Abstract
SCFCdc4 (Skp1, Cdc53/cullin, F-box protein) defines a family of modular ubiquitin ligases (E3s) that regulate diverse processes including cell cycle, immune response, and development. Mass spectrometric analysis of proteins copurifying with Cdc53 identified the RING–H2 finger protein Hrt1 as a subunit of SCF. Hrt1 shows striking similarity to the Apc11 subunit of anaphase-promoting complex. Conditional inactivation of hrt1ts results in stabilization of the SCFCdc4 substrates Sic1 and Cln2 and cell cycle arrest at G1/S. Hrt1 assembles into recombinant SCF complexes and individually binds Cdc4, Cdc53 and Cdc34, but not Skp1. Hrt1 stimulates the E3 activity of recombinant SCF potently and enables the reconstitution of Cln2 ubiquitination by recombinant SCFGrr1. Surprisingly, SCF and the Cdc53/Hrt1 subcomplex activate autoubiquitination of Cdc34 E2 enzyme by a mechanism that does not appear to require a reactive thiol. The highly conserved human HRT1 complements the lethality of hrt1Δ, and human HRT2 binds CUL-1. We conclude that Cdc53/Hrt1 comprise a highly conserved module that serves as the functional core of a broad variety of heteromeric ubiquitin ligases.
Keywords: Roc1, Rbx1, cullin, ubiquitin, RING finger
Proteolysis in the eukaryotic cytosol is triggered by the assembly of a substrate-linked multiubiquitin chain, which targets the protein for degradation by the 26S proteasome (Banerjee et al. 1993). Ubiquitin is activated for transfer to substrate through the ATP-dependent formation of a thioester bond between the carboxyl terminus of ubiquitin and the ubiquitin-activating enzyme, E1. Ubiquitin is transferred subsequently to a member of a family of ubiquitin-conjugating (E2) enzymes. Finally, thioesterified ubiquitin is transferred from E2 to a lysine residue of the target protein, either directly or with the assistance of a ubiquitin ligase (E3). E3s bind substrate directly, suggesting that they provide specificity in ubiquitination reactions. Whereas the budding yeast genome encodes 13 E2-like proteins, it is not known how many E3s exist, because these enzymes do not share a common sequence motif. So far, four different classes of E3 enzymes have been defined: Ubr1 (Bartel et al. 1990), the HECT (homology to E6-AP carboxyl terminus) domain family (Huibregtse et al. 1995), of which Mdm2 has been proposed to be a member (Honda et al. 1997), the SCF family (Feldman et al. 1997; Skowyra et al. 1997), and the anaphase-promoting complex/cyclosome (APC/C) (King et al. 1995).
SCFCdc4, the prototype of the SCF (for Skp1, Cdc53/Cullin, F-box receptor) family of ubiquitin ligases, was first defined by in vitro reconstitution (Feldman et al. 1997; Skowyra et al. 1997; Verma et al. 1997b). Identification of SCFCdc4 stemmed from a genetic analysis of the G1/S transition (Schwab et al. 1994). Budding yeast cells harboring temperature-sensitive mutations in either the Cdc34 E2 enzyme or in any one of the subunits of SCFCdc4 fail to enter S phase at the nonpermissive temperature because they fail to degrade the S phase cyclin/cyclin-dependent kinase (CDK) inhibitor Sic1 (Bai et al. 1996; Schwab et al. 1994). In wild-type cells, G1 cyclin/CDK phosphorylates Sic1 at the end of G1 phase, thereby targeting it for Cdc34-dependent ubiquitination and degradation (Verma et al. 1997a). Phosphorylated Sic1, but not the unmodified protein or a phosphorylation-site mutant, binds tightly to the Cdc4 subunit of SCFCdc4 (Feldman et al. 1997; Skowyra et al. 1997). Cdc34 is also recruited to SCFCdc4, possibly via interaction with the Cdc53 subunit (Willems et al. 1996). Upon assembly with the SCFCdc4 complex, phosphorylated Sic1 is extensively ubiquitinated as a prelude to its degradation.
Besides SCF, the APC/C ubiquitin ligase also plays a key role in cell-cycle control. APC/C controls the separation of sister chromatids and the exit from mitosis by specifying the destruction of Pds1 and cyclin B, respectively (King et al. 1996). Although SCF and APC/C are distinctly regulated and differ greatly in size, subunit composition and apparent complexity, APC/C contains a subunit (Apc2) that bears homology to the carboxy-terminal half of Cdc53/cullin (Yu et al. 1998; Zachariae et al. 1998).
A remarkable property of SCF ubiquitin ligases is that their specificity can be programmed via an array of distinct substrate receptors (Patton et al. 1998b; Skowyra et al. 1997). These receptors are recruited to SCF by the conserved F-box domain they share. The discovery of the F box stemmed from the observation that human cyclin F and Cdc4 share a conserved sequence that mediates their interaction with Skp1 (Bai et al. 1996). The budding-yeast genome encodes 17 proteins that contain an obvious F-box motif (Patton et al. 1998b), and at least three of these proteins assemble into distinct SCF complexes in vivo (Patton et al. 1998a).
Over the past few years, F-box proteins and other components of the SCF/Cdc34 pathway have been identified in many eukaryotes and linked to diverse cellular processes, ranging from the auxin response in plants (Ruegger et al. 1998), withdrawal from the cell cycle in nematodes (Kipreos et al. 1996), signaling via the Wnt and Hedgehog pathways in Drosophila (Jiang and Struhl 1998), and activation of the immunoregulatory protein NF-κB in human cells (Yaron et al. 1998). Although SCF ubiquitin ligases clearly regulate diverse physiological pathways in a variety of eukaryotes, little is known about their structure, regulation, or mechanism of action.
The HECT domain proteins are the only ubiquitin ligases whose mechanism of action is understood. The charter member of the HECT domain family of proteins, E6-associated protein (E6-AP), forms a thioester bond with ubiquitin as an intermediate in the transfer of ubiquitin from E2 to substrate (Scheffner et al. 1995). Mutation of the thioester-forming cysteine of E6-AP destroys its E3 activity. It remains unclear whether other (non-HECT) E3 enzymes catalyze substrate ubiquitination via a similar thioester cascade.
We report the discovery of a heretofore undetected fourth essential subunit of the SCF complex. This new subunit, Hrt1, is a member of the RING–H2 family of proteins (Suarin et al. 1996). Hrt1 is required for degradation of SCF substrates in vivo, and stimulates the ubiquitin ligase activity of both SCFCdc4 and SCFGrr1 complexes in vitro. A Cdc53/Hrt1 subcomplex potently activates autoubiquitination of the Cdc34 E2 enzyme, suggesting that these two proteins define a minimal ubiquitin ligase. Based on analysis of Cdc34 autoubiquitination, we suggest that Cdc53/Hrt1 promotes substrate ubiquitination by a novel mechanism. Remarkably, the SCF and APC/C ubiquitin ligase complexes both contain essential subunits with cullin homology and RING–H2 domains. We propose that these seemingly unrelated ubiquitin ligases define a broad family of cullin-containing E3s that share a common enzymatic mechanism.
Results
Purification of SCF complexes from yeast cells
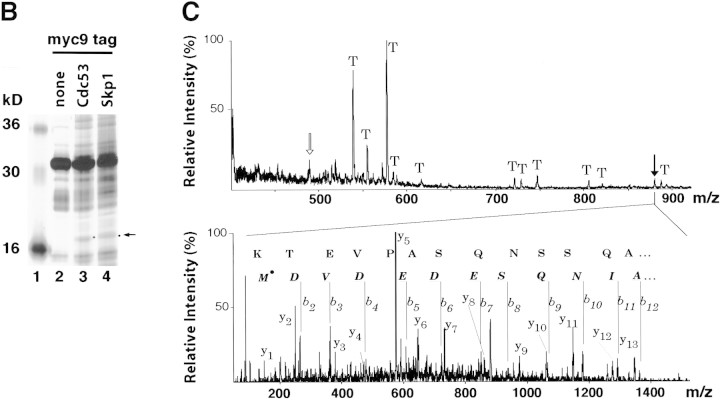
To determine if SCF contains subunits other than the minimal set that suffices for reconstitution of ubiquitin ligase activity, we investigated its native composition in yeast . The chromosomal copies of CDC53 and SKP1 were modified by a PCR-based tagging procedure (see Materials and Methods) to encode proteins with either six or nine copies of the myc epitope (myc6, myc9) appended to their carboxyl termini. CDC53myc6 and control untagged cells were pulse radiolabeled with [35S]-methionine, and proteins bound specifically to Cdc53myc6 were isolated on an α-myc 9E10 monoclonal antibody matrix (Fig. 1A). Among other proteins, Cdc53myc6 immunoprecipitates contained a prominent ∼17-kD band (lane 2) that was not detected in affinity-purified APC/C (lane 3). To isolate SCF-associated proteins in sufficient scale for protein sequencing, Skp1myc9 and Cdc53myc9 were affinity purified on 9E10 beads and associated proteins were resolved by SDS-PAGE and stained with silver (Fig. 1B). Although several proteins were identified in both preparations, Cdc53, YJL149w (an F-box protein), and the 17-kD polypeptide were the only species that were detected consistently in both Skp1myc9 and Cdc53myc9 immunoprecipitates (J.H. Seol and A. Sherchenko, unpubl.). The 17-kD protein was identified as the product of the HRT1 (YOL133w) locus (Fig. 1C) by a combination of high-mass-accuracy MALDI (matrix-assisted laser desorption ionization mass spectrometry) peptide mapping and nanoelectrospray tandem mass spectrometric sequencing (Shevchenko et al. 1996a). HRT1 was first identified as a gene whose high-level expression reduces transposition by the retrotransposon Ty3 (cited in the Saccharomyces Genome Database at http://genome-www.stanford.edu/Saccharomyces/; J. Claypool and S. Sandmeyer, in prep.)
Figure 1.
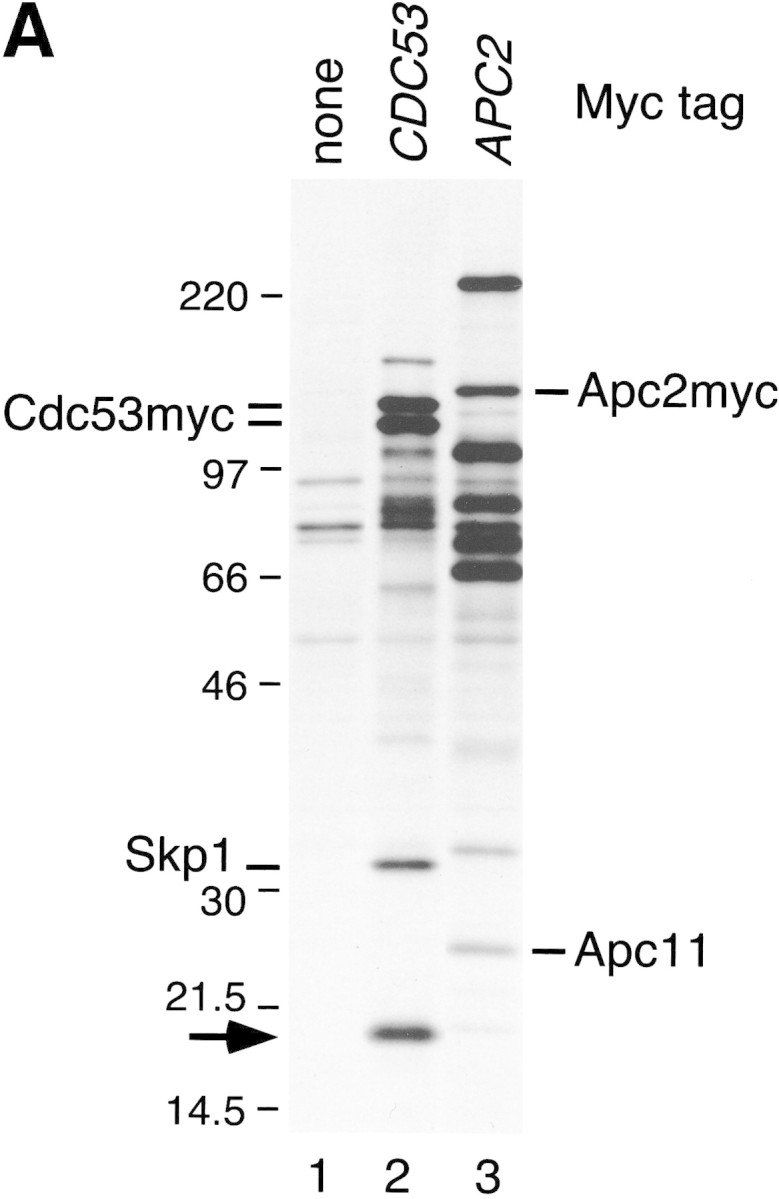
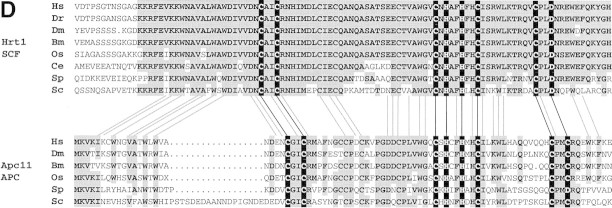
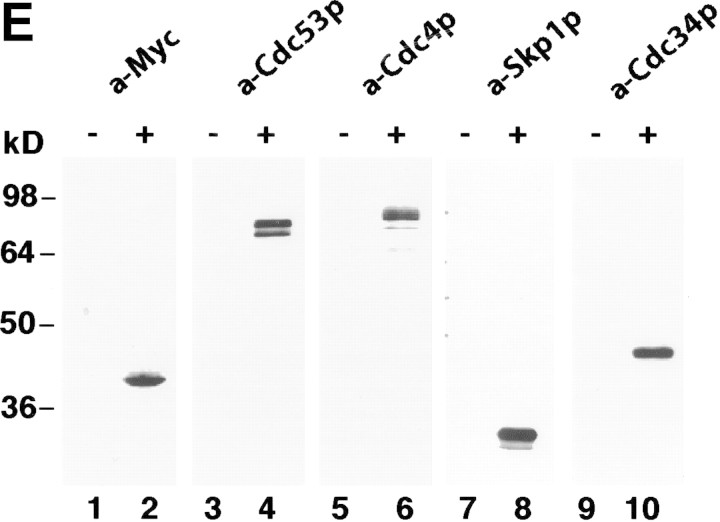
Identification of Hrt1 as a Cdc53-associated protein. (A) [35S]Methionine-labeled proteins that specifically coimmunoprecipitate with Cdc53myc6 (lane 2) or Apc2myc6 (lane 3) were visualized by SDS-PAGE followed by autoradiography. (B) Proteins coimmunoprecipitating with Cdc53myc9 (lane 3) or Skp1myc9 (lane 4) were separated by SDS-PAGE, and stained with silver. Both preparations consistently contained a 17-kD protein (arrow in A and B). (C) Identifiecation of YOL133w by nanoelectrospray tandem mass spectrometry (Nano ES MS/MS). (Top) Partial mass spectrum of a pool of tryptic peptides recovered after in gel digestion of 17-kD band. (T) Peaks identified as trypsin autolysis products. Tandem mass spectrum acquired from the ion with m/z 488.2 (open arrow) identified peptide AFLIEEQK from ribosomal protein RPL34B (data not shown). Tandem mass spectrum (bottom) of the ion with m/z 880.4 (solid arrow) revealed two nonoverlapping series of fragment y-ions and b-ions (Biemann 1988) that both identified the same Hrt1-derived peptide M*DVDEDESQNIAQSSNQSAPVETK (M* stands for methionine sulfoxide). Carboxy-terminal part of the peptide sequence (...AQSSNQSAPVETK) is covered by continuous series of y-ions. Amino-terminal part of the sequence (M*DVDEDESQNIA) is covered by b-ion series. As every amino acid residue in the peptide sequence can be accounted for, identification of Hrt1 is certain despite the complexity of the spectrum and that only a single peptide was identified. (D) Hrt1 is highly conserved between species and related to the APC subunit Apc11. Budding yeast Hrt1 and Apc11 were each aligned with related sequences from different species. Putative zinc-coordinating residues are highlighted black. Residues identical in at least 4 (Apc11) or 5 (Hrt1) sequences are shaded. Lines connecting the alignments indicate residues conserved between Apc11 and Hrt1. (Hs) Homo sapiens, (Dm) Drosophila melanogaster; (Dr) Danio rerio; (Bm) Bombyx mori; (Os) Oryza sativa; (Ce) Caenorhabditis elegans; (Sp) Schizosaccharomyces pombe; (Sc) Saccharomyces cerevisiae. (E) Hrt1 is an authentic subunit of SCF. Crude extracts from untagged control (lane 1) and HRT1myc9 (lane 2) strains were evaluated in parallel by immunoblotting with 9E10 antibodies. Extracts from untagged (odd-numbered lanes) or HRT1myc9 (even-numbered lanes) cells were immunoprecipitated with 9E10 antibodies fractionated by SDS-PAGE, and immunoblotted with α-Cdc53, α-Cdc4, α-Skp1, or α-Cdc34 as indicated to reveal the corresponding protein.
Interestingly, the yeast protein most closely related to Hrt1 was the Apc11 subunit of APC/C (Fig. 1D) (Zachariae et al., 1998). These proteins are 51% related and 28% identical to each other, and both contain highly related RING–H2 domains (Suarin et al. 1996). Hrt1 has even closer relatives in other organisms, including humans. The human hHRT1 (GenBank accession no. D52876) and hHRT2 (TIGR accession no. HGI THC257076) genes are 62.5% identical (79% similar) and 41% identical (63% similar) to yeast HRT1, respectively.
To verify the interaction between Hrt1 and SCF, we modified the chromosomal HRT1 locus to encode HRT1myc9. The resulting strain specifically expressed a 9E10-reactive polypeptide of the expected molecular weight (Fig. 1E, lane 2) and was fully viable at 25°C, but not 37°C (see next section). In coimmunoprecipitation experiments, Cdc53 (lane 4), Cdc4 (lane 6), Skp1 (lane 8), and Cdc34 (lane 10) were detected specifically in 9E10 immunoprecipitates prepared from HRT1myc9 cells, but not in those prepared from a control untagged strain (odd-numbered lanes).
HRT1 is required for SCF activity in vivo
To evaluate the role of Hrt1 in SCF function, an HRT1/hrt1::HIS3 heterozygote was constructed, sporulated, and dissected into tetrads. Only two viable, His− spores were recovered from each tetrad on rich medium (Fig. 2A, top). However, HRT1 overlaps extensively with the YOL134c locus on the opposite strand, such that hrt1::HIS3 disrupted both genes. Two lines of evidence indicate that HRT1 is essential, and YOL134c is dispensable. First, cosegregation of a GAL–HRT1 plasmid complemented hrt1::HIS3 on either galactose medium (second panel) or glucose medium (not shown) as evidenced by the appearance of tetrads with more than two viable spores, at least one of which was His+. Second, insertion of a myc9 epitope immediately upstream of the HRT1 stop codon yielded a temperature-sensitive growth phenotype, whereas insertion of the same epitope immediately downstream of the stop codon (HRT1–stop–myc9) had no effect on growth (e.g., Fig. 2B). A similar effect of the myc9 epitope was observed with APC11 (Zachariae et al. 1998). hHRT1 expressed from the GAL1 promoter also complemented the lethality of hrt1::HIS3 on galactose medium (third panel), but not on glucose medium (data not shown). Thus, human and yeast Hrt1 most likely perform the same biochemical function. In contrast, the more distantly related hHRT2 did not complement hrt1::HIS3 on galactose medium (bottom panel). Nevertheless, hHRT2 is likely to perform a function related to that of hHRT1, because a cDNA encoding amino acids 9–112 of hHRT2 was recovered in a two-hybrid screen for CUL-1-interacting proteins (data not shown).
Figure 2.
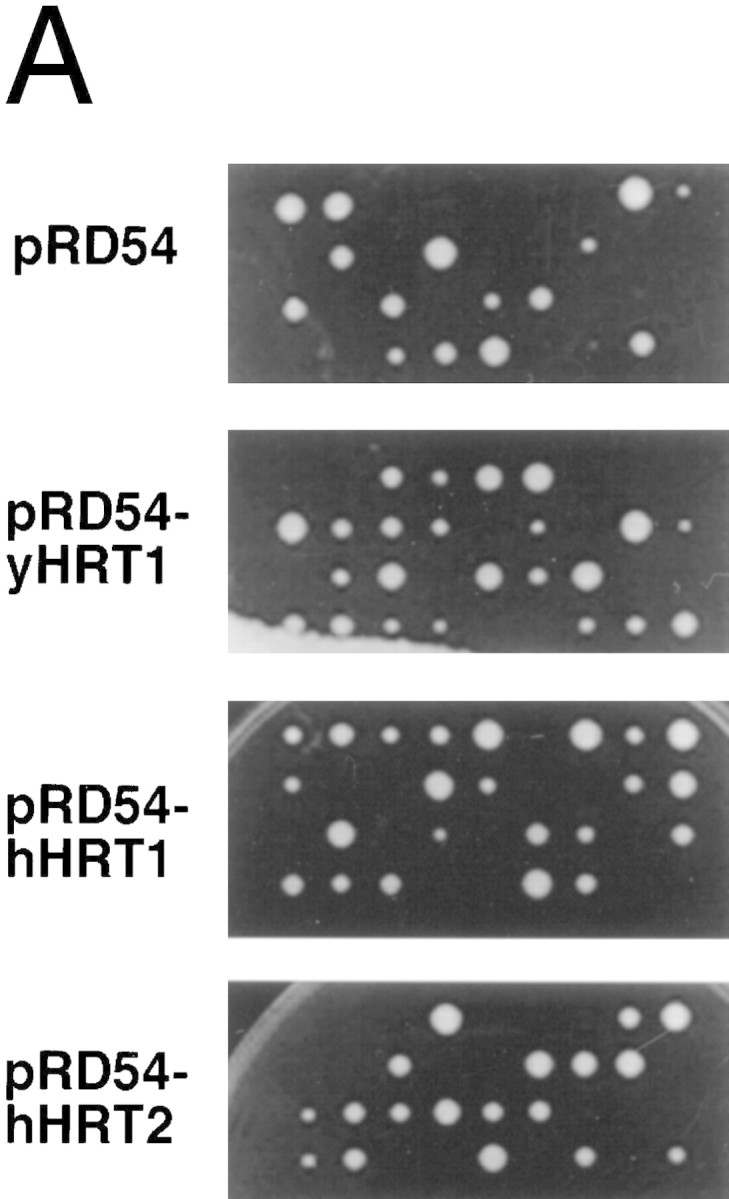
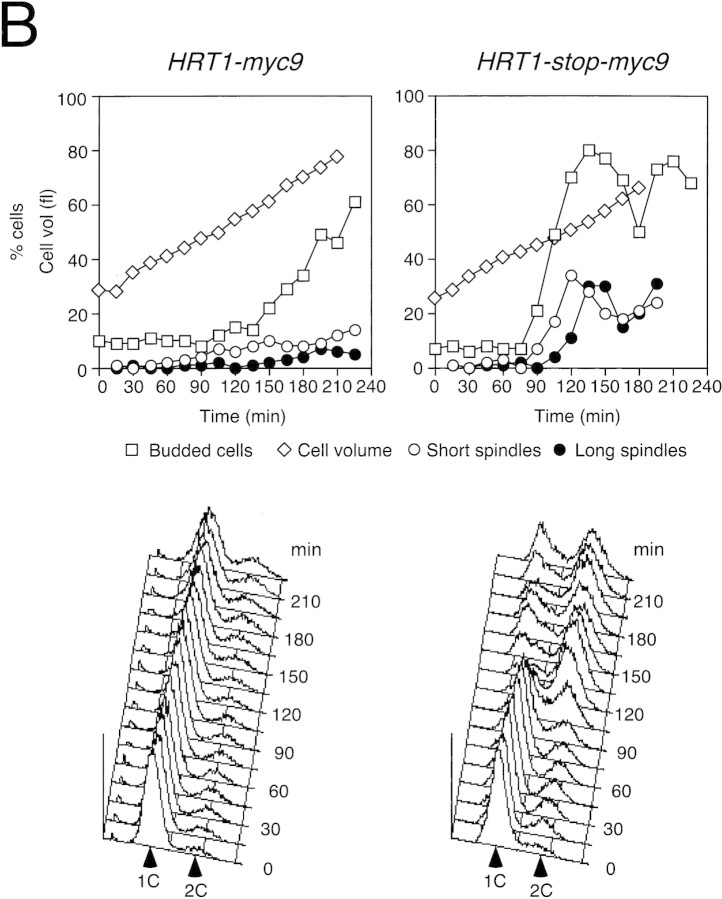
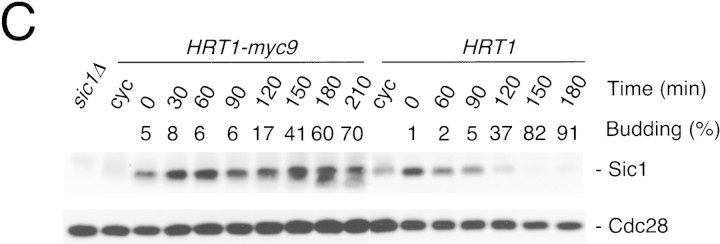
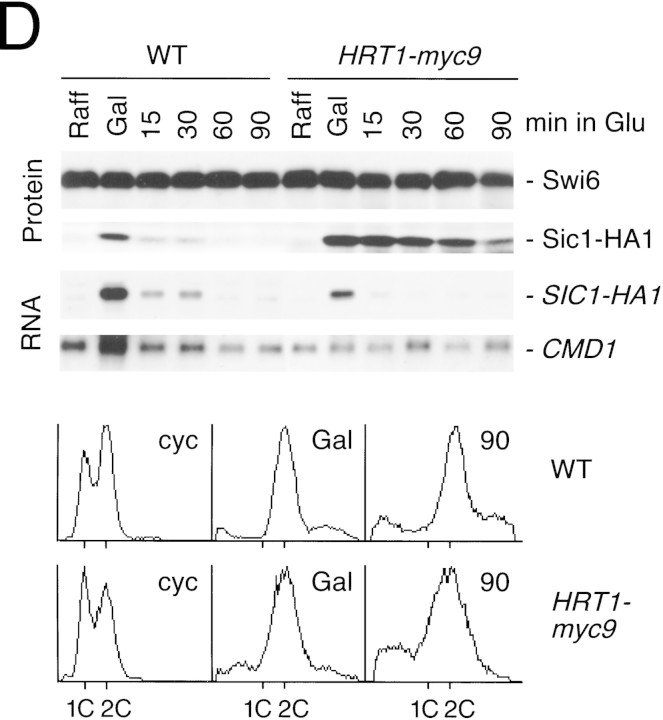
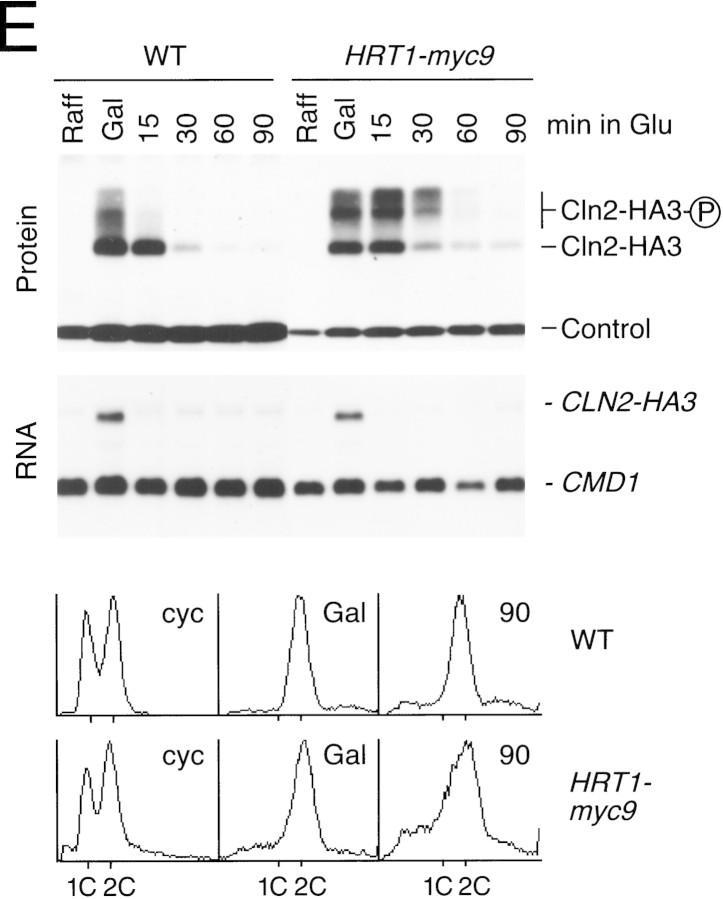
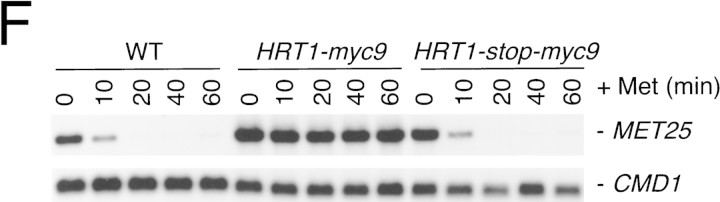
(A) HRT1 is essential. A diploid hrt1::HIS3/HRT1 strain was transformed with pRD54-based plasmids (pRS316 CEN, URA3) containing either yeast HRT1, human HRT1, or human HRT2 under the control of the galactose-inducible GAL1 promoter. Sporulated diploids were dissected on complete galactose medium, followed by incubation at 24°C for 5 days. (B,C) HRT1 is required for both elimination of Sic1 and for S phase. A myc9 epitope cassette was integrated at HRT1 either immediately upstream (HRT1–myc9, left) or downstream (HRT1–stop–myc9, right) of the stop codon. Small G1 cells were isolated by centrifugal elutriation from cultures grown in YPD at 25°C and inoculated into fresh medium at 37°C at time 0. In B, mean cell volume (fl), budding index, formation of mitotic spindles (top panels), and DNA contents (bottom panels) were assessed at the indicated times. In C, budding index, Sic1 levels, and Cdc28 levels were monitored. (D) HRT1 is required for proteolysis of Sic1. HRT1 cells containing five integrated copies of GAL–SIC1–HA1 and HRT1–myc9 GAL–SIC1–HA1 (one copy) cells were grown in YP–raffinose at 25°C (cyc) and arrested in mitosis with nocodazole (Raff). After shifting the cultures to 32°C, expression from the GAL promoter was induced with galactose (Gal) and then repressed by transferring the cells to YP–glucose. Samples were withdrawn at the indicated time points and assayed for: Swi6 and Sic1-HA1 protein levels (top panels); calmodulin and SIC1–HA1 mRNA levels (middle panels); and cellular DNA content (bottom panels). (E) HRT1 is required for rapid degradation of Cln2. Same as D, except CLN2–HA3 expression was induced with galactose at 35°C, and cells were shifted to 37°C upon transfer to YP–glucose. The band labeled control is an unknown polypeptide recognized by 12CA5. (F) HRT1 is required for methionine-dependent repression of MET25 mRNA. Yeast strains indicated were grown in complete medium containing glucose and lacking methionine at 25°C and were then shifted to 37°C for 2 hr. Methionine was added to the medium and samples were withdrawn at the indicated times. MET25 and CMD1 mRNA levels were assayed by Northern blotting.
The fortuitous temperature-sensitive growth phenotype of HRT1myc9 strains allowed us to evaluate the roles of Hrt1 in cell division and degradation of SCF substrates. Cells with temperature-sensitive mutations in SCF/Cdc34 pathway components are sensitive to overexpression of SCF substrates (Deshaies et al. 1995; Bai et al. 1996; Willems et al. 1996). Overproduction of either SIC1HA or CLN2HA3 from the GAL promoter prevented growth at 25°C of HRT1myc9 cells, but not wild-type or HRT1–stop–myc9 cells (data not shown). A second phenotype of cells harboring temperature-sensitive mutations in SCF/Cdc34 pathway components is that they bud but fail to initiate DNA replication at the nonpermissive temperature (Schwab et al. 1994; Bai et al. 1996). HRT1myc9 and HRT1–stop–myc9 cells grown in YP-glucose medium were elutriated to obtain a population of small, unbudded G1 cells, which were reinoculated into fresh YP-glucose medium at 37°C. Cells harvested at 15-min intervals were evaluated for DNA content, cell volume, and budding index (Fig. 2B). Whereas most HRT1–stop–myc9 cells initiated budding and DNA replication at ∼90–105 min, HRT1–myc9 cells did not enter S phase or form mitotic spindles, and exhibited a strong delay in bud emergence. Like HRT1myc9, cdc34ts cells are also delayed in bud emergence (Schwab et al. 1994), and arrested cultures of skp1-11 mutants contain unbudded cells (Bai et al. 1996). These observations suggest that SCF antagonizes an inhibitor of bud emergence. Consistent with their failure to duplicate DNA, Sic1 persisted at high levels in elutriated HRT1myc9 cells, but was degraded concomitant with bud emergence in HRT1 cells (Fig. 2C).
To investigate the role of HRT1 in turnover of substrates independent of its effects on cell-cycle progression, SIC1 and CLN2 expression were induced transiently in G2-arrested wild-type and HRT1myc9 cells incubated at a semirestrictive temperature, and the decline in Sic1 (Fig. 2D) and Cln2 (Fig. 2E) levels were monitored by immunoblotting. Comparison of protein levels (Fig. 2D,E, top panels) at different time points after repression of transcription (middle panels) indicates that both Sic1 and Cln2 were stabilized in HRT1myc9 cells. For unknown reasons, HRT1myc9 had a stronger effect on Sic1 turnover than on Cln2 turnover. Densitometric scanning of the experiment in Figure 2E revealed that the half-life of Cln2 was 8 min in wild-type cells and 19 min in mutant cells. Flow cytometry confirmed that both wild-type and mutant cells remained arrested in G2/M throughout the duration of the time courses (bottom panels). Although a protein target of the SCFMet30 complex has not been described yet, mutations in CDC53, SKP1, and MET30 prevent repression of MET25 mRNA in response to high extracellular methionine (Patton et al. 1998a). Likewise, HRT1myc9 cells were also deficient in MET25 repression (Fig. 2F). Thus, the activities of three distinct SCF complexes—SCFCdc4, SCFGrr1 and SCFMet30—were diminished in HRT1myc9 cells.
Assembly of recombinant Hrt1 with SCFCdc4 subunits
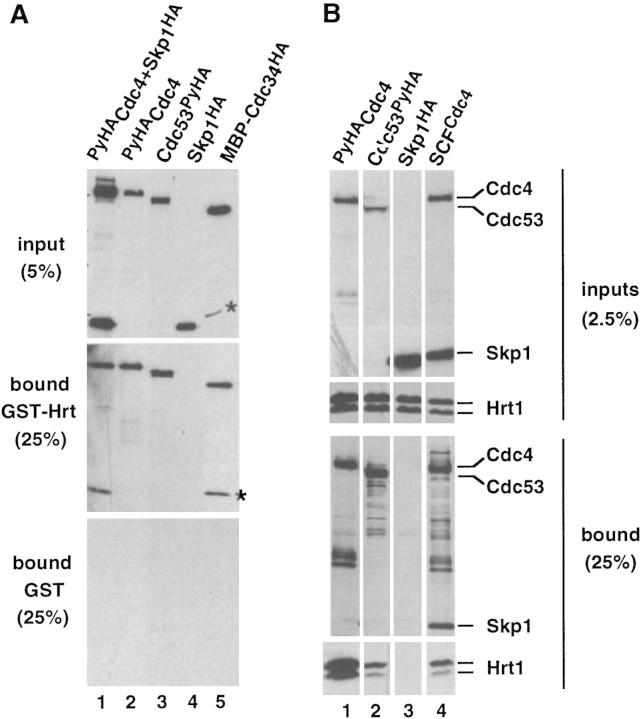
To evaluate the assembly of Hrt1 with SCF subunits in vitro, glutathione S-transferase–Hrt1 (GST–Hrt1) purified from Escherichia coli was mixed with insect cell lysates containing recombinant SCFCdc4 subunits, and proteins recovered on glutathione–agarose were detected by immunoblotting (Fig. 3A). GST–Hrt1 presumably folded correctly, as it stimulated SCFCdc4 E3 activity (Fig. 4A). Whereas Skp1 (Fig. 3A, lane 4) did not bind GST–Hrt1, ∼20% of PyHACdc4 (lanes 1,2) and ∼20% of Cdc53PyHA (lane 3) bound to GST–Hrt1 (middle panel), but not GST (bottom panel). The high efficiencies of binding suggest that interaction of GST–Hrt1 with Cdc4 and Cdc53 was direct and did not require intermediary insect cell proteins. MBP–Cdc34ΔCHA purified from E. coli also bound purified GST–Hrt1 (lane 5), but not GST. Similar protein interactions were observed upon coexpression of Hrt1 with SCFCdc4 subunits in insect cells (Fig. 3B). Hrt1 was retrieved on an antipolyoma antibody matrix (α-Py beads) from insect cell lysates containing coexpressed SCFCdc4 (lane 4; Cdc4 was tagged with PyHA epitope), PyHACdc4 (lane 1) or Cdc53PyHA (lane 2). Unexpectedly, recovery of Hrt1 bound to PyHACdc4 was diminished upon coexpression of Skp1, suggesting that Skp1 and Hrt1 may compete for binding to overlapping sites on Cdc4. Hrt1 was not recovered on α-Py beads upon coexpression with an untagged control protein (Skp1HA; lane 3).
Figure 3.
Hrt1 binds Cdc4, Cdc53, and Cdc34. (A) (Lanes 1–4) GST–Hrt1 (middle panel) or GST (bottom panel) purified from E. coli were mixed with crude insect cell lysates (top panel) that contained the proteins indicated above each lane. Proteins retrieved on glutathione-agarose were fractionated by SDS-PAGE and revealed by immunoblotting with α-HA. (Lane 5) Same as lanes 1–4, except MBP–Cdc34ΔCHA purified from E. coli was evaluated for its ability to bind GST-Hrt1. The asterisks denote a proteolytic fragment of MBP–Cdc34ΔCHA. Input lanes contain 5%; bound lanes contain 25% of the material from a single binding reaction. (B) Lysates from Sf9 insect cells that coexpressed Hrt1 plus PyHACdc4 (lane 1), Cdc53PyHA (lane 2), Skp1HA (lane 3), or PyHACdc4/Cdc53/Skp1HA (lane 4) were adsorbed to α-Py beads, and bound proteins were fractionated by SDS-PAGE and revealed by immunoblotting with α-HA (third row of panels) or α-Hrt1 (bottom row of panels) antibodies. Unfractionated insect cell lysates were directly immunoblotted to evaluate relative expression of the individual proteins (top row of panels, α-HA blot; second row of panels, α-Hrt1 blot).
Figure 4.
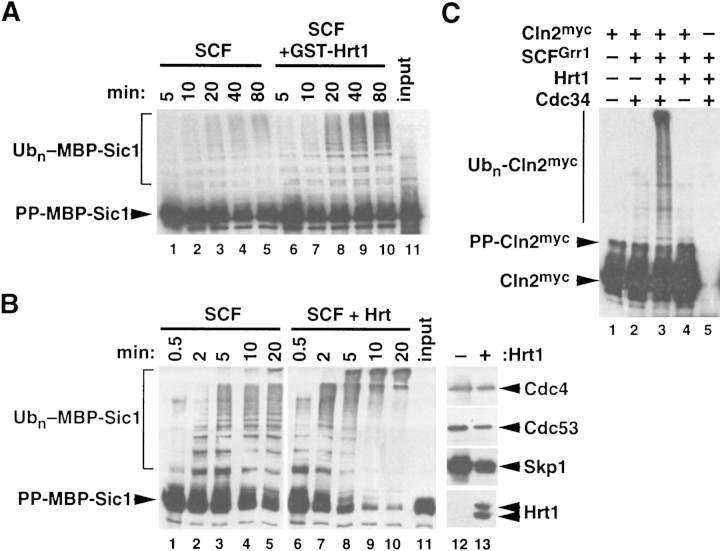
Hrt1 potently stimulates ubiquitin ligase activity of SCFCdc4. (A) Recombinant SCFCdc4 was produced in Sf9 insect cells upon coinfection with baculovirus vectors that express Skp1, Cdc53PyHA, and PyHACdc4, and the complex was purified by adsorption to a protein A resin containing covalently linked antipolyoma monoclonal antibody (α-Py beads). Parallel samples were supplemented with E1 enzyme, Cdc34, ATP, ubiquitin, purified phosphorylated maltose binding protein–Sic1 chimera (PP–MBP–Sic1), and either GST (lanes 1–5) or GST–Hrt1 (lanes 6–10). At the indicated times, samples were evaluated for ubiquitination of phospho–MBP–Sic1 by SDS-PAGE followed by immunoblotting with α-Sic1. (B) (Lanes 1–11) Same as A, except that recombinant SCFCdc4 complexes that either contained (lanes 6–10) or lacked (lanes 1–5) yeast Hrt1 were generated by either triple or quadruple infection of insect Sf9 cells (quadruple infection contained an Hrt1-expressing baculovirus). Input substrate is shown in lane 11. (Lanes 12,13) Levels of SCF subunits in each preparation were evaluated by immunoblotting. (C) Same as B, except that recombinant SCFGrr1 complexes containing (lane 3) or lacking (lane 2) Hrt1 were expressed in insect cells, purified, and mixed with 1/20 volume insect cell lysate that contained recombinant Cln2myc/GST–Cdc28/Cks1 complexes (Cln2myc). In lanes 3 and 4, Cdc34 and substrate, respectively, were omitted from the final reaction. PP–Cln2myc refers to the phosphorylated substrate.
Hrt1 stimulates SCFCdc4 activity in vitro
Ubiquitin ligase activity specific for phosphorylated Sic1 can be reconstituted with recombinant Skp1, Cdc53, and Cdc4 (Feldman et al. 1997; Skowyra et al. 1997). The identification of Hrt1 as a fourth essential subunit of the SCF complex required for normal rates of Sic1 and Cln2 proteolysis in vivo raised the question as to whether Hrt1 functions in substrate ubiquitination. To address this issue, we evaluated the effect of recombinant Hrt1 on SCFCdc4-dependent ubiquitination of Sic1 in vitro. Sf9 cells were infected with baculoviruses encoding Skp1, Cdc53, and Cdc4, and recombinant SCF complexes were retrieved on α-Py beads. A deliberate excess of phosphorylated MBP–Sic1 substrate was incubated with immobilized SCFCdc4 in the presence of E1 enzyme, Cdc34, ubiquitin, ATP, and either GST or GST–Hrt1 purified from E. coli. As shown in Figure 4A, GST–Hrt1 potently stimulated ubiquitination of phospho–MBP–Sic1. A similar result was obtained by evaluating SCFCdc4 activity obtained from insect cells coexpresseing Hrt1 and SCF subunits (Fig. 4B). The potent (>fivefold) stimulatory effect of Hrt1 upon Sic1 ubiquitination was not because of increased expression of SCFCdc4 subunits (lanes 12,13). Notably, Hrt1 augmented both the rate (as measured by percent conversion) and extent (as measured by molecular weight of ubiquitin conjugates) of ubiquitination.
The F-box protein Grr1 is required for Cln1 and Cln2 proteolysis (Barral et al. 1995; Patton et al. 1998a). Whereas recombinant SCF complexes assembled with Grr1 (SCFGrr1) bind phospho-Cln1, they do not support its ubiquitination (Skowyra et al. 1997). As Hrt1 is required for SCF function in vivo and stimulates SCFCdc4 activity in vitro, we reasoned that Hrt1 might be essential for SCFGrr1 function. Recombinant SCFGrr1 complexes that either contained (Fig. 4C, lane 3) or lacked (lane 2) yeast Hrt1 were expressed in insect cells. Complexes retrieved on α-Py beads were then mixed with Sf9 cell lysate that contained Cln2myc/GST–Cdc28/Cks1 substrate. Whereas SCFGrr1 complexes were essentially inactive (lane 2), those that contained Hrt1 sustained Cdc34-dependent ubiquitination of Cln2 (lanes 3,4). Taken together with the previous data, these observations suggest that Hrt1 is an essential core component that contributes to the E3 activity of all SCF complexes.
Cdc53/Hrt1 defines a novel ubiquitin ligase module
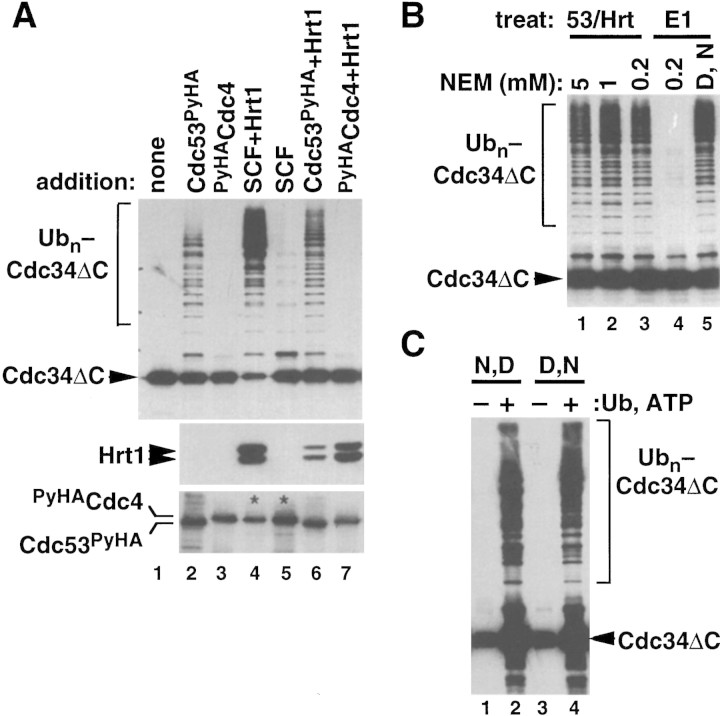
To characterize the mechanism of action of Hrt1/SCFCdc4, we sought to identify a minimal ubiquitin ligase domain. Purified Cdc34 ubiquitinates itself upon incubation with E1 and ubiquitin, but this reaction proceeds slowly (Banerjee et al. 1993). Cdc34 lacking 24 residues from the carboxyl terminus (Cdc34ΔC) no longer detectably ubiquitinates itself (R.M.R. Feldman, unpubl.), but nevertheless complements cdc34Δ (Kolman et al. 1992). We reasoned that the ubiquitin ligase activity of SCF might promote autoubiquitination of either intact Cdc34 or Cdc34ΔC; this simplified system would allow us to monitor SCFCdc4 ubiquitin ligase activity independently of substrate targeting requirements. Strikingly, SCFCdc4 complexes reconstituted with yeast Hrt1 potently activated Cdc34 (data not shown) and Cdc34ΔC (Fig. 5A, top panel, lane 4) autoubiquitination, whereas SCFCdc4 complexes lacking yeast Hrt1 had much less activity (lane 5). Hrt1 did not act by simply enhancing the levels of SCFCdc4 subunits recovered from insect cells (Fig. 4B, lanes 12,13). To determine if this activity of SCFCdc4 resides in a subdomain of the complex, individual subunits were affinity purified from baculovirus-infected Sf9 cells and tested for activity. Cdc53PyHA (Fig. 5A, lane 2), but not equivalent amounts of PyHACdc4 (lane 3) or Skp1HA (data not shown), promoted Cdc34ΔC autoubiquitination. The activity (top panel), but not the yield (bottom panel) of Cdc53PyHA was substantially enhanced by assembly with Hrt1 (lane 6). Hrt1 bound to PyHACdc4, however, was not sufficient to catalyze Cdc34ΔC autoubiquitination (top panel, lane 7) even though it was present in large amounts (middle panel). The activity observed for Cdc53/Hrt1 subcomplexes was consistently less than that of Hrt1-containing SCFCdc4 complexes. Thus, although the ubiquitin ligase activity of SCFCdc4 is intrinsic to the Cdc53/Hrt1 subcomplex, this activity may be enhanced upon assembly of Cdc53/Hrt1 with Cdc4/Skp1.
Figure 5.
Cdc53/Hrt1 comprise an NEM-resistant ubiquitin ligase module. (A) The proteins indicated were retrieved from baculovirus-infected insect cells on α-Py beads, and mixed with E1, Cdc34ΔC, ubiquitin, and ATP. Following a 60-min incubation at 20°C, reactions were evaluated for Cdc34ΔC autoubiquitination (top panel), levels of Hrt1 (middle panel), and levels of Cdc53PyHA or PyHACdc4 (bottom panel) by SDS-PAGE following by immunoblotting. SCFCdc4 complexes (lanes 4,5) contained PyHACdc4 and untagged Cdc53. Thus, only PyHACdc4 is visible in these lanes in the α-HA blot (bottom panel; see asterisks). (B) Either Cdc53PyHA/Hrt1 (lanes 1–3) or E1 enzyme (lane 4) was pretreated with the indicated amount of NEM for 10 min at 20°C. NEM was quenched with DTT, and complete reactions containing E1, Cdc34ΔC, ubiquitin, ATP, and α-Py bead-bound Cdc53PyHA/Hrt1 were incubated for 60 min at 20°C. For the sample shown in lane 5, DTT was mixed with NEM prior to incubation with E1 to confirm the efficacy of the DTT quench. All reactions were subjected to SDS-PAGE followed by immunoblotting with α-Cdc34. (C) Same as B, except that complete reactions were first assembled and then treated with 5 mm NEM followed by 10 mm DTT (lanes 1,2) or vice versa (lanes 3,4). Afterwards, α-Py bead-bound Cdc53PyHA/Hrt1 was recovered, washed, and supplemented with either buffer (lanes 1,3) or fresh E1, Cdc34ΔC, ubiquitin and ATP (lanes 2,4). The signal in lanes 1 and 3 represents Cdc34ΔC from the first stage incubation that was retained on the Cdc53PyHA/Hrt1 matrix.
E3 enzymes of the HECT domain family catalyze substrate ubiquitination by providing a catalytic cysteine residue that accepts ubiquitin from E2 enzyme, and subsequently transfers the thioesterified ubiquitin to the ε-amino group of a substrate lysine residue (Scheffner et al. 1995). To address whether the Cdc53/Hrt1 ligase domain of SCFCdc4 operates by a similar mechanism, we preincubated PyHACdc53/Hrt1 with increasing amounts of the sulfhydryl-modifying reagent NEM, added DTT to quench further cysteine modification, and mixed the resulting samples with Cdc34, E1, ubiquitin, and ATP. As a control, we evaluated N-ethylmaleimide (NEM) sensitivity of the E1 enzyme. Whereas E1 was inactivated completely by pretreatment with 0.2 mm NEM [but not when the NEM was prequenched with dithiothreitol (DTT), cf. lanes 4 and 5 of Fig. 5B], PyHACdc53/Hrt1 was resistant to pretreatment with up to 5 mm NEM. Similarly, ubiquitin ligase activity of intact SCFCdc4 towards either Cdc34ΔC or phosphorylated Sic1 was not affected by NEM (R.M.R. Feldman, unpubl.). To test the possibility that exposure of the putative thioester-forming cysteine of Cdc53/Hrt1 is gated by association of Cdc53/Hrt1 with E2 enzyme, we mixed together E1, Cdc34, ubiquitin, ATP, and either mock beads or beads containing PyHACdc53/Hrt1. After a brief incubation, NEM was added to derivitize reactive cysteines, followed by DTT to quench the NEM. Treated beads were sedimented, washed, and mixed with fresh E1, Cdc34, ubiquitin, and ATP. As a control, a parallel two-stage reaction was carried out in which DTT was added prior to NEM during the first stage. As shown in Figure 5C, PyHACdc53/Hrt1 beads retained high autoubiquitination-promoting activity if incubated with NEM followed by DTT (lane 2), or vice versa (lane 4) during the first stage.
In the course of this work, we made the unexpected observation that polycations influence Cdc34 activity. Remarkably, polylysine (Fig. 6A, lane 5), but not PEG (lanes 2–4) or lysine (lane 1) markedly stimulated Cdc34ΔC autoubiquitination. As an independent test, we evaluated the effect of polycations on Gcn4 ubiquitination. Gcn4 is stabilized in cdc34ts cells (Kornitzer et al. 1994), and Gcn4 ubiquitination can be reconstituted in vitro with recombinant SCFCdc4 and Cdc34 (Y.Chi, unpubl.). Gcn4 was not modified detectably in the presence of E1, Cdc34, ATP, and ubiquitin (Fig. 6B, lane 2), but high-molecular-mass K48-linked multiubiquitin chains were assembled on Gcn4 upon inclusion of polyethyleneimine (lanes 6,7). Thus, polyethyleneimine acts like a classic E3. The ability of polycations to substitute for SCF strongly suggests that an E3-linked ubiquitin thioester intermediate is not required to evoke the intrinsic ability of Cdc34 to assemble K48-linked multiubiquitin chains.
Figure 6.
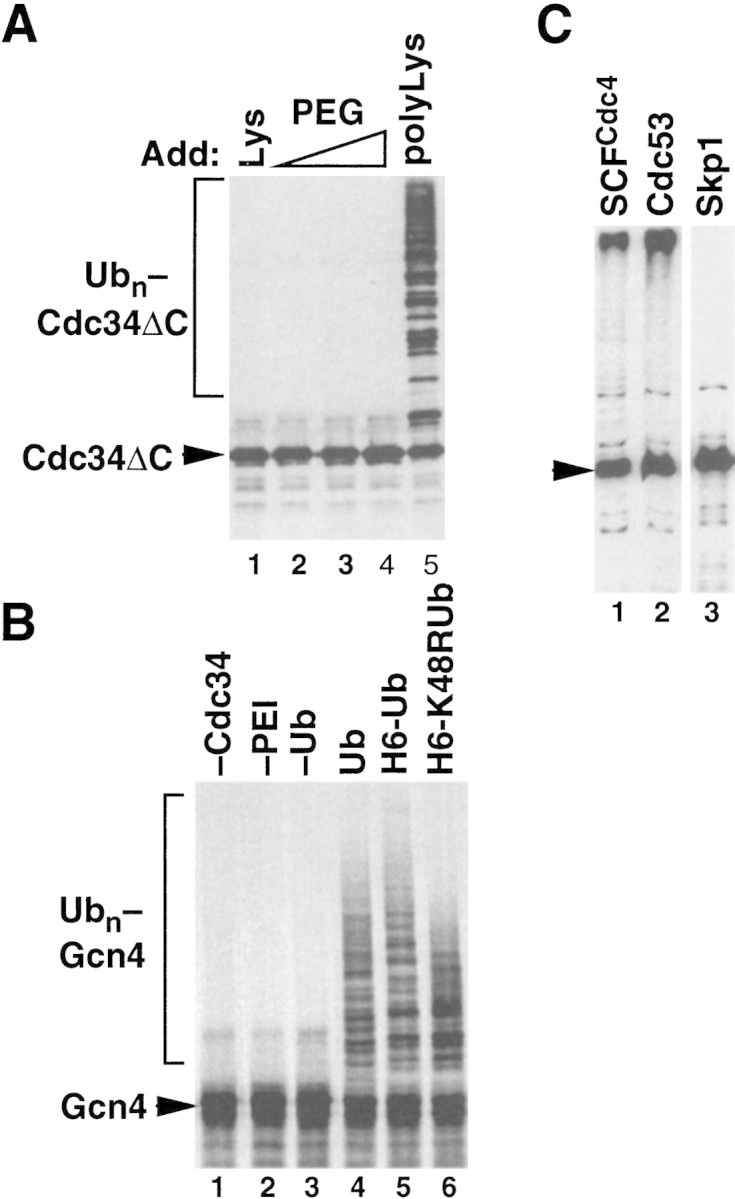
Polycations can substitute for SCF. (A) Polylysine activates Cdc34ΔC autoubiquitination. Reactions containing Cdc34ΔC, ATP, ubiquitin, and E1 enzyme were adjusted to either 10 mm lysine (lane 1), 0.01, 0.1, or 1% polyethylene glycol (PEG, lanes 2–4), or 0.01% (∼300 nm) polylysine (lane 5), and incubated at 20°C for 60 min. Reactions were evaluated as described for Fig. 5A. (B) Polyethyleneimine is an E3 for Gcn4. 35S labeled Gcn4 produced by in vitro translation in reticulocyte lysate and enriched by DEAE chromatography was mixed with E1, Cdc34, ATP, the indicated ubiquitin derivatives (lanes 4–6; H6 stands for His6) and 0.0005% polyethyleneimine (PEI), and incubated at 25°C for 60 min. Reactions conducted in the absence of either Cdc34, PEI, or ubiquitin are depicted in lanes 1–3. Gcn4 was visualized by SDS-PAGE followed by autoradiography. (C) Fusion of Sic1 to Cdc34 does not activate its ubiquitination. Cdc34ΔC–Sic1 (arrowhead) was mixed with E1, ubiquitin, and ATP in the presence of immobilized SCFCdc4 (lane 1), Cdc53PyHA (lane 2), or (as a negative control) Skp1HA (lane 3). Samples were incubated and processed as described for Fig. 5A.
We considered two possible artifactual roles of polycations. First, it is unlikely that polycations serve simply as macromolecular crowding reagents, as their stimulatory effects were observed at relatively low concentrations (300 nm for polylysine and 0.0005% for polyethyleneimine), and were not mimicked by PEG (Fig. 6A). An alternative possibility is that polycations serve to link together ubiquitin donor and ubiquitin acceptor molecules. We sought to address this by fusing the ubiquitin-acceptor domain of Sic1 to the carboxyl terminus of Cdc34ΔC. The rationale underlying this experiment is that if SCFCdc4 or E3 mimetics simply bring Sic1 and Cdc34 into close proximity, then direct fusion of substrate to E2 might bypass the requirement for E3. As shown in Figure 6C, both SCFCdc4 and Cdc53PyHA potently stimulated Cdc34ΔC–Sic1 ubiquitination, whereas Skp1HA had no effect beyond what is observed in the presence of E1, ubiquitin, and Cdc34ΔC (data not shown). Thus, bringing a substrate into close proximity to Cdc34 is not sufficient to activate its ubiquitination.
Discussion
Hrt1, a new subunit of SCF ubiquitin ligase complexes
We have identified by mass spectrometric sequencing of proteins that coimmunoprecipitate with Cdc53 a new subunit of the SCF ubiquitin ligase complex, Hrt1. Hrt1 and the homologous Apc11 protein of APC/C both contain RING–H2 domains (Zachariae et al. 1998). HRT1 is essential, and hrt1ts mutants—like cdc34ts, cdc4ts, cdc53ts, and skp1ts mutants—fail to (1) tolerate overexpression of Cln2 and Sic1, (2) transit from G1 to S phase, (3) properly degrade the SCFCdc4 substrate Sic1 and the SCFGrr1 substrate Cln2, and (4) regulate properly the expression of MET25 mRNA. hHRT1 complements the lethality of hrt1Δ, and human HRT2 binds human CUL-1 in a two-hybrid assay. Hrt1 independently binds to Cdc4, Cdc53, and Cdc34, but not to Skp1. SCFCdc4 complexes assembled with yeast Hrt1 have much higher ubiquitin ligase activity towards phospho-Sic1. Moreover, SCFGrr1 complexes containing yeast Hrt1, but not those lacking the protein, promote ubiquitination of the G1 cyclin Cln2.
SCFCdc4 ubiquitin ligase activity toward phospho-Sic1 can be reconstituted upon expression of the yeast Skp1, Cdc53, and Cdc4 proteins in insect cells (Feldman et al. 1997; Skowyra et al. 1997). This raises a simple question: Is Hrt1 absolutely required for SCFCdc4 function, or does it merely stimulate the basal activity of the heterotrimeric complex? Unambiguous resolution of this question will require the expression of SCF subunits in a system that lacks Hrt1 homologs (e.g., E. coli), or assembly of SCF complexes with mutant subunits that are selectively defective in their ability to recruit Hrt1. We favor the notion that Hrt1 is essential for the E3 activity of all SCF complexes; an insect cell homolog of the highly conserved Hrt1 may weakly substitute for the yeast protein in the SCFCdc4 complex, but perhaps not in SCFGrr1.
Cdc53 and Hrt1 comprise a ubiquitin ligase module
To identify a minimal ubiquitin ligase module in SCFCdc4, we exploited the observation that Cdc34 can conjugate ubiquitin to itself (Banerjee et al. 1993). This reaction is not thought to be of physiological significance (Goebl et al. 1994). Nevertheless, we reasoned that Cdc34 autoubiquitination might serve as a surrogate to monitor the ubiquitin ligase activity of SCF in the absence of substrate targeting requirements. Although it is formally possible that Cdc34 autoubiquitination does not faithfully mimic the ubiquitin ligase activity of SCF towards natural substrates, this seems unlikely given the potent effects of SCF and Cdc53/Hrt1 in this assay. Furthermore, activated Cdc34 assembles multiubiquitin chains upon itself with the appropriate K48 linkage (R.M.R. Feldman, unpubl.). Whereas efficient transfer of ubiquitin from Cdc34 to phospho-Sic1 requires all four subunits of SCFCdc4, either Cdc53 or a subcomplex of Cdc53/Hrt1 can partially substitute for intact SCFCdc4 to promote assembly of multiubiquitin chains on Cdc34; in contrast, Hrt1 retrieved in a complex with Cdc4 is inactive.
How does the Cdc53/Hrt1 ubiquitin ligase module work?
Four distinct classes of ubiquitin ligases have been defined; the mechanism of action of only one—the HECT domain proteins—has been described (Scheffner et al. 1995). The HECT domain ubiquitin ligase E6–AP collaborates with a member of the UbcH5 family of E2s and human papillomavirus E6 protein to specify ubiquitination of p53. Ubiquitin destined for attachment to p53 is transferred from E2 to the conserved cysteine 833 residue of E6–AP (Scheffner et al. 1995). The ubiquitin linked to E6–AP is transferred subsequently to a lysine residue of p53. Mutation of cysteine 833 destroys E6–AP’s E3 activity, indicating that the E6-AP∼ubiquitin thioester is an essential intermediate in the transfer of ubiquitin from E2 to p53. Because the SCF, APC, and Ubr1 E3s do not contain obvious HECT domains, it is unclear whether they catalyze ubiquitination by a similar mechanism.
Three lines of evidence lead us to suggest that SCF promotes Cdc34-dependent ubiquitination of substrates by a novel mechanism that does not involve an E3∼ubiquitin thioester intermediate. First, Cdc34 is able to assemble K48-linked multiubiquitin chains in the absence of an E3 (Banerjee et al. 1993). Thus, the active site of Cdc34 is inherently capable of assembling appropriate multiubiquitin chains in the absence of an additional catalytic center. Second, whereas enzymes that use cysteine residues for catalysis (including ubiquitin pathway components E1 and E2) are typically sensitive to alkylating agents such as NEM and iodoacetamide (Hershko et al. 1983), Cdc53/Hrt1-dependent autoubiquitination of Cdc34 is indifferent to NEM. The NEM resistance of Cdc53/Hrt1 is consistent with previous studies showing that mutation of all cysteine residues within Cdc53 does not impair its biological function (Patton et al. 1998a). Third, compounds lacking sulfhydryls—polylysine and polyethyleneimine—can substitute for SCFCdc4 and promote the assembly of K48 linked multiubiquitin chains on either Cdc34 or the SCFCdc4/Cdc34 pathway substrate Gcn4.
If Cdc53/Hrt1 does not activate ubiquitination of Cdc34 substrates by providing a catalytic thiol residue, how does it promote ubiquitin transfer? We propose that Cdc53/Hrt1 stabilizes the oxyanion that is formed in the transition state as the carboxyl terminus of ubiquitin is transferred from Cdc34 to the ε-amino group of a substrate lysine. Cdc53/Hrt1 (or polycations) may directly provide a positive charge to neutralize the oxyanion, or may elicit a favorable conformational change in Cdc34. Hrt1 binds both Cdc34 and components of the SCF complex directly, placing it in an optimal position to coordinate the transfer of ubiquitin from Cdc34 to substrate.
SCF and APC/C: archetypes of a family of cullin-based ubiquitin ligases
SCF complexes can be subdivided into three distinct functional domains: the F-box subunit recruits substrate (Feldman et al. 1997; Skowyra et al. 1997); the Skp1 subunit links the F-box substrate receptor to the remaining subunits; and Cdc53 (Willems et al. 1996) and Hrt1 subunits recruit the E2 enzyme and comprise the ubiquitin ligase core. Of the known members of the cullin family, the Apc2 subunit of APC/C is the most diverged from Cdc53 (Yu et al. 1998; Zachariae et al. 1998). As APC/C nevertheless also contains an Hrt1-like protein (Apc11) (Zachariae et al. 1998), we suggest that SCF and APC/C define an extended family of multisubunit E3s that are elaborated from a basic ubiquitin ligase module comprised of a cullin (CUL-1 through CUL-5, APC2) and a RING–H2 finger protein (HRT1, HRT2, or APC11). The RING–H2/cullin module in turn can be linked to families of putative substrate receptors, three of which have been defined so far: the F-box proteins for SCF (Bai et al. 1996), the SOCS box proteins for the presumed CUL-2-based ubiquitin ligases (Kamura et al. 1998), and the WD-40 repeat proteins Cdc20 and Hct1/Cdh1 for APC/C (Schwab et al. 1997; Visintin et al. 1997; Fang et al. 1998). Thus, many distinct ubiquitin ligase complexes can be assembled from the RING–H2/cullin ubiquitin ligase module.
Several other proteins implicated in ubiquitin-dependent proteolysis, including SINA (Hu and Fearon 1999), Der3/Hrd1 (Hampton et al. 1996; Bordallo et al. 1998), and the ubiquitin ligases Ubr1 and Mdm2, contain a putative RING finger (Kwon et al. 1998; Suarin et al. 1996). Both Ubr1 (F. Du and A. Varshavsky, pers. comm.) and Mdm2 (Honda et al. 1997) are thought to function without benefit of a cullin subunit. Intriguingly, E3a, which is the mammalian homolog of Ubr1, is resistant to NEM treatment (Hershko et al. 1983). Although Cys-464 has been proposed to define a divergent HECT-like catalytic center within Mdm2 (Honda et al. 1997), this residue also occupies a conserved position within Mdm2’s putative RING finger (Suarin et al. 1996). Without a definitive demonstration that ubiquitin thioesterified to Mdm2 can be chased to substrate, it is judicious to keep open the possibility that Mdm2 is in fact a member of a distinct family of RING-containing ubiquitin ligases. The RING finger may have been recruited early during eukaryotic evolution to serve as a basis for the elaboration of a broad range of cullin-dependent and -independent ubiquitin ligases.
Materials and methods
Strains, media, and reagents
All yeast strains were derivatives of W303 (HMLα or HMRa ho ade2-1 trp1-1 can1-100 leu2-3, 112 his3-11, 15 ura3-1 ssd1). Yeast strains were grown in YEP medium (1% yeast extract, 2% Bacto-Peptone, 50 mg/liter adenine) supplemented with 2% of glucose (YPD), raffinose, or raffinose plus galactose. Nocodazole (15 μg/ml) was added to arrest cells in mitosis. Rabbit polyclonal antisera against Sic1 (Mike Tyers, Samuel Lunenfeld Research Institute, Toronto, Canada), Cdc4 and Cdc34 (Zach Pitluck), and rat α-tubulin monoclonal antibody YOL1/34 (John Kilmartin, MRC, Cambridge, UK) were generously provided by the indicated investigators. Rabbit polyclonal α-Skp1 and α-Hrt1 antibodies were raised against bacterially expressed GST–Skp1 and His6–Hrt1, respectively. Production of polyclonal α-Cdc53 and cross-linked α-polyoma monoclonal antibody beads were described previously (Feldman et al. 1997). Methyl ubiquitin was purchased from Boston Biochem.
Strain and plasmid constructions
To epitope tag yeast genes, cassettes encoding an epitope tag, a 3′ UTR, and a selectable marker were amplified by PCR with target-gene-specific primers and integrated at the respective genomic locus; detailed descriptions of the tagging cassettes are available upon request. Strains containing one or five copies of a GAL promoter–SIC1–HA1 fusion integrated at the ura3 locus were generously provided by Etienne Schwab (CNRS, Montpellier, France). GAL–SIC1–HA1 HRT1–myc9 and GAL–SIC1–HA1 HRT1–stop–myc9 strains were obtained by genetic crosses.
To generate plasmids expressing yeast or human Hrt1 in bacteria, yeast, or insect cells, the appropriate open reading frames were amplified by PCR and inserted into expression vectors for E. coli (pGEX-4T-1 or pMAL-c2), Saccharomyces cerevisiae (pRD54), or a baculovirus transfer vector (pFastBac1). Cdc34ΔC and MBP–Cdc34HA were created by replacing the 24 carboxy-terminal residues of Cdc34 with six histidines or the HA epitope and cloning the resulting reading frames into pET11b or pMAL-c2, respectively. A baculovirus expressing PyHA epitope-tagged Grr1 was obtained by annealing a set of four oligos containing a Drosophila codon-optimized sequence encoding the amino-terminal region of Grr1. This segment was followed by the remainder of the yeast GRR1 sequence and the entire gene cloned into pFastbac. Sequences of the oligos and the details of cloning strategies used are available upon request.
Preparation of yeast cell extracts and immunoprecipitations
Exponentially growing yeast cells were pelleted, washed with water, and resuspended in lysis buffer (50 mm Tris-HCl at pH 7.8, 100 mm NaCl, 0.1 mm EDTA, 0.2% Triton X-100) containing protease inhibitors (1 mm PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml aprotinin). Cells were lysed with glass beads by 10 cycles of vortexing for 40 sec followed by a 1 min incubation on ice. After centrifugation (30 min at 100,000g), extracts (7 mg) were incubated with monoclonal antibody 9E10 cross-linked to protein A–Sepharose (9E10 beads, 20 μl) for 90 min at 4°C. Beads were washed four times with 1 ml of lysis buffer lacking protease inhibitors, and once with 1 ml of 50 mm Tris-HCl at pH 7.8. Proteins were eluted from the beads in a minimal amount of 50 mM Tris-HCl at pH 7.8 plus 0.2% SDS, lyophilized, resuspended in SDS-PAGE sample buffer, and applied to a 8%–16% gradient SDS–polyacrylamide gel. Protein bands were visualized by silver staining (Harlow and Lane 1988). The larger-scale prep used to obtain Hrt1 for mass spectrometry-based sequencing was performed as described above, except that 600 mg of crude extract protein from CDC53myc9 cells was applied to a 30-ml DEAE column, and the 0.35 m NaCl eluate was collected essentially as described (Deshaies et al. 1995). The resulting fraction II (100 mg) was mixed with 250 μl of 9E10 beads, and the immunoprecipitates were processed as described above except that proteins were visualized by Coomassie Blue stain following SDS-PAGE.
Mass spectrometry
A 17-kD Coomassie-stained band present in Cdc53myc9 immunoprecipitates was excised and in-gel digested with trypsin (Boehringer Mannheim, unmodified, sequencing grade) as described (Shevchenko et al. 1996b). MALDI mass spectrometry (MS) analysis of an aliquot withdrawn from the digest failed to identify proteins and therefore the recovered pool of tryptic peptides was analyzed subsequently by nanoelectrospray tandem MS (Nano ES MS/MS) (Wilm et al. 1996). Nano ES MS/MS was performed on a API III triple quadrupole mass spectrometer (PE Sciex, Ontario, Canada) equipped with a nanoelectrospray ion source (Wilm and Mann 1996) developed at European Molecular Biology Laboratory (EMBL). Short sequence stretches were determined from tandem mass spectra of individual tryptic peptides and, in combination with the masses of the intact peptides, were assembled into peptide sequence tags (Mann and Wilm 1994) and used to search a comprehensive sequence database with PeptideSearch version 3.0 software developed at EMBL.
Analysis of Hrt1 function in vivo
Small G1 cells were isolated by centrifugal elutriation from cultures grown at 25°C in YPD medium as described (Schwab and Nasmyth 1993). A Becton-Dickinson FACScan was used for flow cytometric measurement of cellular DNA content (Epstein and Cross 1992). Spindles were detected by indirect immunofluorescence using anti-tubulin antibodies (YOL1/34) and FITC-conjugated secondary antibodies. To measure cell volume, cells were fixed in YPD medium containing formaldehyde (3.8%) for 4 hr on ice. About 20,000 cells were counted with a CASY1 Cell Counter Plus Analyser (Model TTC, Schaerfe System, Reutlingen, Germany) and peak cell volume was measured in triplicate.
Swi6 and Sic1–HA1 were detected with rabbit polyclonal antibodies. Detection with monoclonal α-HA antibodies gave similar results for Sic1–HA1. RNA isolation and Northern blot analysis were performed as described (Cross and Tinkelenberg 1991; Price et al. 1991). Fragments from the coding regions of SIC1 and CMD1 were used as probes.
Protein expression
GST–Hrt1, Cdc34ΔCHis6, and MBP–Cdc34ΔCHA proteins were expressed in E. coli BL21(DE3)+pLysS cells and purified by glutathione, Ni-NTA, and amylose affinity chromatography as directed by the resin suppliers, respectively. MBP–Cdc34ΔCHA formed thioesters with ubiquitin, served as an E2 in Sic1 ubiquitination reactions, and could autoubiquitinate in the presence of Cdc53 (R.M.R. Feldman, unpubl.). Baculoviral expression of SCF subunits was as described (Feldman et al. 1997). Cln2myc/GST–Cdc28/Cks1/Cak1 proteins were expressed in Sf9 insect cells infected with the respective recombinant baculoviruses in a 10:5:3:1 ratio.
GST pull-downs, α-Py IP from insect cells
GST pull-downs contained 2.5 μg of either GST or GST–Hrt1, 50 μl (∼200 μg) lysate from insect cells infected with the indicated baculoviruses, and 100 μl of BVLB (30 mm HEPES at pH 7.4, 150 mm NaCl, 0.1% Triton X-100) containing 1 mg/ml BSA. Reactions were incubated 15 min at 4°C, supplemented with 7 μl of glutathione–agarose beads (Sigma), and incubated a further 15 min at 4°C. Beads were collected by brief centrifugation and washed four times in BVLB fortified with 100 mm NaCl (250 mm final). After washing, beads were aspirated dry and resuspended in 1× SDS-PAGE buffer, boiled, and analyzed by SDS-PAGE and Western blotting.
Experiments utilizing baculovirus-expressed Hrt1 (Fig. 3B) contained 100 μl of lysate from insect cells coinfected with Hrt1-expressing and other viruses as indicated. Lysates were incubated with 7 μl α-Py beads for 20 min at 4°C, and were washed and analyzed as described above.
In vitro assays
To analyze Cdc34ΔCHis6 autoubiquitination, 200 μl of lysate (∼4 mg/ml) from insect cells infected with the indicated baculovirus (es) was incubated with 7 μl of α-Py beads for 1 hr at 4°C. The beads were then washed three times in BVLB, three times in KAB (50 mm HEPES at pH 7.4, 50 mm NaCl, 5 mm MgCl2, 1 mm EDTA), aspirated to dryness, and resuspended in reaction buffer (30 mm Tris at pH 7.4, 5 mm MgCl2, 2 mm ATP) supplemented with 100 ng of Uba1 (Feldman et al. 1997), 5 μg ubiquitin and 600 ng of Cdc34ΔHis6 (20 μl total volume). Reactions were incubated at 24°C for 1 hr with gentle mixing, and were stopped by addition of an equal volume of 2× SDS-PAGE sample buffer and boiled for 3 min. Samples were analyzed by SDS-PAGE and Western blotting using α-Cdc34 followed by HRP-conjugated α-rabbit secondary antibodies.
Sic1 ubiquitination reactions were performed as described (Feldman et al. 1997), except that 100 ng of phosphorylated MBP–Sic1 was used per reaction, and 200 μl (∼ 800 μg) of insect cell lysate that contains polyoma-tagged SCF was immunoprecipitated with 7 μl Py beads and washed as described above, typically yielding 150–200 ng of SCF components per sample as estimated from Coomassie-stained gels. For Figure 4A, 2.5 μg of purified GST or GST–Hrt1 was added to reactions that contained 4 μg of phospho-MBP–Sic1.
To test NEM sensitivity, α-Py immunoprecipitates were prepared as described above. NEM (5 mm unless otherwise indicated) was added to beads (or to Uba1 in the presence of an equal amount of beads) and incubated at 24°C for 10 min, at which point a twofold molar excess of DTT was added and the incubation was continued for 10 min. As a control, beads were treated with NEM that had been premixed with twofold excess DTT. After incubations, beads were washed with KAB and used in reactions as described; Uba1 treated in the presence of beads was used as is.
Sequence analysis
Databases were searched with Gapped BLAST (Altschul et al. 1997). ESTs were assembled using AssemblyLIGN (Oxford Molecular, Oxford, UK), and protein sequences were aligned with CLUSTAL W (Thompson et al. 1994) using the Blosum 62 matrix with default parameters.
Acknowledgments
We thank S. Elledge, J. Kilmartin, Z. Pitluck, E. Schwab, and M. Tyers for supplying strains, baculoviruses, antibodies, libraries, and plasmids. We thank Y. Xiong for communicating unpublished data and V. Chau for stimulating discussion regarding the mechanism of SCF action. This work was supported by a grant from the National Institutes of Health (GM52466-01, R.J.D.), a Burroughs-Wellcome New Investigator Award in Basic Pharmacological Sciences (R.J.D.), a Howard Hughes Medical Institute Predoctoral Fellowship (R.M.R.F.), and a Leukemia Society Postdoctoral Fellowship (C.C.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Note
While this manuscript was under review, Ohta et al. (1999), Tan et al. (1999), Kamura et al. (1999), and Skowyra et al. (1999) reported the identification of yeast and human HRT1.
Footnotes
E-MAIL deshaies@cco.caltech.edu; FAX (626) 449-0756.
References
- Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Gregori L, Xu Y, Chau V. The bacterially expressed yeast CDC34gene product can undergo autoubiquitination to form a multiubiquitin chain-linked protein. J Biol Chem. 1993;268:5668–5675. [PubMed] [Google Scholar]
- Barral Y, Jentsch S, Mann C. G(1) cyclin turnover and nutrient-uptake are controlled by a common pathway in yeast. Genes & Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- Bartel B, Wunning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemann K. Contributions of mass spectrometry to peptide and protein structure. Biomed Environ Mass Spectrom. 1988;16:99–111. doi: 10.1002/bms.1200160119. [DOI] [PubMed] [Google Scholar]
- Bordallo J, Plemper RK, Finger A, Wolf DH. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR, Tinkelenberg AH. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Chau V, Kirschner M. Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 1995;14:303–312. doi: 10.1002/j.1460-2075.1995.tb07004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CB, Cross FR. CLB5: A novel B cyclin from budding yeast with a role in S phase. Genes & Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- Fang GW, Yu HT, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Feldman RMR, Correll C, Kaplan K, Deshaies R. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Goebl MG, Goetsch L, Byers B. The Ubc3 (Cdc34) ubiquitin-conjugating enzyme is ubiquitinated and phosphorylated in vivo. Mol Cell Biol. 1994;14:3022–3029. doi: 10.1128/mcb.14.5.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Gardner RG, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system: Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Hu G, Fearon E. Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol Cell Biol. 1999;19:724–732. doi: 10.1128/mcb.19.1.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin protein ligase. Proc Natl Acad Sci. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F- box/WD40-repeat protein Slimb. Nature. 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- Kamura T, Sato S, Haque D, Liu L, Kaelin WG, Conaway RC, Conaway JW. The elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin families. Genes & Dev. 1998;12:3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Jr, Elledge SJ, Conaway RC, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peter JM, Kirschner M. How proteolysis controls cell division. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:1–20. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- Kolman CJ, Toth J, Gonda DK. Identification of a portable determinant of cell cycle function within the carboxyl-terminal domain of the yeast CDC34 (UBC3) ubiquitin conjugating (E2) enzyme. EMBO J. 1992;11:3081–3090. doi: 10.1002/j.1460-2075.1992.tb05380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornitzer D, Raboy B, Kulka RG, Fink GR. Regulated degradation of the transcription factor GCN4. EMBO J. 1994;13:6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y, Reiss Y, Fried V, Hershko A, Yoon J, Gonda D, Sangan P, Copeland N, Jenkins N, Varshavsky A. The mouse and human genes encoding the recognition component of the N-end rule pathway. Proc Natl Acad Sci. 1998;95:7898–7903. doi: 10.1073/pnas.95.14.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M, Wilm M. Error tolerant identification of peptides in sequence databases by peptide sequence tags. Anal Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquiting ligase activity. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Sa D, Kuras L, Thomas D, Craig KL, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes & Dev. 1998a;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: Don’t Skp the F-box hypothesis. Trends Genet. 1998b;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- Price C, Nasmyth K, Schuster T. A general approach to the isolation of cell cycle-regulated genes in the budding yeast Saccharomyces cerevisiae. J Mol Biol. 1991;218:543–556. doi: 10.1016/0022-2836(91)90700-g. [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes & Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- Schwab E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes & Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- Schwab E, Böhm T, Mendenhall M, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1/S transition in Saccharomyces cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulators of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Jensen O, Podtelejnikov A, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: Large scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci. 1996a;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal Chem. 1996b;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig K, Tyers M, Elledge S, Harper J. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Suarin A, Borden K, Boddy M, Freemont P. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan ZQ. Recruitment of a ROC1-Cul1 ubiquitin ligase by Skp1 and HOS to catalyze the ubitquitination of I kappa B alpha. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- Thompson J, Higgins D, Gibson T. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalities and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997a;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- Verma R, Feldman R, Deshaies RJ. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol Biol Cell. 1997b;8:1427–1437. doi: 10.1091/mbc.8.8.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: A family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Willems AR, Lanker S, Patton EE, Craig KL, Nason TF, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- Wilm M, Mann M. Analytical properties of the nano electrospray ion source. Anal Chem. 1996;66:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nanoelectrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y. Identification of the receptor component of the IκBα-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- Yu H, Peters JM, King RW, Page AM, Hieter P, Kirschner MW. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science. 1998;279:1219–1222. doi: 10.1126/science.279.5354.1219. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Shevchenko A, Andrews PD, Ciosk R, Galova M, Stark MJ, Mann M, Nasmyth K. Mass spectrometric analysis of the anaphase-promoting complex from yeast: Identification of a subunit related to cullins. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]