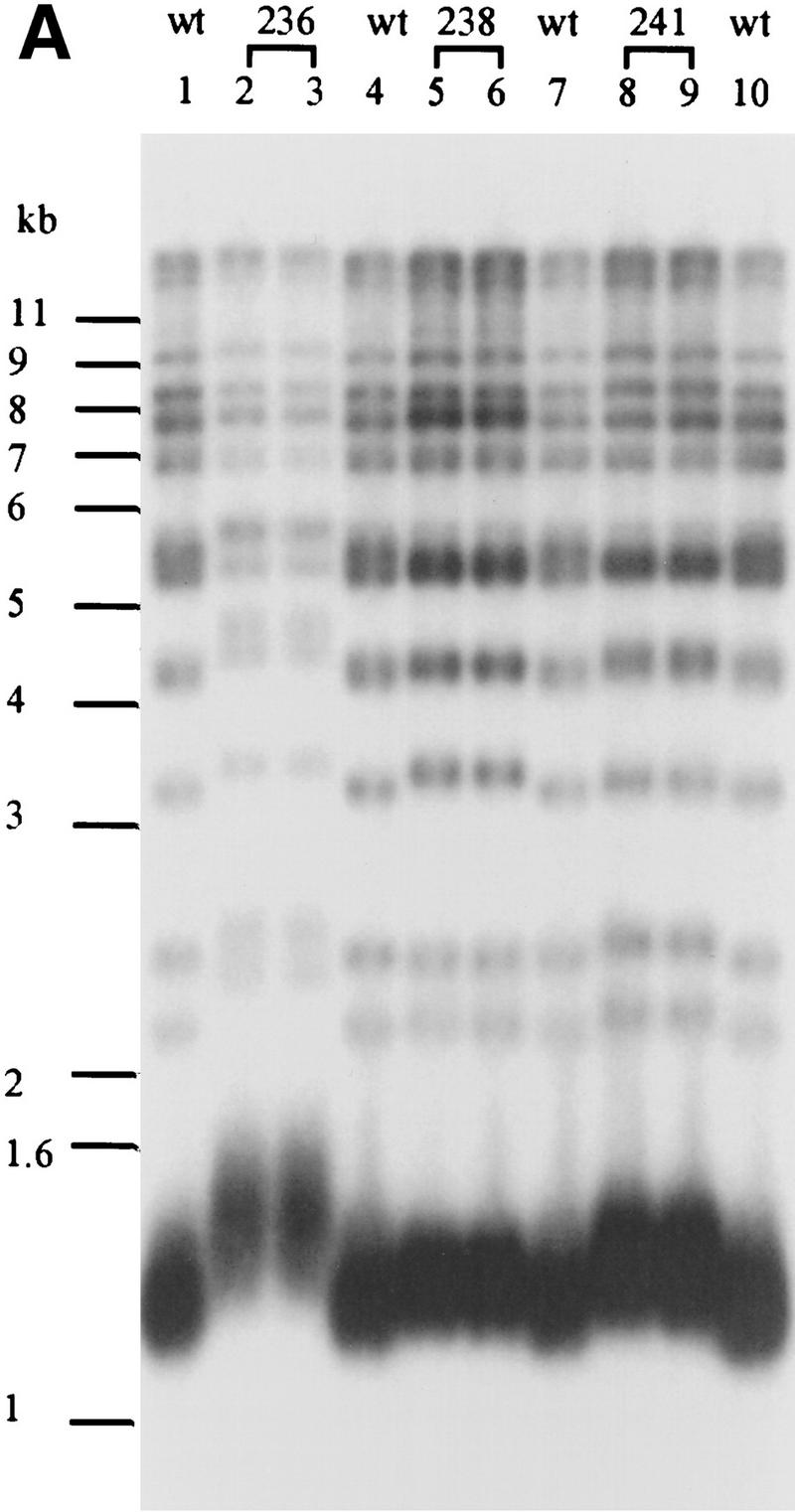
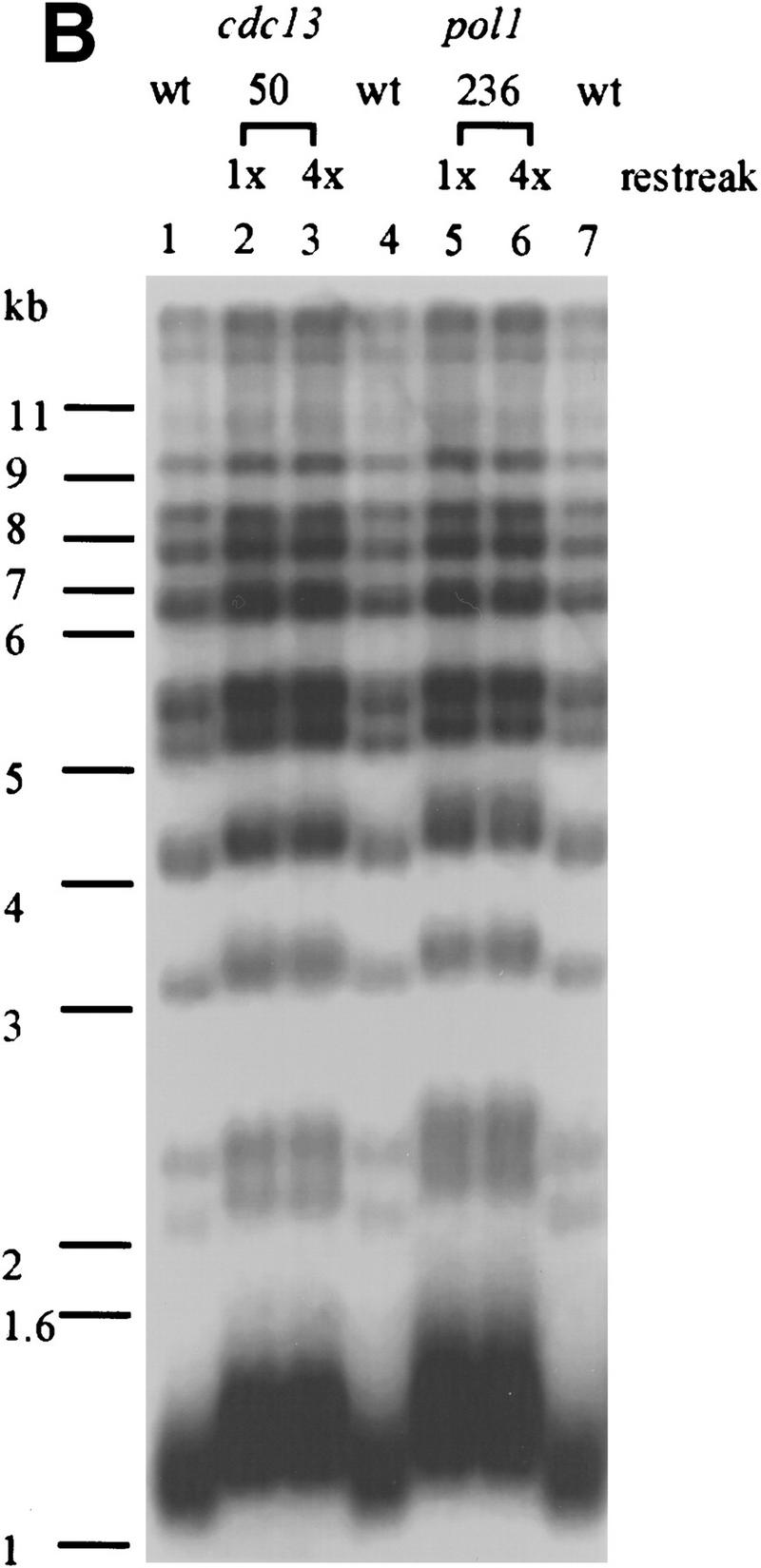
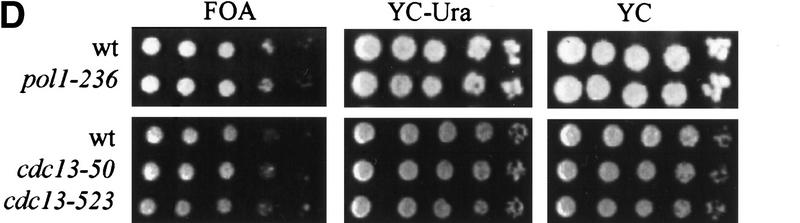
The pol1 alleles that disrupt Pol1p-Cdc13p interaction cause telomere lengthening. (A) Genomic DNA was isolated from wild type and two independent isolates for each of the three pol1 alleles (allele numbers are above the lanes), digested with XhoI and analyzed by Southern hybridization using a telomeric probe. (Lanes 2, 3) pol1-236 cells; (lanes 5, 6) pol1-238 cells; (lanes 8, 9) pol1-241 cells. (Lanes 1, 4, 7, 10) DNA from an otherwise isogenic wild-type strain. (B) Cells of the indicated genotype were streaked on complete media right after meiosis and allowed to form colonies. DNA was prepared from colonies after the first restreak (∼55 divisions postgermination) and after four restreaks (∼130 generations). DNA was analyzed as described for panel A. DNA is from wild type (lanes 1, 4, 7), cdc13-50 cells (lane 2, 55 divisions; lane 3, 155 divisions), and pol1-236 cells (lane 5, 55 divisions; lane 6, 155 divisions). (C) Western analysis was carried out using DNA from POL1 or pol1-236, -238, and -241 strains expressing 3XMYC- Cdc13p (mutant allele indicated above the lanes; extracts from two independent mutant isolates are shown for each mutant allele). Extracts were analyzed by SDS–PAGE and Western blotting using the anti-Pol1p serum (top) or anti-myc serum (lower panel). The minor differences in protein levels were not reproducible. (D) To measure telomere position effect, ten-fold serial dilutions of otherwise isogenic strains that were either wild type (wt) or contained the indicated mutations and having URA3 next to the left telomere of chromosome VII were spotted onto plates containing complete medium plus FOA, complete medium lacking uracil (YC-uracil), or complete medium (YC).
Telomere position effect, (TPE), refers to the fact that genes placed near a yeast telomere are transcriptionally repressed (Gottschling et al. 1990). Certain POL1 alleles that affect the catalytic activity of polymerase α cause telomere lengthening, loss of TPE, and altered G-tail metabolism (Adams-Martin et al. 2000). To monitor TPE, the URA3 gene was placed next to the left telomere of chromosome VII in the pol1-236 strain and the fraction of cells able to grow on FOA medium determined. As the pol1-236 strain had the same fraction of FOAR cells as an isogenic wild-type control (Fig. 4D), TPE was not affected by the pol1-236 mutation. Long TG1-3 tails are detected on yeast telomeres at the end of S phase (Wellinger et al. 1993b) but not in DNA from log phase cultures except in strains that are defective in their processing, as occurs in cdc13-1 cells growing at high temperatures (Gravel et al. 1998) or hdf1 strains that lack the Ku heterodimeric complex (Gravel et al. 1998; Polotnianka et al. 1998). Using the nondenaturing hybridization method described in Dionne and Wellinger (1996), G-tails were not detected at pol1-236 telomeres whereas G-tails were readily detected on cdc13-1 and hdf1Δ telomeres (data not shown). Thus, by the limits of this assay, a mutation that disrupted Cdc13p–Pol1p interaction did not affect G-tail structure.
The pol1-236 mutation eliminated interaction of Pol1Np with Cdc13Np, as monitored by the two-hybrid assay (Fig. 3C). To determine if this mutation eliminated the interaction of full-length Pol1p and full-length Cdc13p, we carried out the same experiment used to detect the interaction of these proteins in wild-type cells (Fig. 2B). A protein extract was prepared from pol1-236 cells expressing a MYC3-tagged Cdc13p. The extract was immunoprecipitated with anti-MYC antibodies and analyzed by Western blotting using both anti-MYC and anti-Pol1p antibodies. The amount of Pol1–236p in the immunoprecipitate was reduced substantially but not eliminated (Fig. 2B, IP panel, lane 2). The residual interaction between Cdc13p and Pol1–236p suggests that the Pol1p–Cdc13p interaction is mediated by more than one region in Pol1p and/or that other subunits of polymerase α contribute to its interaction with Cdc13p.
Mutations in CDC13 that disrupt the Cdc13Np–Pol1Np interaction cause long telomeres
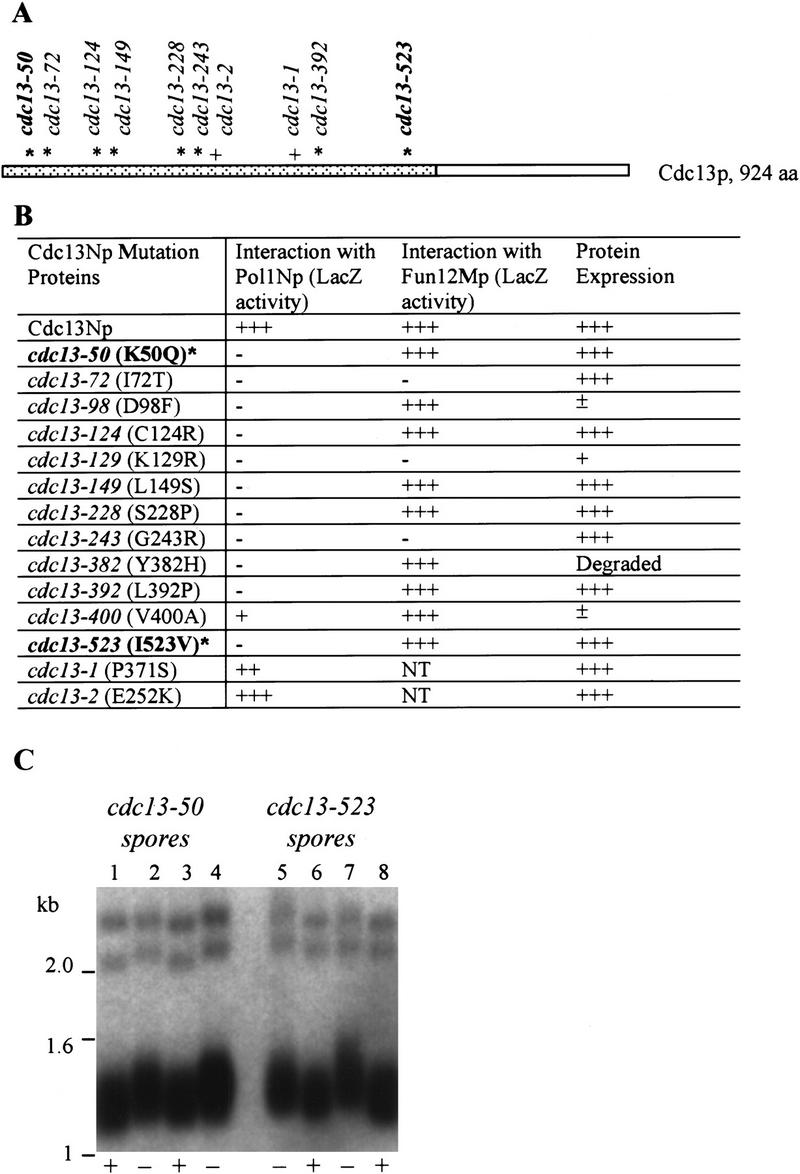
To identify mutations in CDC13 that disrupt the Cdc13Np–Pol1Np interaction, the 600 amino acid fragment of Cdc13p used in the initial two-hybrid screen (Fig. 1) was mutagenized by PCR. The mutagenized segments were reintroduced into the yeast two hybrid strain, and mutants that failed to interact with Pol1Np were identified by their generating white colonies in the lacZ filter assay. Of the 14 candidate mutations that produced full-length protein, DNA sequencing revealed that 12 contained a single amino acid mutation, and two contained multiple point mutations. The single mutations were spread throughout the 600 amino acids of Cdc13Np (Fig. 5A). As with the pol1 mutations, these alleles were named according to the mutated amino acid residue. Eight of the 12 mutant polypeptides were expressed at wild-type levels, and six of these still interacted with Fun12Mp in the two hybrid assay, one of three polypeptides identified as interacting with wild-type Cdc13Np (summarized in Fig. 5B). Mutants cdc13-50 (K50Q), cdc13-124 (C124R), cdc13-129 (L129S), cdc13-228 (S228P), cdc13-392, (L392P) and cdc13-523 (I523V) were defective specifically in their ability to interact with Pol1Np, and these defects were not due to reduced protein expression.
Figure 5.
Alleles of CDC13 that disrupt interactions with POL1. (A) The portion of Cdc13p that was used as bait in the two-hybrid screen is represented by the dotted region. Asterisks mark the sites of single amino acid substitutions that reduced the interaction of Cdc13Np with Pol1Np by two hybrid criteria. The alleles in bold are those used for phenotypic analyses. The locations of the cdc13-1 and cdc13-2 alleles are also indicated (+) (Lin and Zakian 1996; Nugent et al. 1996). (B) Alleles for cdc13 polypeptides that lost interaction with Pol1Np are named by the number of the mutated residue. The amino acid change in each allele is noted in parentheses. Each mutant allele was checked for its ability to interact with both Pol1Np and Fun12Mp in the two hybrid assay using the LacZ filter assay. Western blotting was used to determine if strains carrying the mutant allele made wild-type levels of Cdc13p. (NT) Not tested (+++) wild-type levels of interaction or protein expression, (−) no interaction or protein expression; (±, +, and ++) intermediate levels of interaction or protein expression. (C) XhoI digested DNA from wild-type strain or cdc13Δ strains carrying a centromere plasmid with mutant cdc13 alleles (cdc13-50 and cdc13-523) that disrupted interaction of Cdc13Np with Pol1Np was analyzed by Southern blotting. Only the lower portion of the gels is shown. (+) Wild type (−) mutant cdc13 alleles.
To determine the phenotypes of cdc13 alleles that reduced the Cdc13Np–Pol1Np interaction, we introduced two of the mutations, cdc13-50 and cdc13-523, on a centromere plasmid into a cdc13Δ strain such that the mutant allele was the only copy of CDC13 in the strain. These alleles were chosen because they produced stable protein and had little or no interaction with Pol1Np by two hybrid criteria, yet continued to interact with Fun12Mp (Fig. 5B). The growth of cdc13-50 and cdc13-523 cells was comparable to that of wild-type cells at 23°C, 30°C, and 37°C (data not shown), but cdc13-50 (Fig. 5C, lanes 2,4) and cdc13-523 (Fig. 5C, lanes 5,7) cells had telomeres that averaged 50 bp longer than wild type (Fig. 5C, lanes 1,3,6,8). As with the pol1 alleles, this lengthening was not progressive (Fig. 4B, cdc13-50; cf. lanes 2 and 3). Cells carrying the cdc13-50 and cdc13-523 alleles had wild-type levels of TPE (Fig. 4D).
Cdc13Np interacts with Est1Cp by two-hybrid, genetic, and biochemical criteria
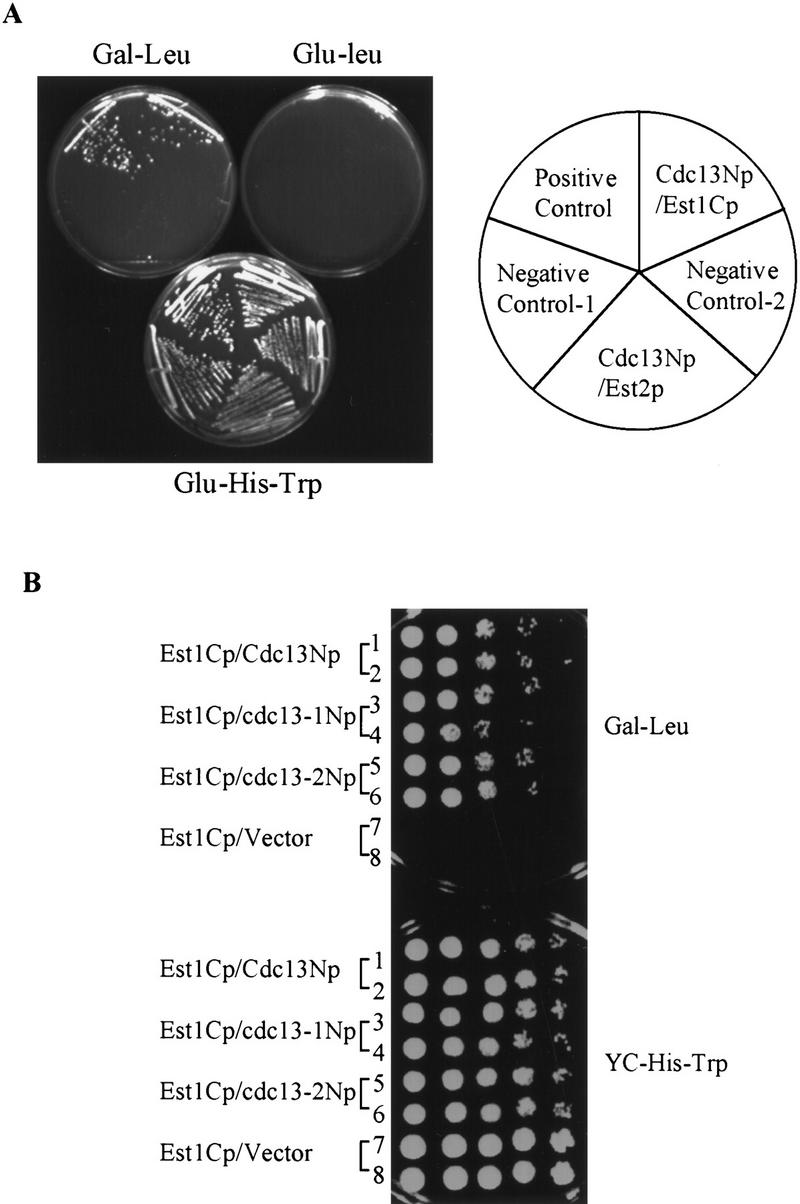
Telomere-bound Cdc13p has been proposed to function by recruiting telomerase to the telomere (Evans and Lundblad 1999; Zhou et al. 2000). As both Est1p (Lin and Zakian 1995; Steiner et al. 1996) and Est2p (Counter et al. 1997; Lingner et al. 1997a,b) are associated with TLC1 telomerase RNA, we tested the ability of Cdc13Np to interact with both Est1p and Est2p in the two-hybrid assay. Full-length Est2p or the carboxy-terminal region of Est1p (Est1Cp) were cloned into the prey vector (Fig. 1). These plasmids were introduced into the two hybrid reporter strain expressing Cdc13Np. Cells expressing the Est2p fusion protein did not interact with Cdc13Np in the two-hybrid assay (Fig. 6A), although Western analysis showed that the Est2p fusion protein was expressed (data not shown). In contrast, Est1Cp, which contained the carboxy-terminal 200 amino acids of the 699 amino acid Est1p, did interact with Cdc13Np as demonstrated by the growth of cells expressing Est1Cp and Cdc13Np on plates lacking leucine (Fig. 6A,B). However, this association was weaker than the Cdc13Np–Pol1Np interaction, as expressing Cdc13Np and Est1Cp did not activate the LacZ reporter gene (data not shown). By the two hybrid assay, Est1Cp also interacted with mutant proteins Cdc13-1Np and Cdc13-2Np (Fig. 6B).
Figure 6.
Cdc13Np interacts with Est1Cp in a two-hybrid assay. (A) Two hybrid analysis demonstrates that Cdc13Np interacted with Est1Cp but not with full-length Est2p. Positive and negative controls are the same as in Figure 2A. (B) Ten-fold serial dilutions of each strain were plated onto media selecting for the two-hybrid interaction (Gal - Leu) or selecting for the two plasmids (YC-His-Trp). The terminal 600 amino acids from CDC13, cdc13-1, or cdc13-2 were tested for their ability to interact with Est1Cp.
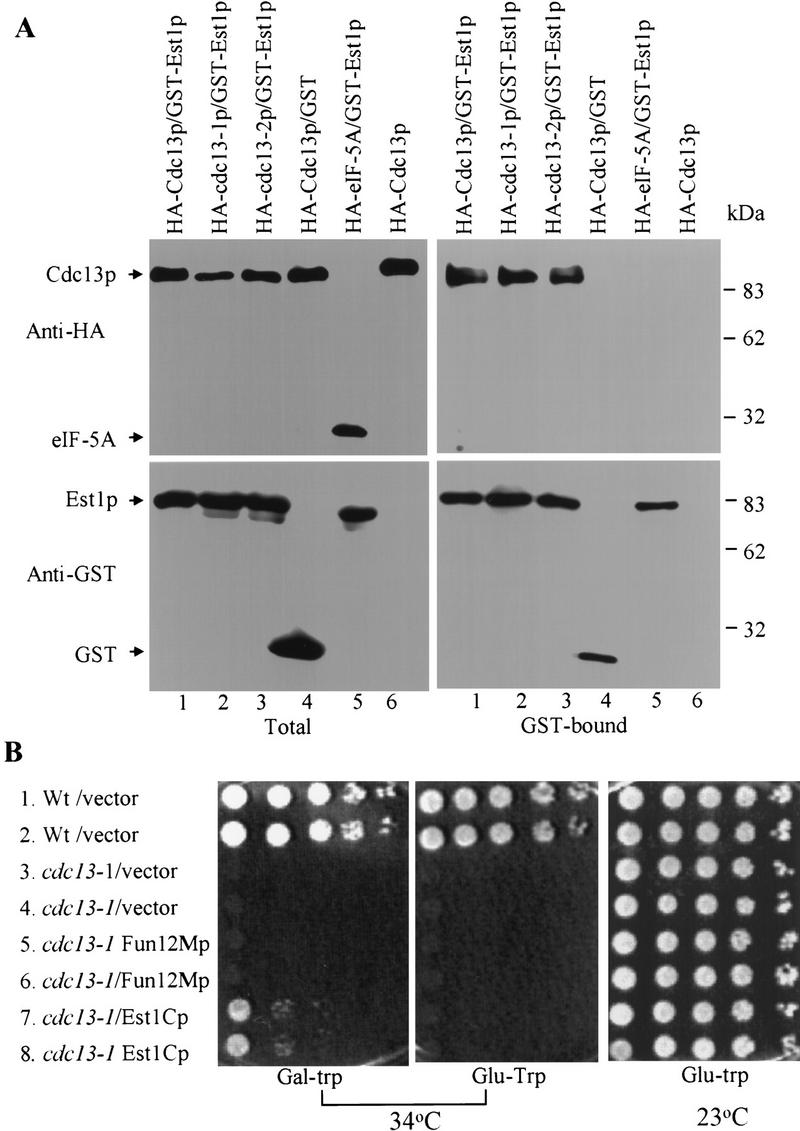
Using the biochemical approach that detected interaction of Cdc13p and Pol1p (Fig. 2B), we were unable to detect association of endogenous Cdc13p and Est1p (data not shown), perhaps because both proteins are rare and/or because their interaction within cells occurs only in a transient manner. To demonstrate the interaction biochemically, we prepared extracts from cells that overexpressed both proteins. GST–Est1p was expressed from the centromere plasmid pKT/EST1 (Mitchell et al. 1993) using the inducible GAL1 promoter, and HA-tagged Cdc13p was expressed from the PGK1 promoter on the 2 μm-based plasmid pTHA/CDC13 (Lin and Zakian 1996). Although the GST–Est1p lacked the first 31 amino acids of Est1p, it complemented an est1 strain (Y. Yamashita, unpubl.). Extracts were incubated with glutathione sepharose (GS) beads. After extensive washing, the material bound to the beads was eluted and analyzed by Western blotting for the presence of GST–Est1p and HA–Cdc13p (Fig. 7A). HA–Cdc13p was bound to the beads in extracts from cells expressing GST–Est1p (Fig. 7A, upper right panel, lane 1) but not from cells expressing just the GST protein (Fig. 7, upper right panel, lane 4) or no GST protein at all (Fig. 7, lane 6). The telomere-unrelated protein HA-eIF-5Ap did not interact with GST–Est1p (Fig. 7, lane 5). HA-Cdc13-1p (Fig. 7, lane 2) and HA-Cdc13-2p (Fig. 7, lane 3) also interacted with GST–Est1p in this assay.
Figure 7.
The interaction of Cdc13p and Est1p can be detected biochemically and has functional significance. (A) Extracts were prepared from cells expressing HA-tagged wild type (lanes 1,4,6) or mutant Cdc13p (lane 2, cdc13-1; lane 3, cdc13-2) or HA-tagged eiF-5Ap (lane 5). Cells also expressed GST-fused Est1p (lanes 1,2,3,5), the GST polypeptide (lane 4) or no GST protein (lane 6). The total extract was analyzed by Western blotting (left panels) with an anti-HA (top) or anti-GST sera (bottom). The extracts were incubated with glutathione sepharose (GS) beads, and the bound proteins eluted (right panels) and analyzed by Western blotting as described for the unfractionated extract. (B) To determine the effect of Est1Cp overexpression on growth, ten-fold serial dilutions of the CDC13 strain carrying pJG4-5 vector (lines 1,2); cdc13-1 carrying pJG4-5 vector (lines 3,4); cdc13-1 expressing Fun12Mp from the pJG4-5 vector (lines 5,6); cdc13-1 expressing Est1Cp from the pJG4-5 vector (lines 7,8) were plated on galactose minus tryptophan medium and grown at 34°C (left) or glucose minus tryptophan medium and grown at 34°C (middle) or glucose minus tryptophan medium and grown at 25°C (right). Expression of Est1Cp and Fun12Mp was under control of the galactose inducible GAL1 promoter.
To assess if the Cdc13Np–Est1Cp interaction has in vivo significance, we asked if high-level expression of Est1Cp could rescue the temperature-sensitive lethality of cdc13-1 cells (Fig. 7B). The maximum permissive temperature for cells carrying the cdc13-1 allele was 28°C although the isogenic wild-type strain grew well at 37°C. Ten-fold serial dilutions of wild-type or cdc13-1 cells were spotted on galactose (gal) or glucose (glu) plates lacking tryptophan (galactose induces expression of the fusion proteins; no tryptophan selects for the prey plasmid) and the plates incubated at different temperatures (Fig. 7B). The cdc13-1 cells carried either prey vector alone, the prey vector expressing Fun12Mp, or the prey vector expressing Est1Cp. All strains grew on glucose plates at 25° (Fig. 7B, right panel); wild-type cells grew on both glucose (Fig. 7B, middle panel) and galactose (Fig. 7B, left panel) plates at 34°. However, cdc13-1 cells did not grow at 34° unless they expressed Est1Cp: Cells with Est1Cp did not grow at 34° on glucose medium where Est1Cp was not expressed (Fig. 7B, middle panel) nor on galactose plates when expressing Fun12Mp (Fig. 7B, left panel). Thus, high-level expression specifically of Est1Cp suppressed the temperature sensitivity of cdc13-1 cells, suggesting that the Est1Cp–Cdc13Np interaction was significant. The suppression of the cdc13-1 growth defect was partial: cdc13-1 cells expressing Est1Cp did not grow to the same dilution at 34° as the same strain at 25°. Also, the expression of Est1Cp did not allow cdc13-1 cells to grow at 37° (data not shown).
Discussion
Two-hybrid analysis demonstrated that the amino-terminal two-thirds of Cdc13p interacts with the amino terminus of Pol1p, the catalytic subunit of DNA polymerase α (Fig. 2A). This interaction was confirmed by demonstrating that full-length Cdc13p and Pol1p could be coimmunoprecipitated (Fig. 2B). The interaction could be direct or indirect, mediated by one or more as yet unidentified proteins. Whether direct or not, this interaction is satisfying because polymerase α, by virtue of its primase association, is the only eukaryotic polymerase that can initiate DNA replication (Waga and Stillman 1998) and is hence the best candidate for the polymerase that resynthesizes C-strand DNA. POL1 mutations that reduced this interaction mapped to a negatively charged, five amino acid patch within the amino-terminal 379 amino acid segment of Pol1p, identified by two-hybrid experiments (Fig. 3A). Although the catalytic subunit of DNA polymerase α is conserved among eukaryotes, its amino-terminal portion is not conserved and no function had been mapped previously to this region (Fig. 3A).
Mutations in either POL1 (Fig. 4A) or CDC13 (Fig. 5C) that reduced the Pol1p–Cdc13p interaction caused telomerase-mediated telomere lengthening. For the POL1 mutations, this TLC1-dependent length increase was greatest in the mutant with the most severe effect on the Pol1p–Cdc13p interaction (Figs. 3C and 4A). These genetic results argue strongly that the Pol1p–Cdc13p interaction detected by both two-hybrid (Fig. 2A) and biochemical (Fig. 2B) criteria is important for telomere maintenance. Others have shown that certain pol1 alleles show telomerase-mediated telomere lengthening (Carson and Hartwell 1985; Adams and Holm 1996). However, in these cases the mutations are temperature sensitive, affect the catalytic activity of Pol1p, and have global effects on both DNA replication and cell viability. In contrast, the pol1 mutations studied here occurred in a region that is thought to be dispensable for catalysis, and these mutations had no effect on cell growth or TPE (Fig. 4D). Because these mutations disrupted interaction with Cdc13p (Figs. 2B and 3C), an in vivo telomere binding protein (Bourns et al. 1998), their effects on telomere length were likely direct. Based on these data, we propose that telomere-bound Cdc13p recruits polymerase α to the telomere and that this interaction promotes C-strand resynthesis.
If the Pol1p–Cdc13p interaction is essential for C-strand resynthesis, mutations like pol1-236 that eliminated this interaction by two-hybrid criteria (Fig. 3C) should be lethal due to loss of telomeric DNA and/or activation of DNA damage checkpoints by long ss G-tails (Lee et al. 1998). However, biochemical experiments demonstrated that there is residual interaction between Cdc13p and Pol1p even in the pol1-236 strain (Fig. 2B), suggesting that other parts of Pol1p might also interact with Cdc13p. Alternatively or in addition, other telomere proteins (such as Stn1p) whose loss has a similar effect on C-strand degradation as the cdc13-1 mutation (Grandin et al. 1997), or the Ku heterodimer, whose loss results in G-tails throughout the cell cycle (Gravel et al. 1998; Polotnianka et al. 1998), might be partially redundant with Cdc13p and help recruit Pol1p to the telomere.
We also report that the amino terminus of Cdc13p interacted with the carboxyl third of Est1p by two-hybrid analysis (Fig. 6A). Biochemical experiments confirmed this interaction, demonstrating that full-length Cdc13p interacted in vivo with a close to full-length, functional Est1p (Fig. 7A). Again this interaction could be direct or mediated by another protein. As overexpression of the carboxy- terminal 200 amino acid segment of Est1p partially suppressed the temperature sensitive lethality of cdc13-1 cells whereas overexpression of another Cdc13p interacting polypeptide, Fun12Mp, did not (Fig. 7B), the Cdc13p–Est1p interaction is likely to be important for the essential function of Cdc13p.
Because Est1p is associated with telomerase RNA (Lin and Zakian 1995; Steiner et al. 1996), a Cdc13p–Est1p interaction could recruit telomerase to telomeres, thereby explaining the senescence phenotype of cdc13-2 cells. That Cdc13p functions to recruit telomerase is also supported strongly by the demonstration that expression of a Cdc13p–Est1p or a Cdc13p–Est2p fusion protein supplants the need for Est1p in telomere maintenance (Evans and Lundblad 1999). However, as these fusion proteins might bypass the normal role of Est1p, these data do not address whether Cdc13p and Est1p interact in vivo, whereas our data provide strong support for this possibility.
If Cdc13p recruits telomerase by virtue of its ability to interact with Est1p, the phenotype of cdc13-2 cells could be explained if Cdc13-2p were unable to interact with Est1p. However, Est1p interacted with Cdc13-2p as assayed by both two-hybrid (Fig. 6B) and biochemical (Fig. 7A) criteria. One explanation for this result is trivial: Est1p might be able to interact with Cdc13-2p when both proteins are overproduced as they were in both the two-hybrid and biochemical assays, but not when the proteins are expressed at physiological levels. This possibility predicts that overexpression of Est1p would suppress the telomere defects of a cdc13-2 strain. However, overexpression of a GST–EST1p fusion protein that contained all but the most amino-terminal 31 amino acids of Est1p, and which complemented an est1 strain or overexpression of the carboxy-terminal portion of Est1p, did not suppress the telomere length defect of cdc13-2 cells (data not shown). Alternatively, interaction of Cdc13p with Est1p might trigger a conformational change in Cdc13p, such as phosphorylation, that makes it better able to promote the telomerase pathway: Cdc13-2p might be able to interact with Est1p but not undergo the subsequent conformational change.
In the ciliate Euplotes, aphidicolin, an inhibitor of conventional DNA polymerases, causes changes in the length of both the C- and G-strands of telomeric DNA. This result led to the proposal that G-strand lengthening and C-strand synthesis are coordinately regulated (Fan and Price 1997). In Saccharomyces, addition of duplex telomeric DNA to a double-strand break requires not only telomerase but also DNA primase and two of the three conventional DNA polymerases, polymerase α and polymerase β (Diede and Gottschling 1999). These data also support a model of coordinate regulation of C- and G-strand synthesis. Cdc13p, by virtue of its ability to interact with both the conventional replication apparatus (Fig. 2A) and a telomerase component (Fig. 6A), could play a critical role in this coordination.
Alternatively, there might be competition between C-strand resynthesis by polymerase α and G-strand lengthening by telomerase. In support of this model, when the interaction between Cdc13p and Pol1p was weakened, telomerase-mediated lengthening of telomeres increased (Fig. 4A). By two hybrid criteria, Cdc13p interacted more strongly with Pol1p than it did with Est1p. Also, polymerase α is much more abundant than telomerase. Based on these considerations, a competition model predicts that recruitment by Cdc13p of polymerase α to ss TG1-3 tails is much more likely than recruitment of telomerase. As short telomeres are preferentially lengthened by telomerase (Marcand et al. 1999), telomere length may influence this competition.
Materials and methods
The yeast two-hybrid strain YEM1α (MATa his3-11 trp1-1 LEU2::pLexAop-LEU2 ura3-1:: URA3-pLexAop-Gal1-lacZ) was constructed by E. Monson in our lab using reagents described in Gyuris et al. (1993). The protease-deficient yeast strain BJ2168 (Jones 1991) was used to detect Est1p–Cdc13p interaction. YPH499, YPH500, and YPH501 (Sikorski and Hieter 1989) were used for analysis of pol1 and cdc13 mutant phenotypes. CDC13 deletion in YPH501 was created by J.J. Lin (Lin and Zakian 1996). Cdc13p was tagged at its carboxyl terminus with three MYC epitopes (Schneider et al. 1995) in strain VPS106 (V. Schulz and V. Zakian 1994) and used for immunoprecipitation. CDC13 was also tagged with nine MYC epitopes at its carboxyl terminus in strain YPH499 (A. Taggart and V.A. Zakian, unpubl.) as in Zachariae et al. (1996). Cells containing either 3-myc-tagged or 9-myc-tagged CDC13 had normal cell growth and telomere length.
The two-hybrid assay was done essentially as described (Gyuris et al. 1993) using components generously provided by Dr. R. Brent and colleagues. The prey library (pJG4-5/X) (Gyuris et al. 1993; Golemis et al. 1994) was transformed into YEM1α strain containing the CDC13N bait plasmid (pEG202/CDC13-B; constructed by J.-J. Lin) (Fig. 1). HA3-tagged CDC13 from pTHA/CDC13 (Lin and Zakian 1996) was inserted into BamHI–SalI digested pVZ1, then BglII digested, followed by self-ligation to delete the BglII fragment within CDC13. This introduces a frameshift at the ligation site such that only the first 600 amino acids of Cdc13p are produced. The HA-tagged amino-terminal region of CDC13 was cloned into pEG202 generating a LexA DNA binding domain–Cdc13Np fusion protein expressed from the ADH1 promoter. The library proteins were also HA tagged and expressed from a GAL1 promoter. Transformants were harvested and replated to 3% galactose YC plates minus leucine. Leu+ colonies were transferred to nitrocellulose filters (Schleicher & Schuell) for the colony lacZ filter assay (Breeden and Nasmyth 1985). Liquid lacZ assays were done as in Miller (1972) and Guarente (1983). Leu+ LacZ+ colonies were purified and confirmed by rescuing the plasmids into E. coli and retransforming them into the two-hybrid yeast strain. Prey plasmids that retested positively were sequenced with the amplitaq FS dye terminator cycle sequencing kit (ABI).
The plasmid pJG4-5/EST1C was constructed by Y. Yamashita and contained amino acids 499–699 of Est1p fused in-frame with the B42 trans-activation domain and an HA epitope tag in the prey vector pJG4-5. Full-length EST2 was PCR amplified from YPH499 genomic DNA and cloned into pJG4-5 to create pJG4-5/EST2 (made by S-C. Teng). This plasmid complements an est2Δ strain. The cdc13-1 mutation P371S creates an additional EcoRI site (Lin and Zakian 1996) and cdc13-2 mutation E252K eliminates the first EcoRI site in CDC13 (Nugent et al. 1996). pEG202/cdc13-1B and pTHA/cdc13-1 were created by site-directed mutagenesis (QuikChange Site-Directed Mutagenesis Kit, Stratagene) from pEG202/CDC13N and pTHA/CDC13, respectively. pEG202/cdc13-2B and pTHA/cdc13-2 were created from pEG202/CDC13N and pTHA/CDC13, respectively. The mutated plasmids were introduced into E. coli, purified, and then sequenced to confirm that the correctmutation was made. Plasmids producing LexA::cRafp or LexA::Krev1p fusion protein were gifts from E. Golemis.
Immunoprecipitation to detect Cdc13p–Pol1p interaction was done in yeast strain VPS106 containing 3XMYC-CDC13 as in Garrett et al. (1991), except that cells were lysed in a homogenizer (Avestin's High Pressure Homogenizer EmulsiFlexC-5) in 40 mm Tris (pH 8.0), 75 mm NaCl, and 20 mm KOAc, 1 mm 2-mercaptoethanol, 0.01% NP-40, 1 mm phenylmethylsulfonyl fluoride (PMSF), 10 μg/ml pepstain A, 5 μg/ml leupeptin, 10 μg/ml aprotinin, 10 mm benzamidine, 250 μg/ml soybean trypsin inhibitor, and 10% glycerol. Monoclonal anti-MYC 9E10 (Princeton University) was used for the immunoprecipitation. Monoclonal anti-Pol1p antibody (from P. Plevani), anti-MYC antibody, and ECL (Amersham Life Science) were used to visualize the proteins.
The GST (glutathione S transferase) assay was conducted essentially as described in Mitchell et al. (1993). pEGKT/EST1, a plasmid that produces a GST::Est1p fusion protein under the control of a GAL1 promoter, was constructed and generously provided by Y. Yamashita. In brief, the HincII fragment from EST1, which contains amino acids 31–699 of Est1p, was cloned into pEGKT (Mitchell et al. 1993). Expression of GST::Est1p from this plasmid complements an est1Δ mutant (data not shown). Full-length Cdc13p tagged with 3 × HA at its amino terminus was expressed from a PGK1 promoter on plasmid pTHA/CDC13 (Lin and Zakian 1996). The two proteins were coexpressed in strain BJ2168. A similarly HA-tagged version of the yeast protein eIF-5A (Kang and Hershey 1994) was constructed by J-J. Lin and used as a negative control. Raffinose-grown log phase cells were transferred to 3% galactose medium and incubated at room temperature overnight to induce expression of GAL1-regulated genes. Cells were lysed in 20 mm Tris (pH 8.0), 200 mm NaCl, 1 mm EDTA, 1 mm DTT, 0.01% NP-40, 10% glycerol with protease inhibitors plus an equal volume of glass beads by vortexing in mini-beadbeater-8 (Biospec Products). NP-40 and Triton X-100 were added to the soluble protein fractions to final concentrations of 0.5% and 0.1%, respectively. GST-Sepharose 4B beads (Pharmacia Biotech) were then added. After incubation at 4°C, beads were collected by centrifugation and then washed once with the lysis buffer containing 1% NP-40 0.1% Triton X-100 and once with lysis buffer containing 450 mm NaCl. The beads were treated with 0.1 mg/ml DNase I in lysis buffer plus 1 mm MgCl2, then washed with lysis buffer containing 450 mm NaCl and 350 mm KOAC. Proteins were visualized by ECL Western using anti-HA antibody 12CA5 (Princeton University) and rabbit anti-GST antibodies (from Jin-Qiu Zhou).
PCR mutagenesis was used to generate POL1 and CDC13 mutations (Grandin et al. 1997). Two DNA oligonucleotides that were complementary to the sequences from the prey vector pJG4-5 (for POL1 mutations) or to the bait vector pEG202 (for CDC13) were used. A modified random PCR mutagenesis (Leung et al. 1989; Zhou et al. 1991) was done in a reaction mixture containing 100 μm dNTP, 200 νm oligonucleotides, 1× PCR buffer with 3 mm MgCl2, 1–10 ng/100 μl plasmid DNA template, and 1 unit Taq polymerase (Roche Molecular Biochemicals). PCR products were cotransformed into the two-hybrid strain containing bait or prey plasmids with XhoI linearized pJG4-5 or pEG202 vector. Pools of the PCR mutagenized POL1 segments were introduced by transformation into the yeast strain containing both the two-hybrid reporter genes and expressing Cdc13Np (Grandin et al. 1997). Transformants were selected by their ability to grow on media lacking histidine and tryptophan, thus selecting for both the prey and bait plasmids but not for the two-hybrid interaction. Transformants were replica plated to galactose plates then transferred to nitrocellulose membranes for LacZ assays.
The pol1-236, pol1-238, and pol1-241 mutant alleles were introduced into the POL1 integration plasmid pCM1 (Lucchini et al. 1988) using the QuickChange site-directed Mutagenesis Kit (Stratagene). Silent mutations that created an AclI site in pol1-236 (T708→C and G710→T in addition to the D236N mutation, G706→A) or an additional EcoRI site in pol1-238 and 241 (A713→G in addition to the E238K mutation, G711→A or P241T, C720→A) were also introduced to allow identification of mutant alleles by restriction enzyme digestion. The mutagenized plasmids were linearized by BstXI digestion and integrated into the POL1 locus of diploid strain YPH501 as in Lucchini et al. (1988). Transformants were selected by their Ura+ phenotype and verified by Southern. The diploid cells were sporulated; spores containing the mutant POL1 alleles were identified by Ura+ phenotype. Cells that excised URA3, leaving behind an intact but mutated POL1 gene, were selected on plates containing 5-fluoro orotic acid.
Wild-type CDC13 with its own promoter and terminator sequences was released from YEP24/CDC13 using ApaI digestion (Garvik et al. 1995) and cloned into the ApaI-digested CEN plasmid pRS314. The pRS314 /CDC13 was mutagenized using the QuickChange mutagenesis kit. To facilitate identification of mutations, an AvrII site was created near K50Q in pRS/cdc13-50 and the first HindIII site of CDC13 was eliminated at I523V in pRS314/cdc13-523 without changing additional amino acid residues. The mutated plasmids were introduced into a derivative of YPH 501 that had one copy of CDC13 and one copy of cdc13::HIS3 (Lin and Zakian 1996). After sporulation and dissection, haploid cdc13::HIS3 spores carrying the mutated cdc13 plasmid were selected by their His+ Trp+ phenotype.
Acknowledgments
We thank R. Brent and his lab, especially E. Golemis, for advice and reagents and P. Plevani for his generous gift of anti-Pol1p serum. We thank past and current members of our lab, J.-J. Lin, E. Monson, A. Taggart, S.-C. Teng, and Y. Yamashita, for unpublished strains or plasmids. We also thank R. Jiang, E. Monson, A. Taggart, S.-C. Teng, Y. Tsukatmoto, and J.-Q. Zhou for comments on the manuscript. This work was supported by grant GM RO1 43265 from the National Institutes of Health and a U.S. Army Breast Cancer postdoctoral fellowship to H.Q.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL vzakian@molbio.princeton.edu; FAX (609) 258-1701.
References
- Adams AK, Holm C. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4614–4620. doi: 10.1128/mcb.16.9.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams-Martin A, Dionne I, Wellinger RJ, Holm C. The function of DNA polymerase alpha at telomeric G tails is important for telomere homeostasis. Mol Cell Biol. 2000;20:786–796. doi: 10.1128/mcb.20.3.786-796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourns BD, Alexander MK, Smith AM, Zakian VA. Sir proteins, Rif proteins and Cdc13p bind Saccharomyces telomeres in vivo. Mol Cell Biol. 1998;18:5600–5608. doi: 10.1128/mcb.18.9.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- Carson MJ, Hartwell L. CDC 17: An essential gene that prevents telomere elongation in yeast. Cell. 1985;42:249–257. doi: 10.1016/s0092-8674(85)80120-3. [DOI] [PubMed] [Google Scholar]
- Choi SK, Lee JH, Zoll WL, Merrick WC, Dever TE. Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science. 1998;280:1757–1760. doi: 10.1126/science.280.5370.1757. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Wright JH, Wolf AJ, Zakian VA. RAP1 protein interacts with yeast telomeres in vivo: Overproduction alters telomere structure and decreases chromosome stability. Cell. 1990;63:739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- Counter CM, Meyerson M, Eaton EN, Weinberg RA. The catalytic subunit of yeast telomerase. Proc Natl Acad Sci. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- Dionne I, Wellinger RJ. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc Natl Acad Sci. 1996;93:13902–13907. doi: 10.1073/pnas.93.24.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- Fan X, Price CM. Coordinate regulation of G- and C-strand length during new telomere synthesis. Mol Biol Cell. 1997;8:2145–2155. doi: 10.1091/mbc.8.11.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song O-K. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Garrett S, Menold MM, Broach JR. The Saccharomyces cerevisiae YAK1 gene encodes a protein kinase that is induced by arrest early in the cell cycle. Mol Cell Biol. 1991;11:4045–4052. doi: 10.1128/mcb.11.8.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis EA, Gyuris J, Brent R. Interaction trap/ two-hybrid system to identify interacting proteins. In: Ausubel FM, Brent R, Kingston R, Moore D, Seidman J, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1994. pp. 20.0.1–20.1.28. [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Grandin N, Reed SI, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes & Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- Gravel S, Larrivee M, Labrecque P, Wellinger RJ. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101C:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- Jones E. Tackling the Protease Problem in Saccharomyces cerevisiae. In: Guthrie C, Fink G, editors. Guide to Yeast Genetics and Molecular Biology. San Diego, California: Academic Press; 1991. p. 933. [Google Scholar]
- Kang HA, Hershey JW. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J Biol Chem. 1994;269:3934–3940. [PubMed] [Google Scholar]
- Khazak V, Sadhale PP, Woychik NA, Brent R, Golemis EA. Human RNA polymerase II subunit hsRPB7 functions in yeast and influences stress survival and cell morphology. Mol Biol Cell. 1995;6:759–775. doi: 10.1091/mbc.6.7.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Leung D, Chen E, Goeddel D. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- Lin J-J, Zakian VA. An in vitro assay for Saccharomyces telomerase requires EST1. Cell. 1995;81:1127–1135. doi: 10.1016/s0092-8674(05)80017-0. [DOI] [PubMed] [Google Scholar]
- ————— The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR, Hughes TR, Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci. 1997a;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997b;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Lucchini G, Mazza C, Scacheri E, Plevani P. Genetic mapping of the Saccharomyces cerevisiae DNA polymerase I gene and characterization of a pol1 temperature-sensitive mutant altered in DNA primase-polymerase complex stability. Mol Gen Genet. 1988;212:459–465. doi: 10.1007/BF00330850. [DOI] [PubMed] [Google Scholar]
- Lucchini G, Falconi MM, Pizzagalli A, Aguilera A, Klein HL, Plevani P. Nucleotide sequence and characterization of temperature-sensitive pol1 mutants of Saccharomyces cerevisiae. Gene. 1990;90:99–104. doi: 10.1016/0378-1119(90)90444-v. [DOI] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Gilson E. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 1999;18:3509–3519. doi: 10.1093/emboj/18.12.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Replica plating. In: Miller JH, editor. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. pp. 56–59. [Google Scholar]
- Mitchell DA, Marshall TK, Deschenes RJ. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–723. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- Nugent CI, Lundblad V. The telomerase reverse transcriptase: Components and regulation. Genes & Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- Pizzagalli A, Valsasnini P, Plevani P, Lucchini G. DNA polymerase I gene of Saccharomyces cerevisiae: Nucleotide sequence, mapping of a temperature-sensitive mutation, and protein homology with other DNA polymerases. Proc Natl Acad Sci. 1988;85:3772–3776. doi: 10.1073/pnas.85.11.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polotnianka RM, Li J, Lustig AJ. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol. 1998;8:831–834. doi: 10.1016/s0960-9822(98)70325-2. [DOI] [PubMed] [Google Scholar]
- Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Schulz VP, Zakian VA. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner BR, Hidaka K, Futcher B. Association of the Est1 protein with telomerase activity in yeast. Proc Natl Acad Sci. 1996;93:2817–2821. doi: 10.1073/pnas.93.7.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virta-Pearlman V, Morris DK, Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes & Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- Watt PM, Louis EJ, Borts RH, Hickson ID. Sgs1: A eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–60. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Wolf AJ, Zakian VA. Origin activation and formation of single-strand TG1-3 tails occur sequentially in late S phase on a yeast linear plasmid. Mol Cell Biol. 1993a;13:4057–4065. doi: 10.1128/mcb.13.7.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell. 1993b;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Ethier K, Labrecque P, Zakian VA. Evidence for a new step in telomere maintenance. Cell. 1996;85:423–433. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]
- Wright JH, Zakian VA. Protein–DNA interactions in soluble telosomes from Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:1454–1460. doi: 10.1093/nar/23.9.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JH, Gottschling DE, Zakian VA. Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes & Dev. 1992;6:197–210. doi: 10.1101/gad.6.2.197. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Shin TH, Galova M, Obermaier B, Nasmyth K. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science. 1996;274:1201–1204. doi: 10.1126/science.274.5290.1201. [DOI] [PubMed] [Google Scholar]
- Zhou J, Hidaka K, Futcher B. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol Cell Biol. 2000;20:1947–1955. doi: 10.1128/mcb.20.6.1947-1955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YH, Zhang XP, Ebright RH. Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res. 1991;19:6052. doi: 10.1093/nar/19.21.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]