Abstract
The initiation of DNA synthesis is an important cell cycle event that defines the beginning of S phase. This critical event involves the participation of proteins whose functions are regulated by cyclin dependent protein kinases (Cdks). The Mcm2–7 proteins are a family of six conserved proteins that are essential for the initiation of DNA synthesis in all eukaryotes. In Saccharomyces cerevisiae, members of the Mcm2–7 family undergo cell cycle-specific phosphorylation. Phosphorylation of Mcm proteins at the beginning of S phase coincides with the removal of these proteins from chromatin and the onset of DNA synthesis. In this study, we identified DBF4, which encodes the regulatory subunit of a Cdk-like protein kinase Cdc7–Dbf4, in a screen for second site suppressors of mcm2-1. The dbf4 suppressor mutation restores competence to initiate DNA synthesis to the mcm2-1 mutant. Cdc7–Dbf4 interacts physically with Mcm2 and phosphorylates Mcm2 and three other members of the Mcm2–7 family in vitro. Blocking the kinase activity of Cdc7–Dbf4 at the G1-to-S phase transition also blocks the phosphorylation of Mcm2 at this defined point of the cell cycle. Taken together, our data suggest that phosphorylation of Mcm2 and probably other members of the Mcm2–7 proteins by Cdc7–Dbf4 at the G1-to-S phase transition is a critical step in the initiation of DNA synthesis at replication origins.
Keywords: DNA replication, Mcm proteins, Cdc7–Dbf4 kinase
The initiation of DNA replication in eukaryotic cells is a tightly regulated process that is strictly coupled to the progression of the cell cycle. At the end of mitosis, DNA replication proteins such as the origin recognition complex (ORC) (Bell and Stillman 1992), Cdc6 (Cocker et al. 1996), and the Mcm proteins (Tye 1994) are assembled at replication origins to form the pre-replicative chromatin (Coleman et al. 1996; Nasmyth 1996; Stillman 1996). In the yeast Saccharomyces cerevisiae, this competent state of chromatin is marked by a distinctive protection pattern against DNase I digestion on DNA replication origins (Diffley et al. 1994; Diffley 1996). After cells pass Start, the prereplicative chromatin is activated and DNA synthesis is initiated at replication origins. This process defines the beginning of S phase. As S phase progresses, the prereplicative chromatin shifts to a post-replicative state that is no longer competent to initiate another round of DNA synthesis. Recently, it has become clear that the assembly and activation of competent chromatin are under the control of cell cycle-dependent kinase activities (Nasmyth 1996; Stillman 1996). It remains largely unknown, however, which replication protein, at each of the major control points, is the specific in vivo target of a certain known kinase.
The Mcm2–7 proteins are a family of six conserved proteins (Hennessy et al. 1991; Yan et al. 1991; Merchant et al. 1997), that are essential for the initiation of DNA synthesis at replication origins in S. cerevisiae (Yan et al. 1993). Each of these proteins has a corresponding homolog in all eukaryotes examined so far (Tye 1994; Kearsey et al. 1995), suggesting that their role in DNA replication initiation is universal. Studies in S. cerevisiae indicate that lesions in members of the Mcm2–7 protein family prevent initiation of DNA synthesis at replication origins in plasmids and in chromosomes (Maine et al. 1984; Hennessy et al. 1991; Yan et al. 1993). The replication defects caused by mutations in the MCM genes are origin specific: For a certain mcm mutation, the defect at some replication origins is much more severe than at others (Yan et al. 1993), suggesting that Mcm proteins are directly involved in the initiation of DNA synthesis at origins. Studies with in vitro-assembled nuclei in Xenopus egg extracts showed that the XMcm proteins are essential components of the DNA-replication activity, which is competent for a single round of DNA synthesis (Chong et al. 1995; Kubota et al. 1995; Madine et al. 1995). Several reports showed that the Mcm2–7 proteins interact with each other to form multimeric complexes (Chong et al. 1995; Kubota et al. 1995; Lei et al. 1996; Thommes et al. 1997). The Mcms are abundant proteins in proliferating cells (Treisman et al. 1995; Lei et al. 1996; Donovan et al. 1997; Young and Tye 1997). In S. cerevisiae, these proteins are constitutively present in the cytoplasm and in the nucleus (Young and Tye 1997). A fraction of the nuclear Mcm proteins is associated with chromatin in a cell cycle-specific manner: they are chromatin bound in the G1 phase, but are removed from the chromatin as S phase progresses (Todorov et al. 1995; Coue et al. 1996; Donovan et al. 1997; Young and Tye 1997). Binding of the Mcm proteins to chromatin appears to be essential for the transition of chromatin from the replication-incompetent to the replication-competent state (Kubota et al. 1995). Recent studies suggest that the binding of the Mcm proteins to the chromatin, as a step of the ordered assembly of prereplication complex (pre-RC), is dependent on the binding of ORC and Cdc6 protein to chromatin (Coleman et al. 1996). Recent protein–DNA cross-linking studies show that the Mcm proteins are localized at replication origins during G1 phase (Aparicio et al. 1997; Tanaka et al. 1997). These properties of the Mcm proteins suggest that they may play a regulatory role in restricting DNA synthesis to once per cell cycle and in determining the competence of replication origins.
Several lines of evidence suggest that the cell cycle-specific functions of the Mcm proteins are regulated by changes in the phosphorylation states of these proteins. Mcm proteins are phosphoproteins that undergo cell cycle-specific phosphomodifications (Todorov et al. 1995; Coue et al. 1996; Young and Tye 1997). Both Mcm2 and Mcm3 are converted from hypophosphorylated to hyperphosphorylated isoforms during the G1- to S-phase transition, which coincides with the gradual dissociation of Mcm proteins from the chromatin and the beginning of DNA synthesis (Young and Tye 1997). Recently, it was shown that XMcm4 is phosphorylated by Cdc2-cyclin B kinases, and the phosphorylated XMcm4 has reduced affinity to chromatin (Hendrickson et al. 1996). These studies suggest that protein kinases are involved in the regulation of the function of Mcm proteins during the transition from G1 to S phase.
Cdc7–Dbf4 is a Cdk-like serine/threonine protein kinase that is required for the onset of DNA synthesis (Hollingsworth and Sclafani 1992; Jackson et al. 1993). Cdc7, the catalytic subunit, is conserved in Schizosaccharomyces pombe and mammals (Masai et al. 1995; Sato et al 1997) and is maintained at a constant protein level throughout the cell cycle. Its protein kinase activity, however, is activated only during the G1-to-S-phase transition by the regulatory subunit Dbf4 (Jackson et al. 1993; Kitada et al. 1993). Dbf4, like cyclins, is expressed periodically during the cell cycle (Jackson et al. 1993). Mutations in either CDC7 or DBF4 block cell cycle progression at a point after Start, but before the hydroxyurea block (Hereford and Hartwell 1974; Hartwell 1976; Pringle and Hartwell 1981; Johnston and Thomas 1982). In vivo footprinting studies indicate that cells blocked at the G1-to-S-phase transition by mutations in CDC7 still maintain the prereplicative chromatin state (Diffley et al. 1994). Physical and functional interactions between Cdc7–Dbf4 and replication proteins or replication origins have been observed. Genetic studies suggested that Cdc7 interacts with one of the ORC subunits, Orc2 (Hardy and Pantz 1996). Dbf4 was shown to be associated with the A domain of replication origins through other origin-binding proteins (Dowell et al. 1994). The bob1 mutation, which was recently identified as a mutant allele of MCM5/CDC46 (Hardy et al. 1997), bypasses the requirement for CDC7 entirely during mitotic growth (Jackson et al. 1993). These findings strongly suggest a direct regulatory role for Cdc7–Dbf4 in the activation of the initiation of DNA replication, presumably by phosphorylating replication proteins at replication origins. In vivo substrates for this kinase, however, have not been identified so far. In addition to its essential role in the initiation of DNA replication, Cdc7 is known to be involved in other important cellular functions such as meiotic recombination (Simchen 1974; Schild and Byers 1978), replication dependent DNA repair (Hollingsworth and Sclafani 1992) and transcriptional silencing (Axelrod and Rine 1991).
In this study, we identify a new dbf4 mutant allele that suppresses the replication initiation defect of mcm2-1. On the basis of genetic analyses, physical interaction studies, in vitro and in vivo phosphorylation assays, we show that Mcm2 is a target of regulation by Cdc7–Dbf4 during the initiation of DNA synthesis in S. cerevisiae.
Results
Identification of DBF4 as a suppressor gene of mcm2-1
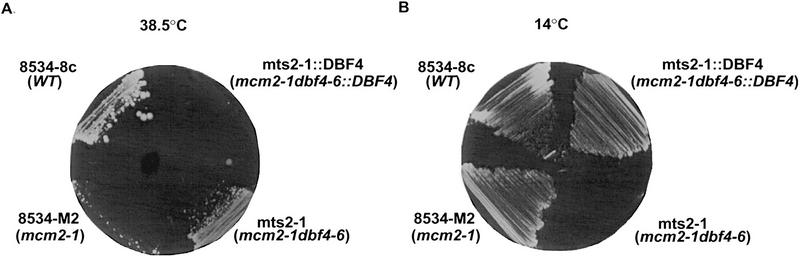
Mcm2 plays a critical role in the regulation of DNA replication initiation (Tye 1994). A mutation in the MCM2 gene, mcm2-1, reduces the frequency of DNA-replication initiation at replication origins and results in a defect in minichromosome maintenance (Yan et al. 1991, 1993). This mutant allele is conditionally lethal at 38.5°C (Fig. 1A). To understand the molecular mechanism that regulates the function of Mcm2 and other Mcm proteins, we sought to identify genes that functionally interact with MCM2. We carried out a genetic screen to isolate spontaneous second-site mutations that suppress the temperature-sensitive growth defect of the mcm2-1 mutant. From four independent pools of cells (∼2.5 × 108 cells in total), we isolated six temperature-sensitive suppressors that are also conditionally lethal at 14°C. Diploid strains resulting from a cross between these suppressor strains and a mcm2-1 strain, M46Y1332A (Table 1), were all temperature-sensitive at 38.5°C, and viable at 14°C (data not shown). Therefore, both the temperature-sensitive suppression and the cold-sensitive growth defect of these suppressors are recessive. To identify the suppressor genes, one approach we took was to transform the suppressor strains with plasmids that harbor genes known to be involved in the control of DNA replication. When a centromeric plasmid containing the DBF4 gene [pKK823, (Kitada et al. 1992)] was transformed into each of the six suppressor strains, the cold-sensitive phenotype of one of them, mts2-1 (mcm temperature-sensitive suppressors), was complemented at 14°C. The transformant also became temperature-sensitive at 38.5°C. We then integrate the DBF4 gene at the dbf4 locus in the mts2-1 strain. Whereas the suppressor strain, mts2-1, is viable at 38.5°C and cold-sensitive at 14°C, the integrant, mts2-1::DBF4, is temperature-sensitive at 38.5°C and viable at 14°C (Fig.1A,B), indicating that both the temperature-sensitive suppression and the cold-sensitive phenotypes of mts2-1 are complemented by DBF4. Tetrad analysis of a diploid strain, mts2-1::DBF4/M46Y1332A (mts2-1::DBF4 mcm2-1/MTS2 mcm2-1) showed that all four spores from each of the nine sets of tetrads examined are temperature-sensitive at 38.5°C (data not shown), indicating that MTS2 is tightly linked to DBF4. These results together indicate that MTS2 is identical to DBF4. The mts2-1 mutation will be referred to as dbf4-6 hereafter, and the other five suppressors will be described elsewhere. Tetrad analysis of a diploid strain mts2-1/DBY2065 (mcm2-1 dbf4-6/MCM2 DBF4) showed that spores with dbf4-6 in a wild-type MCM2 background were inviable (data not shown), suggesting that dbf4-6 by itself is lethal for growth and that viability of the mts2-1 strain at 38.5°C is the result of cosuppression of mcm2-1 and dbf4-6 (Adams et al. 1989).
Figure 1.
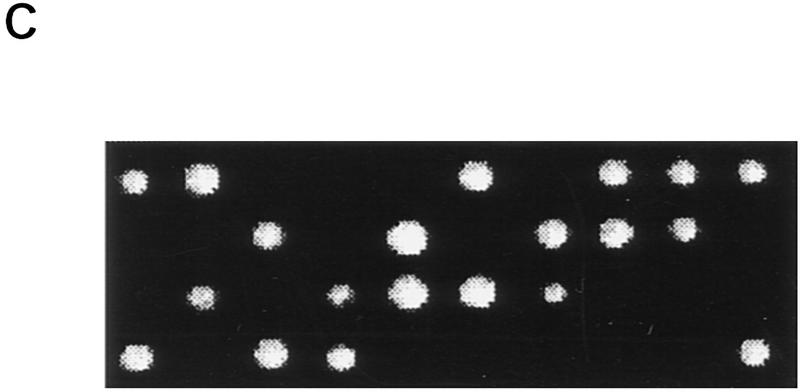
The DBF4 gene complements both the heat-sensitive suppression and the cold-sensitive growth phenotype of mts2-1. mts2-1::DBF4 contains a wild-type DBF4 gene integrated at the dbf4 locus of mts2-1. Yeast cells were streaked onto YEPD plates and grown at 38.5°C for 3 days (A), or at 14°C for 7 days (B). (C) The result of the tetrad analysis of a diploid strain that is mcm2-1 homozygous and dbf4-3 heterozygous (mcm2-1 dbf4-3/mcm2-1 DBF4; see Materials and Methods for construction of the strain). Tetrads were dissected on YEPD plates and grown at 30°C for 3 days.
Table 1.
Plasmids and strains
| Plasmids/strains
|
Description
|
Source or reference
|
|---|---|---|
| pRS306.DBF4 | URA3 DBF4 | this study |
| pKK823 | ARS1 CEN4 LEU2 DBF4 | Kitada et al. 1992 |
| pMcm2-1.s | YIp56 with mcm2-1 | Yan et al. 1991 |
| pHY1 | pSE355 with mcm2-1 | Yan et al. 1991 |
| pBTM116 | 2μ TRP1 lexA(1–202) | S. Fields (Washington University, St. Louis, MO) |
| pBTM116.MCM2 | 2μ TRP1 lexA–mcm2 | Lei et al. 1996 |
| pBTM116.MCM2-1 | 2μ TRP1 lexA–mcm2-1 | this study |
| pKH125 | 2μ TRP1 lexA–DBF4(52–695) | Jackson et al. 1993 |
| pGAD2F | 2μ LEU2 GAL4(768–881) | S. Fields |
| pC2 | 2μ LEU2 GAL4–DBF4(4–416) | Dowell et al. 1994 |
| pKH122 | 2μ LEU2 GAL4–CDC7(1–507) | Jackson et al. 1993 |
| pEG(KT) | 2μ URA3 GAL1–GST | D. Mitchell (University of Iowa, Iowa City) |
| pEG.MCM2 | 2μ URA3 GAL1–GST–mcm2 | Lei et al. 1996 |
| pEG.MCM3 | 2μ URA3 GAL1–GST–mcm3 | Lei et al. 1996 |
| pEG.MCM4 | 2μ URA3 GAL1–GST–mcm4 | Merchant et al. 1997 |
| pEG.MCM5 | 2μ URA3 GAL1–GST–mcm5 | Merchant et al. 1997 |
| pEG.MCM6 | 2μ URA3 GAL1–GST–mcm6 | Merchant et al. 1997 |
| pEG.MCM7 | 2μ URA3 GAL1–GST–mcm7 | Merchant et al. 1997 |
| pEG.MCM2-1 | 2μ URA3 GAL1–GST–mcm2-1 | this study |
| 8534-8c | α ura3 his3 leu2 | Yan et al. 1993 |
| 8534-M2 | α ura3 his3 leu2 mcm2-1 | Yan et al. 1993 |
| DBY2065 | a lys | D. Botstein (Stanford University, Stanford, CA) |
| L128-2D | a dbf4-1 ura3 trp1 ade2 leu2 | Johnston and Thomas 1982 |
| KKY743 | α dbf4-3 ura3-52 leu2-3,112 trp1-289 ade5 his7-2 | Kitada et al. 1992 |
| KKY744 | α dbf4-4 ura3-52 leu2-3,112 trp1-289 ade5 his7-2 | Kitada et al. 1992 |
| KKY745 | α dbf4-5 ura3-52 leu2-3,112 trp1-289 ade5 his7-2 | Kitada et al. 1992 |
| M46Y1332A | α ura3 his3 ade2-101 mcm2-1 | Gibson 1989 |
| M46Y1333B | α ura3 his3 ade2-101 mcm2-1 | Gibson 1989 |
| mts2-1 | α ura3 his3 leu2 mcm2-1 dbf4-6 | this study |
| mts2-1∷DBF4 | α ura3 his3 leu2 mcm2-1 dbf4-6∷DBF4 | this study |
To investigate whether the dbf4-6 suppression of mcm2-1 is allele specific, we constructed double mutants of mcm2-1 with four temperature-sensitive dbf4 alleles, dbf4-1, -3, -4, -5 (Johnston and Thomas 1982; Kitada et al. 1992). Diploid strains that are mcm2-1 homozygous and dbf4 heterozygous were constructed as described in Materials and Methods. Tetrad analysis of all four of these diploid strains showed that each tetrad has two live and two dead spores, suggesting that mcm2-1 is synthetically lethal with each of the dbf4-1, -2, -3, -4 alleles. Figure 1C is a prototypical result of these tetrad analyses. The allele-specific suppression of mcm2-1 by dbf4-6 and the synergistic effect of mcm2-1 with other dbf4 alleles suggest that Mcm2 and Cdc7–Dbf4 kinase are involved in the same regulatory pathway, and that Cdc7–Dbf4 may regulate the function of Mcm2 by direct physical interaction.
dbf4-6 suppresses the replication initiation defect of mcm2-1
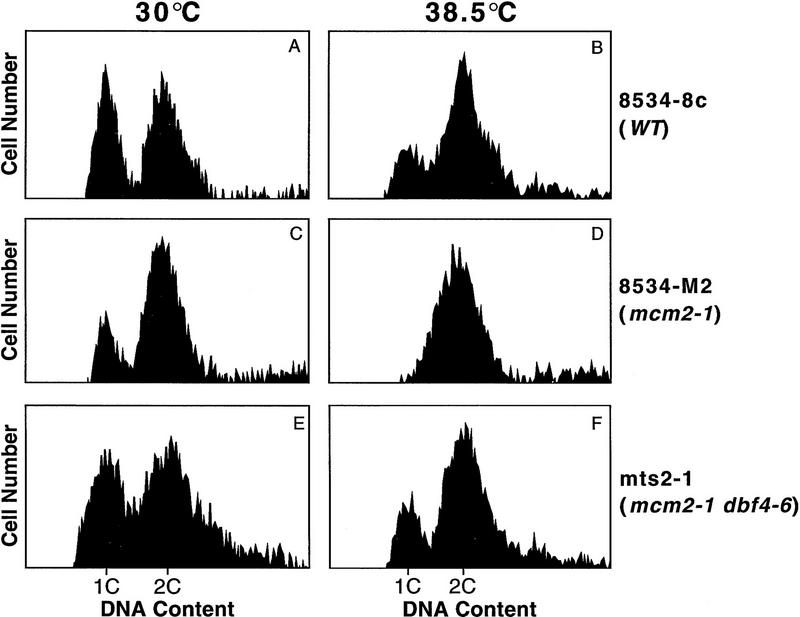
When yeast cells are depleted of Mcm2, cells arrest with a large bud and an undivided nucleus with almost 2C DNA caused by incomplete replication (Yan et al. 1991). At 38.5°C, the mcm2-1 mutant cells have a similar arrest phenotype (data not shown), which is corrected in the suppressor strain, mts2-1 (Fig. 1). The DNA content of the wild-type, mcm2-1 and suppressor cells growing at 30°C or 38.5°C was analyzed by flow cytometry (Fig. 2). At 38.5°C, mcm2-1 cells are arrested with a DNA content close to 2C (Fig. 2D), similar to that observed in cells depleted of Mcm2 (Yan et al. 1991). This arrest phenotype is reversed in the mts2-1 strain, as shown by the two distinct populations of cells containing 1C or 2C DNA (Fig. 2F), similar to wild-type cells (Fig. 2B).
Figure 2.
Growth arrest of mcm2-1 cells with nearly 2C DNA at 38.5°C is reversed by the dbf4-6 mutation. 8534-8c (WT), 8534-M2 (mcm2-1) and mts2-1 (mcm2-1 dbf4-6) cells grown at 30°C or 38.5°C were stained with propidium iodide, and the DNA content of the cells were analyzed by flow cytometry. (A,C,E) DNA content profiles of the wild-type, mutant and suppressor strains at 30°C. (B,D,F) DNA content profiles of the three strains at 38.5°C.
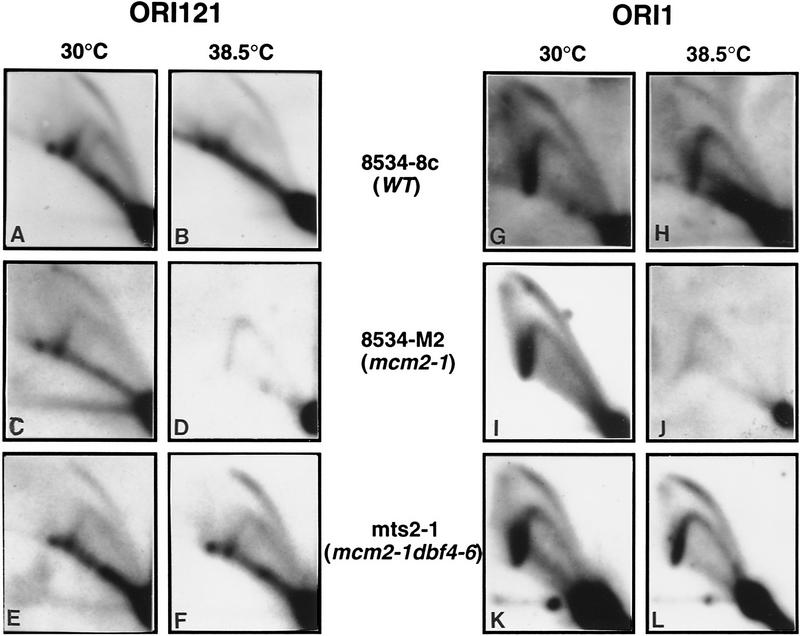
The mcm2-1 mutant is defective in the initiation of DNA synthesis at replication origins (Yan et al. 1993). To investigate whether the suppressor restores competence for initiating DNA synthesis at replication origins, we examined the occurence of replication initiation events at ORI121 and ORI1 by two-dimensional DNA gel analysis (Fig. 3). In the wild-type cells, replication initiation is detected in the form of bubble arcs at both ORI121 and ORI1 at 30°C and 38.5°C (Fig. 3A,B,G,H). In the mcm2-1 mutant cells, however, no bubble arc was detected at 38.5°C, indicating that initiation at ORI1 and ORI121 is greatly reduced (Fig. 3C,D,I,J). The Y arcs detected result from elongation forks that emanate from initiation events at other origins. In the mts2-1 strain, initiation at ORI121 and ORI1 is restored to near wild-type level at 38.5°C, as shown by the strong bubble arcs (Fig. 3E,F,K,L). This result suggests that the dbf4-6 mutation can restore the function of mcm2-1 such that the initiation of DNA synthesis is returned to normal at replication origins.
Figure 3.
Two-dimensional DNA gel analysis of the initiation of DNA synthesis at replication origins in 8534-8c (WT), 8534-M2 (mcm2-1) and mts2-1 (mcm2-1 dbf4-6) cells. (A–F) Analysis of replication events at ORI121. (G–L) Analysis of replication events at ORI1.
Cdc7–Dbf4 physically interacts with Mcm2
The allele-specific suppression of mcm2-1 by dbf4-6 strongly suggests a direct physical interaction between the Mcm2 and Dbf4 proteins. Therefore, we decided to investigate whether Mcm2 and Dbf4–Cdc7 are physically associated.
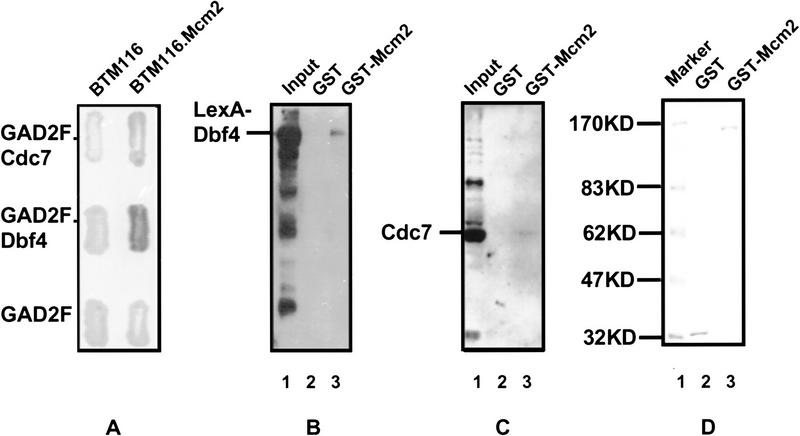
The yeast two-hybrid system (Fields and Song 1989) was used to examine the interaction between Cdc7–Dbf4 and Mcm2. In this assay, both Dbf4 and Cdc7 interact with Mcm2, although the interaction between Cdc7 and Mcm2 appears to be much weaker than that detected for Dbf4 (Fig. 4A). The mcm2-1 mutation significantly affects the interaction between Mcm2 and Dbf4. When measured by β-galactosidase activities in the two-hybrid assay, the interaction between Dbf4 and Mcm2 (175.5±8.5 miller units) is ∼2.5-fold stronger than that between Dbf4 and Mcm2-1 (70±2.3 miller units). Physical interactions between Cdc7–Dbf4 and Mcm2 were also investigated by use of affinity chromatography. GST and GST–Mcm2 fusion proteins expressed in yeast were conjugated to glutathione-Sepharose resin and incubated with yeast protein extracts. For the interaction between Cdc7 and Mcm2, protein extracts from the protease deficient yeast strain, BJ2168, were used. For the interaction between Dbf4 and Mcm2, protein extracts from BJ2168 cells carrying a plasmid (pKH125) that produces LexA–Dbf4 were used. The LexA–Dbf4 fusion protein complements the temperature-sensitive growth defect of dbf4-1 cells at 37°C (data not shown). After extensive washing, proteins bound to GST and GST–Mcm2 were eluted from the resin. The eluent was analyzed by SDS-PAGE and probed with either anti-LexA or anti-Cdc7 antibodies in the Western blots. The result (Fig. 4B) shows that LexA–Dbf4 is retained by GST–Mcm2 (lane 3) but not by GST (lane 2). LexA alone was not retained by GST or GST–Mcm2 fusion protein (data not shown). Figure 4C shows that Cdc7 is retained by GST–Mcm2 (lane 3) but not GST (lane 2). GST and GST–Mcm2 coupled to the resin were then released by 10 mm glutathione. The concentrations of the released proteins were compared in SDS-PAGE. Figure 4D shows that about equal amounts of GST and GST–Mcm2 proteins were coupled to the resin in this experiment. These results indicate that Mcm2 is physically associated with the Cdc7–Dbf4 kinase.
Figure 4.
Physical interactions between Cdc7–Dbf4 and Mcm2. (A) Two-hybrid analysis. BTM116 and BTM116.Mcm2 denote plasmids that express LexA and LexA–Mcm2 proteins, respectively. GAD2F, GAD2F.Cdc7, and GAD2F.Dbf4 denote plasmids that express GAL4, GAL4–Cdc7, and GAL4–Dbf4 proteins, respectively. Yeast transformants carrying each pair of the plasmids and a LexAop–lacZ reporter gene were patched on 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal) plates. The expression of β-galactosidase is shown by the blue color of colonies. (B,C) GST–Mcm2 fusion affinity column chromatography. (B) Western blot with anti-LexA antibodies. (Lane 1) Total yeast protein extract from BJ2168 cells carrying pKH125; (lane 2,3) proteins eluted from glutathione Sepharose 4B beads coupled with GST (lane 2) and GST.Mcm2 (lane 3). (C) Western blot with anti-Cdc7 antibodies. (Lane 1) Total yeast protein extract from BJ2168 cells; (lanes 2,3) proteins eluted from glutathione Sepharose 4B beads coupled with GST (lane 2) and GST.Mcm2 (lane 3). (D) Coomassie Brilliant Blue staining of a SDS-PAGE showing the GST or GST–Mcm2 released from the beads by 10 mm glutathione after elution of the interacting proteins.
Mcm2, Mcm3, Cdc54/Mcm4, and Mcm6 are phosphorylated by Cdc7–Dbf4 in vitro
The interactions between Cdc7–Dbf4 and Mcm2 led us to examine whether Mcm2 is a substrate of the Cdc7–Dbf4 kinase. Because members of the Mcm2–7 family interact with one another and form complexes (Lei et al. 1996; Thommes et al. 1997), we also examined if other members of the Mcm2–7 family are phosphorylated by Cdc7–Dbf4. Cdc7–Dbf4 kinase purified from Sf9 insect cells coexpressing the yeast CDC7 and DBF4 genes (M. Kihara, K. Kitada, and A. Sugino, unpubl.) was used in the kinase assay (Fig. 5A). Approximately equal amounts of GST–Mcm2–7 fusion proteins purified from yeast (Lei et al. 1996) were used as substrates (see Fig. 5B). The results indicate that GST–Mcm2 (Fig. 5A, lane 1), GST–Mcm3 (lane 2), GST–Cdc54/Mcm4 (lane 3), and GST–Mcm6 (lane 5) are phosphorylated by Cdc7–Dbf4, but GST–Cdc46/Mcm5 (lane 4) and GST–Cdc47/Mcm7 (lane 6) are not. Of the four Mcm proteins that are phosphorylated, Mcm2 and Mcm6 are phosphorylated to the greatest extent.
Figure 5.
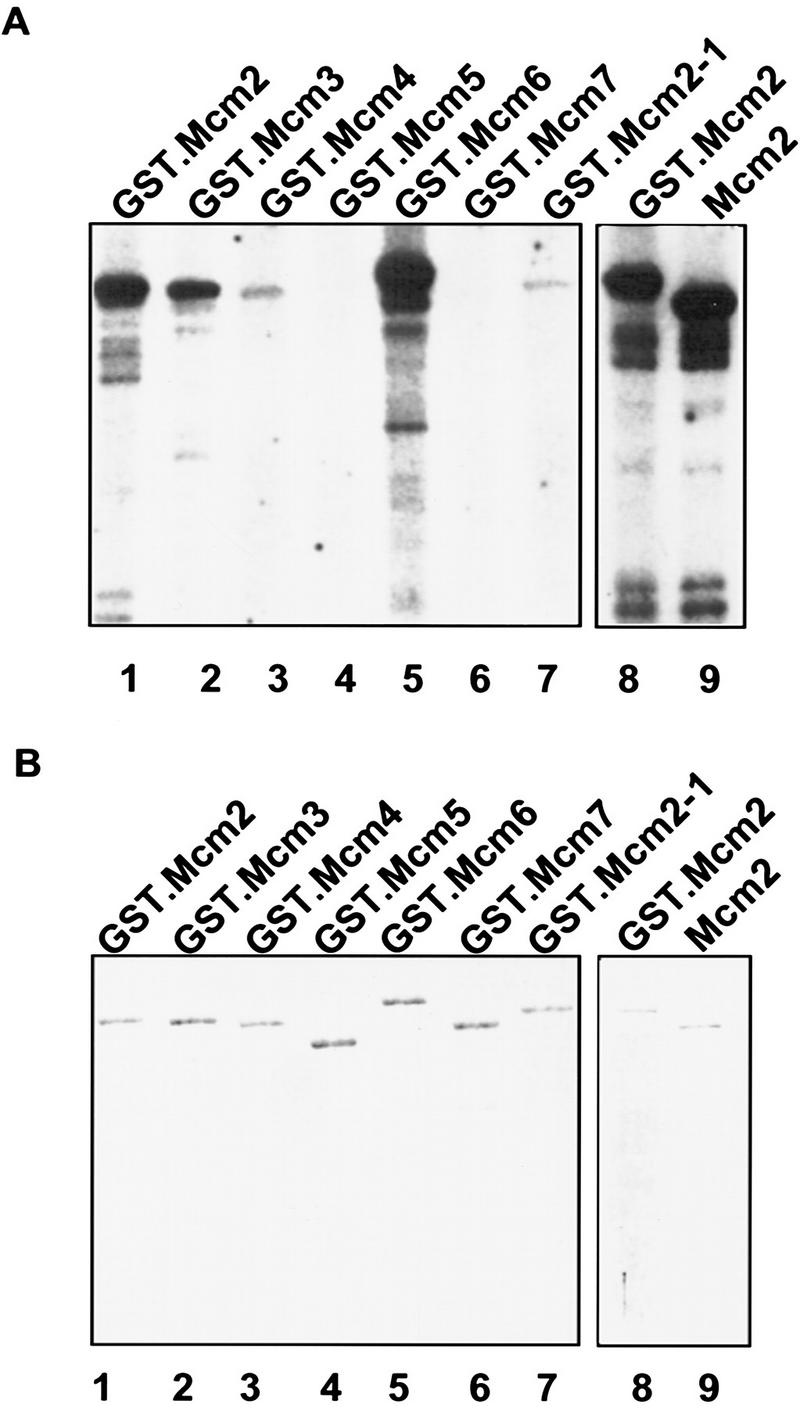
Mcm2, Mcm3, Mcm4, and Mcm6 are substrates of the Cdc7–Dbf4 kinase in vitro. The kinase assay was performed with Cdc7–Dbf4 protein kinase purified from Sf9 cells, and GST–Mcm proteins purified from yeast. The reactions were resolved in SDS–polyacrylamide gels for autoradiography (A), or for Coomassie brilliant blue staining (B). GST.Mcm2, (lane 1); GST.Mcm3, (lane 2); GST.Mcm4, (lane 3); GST.Mcm5, (lane 4); GST.Mcm6, (lane 5); GST.Mcm7, (lane 6); GST.Mcm2-1, (lane 7); GST.Mcm2, (lane 8); Mcm2, (lane 9).
Previously, we have characterized several point mutations in MCM2 (Yan et al. 1991). The mcm2-1 mutant allele (Glu-392 to Lys) has a dramatic mcm defect that reduces the frequency of initiation at replication origins (Fig. 3; Yan et al. 1993). When GST–Mcm2-1 was purified and used as substrates in the in vitro assay, it was phosphorylated to a much lower level than was the wild-type Mcm2 protein (Fig. 5A, lane 7). This result suggests a correlation between the function of Mcm2 and its phosphorylation by Cdc7–Dbf4. The GST tag in the fusion protein does not have an obvious effect on the phosphorylation of Mcm2 by Cdc7–Dbf4. When the tag was cleaved from GST–Mcm2, the Mcm2 protein was phosphorylated to a similar level as GST–Mcm2 (Fig. 5A, lanes 8,9).
A lesion in Dbf4 prevents phosphorylation of the Mcm2 protein at the G1-to-S-phase transition
Mcm2 and Mcm3 undergo phosphorylation during the G1-to-S-phase transition. The conversion of hypophosphorylated isoforms to hyperphosphorylated isoforms during this process was shown previously by two-dimensional protein gel analysis (Young and Tye 1997). The appearance of the hyperphosphorylated forms coincides with the execution point of the kinase activity of Cdc7–Dbf4 (Jackson et al. 1993). Because Mcm2 and three other Mcm proteins are specific substrates of the Cdc7–Dbf4 kinase in vitro, we investigated whether the Cdc7–Dbf4 kinase indeed phosphorylates Mcm2 in vivo. As a comparison, we also examined the effect of Cdc7–Dbf4 on the phosphorylation of Mcm5, which is not a substrate for the Cdc7–Dbf4 kinase in vitro.
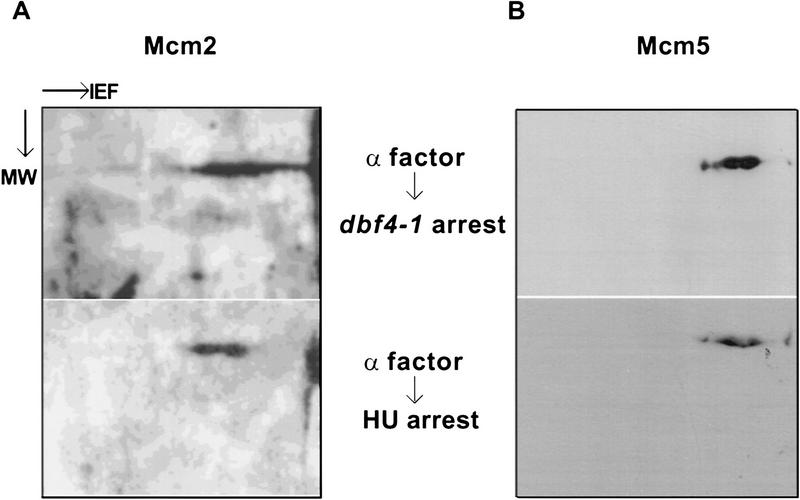
The dbf4-1 mutant strain (L128-2D, Table 1) was used in this experiment. At the nonpermissive temperature, the Cdc7–Dbf4 kinase activity in the mutant cells is inactivated and the cell-cycle progression is blocked at the G1-to-S-phase transition (Jackson et al. 1993; Kitada et al. 1993). The dbf4-1 cells arrested at the G1 phase with α factor were released from the G1 arrest, and shifted to either 37°C or to YEPD medium containing hydroxyurea (HU). HU blocks cell cycle progression at a very early point in the S phase, after the execution point of the Cdc7–Dbf4 kinase. Protein extracts from these arrested cells were analyzed by two-dimensional gel electrophoresis. Western blots of the two-dimensional gels were probed with antibodies specific to Mcm2 or Mcm5 to visualize the isoforms of these proteins at the G1-to-S-phase transition (dbf4-1 arrest), and the beginning of S-phase (HU arrest).
When the Cdc7–Dbf4 kinase is inactivated at the G1-to-S-phase transition, the Mcm2 protein is distributed on the two-dimensional gel as a heterogeneous array of isoforms that have a similar molecular weight but very different isoelectric points (Fig. 6A, top). This display of multiple isoforms is consistent with previous reports that the Mcm2 protein is distributed in different cellular compartments and, therefore, may assume different phosphorylation states (Young and Tye 1997). When cells are allowed to pass through the execution point of Cdc7–Dbf4 and blocked in S phase by hydroxyurea, the more basic, hypophosphorylated isoforms are converted to the more acidic, hyperphosphorylated isoforms (Fig. 6A, bottom panel). Because the dbf4-1 arrest and the HU arrest demarcate the narrowest time window that can be genetically or chemically defined during the G1-to-S-phase transition, these results suggest that Cdc7–Dbf4 is likely to be responsible for the phosphorylation of Mcm2 at the G1-to-S-phase transition. In comparison, the activity of Cdc7–Dbf4 appears to have very little effect on the phosphorylation states of the Mcm5 protein during the same time frame (Fig. 6B). The results from in vivo studies with the dbf4-1 allele are consistent with the results from in vitro studies, suggesting that Mcm2, but not Mcm5, is phosphorylated by the Cdc7–Dbf4 kinase at the G1-to-S-phase transition. Further studies to map and mutagenize the phosphorylation sites in Mcm2, however, are needed to conclude whether Mcm2 is directly phosphorylated by Cdc7–Dbf4 in vivo.
Figure 6.
Phosphorylation of Mcm2 at the G1-to-S-phase transition is blocked when Cdc7–Dbf4 kinase is inactivated. The isoform distributions of Mcm2 (left) and Mcm5 (right) were analyzed by immunoblots of two-dimensional protein gels with monospecific antibodies. dbf4-1 cells used in this assay were first arrested at Start by α factor at 30°C. Proteins were isolated from cells released from the α factor block and then arrested at G1-to-S-phase transition by shifting to 37°C (top) and from cells released from the α factor block and then arrested at early S by adding HU to the culture (bottom). In the first dimension gel, proteins are separated by isoelectrofocusing (IEF). In the second dimension gel, proteins are separated by molecular weight (MW). The two-dimensional gels were aligned with chemiluminescence signals from protein aggregates that did not enter the first dimension gel (Young and Tye 1997).
Discussion
Cdc7–Dbf4 is a Cdk-like protein kinase that plays an essential role in the initiation of DNA synthesis. The Mcm2–7 proteins are a family of six conserved proteins that are required for the initiation of DNA synthesis at replication origins. We isolated a new mutant allele of dbf4 that specifically suppresses the replication defects of mcm2-1 mutation. This allele-specific suppression suggests that Mcm2 is a target of regulation by Cdc7–Dbf4 during the initiation of DNA synthesis. We further showed that Mcm2, Mcm3, Mcm4, and Mcm6 are substrates for phosphorylation by Cdc7–Dbf4 in vitro, and that Mcm2 is a likely target for phosphorylation in vivo by Cdc7–Dbf4 during the G1-to-S-phase transition.
The Mcm proteins are essential components of the pre-replication complex assembled at replication origins (Diffley et al. 1994; Coleman et al. 1996). They are positioned at replication origins during G1 phase (Aparicio et al. 1997; Tanaka et al. 1997). Previous work with a reporter gene assay suggests that Dbf4 is recruited to the essential A element of replication origins (Dowell et al. 1994). Together, these studies argue strongly that the phosphorylation of Mcm proteins by Cdc7–Dbf4 is likely to take place at replication origins, leading to the initiation of DNA synthesis.
The mcm2-1 mutation is a single amino acid change (Glu-392 to Lys), 25 residues carboxy-terminal to the zinc finger motif, in a conserved region of Mcm2 (Yan et al. 1991). We showed that this mutation significantly affects the interaction of Mcm2 with Dbf4 and the phosphorylation of Mcm2 by Cdc7–Dbf4. The compromised interaction between Mcm2-1 and Dbf4, and the decreased phosphorylation of Mcm2-1 by Cdc7–Dbf4 are likely causes for the replication initiation defect of Mcm2-1. The replication initiation defect of Mcm2-1 is corrected by the dbf4-6 mutation, restoring viability to the mutant at 38.5°C by allowing initiation of DNA synthesis to occur at replication origins. The molecular mechanism of the suppression is not clear at this time, though the allele-specificity of the suppression suggests that dbf4-6 may be a compensatory mutation of mcm2-1 that restores physical interactions between the Mcm2 and Dbf4 proteins.
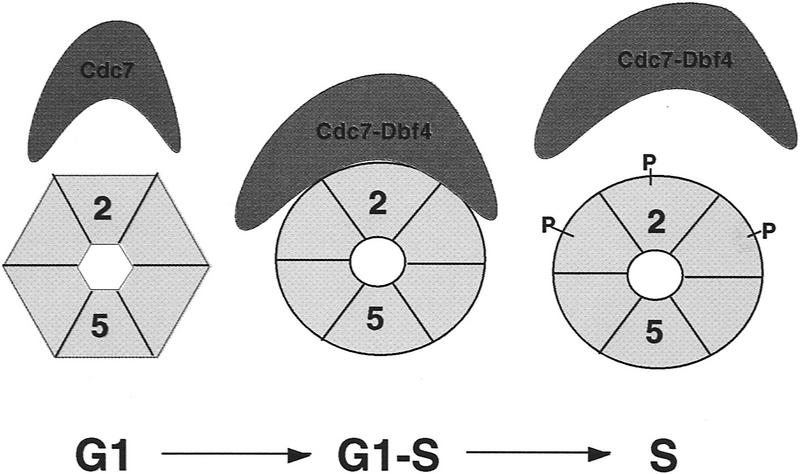
We were surprised that Mcm5 did not serve as a substrate for Cdc7–Dbf4 in our assays because recent studies suggest that the roles of Mcm5 and Cdc7–Dbf4 in DNA replication are intimately related. A mutation, bob1, which bypasses the essential function of Cdc7–Dbf4 (Jackson et al. 1993), was recently identified as an allele of mcm5 (Hardy et al. 1997). The molecular mechanism of the bypass phenotype of bob1 has yet to be determined. Given that the requirement of Mcm2 phosphorylation by Cdc7–Dbf4 is apparently bypassed in the mcm5–bob1 mutant, we envision that the mcm5–bob1 mutation may induce a conformational change in the Mcm complex similar to that induced by the phosphorylation of Cdc7–Dbf4 (Fig. 7). When such a conformational change in the Mcm complex is attained via the mcm5–bob1 mutation, without the phosphorylation by Cdc7–Dbf4, the essential role of Cdc7–Dbf4 in DNA replication initiation becomes dispensable.
Figure 7.
A model for the regulation of Mcm2 by the Cdc7–Dbf4 kinase in the initiation of DNA synthesis at replication origins. Cdc7–Dbf4 interacts with and phosphorylates Mcm2, and likely several other members of the Mcm2–7 proteins. We propose that during the G1-to-S-phase transition, Cdc7–Dbf4 interacts with Mcm2, which is in a complex with other members of the Mcm2–7 family. Phosphorylation of Mcm2 and other subunits in this complex drives the complex into an active conformation that can also be achieved by a single base change mutation in MCM5 (Hardy et al. 1997). This active conformation signals subsequent steps in the replication initiation process.
Cell cycle-regulated protein kinases play major roles in restricting DNA synthesis to once per cell cycle in eukaryotic cells. In addition to Cdc7–Dbf4, which plays a critical role in the entry of S phase, the cyclin-dependent kinase Cdc28, together with various B type cyclins, controls the assembly as well as the activation of the pre-replication complex, and prevents the reassembly of pre-RC before the completion of mitosis (Nasmyth 1996; Stillman 1996). Determining the specific enzyme–substrate relationship between the various protein kinases and replication proteins is undoubtedly a critical step toward unveiling the mechanisms that control DNA replication.
Our work presents the first example of identifying a family of replication proteins as the substrates of Cdc7–Dbf4, a kinase that plays a key role in regulating DNA replication. Cdc7–Dbf4 may also regulate the function of other replication proteins. Physical associations between Cdc7–Dbf4 and ORC have been reported (Dowell et al. 1994; Hardy and Pautz 1996). It is obvious that Cdc7–Dbf4 is not the only cell cycle-specific protein kinase that regulates replication proteins by phosphorylation. Multiple distinct isoforms of the Mcm proteins detected at other stages of the cell cycle suggest that Mcm proteins are modified by other protein kinases or phosphatases (Young and Tye 1997). Recent results suggest that Mcm3 is the substrate of Cdc28–cyclin B kinases (Young et al. 1997). Recent studies suggest that XMcm4 is phosphorylated by Cdc28–cyclin B kinases (Hendrickson et al. 1996). Cdc28–cyclin B kinases also interact with ORC (Leatherwood et al. 1996), and phosphorylate Cdc6 in vitro (Elsasser et al. 1996). These observations put together an emerging picture that the control of DNA replication by cell cycle regulated protein kinases is accomplished through coordinated, multiple phosphomodifications on many of the replication proteins.
Materials and methods
Second-site suppressor screening
8534-M2 (mcm2-1) cells were grown to log phase at 30°C, plated on YEPD agar plates, and incubated at 38.5°C for 3 days. From a total of 2.5 × 108 cells in four independent pools, we obtained ∼2500 temperature-sensitive suppressors. These temperature-sensitive suppressors were replica-plated and incubated at 14°C for 7 days. Colonies that grew at 38.5°C but not at 14°C were further analyzed. Six temperature-sensitive suppressors that are also conditionally lethal at 14°C (cold-sensitive) were isolated. The mts2-1::DBF4 strain was generated by integrating MluI-digested pRS306. DBF4 plasmid at the dbf4 locus in the mts2-1 strain.
Construction of double mutants
Plasmid pMcm2-1.s (Yan et al. 1991), which contains part of the mcm2-1 mutant allele and URA3 gene, was digested with BglII and integrated into dbf4-1, dbf4-3, dbf4-4, and dbf4-5 mutant strains, respectively, at the MCM2 locus. The integrated strains were crossed with M46Y1333B (α ura3 his3 ade2–101 mcm2-1) or M46Y1332A (α ura3 his3 ade2–101 mcm2-1), respectively. The resulting diploid strains were then grown on 5′-FOA plates to yield diploids that are either heterozygous or homozygous for mcm2-1. The mcm2-1 homozygotes were selected by their temperature-sensitive growth phenotype at 38.5°C. Sporulation and tetrad dissection were carried out following standard procedures (Sherman and Wakem 1991). Spores were germinated at 30°C on YEPD plates.
Flow cytometry
Yeast cells were grown in YEPD medium at 30°C until early log phase (OD600 ∼ 1, 107 cells/ml). An aliquot of the culture was harvested, and the remaining culture was shifted to 38.5°C for 3 hr prior to harvest. Cells were fixed in 70% ethanol, treated with 0.5 mg/ml of RNaseA at 37°C for 1.5 hr, then stained with 50 μg/ml of propidium iodide. The analysis was performed on EPICS Profile (Hutter and Eipel 1978).
Plasmid construction
pRS306.DBF4: A BamHI–HindIII fragment containing the DBF4 gene was isolated from pKK823 (Kitada et al. 1992), and cloned into pRS306 at the BamHI–HindIII sites. pBTM116.MCM2-1: The mcm2-1 mutant allele was PCR amplified as a BamHI fragment by use of pHY1 (Yan et al. 1991) as template, and cloned into pBTM116 at the BamHI site. pEG.MCM2-1: The BamHI fragment containing the mcm2-1 mutant allele was isolated from pBTM.MCM2-1 and cloned into pEG(KT) at the BamHI site.
Two-dimensional DNA gel analysis
Yeast cells were grown in YEPD medium at 30°C until early log phase (OD600∼0.9). An aliquot of the culture was harvested, and the remaining culture was shifted to 38.5°C for 3 hr prior to harvest. Genomic DNA was prepared according to Merchant et al. (1997). Replication intermediates were enriched by BND cellulose column chromatography as described (Dijkwel et al. 1991). Two-dimensional DNA gel analysis (Brewer and Fangman 1987) was carried out as described (Merchant et al. 1997). For the analysis of ORI1, genomic DNA was digested with NcoI. For the analysis of ORI121, genomic DNA was digested with EcoRI–BamHI. The blots were hybridized with probes labeled with [α-32P]dATP by random priming.
Two-hybrid analysis and affinity chromatography
Two-hybrid analysis and the β-galactosidase activity assay were performed as described (Lei et al. 1996). Glutathione S-transferase (GST) and GST–Mcm2 fusion proteins expressed in yeast cells were prepared and conjugated to glutathione–Sepharose 4B (Pharmacia) as described (Lei et al. 1996). The GST or GST–Mcm2 conjugated resin (0.1 ml) was then recovered by centrifugation and incubated at 4°C for 2 hr with soluble proteins extracted from 100 ml of log phase BJ2168 cells or BJ2168 cells containing pKH125. The resin was washed with 5 × 1 ml of buffer B (Lei et al. 1996) containing 0.1 m NaCl. Specifically bound proteins were then eluted from the resin with 2 × 0.1 ml of buffer B containing 0.5 M NaCl. Finally, GST and GST–Mcm2 were released from the resin with 0.2 ml buffer B containing 10 mm glutathione. The relative concentrations of the GST and GST–Mcm2 fusion proteins were compared in Coomassie brilliant blue-stained SDS–polyacrylamide gels. The Western blots were probed with polyclonal anti-LexA or anti-Cdc7 antisera, respectively.
In vitro kinase assay
GST-Mcm fusion proteins were purified as previously described (Lei et al. 1996). To obtain GST-free Mcm2 protein, GST–Mcm2-coupled glutathione–Sepharose resin was treated with thrombin (50 μg/ml) for 2 hr at 23°C and the supernatant was recovered. The purified proteins were heated at 65°C for 10 min to inactivate the endogenous kinase activity before the kinase assay. For a 20 μl reaction, 5 pmoles of substrate proteins were mixed with 10 μl of the reaction buffer (25 mm HEPES at pH 7.4, 10 mm MgCl2, 100 mm NaCl, 1 mm DTT, 20 mm ATP), 20 μCi of [γ-32P]ATP and the Cdc7–Dbf4 protein kinase. The reaction was incubated at 30°C for 10 min, then stopped by adding SDS sample buffer and incubated in a boiling water bath for 3 min. Samples were then applied to an 8% SDS–polyacrylamide gel. The gels were stained with Coomassie brilliant blue and dried on filter paper for autoradiograph.
Two-dimensional protein gel analysis
dbf4-1 cells growing at 30°C in YEPD media were arrested by adding α-factor to a final concentration of 10 μg/ml. The α-factor-arrested cells were washed three times with YEPD and divided into two aliquots. One aliquot of the cells was resuspended in YEPD and shifted to 37°C for 3 hr. The other aliquot was resuspended in YEPD containing 0.2 m HU and incubated at 30°C for 3 hr. Cells were then harvested by centrifugation. Pelleted cells were resuspended in 500 μl breaking buffer plus protease inhibitors and phosphatase inhibitors (Young and Tye 1997), then disrupted by vortexing for six 1-min cycles, at 4°C, in the presence of glass beads. Two-dimensional protein gel analysis was performed as described (Young and Tye 1997) with one modification: the ampholyte mixture for first dimension isoelectrofocusing was 3 parts pH 5–7 ampholytes (Bio-Rad) to 1 part pH 4–6 ampholytes (Pharmacia).
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant GM34190 to BKT, and by grant-in-aid for International Scientific Research (Joint Research) of the Ministry of Education, Science, Sports and Culture of Japan to A.S. We thank Dr. Eric Alani and Dr. Tim Huffaker for critical reading of this manuscript. Y.K. was a recipient of a postdoctoral fellowship from the Human Frontier Science Program. M.R.Y. was a recipient of a postdoctoral fellowship from NIH.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bt16@cornell.edu; FAX (607) 258-2428.
References
- Adams AEM, Botstein D, Drubin DG. A yeast actin-binding protein is encoded by SAC6, a gene found by suppression of an actin mutation. Nature. 1989;243:231–233. doi: 10.1126/science.2643162. [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Axelrod A, Rine J. A role for CDC7 in repression of transcription at the silent mating-type locus HMR in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1080–1091. doi: 10.1128/mcb.11.2.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Chong J, Mahbubani HM, Khoo CY, Blow JJ. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature. 1995;375:418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JFX. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 Protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Coue M, Kearsey SE, Mechali M. Chromatin binding, nuclear localization and phosphorylation of Xenopus cdc21 are cell-cycle dependent and associated with the control of initiation of DNA replication. EMBO J. 1996;15:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Diffley JFX. Once and only once upon a time: Specifying and regulating origins of DNA replication in eukaryotic cells. Genes & Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- Diffley JFX, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Dijkwel PA, Vaughn JP, Hamlin JL. Mapping of replication initiation sites in mammalian genomes by two-dimensional gel analysis: Stabilization and enrichment of replication intermediates by isolation on the nuclear matrix. Mol Cell Biol. 1991;11:3850–3859. doi: 10.1128/mcb.11.8.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JFX. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell SJ, Romanoski P, Diffley JFX. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science. 1994;265:1243–1246. doi: 10.1126/science.8066465. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Lou F, Wang B, Campbell J, Jong A. Interaction between yeast Cdc6 protein and B-type cyclin/Cdc28 kinases. Mol Biol Cell. 1996;7:1723–1735. doi: 10.1091/mbc.7.11.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song OK. A novel genetic system to detect protein-protein interaction. Proc Natl Acad Sci. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Gibson SI. “Analysis of Mcm3, a minichromosome maintenance mutant of yeast with a cell division cycle arrest phenotype,” Ph.D. thesis. Ithaca, NY: Cornell University; 1989. [Google Scholar]
- Hardy CFJ, Pautz A. A novel role for Cdc5p in DNA replication. Mol Cell Biol. 1996;16:6775–6782. doi: 10.1128/mcb.16.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CFJ, Dryga O, Pahl PMB, Sclafani RA. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol. 1976;104:803–817. doi: 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- Hendrickson M, Madine M, Dalton S, Gautier J. Phosphorylation of MCM4 by cdc2 proteins kinase inhibits the activity of the minichromosome maintenance complex. Proc Natl Acad Sci. 1996;93:12223–12228. doi: 10.1073/pnas.93.22.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy KM, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes & Dev. 1991;5:958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- Hereford L, Hartwell L. Sequential gene function in the initiation of S. cerevisiae DNA synthesis. J Mol Biol. 1974;84:445–461. doi: 10.1016/0022-2836(74)90451-3. [DOI] [PubMed] [Google Scholar]
- Hollingsworth RE, Sclafani R. Molecular genetic studies of the Cdc7 protein kinase and induced mutagenesis in yeast. Genetics. 1992;132:53–62. doi: 10.1093/genetics/132.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter KJ, Eipel HE. Flow cytometric determinations of cellular substances in algae, bacteria, molds and yeasts. J Microbiol Ser. 1978;44:269–282. doi: 10.1007/BF00394305. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Pahl PMB, Harrison K, Rosamond J, Sclafani RA. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol Cell Biol. 1993;13:2899–2908. doi: 10.1128/mcb.13.5.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LH, Thomas APM. The isolation of new DNA synthesis mutants in the yeast Saccharomyces cerevisiae. Mol & Gen Genet. 1982;186:445–448. doi: 10.1007/BF00729466. [DOI] [PubMed] [Google Scholar]
- Kearsey SE, Maiorano D, Holmes EC, Todorov I. The role of MCM proteins in the control of genome duplication. BioEssays. 1995;18:183–189. doi: 10.1002/bies.950180305. [DOI] [PubMed] [Google Scholar]
- Kitada K, Johnston L, Sugino T, Sugino A. Temperature-sensitive cdc7 mutations of Saccharomyces cerevisiae are suppressed by the DBF4 gene, which is required for the G1/S cell cycle transition. Genetics. 1992;131:21–29. doi: 10.1093/genetics/131.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada K, Johnson AL, Johnston LH, Sugino A. A multipcopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is identified as CDC5. Mol Cell Biol. 1993;13:4445–4457. doi: 10.1128/mcb.13.7.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S-i, Takisawa H, Nojima H. Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell. 1995;81:601–610. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- Leatherwood J, Lopez-Girona A, Russell P. Interaction of Cdc2 and Cdc18 with a fission yeast ORC2-like protein. Nature. 1996;379:360–363. doi: 10.1038/379360a0. [DOI] [PubMed] [Google Scholar]
- Lei M, Kawasaki Y, Tye BK. Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in S. cerevisiae. Mol Cell Biol. 1996;16:5081–5090. doi: 10.1128/mcb.16.9.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madine MA, Khoo C-Y, Mills AD, Laskey RA. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature. 1995;375:421–424. doi: 10.1038/375421a0. [DOI] [PubMed] [Google Scholar]
- Maine GT, Sinha P, Tye BK. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics. 1984;106:365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Miyake T, Arai K. Hsk1(+), a Sachizosaccharomyces pombe gene related to Saccharomyces cerevisiae Cdc7, is required for chromosomal replication. EMBO J. 1995;14:3094–3104. doi: 10.1002/j.1460-2075.1995.tb07312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant AM, Kawasaki Y, Chen Y, Lei M, Tye BK. A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in S. cerevisiae. Mol Cell Biol. 1997;17:3261–3271. doi: 10.1128/mcb.17.6.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Viewpoints: Putting the cell cycle in order. Science. 1996;274:1643–1645. doi: 10.1126/science.274.5293.1643. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Hartwell LH. The Saccharomyces cerevisiae cell cycle. In: Strathern JN, Jones EW, Broach JR, editors. the molecular biology of the yeast Saccharomycesi. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1981. pp. 97–142. [Google Scholar]
- Sato N, Arai K-i, Masai H. Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: In vitro phosphorylation of MCM subunits by a putative human homologue of Cdc7. EMBO J. 1997;16:4340–4351. doi: 10.1093/emboj/16.14.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D, Byers B. Meiotic effects of DNA-defective cell division cycle mutations of Saccharomyces cerevisiae. Chromosoma. 1978;70:109–130. doi: 10.1007/BF00292220. [DOI] [PubMed] [Google Scholar]
- Sherman F, Wakem P. Mapping yeast genes. Meth Enzymol. 1991;194:38–57. doi: 10.1016/0076-6879(91)94006-x. [DOI] [PubMed] [Google Scholar]
- Simchen G. Are mitotic functions required in meiosis? Genetics. 1974;114:753–767. doi: 10.1093/genetics/76.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1663. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Thommes P, Kubota Y, Takisawa H, Blow JJ. The RLF-M component of the replication licensing system forms complexes containing all six MCM/P1 polypeptides. EMBO J. 1997;16:3312–3319. doi: 10.1093/emboj/16.11.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov IT, Attaran A, Kearsey SE. BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995;129:1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman JE, Follette PJ, O’Farrell PH, Rubin GM. Cell proliferation and DNA replication defects in a Drosophila MCM2 mutant. Genes & Dev. 1995;9:1709–1715. doi: 10.1101/gad.9.14.1709. [DOI] [PubMed] [Google Scholar]
- Tye BK. The Mcm2-3-5 proteins: Are they replication licensing factors? Trends Cell Biol. 1994;4:160–166. doi: 10.1016/0962-8924(94)90200-3. [DOI] [PubMed] [Google Scholar]
- Yan H, Gibson S, Tye BK. Mcm2 and Mcm3, two proteins important for ARS activity, are related in structure and function. Genes & Dev. 1991;5:944–957. doi: 10.1101/gad.5.6.944. [DOI] [PubMed] [Google Scholar]
- Yan H, Merchant AM, Tye BK. Cell cycle-regulated nuclear localization of MCM2 and MCM3, which are required for the initiation of DNA synthesis at chromosomal replication origins in yeast. Genes & Dev. 1993;7:2149–2160. doi: 10.1101/gad.7.11.2149. [DOI] [PubMed] [Google Scholar]
- Young M, Tye BK. Mcm2 and Mcm3 are constitutive nuclear proteins which exhibit distinct isoforms and bind chromatin during specific cell cycle stages of S. cerevisiae. Mol Biol Cell. 1997;8:1587–1601. doi: 10.1091/mbc.8.8.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, M., K. Suzuki, H. Yan, S. Gibson, and B.K. Tye. 1997. Nuclear accumulation of S. cerevisiae Mcm3 is dependent on its nuclear localization sequence. Genes Cells (in press). [DOI] [PubMed]