Abstract
Kinesin-1 is an ATP-driven, processive motor that transports cargo along microtubules in a tightly regulated stepping cycle. Efficient gating mechanisms ensure that the sequence of kinetic events proceeds in proper order, generating a large number of successive reaction cycles. To study gating, we created two mutant constructs with extended neck-linkers and measured their properties using single-molecule optical trapping and ensemble fluorescence techniques. Due to a reduction in the inter-head tension, the constructs access an otherwise rarely populated conformational state where both motor heads remain bound to the microtubule. ATP-dependent, processive backstepping and futile hydrolysis were observed under moderate hindering loads. Based on measurements, we formulated a comprehensive model for kinesin motion that incorporates reaction pathways for both forward and backward stepping. In addition to inter-head tension, we find that neck-linker orientation is also responsible for ensuring gating in kinesin.
Kinesin-1 is a motor protein that proceeds unidirectionally towards the plus end of the microtubule in discrete, 8.2-nm steps1 by alternately advancing each of its two heads, walking in an asymmetric, hand-over-hand fashion2–4. Kinesin motion is remarkably processive, with single motors typically taking ~100 steps before detaching from the microtubule5. The processivity and thermodynamic efficiency of kinesin-1 arise from a coordinated mechanochemical cycle in which a single ATP molecule is hydrolyzed per mechanical step6–8. Deviations from that cycle, such as those resulting in backsteps towards the microtubule minus end, occur only rarely, or in response to external loads that approach or exceed the stall force9. Therefore, the kinesin molecule must employ gating mechanisms to coordinate its catalytic heads and move efficiently10.
Mechanical strain developed within the kinesin molecule is thought to underlie the mechanism by which kinesin gates the enzymatic activity of its heads11–13. Within the crystal structure of the kinesin dimer14, the microtubule-binding domains of the twin heads are separated by ~5 nm. When both heads bind strongly to the microtubule, they become separated by a distance corresponding to the tubulin-dimer lattice spacing of 8.15 nm. Substantial inter-head tension is consequently thought to be developed between the heads via the neck-linker regions: molecular dynamics simulations suggest that 15–35 pN of tension may be produced15,16. There are two competing models explaining the roles that tension and conformational changes domains play in gating. Generally speaking, front head-gated models propose that tension reduces the binding affinity of the front head for ATP until the rear head has detached from the microtubule12,17,18. Rear head-gated models propose that tension from the front head accelerates the detachment of the rear head13,19,20. These models are not mutually exclusive, and kinesin may utilize both mechanisms to maintain coordination during processive stepping. Loss of coordination between the heads can lead to conformational states that promote detachment from the microtubule, backward steps, and/or futile hydrolysis cycles. Investigations into the prevalence of backward steps near the stall force21 and beyond22 have demonstrated that ATP binding, but not necessarily hydrolysis, is required to backstep. Various kinetic models have been advanced to explain this process22–24, but the rate equations implied by such models have not yet been shown to describe quantitatively the detailed data available from single-molecule studies, such as velocities under load, ratios of forward to backward steps, and randomness parameters.
The neck-linker consists of a short, largely unstructured ~14 amino acid sequence that connects each of the kinesin heads to the common stalk, and it has been implicated in the conformational change thought to drive motility25. To better understand gating mechanisms and kinetics in the kinesin enzymatic cycle, we lengthened the neck-linker, introducing six additional amino acids with a net neutral charge (AEQKLT) into the C-terminal portion of the linker region in a truncated construct based on the human kinesin heavy chain, expressed in bacteria. When hindering loads were applied to this construct (Kin6AA) in a single-molecule optical-trapping assay, the motion was found to consist of an admixture of forward and backward steps, in a proportion that was strongly force dependent. Essentially identical results were obtained with a construct carrying a 9-residue insert with 6 adjacent prolines in its linker (Kin6P). In contrast to wild-type (WT) kinesin, the stall force of the mutant constructs varied greatly with the ATP concentration, and the average velocity smoothly reversed and became negative as the hindering load was increased beyond stall. Comparisons of data from the mutant and WT kinesin molecules led to the development of a unified, five-state minimal kinetic model that accounts quantitatively for both forward and backward stepping by kinesin.
RESULTS
Kin6AA moves backwards processively
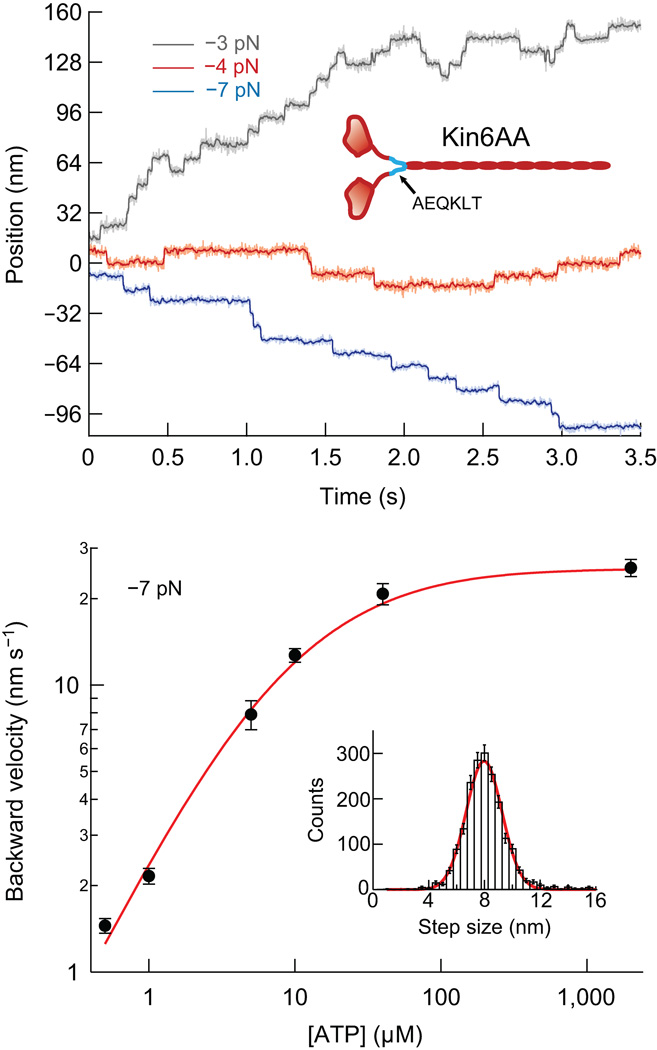
The stepping dynamics of Kin6AA were studied using an optical force clamp, which employs feedback to servo the optical trap to a fixed distance from the bead center, maintaining constant force, of either sign, during subsequent motion26,27. Representative displacement records for Kin6AA under different loads are presented in Figure 1a. The traces display a striking difference from those obtained with WT kinesin with respect to the frequency of backstepping. Whereas WT kinesin backsteps only rarely, with 2–4% probability when subjected to ~3 pN hindering load8,21,22,28, Kin6AA backsteps ~20% of the time under comparable forces. The prevalence of backsteps in the mutant gives experimental access to kinetic states that are too rare or too transient in the WT for practical study.
Figure 1.
Single-molecule records and backstepping velocity for Kin6AA. (a) Representative traces obtained under constant force at loads of −3 pN (gray), −4 pN (red), and −7 pN (blue), at 2 mM ATP. Light traces are unfiltered, darker traces are median-filtered. The records show clear 8-nm forward and backward steps. (b) Double-log plot of backstepping velocity vs. ATP concentration under −7-pN load (black dots; mean ± s.e.m.; N = 21−62). The solid line (red) shows the global fit to all single-molecule data for the model described in the text. Inset, histogram of the backward step size at a −7-pN load for all ATP concentrations (8.12 ± 1.72 nm, mean ± s.d., N = 1,906) and Gaussian fit (red line), centered at 7.97 nm.
The differences between Kin6AA and WT kinesin became more dramatic as load was increased. At forces approaching stall with WT kinesin, processivity is lost and motors typically display small numbers of forward and backward steps before detaching from the microtubule21. By contrast, Kin6AA motors approaching stall remained bound to the microtubule while undertaking numerous forward and backward steps with equal probability, leading to a large mean-squared displacement but no net progress (Fig. 1a, red trace). As the force was increased beyond stall, Kin6AA began to step rearwards processively, with a slow velocity that was strongly dependent upon the ATP concentration, but comparatively insensitive to load (Figs. 1b and 2a). The velocities for processive backstepping were broadly consistent with rates previously measured for WT kinesin subjected to super-stall loads22, supporting the notion that backsteps in Kin6AA and WT kinesin arise from a common molecular mechanism. The backward step size for Kin6AA averaged 8.12 nm, indicating that the motor accesses the same microtubule binding sites as during forward stepping (Fig. 1b, inset). In contrast to one study that employed much longer inserts in the neck-linker13, steps that were integral multiples of the 8 nm spacing were not prevalent for Kin6AA. Although the velocity data (Fig. 1b) displayed ATP dependence suggestive of Michaelis-Menten kinetics, these and other single-molecule data (Figs. 1 and 2) were globally fit to a more detailed five-state model.
Figure 2.
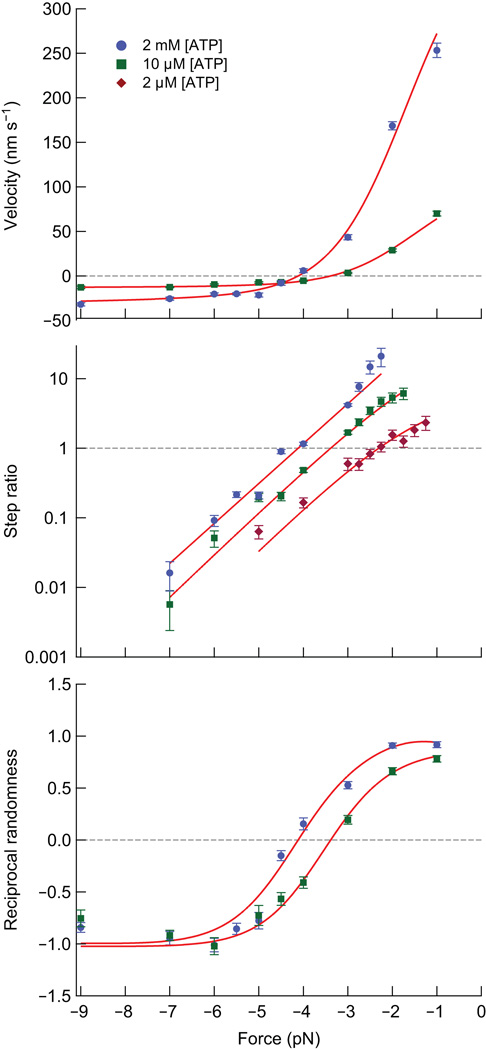
Bi-directionality of Kin6AA as a function of load and ATP concentration. Solid lines (red) show the global fits to all single-molecule data for the model described in the text. (a) Velocity, v (mean ± s.e.m.) vs. force for 2 mM ATP (blue circles; N = 25−164) and 10 µM ATP (green squares; N = 18−74). Stall occurs where the fit data cross the horizontal dashed line (grey) at v = 0. (b) Step ratio, SR (mean ± s.e.m.; ratio of number of forward to backward steps) vs. force at 2 mM ATP (blue circles; N = 264−3,331), 10 µM ATP (green squares; N = 235−1,412), and 2 µM ATP (purple diamonds; N = 90−368). Stall occurs where the fit data cross the horizontal dashed line (grey) at SR = 1. (c) Reciprocal randomness, r−1, (mean ± s.e.m.), color-coded as in (a). Note that the fit data cross r−1 = 0 at the stall forces for the data in (a).
The KIN6AA stall force is ATP-dependent
The neck-linker insert exerted a substantial effect on the stall force. In contrast to WT kinesin, the stall force for Kin6AA depended sharply upon the ATP concentration, as evidenced by plots of the velocity and forward/backward step ratio (Fig. 2a,b). Here, a stall was operationally defined as either the force where the mean velocity, v, dropped to zero or the force where the ratio of forward to backward steps (the step ratio, SR) became unity. These metrics were mutually consistent (Fig. 2a,b), with v = 0 and SR = 1 occurring at the identical force for each ATP concentration.
The randomness constrains possible models
In addition to v and SR, we computed the randomness parameter, r, which is a statistical measure of the regularity of steps8,29–31. For a clock-like process with identical stepping times, r = 0. A Poisson process with exponentially distributed times (and no backsteps) produces a value of r = 1. The reciprocal randomness, r−1, has been used to distinguish between competing kinetic pathways for kinesin8,32. In the case of an unbranched kinetic cycle, the maximal value of r−1 supplies the minimum number of rate-limiting transitions. In practice, distributions of dwell times can vary enormously for different kinetic models with otherwise identical cycle times, requiring a metric such as randomness to differentiate among them33. A plot of r−1 versus load for two different ATP concentrations is shown in Figure 2c. Because r−1 is proportional to the mean velocity, the randomness value changes sign and crosses the abscissa at stall.
Mechanochemistry: The forward stepping pathway
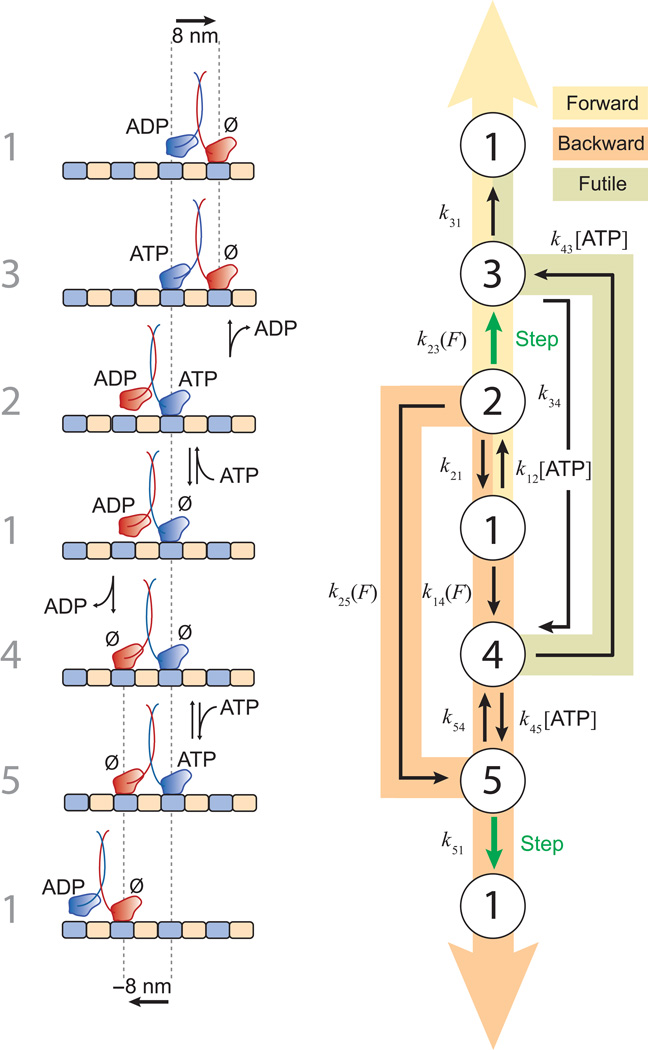
Five states proved necessary to account for the range of single-molecule data acquired in these experiments. The data (Figs. 1 and 2) were globally fit by a model that incorporates explicit reaction pathways for both forward and backward steps. A block diagram of the kinetic scheme, together with the molecular configurations presumed to correspond to these states, is shown inFigure 3. The pathway begins at an arbitrary starting point, labeled State {1}, which represents the so-called ‘ATP waiting state’34–36. At low ATP concentrations, WT kinesin is found predominantly in this state, where the leading head is without nucleotide and strongly bound to the microtubule, while the trailing head, carrying ADP, remains detached from the microtubule34, or is perhaps transiently (weakly) associated36,37. Because Kin6AA carries an extended linker designed to relieve inter-head tension, it is possible, in principle, for its trailing head to bind and unbind the microtubule transiently from this state, provided that this conformation is not excessively prone to releasing its bound ADP. This characteristic permits State {1} to serve a common starting point for both forward and backward step cycles. The choice of a common starting point is consistent with a Markov property of stepping, which was confirmed experimentally: the probability of a step in any direction is uncorrelated with the direction of the preceding step (Supplementary Fig. 1). In the forward-stepping pathway (Fig. 3b, yellow), ATP binding to the leading head advances the motor from State {1} to State {2}. Next, a largely irreversible, load-dependent forward step occurs between States {2} and {3} as the leading and trailing heads swap positions and ADP is released. The motor returns to State {1} and completes its cycle after subsequent ATP hydrolysis followed by Pi release. Both here and elsewhere, multiple biochemical transitions occurring in rapid succession have been lumped into single states to form a minimal model. Similar, unbranched reaction pathways for forward steps have been proposed38 based on the available mechanochemical, biochemical, and structural data, and an analogous 3-state cycle was used to model single-molecule data32. However, these pathways do not account for backstepping and futile hydrolysis.
Figure 3.
Model for stepping by kinesin dimers, showing forward stepping, backward stepping, and futile hydrolysis pathways. See text. (a) Left, the numbers assigned to each of the five states. The molecular configurations of the kinesin dimer on the microtubule thought to correspond to each of the states are illustrated, along with any nucleotides bound. No particular docking state of the neck-linker is implied in this diagram. Kinesin heads are color-coded (red, blue). Starting from State {1} (middle row), forward steps are accomplished by ascending the diagram; backward steps by descending. (b) Reaction diagram for the model, displaying the transition rates among states. Load- and ATP-dependent transitions are indicated. Three main pathways are shaded: forward stepping (yellow), backward stepping (orange), and futile hydrolysis (light green). Largely irreversible transitions between states that produce ±8-nm displacements are shown (green arrows). In this minimal model, fast transitions occurring in rapid succession were combined to generate composite states in several instances. Note: The transition from State {4} to State {3} in the futile hydrolysis pathway involves ATP binding to the rear head, but, unlike the stepping pathways, heads do not swap positions and no step is taken.
Mechanochemistry: The backward stepping pathway
Unlike forward steps, backward steps may be completed following alternative, branched kinetic paths (Fig. 3b, orange). Beginning from State {1}, two general properties must be satisfied to generate a successful backstep. First, the trailing head must be able to bind the microtubule to generate a two-heads-bound state, leading to release of ADP from the rear head. Second, the backward cycle must incorporate an ATP binding event to account for the ATP dependence of processive backstepping (Fig. 1b). In the model, reversible ATP binding can occur in two ways: either prior to the rear head binding the microtubule, following the sequence {1}→{2}→{5}→{1}, or afterwards, following the sequence {1}→{4}→{5}→{1}. The existence of alternative backstepping pathways results in a forward/backward step ratio exhibiting ATP dependence (Fig. 2b). Unbranched cycles, which lack this property, have been proposed to explain the much weaker ATP dependence of the step ratio for WT kinesin22,23, which, in the context of this model, predominantly follows the sequence {1}→{2}→{5}→{1}. From State {5}, which is a state unique to the backstepping pathway, we propose that ATP hydrolysis by the front head leads to a weakly-bound ADP state and head detachment, and thereby promotes backstepping, leading to a return to State {1}.
Inclusion of futile hydrolysis
The model also incorporates a pathway for futile hydrolysis, where ATP binds the trailing head, following the sequence {4}→{3}→{1} (Fig. 3b). Absent such a pathway, the only route of departure from State {4} is via an obligatory backstep, which has a maximal rate of ~3 s−1 under saturating ATP conditions (Fig. 1b). Without futile hydrolysis, the slow backstepping rate would result in extended dwell times, yielding small values of r−1 at low forces, which were not observed (Fig. 2c). The inclusion of a futile hydrolysis cycle leads to an accurate fit of the randomness data. Direct evidence for futile hydrolysis by Kin6AA is provided by the ensemble ATPase data, where the measured rate of ATP hydrolysis was 143 s−1 molecule−1 (Supplementary Fig. 2). Because the unloaded velocity of Kin6AA is 40 steps s−1 under saturating ATP conditions, an average of 3.5 ATP molecules are hydrolyzed per forward step. Futile hydrolysis is suppressed in the WT, leading to greater efficiency and ~1 ATP per forward step6–8.
Analytical expressions for the velocity, randomness, and step ratio were developed from the cycle of Figure 3b following the approach of Chemla et al.31 Equations for v, r, and SR in terms of transition rates between the states, kij, and the ATP concentration are found in Supplementary Methods. A global fit to the data (Figs. 1 and 2) was carried out to determine all transition rates. The three transition rates that involve taking a forward step (k23) or rebinding the rear head to the microtubule during a backstep (k14, k25) were found to carry load dependence an"d were modeled by Boltzmann expressions, , where δij is a characteristic distance. This distance was a free parameter in the fits. Values for the rates and distances are found in Table 1. We note that a state where both heads bind ATP and remain bound to the microtubule was not included in the model. The existence of such a state would introduce one additional pathway for futile hydrolysis as well as one for backstepping, increasing the model complexity but without adding any new observable features. Additional states or transitions can always be added, in principle, but the sequence of Figure 3 is a minimal scheme that effectively captures the experimental data for processive stepping. We have not, however, modeled any branching reaction paths that lead to loss of processivity through head release and microtubule dissociation.
Table 1.
Kinetic parameters, measured and fit
| Parameter | Description | Global fit value for model |
Ensemble measurement |
Comment |
|---|---|---|---|---|
| k12, k45 | ATP binding rates to front head | 3.7 ± 0.5 µM−1s−1 | 2.6 ±0.3 µM−1 s−1 | ATP binding rate, average of both headsa |
| k43 | ATP binding rate to rear head | 4.0 ± 0.8 µM−1s−1 | ND | Transition leading to futile hydrolysis |
| k21, k54 | ATP dissociation rates from front head | 68 ± 10 s−1 | 55–135 s−1 | ATP dissociation rate, average of both headsa,b |
| k34 | ATP dissociation rate from rear head | 12 ± 7 s−1 | ND | ATP unbinding rate from read head |
| k23(F) | Forward step and ADP release | 570 ± 90 s−1 | ND | Load-dependent step transition; value for F = 0 |
| δ23 | Characteristic distance for k23(F) | 4.3 ± 0.2 nm | ND | Transition distance leading to a forward step |
| k31 | ATP hydrolysis, Pi release, and dissociation of the rear head | 57 ± 5 s−1 | 79 ± 4 s−1 | Head dissociation ratec |
| k51 | ATP hydrolysis, Pi release, front head | 6.5 ± 0.4 s−1 | ND | ATP hydrolysis and backstep |
| k14(F), k25(F) | Rates for rear head rebinding to microtubule | 14 ± 3 s−1 | 11 ± 4 s−1 | Load-dependent head rebinding ratesc; value for F = 0 |
| δ14, δ25 | Characteristic distances for k14(F), k25(F) | −1.0 ± 0.2 nm | ND | Transition distance leading to a backward step or futile hydrolysis |
Ensemble values were derived from afit to 2′dmT data, initial exponential increase (Fig. 5b) and cfit to the concentration dependence of initial TMR decay (Fig. 5c). Rates that describe comparable processes (k12 and k45, k21 and k54, k14 and k25) were set to be equal during global fitting.
The range of values for the ATP dissociation rate constant based on ensemble measurements was calculated as described in Supplementary Methods. ND, Not Determined.
Ensemble experiments
Results from fits to the model in Figure 3 supply predictions about molecular configurations and transitions between states. To test such predictions independently, we performed ensemble kinetic experiments using a stopped-flow fluorimeter. One of the key features distinguishing the present model from earlier representations of the kinesin cycle is State {4}, where both heads bind the microtubule in rigor. It has long been known39 that kinesin dimers labeled with a fluorescent ADP analog will release just one ADP molecule upon binding the microtubule, while release of the second ADP requires the further addition of ATP39,40. However, when Kin6AA with 2′–deoxy 3′–mant ADP (2′dmD) bound was rapidly mixed with microtubules, both bound nucleotides were released together (Supplementary Fig. 3). This finding supports a two-heads-bound state. The simultaneous release of both nucleotides, with consequent support for a two-heads-bound state, was previously reported by Hackney and coworkers for a different mutant construct that also carried an extended neck-linker.41
Two-heads-bound state studied using TMR
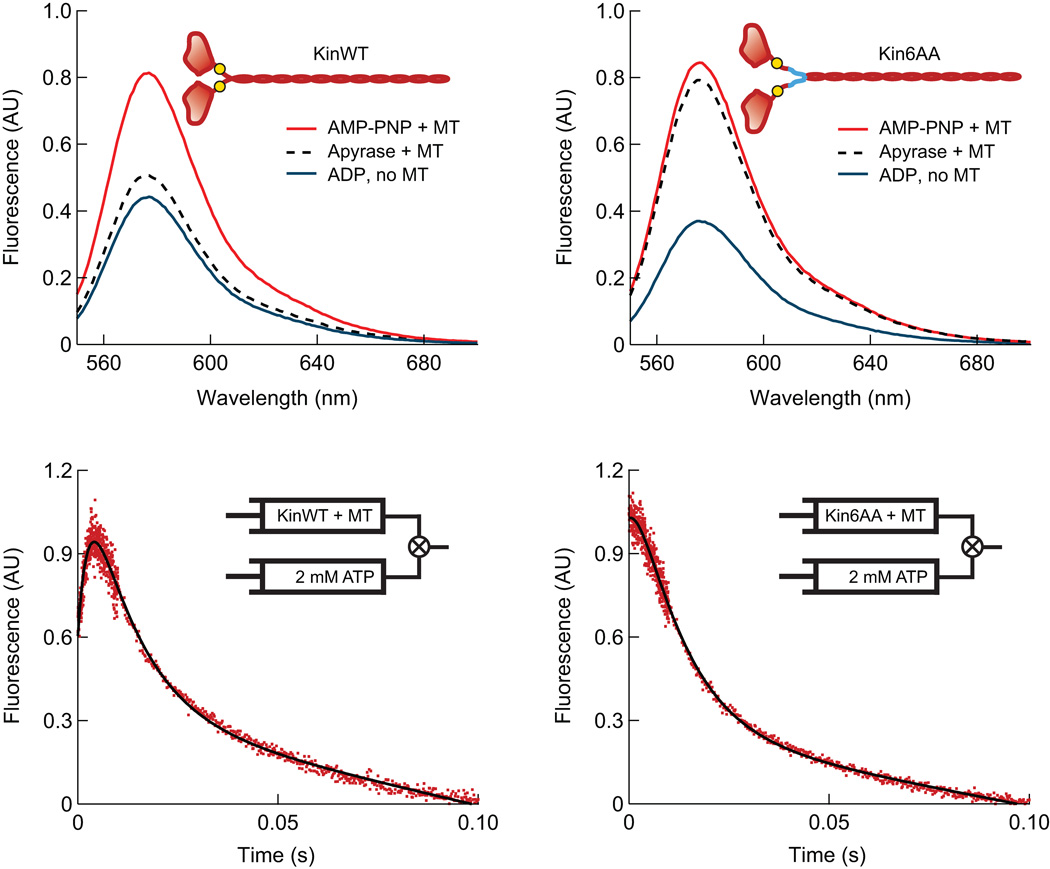
Further structural and kinetic evidence for State {4} came from steady-state and kinetic ensemble experiments employing tetramethylrhodamine (TMR) labels (Fig. 4). TMR dyes were covalently linked to a cysteine residue introduced at amino-acid position 333, in the middle of the neck-linker, in both Kin6AA and KinWT constructs (cys-light versions of human kinesin 1; Supplementary Methods). TMR dyes positioned in sufficiently close proximity42 (~1 nm) interact in an orientation that quenches fluorescence. We had previously shown that separation of these dyes occurs when WT kinesin takes a forward step, and results in a large fluorescence enhancement12. Previous experiments with kinesin have shown that both motor domains bind tightly to the microtubule in the presence of the non-hydrolyzable analog, AMP–PNP43. TMR-labeled KinWT produced a large fluorescence signal when incubated with AMP–PNP and microtubules (Fig. 4a), indicating that its neck-linkers separate when the motor binds the microtubule. An intermediate-level signal was measured for KinWT in the absence of nucleotide, however, indicating a closer average neck-linker separation. This finding is consistent with a single head being strongly bound to the microtubule while its partner remains tethered, or alternatively, with an admixture of singly- and doubly-bound head states43,44. By contrast, Kin6AA not only permits neck-linker separation on microtubules in the presence of AMP–PNP, but also in the absence of nucleotide (Fig. 4b).
Figure 4.
Fluorescence data for Kin6AA and KinWT with TMR probes attached to both neck-linkers. (a, b) Steady-state TMR fluorescence emission spectra for KinWT (a) and Kin6AA (b), which monitors neck-linker separation under the following conditions: microtubules plus 2 mM AMP–PNP (red), microtubules plus apyrase to remove nucleotides (black, dashed), and 2 mM ADP without microtubules (blue). The large signal increase in the absence of nucleotide (apyrase present) for Kin6AA is consistent with neck-linker separation. In the inset cartoons, approximate locations of the TMR probes (at position 333) are indicated (yellow circles), as well as the neck-linker inserts (blue lines). (c, d) Pre-steady-state TMR kinetic records for KinWT and Kin6AA. TMR-labeled motors complexed to a five-fold excess of microtubules and treated with apyrase were mixed with 2 mM ATP. The initial increase in fluorescence seen for KinWT is absent for Kin6AA, indicating that prior to mixing with ATP, both heads of Kin6AA are bound to the microtubule, and consequently, the neck-linkers of this mutant are separated.
Transient-state kinetic studies of KinWT and Kin6AA (Fig. 4c,d) also support the existence of a two-heads-bound state. TMR-labeled KinWT or Kin6A were complexed with an excess of microtubules and rapidly mixed with ATP. When this procedure was performed using KinWT, a rapid initial rise in fluorescence was observed, followed by a slower exponential fall (Fig. 4c). The initial increase reflects the separation of the neck-linkers, and its kinetics are consistent with ATP-induced forward stepping12,37. The subsequent decay occurs in two phases. The faster phase constitutes 80–85% of the total amplitude of the decay phase, and it has kinetics consistent with the dissociation of the new trailing head from the microtubule to regenerate a singly-attached species. That species consists of a strongly-bound leading head and a trailing head that is unbound and mobile, or possibly in an admixture of unbound and weakly-attached states that interconvert. The slower phase has kinetics consistent with the subsequent dissociation of the motor from the microtubule, because this phase is absent when the experiment is repeated in low ionic-strength buffer12. The data also suggest that the TMR dyes are in rapid equilibrium between quenched and unquenched states prior to the addition of ATP, as anticipated for a motor in a one-head, strongly-bound state. Notably, the trace for Kin6AA (Fig. 4d) lacked any initial rising phase, indicating that prior to ATP binding, its neck-linkers were fully separated. Combined with the finding that both heads are nucleotide-free in this state (Supplementary Fig. 3), this result suggests that both heads are bound to the microtubule in rigor. After a delay of ~2 ms, fluorescence was quenched at an initial rate that depended upon ATP concentration. Fitting the concentration dependence of this decay to a hyperbola generates a curve with a y-intercept of 11 ± 4 s−1 (Fig. 5), which we interpret as the rate at which the trailing head rebinds the microtubule.
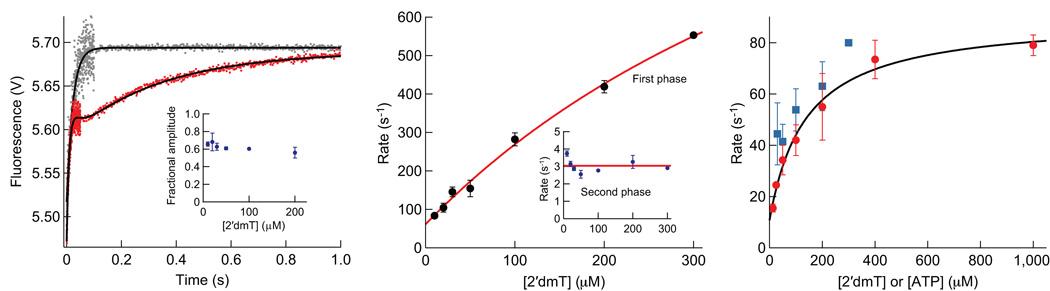
Figure 5.
Binding of 2′dmT to Kin6AA. (a) A complex of Kin6AA and microtubules was pre-formed and mixed in a stopped-flow apparatus with 2′dmT (Supplementary Methods). The resulting fluorescence signal (red) consisted of three sequential phases: a first phase of increasing fluorescence, a lag phase, and a second phase of increasing fluorescence. Fitting this signal required three exponential terms (black curve). Two terms, corresponding to the phases of increasing fluorescence, were associated with rate constants of 81.5 ± 21.0 s−1 and 3.0 ± 0.1 s−1. The third term was associated with a low-amplitude, decreasing phase, consistent with a lag, and a rate constant of 55.6 ± 24.8 s−1. The same experiment in the absence of microtubules (grey) produced a fluorescence increase with a single exponential phase with a rate constant of 45.0 ± 1.0 s−1. Inset: Fractional amplitude of the first phase vs. [2′dmT]. (b) Rate constant for the first phase of fluorescence increase vs. [2′dmT]. Data (black dots; mean ± s.e.m.) were fit to a hyperbola (red curve) that extrapolates to 61 ± 12 s−1 at zero [2′dmT] and is associated with a second-order rate constant of 2.6 ± 0.3 µM−1 s−1. Inset: Rate constant for the second phase of fluorescence vs. [2′dmT], which averages 3.0 ± 0.4 s−1 (red line) (c) Rate of initial decay of TMR fluorescence as a function of [ATP], compared to the rate of the lag phase in (a). Data (mean ± s.e.m.; N = 18–35; red dots) were fit to a rectangular hyperbola (black curve); the asymptotic rate at saturating ATP was 90 ± 4 s−1. The y-intercept of the fit at 11 ± 5 s−1 is interpreted as the rate at which a head rebinds to the microtubule. Rate constant for the lag phase vs. [2′dmT] (mean ± s.e.m.; N = 10–20; blue squares).
Binding of a fluorescent ATP analog suggests Kin6AA is gated
We monitored the binding of 2′–deoxy 3′–mant ATP (2′dmT) to investigate the binding and release of ATP to the heads. FRET between intrinsic tryptophan residues (in the motor domain and microtubule) and the mant fluorophore in 2′dmT produces fluorescence when binding occurs. Figure 5a displays the signal produced when Kin6AA was complexed with an excess of microtubules and rapidly mixed with 2′dmT in the stopped-flow spectrophotometer. The fluorescence changes progress in three phases. A rapid initial increase could be fit to a single exponential process, with a rate constant that displayed a hyperbolic dependence on 2′dmT concentration (Fig. 5b). Next came a brief lag phase, followed in turn by a second, slower exponential increase with a rate constant of ~3 s−1 that displayed little 2′dmT concentration-dependence (Fig. 5b, inset). Both phases of increasing fluorescence were of similar magnitude for all 2′dmT concentrations (Fig. 5a, inset). These results suggest that nucleotide binding occurs sequentially to the two heads when these are bound to the microtubule. This interpretation is bolstered by the observation that mixing Kin6AA with 2′dmT in the absence of microtubules produces a rapid increase in fluorescence consisting of a solitary phase, with amplitude similar to that in the presence of microtubules (Fig. 5a, grey). Furthermore, the rate constant associated with the lag phase (Fig. 5c, blue) closely matches the corresponding rate constant for dissociation of the trailing head, as measured by the TMR transient (Fig. 5c, red). Because both TMR- and 2′dmD-release experiments indicate that Kin6AA starts from a two-heads-bound state prior to addition of nucleotide (State {4}), the results from this experiment imply that the Kin6AA heads continue to be gated, despite any possible reduction in inter-head tension due to the neck-linker extension.
Additional tests of the model
All rates and load dependencies for the five-state model were determined by a global fit to the single-molecule data for the backward velocity, step ratio, and randomness as functions of load and ATP concentration for Kin6AA (Figs. 1 and 2 and Table 1). In all, ten parameters were fit. Direct, independent measurements of four of these rates were obtained from the ensemble kinetic measurements of Kin6AA, and these were in close agreement with the globally fit values (Table 1). In addition, the single-molecule dataset for the unloaded forward velocity of Kin6AA as a function of ATP was not used to fit the parameters of the model. The model should therefore predict this function independently, with no adjustable parameters: the agreement between the data and prediction was excellent (Supplementary Fig. 4).
DISCUSSION
Kinesin is gated by strain and by steric effects
The kinetics implied by the mechanochemical model and associated single-molecule data (Fig. 3), the TMR data, and the 2′dmT binding experiments all suggest that kinesin employs multiple gating strategies to maintain coordination between its heads during a processive run. The TMR data provide direct evidence that extending the neck-linkers allows both Kin6AA heads to attach stably to the microtubule in rigor (State {4}). This conformational state is partially suppressed in WT kinesin, which tends to adopt a configuration where just one head is strongly attached to the microtubule39,40, while the partner head, carrying ADP, is more-or-less free of the microtubule34–36, particularly at low ATP concentrations. The inhibition of the rear head from binding leads to efficient, unidirectional forward motion by reducing backstepping, futile hydrolysis, and access to states where both heads are released, leading to loss of processivity34. The experiments with Kin6AA demonstrate that moderately lengthening the neck-linker allows the trailing head to bind strongly to the microtubule and release ADP, yet retain substantial aspects of gating. Indeed, the processivity of Kin6AA exceeds that of the KinWT construct, supporting long runs that average over 150 steps (Supplementary Fig. 5). However, a price is clearly paid, through increased backstepping under load and marked reductions in both speed and stall force, along with energy efficiency. Our results do, however, support a mechanism whereby attachment of the trailing head is strain-based, with the accessibility of this state being promoted by a reduction in inter-head tension.
Neck-linker orientation also gates kinesin
Although an extended neck-linker is thought to mitigate the effectiveness of strain-based gating mechanisms13,41, our results clearly indicate that Kin6AA is still well gated, and therefore support the notion that there is more to gating than the tension exerted between heads13,34. The 2′dmT binding data reveal that even when both heads bind the microtubule, nucleotide binding is sequential. The rates fit to the five-state model also show that heads maintain excellent coordination. From the data in Table 1, the average rate of ATP hydrolysis by the trailing head is nearly 9-fold faster than the corresponding rate for the leading head. Similarly, the model predicts that the average rate of ATP unbinding is almost 6-fold faster in the front than the rear. The picture that emerges is that when the neck-linker is in its forward (docked) orientation, ATP hydrolysis is fast and ATP unbinding is slow. Conversely, when the neck-linker is in a rearward orientation, ATP hydrolysis is slow and ATP unbinding is fast. In effect, some conformational change associated with neck-linker docking (or simply orientation) ‘locks’ ATP in place in the rear head and efficiently hydrolyses it before ATP tightly binds to the front head. We emphasize that this gating mechanism does not require substantial inter-head tension to function, and is most easily understood in a two-heads-bound state, where the neck-linker of the trailing head is free to dock, but the neck-linker of the leading head is restricted from so doing, because it is oriented backwards (i.e., unfavorably) by the trailing head.
Such a mechanism is fully consistent with a model conjectured previously to explain gating in WT kinesin at low ATP concentrations, where only one head is generally bound34. It is also consistent with the kinetics of 2′dmT binding to Kin6AA (Fig. 5), as compared to KinWT12. The binding of 2′dmT to Kin6AA occurs in two steps, with similar amplitudes that are separated by a lag (Fig. 5a). The kinetics of the lag (Fig. 5c, blue) strongly suggests that it corresponds to the dissociation of one of the two heads, as monitored by TMR fluorescence (Fig. 5c, red). This result implies that binding of 2′dmT to the second head can only occur after dissociation of the first head, and the concomitant reduction of any inter-head tension imposed by microtubule binding. In Kin6AA, the reorientation of the head with a lengthened neck-linker (in the absence of any external load) is a comparatively slow process, (~3 s−1, Fig. 5b inset) and it is this process, and not the relief of inter-head tension, per se, that gates ATP binding. We had previously noted that in KinWT, by contrast, the change in neck-linker orientation associated with head repositioning is at least two orders of magnitude faster37. We therefore propose that it is the orientation of the neck-linker, and not inter-head tension, that forms the basis for gating in kinesin. This proposal is supported by recent single-molecule and ensemble-fluorescence measurements37 indicating that the gating of ATP binding to the leading head requires neck-linker separation, but does not require strong binding of both heads to the microtubule. It is also supported by results showing that in the ATP-waiting state, the trailing head carrying ADP is mobile and free of the microtubule34, and therefore unable to exert substantial tension on the leading head. A neck-linker orientation-based gating mechanism can explain the large processivity of native kinesin, even under force45, since it reduces the probability of ATP hydrolysis until a mechanical step has been completed.
When kinesin binds a non-hydrolyzable ATP analog, such as AMP–PNP or ADP–BeFx, its motion becomes arrested, and an obligatory backstep becomes necessary to unbind the analog before allowing any forward motion to continue in the presence of ATP17. It was concluded that the analog could only be released from the leading head, and not the trailing one, necessitating a backstep to interchange head positions whenever the (former) trailing head bound analog. Because such analogs are preferentially released from the leading head, these results were interpreted as evidence for front-head gating. Our results are fully consistent with that interpretation, since the predicted rate of nucleotide unbinding is almost six-fold faster in the leading than the trailing head. Therefore, a backstep should efficiently release the bound nucleotide. Although inter-head tension was invoked to explain why the front head had a reduced nucleotide affinity, we note that these findings are equally consistent with a mechanism involving gating through neck-linker orientation.
A previous study of kinesin mutants with a series of neck-linker extensions was carried out by Yildiz et al.13. In that work, 2–26 additional prolines were inserted into the neck-linkers of kinesin, including one mutant with an extra six prolines, and the behavior was characterized by single-molecule, optical trapping assays. It was discovered that the neck-linker mutants moved uniformly slower along microtubules than wild-type kinesin, but that their velocities could be increased substantially via the application of assisting load. Based on this property, it was conjectured that additional inter-head tension, due to ATP-induced neck-linker docking, increased the rate of rear-head detachment and was largely responsible for coordination between the heads. To compare directly with our results, we prepared an analogous construct with six additional prolines (nine extra amino acids), identical in linker sequence to one of the mutants studied by Yildiz and coworkers. We confirmed that this construct stepped backwards processively in the presence of ATP, at a velocity comparable to Kin6AA, for loads beyond stall (Supplementary Fig. 6). Kin6AA and the six-proline construct displayed similar unloaded run lengths (Supplementary Fig. 5). High processivities are predicted by the model, since the mutants spend a greater portion of the reaction cycle with both heads attached to the microtubule, which reduces the chance of detachment from a one-head-bound state. Based on our results, we would interpret the decrease in velocity for the neck-linker mutants to be a consequence of their increased propensity to enter a doubly-bound state on the microtubule, rather than to a large modulation in the rear-head detachment rate, especially under hindering loads. The only escape from a doubly-bound state (Fig. 3b) is via futile hydrolysis or a backstep, both of which act to decrease the velocity and efficiency. The application of an assisting load decreases the probability that the trailing head will bind tightly to the microtubule, due to the force dependence of the rebinding rates (k14, k25), which effectively increases the average velocity: this supplies an alternate explanation for the earlier results, but without invoking inter-head tension as a gating mechanism.
The minimal (five-state) reaction cycle for kinesin stepping presented here can account successfully for a variety of single-molecule and ensemble kinetic measurements. It incorporates explicit pathways for both forward and backward stepping, as well as for futile hydrolysis. It fits well to the experimentally determined force-velocity data, as well as to the randomness and the stepping ratios as functions of load, while capturing the apparent Michaelis-Menten behavior of velocity as a function of ATP concentration. Although some biochemical states known to be distinct were modeled as composites (e.g., ADP and Pi release), the rate constants associated with such a minimal model represent the ones most directly responsible for the mechanochemistry. Presumably, the dynamics of any kinesin dimer that steps discretely along a microtubule may be modeled in similar fashion, albeit with different rates. The values for the reaction rates obtained here suggest how exquisitely tuned head coordination is in kinesin, and shed additional light on the mechanisms responsible for gating.
METHODS
Single-molecule experiments
Assays
Experimental flowcells (volume, ~10 µl) were constructed by attaching APTES (Sigma)-coated coverslips to glass microscope slides using pieces of double-sided tape (3M). Microtubules were chemically attached to the coverslip surface by crosslinking with glutaraldehyde before the surface was passivated using bovine serum albumin (BSA, Sigma). Polystyrene beads (0.44 µm diameter, Spherotech) were functionalized with penta-His antibody (Qiagen) and incubated with the appropriate kinesin construct at 4°C for a minimum of 3 hr. The motor protein-bead ratio was adjusted so that fewer than 25% of kinesin-coated beads captured by the optical trap and tested on microtubules showed motility. The buffer used for motility assays consisted of 80 mM PIPES (pH 6.9), 1 mM EGTA, 4 mM MgCl2, 50 mM potassium acetate, 2 mM dithiothreitol, 10 µM Taxol (Paclitaxel), 2 mg ml−1 BSA, and the desired concentration of ATP. Immediately prior to performing measurements, an oxygen-scavenging system was added to the buffer, consisting of 50 µg ml−1 glucose oxidase (Calbiochem), 12 µg ml−1 catalase (Sigma), and 1 mg ml−1 glucose.
Instrumentation and analysis
Loads were applied to kinesin-coated beads by an optical trapping apparatus previously described27. Data were filtered at 1 kHz, acquired at 20 kHz, decimated, and then recorded at 2 kHz. The velocities, v, were calculated from the slopes of straight line fits to single-molecule traces. Average velocities, 〈v〉, were computed as the statistical mean of all individual velocities and associated standard errors. To determine randomness, the individual variance was calculated for each run, σ2(Δt) = {x(t+Δt) − (x(t)+〈v〉Δt)}2, as a function of Δt. The individual variances were used to create an average variance, 〈σ2(Δt)〉, to which a straight line was fit over the first 20 nm 〈v〉−1, neglecting the first 3.5 ms of each record, which is dominated by the viscous relaxation time of the bead. The randomness, r, was calculated from the slope of the linefit to the average variance, divided by d〉v〉. Randomness error was propagated from the error in determining the average velocity. The step ratio, SR, was calculated by counting the number of forward steps, n+, and backward steps, n−, at a given force and ATP concentration, and determining the ratio (n+/n−). The estimated errors in the numbers of steps were taken to be the statistical errors (square root of the numbers of events). All data for forces exceeding −3 pN were obtained under force clamped conditions using a feedback-based optical trapping system. However, for small forces below −3 pN, data were collected without force feedback, leading to a minor change in force during the course of each step. In such cases, step ratio was determined by collecting n+ and n− in force bins of width 0.25 pN, a bin size chosen to correspond approximately to the change in force experienced by a motor as it undertakes an 8-nm step in a trap with average stiffness 0.03 pN nm−1.
Global Curve Fitting
Analytical expressions for the velocity (v), step ratio (SR) and randomness (r) in terms of the model parameters of Fig. 3 and Table 1, along with a description of the methods used for the global fits, may be found in theSupplementary Methods.
Ensemble fluorescence experiments
Engineered Constructs
The template for a ‘wild-type’ kinesin, KinWT, was based on a 560-residue (truncated), cysteine-light version of the human ubiquitous kinesin-1 gene, an extended version of a 413-residue construct described previously46. To generate Kin6AA, an insert consisting of six amino acids residues with net neutral charge (AEQKLT) was introduced in the C-terminal end of the neck linker region (after L335), N-terminal to the start of the dimerization domain of the stalk. Previous work involving constructs with insertions in the neck linker region13 focused instead on the addition of prolines to ensure that no protein secondary structure would form. For comparison purposes (see main text), we therefore generated and tested one additional construct, based on KinWT, carrying a 9-residue insert in its neck-linker region with six adjacent prolines, designated as the Kin6P construct. The Kin6P construct carries its insert in the identical location as Kin6AA, with the sequence KKPPPPPPG. The basic rationale for this particular sequence was furnished by Yildiz et al.13 In brief, prolines have restricted dihedral angles that present an entropic barrier to conformational changes. To ameliorate this concern, a glycine ‘swivel’ residue was introduced C-terminal to the prolines, adjacent to the dimerization domain of the stalk. In addition, wild-type kinesin is believed to form an electrostatic interaction between positively-charged lysines found in its proximal dimerization domain and C-terminus of tubulin, which carries a complimentary negative charge. To allow for such a possibility, two adjacent lysine residues were incorporated adjacent to the 6-proline insert (above), designed to mimic the charged residues found in the dimerization domain. The KinWT, Kin6AA, and Kin6P constructs all formed fully functional, processive motors when expressed in bacteria and subsequently purified.
Protein expression and purification
The kinesin constructs were expressed and purified as described47. Briefly, the DNA encoding the constructs was transformed into BL21 cells (Stratagene), which were grown in LB medium (2% (w/v) tryptone, 1% (w/v) yeast extract, 0.5% (w/v) NaCl, 0.2% (w/v) glycerol, 50 mM Na2HPO4, 50 mM K2HPO4, 50 mg L−1 ampicillin, 50 mg L−1 chloramphenicol) to O.D. 1.0 at wavelength 595 nm, then induced using IPTG (isopropyl-beta-D-thiogalactopyranoside). The proteins were expressed for 48 hours at 18°C before harvesting and lysing. A batch method was used to purify proteins over Ni-Sepharose high-performance beads (GE Healthcare). The elutant was then dialyzed overnight at 4°C in low ionic strength buffer (25 mM HEPES (pH 7.5), 10 mM KCl, 2 mM MgCl2, 1 M DTT, 1 mM ATP) and centrifuged at 25,000 RCF to remove any aggregated proteins.
Experiments with 2′dmT and 2′dmD
Fluorescently labeled 2′–deoxy 3′–mant ATP (2′dmT) and 2′–deoxy 3′–mant ADP (2′dmD) were synthesized by reacting the corresponding 2′–deoxyadenosine precursors (Sigma-Aldrich) with N-methylisatoic anhydride (Invitrogen). The 2′dmD and 2′dmT reaction products were purified by chromatography over a Sephadex LH20 column to >98% purity, determined from the relative absorbances at 255 and 356 nm, as described48. Measurements of the kinetics of 2′dmD release were carried out by incubating the kinesin constructs overnight with a slight excess of 2′dmD. The sample (1.0 µM) was then mixed in a stopped-flow fluorimeter with microtubules (10.0 µM polymerized tubulin), and the 2′dmD or 2′dmT fluorescence was monitored through FRET from protein tryptophans excited at 290 nm. The sensitized emission was observed at 90° from the excitation by using a broad bandpass filter centered at 450 nm. For 2′dmT-binding experiments, Kin6AA and KinWT were incubated with a 5– to 10–fold excess of microtubules and treated with 0.2 U ml−1 apyrase for 20 min. The complex was then mixed with 2′dmT in the stopped-flow spectrometer, and the sensitized fluorescence emission was observed as in the 2′dmD-release measurements. All kinetic experiments were performed at 20°C in 100 mM KCl, 25 mM HEPES, 2 mM MgCl2, pH 7.50.
Additional Methods
Further details of the methods used to determine ATPase activity, the generation of the mutant constructs, and a derivation of the dissociation rate constant for ATP release measured in ensemble experiments may be found in the Supplementary Methods.
Supplementary Material
ACKNOWLEDGEMENTS
SMB acknowledges support from grant GM51453 and SSR acknowledges support from grant AR048565 from the National Institutes of Health.
Footnotes
AUTHOR CONTRIBUTIONS. BEC, WMB-P, and JA designed and carried out experiments, collected and analyzed data, and co-wrote the paper. BEC and JA carried out modeling and fits. SMB and SSR helped to design experiments, analyze results and co-wrote the paper.
MAIN REFERENCES
- 1.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 2.Asbury CL, Fehr AN, Block SM. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaseda K, Higuchi H, Hirose K. Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat Cell Biol. 2003;5:1079–1082. doi: 10.1038/ncb1067. [DOI] [PubMed] [Google Scholar]
- 4.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 5.Block SM, Goldstein LS, Schnapp BJ. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348:348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- 6.Coy DL, Wagenbach M, Howard J. Kinesin takes one 8-nm step for each ATP that it hydrolyzes. J Biol Chem. 1999;274:3667–3671. doi: 10.1074/jbc.274.6.3667. [DOI] [PubMed] [Google Scholar]
- 7.Hua W, Young EC, Fleming ML, Gelles J. Coupling of kinesin steps to ATP hydrolysis. Nature. 1997;388:390–393. doi: 10.1038/41118. [DOI] [PubMed] [Google Scholar]
- 8.Schnitzer MJ, Block SM. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388:386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 9.Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 10.Block SM. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys J. 2007;92:2986–2995. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guydosh NR, Block SM. Not so lame after all: kinesin still walks with a hobbled head. J Gen Physiol. 2007;130:441–444. doi: 10.1085/jgp.200709902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenfeld SS, Fordyce PM, Jefferson GM, King PH, Block SM. Stepping and stretching. How kinesin uses internal strain to walk processively. J Biol Chem. 2003;278:18550–18556. doi: 10.1074/jbc.M300849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozielski F, et al. The crystal structure of dimeric kinesin and implications for microtubuledependent motility. Cell. 1997;91:985–994. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- 15.Hyeon C, Onuchic JN. Internal strain regulates the nucleotide binding site of the kinesin leading head. Proc Natl Acad Sci U S A. 2007;104:2175–2180. doi: 10.1073/pnas.0610939104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hariharan V, Hancock WO. Insights into the mechanical properties of the kinesin neck-linker domain from sequence analysis and molecular dynamics simulations. Cellular and Molecular Bioengineering. 2009;2:177–189. doi: 10.1007/s12195-009-0059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guydosh NR, Block SM. Backsteps induced by nucleotide analogs suggest the front head of kinesin is gated by strain. Proc Natl Acad Sci U S A. 2006;103:8054–8059. doi: 10.1073/pnas.0600931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klumpp LM, Hoenger A, Gilbert SP. Kinesin's second step. Proc Natl Acad Sci U S A. 2004;101:3444–3449. doi: 10.1073/pnas.0307691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crevel IM, et al. What kinesin does at roadblocks: the coordination mechanism for molecular walking. EMBO J. 2004;23:23–32. doi: 10.1038/sj.emboj.7600042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schief WR, Clark RH, Crevenna AH, Howard J. Inhibition of kinesin motility by ADP and phosphate supports a hand-over-hand mechanism. Proc Natl Acad Sci U S A. 2004;101:1183–1188. doi: 10.1073/pnas.0304369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishiyama M, Higuchi H, Yanagida T. Chemomechanical coupling of the forward and backward steps of single kinesin molecules. Nat Cell Biol. 2002;4:790–797. doi: 10.1038/ncb857. [DOI] [PubMed] [Google Scholar]
- 22.Carter NJ, Cross RA. Mechanics of the kinesin step. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 23.Hyeon C, Klumpp S, Onuchic JN. Kinesin's backsteps under mechanical load. Phys Chem Chem Phys. 2009;11:4899–4910. doi: 10.1039/b903536b. [DOI] [PubMed] [Google Scholar]
- 24.Liepelt S, Lipowsky R. Kinesin's network of chemomechanical motor cycles. Phys Rev Lett. 2007;98:258102. doi: 10.1103/PhysRevLett.98.258102. [DOI] [PubMed] [Google Scholar]
- 25.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 26.Visscher K, Schnitzer MJ, Block SM. Single kinesin molecules studied with a molecular force clamp. Nature. 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- 27.Valentine MT, et al. Precision steering of an optical trap by electro-optic deflection. Opt Lett. 2008;33:599–601. doi: 10.1364/ol.33.000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coppin CM, Pierce DW, Hsu L, Vale RD. The load dependence of kinesin's mechanical cycle. Proc Natl Acad Sci U S A. 1997;94:8539–8544. doi: 10.1073/pnas.94.16.8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svoboda K, Mitra PP, Block SM. Fluctuation analysis of motor protein movement and single enzyme kinetics. Proc Natl Acad Sci U S A. 1994;91:11782–11786. doi: 10.1073/pnas.91.25.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaevitz JW, Block SM, Schnitzer MJ. Statistical kinetics of macromolecular dynamics. Biophys J. 2005;89:2277–2285. doi: 10.1529/biophysj.105.064295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chemla YR, Moffitt JR, Bustamante C. Exact solutions for kinetic models of macromolecular dynamics. J Phys Chem B. 2008;112:6025–6044. doi: 10.1021/jp076153r. [DOI] [PubMed] [Google Scholar]
- 32.Valentine MT, Fordyce PM, Krzysiak TC, Gilbert SP, Block SM. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat Cell Biol. 2006;8:470–476. doi: 10.1038/ncb1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moffitt JR, Chemla YR, Bustamante C. Methods in statistical kinetics. Methods Enzymol. 475:221–257. doi: 10.1016/S0076-6879(10)75010-2. [DOI] [PubMed] [Google Scholar]
- 34.Guydosh NR, Block SM. Direct observation of the binding state of the kinesin head to the microtubule. Nature. 2009;461:125–128. doi: 10.1038/nature08259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asenjo AB, Sosa H. A mobile kinesin-head intermediate during the ATP-waiting state. Proc Natl Acad Sci U S A. 2009;106:5657–5662. doi: 10.1073/pnas.0808355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mori T, Vale RD, Tomishige M. How kinesin waits between steps. Nature. 2007;450:750–754. doi: 10.1038/nature06346. [DOI] [PubMed] [Google Scholar]
- 37.Toprak E, Yildiz A, Hoffman MT, Rosenfeld SS, Selvin PR. Why kinesin is so processive. Proc Natl Acad Sci U S A. 2009;106:12717–12722. doi: 10.1073/pnas.0808396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valentine MT, Gilbert SP. To step or not to step? How biochemistry and mechanics influence processivity in Kinesin and Eg5. Curr Opin Cell Biol. 2007;19:75–81. doi: 10.1016/j.ceb.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hackney DD. Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc Natl Acad Sci U S A. 1994;91:6865–6869. doi: 10.1073/pnas.91.15.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alonso MC, et al. An ATP gate controls tubulin binding by the tethered head of kinesin-1. Science. 2007;316:120–123. doi: 10.1126/science.1136985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hackney DD, Stock MF, Moore J, Patterson RA. Modulation of kinesin half-site ADP release and kinetic processivity by a spacer between the head groups. Biochemistry. 2003;42:12011–12018. doi: 10.1021/bi0349118. [DOI] [PubMed] [Google Scholar]
- 42.Blackman MJ, et al. Structural and biochemical characterization of a fluorogenic rhodamine-labeled malarial protease substrate. Biochemistry. 2002;41:12244–12252. doi: 10.1021/bi0263661. [DOI] [PubMed] [Google Scholar]
- 43.Kawaguchi K, Ishiwata S. Nucleotide-dependent single- to double-headed binding of kinesin. Science. 2001;291:667–669. doi: 10.1126/science.291.5504.667. [DOI] [PubMed] [Google Scholar]
- 44.Gutierrez-Medina B, Fehr AN, Block SM. Direct measurements of kinesin torsional properties reveal flexible domains and occasional stalk reversals during stepping. Proc Natl Acad Sci U S A. 2009;106:17007–17012. doi: 10.1073/pnas.0907133106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnitzer MJ, Visscher K, Block SM. Force production by single kinesin motors. Nat Cell Biol. 2000;2:718–723. doi: 10.1038/35036345. [DOI] [PubMed] [Google Scholar]
Additional References for Methods
- 46.Rosenfeld SS, Xing J, Jefferson GM, Cheung HC, King PH. Measuring kinesin's first step. J Biol Chem. 2002;277:36731–36739. doi: 10.1074/jbc.M205261200. [DOI] [PubMed] [Google Scholar]
- 47.Behnke-Parks WM, et al. Loop L5 acts as a conformational latch in the mitotic Kinesin Eg5. J Biol Chem. 286:5242–5253. doi: 10.1074/jbc.M110.192930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hiratsuka T. New ribose-modified fluorescent analogs of adenine and guanine nucleotides available as substrates for various enzymes. Biochim Biophys Acta. 1983;742:496–508. doi: 10.1016/0167-4838(83)90267-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.