Abstract
The eukaryotic cell cycle is overseen by regulatory mechanisms, termed checkpoints, that respond to DNA damage, mitotic spindle defects, and errors in the ordering of cell cycle events. The DNA replication and DNA damage cell cycle checkpoints of the fission yeast Schizosaccharomyces pombe require the hus1+ (hydroxyurea sensitive) gene. To determine the role of the mouse homolog of hus1+ in murine development and cell cycle checkpoint function, we produced a targeted disruption of mouse Hus1. Inactivation of Hus1 results in mid-gestational embryonic lethality due to widespread apoptosis and defective development of essential extra-embryonic tissues. DNA damage-inducible genes are up-regulated in Hus1-deficient embryos, and primary cells from Hus1-null embryos contain increased spontaneous chromosomal abnormalities, suggesting that loss of Hus1 leads to an accumulation of genome damage. Embryonic fibroblasts lacking Hus1 fail to proliferate in vitro, but inactivation of p21 allows for the continued growth of Hus1-deficient cells. Hus1−/−p21−/− cells display a unique profile of significantly heightened sensitivity to hydroxyurea, a DNA replication inhibitor, and ultraviolet light, but only slightly increased sensitivity to ionizing radiation. Taken together, these results indicate that mouse Hus1 functions in the maintenance of genomic stability and additionally identify an evolutionarily-conserved role for Hus1 in mediating cellular responses to genotoxins.
Keywords: Apoptosis, cell cycle checkpoints, genomic instability, DNA damage
Cell cycle checkpoints are surveillance mechanisms that monitor the cell cycle and protect genome integrity by inducing cell cycle arrest or programmed cell death in response to DNA damage, mitotic spindle defects, or errors in the ordering of cell cycle events (Hartwell and Weinert 1989; Elledge 1996). Among these regulatory pathways are the DNA replication checkpoint, which couples mitosis to the completion of DNA replication, and the DNA damage checkpoint, which promotes the repair of DNA lesions before DNA replication or cell division. In addition to overseeing progression of the mitotic cell cycle, checkpoint proteins also act to maintain telomeres (Dahlen et al. 1998; Ahmed and Hodgkin 2000), to monitor meiosis (for review, see Page and Orr-Weaver 1997), and to regulate dNTP biosynthesis and maintain the cellular capacity for DNA synthesis (Desany et al. 1998; Zhao et al. 1998). Defects in checkpoint pathways can result in cell death or genomic instability. In mammals, genomic instability is thought to promote tumorigenesis (Hartwell and Kastan 1994; Lengauer et al. 1998), and mutations in the checkpoint genes p53 (for review, see Levine 1997), ATM (for review, see Rotman and Shiloh 1999), and hCHK2 (Bell et al. 1999) are associated with familial cancer syndromes. In addition, cellular responses to anticancer drugs, many of which act by damaging DNA or inhibiting DNA replication, may depend largely on the activity of cell cycle checkpoints (Hartwell and Kastan 1994).
Recent studies indicate that several features of cell cycle checkpoints are conserved throughout evolution. As a result, knowledge of cell cycle checkpoints in the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe provides a framework for developing a further understanding of checkpoint pathways in higher eukaryotes. In fission yeast six non-essential genes (hus1+, rad1+, rad3+, rad9+, rad17+, and rad26+), known as the checkpoint rad genes, are required for both the DNA replication and DNA damage checkpoints (Al-Khodairy and Carr 1992; Enoch et al. 1992; Rowley et al. 1992; Al-Khodairy et al. 1994). With the possible exception of rad26+, each of the checkpoint rad genes is represented in mammals by a closely related homolog (for review, see Caspari and Carr 1999; Dasika et al. 1999). Evidence that this sequence homology reflects conserved protein function is provided by the rad3+-related gene ATM, which encodes a bona fide mammalian checkpoint protein. In addition to the checkpoint rad genes, other fission yeast genes that function in checkpoint responses, such as chk1+ and cds1+, also appear to be retained in higher eukaryotes (for review, see Caspari and Carr 1999; Dasika et al. 1999). Taken together, these evolutionarily-conserved checkpoint genes may represent a core checkpoint apparatus, analysis of which will likely provide considerable insight into mammalian DNA damage responses.
Fission yeast hus1+ (hydroxyurea sensitive) was originally identified in a screen for genes that, when inactivated, conferred sensitivity to the DNA replication inhibitor hydroxyurea (HU) (Enoch et al. 1992). Fission yeast lacking hus1+ appear normal under standard growth conditions, but fail to arrest the cell cycle after DNA damage or blockage of DNA synthesis (Enoch et al. 1992; Kostrub et al. 1997). Instead, hus1− strains proceed into mitosis with damaged or incompletely replicated genomes. In addition, hus1+ plays a role in the poorly understood process of recovery from S-phase arrest, as HU-treated hus1− yeast become irreversibly damaged during interphase, before undergoing premature cell division (Enoch et al. 1992). Although the Hus1p polypeptide lacks domains that might indicate its biochemical function, sequence analyses suggest that Hus1p may be structurally similar to the DNA sliding clamp protein PCNA, which forms a trimer that encircles DNA (Aravind et al. 1999; Caspari et al. 2000). Hus1p is phosphorylated after DNA damage, and this modification requires the other checkpoint rad proteins, including the ATM-related kinase Rad3p (Kostrub et al. 1998). In addition, Hus1p physically associates with two other checkpoint polypeptides, Rad1p and Rad9p (Kostrub et al. 1998; Caspari et al. 2000).
Homologs of hus1+ have been identified in a number of higher eukaryotes, including Mus musculus and Homo sapiens (Dean et al. 1998; Kostrub et al. 1998). The mammalian Hus1 proteins are similar in length to Hus1p and share ∼30% amino acid identity with the S. pombe protein. The notion that these homologs are functionally equivalent to their fission yeast counterpart is strengthened by the finding that the human HUS1 protein interacts with human RAD1 and RAD9 polypeptides (St. Onge et al. 1999; Volkmer and Karnitz 1999). Mouse Hus1 is a single copy gene located at the proximal end of chromosome 11 (Weiss et al. 1999). It is expressed throughout embryonic development and displays a ubiquitous expression pattern in adult tissues. To determine the role of Hus1 in murine development and cell cycle checkpoint function, we produced a targeted disruption of Hus1. Here, we report that Hus1 inactivation causes mid-gestational embryonic lethality. Hus1-null embryos show transcriptional induction of DNA damage-responsive genes and yield cells that contain increased spontaneous chromosomal abnormalities, suggesting that Hus1 is required for the maintenance of genomic stability. In addition, analysis of Hus1-deficient fibroblast cultures indicates that mouse Hus1, like S. pombe hus1+, has a role in cellular responses to genotoxic stress.
Results
Hus1-deficiency results in mid-gestational embryonic lethality
Mouse Hus1 was disrupted by homologous recombination in embryonic stem (ES) cells using a targeting vector in which 1 kb of genomic sequence from the Hus1 locus was deleted (Fig. 1A). Because the deleted region included Hus1 exon one and the translation initiation codon, homologous recombination of the targeting construct was predicted to produce a null Hus1 allele. The targeting construct was electroporated into TC-1 ES cells (Deng et al. 1996) and, of 120 G418- and FIAU-resistant clones screened by Southern blot hybridization, eight were found to be correctly targeted (Fig. 1B). Five of the targeted ES cell clones were micro-injected into donor blastocysts, and chimeric males derived from three of the clones transmitted the targeted allele through the germline when mated with 129S6 and Black Swiss females. Mice heterozygous for the targeted allele (Hus1+/−) were normal and fertile, and no overt phenotypes have been observed in Hus1+/− mice at up to one year of age.
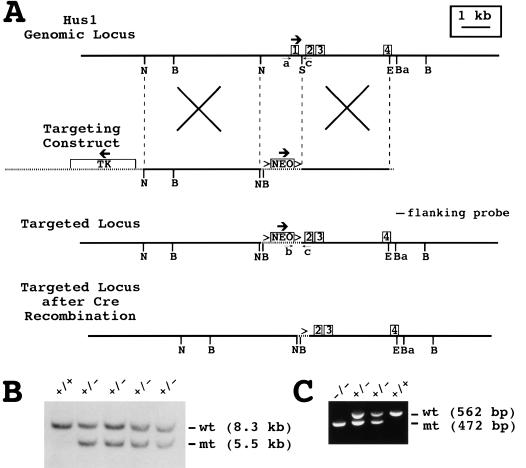
Figure 1.
Targeted disruption of the mouse Hus1 gene. (A) Restriction maps of the Hus1 genomic locus, targeting construct, targeted locus, and targeted locus following cre-mediated recombination. The first four Hus1 exons are shown as boxes. Vector-derived sequences are shown as stippled lines, and thick arrows indicate the direction of transcription for the Hus1, neomycin resistance (Neo), and thymidine kinase (Tk) genes. LoxP sites are represented by the (>) symbols. The positions of the flanking probe and PCR primers a, b, and c used for genotyping are indicated. Homologous recombination of the targeting construct is depicted by the large Xs. (B) BglI, (Ba) BamHI, (E) EcoRV, (N) NheI, (S) SmaI. (B) Southern blot analysis of ES cell genomic DNA. Genomic DNA from wild-type TC-1 cells (+/+) or four independent targeted ES cell clones (+/−) was digested with BglI and hybridized with the flanking probe. The positions of the bands corresponding to the wild-type (wt) and targeted mutant (mt) Hus1 alleles are indicated. (C) Representative PCR analysis of yolk sac genomic DNA obtained from progeny of a Hus1 heterozygote intercross using the primers shown in A. The positions of PCR products from the wild-type (wt) and targeted mutant (mt) Hus1 alleles are indicated.
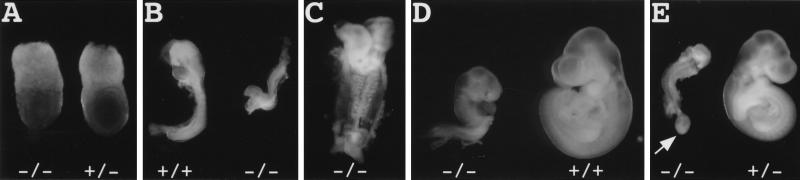
Hus1+/− mice were intercrossed in order to produce Hus1−/− mice. However, no nullizygous mice were detected among 243 offspring, indicating that loss of Hus1 caused recessive embryonic lethality (Table 1). To characterize the timing and nature of the embryonic lethality, we analyzed the morphology of embryos from timed Hus1 heterozygote intercrosses at different stages of gestation. Embryos were genotyped by PCR (Fig. 1C). At 7.5 dpc (days post coitum) Hus1−/− embryos appeared grossly normal but were on average slightly smaller than Hus1+/+ or Hus1+/− littermates (Table 1; Fig. 2A). Examination of histological sections from 7.5 dpc Hus1 embryos confirmed these observations and additionally revealed that Hus1−/− embryos gastrulated normally (data not shown). By 8.5 dpc, Hus1−/− embryos showed atypical small size and occasional morphological defects (Table 1; Fig. 2B). In addition, Hus1-deficient embryos at 8.5 dpc were developmentally delayed, possessing fewer somites than normal embryos and failing to initiate axial rotation. At 9.5 dpc, Hus1−/− embryos were invariably abnormal, showing small size, severe developmental delay, and numerous morphological abnormalities (Table 1; Fig. 2C–E). Hus1-null embryos at 9.5 dpc failed to undergo axial rotation, whereas normal embryos had completely turned by this time. The posterior of Hus1−/− embryos was severely underdeveloped and limb bud formation could not be detected. Hus1−/− embryos had eight to 13 somites at this stage, compared to the 20–24 somites of normal embryos, and the somites of Hus1-null embryos often appeared misshapen (Fig. 2C). The neural tube of Hus1−/− embryos showed extensive kinking (Fig. 2C), and the head folds of nullizygous embryos usually failed to close (Fig. 2C,E), although a few Hus1−/− embryos showed more advanced development of cranial structures (Fig. 2D). In 17% (4 of 23) of Hus1-null embryos, the allantois failed to fuse to the chorion by 9.5 dpc and instead appeared as a swollen, bulbous structure at the posterior of the embryo (Fig. 2E), a phenotype also observed in irradiated rat embryos (Kuznetsova 1957; Walshtrem 1960). Approximately one-half of Hus1−/− embryos possessed a beating heart at 9.5 dpc, but only a single live Hus1−/− embryo was identified at 10.5 dpc. No additional development of Hus1−/− embryos beyond that observed at 9.5 dpc was detected at 10.5 dpc, and intact Hus1−/− embryos were not detected at latter stages of gestation (Table 1). Similar results were obtained from the analysis of mice derived from three independent ES cell clones. In a pure 129S6 genetic background, the phenotype of Hus1−/− embryos was similar but more severe than that described above for the mixed (129S6 × Black Swiss) background, resulting in the death of all Hus1−/− embryos by 9.5 dpc (data not shown). In addition, the Hus1−/− phenotype was unchanged following cre-mediated recombination at the targeted allele (Fig. 1A; data not shown), indicating that embryonic lethality did not result from interference with neighboring gene function by the Neo cassette.
Table 1.
Genotype and phenotype analysis of offspring from Hus1 heterozygote matingsa
| Stage
|
Genotype
|
Resorptions
|
||
|---|---|---|---|---|
| +/+
|
+/−
|
−/−
|
||
| 7.5 dpc | 23 | 38 | 18 | 1 |
| 8.5 dpc | 9 | 24 (1A)b | 16 (15A) | 1 |
| 9.5 dpc | 21 | 35 (1A) | 23 (23A) | 4 |
| 10.5 dpc | 16 (1A) | 28 (1A) | 9 (9A) | 7 |
| 11.5–13.5 dpc | 10 | 25 | 0 | 16 |
| Weanling | 83 | 160 | 0 | — |
For each developmental stage, the number of embryos of each genotype and the number of resorptions is indicated. The number of phenotypically abnormal embryos is noted in parentheses.
(A) Abnormal embryo defined by small size, developmental delay, or morphological defects; see text for detailed description of mutant embryo phenotypes.
Figure 2.
Morphology of Hus1 embryos. Embryos from timed Hus1 heterozygote intercrosses were photographed at the time of dissection and genotyped thereafter. (A) 7.5 dpc, (B) 8.5 dpc, (C–E) 9.5 dpc. Arrow in E indicates hydropic allantois.
Extra-embryonic defects in Hus1-null embryos
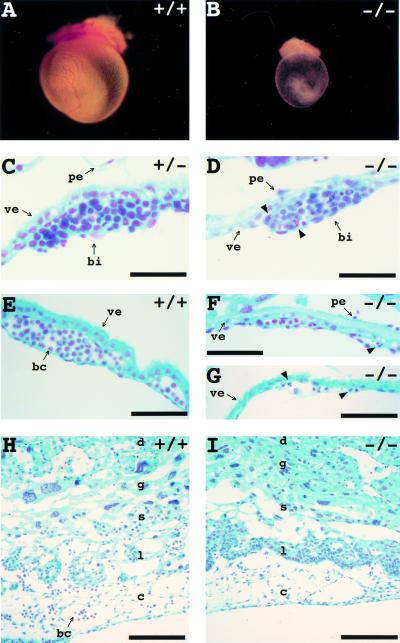
In several mouse models, the occurrence of embryonic lethality between 10.5 and 12.5 dpc is due to defective development of the yolk sac and/or placenta, which become essential at this stage of gestation (for review, see Copp 1995). Because loss of Hus1 caused a similar mid-gestational embryonic lethality, we investigated the development of extra-embryonic tissues in Hus1-null embryos. The yolk sac transports nutrients and gases from the surrounding maternal deciduum to the embryo (for review, see Cross et al. 1994; Rinkenberger et al. 1997). By gross inspection, the yolk sacs of 9.5 dpc Hus1+/+ and Hus1+/− embryos were well-vascularized and contained numerous blood-filled vitillene vessels (Fig. 3A). In contrast, the yolk sacs of Hus1−/− embryos appeared thin and fragile, and lacked prominent vasculature, although some blood cells could be detected (Fig. 3B). These observations prompted a histological analysis of yolk sac development in Hus1 embryos. The yolk sac circulation derives from extra-embryonic mesodermal aggregates termed blood islands (for review, see Cross et al. 1994; Rinkenberger et al. 1997). Blood islands formed by 8.0 dpc in Hus1 embryos of all genotypes, but the blood islands of Hus1−/− embryos contained cells with abnormal pyknotic nuclei which were not observed in blood islands from Hus1+/+ or Hus1+/− embryos (Fig. 3C,D). Furthermore, whereas the blood islands of Hus1+/+ and Hus1+/− embryos gave rise to numerous vessels filled with primitive, nucleated blood cells by 9.5 dpc (Fig. 3E), blood cells were scarce in sections of yolk sac from Hus1−/− littermates and atypical cells with pyknotic nuclei were apparent (Fig. 3F,G).
Figure 3.
Abnormal development of yolk sac and placenta in Hus1−/− embryos. (A,B) Hus1 embryos at 9.5 dpc were photographed following removal of deciduum, trophoblast giant cells, and Reichert's membrane. (C–I) Sections of embryos from timed Hus1 heterozygote intercrosses were stained by the Feulgen method. (C,D) 8.5 dpc blood islands; (E–G) 9.5 dpc yolk sacs; (H,I) 9.5 dpc placentas. Arrowheads indicate some of the cells with pyknotic or otherwise abnormal nuclei. Abbreviations: (bc) blood cell, (bi) blood island, (c) chorionic plate, (d) maternal decidua, (g) trophoblast giant cell layer, (l) labyrinthine layer, (pe) parietal endoderm, (s) spongiotrophoblast layer, (ve) visceral endoderm. Size bars: (C,D) 31 μm; (E–G) 62.5 μm; (H,I) 125 μm.
Development of the chorioallantoic placenta was also examined. As described earlier, the allantois failed to fuse with the chorion in a subset of Hus1-deficient embryos. However, the morphology of Hus1−/− embryos was the same regardless of whether chorioallantoic fusion had occurred, suggesting that the fusion events that did happen were not productive. When chorioallantoic fusion did occur, embryonic blood cells were nevertheless absent from the primitive umbilicus of Hus1−/− embryos, unlike Hus1+/+ or Hus1+/− embryos (data not shown). In addition, although all four embryonic layers of the placenta were present in Hus1−/− embryos, the labyrinthine layer was abnormally compact and avascular (Fig. 3H,I). Furthermore, numerous apoptotic cells were detected throughout the chorionic plate, and embryonic blood cells were notably absent from this region. These data suggest that the death of Hus1−/− embryos is due in part to defective development of essential extra-embryonic tissues.
Increased apoptosis but normal cell proliferation in Hus1−/− embryos
The two primary cellular responses to genotoxic stress in mammals are apoptosis and cell cycle arrest. We reasoned that, in the absence of Hus1, these processes might be deregulated and that increased apoptosis or reduced cell proliferation could account for the small size and developmental delay observed in Hus1−/− embryos. Evaluations of histological sections from Hus1 embryos indicated that Hus1−/− embryos contained increased numbers of cells with pyknotic nuclei, a characteristic of apoptotic cells. Therefore, the occurrence of apoptosis in embryo sections was assessed more rigorously by TUNEL (TdT-mediated dUTP nick end labeling) staining, a method for detecting apoptosis based on the presence of fragmented DNA in apoptotic cells. As expected, few TUNEL-positive cells were observed in Hus1+/+ and Hus1+/− embryos (Fig. 4A–D), and none were detected in control sections on which mock TUNEL staining was performed without TdT enzyme (data not shown). In contrast, TUNEL staining of Hus1−/− embryo sections at 7.5 dpc identified multiple apoptotic cells within the amniotic cavity and scattered throughout the embryo (Fig. 4F). Increased numbers of TUNEL-positive apoptotic cells were also observed in Hus1−/− embryos at 8.0 and 8.5 dpc (Fig. 4G–I). Cell death was widespread in Hus1−/− embryos and appeared to occur in all cell types. Quantitation of TUNEL-positive cells in embryonic ectoderm revealed an ∼5-fold elevation in apoptosis in Hus1-deficient embryos at 7.5, 8.0, and 8.5 dpc.
Figure 4.
Increased apoptosis but normal cell proliferation in Hus1−/− embryos. (A–D,F–I) Sections of Hus1 embryos were stained by the TUNEL method. (A,F) 7.5 dpc, (B,G) 8.0 dpc, (C,H) higher magnification view of boxed region in B and G, (D,I) 8.5 dpc. Arrows in A, C, F, and H indicate some of the TUNEL-positive apoptotic cells, which are darkly-stained in contrast to the methyl green counterstain. (E,J) Sections of BrdU-labeled Hus1 embryos at 7.5 dpc were stained with an anti-BrdU antibody. BrdU-positive cells are darkly-stained in contrast to the hematoxylin (blue) counterstain. Size bars: (A,D,F,I) 125 μm; (B,G) 250 μm; (C,E,H,J) 62.5 μm.
Further examination of histological sections from Hus1 embryos indicated that the mitotic index of Hus1−/− embryos was normal (data not shown). To confirm that cell proliferation was unaffected by loss of Hus1, we examined the ability of Hus1 embryos to incorporate BrdU, an indication of DNA synthesis and thus a marker for S-phase of the cell cycle. Embryos were labeled with BrdU in utero, sectioned, and stained with an anti-BrdU monoclonal antibody. Normal embryos at 7.5 dpc are characterized by rapid cell proliferation, and ∼85% of cells in the epiblast of Hus1+/+ or Hus1+/− embryos were BrdU-positive (Fig. 4E). A similar frequency of BrdU incorporation was detected in Hus1−/− embryos (Fig. 4J). Furthermore, regardless of genotype, ∼75% of embryonic ectodermal cells from 8.5 dpc embryos stained positive for BrdU incorporation (data not shown). Thus, cells of Hus1-deficient embryos appear to proliferate normally but undergo apoptosis at a relatively high frequency.
Increased expression of DNA damage-responsive genes in Hus1-deficient embryos
Given the role of cell cycle checkpoints in maintaining the integrity of the genome, genomic instability and DNA damage accumulation were possible consequences of Hus1 inactivation. As a readout for the presence of DNA damage in Hus1 embryos, the expression levels of p21 and other DNA damage-responsive genes were examined by Northern blot hybridization. RNA was prepared from individual Hus1 embryos at 8.5 dpc, a stage at which Hus1−/− embryos were consistently abnormal morphologically but did not show severe defects that might indirectly affect the expression of many genes. Contaminating genomic DNA in the RNA samples permitted genotyping by PCR (data not shown). The RNAs were first hybridized to a probe encompassing the entire Hus1 open reading frame to verify the genotypes obtained by PCR and to determine whether any Hus1 transcripts were produced from the targeted allele. Importantly, no Hus1 transcripts were detected in Hus1−/− embryos, confirming that the targeted disruption of Hus1 resulted in a null allele (Fig. 5A). As expected, reduced levels of Hus1 mRNA were detected in Hus1+/− embryos relative to Hus1+/+ embryos.
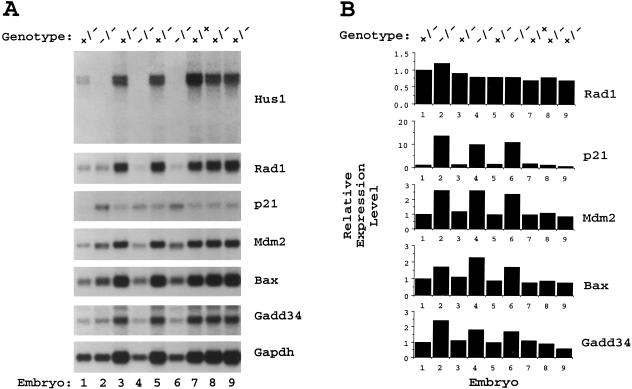
Figure 5.
Lack of detectable Hus1 transcripts but elevated expression of DNA damage-inducible genes in Hus1−/− embryos. (A) Northern blot analysis of 8.5 dpc embryos from a timed Hus1 heterozygote intercross. Total RNA was prepared from individual embryos and subjected to Northern blot hybridization with the indicated 32P-labeled cDNA probes. Note that Hus1−/− embryos are smaller than normal embryos at 8.5 dpc and therefore appear to be under-loaded. A portion of the RNA from embryo number one was lost during sample preparation and thus approximately equal amounts of RNA were obtained from this Hus1+/− embryo and the Hus1−/− embryos. (B) Quantitative analysis of gene expression levels. Radioactive signal intensity was quantitated with a PhosphorImager. Values were normalized based on results for the loading control Gapdh, and for each cDNA probe the expression level in each embryo is shown relative to the signal for embryo one, which was assigned an arbitrary expression level of one.
The same Northern blot was then sequentially hybridized to a series of cDNA probes for genes involved in cellular DNA damage responses. Because less total RNA was obtained from the undersized Hus1−/− embryos, results were quantitated on a PhosphorImager and values were normalized based on results for the loading control, Gapdh (Fig. 5B). First to be assessed was the p53-regulated, DNA damage-inducible gene p21 (for review, see Ko and Prives 1996). A striking increase in p21 mRNA was detected in all Hus1−/− embryos (mean induction of 9.9-fold in Hus1−/− versus Hus1+/+ and Hus1+/− embryos). The expression levels of Mdm2 and Bax, two additional p53-regulated genes that are induced following DNA damage (for review, see Ko and Prives 1996), were also elevated in the absence of Hus1 (mean induction of 2.6- and 2.1-fold for Mdm2 and Bax, respectively). Also examined was the expression of Gadd34, a DNA damage-responsive gene that is regulated in a p53-independent manner (Hollander et al. 1997). A modest increase in Gadd34 expression (mean induction of 2.1-fold) was detected in Hus1-null embryos. To determine the specificity of the changes in gene expression observed in Hus1−/− embryos, we examined the expression of Rad1, the human homolog of which encodes a HUS1-interacting protein (St. Onge et al. 1999; Volkmer and Karnitz 1999) that is not induced by DNA damage (Freire et al. 1998; Parker et al. 1998). Consistent with the observation that fission yeast lacking hus1+ show no alterations in rad1+ expression (Kostrub et al. 1998), Rad1 expression was found to be essentially unchanged in Hus1−/− embryos (mean induction of 1.2-fold). Thus, loss of Hus1 specifically results in transcriptional upregulation of p21 and, to lesser extents, other DNA damage-inducible genes.
Chromosomal abnormalities in Hus1−/− embryos
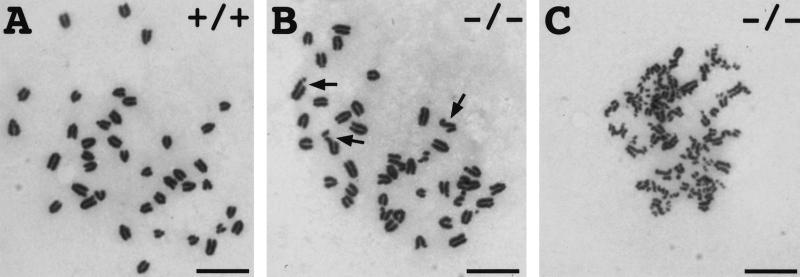
The elevated expression of p21 and other DNA damage-responsive genes observed in Hus1−/− embryos suggested that Hus1-deficient embryos might contain increased DNA damage. To directly assess genome damage in Hus1-null embryos, we examined the frequency of chromosomal abnormalities in metaphase spreads prepared from primary Hus1 embryo cultures. A total of 9 Hus1+/+, 26 Hus1+/−, and 14 Hus1−/− embryos, from five litters, were assessed. Representative metaphases are shown in Figure 6. A significant increase in chromosomal abnormalities was detected in Hus1−/− embryos, as 55% (45 of 82) of metaphases from Hus1−/− embryos contained at least one chromosomal abnormality, compared to only 19% (24 of 130) of metaphases from Hus1+/+ or Hus1+/− embryos (χ2 = 30.37, P < 0.001). Chromatid breaks were the most common abnormality in Hus1−/− embryos, and this type of aberration was observed in 43 of 82 Hus1−/− metaphases. Also detected were chromatid gaps (4 of 82), terminal deletions (5 of 82), acentric fragments (4 of 82), and chromatid exchanges (11 of 82). A similar distribution of abnormalities was observed at reduced frequency in Hus1+/+ and Hus1+/− embryos, and included chromatid breaks (21 of 130), chromatid gaps (2 of 130), acentric fragments (1 of 130), and chromatid exchanges (3 of 130). More striking than the increased occurrence of abnormal metaphases in Hus1−/− cells was the frequency of metaphases containing multiple chromosomal abnormalities. Forty-four percent (36 of 82) of metaphases from Hus1−/− embryos contained more than one aberration and 27% (22 of 82) contained greater than five, whereas only 4% (5 of 130) of metaphases from Hus1+/+ or Hus1+/− embryos contained more than one abnormality and none had greater than four abnormalities. Moreover, some metaphases from Hus1−/− cells contained such extensive chromosome damage that the abnormalities could not be enumerated (Fig. 6C). These data suggest an important role for Hus1 in the maintenance of genomic stability.
Figure 6.
Chromosomal abnormalities in primary cultures of Hus1−/− embryos. Metaphase spreads were prepared from primary cultures of 9.5 dpc Hus1 embryos and stained with Giemsa. Shown are: (A) A normal Hus1+/+ metaphase, (B) a Hus1−/− metaphase containing multiple chromatid breaks, and (C) a Hus1−/− metaphase with extensive chromosome damage. Arrows in B indicate some of the chromatid breaks. Size bar: 12.5 μm.
Increased sensitivity of Hus1-deficient cells to genotoxic stress
To facilitate additional studies of Hus1 cellular functions, we attempted to culture mouse embryonic fibroblasts (MEFs) from 9.5 dpc Hus1 embryos. Whereas Hus1+/+ and Hus1+/− MEF cultures grew well and could be serially passaged, Hus1−/− MEFs failed to proliferate (data not shown). In light of the increased chromosomal abnormalities in primary Hus1-null cells, it seemed plausible that spontaneous genome damage in Hus1−/− MEFs might trigger a Hus1-independent checkpoint, resulting in cell cycle arrest. A likely candidate for enforcing such a growth arrest was p21, given its increased expression in Hus1-null embryos. Therefore, the targeted Hus1 allele was bred onto a p21−/− background, and MEFs were generated from Hus1+/−p21−/− intercrosses at 9.5 dpc to determine if deletion of p21 might enable Hus1-deficient cells to continue proliferating. Whereas Hus1−/− MEFs failed to proliferate under the same conditions, cells from three of 13 Hus1−/−p21−/− embryos placed into culture continued to divide and could be continuously passaged. It is unclear whether the low efficiency of establishing Hus1−/−p21−/− cultures is due to a requirement for additional genetic changes or is a reflection of technical difficulties associated with culturing abnormal embryos at an early developmental stage. Northern blot analyses confirmed that the Hus1−/−p21−/− MEFs did not contain wild-type Hus1 transcripts but revealed aberrant Hus1 transcripts that were not detected in Hus1−/− embryos (data not shown). Because the targeted allele lacks Hus1 exon one and the translation initiation codon, it is unlikely that these mRNAs give rise to functional Hus1 protein, although we cannot rule out the possibility that truncated Hus1 proteins are produced. The Hus1−/−p21−/− cells showed growth rates similar to those of Hus1+/+p21−/− cells, despite the presence of increased chromosomal abnormalities (data not shown).
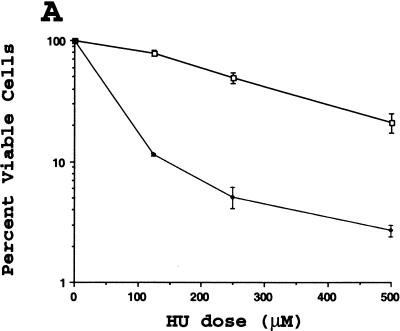
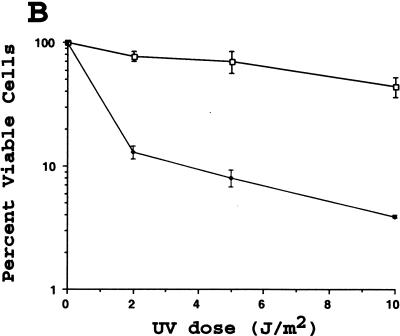
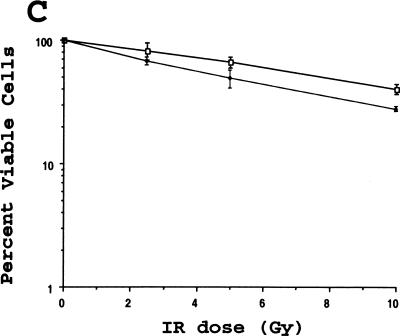
Fission yeast lacking hus1+ are highly sensitive to DNA replication inhibitors as well as various DNA damaging agents, including ultraviolet light (UV) and the radiomimetic drug bleomycin (Enoch et al. 1992; Kostrub et al. 1998). To test whether mouse Hus1 was likewise required for proper cellular responses to genotoxic stress, we examined the viability of Hus1−/−p21−/− and Hus1+/+p21−/− cells after treatment with increasing doses of HU, UV, and ionizing radiation (IR). Hus1−/− p21−/− MEFs were 5- to 10-fold more sensitive to HU than Hus1+/+p21−/− cells, suggesting that Hus1 participates in cellular responses to DNA replication blocks (Fig. 7A). The Hus1−/− p21−/− cells also showed an increased sensitivity to UV (Fig. 7B). Surprisingly, however, the Hus1−/−p21−/− cells did not display a similarly heightened sensitivity to IR. In some experiments, the Hus1−/−p21−/− cells showed a less than twofold increase in sensitivity to IR (Fig. 7C), whereas in other experiments even this slight difference in IR sensitivity was not observed. Therefore, we conclude that loss of Hus1 results in significantly increased sensitivity to HU and UV, but has a relatively minor effect on IR sensitivity.
Figure 7.
Increased sensitivity of Hus1-deficient cells to certain genotoxins. Hus1+/+p21−/− (□) or Hus1−/−p21−/− (♦) MEFs were plated in 6-well culture dishes and treated with the indicated doses of (A) HU, (B) UV, or (C) IR as described in Materials and Methods. Cell viability was assessed 72 hr after treatment and is plotted as the percent viable cells relative to results for untreated control cultures. Each point represents the mean of three samples, with error bars showing standard deviation. The plots are representative of results obtained for three independent Hus1−/−p21−/− and matched control Hus1+/+p21−/− MEF cultures.
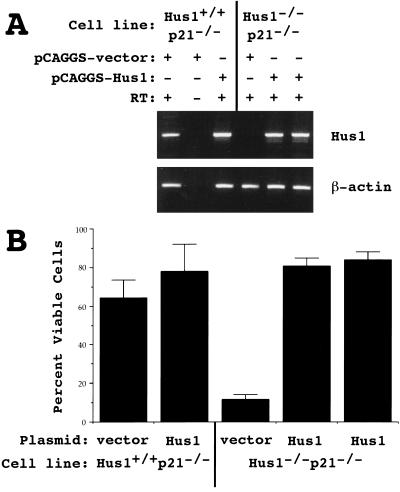
Although three independent Hus1−/−p21−/− cultures gave results similar to those shown in Figure 7, it was possible that these findings were affected by secondary mutations that might have arisen during the culturing of Hus1–null cells. To confirm that the increased sensitivity of Hus1−/−p21−/− cells to genotoxins was due to the absence of Hus1, we tested whether restoring Hus1 expression would rescue the UV sensitivity of Hus1−/−p21−/− cells. Hus1−/−p21−/− and control Hus1+/+ p21−/− cells were stably transfected with either an expression plasmid encoding Hus1 (pCAGGS–Hus1) or the empty expression plasmid vector (pCAGGS-vector). Individual clones were analyzed for Hus1 expression by RT–PCR, using primers spanning the entire Hus1 ORF (Fig. 8A). As expected, Hus1 expression was evident in Hus1+/+p21−/− cells containing either pCAGGS-vector or pCAGGS-Hus1. Hus1 expression was not detected in Hus1−/−p21−/− clones containing pCAGGS–vector, but was restored in Hus1–null cells containing pCAGGS–Hus1. The UV sensitivity of the stable clones was then tested, and representative results are shown in Figure 8B. Clones containing pCAGGS-vector showed UV sensitivities similar to those of the parental cultures, with Hus1–null clones displaying increased UV sensitivity. Significantly, Hus1 overexpression fully complemented the UV sensitivity of Hus1−/−p21−/− cells, as four of four Hus1−/−p21−/− clones overexpressing Hus1 survived UV treatment as well as Hus1+/+p21−/− cells (Fig. 8B; data not shown). The UV sensitivity of Hus1+/+p21−/− cells was not significantly affected by Hus1 overexpression. These results indicate that the heightened DNA damage sensitivity of Hus1−/−p21−/− cells is due at least in part to the absence of Hus1.
Figure 8.
Restoration of Hus1 expression complements the UV sensitivity of Hus1-deficient cells. Hus1−/−p21−/− and Hus1+/+p21−/− MEFs were stably transfected with either an empty expression plasmid vector (pCAGGS–vector) or an expression plasmid encoding Hus1 (pCAGGS–Hus1). (A) Gene expression in individual stable clones was analyzed by RT–PCR, with primers specific for Hus1 (top) or β-actin (bottom). (B) The indicated cell lines were treated with 5 J/m2 of UV and cell viability was assessed 72 hr after treatment. Values are the mean of three samples, with error bars showing standard deviation.
Discussion
Fission yeast hus1+ is a component of the cellular machinery that responds to stalled DNA replication and DNA damage. In the present study, we examined the functions of the mouse homolog of S. pombe hus1+ by assessing the consequences of a targeted disruption of mouse Hus1. Our findings identify an important role for Hus1 in the maintenance of genomic stability, as inactivation of Hus1 in mouse embryos resulted in an accumulation of spontaneous genome damage, widespread cell death, and ultimately embryonic lethality. In addition, Hus1-null cells were found to be highly sensitive to the DNA replication inhibitor hydroxyurea and DNA damaging ultraviolet light, indicating that mouse Hus1, like its fission yeast counterpart, participates in cellular responses to genotoxic stress.
Mouse Hus1 is essential for proper embryonic development, whereas fission yeast lacking hus1+ appear normal under standard growth conditions (Enoch et al. 1992). This may indicate that mouse Hus1 mediates additional functions not performed by its fission yeast homolog, or that fundamental physiological differences between yeast and mammalian cells create a greater requirement for mouse Hus1. Yet, Hus1 is not absolutely required for cell survival, as Hus1−/−p21−/− MEFs with normal growth capacities could be generated. In Drosophila, homologs of the fission yeast checkpoint proteins chk1+ (grapes) and rad3+ (mei-41) are essential specifically during embryonic development, but are dispensable in the adult fly (Hari et al. 1995; Fogarty et al. 1997; Sibon et al. 1997, 1999). Mouse embryogenesis may similarly create a strict requirement for Hus1. Cell division can occur as rapidly as every 2–3 hr in some regions of gastrulating mouse embryos (Snow 1977), and these extraordinary circumstances might necessitate regulatory mechanisms to ensure the correct ordering of cell cycle events or the repair of DNA damage. Conditional gene targeting should reveal the role of Hus1 in adult tissues.
Inactivation of Hus1 results in genomic instability, as evidenced by the increased expression of DNA damage-inducible genes in Hus1−/− embryos and the presence of increased chromosomal abnormalities in primary Hus1−/− embryo cultures. The identification of a role for Hus1 in genome maintenance raises the possibility that Hus1 is a tumor suppressor gene, as genetic instability may be an important driving force in tumorigenesis (Hartwell and Kastan 1994; Lengauer et al. 1998). Although it has not been determined whether human HUS1 is mutated in any malignancies, the HUS1 locus resides in a genomic region reported to be altered in choriocarcinomas and ovarian cancers (Dean et al. 1998).
The molecular basis for the abnormalities that arise in the absence of Hus1 remains unknown. The chromosomal aberrations visible in metaphase spreads from Hus1−/− embryos, mostly chromatid breaks, are similar to those that occur in normal embryos and therefore may represent spontaneous DNA damage that would ordinarily be repaired in a Hus1-dependent manner. On the other hand, loss of Hus1 may promote the occurrence of DNA damage. Premature progression of cells with incompletely-replicated chromosomes into mitosis due to the absence of a Hus1-mediated checkpoint could result in significant genome damage. Alternatively, the genome damage observed in Hus1-null cells could be the consequence of a failure to properly recover from transient S-phase arrest, as S. pombe hus1+ plays an important role during S-phase in the response to stalled DNA replication (Enoch et al. 1992). Further analysis of the Hus1-deficient cells should reveal which of these possible defects is responsible for the genome damage that occurs in the absence of Hus1.
Hus1−/− embryonic fibroblasts fail to proliferate and cannot be serially passaged. However, inactivation of p21 allowed for the continued growth of Hus1-deficient MEFs. The poor growth of Atm-null MEFs in vitro can similarly be rescued by deletion of p21 (Wang et al. 1997; Xu et al. 1998). We suspect that genome damage in Hus1−/− MEFs triggers cell cycle arrest and that removal of p21 enables the damaged cells to continue cycling. Consistent with this possibility, Hus1−/−p21−/− MEFs proliferate despite numerous chromosomal abnormalities (data not shown). That genome damage in Hus1−/− embryos is accompanied by widespread apoptosis, rather than growth arrest, may be a reflection of tissue-specific DNA damage responses (for review, see Evan and Littlewood 1998; Bates and Vousden 1999). It should be noted that loss of p21 does not allow for the survival of Hus1-null mice; whether it partially rescues the developmental defects of Hus1−/− embryos is under investigation.
Hus1−/−p21−/− cells are highly sensitive to HU and UV, but show little increase in IR sensitivity compared to Hus1+/+p21−/− cells. Although we cannot rule out a contribution from p21-deficiency, these phenotypes are due at least in part to the absence of Hus1, as restoration of Hus1 expression fully complements the UV sensitivity of Hus1−/−p21−/− cells. These results establish a role for Hus1 in the response to DNA replication blocks and certain forms of DNA damage. Considering that UV and IR cause different types of DNA lesions, a pathway involving Hus1 might recognize UV photoproducts, but not strand breaks caused by IR. The rad3+-related checkpoint gene Atm provides a precedent for such specialization in mammalian DNA damage responses. Whereas fission yeast lacking rad3+ are sensitive to both IR and UV (Al-Khodairy and Carr 1992; Jimenez et al. 1992), Atm-deficient cells are sensitive to IR, but not UV (for review, see Rotman and Shiloh 1999). Thus, in mammalian cells Hus1 might respond specifically to DNA damage caused by UV. Alternatively, the increased UV sensitivity of Hus1−/−p21−/− cells might reflect a deficiency in an S-phase arrest response, rather than a DNA damage checkpoint defect, as most DNA polymerases are unable to replicate templates containing UV-induced DNA lesions (for review, see Naegeli 1994). Finally, although the Hus1−/−p21−/− cells show only slightly increased IR sensitivity, a role for Hus1 in responding to IR-induced DNA damage cannot be excluded. An evolutionarily conserved function for the Hus1/Rad1/Rad9 complex in responding to strand breaks is suggested by the finding that the Caenorhabditis elegans homolog of S. pombe rad1+ (mrt-2) is required for IR-induced apoptosis in germ cells (Gartner et al. 2000). A similar function for mouse Hus1 might be masked by redundant pathways that can respond to the same damage. Because primary Hus1−/− cells contain numerous chromatid breaks, it also remains possible that the Hus1−/−p21−/− cells have adapted to this form of DNA damage and therefore are less sensitive to IR.
This analysis of Hus1-deficient embryos and cells implicates Hus1 in a subset of mammalian checkpoint pathways. An important component of many mammalian DNA damage responses is the transcription factor p53, which becomes activated in response to genotoxic stress and induces the expression of genes involved in cell cycle control, apoptosis, and DNA repair (for review, see Levine 1997). Hus1−/− embryos show elevated expression of p53 target genes p21, Mdm2, and Bax, and appear proficient for IR-induced apoptosis (R. Weiss and P. Leder, unpubl.), another process largely dependent on p53 (Norimura et al. 1996). Although direct analysis of p53 in Hus1-deficient cells is still necessary, these data suggest that Hus1 may not be required for p53 activation. In fission yeast, hus1+ acts in the same pathway as rad3+ (for review, see Caspari and Carr 1999). Whereas the mammalian checkpoint genes Atm and Atr are both related to rad3+, several lines of evidence suggest that mouse Hus1 acts in conjunction with Atr, rather than Atm. Like Hus1, Atr is essential, and both Atr−/− and Hus1−/− embryos show spontaneous chromosomal abnormalities and extensive apoptosis (Brown and Baltimore 2000). In contrast, Atm is not an essential gene (for review, see Rotman and Shiloh 1999). Furthermore, cells overexpressing dominant-negative Atr, like Hus1-null cells, show increased sensitivity to UV and HU (Cliby et al. 1998; Wright et al. 1998), whereas Atm−/− cells are not hypersensitive to UV or HU. Because Atr also appears to act in response to IR and Atr-null embryos die earlier in development than Hus1−/− embryos, Atr likely has additional Hus1-independent functions. The Hus1-deficient cells described in this study should be a useful tool for assessing the possible functional interactions between Hus1 and Atr, as well as other checkpoint components. The strong links between cell cycle checkpoints and cancer prevention underscore the importance of such efforts to develop a detailed understanding of mammalian checkpoint pathways.
Materials and methods
Plasmids
A Hus1 targeting vector was constructed in two steps in the OS.DUP/DEL vector (a gift of O. Smithies), which contains a loxP-flanked MC1-Neo expression cassette for positive selection and a PGK-TK expression module for negative selection. First, an ∼4-kb NheI fragment from plasmid pMH1 (Weiss et al. 1999) was subcloned into the NheI site of OS.DUP/DEL, producing pOS–HKO5′. Next, an ∼3.5-kb SmaI–EcoRV fragment was removed from plasmid pMH1, blunt-ended, and cloned into a blunt-ended BamHI site of pOS–HKO5′, generating the final targeting construct, pOS–HKO (Fig. 1A). After generating Hus1+/− mice using pOS-HKO, we were notified that the Neo cassette in OS.DUP/DEL does not contain a polyA signal sequence (O. Smithies, pers. comm.). However, in the case of Hus1 targeting, this did not appear to compromise Neo function, as evidenced by the appearance of typical numbers of G418-resistant colonies (data not shown), or lead to the generation of detectable read-through transcripts in embryos (see Fig. 5A). The Hus1 expression plasmid pCAGGS–Hus1 was generated by cloning the Hus1 ORF into the pCAGGS vector (Niwa et al. 1991).
ES cell culture, gene targeting, and generation of Hus1 mice
TC-1 ES cells were electroporated with NotI-linearized pOS-HKO and selected in G418 and FIAU as described previously (Deng et al. 1996). Genomic DNA from 120 drug-resistant colonies was screened for homologous recombination at the Hus1 locus by Southern blot hybridization with an external probe, and eight correctly-targeted clones were identified. The correct targeting of the heterozygous clones was verified by additional Southern blot analyses using alternative restriction enzyme digests, as well as an internal probe. ES cells from correctly-targeted clones were micro-injected into C57BL/6 (Taconic) blastocysts, which were then transferred into pseudopregnant Swiss Webster (Taconic) foster mothers. Chimeric male mice, identified by the presence of agouti coat color, were mated to 129S6 (Taconic) and Black Swiss (Taconic) females, and transmission of the targeted allele through the germline was determined by coat color and Southern blot analyses. Mice heterozygous for the targeted allele were intercrossed and also mated to EIIa-cre transgenic mice (a gift of F. Alt) for cre-mediated recombination and excision of the Neo cassette (Lakso et al. 1996). For timed matings, mice were mated overnight and females were inspected for vaginal plugs the following morning. Noon on the day of vaginal plug detection was defined as day 0.5 of gestation.
Genotyping by Southern blot and PCR analyses
Genomic DNA was isolated from ES cells and tail biopsy fragments by proteinase K digestion and precipitation with ethanol. Whole-embryo and yolk sac genomic DNA was prepared by incubating samples in digestion buffer (500 mm Tris at pH 9.5, 10 mm NaCl, 20 mm EDTA, 3.0% Tween-20, 0.5% xylene cyanol, 1.5 mg/ml proteinase K) at 50°C overnight, diluting the sample fivefold with water, and heating at 100°C for 10 min. Genomic DNA was obtained from embryo sections as described (Zeitlin et al. 1995). For Southern blot analyses, approximately 10 μg of genomic DNA was digested with BglI, resolved on a 0.8% agarose gel, transferred to a nylon membrane, and hybridized with 32P-labeled probe. To generate an external probe, we first subcloned a blunt-ended BglI–BamHI fragment from plasmid pMH2 (Weiss et al. 1999) into the SmaI site of pBluescript-SK, producing pBS–Int4. The flanking probe (Fig. 1A) was a SspI–PstI fragment from pBS–Int4. For PCR analysis, 0.5–2.0 μl of genomic DNA was PCR amplified with Taq DNA polymerase (Boehringer Mannheim) under standard reaction conditions in a total volume of 50 μl with the three oligonucleotide primers depicted in Figure 1A: (a) 5′-CCGTCGGCCTGGTATCCGCCATGA-3′; (b) 5′-GGTATCGCCGCTCCCGATTCGCAG-3′; (c) 5′-GGGCTGATGCGGAGGGTGCAGGTT-3′. After denaturation at 94°C for 5 min, 35 cycles of amplification (94°C, 20 sec; 60°C, 30 sec; 72°C, 1 min) were performed, followed by a final extension step of 72°C for 10 min.
Histological analyses
Decidua from timed Hus1 heterozygote matings were isolated at 7.5–9.5 dpc. Following fixation for 2–4 hr at 4°C in 4% paraformaldehyde, 1× PBS (137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, 1.4 mm KH2PO4), decidua were sequentially dehydrated in 50%, 70%, 95%, and 100% ethanol, cleared in xylenes, and embedded in paraffin. Paraffin blocks were sectioned at 5 μm. Staining of sections by the Feulgen reaction was performed as described previously (Chester et al. 1998). TUNEL staining was performed with the ApopTag peroxidase in Situ apoptosis detection kit (Intergen Co.) according to the manufacturer's directions. VIP (Vector Laboratories) was used as the peroxidase substrate, and counterstaining was performed with 0.5% (wt/vol) methyl green in 0.1 m sodium acetate (pH 4.0). To analyze BrdU incorporation, we injected BrdU (100 μg per gram of body weight) intraperitoneally into pregnant females. One hour after BrdU injection, decidua were isolated, processed, and sectioned as described above. BrdU staining was performed with a biotinylated mouse anti-BrdU antibody using a BrdU staining kit (Zymed Laboratories, Inc.) according to the manufacturer's directions, with the exception that 5 μg/ml proteinase K was used in place of trypsin for antigen retrieval. DAB was used as the peroxidase substrate and counterstaining was performed with hematoxylin.
Northern blot hybridizations and RT–PCR
Embryos from timed Hus1 heterozygote matings at 8.5 dpc were dissected free of the decidua. Total RNA was prepared from individual embryos, including yolk sac and amnion, or MEF cultures using the RNA STAT-60 reagent (Tel-Test, Inc.). A fraction of each RNA sample was reserved for genotyping by PCR and the remainder was subjected to Northern blotting as described previously (Weiss et al. 1999). For RT–PCR, cDNA was prepared from 5 μg of DNase-treated total RNA by random priming using the SuperScript preamplification system (Life Technologies, Inc.). PCR amplification of cDNAs was done using primers specific for Hus1 (5′-ATGAAGTTTCGCGCCAAGATCGTGGACC-3′ and 5′-CTAGGACAAGGCTGGGATGAAATACTG-3′) or β-actin (5′-TGTGATGGTGGGAATGGGTCAG-3′ and 5′-TTTGATGTCACGCACGATTTCC-3′).
Chromosome preparations
Embryos from Hus1 heterozygote intercrosses at 9.5 dpc were dissected from the deciduum in Hanks' Balanced Salt Solution, mechanically disrupted in 100 μl of 0.25% trypsin, 1.0 mm EDTA, and placed into individual wells of gelatinized 24-well culture dishes in culture medium (DMEM supplemented with 20% fetal bovine serum, 1.0 mm l-glutamine, 0.1 mm MEM non-essential amino acids, 100 μg/ml of streptomycin sulfate, and 100 U/ml of penicillin). Yolk sacs were reserved for genotyping. The following day, the embryo cultures were incubated in culture medium containing 0.1 μg/ml of colcemid for 2 hr. Cells were then harvested by trypsinization, swollen for 10 min at 37°C in 0.075 m KCl, and fixed for at least 20 min at 4°C in 75% methanol, 25% acetic acid. Cells in fixative were then spotted onto microscope slides and stained with 2.0% Giemsa in phosphate buffer (pH 6.8). Chromosomal abnormalities were scored based on guidelines in the International System for Human Cytogenetic Nomenclature (Mitelman 1995). Statistical comparison of the frequency of chromosomal abnormalities was done by Pearson Chi-square.
Hus1 MEF cultures, cell survival assays, and stable transfections
Hus1+/− mice were bred with p21−/− mice (Deng et al. 1995), and individual embryos from Hus1+/−p21−/− intercrosses at 9.5 dpc were placed into culture as described above. After the cultures expanded to fill a 10-cm dish, the culture medium serum content was reduced to 10%, and the cells were subsequently maintained on a 3T3 culture protocol in which 106 cells were passed onto a new gelatinized 10-cm dish every three days. Sensitivity to genotoxins was assessed by seeding 105 cells into individual gelatinized wells of a six-well dish in triplicate and treating the cultures the following day. For UV treatment, the culture medium was removed, cells were exposed to 254 nm UV light in a XL-1500 Spectrolinker (Spectronics Corp), and fresh medium was placed onto the cells. For IR treatment, cells were irradiated with a 137Cs source (Mark 1 Irradiator, J.L. Sheperd and Sons) at a dose rate of 2.0 Gy/min. For HU treatment, cells were cultured in medium containing HU (Sigma) for 24 hr, and then the medium was removed and replaced with culture medium lacking HU. Three days after treatment, cells were harvested by trypsinization, incubated with trypan blue dye, and counted. For restoration of Hus1 expression, MEFs were transfected with either 2.0 μg pCAGGS-vector or 2.0 μg of pCAGGS-Hus1 and 0.2 μg of PGK-puro DNA using the Lipofectamine Plus transfection reagent (Life Technologies, Inc.) according to manufacturer's directions. Transfected cells were selected in 2.5 μg/ml of puromycin, and individual clones were isolated, expanded, and analyzed.
Acknowledgments
We thank Fred Alt, Nick Chester, Jan Pinkas, and Oliver Smithies for reagents; Cynthia Morton and Charles Lee for advice on chromosome analysis; Anne Harrington for performing blastocyst microinjections; Cathie Daugherty for assistance with ES cell culture; Rick Van Etten for assistance with cell irradiation; and Mark Bedford, Nick Chester, Liz Harrington, Boris Reizis, Yaoqi Wang, and Tom Wolkow for helpful discussions and for critically reading the manuscript. R.S.W. is supported by a postdoctoral fellowship (PF-98-131-01) from the American Cancer Society, T.E. is supported by NIH grant number GM50015, and P.L. is a senior investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL leder@rascal.med.harvard.edu; FAX (617) 432-7663.
References
- Ahmed S, Hodgkin J. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature. 2000;403:159–164. doi: 10.1038/35003120. [DOI] [PubMed] [Google Scholar]
- Al-Khodairy F, Carr AM. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khodairy F, Fotou E, Sheldrick KS, Griffiths DJ, Lehmann AR, Carr AM. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, Vousden KH. Mechanisms of p53-mediated apoptosis. Cell Mol Life Sci. 1999;55:28–37. doi: 10.1007/s000180050267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DW, Varley JM, Szydlo TE, Kang DH, Wahrer DC, Shannon KE, Lubratovich M, Verselis SJ, Isselbacher KJ, Fraumeni JF, et al. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science. 1999;286:2528–2531. doi: 10.1126/science.286.5449.2528. [DOI] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes & Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- Caspari T, Carr AM. DNA structure checkpoint pathways in Schizosaccharomyces pombe. Biochimie. 1999;81:173–181. doi: 10.1016/s0300-9084(99)80050-9. [DOI] [PubMed] [Google Scholar]
- Caspari T, Dahlen M, Kanter-Smoler G, Lindsay HD, Hofmann K, Papadimitriou K, Sunnerhagen P, Carr AM. Characterization of Schizosaccharomyces pombe Hus1: A PCNA-related protein that associates with Rad1 and Rad9. Mol Cell Biol. 2000;20:1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester N, Kuo F, Kozak C, O'Hara CD, Leder P. Stage-specific apoptosis, developmental delay, and embryonic lethality in mice homozygous for a targeted disruption in the murine Bloom's syndrome gene. Genes & Dev. 1998;12:3382–3393. doi: 10.1101/gad.12.21.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliby WA, Roberts CJ, Cimprich KA, Stringer CM, Lamb JR, Schreiber SL, Friend SH. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp AJ. Death before birth: Clues from gene knockouts and mutations. Trends Genet. 1995;11:87–93. doi: 10.1016/S0168-9525(00)89008-3. [DOI] [PubMed] [Google Scholar]
- Cross JC, Werb Z, Fisher SJ. Implantation and the placenta: Key pieces of the development puzzle. Science. 1994;266:1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- Dahlen M, Olsson T, Kanter-Smoler G, Ramne A, Sunnerhagen P. Regulation of telomere length by checkpoint genes in Schizosaccharomyces pombe. Mol Biol Cell. 1998;9:611–621. doi: 10.1091/mbc.9.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasika GK, Lin SC, Zhao S, Sung P, Tomkinson A, Lee EY. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene. 1999;18:7883–7899. doi: 10.1038/sj.onc.1203283. [DOI] [PubMed] [Google Scholar]
- Dean FB, Lian L, O'Donnell M. cDNA cloning and gene mapping of human homologs for Schizosaccharomyces pombe rad17, rad1, and hus1 and cloning of homologs from mouse, Caenorhabditis elegans, and Drosophila melanogaster. Genomics. 1998;54:424–436. doi: 10.1006/geno.1998.5587. [DOI] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes & Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: Preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Enoch T, Carr AM, Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes & Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- Evan G, Littlewood T. A matter of life and cell death. Science. 1998;281:1317–1322. doi: 10.1126/science.281.5381.1317. [DOI] [PubMed] [Google Scholar]
- Fogarty P, Campbell SD, Abu-Shumays R, Phalle BS, Yu KR, Uy GL, Goldberg ML, Sullivan W. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr Biol. 1997;7:418–426. doi: 10.1016/s0960-9822(06)00189-8. [DOI] [PubMed] [Google Scholar]
- Freire R, Murguia JR, Tarsounas M, Lowndes NF, Moens PB, Jackson SP. Human and mouse homologs of Schizosaccharomyces pombe rad1+ and Saccharomyces cerevisiae RAD17: Linkage to checkpoint control and mammalian meiosis. Genes & Dev. 1998;12:2560–2573. doi: 10.1101/gad.12.16.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- Hari KL, Santerre A, Sekelsky JJ, McKim KS, Boyd JB, Hawley RS. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell. 1995;82:815–821. doi: 10.1016/0092-8674(95)90478-6. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: Controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hollander MC, Zhan Q, Bae I, Fornace AJ., Jr Mammalian GADD34, an apoptosis- and DNA damage-inducible gene. J Biol Chem. 1997;272:13731–13737. doi: 10.1074/jbc.272.21.13731. [DOI] [PubMed] [Google Scholar]
- Jimenez G, Yucel J, Rowley R, Subramani S. The rad3+ gene of Schizosaccharomyces pombe is involved in multiple checkpoint functions and in DNA repair. Proc Natl Acad Sci. 1992;89:4952–4956. doi: 10.1073/pnas.89.11.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: Puzzle and paradigm. Genes & Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Kostrub CF, Al-Khodairy F, Ghazizadeh H, Carr AM, Enoch T. Molecular analysis of hus1+, a fission yeast gene required for S-M and DNA damage checkpoints. Mol Gen Genet. 1997;254:389–399. doi: 10.1007/pl00008606. [DOI] [PubMed] [Google Scholar]
- Kostrub CF, Knudsen K, Subramani S, Enoch T. Hus1p, a conserved fission yeast checkpoint protein, interacts with Rad1p and is phosphorylated in response to DNA damage. EMBO J. 1998;17:2055–2066. doi: 10.1093/emboj/17.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova MN. Histopathology of the placenta in radiation sickness. Akush Ginekol (Mosc) 1957;33/4:50–55. [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Mitelman F. ISCN (1995): An International System for Human Cytogenetic Nomenclature. Basel: S. Karger; 1995. [Google Scholar]
- Naegeli H. Roadblocks and detours during DNA replication: Mechanisms of mutagenesis in mammalian cells. BioEssays. 1994;16:557–564. doi: 10.1002/bies.950160809. [DOI] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Norimura T, Nomoto S, Katsuki M, Gondo Y, Kondo S. p53-dependent apoptosis suppresses radiation-induced teratogenesis. Nat Med. 1996;2:577–580. doi: 10.1038/nm0596-577. [DOI] [PubMed] [Google Scholar]
- Page AW, Orr-Weaver TL. Stopping and starting the meiotic cell cycle. Curr Opin Genet Dev. 1997;7:23–31. doi: 10.1016/s0959-437x(97)80105-0. [DOI] [PubMed] [Google Scholar]
- Parker AE, Van de Weyer I, Laus MC, Oostveen I, Yon J, Verhasselt P, Luyten WH. A human homologue of the Schizosaccharomyces pombe rad1+ checkpoint gene encodes an exonuclease. J Biol Chem. 1998;273:18332–18339. doi: 10.1074/jbc.273.29.18332. [DOI] [PubMed] [Google Scholar]
- Rinkenberger JL, Cross JC, Werb Z. Molecular genetics of implantation in the mouse. Dev Genet. 1997;21:6–20. doi: 10.1002/(SICI)1520-6408(1997)21:1<6::AID-DVG2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Rotman G, Shiloh Y. ATM: A mediator of multiple responses to genotoxic stress. Oncogene. 1999;18:6135–6144. doi: 10.1038/sj.onc.1203124. [DOI] [PubMed] [Google Scholar]
- Rowley R, Subramani S, Young PG. Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J. 1992;11:1335–1342. doi: 10.1002/j.1460-2075.1992.tb05178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibon OC, Laurencon A, Hawley R, Theurkauf WE. The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr Biol. 1999;9:302–312. doi: 10.1016/s0960-9822(99)80138-9. [DOI] [PubMed] [Google Scholar]
- Sibon OC, Stevenson VA, Theurkauf WE. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature. 1997;388:93–97. doi: 10.1038/40439. [DOI] [PubMed] [Google Scholar]
- Snow MHL. Gastrulation in the mouse: Growth and regionalization of the epiblast. J Embryol Exp Morph. 1977;42:293–303. [Google Scholar]
- St. Onge RP, Udell CM, Casselman R, Davey S. The human G2 checkpoint control protein hRAD9 is a nuclear phosphoprotein that forms complexes with hRAD1 and hHUS1. Mol Biol Cell. 1999;10:1985–1995. doi: 10.1091/mbc.10.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmer E, Karnitz LM. Human homologs of Schizosaccharomyces pombe Rad1, Hus1, and Rad9 form a DNA damage-responsive protein complex. J Biol Chem. 1999;274:567–570. doi: 10.1074/jbc.274.2.567. [DOI] [PubMed] [Google Scholar]
- Walshtrem EA. Pathogenesis of irradiation hazards and reparative processes in rat embryos after x-raying of rat females on the 10th day of pregnancy. Arkh Anat Gistol Embriol. 1960;38:72–79. [PubMed] [Google Scholar]
- Wang YA, Elson A, Leder P. Loss of p21 increases sensitivity to ionizing radiation and delays the onset of lymphoma in atm-deficient mice. Proc Natl Acad Sci. 1997;94:14590–14595. doi: 10.1073/pnas.94.26.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RS, Kostrub CF, Enoch T, Leder P. Mouse Hus1, a homolog of the Schizosaccharomyces pombe hus1+ cell cycle checkpoint gene. Genomics. 1999;59:32–39. doi: 10.1006/geno.1999.5865. [DOI] [PubMed] [Google Scholar]
- Wright JA, Keegan KS, Herendeen DR, Bentley NJ, Carr AM, Hoekstra MF, Concannon P. Protein kinase mutants of human ATR increase sensitivity to UV and ionizing radiation and abrogate cell cycle checkpoint control. Proc Natl Acad Sci. 1998;95:7445–7450. doi: 10.1073/pnas.95.13.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Yang EM, Brugarolas J, Jacks T, Baltimore D. Involvement of p53 and p21 in cellular defects and tumorigenesis in Atm−/− mice. Mol Cell Biol. 1998;18:4385–4390. doi: 10.1128/mcb.18.7.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]