Abstract
Patched (Ptc), initially identified in Drosophila, defines a class of multipass membrane proteins that control cell fate and cell proliferation. Biochemical studies in vertebrates indicate that the membrane proteins Ptc and Smoothened (Smo) form a receptor complex that binds Hedgehog (Hh) morphogens. Smo transduces the Hh signal to downstream effectors. The Caenorhabditis elegans genome encodes two Ptc homologs and one related pseudogene but does not encode obvious Hh or Smo homologs. We have analyzed ptc-1 by RNAi and mutational deletion and find that it is an essential gene, although the absence of ptc-1 has no detectable effect on body patterning or proliferation. Therefore, the C. elegans ptc-1 gene is functional despite the lack of Hh and Smo homologs. We find that the activity and expression of ptc-1 is essentially confined to the germ line and its progenitors. ptc-1 null mutants are sterile with multinucleate germ cells arising from a probable cytokinesis defect. We have also identified a surprisingly large family of PTC-related proteins containing sterol-sensing domains, including homologs of Drosophila dispatched, in C. elegans and other phyla. These results suggest that the PTC superfamily has multiple functions in animal development.
Keywords: Multinucleate, cytokinesis, Hedgehog, signaling, C. elegans, cholesterol
Intercellular signaling has a central role in specifying cell fate and establishing body pattern. In recent years, the Hedgehog (Hh) morphogen (Hammerschmidt et al. 1997) and the Patched (Ptc) receptor have been shown to control many aspects of pattern formation and cell proliferation in Drosophila and vertebrates (Ingham 1998; Johnson and Scott 1998). The Caenorhabditis elegans ptc-1 gene is the focus of this study: C. elegans ptc genes have not been studied previously despite the availability of a complete genome sequence.
The ptc and hh genes were originally identified in Drosophila because of their roles in specifying anterior/posterior polarity in body segments and limbs (Ingham 1998; Johnson and Scott 1998). Hh acts as a short-range signal originating from posterior cells that induces anterior cells to secrete other signaling molecules, such as Wingless (Wg) or Decapentaplegic (Dpp) (Hidalgo and Ingham 1990; Ingham et al. 1991). The membrane protein Ptc appears to transduce the Hh signal and also limit its movement (Chen and Struhl 1996). The absence of ptc results in a phenotype that might be expected if there was uniform reception of the Hh signal in the anterior compartment.
The Drosophila ptc gene encodes a membrane protein with 12 predicted transmembrane (TM) domains (Hooper and Scott 1989; Nakano et al. 1989). The TM domains appear to be arranged in two sets of (1 + 5) membrane-spanning domains reminiscent of transporters; in particular, the TM topology is similar to that of the resistance/nodulation/cell division (RND) family of prototypic bacterial multidrug efflux pumps (Paulsen et al. 1996). Ptc also contains a sterol-sensing domain (SSD), which is associated with proteins involved in cholesterol homeostasis (Lange and Steck 1998; Osborne and Rosenfeld 1998). The presence of an SSD became apparent after the cloning of NPC1, a gene responsible for the neurovisceral disorder Niemann Pick type-C (NPC) disease, which affects cholesterol trafficking. There is increasing evidence that cholesterol may regulate the release of Hh from signaling cells and also help to target Hh to receiving cells expressing Ptc (Porter et al. 1996a,b; Cooper et al. 1998; Burke et al. 1999). It has been shown recently that Dispatched (Disp), which carries an SSD and shares sequence similarity to Ptc, may facilitate Hh release from signaling cells (Burke et al. 1999). Two Disp homologs are found in C. elegans (Burke et al. 1999; this paper).
In vertebrates, PTCH not only specifies cell fate but also regulates cell proliferation. Mutations in PTCH are associated with Gorlin's disease, a rare inherited disease causing cancer and developmental abnormalities; sporadic mutations in PTCH can lead to basal cell carcinomas (Hahn et al. 1996; Johnson et al. 1996). PTCH has the properties of a classic tumor suppressor. In contrast to Drosophila, vertebrate genomes encode multiple Hh proteins (Hammerschmidt et al. 1997) and at least two PTCH proteins (Concordet et al. 1996; Carpenter et al. 1998; Motoyama et al. 1998).
Our present understanding of the signaling pathway involving Hh and Ptc is derived from genetic studies in Drosophila and biochemical studies in vertebrates (Ingham 1998; Johnson and Scott 1998). Biochemical studies have shown that Ptc complexes with the serpentine membrane protein Smoothened (Smo) (Marigo et al. 1996; Stone et al. 1996). It is proposed that Ptc inhibits Smo and that Hh relieves Smo from this inhibition by binding Ptc. In turn, Smo promotes Cubitus interruptus (Ci) to up-regulate expression of ptc and the growth factor genes wg and dpp. The mechanism by which these downstream signaling events are mediated is not understood; however, a cytoplasmic complex composed of the Fused kinase, the kinesin-related protein Costal2, and Ci has been implicated in regulating the entry of Ci into the nucleus (Ingham 1998; Johnson and Scott 1998).
The completion of the C. elegans genome sequence means that most nematode genes have been identified at the sequence level (Consortium 1998). We observed that the worm had at least one gene encoding a PTC protein but no genes encoding obvious Hh or Smo homologs. Despite the absence of a Hh homolog with sequence similarity to the signaling domain of Hh, the C. elegans genome encodes a large family of Hh-related proteins, which are so far found in other nematodes but not in other phyla (Aspöck et al. 1999). Some of these predicted proteins may have signaling activities that are potentially homologous in function to Hh (Bürglin 1996; Porter et al. 1996b; Hall et al. 1997; Aspöck et al. 1999). Given the importance of Hh and Smo in controlling the activity of PTC in other organisms, the conspicuous absence of these genes in C. elegans led us to ask whether the activity of Ce-PTC-1 was conserved. From an evolutionary standpoint, a study of the C. elegans ptc genes might shed light on the ancestral roles of PTC proteins or uncover new functions for these proteins.
Here we report that the C. elegans genome encodes three PTC homologs and a large number of PTC-related proteins. We show that PTC-1 is functional despite the surprising absence of Hh and Smo homologs. Inactivation of the C. elegans ptc-1 gene by RNA-mediated interference (RNAi) or gene deletion reveals that ptc-1 is needed for cytokinesis in the germ-line syncytium but is not essential for zygotic development. These findings expand the repertoire of functions for PTC proteins and indicate that their activities and control have diverged or have been elaborated upon during evolution.
Results
The genome of C. elegans encodes multiple ptc genes
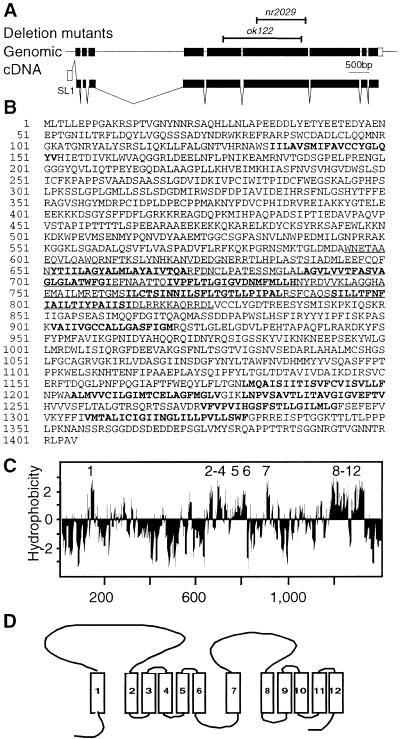
We began a characterization of the ptc-1 gene found on cosmid ZK675 (LGII) (Consortium 1998) by obtaining a full-length ptc-1 cDNA sequence, which carries the trans-spliced SL1 sequence, from clones generated by RT–PCR. A single ptc-1 4.3-kb transcript was detected by Northern analysis (data not shown). The intron/exon organization of the ptc-1 locus was deduced by comparing cDNA to genomic sequence (Fig. 1A). Ce-ptc-1 encodes a predicted protein of 1405 amino acids (Fig. 1B), named PTC-1, which shares 50% similarity (29% identity) to Drosophila Ptc and 46% similarity (30% identity) to human PTCH1. The PTC-1 protein contains a putative SSD found in other PTC and NPC proteins (Carstea et al. 1997; Loftus et al. 1997). Hydropathy analysis indicates that PTC-1 has 12 potential membrane-spanning domains, which can assume a topology similar to that predicted for Drosophila and vertebrate PTC proteins (Fig. 1C,D). The predicted topology of PTC-1 deviates from that of PTCH1 because it has a shorter carboxy-terminal intracellular tail, a 65-amino-acid shorter intracellular loop between TM6 and TM7, and a 130-amino-acid-longer extracellular loop between TM1 and TM2.
Figure 1.
Organization of the C. elegans ptc-1 gene. (A) The ptc-1 cDNA sequence was obtained by sequencing RT–PCR fragments. ptc-1 mRNA is at least 4.24 kb in length, composed of 9 exons, and carries the SL1 trans-splice leader sequence. Overhead bars indicate the extent of the ptc-1 sequence removed by the deletion alleles ok122 and nr2029. (B) The PTC-1 protein sequence. Amino acids in bold correspond to the 12 predicted TM domains. The underlined sequence corresponds to a predicted SSD (Carstea et al. 1997; Loftus et al. 1997). (C) Hydropathy profile of the PTC-1 protein using a window size of 9 amino acids (Kyte and Doolittle 1982). The 12 numbered peaks represent predicted TM domains. (D) Predicted topology of PTC-1.
Surprisingly, BLAST searches also revealed that cosmid F21H12 (LGII) carries three predicted ORFs with >95% nucleotide identity to ptc-1. Collectively, the locus carrying these ORFs has been designated ptc-2, although not all ORFs are transcribed from the same strand. All three predicted PTC-2 isoforms are severely truncated, and attempts to generate F21H12.4-specific cDNA sequences by RT–PCR have been unsuccessful. Taken together, these data indicate that ptc-2 is likely to be a pseudogene that arose through a recent gene duplication.
Further database searches using the ptc-1 cDNA sequence identified two ESTs, yk68c12 and yk92c4 (kindly provided by Y. Kohara, National Institute of Genetics, Mishima, Japan). We sequenced these cDNAs and found that they defined a third ptc gene, named ptc-3; the complete ptc-3 cDNA sequence was obtained by RT–PCR (P. Kuwabara, unpubl.). The predicted PTC-3 is composed of 1358 amino acids with 61% similarity (44% identity) to PTC-1; PTC-3 is also likely to share the same topology as PTC-1.
A new C. elegans PTC superfamily: identification of ptc-related genes
We identified 23 additional C. elegans ORFs with significant sequence similarity to PTC using BLAST and sensitive hidden Markov models (HMMs); several of these ORFs were noted previously (Johnson and Scott 1997). The genes encoding these proteins have been named ptc-related (ptr); ESTs are associated with many of these genes (Table 1). All of the predicted ptr genes encode proteins with significant similarity to a protein encoded by a 131-kb unannotated genomic sequence from human chromosome 10p12.1 (AC005875) and to human NPC1 and PTCH. The predicted Drosophila protein closest in similarity to the PTR proteins is encoded by a gene located at 42A10 (AAF57274). Two other Ce-ptr members have been named ptd-1 and ptd-2 (patched-related, dispatched-like) because they encode proteins with similarity to Drosophila Disp (Burke et al. 1999). Hydropathy analyses indicate that PTR proteins are likely to encode TM proteins with topologies similar to PTC-1 and PTC-3 (Table 1). Clustal W analysis shows that PTR proteins cluster separately from PTC and NPC proteins (Fig. 2) and confirms that PTC-1 and PTC-3 are the closest C. elegans relatives of the Drosophila and vertebrate PTC proteins.
Table 1.
PTC superfamily
| Gene
|
Cosmida
|
LG
|
Amino acids
|
BLAST Pb
|
SSD Ec
|
PND Ea
|
TM summaryd
|
TMs
|
EST hits
|
|---|---|---|---|---|---|---|---|---|---|
| tra-2 | C15F1.3 | II | 1147 | N.S. | N.S. | N.S. | 1,3,2,1,3,2 | 12 | 4 |
| ptr-1 | C24B5.3 | V | 956 | 2.4E-22 | 7.1E-09 | 1.30E-24 | 1,3,2,1,3,2 | 12 | 0 |
| ptr-2 | C32E8.8 | I | 933 | 7.6E-26 | 1.2E-28 | 1.80E-45 | 1,3,2,1,3,2 | 12 | 16 |
| ptr-3 | C41D7.2 | II | 871 | 2.1E-09 | 7.1E-08 | 5.40E-14 | 3,2,1,3,2 | 11 | 6 |
| ptr-4 | C45B2.7 | X | 1037 | 1.7E-22 | 1.7E-09 | 8.80E-21 | 1,3,2,1,3,2 | 12 | 21 |
| ptr-5 | C53C11.3 | X | 845 | 2.6E-19 | 3.1E-09 | 2.30E-13 | 3,2,1,3,2 | 11 | 11 |
| ptr-6 | C54A12.1 | II | 1015 | 1.8E-33 | 7.4E-14 | 3.60E-32 | 1,3,2,1,3,2 | 12 | 4 |
| scp-1 | D2013.8 | II | 1032 | 1.5E-03 | 3.5E-78 | N.S. | 1,3,2,1,1 | 8 | 20 |
| npc-1 | F02E8.6 | X | 1456 | 2.4E-14 | 2.8E-10 | 3.10E-96 | 1,1,3,2,1,3,2 | 13 | 6 |
| ptd-2 | F07C3.1 | V | 932 | 1.9E-04 | 6.7E-02 | 7.40E-292 | 1,3,2,1,3,2 | 12 | 2 |
| npc-2 | F09G8.4 | III | 915 | 4.5E-07 | 1.8E-06 | 4.80E-23 | 1,1,3,2,1,3,2 | 13 | 2 |
| ptc-2 | F21H12.4 | II | 714 | 0.0 | 2.4E-67 | 7.90E-243 | 3,2,1,3 | 9 | 0 |
| ptr-7 | F31F6.5 | X | 955 | 4.5E-21 | 6.8E-10 | 4.50E-23 | 1,3,2,1,3,2 | 12 | 2 |
| ptr-8 | F44F4.4 | II | 890 | 6.6E-10 | 5.6E-08 | 5.30E-12 | 1,3,2,1,3,2 | 12 | 10 |
| ptr-9 | F54G8.5 | III | 413 | 2.1E-10 | 1.2E-02 | 1.10E-15 | 2,1,3 | 6 | 0 |
| ptr-10 | F55F8.1 | I | 889 | 7.8E-27 | 7.4E-14 | 4.70E-28 | 1,3,2,1,3,2 | 12 | 4 |
| ptr-11 | F56C11.2 | I | 820 | 8.0E-24 | 6.1E-10 | 1.80E-20 | 1,3,2,1,3,2 | 12 | 4 |
| ptd-1 | F45H1.1 | I | 969 | 1.9E-06 | 1.9E-05 | 1.10E-282 | 1,3,2,1,3,2 | 12 | 3 |
| ptr-12 | K07A3.2 | I | 1110 | 4.0E-09 | 2.7E-07 | 6.90E-09 | 1,1,3,2,1,2,2,1 | 13 | 1 |
| ptr-13 | K07C10.1 | II | 690 | 1.9E-17 | 6.9E-11 | 1.60E-21 | 3,2,1,3,2 | 11 | 0 |
| ptr-14 | R09H10.4 | IV | 877 | 5.8E-20 | 2.2E-12 | 2.10E-18 | 1*,3,2,1,3,2 | 12 | 11 |
| ptr-15 | T07H8.6 | V | 880 | 1.1E-21 | 4.4E-15 | 3.70E-32 | 1,3,2,1,3,2 | 12 | 0 |
| ptr-16 | T21H3.2 | V | 881 | 4.8E-17 | 3.6E-21 | 7.50E-21 | 1,3,2,1,3,2 | 12 | 0 |
| ptc-3 | Y110A2AL.8 | II | 1314 | 1.4E-223 | 2.8E-96 | 0 | 1,3,2,1,3,2 | 12 | 12 |
| ptr-17 | Y18D10A.7 | I | 936 | 7.2E-24 | 5.1E-15 | 8.20E-31 | 1,3,2,1,3,2 | 12 | 0 |
| ptr-18 | Y38F1A.3 | II | 800 | 5.1E-21 | 2.4E-22 | 8.20E-32 | 3,2,1,3,2 | 11 | 4 |
| ptr-19 | Y39A1B.2 | III | 1003 | 2.0E-25 | 3.1E-13 | 2.40E-29 | 1,3,2,1,3,2 | 12 | 0 |
| ptr-20 | Y53F4B.28 | II | 860 | 6.9E-15 | 9.2E-19 | 3.80E-27 | 1,3,2,1,3,2 | 12 | 7 |
| ptr-21 | Y65B4BR.3 | I | 909 | 6.3E-13 | 1.8E-08 | 2.60E-05 | 1,3,2,1,3,2 | 12 | 0 |
| ptr-22 | Y80D3A.7 | V | 859 | 2.9E-16 | 1.4E-08 | 2.40E-20 | 1,3*,2,1,3,2 | 12 | 0 |
| ptr-23 | ZK270.1 | I | 983 | 1.1E-13 | 1.5E-08 | 4.30E-10 | 1,3,2,1,3,2 | 12 | 24 |
| ptc-1 | ZK675.1 | II | 1405 | 0.0 | 1.8E-93 | 0 | 1,3,2,1,3,2 | 12 | 2 |
(LG) Linkage group. (*) An uncertainty in assignment. (N.S.) Not significant.
A profile HMM was constructed using HMMER 2.1.1 (http://www.hmmer.wustl.edu/); the seed alignment for the HMM was a Clustal W alignment of all currently identified PTC, NPC1, and DISP homologs (http://cmgm.stanford.edu/∼jefferis/paperdata/PND.aln). PND E-values were derived by using this calibrated HMM (http://cmgm.stanford.edu/∼jefferis/paperdata/PND.shmm) to search the Wormpep 20 database (http://www.sanger.acu.uk/Projects/C_elegans/wormpep/) in best local alignment mode (http://cmgm.stanford.edu/∼jefferis/paperdata/PND.shmms).
BLAST P represents the probability of a match using PTC-1 to search Wormpep 10 using BLASTP algorithm with default parameters.
A profile HMM was constructed for the SSD motif (Carstea et al. 1997) using a Clustal W alignment of all currently identified PTC, NPC1, and SCAP orthologs and a representative selection of SSD containing HMGCoAR proteins. This calibrated HMM was used to search Wormpep 20, and expectation values are tabulated as SSD E.
The TM domains in this family are arranged in a double repeat of the form 2(1,3,2), where each number represents a closely spaced group of TM domains and the commas represent extramembranous regions.
Figure 2.
Clustal W alignment of SSD-containing proteins including C. elegans (Ce) PTC, NPC, and PTR proteins with key representatives from Drosophila melanogaster (Dm), Dictyostelium discoideum (Dd), Danio rerio (Dr), Homo sapiens (Hs), and Rattus norvegicus (Rn). CePTR-10 was omitted from the analysis because of its length.
The role of ptc-1 during C. elegans development
To understand the role that ptc-1 has in C. elegans development, RNAi using double-stranded RNA (dsRNA) was performed to disrupt its activity (Guo and Kemphues 1995; Fire et al. 1998). We found that the F1 progeny of ptc-1(RNAi) mothers showed no obvious somatic patterning defects comparable to those described in Drosophila ptc or other Hh pathway mutants (Table 2). Instead, Nomarski DIC microscopy revealed that F1 progeny (37%) developed into sterile adults with abnormal multinucleate oocytes. We also observed that injected mothers developed germ-line anatomical defects and became sterile like their sterile F1 progeny. In addition, 30% of the F1 progeny arrested as abnormal embryos, probably because they developed from already defective oocytes. The primary target for ptc-1(RNAi) thus appears to be the germ line. When RNAi was performed using a sequence common to both ptc-1 and ptc-2, a more severe additive phenotype was not observed, suggesting further that ptc-2 is a pseudogene. A more extensive discussion of the ptc-1 mutant phenotype is provided below. A description of ptc-3 RNAi and mutant phenotype will be presented elsewhere (P. Kuwabara, unpubl.).
Table 2.
Inactivation of ptc-1 activity by RNAi
| dsRNA injected
|
|
Phenotype of progeny
|
|||
|---|---|---|---|---|---|
| dead eggs
|
larval arrest
|
sterile adults
|
fertile adultsa
|
||
| ptc-1 (3.6 mg/ml) | average | 30% (13.7) | 2% (1.0) | 37% (17.1) | 31% (14.4) |
| Po = 7 | totalb | 96 | 7 | 120 | 101 |
| range | 6–27 | 0–4 | 6–28 | 13–18 | |
Wild-type adults were produced at the beginning of the brood and probably represent animals that were fertilized prior to the effects of dsRNA injection.
Total brood counts are low because RNAi disrupted the fertility of injected parents.
ptc-1 mRNA is localized to the germ line and germ-line progenitor blastomeres
The mutant phenotypes observed in ptc-1(RNAi) animals suggested that ptc-1 activity is required primarily by the germ line and not the soma. To examine whether ptc-1 expression is similarly localized, in situ hybridization was performed on embryos and gonad arms dissected from wild-type hermaphrodites. We found that ptc-1 mRNA is maternally transcribed: ptc-1 mRNA was initially detected in the pachytene region of the germ line and increased in steady-state levels as germ cells progressed to fully grown oocytes (Fig. 3, top). After fertilization, maternal ptc-1 mRNA was evenly distributed in early two- and four-cell embryos (Fig. 3, bottom). However, as P-cell divisions continued, ptc-1 mRNA localized to germ-line blastomeres and disappeared from somatic blastomeres (Fig. 3, bottom right).
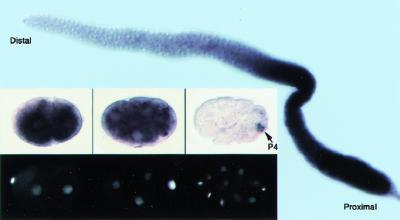
Figure 3.
Tissue-specific localization of ptc-1 mRNA to the C. elegans germ line and its progenitors by in situ hybridization. A dissected adult hermaphrodite gonad revealing that purple staining ptc-1 mRNA is first detected at meiotic prophase and increases progressively in steady-state levels with germ cell maturation. ptc-1 mRNA accumulation during early embryogenesis: (top panels) purple ptc-1 staining; (bottom panels) the same embryos as in the top, stained with DAPI to visualize DNA. Panels show (left) 2-cell stage; (middle) 4-cell stage; (right) 14-cell stage, with germ-line blastomere P4 indicated (arrow). Scale bar, 20 μm.
Characterization of ptc-1 deletion mutants
Two heterogeneous populations of worms carrying ptc-1 deletion alleles, ok122 (kindly provided by G. Moulder and R. Barstead, Oklahoma Medical Research Foundation, Oklahoma City) and nr2029 (kindly provided by the NemaPharm Group at Axys Pharmaceuticals), were obtained to corroborate the ptc-1(RNAi) phenotype and to understand the basis of the sterility. Prior to analyzing these mutants, we isolated and extensively outcrossed each mutant, tracking the deletion allele by PCR. We found by sequencing deletion breakpoints that ok122 deleted 1.708 kb and nr2029 deleted 0.96 kb of the ptc-1 locus; both deletions removed extensive blocks of ptc-1 coding sequence and produced frameshifts that introduced termination codons close to the deletion breakpoints (Fig. 1A). Only the phenotype of ptc-1(ok122) is reported because the two alleles showed no discernible phenotypic differences.
We examined 75 individual ptc-1 (m+ z−; refers to the presence or absence of maternal or zygotic gene activity) mutant hermaphrodites and found that 72% were sterile whereas the remaining 28% produced one or two escaper progeny that developed into sterile adults (m− z−) (Table 3). The detection of escaper progeny (m− z−) indicates that ptc-1 is not essential for embryonic or larval development and that the embryonic lethality observed for ptc-1(RNAi) is likely to result from defective oogenesis. Examination of ptc-1 L4 and adult hermaphrodites by Nomarski DIC microscopy revealed no obvious somatic defects, whereas the germ lines contained multinucleate oocytes, displayed a general lack of organization of nuclei, and were underproliferated. The multinucleate oocyte phenotype caused by the absence of ptc-1 will be referred to as the Moo (multinucleate oocyte) phenotype.
Table 3.
The effects of ptc-1 on fertility
| Genotype
|
Po Number fertile/total examined
|
|
F1 Progeny
|
||
|---|---|---|---|---|---|
| dead eggs
|
larval arrest
|
adults
|
|||
| ptc-1 | total | 20 | 2 | 11 | |
| Self-progenya | 21/75 | range | 0–2 | 0–1 | 0–2 |
| meanb | 1.0 | 0.1 | 0.5 | ||
| ptc-1 | total | 467 | 34 | 129 | |
| Cross-progenyc | 35/35 | range | 0–44 | 0–5 | 0–14 |
| mean | 13.3 | 1.0 | 3.7 | ||
These animals were identified as uncoordinated progeny of unc-4 ptc-1/mnC1 hermaphrodites, using unc-4 to mark ptc-1 in cis.
Mean is calculated only for those animals displaying fertility.
Cross-progeny produced by mating L4-staged XX unc-4 ptc-1 hermaphrodites to XO wild-type males. Cross-progeny can be distinguished from self-progeny because they are non-Unc in phenotype.
C. elegans ptc-1 functions in germ-line cytokinesis
To understand the genesis of the Moo phenotype we examined ptc-1 germ lines in dissected gonads stained with DAPI to visualize DNA, anti-syntaxin-4 (SYN-4) to outline cell membranes (Jantsch-Plunger and Glotzer 1999), and anti-phosphohistone H3 (Hendzel et al. 1997) to reveal mitotic nuclei (M phase). A description of the germ-line anatomy of a single gonad arm from a wild-type adult hermaphrodite is illustrated schematically in Figure 4A and in cross section at the ultrastructural level by transmission electron microscopy (Fig. 4B). In the C. elegans germ line, plasma membranes surround but do not fully enclose each nucleus so that they are interconnected as a syncytium. By convention, a nucleus and its surrounding cytoplasm and membranes are referred to as a germ cell. At the distal end, there is a stem cell population of mitotically proliferating germ cells that initiate meiotic development in the transition zone before progressing through an extended pachytene stage. At the loop region, germ cells transit from pachytene to diplotene and begin oogenesis. In the proximal region, oocytes enlarge, become fully cellularized, and are in diakinesis.
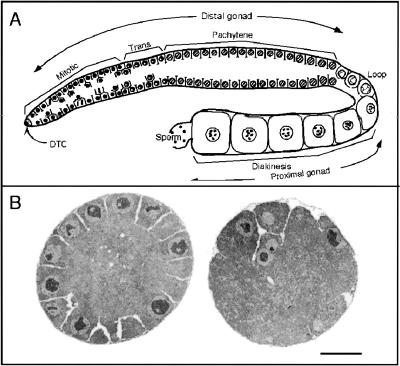
Figure 4.
Organization of the adult hermaphrodite gonad arm. (A) Schematic diagram of a single wild-type adult hermaphrodite gonad arm, interior view. The tube-shaped distal gonad arm contains nuclei circumferentially arranged on the surface and a central rachis or cytoplasmic core that is essentially devoid of nuclei and plasma membranes (Hirsh et al. 1976; Hall et al. 1999). Each nucleus is partially enclosed by plasma membrane; although this is a syncytium, each nucleus and its surrounding cytoplasm and membrane are called a germ cell. At the distal end, adjacent to the somatic distal tip cell (DTC), a population of mitotically proliferating germ cells begin meiotic development in the transition zone (trans) and then progress through an extended pachytene region. From the loop into the proximal gonad, nuclei progress from pachytene to diakinesis; oocytes become more fully cellularized and increase in both nuclear and cytoplasmic volume (Hirsh et al. 1976; Strome 1986; Crittenden et al. 1994; Hall et al. 1999). (B) Low-power electron microscopy cross sections of distal gonad arms dissected from adult hermaphrodites. (Left) Wild-type gonad with germ nuclei positioned radially at the periphery. Germ nuclei are surrounded by membranes on all sides but one, providing cytoplasmic continuity with the central rachis. (Right) Gonad from ptc-1 mutant shows irregular organization and an abnormal distribution of germ nuclei with large acellular patches. Two cells, each containing two nuclei, are positioned centrally and at 5 o'clock. Scale bar, 5 μm.
Multinucleate germ cells are found in ptc-1 hermaphrodites from the L4 stage onward. Fifty percent of the oocytes in ptc-1 mutants are multinucleate (n = 49). Figure 5A shows a ptc-1 adult gonad containing oocytes with three diakinesis nuclei in the proximal gonad and two just distal to the loop. Many abnormal cells with multiple pachytene nuclei are observed, but mononucleated cells are also present (Fig. 5A). Acellular regions on the surface of the gonadal tube are also common and can be viewed more clearly at the ultrastructural level in cross section (Fig. 4B, right). Male germ-line development also requires ptc-1 activity, as mutant males and L4 hermaphrodites undergoing spermatogenesis have multinucleate germ cells (data not shown).
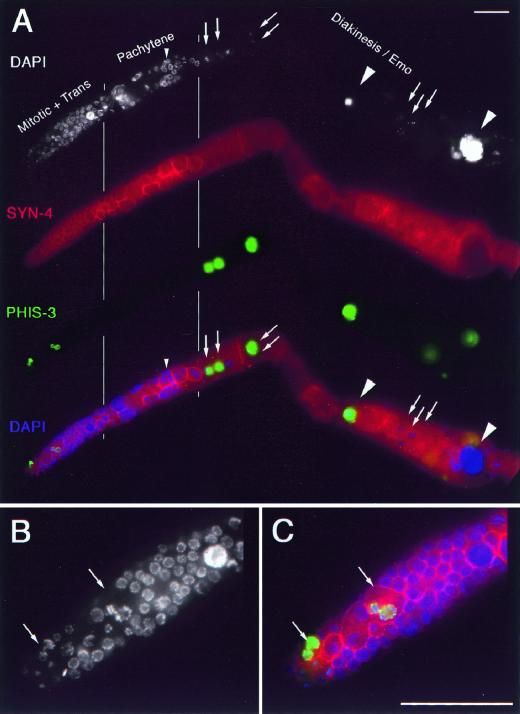
Figure 5.
Multinucleate germ cells in ptc-1 mutant hermaphrodite gonad. For reference, refer to wild-type gonad arm (Fig. 7). Gonad arm from a ptc-1 hermaphrodite (42 hr post-L4) stained with DAPI (white) to visualize DNA, anti-SYN-4 (red) to visualize plasma membranes (Jantsch-Plunger and Glotzer 1999), and anti-PHIS-3 (green) to visualize proliferating germ-line nuclei in M phase and oocyte nuclei in diakinesis (Hendzel et al. 1997). (A) Four panels of the same focal plane showing a surface view of the distal gonad and an interior view of the proximal gonad (left to right). The bottom panel merges the information from the top panels; DAPI fluorescence is false-colored purple. Multinucleate oocytes are indicated by arrows pointing to individual diakinesis nuclei. Many multinucleate meiotic prophase cells in pachytene are visible; a small arrowhead highlights an example. Endomitotic oocytes within the gonad arm (Emo phenotype) are indicated by large arrowheads. Diakinesis chromosomes, in mono- or multinucleated oocytes, are found in the normal six bivalent configuration indicating that ptc-1(ok122) does not disrupt meiotic pairing/recombination. (B,C) Magnified view of the distal region of the germ line. (B) DAPI fluorescence of nuclei. (C) Merged image; DAPI is false-colored purple. Multinucleate germ cells in M phase are indicated by arrows. The left-most M-phase cell has two nuclei; the right-most cell has three nuclei with one partially out of the plane of focus. Note that the right-most M-phase cell has an anuclear patch. Additional M-phase cells are present in this gonad arm, but they are partially out of the plane of focus. Rare (fewer than one per gonad arm) polyploid nuclei are also detected in the distal germ line such as the one shown near the transition zone (B,C, right).
The appearance of multinucleate germ cells in meiotic prophase initially suggested that the partially enclosing plasma membrane separating individual nuclei may have retracted or adjoining cells may have fused. However, the existence of multinucleate proliferative germ cells in the distal-most region of the ptc-1 germ line, some of which were in M phase, indicated instead that the defect giving rise to multinucleate germ cells probably occurred earlier in the pool of mitotically proliferating germ cells undergoing cytokinesis (Fig 5B,C). The simplest interpretation of these results is that ptc-1 functions in cytokinesis; in ptc-1 mutants, germ-line nuclear divisions occur, but cytokinesis is sometimes defective. Because the frequency of multinucleate cells is similar in all regions, we suggest that multinucleate meiotic prophase cells arise principally from multinucleate proliferative cells. The basis of this cytokinesis defect could range from an early defect in cleavage furrow formation to a late defect in furrow stabilization. ptc-1 is apparently not absolutely essential for germ-line cytokinesis, as some proliferative and meiotic prophase cells are mononucleated, nor is ptc-1 essential for cytokinesis in somatic cells.
Sterility and embryonic lethality are due to defects in both male and female germ cells
Other aspects of the ptc-1 mutant phenotype are visible in Figure 5. Most adult hermaphrodite gonads have an Emo phenotype characterized by endomitotic oocytes in the proximal gonad that complete meiotic maturation but cycle mitotically without undergoing karyokinesis and cytokinesis in the absence of ovulation and fertilization (Iwasaki et al. 1996). Defective ptc-1 sperm cells appear to contribute to the Emo phenotype because mating with wild-type males partially suppresses the Emo phenotype of the ptc-1 mother; 19% (7/37) of gonad arms from mated ptc-1 (m+ z−) mutants were Emo, whereas 69% (29/42) of unmated mutants were Emo. These matings also increased fecundity, although ptc-1 (m− z+) cross-progeny continued to suffer from a high incidence of lethality (Table 3); 100% of ptc-1 mothers produced cross-progeny (cf. 28% for unmated), and the average brood size increased to 18 (cf. 1.6 for unmated). Taken together, it appears that the small number of viable escaper progeny (m− z−) from ptc-1 mothers results from the rare union of a haploid sperm with a successfully ovulated oocyte containing a single diakinesis nucleus.
C. elegans ptc-1 maintains cell cycle autonomy among proliferating germ cells
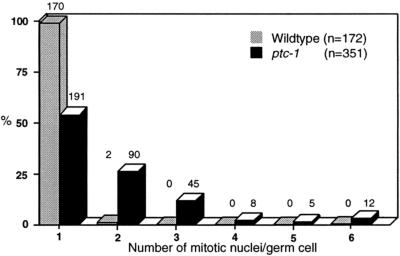
Germ cells in the distal proliferative region are syncytial (Hall et al. 1999), yet they progress through the mitotic cell cycle asynchronously; at any one time, nuclei in M phase are few and randomly scattered (Crittenden et al. 1994; Francis et al. 1995). In contrast, a high degree of mitotic cell cycle synchrony is observed in other syncytia, for example, the early Drosophila embryo before cellularization (Foe and Alberts 1983). These observations have led to the proposal that a germ nucleus in the distal proliferative region of the C. elegans germ line, although syncytial, resides in a cytoplasmic domain that is autonomous with respect to mitotic cell cycle progression. Therefore, the unexpected appearance of M-phase nuclear clusters in the distal proliferative region of the ptc-1 hermaphrodite germ line led us to compare the frequency of these clusters between ptc-1 and wild-type animals (Fig. 5B,C). As shown in Figure 6, the occurrence of clusters of M-phase nuclei normally is rare; only 1% of wild-type M-phase nuclei display this phenotype. In ptc-1 mutants, however, 46% of M-phase nuclei are found in multinucleate germ cells containing two to six nuclei. These results indicate that mitotic cell cycle progression is normally autonomous among distal proliferative nuclei and further suggest that the plasma membranes, which normally separate and partially enclose each nucleus, are important for maintaining this autonomy. When these membranes are absent, such as in ptc-1 mutants, multiple nuclei contained within a single cell acquire the ability to progress through the cell cycle synchronously. We speculate that these membranes may contribute to germ-cell autonomy by localizing or partitioning information affecting cell fate or cell proliferation.
Figure 6.
Loss of germ-cell nuclear autonomy in XX hermaphrodite ptc-1 mutants. Thirty hermaphrodite gonad arms, each from ptc-1 unc-4 mutants and unc-4 (control) hermaphrodites, were examined 18 hr after the L4 stage using anti-PHIS-3 to visualize nuclei in M phase (Hendzel et al. 1997) and anti-SYN-4 to highlight plasma membranes (see Materials and Methods). The graph plots the frequency and range in the number of multiple mitotic nuclei contained within a single germ cell, which is defined by the smallest cubic volume that is surrounded on all sides but one by SYN-4-positive plasma membrane. (Shaded bars) Wild type; (solid bars) ptc-1; (n) number of PHIS-3-positive nuclei scored.
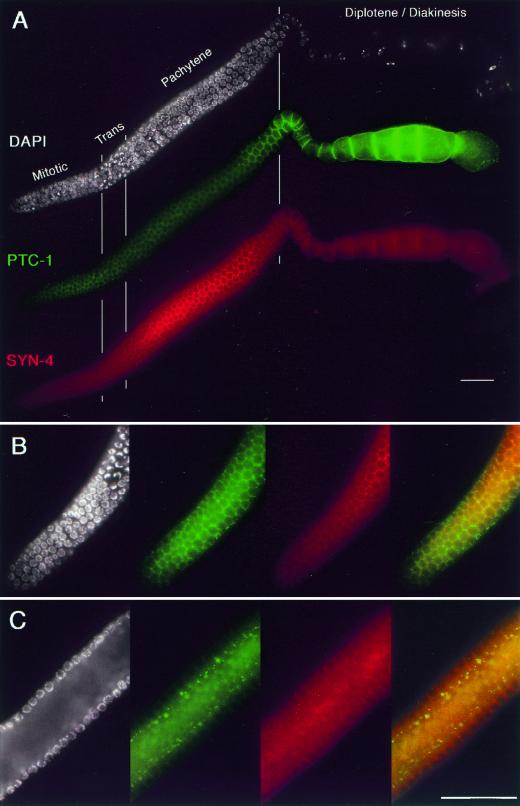
PTC-1 protein is enriched during membrane growth and expansion
The germ-line localization of PTC-1 protein was examined using an affinity-purified anti-PTC-1 polyclonal antibody. We found that anti-PTC-1 highlights plasma membranes of all germ cells in wild-type hermaphrodites (Fig. 7A) and males, with the exception of mature sperm (data not shown). When the fluorescence intensity of PTC-1 was compared with SYN-4, another plasma membrane-associated protein (Jantsch-Plunger and Glotzer 1999), we found that PTC-1 was not distributed uniformly through the germ line. PTC-1 fluorescence was more intense in the distal proliferative region and in oocytes than in the pachytene region (Fig. 7A,B); these zones correspond to regions that would be expected to undergo high levels of membrane growth and expansion. When focusing on a more central focal plane of the pachytene zone (similar to that diagramed in Fig. 4A), we also observed that PTC-1 was more highly concentrated in a punctate pattern at the apices of internal membranes adjacent to the rachis (Fig. 7C).
Figure 7.
PTC-1 localizes to germ-cell membranes. Gonad arm, dissected from a wild-type adult hermaphrodite, stained with DAPI to visualize DNA (white), anti-SYN-4 (red) to visualize plasma membranes, and anti-PTC-1 (green). (A) PTC-1 and SYN-4 colocalize to plasma membranes. Three composite photomicrographs show the same focal plane through an intact gonad: surface view of the distal gonad highlighting the honeycomb arrangement of plasma membranes that surround germ-line nuclei and an interior view of the proximal gonad showing oocyte membrane staining. Spatial distribution of PTC-1 is nonuniform with more intense staining in the distal, proliferative mitotic region and in proximal growing oocytes. The distribution of SYN-4 fluorescence is inverse to PTC-1 (also see B); SYN-4 fluorescence is more intense in the pachytene region and less intense in the mitotic region and in growing oocytes. (B) Surface view of distal proliferative region, magnified. (Panels from left to right) DAPI, anti-PTC-1, anti-SYN-4, merge of anti-PTC-1 and anti-SYN-4. The distal-most region is enriched in PTC-1, with a stronger green signal in the merge (right-most panel); more proximally, PTC-1 and SYN-4 have similar intensities (orange). (C) Interior view of the pachytene region. Panels from left to right are as in B. PTC-1 and SYN-4 both stain lateral plasma membranes. PTC-1 also shows very strong punctate staining at the apparent apices of internal membranes adjacent to the rachis.
Discussion
Here we have shown that C. elegans encodes multiple PTC and PTC-related proteins that share extensive sequence similarity and conserved topology to Ptc proteins found in other animals. We have shown by RNAi and mutational analysis that Ce-ptc-1 has an essential role in nematode development, indicating that ptc-1 is functional despite the apparent absence of other components of the Hh/Ptc signaling pathway. We find that ptc-1 is required for C. elegans germ-line development because the absence of ptc-1 leads to the formation of multinucleate germ cells and sterility. The tissue-specific role of ptc-1 is supported by mRNA and protein localization studies. Our analyses also revealed that the multinucleate germ cells found in ptc-1 mutants probably originated from cytokinesis defects that occurred among the pool of mitotically proliferating, distally situated germ cells. A consequence of this defect is that multiple nuclei within a single cell acquire the ability to progress through mitosis synchronously, indicating that the plasma membrane furrows that normally separate germ nuclei are important for establishing domains within the syncytium that promote nuclear autonomy. An association between the absence of ptc-1 activity and defects in cytokinesis has hitherto not been reported, although mutations in the vertebrate PTCH gene have demonstrated a role for ptc in regulating cell proliferation. The implications of these findings are discussed.
Many components of the Hh/Ptc signaling pathway are not conserved in C. elegans
PTC proteins have been conserved in C. elegans and in other organisms across the animal kingdom; however, this conservation apparently does not extend to other components of the Hh/Ptc signaling pathway in C. elegans. We have been unable to detect Hh and Smo orthologs after exhaustive searches of the essentially complete C. elegans genome sequence (Consortium 1998), although a large number of predicted Hh relatives have been identified (Bürglin 1996; Porter et al. 1996b; Hall et al. 1997; Aspöck et al. 1999). Despite the absence of Hh, the C. elegans genome encodes two homologs each of the Drosophila tout velu and disp proteins, which are postulated to facilitate the movement of Hh (Clines et al. 1997). In addition, the activity of the transcriptional regulator TRA-1, the single C. elegans homolog of Drosophila Ci and human GLI, appears to have been usurped by the sex determination pathway. The absence of ptc-1 activity has no detectable effect on sexual phenotype. Similarly, tra-1 mutations, either loss of function or gain of function, do not show phenotypes that resemble ptc-1 or ptc-3 mutants (Hodgkin 1987; Schedl et al. 1989; P. Kuwabara, unpubl.). Thus, PTC-1 and likely PTC-3 do not appear to mediate transcriptional regulation through TRA-1. Nonetheless, PTC proteins continue to have essential roles in C. elegans development, although they have no obvious role in somatic patterning.
Has the signaling activity of Ce-PTC-1 been conserved despite the apparent absence of Hh and Smo?
We interpret the absence of a C. elegans Hh homolog to indicate that either Hh was lost from the C. elegans genome or that the worm diverged from Drosophila and vertebrates before the acquisition of Hh. The closest C. elegans Hh relatives are those that share similarity to the carboxy-terminal autoprocessing domain (Bürglin 1996; Porter et al. 1996b; Hall et al. 1997; Aspöck et al. 1999); several of these proteins have the potential to be processed and may mediate cell signaling (Porter et al. 1996b). Therefore, if the PTC proteins continue to function in signaling in C. elegans, it is possible that they function without a ligand or that they have acquired new ligands through coevolution with the worm Hh relatives and even new membrane protein partners. For example, although Smo is absent, the C. elegans genome encodes a number of frizzled homologs that are related by sequence to Smo. However, similar to Drosophila, these proteins probably function as Wnt receptors (Wodarz and Nusse 1998).
The C. elegans ptc-1 gene may uncover new functions for PTC proteins
Alternatively, the discovery that mutations in the C. elegans ptc-1 gene disrupt germ-line cytokinesis suggests that we may have uncovered a novel or perhaps an ancestral activity for PTC that is not dependent on Hh or Smo. Comparative studies suggest that the fundamental mechanisms of cytokinesis probably have been conserved among eukaryotes (Field et al. 1999). These observations have led us to postulate that within the C. elegans germ-line syncytium, bipolar cytokinesis involves a contractile-process membrane fusion requiring vesicular transport (Jantsch-Plunger and Glotzer 1999), and stabilization of the incomplete membrane furrows within the syncytium. We suggest that ptc-1 contributes to the process of syncytial cytokinesis by helping to establish or maintain the incomplete plasma membrane furrows separating individual nuclei within the syncytium. Furthermore, it may be found that one or more of the PTR proteins has a role in somatic cytokinesis similar to that of PTC-1 in the germ line.
How might PTC-1 participate in cytokinesis? First, we speculate that because the absence of ptc-1 disrupts either membrane deposition or stabilization during germ-line cytokinesis, PTC-1 may promote vesicle trafficking and membrane fusion events required to complete cytokinesis. It has been shown in C. elegans that membrane fusion during cytokinesis is dependent on the t-SNARE membrane fusion protein SYN-4 (Jantsch-Plunger and Glotzer 1999). Furthermore, there is evidence that lipids and cholesterol have important roles in regulating the sorting of proteins and other lipids during intracellular trafficking (Mukherjee and Maxfield 1999). PTC has the ability to detect or be regulated by sterols because it carries an SSD (Fig. 2; Lange and Steck 1998; Osborne and Rosenfeld 1998). In Drosophila, Hh carries a cholesterol moiety that may help to target it to the SSD of the PTC receptor, although this moiety is not essential for receptor recognition (Marigo et al. 1996; Stone et al. 1996). It has also been suggested that the teratogenic effects of veratrum alkaloids, which block cholesterol biosynthesis and Sonic hedgehog (Shh) signaling, may be mediated through the SSD of Ptc and not by disrupting Hh processing (Cooper et al. 1998). Here we raise the possibility that there is an additional connection between cholesterol homeostasis and Ce-PTC-1 mediated through the SSD, which may affect membrane deposition or stabilization during germ-line cytokinesis. Second, PTC-1 may have an adhesive function. It is not known how the membrane furrows separating individual germ nuclei within the germ-line syncytium retain their integrity; therefore, it is possible that the extracellular domains of PTC-1 interact with an extracellular protein or with each other to stabilize the newly formed furrows. Third, it remains possible that the cytoplasmic domains of PTC-1 interact with the cytoskeleton to promote cytokinesis.
Dual roles for PTC proteins?
We have suggested that the C. elegans PTC-1 might retain a signaling role in the germ line as a receptor for one of the novel Hh-related molecules, have an indirect role in vesicle trafficking and membrane deposition by affecting cholesterol homeostasis, or perhaps assume a more structural role. These roles are not mutually exclusive. One example of a protein with dual roles in cell adhesion and signaling is β-catenin (Wodarz and Nusse 1998). β-Catenin is a structural component of vertebrate adherens junctions that helps to link cadherins to the actin cytoskeleton. In Drosophila, its ortholog Armadillo is a regulator of Wg/Wnt signaling.
C. elegans offers opportunities for exploring the roles of SSD-containing proteins in development and cholesterol homeostasis
Examination of the complete genome sequence of C. elegans has revealed the presence of a large worm PTC superfamily. This is composed of PTC and Disp homologs, as well as novel PTR proteins, all of which are related by a shared 12-pass modular topology and the presence of an SSD. This feature suggests that C. elegans offers opportunities for uncovering developmental processes that may be regulated or influenced by sterols. Such studies are likely to reveal additional novel functions and signaling activities for members of this protein family and may shed further light on mechanisms involved in cholesterol homeostasis.
Materials and methods
Nematode strains and culture
Standard procedures for nematode culture and genetic manipulation were followed, and all genetic characterizations were performed at 20°C (Sulston and Hodgkin 1988). Descriptions of C. elegans genes and alleles can be found elsewhere (Hodgkin 1997). ptc-1 II deletion mutants were outcrossed five times and maintained as ptc-1 unc-4(e120) recombinants balanced by mnC1. The strain ptc-1 unc-4/mnC1 II; him-8(e1489) V was constructed to provide a source of males of the appropriate genotype. Descriptions of the ptc-1 mutant phenotype refer to the ok122 allele.
Molecular cloning and sequencing
Overlapping ptc-1 cDNA fragments were prepared by RT–PCR using Superscript RNaseH(−) reverse transcriptase (Life Technologies) and Expand polymerase (Boehringer-Mannheim), and blunt-end-cloned into the EcoRV site of pBluescript KSII(+). cDNA sequencing was performed on an ABI 377 sequencer. The deletion breakpoint of ptc-1(nr2029) was obtained by amplifying DNA prepared from ptc-1(nr2029) homozygotes with the primers ptc1 (5′-CATACCGGAAGTCTGCTTTCG) and ptc1R2 (5′-CGGAAACATTGCTGCCAATAACTG), and sequencing the product with the ptc1 primer. The deletion breakpoint of ptc-1(ok122) was obtained similarly by amplifying with the primers ptc36 (5′-ACGGACATTGTGAAACTATGG) and ptc4 (5′-ATCATTCTCCACACTTCTCGG) and sequencing with ptc24 (5′-CTGGGCCGGAACTTCCCAG).
BLAST analysis and phylogenetic comparisons
Sequence similarity searches of various public databases were performed using BLAST (Altschul et al. 1997). Profile HMMs constructed with HMMER 2.1.1 (http://hmmer.wustl.edu/) were used to search the Wormpep 20 database (http://www.sanger.ac.uk/Projects/C_elegans/wormpep/). Cluster analysis was performed using Clustal W (Thompson et al. 1997), and output was viewed using TREEVIEW (Page 1996). Tmpred was used to identify potential membrane-spanning domains (Hofmann and Stoffel 1993). MacVector 6.5 (Oxford Molecular) was used for routine sequence management.
RNA microinjection
pPK277, a clone containing 640 bp of 5′ ptc-1 cDNA sequence with no significant stretches of nucleotide identity to ptc-2 or ptc-3, was digested with BssHII, and single-stranded RNA was transcribed from the T3 and T7 RNA polymerase promoters using the Megascript kit (Ambion). After transcription, single-stranded RNA molecules were annealed and prepared for microinjection. Injected animals were transferred to fresh plates every 12–18 hr. Eggs were scored as dead if they failed to hatch within 24 hr after transfer of the mother. Sterility was assessed by examining the germ lines of adult XX hermaphrodites using Nomarski DIC optics.
Electron microscopy
Gonads were dissected in M9 buffer containing 0.2 mm tetramisole and fixed in 0.1 m sodium phosphate buffer (pH 7.4) containing 3% glutaraldehyde for 2 hr and postfixed in 1% osmium tetroxide prior to embedding and sectioning.
Production of anti-PTC-1 antibody
A polyclonal antisera to PTC-1 was generated by immunizing a rabbit with an amino-terminal PTC-1 fragment (amino acids 384–522 fused to GST); this region is absent in ptc-1(ok122) mutants. Affinity-purified antiserum was prepared by coupling the PTC-1–GST fusion protein to Actigel ALD, binding PTC-1 antisera, and eluting with Actisep as recommended by the manufacturer (Sterogene).
Gonad dissection, RNA in situ hybridization, and antibody staining
Dissected gonads from L4 and 18 and 42-hr post-L4 adult hermaphrodites were prepared as described (Francis et al. 1995). For RNA in situ hybridization, dissected gonads were fixed in 0.25% glutaraldehyde/3% formaldehyde, 100 mm K2HPO4 (pH 7.2), and processed as described (Iwasaki et al. 1996; Jones et al. 1996). Whole-mount RNA in situ hybridization of embryos was performed essentially as described by Seydoux and Fire (1994), except that worms were mounted on Superfrost/Plus slides (Fisher Scientific). Sense and antisense hybridization probes were synthesized with digoxigenin-11–dUTP by repeated primer extension (Seydoux and Fire 1994; Jones et al. 1996). A control sense probe gave little or no signal from either dissected gonads or embryos.
For antibody staining, dissected gonads were fixed with 3% formaldehyde, 100 mm K2HPO4 (pH 7.2), for 1 hr and postfixed with cold (−20°C) 100% methanol for 5 min (Francis et al. 1995). Antibody incubations and washes were performed as described (Jones et al. 1996). Anti-phosphohistone H3 antibody (Upstate Biotechnology) was used at a 1:400 dilution to visualize mitotic chromosomes. Plasma membranes were visualized with mouse anti-SYN-4 antibodies (kindly provided by M. Glotzer, Research Institute of Molecular Pathology; Vienna). Affinity-purified rabbit polyclonal anti-PTC-1 antibody was used at a 1:20 dilution. No immunofluorescence was detected in ptc-1 mutants (data not shown). Protocols are available at http://www.genetics.wustl.edu/tslab/protocols.html.
Images were captured using either a Zeiss Axioskop or Axioplan2 equipped with either a SPOT (Diagnostic Instruments, Inc.) or Hamamatsu (Hamamatsu Photonics K.K.) digital CCD and processed with Adobe Photoshop 5.5.
Acknowledgments
We thank Gary Moulder and Bob Barstead for the ptc-1(ok122) deletion mutant; the NemaPharm Group at Axys Pharmaceuticals for the ptc-1(nr2029) deletion mutant; Verena Jantsch-Plunger and Michael Glotzer for SYN-4 antisera; Yuji Kohara for cDNA clones; Douglas Kershaw and Ian Adams for assistance with electron microscopy; Alan Coulson, Ratna Shownkeen, and Stephanie Chissoe for cosmid clones, YAC filters, and genomic sequence information; and Matt Scott for comments on an earlier version of this manuscript. We also thank Carl Johnson and Jonathan Hodgkin for comments and discussions pertaining to this manuscript. This work was supported by the MRC. Work in the Schedl laboratory was supported by NIH RO1 HD25614.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pek@sanger.ac.uk; FAX 44 1223 494919.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspöck G, Kagoshima H, Niklaus G, Bürglin TR. Caenorhabditis elegans has scores of hedgehog-related genes: Sequence and expression analysis. Genome Res. 1999;9:909–923. doi: 10.1101/gr.9.10.909. [DOI] [PubMed] [Google Scholar]
- Bürglin TR. Warthog and Groundhog, novel families related to Hedgehog. Curr Biol. 1996;6:1047–1050. doi: 10.1016/s0960-9822(02)70659-3. [DOI] [PubMed] [Google Scholar]
- Burke R, Nellen D, Bellotto M, Dickson BJ, Basler K. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified Hedgehog from signaling cells. Cell. 1999;99:803–815. doi: 10.1016/s0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- Carpenter D, Stone DM, Brush J, Ryan A, Armanini M, Frantz G, Rosenthal A, De Sauvage FJ. Characterization of two patched receptors for the vertebrate hedgehog protein family. Proc Nat Acad Sci. 1998;95:13630–13634. doi: 10.1073/pnas.95.23.13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstea ED, Morris JA, Coleman KG, Loftus SK, Zhang D, Cummings C, Gu J, Rosenfeld MA, Pavan WJ, Krizman DB, et al. Niemann-Pick C1 disease gene: Homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- Chen Y, Struhl G. Dual roles for patched in sequestering and transducing hedgehog. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- Clines GA, Ashley JA, Shah S, Lovett M. The structure of the human multiple Exostoses 2 gene and characterization of homologs in mouse and Caenorhabditis elegans. Genome Res. 1997;7:359–367. doi: 10.1101/gr.7.4.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet J-P, Lewis K, Moore J, Goodrich LV, Johnson RL, Scott MP, Ingham PW. Spatial regulation of a zebrafish patched homologue reflects the roles of sonic hedgehog and protein kinase A in neural tube and somite patterning. Development. 1996;122:2835–2846. doi: 10.1242/dev.122.9.2835. [DOI] [PubMed] [Google Scholar]
- Consortium, The C. elegans Sequencing. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Cooper MK, Porter JA, Young KE, Beachy PA. Teratogen-mediated inhibition of target tissue response to Shh signaling. Science. 1998;280:1603–1607. doi: 10.1126/science.280.5369.1603. [DOI] [PubMed] [Google Scholar]
- Crittenden SL, Troemel ER, Evans TC, Kimble J. GLP-1 is localized to the mitotic region of the C. elegans germ line. Development. 1994;120:2901–2911. doi: 10.1242/dev.120.10.2901. [DOI] [PubMed] [Google Scholar]
- Field C, Li R, Oegema K. Cytokinesis in eukaryotes: A mechanistic comparison. Curr Opin Cell Biol. 1999;11:68–80. doi: 10.1016/s0955-0674(99)80009-x. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- Francis R, Barton MK, Kimble J, Schedl T. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics. 1995;139:579–606. doi: 10.1093/genetics/139.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden AB, Gillies S, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- Hall DH, Winfrey VP, Blaeur G, Hoffman LH, Furuta T, Rose KL, Hobert O, Greenstein D. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: Relations between the germ line and soma. Dev Biol. 1999;212:101–123. doi: 10.1006/dbio.1999.9356. [DOI] [PubMed] [Google Scholar]
- Hall TMT, Porter JA, Young KE, Koonin EV, Beachy PA, Leahy DJ. Crystal structure of a Hedgehog autoprocessing domain: Homology between Hedgehog and self-splicing proteins. Cell. 1997;91:85–97. doi: 10.1016/s0092-8674(01)80011-8. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M, Brook A, McMahon AP. The world according to hedgehog. Trends Genet. 1997;13:14–21. doi: 10.1016/s0168-9525(96)10051-2. [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, Ingham P. Cell patterning in the Drosophila segment: Spatial regulation of the segment polarity gene patched. Development. 1990;110:291–301. doi: 10.1242/dev.110.1.291. [DOI] [PubMed] [Google Scholar]
- Hirsh D, Oppenheim D, Klass M. Development of the reproductive system of Caenorhabditis elegans. Dev Biol. 1976;49:200–219. doi: 10.1016/0012-1606(76)90267-0. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. A genetic analysis of the sex determining gene, tra-1, in the nematode Caenorhabditis elegans. Genes & Dev. 1987;1:731–745. doi: 10.1101/gad.1.7.731. [DOI] [PubMed] [Google Scholar]
- ————— . Appendix 1. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase—A database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- Hooper JE, Scott MP. The Drosophila patched gene encodes a putative membrane protein required for segmental patterning. Cell. 1989;59:751–765. doi: 10.1016/0092-8674(89)90021-4. [DOI] [PubMed] [Google Scholar]
- Ingham PW. The patched gene in development and cancer. Curr Opin Genet Dev. 1998;8:88–94. doi: 10.1016/s0959-437x(98)80067-1. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Taylor AM, Nakano Y. Role of the Drosophila patched gene in positional signalling. Nature. 1991;355:184–187. doi: 10.1038/353184a0. [DOI] [PubMed] [Google Scholar]
- Iwasaki K, McCarter J, Francis R, Schedl R. emo-1, a Caenorhabditis elegans Sec61p γ homologue, is required for ooocyte development and ovulation. J Cell Biol. 1996;134:699–714. doi: 10.1083/jcb.134.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch-Plunger V, Glotzer M. Depletion of syntaxins in the early Caenorhabditis elegans embryo reveals a role for membrane fusion events in cytokinesis. Curr Biol. 1999;9:738–745. doi: 10.1016/s0960-9822(99)80333-9. [DOI] [PubMed] [Google Scholar]
- Johnson RL, Scott MP. Control of cell growth and fate by patched genes. Cold Spring Harbor Symposia on Quantitative Biology. 1997;LXII:205–215. [PubMed] [Google Scholar]
- Johnson RL, Scott MP. New players and puzzles in the Hedgehog signaling pathway. Curr Opin Genet Dev. 1998;8:450–456. doi: 10.1016/s0959-437x(98)80117-2. [DOI] [PubMed] [Google Scholar]
- Johnson R, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, Quinn AG, Myers RM, Cox DR, Epstein EH, Jr, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- Jones AR, Francis R, Schedl T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev Biol. 1996;180:165–183. doi: 10.1006/dbio.1996.0293. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:133–148. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lange Y, Steck TL. Four cholesterol-sensing proteins. Curr Opin Struct Biol. 1998;8:435–439. doi: 10.1016/s0959-440x(98)80119-x. [DOI] [PubMed] [Google Scholar]
- Loftus SK, Morris JA, Carstea ED, Gu JZ, Cummings C, Brown A, Ellison J, Ohno K, Rosenfeld MA, Tagle DA, et al. Murine model of Niemann-Pick C disease: Mutation in a cholesterol homeostasis gene. Science. 1997;277:232–235. doi: 10.1126/science.277.5323.232. [DOI] [PubMed] [Google Scholar]
- Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. Biochemical evidence that Patched is the Hedgehog receptor. Nature. 1996;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- Motoyama J, Takabatake T, Takeshima K, Hui C-C. Ptch2, a second mouse Patched gene is co-expressed with Sonic hedgehog. Nat Genet. 1998;18:104–106. doi: 10.1038/ng0298-104. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Maxfield FR. Cholesterol: Stuck in traffic. Nat Cell Biol. 1999;1:E37–E38. doi: 10.1038/10030. [DOI] [PubMed] [Google Scholar]
- Nakano Y, Guerrero I, Hidalgo A, Taylor A, Whittle JRS, Ingham PW. A protein with several possible membrane-spanning domains encoded by the Drosophila segment polarity gene patched. Nature. 1989;341:508–513. doi: 10.1038/341508a0. [DOI] [PubMed] [Google Scholar]
- Osborne TF, Rosenfeld JM. Related membrane domains in proteins of sterol sensing and cell signaling provide a glimpse of treasures still buried within the dynamic realm of intracellular metabolic regulation. Curr Opin Lipidol. 1998;9:137–140. doi: 10.1097/00041433-199804000-00010. [DOI] [PubMed] [Google Scholar]
- Page RDM. TREEVIEW: An application to display phylogenetic trees on personal computers. Comp Applic Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Brown MH, Skurray RA. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JA, Young KE, Beachy PA. Cholesterol modification of Hedgehog signaling proteins in animal development. Science. 1996a;274:255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- Porter JA, Ekker SC, Park W-J, von Kessler DP, Young KE, Chen C-H, Ma Y, Woods AS, Cotter RJ, Koonin EV, Beachy PA. Hedgehog patterning activity: Role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell. 1996b;86:21–34. doi: 10.1016/s0092-8674(00)80074-4. [DOI] [PubMed] [Google Scholar]
- Schedl T, Graham PL, Barton MK, Kimble J. Analysis of the role of tra-1 in germline sex determination in the nematode Caenorhabditis elegans. Genetics. 1989;123:755–769. doi: 10.1093/genetics/123.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Fire A. Soma-germline asymmetry in the distribution of maternal and zygotic mRNAs in early C. elegans embryos. Development. 1994;120:2823–2834. doi: 10.1242/dev.120.10.2823. [DOI] [PubMed] [Google Scholar]
- Stone D, Hynes M, Armanini M, Swanson T, Gu Q, Johnson R, Scott MP, Pennica D, Goddard A, Phillips H, et al. The tumor-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- Strome S. Fluorescence visualization of the distribution of microfilaments in gonads and early embryos of the nematode Caenorhabditis elegans. J Cell Biol. 1986;103:2241–2252. doi: 10.1083/jcb.103.6.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Hodgkin J. Methods. In: Wood WB, editor. The nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. pp. 587–606. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Nusse R. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]