Abstract
Telomere proteins protect the chromosomal terminus from nucleolytic degradation and end-to-end fusion, and they may contribute to telomere length control and the regulation of telomerase. The current studies investigate the effect of Oxytricha single-stranded telomere DNA-binding protein subunits α and β on telomerase elongation of telomeric DNA. A native agarose gel system was used to evaluate telomere DNA-binding protein complex composition, and the ability of telomerase to use these complexes as substrates was characterized. Efficient elongation occurred in the presence of the α subunit. Moreover, the α–DNA cross-linked complex was a substrate for telomerase. At higher α concentrations, two α subunits bound to the 16-nucleotide single-stranded DNA substrate and rendered it inaccessible to telomerase. The formation of this α · DNA · α complex may contribute to regulation of telomere length. The α · β · DNA ternary complex was not a substrate for telomerase. Even when telomerase was prebound to telomeric DNA, the addition of α and β inhibited elongation, suggesting that these telomere protein subunits have a greater affinity for the DNA and are able to displace telomerase. In addition, the ternary complex was not a substrate for terminal deoxynucleotidyltransferase. We conclude that the telomere protein inhibits telomerase by rendering the telomeric DNA inaccessible, thereby helping to maintain telomere length.
Keywords: Telomere, telomerase, regulation, telomere DNA-binding proteins, Oxytricha
Telomeres, the protein–DNA complexes at the termini of eukaryotic chromosomes, are vital for the preservation and complete replication of the genome (for reviews, see Blackburn 1991; Zakian 1995; Greider 1996). The past few years have seen explosive growth in our knowledge of telomere DNA-binding proteins and of telomerase, the enzyme that synthesizes telomeric DNA (for review, see Lingner and Cech 1998). Much less is known about the interaction between these two macromolecular complexes. The mechanisms of telomere–telomerase interaction provide the subject of the investigations presented here.
Typically telomeric DNA consists of tandem repeats of a short sequence with the guanine-rich strand oriented 5′ → 3′ toward the chromosome terminus (e.g., Oxytricha telomeres consist of T4G4 repeats; Klobutcher et al. 1981). Although telomere length varies from species to species, the protrusion of the G-rich strand as a single-stranded overhang is a feature conserved among ciliated protozoa (for review, see Zakian 1989, 1995), yeast (Wellinger et al. 1993, 1996; Zakian 1996), and mammals (Makarov et al. 1997; McElligott and Wellinger 1997; Wright et al. 1997). Thus, despite differences in telomeric sequence and length between species, there may be similar functional mechanisms in telomere maintenance.
In addition to the simple DNA repeats, a number of proteins play integral roles in telomere structure and function. Telomere proteins are of two types: those that bind the double-stranded portion of the telomere and those that bind the single-stranded telomeric overhang (for reviews, see Fang and Cech 1995a; Greider 1996; Brun et al. 1997). The best characterized single-stranded telomeric DNA-binding protein is that of the ciliated protozoan Oxytricha nova; the crystal structure of its complex with telomeric DNA has been solved recently (M. Horvath, V. Schweiker, J. Ruggles, J.M. Bevilacqua, and S.C. Schultz, unpubl.). The protein is a heterodimer consisting of a 56-kD α subunit and a 41-kD β subunit (Gottschling and Zakian 1986; Price and Cech 1987). Initial research revealed that the α subunit forms a specific complex with telomeric DNA, whereas the β subunit does not bind to DNA with sequence specificity by itself. The α and β subunits together bind tenaciously to telomeric DNA to form a stable ternary complex (Gray et al. 1991).
The β subunit is capable of chaperoning the formation of G-quartet structures (Fang and Cech 1993a). G-quartets form monovalent cation-induced tetraplex DNA structures (Williamson 1994), which previously were shown to be poor substrates for telomerase (Zahler et al. 1991). The highly charged carboxyl domain of the β subunit mediates G-quartet structure formation in vitro (Fang and Cech, 1993a). The α · β heterodimer does not form on DNA folded into G-quartets, but rather requires an unfolded telomeric DNA substrate (Raghuraman and Cech 1990).
One of the functions of the telomere is to act as a substrate for telomerase, the ribonucleoprotein that catalyzes the synthesis of telomeric DNA repeats (for reviews, see Blackburn 1992; Greider 1996; Lingner and Cech 1998). The regulation of telomere length and telomerase activity appear pivotal for cellular life span (Lundblad and Szostak 1989; Harley and Villeponteau 1995; Bodnar et al. 1998). Telomerase and single-stranded telomere DNA-binding proteins share substrate specificity (the telomeric DNA overhang). Both localize to the Oxytricha replication band during S-phase (Fang and Cech 1995b), and the telomere DNA-binding protein localizes behind telomerase in these analyses. Thus, it is thought that the telomere protein may bind to newly synthesized telomeres for their protection. A similar situation pertains in Euplotes crassus, with the interesting additional feature of a replication-specific version of the telomere protein (Skopp et al. 1996).
The effect of the Oxytricha telomere DNA-binding proteins on telomerase activity was studied initially by Shippen et al. (1994). They reported that the ternary complex was a substrate for telomerase, although not as good a substrate as DNA free in solution. Because these experiments used native proteins that cannot be isolated in large quantities, the integrity of the reconstituted telomere protein–DNA complexes was difficult to evaluate. Moreover, the α · DNA complex, although initially characterized by nitrocellulose filter binding and dimethylsulfate (DMS) protection (Gray et al. 1991; Fang et al. 1993), had not been visualized as a discrete complex with the biological DNA substrate by gel electrophoresis.
The present studies used recombinant Oxytricha telomere DNA-binding proteins in conjunction with native agarose gel electrophoresis (J.M. Bevilacqua and S.C.Schultz, unpubl.) to monitor telomere protein complex formation. The effect of telomere protein subunits on the synthesis of telomeric repeats by telomerase was then analyzed. Telomere DNA-binding protein subunits were found to inhibit telomerase by altering the telomeric DNA substrate accessibility and, therefore, may collaborate with telomerase to maintain telomere length. A model of telomere protein involvement in telomere synthesis and length regulation is presented.
Results
Telomere protein–DNA complex formation
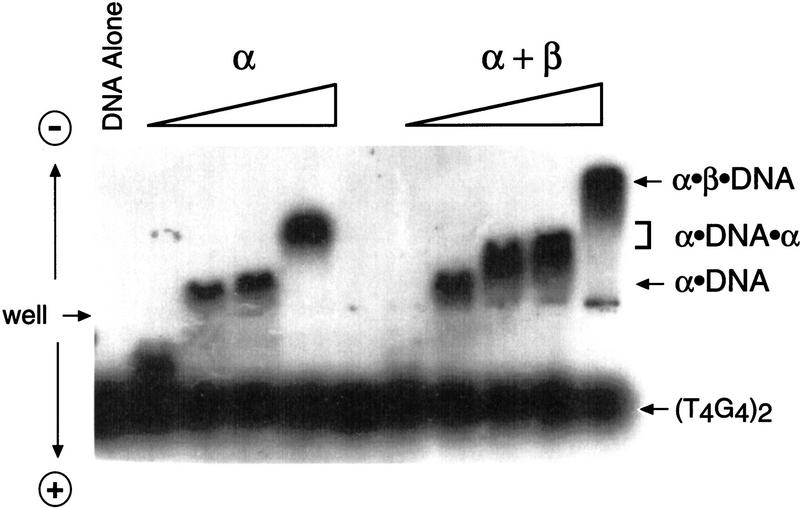
The α and β subunits form a stable ternary complex with telomeric DNA, which has been observed as a discrete band by polyacrylamide gel shift analysis (Fang and Cech 1993b; Fang et al. 1993). In contrast, the less stable α · DNA complex forms smears on polyacrylamide gels and, therefore, has been characterized by nitrocellulose filter binding and DMS protection (Gray et al. 1991; Fang et al. 1993). Many of these previous studies were performed with single-stranded oligonucleotides much larger than the biological substrate to counterbalance the high positive charge inherent to the telomere proteins. To determine which telomere protein · DNA complexes were present and extended by telomerase, a novel agarose gel system was used (J.M. Bevilacqua and S.C.Schultz, unpubl.). Samples are loaded in the center of a horizontal agarose gel, and during electrophoresis, the positively charged DNA · protein complexes migrate in one direction, whereas the negatively charged DNA migrates in the opposite direction. This methodology permits the use of the biological length substrate [i.e., the single-stranded 16-mer (T4G4)2].
Gel-shift analysis resulted in a distinct banding pattern with the addition of increasing concentrations of protein. The addition of the α subunit resulted in formation of α · DNA and subsequently α · DNA · α complexes (Fig. 1), as expected from the previous conclusion that two α subunits can bind a two-repeat DNA in a noncooperative fashion (Gray et al. 1991). The transition between the α · DNA and the α · DNA · α complex began to occur beyond ∼250 nm. When both α and β were added, an additional complex of higher positive charge/mass ratio was formed. The increased mobility of this complex is consistent with the presence of the β subunit, which is highly basic (Hicke et al. 1990). Therefore, this complex is identified as the 1:1:1 complex of α·β·DNA characterized previously (Fang and Cech 1993b).
Figure 1.
Telomere DNA-binding protein complex formation analyzed by agarose gel electrophoresis. The DNA substrate (T4G4)2 was 5′-radiolabeled and was kept at a constant concentration throughout the binding assay (10 nm, submolar to the protein concentration). The α subunit was present at 25 nm, 100 nm, 1 μm, and 2.5 μm; protein concentrations for formation of the α · β · DNA ternary complex were 5 nm α and 5 nm β; 25 nm α and 10 nm β; 250 nm α and 25 nm β; 1 μm α and 50 nm β; 2.5 μm α and 100 nm β. The data are representative of five experiments.
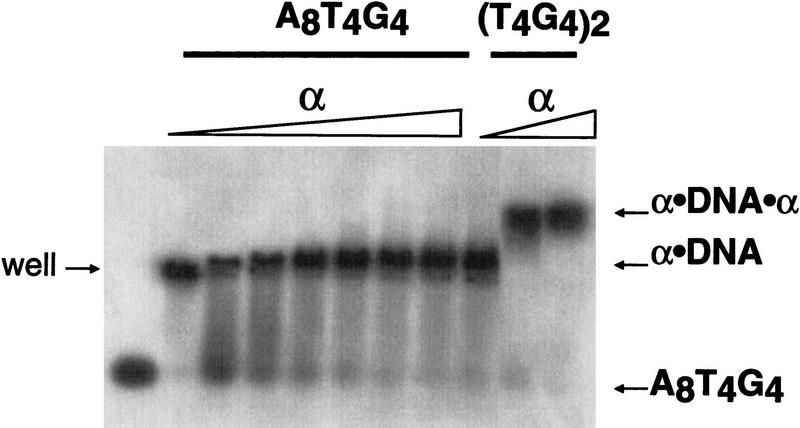
Further evidence for the identification of the α · DNA · α complex was obtained by analyzing binding of the α subunit to oligonucleotides containing one and two telomeric repeats (Fig. 2). Previous DMS protection studies had defined the α subunit binding site as a single T4G4 repeat (Gray et al. 1991). Consistent with this finding, the putative α · DNA · α complex was observed only with the oligonucleotide containing two telomeric repeats.
Figure 2.
An α subunit binds to each T4G4 repeat forming an α · DNA · α complex on the native telomeric sequence. The oligonucleotide A8T4G4, which has only one telomeric-binding register, was used to analyze the binding of the α subunit. The oligonucleotide was present at a constant concentration of 1 nm. The α subunit was present at 10, 25, 50, 100, and 500 nm, and at 1 and 2.5 μm. The three right lanes are binding analyses with the natural sequence oligonucleotide (T4G4)2 at α concentrations of 0.25, 1, and 5 μm.
Agarose gel electrophoresis does not trap the complexes but allows dissociation within the gel matrix. Therefore, the apparent Kd values in the present study may be greater than the actual Kd values. At certain concentrations, kinetic transitions between complexes result in intermediate electrophoretic mobilities. However, the complexes that are stable in these gels will be even more stable in solution, where dissociated DNA cannot electrophorese away from the protein.
From these studies, the complexes present at specific concentrations were identified, and the ability of telomerase to exploit these complexes as substrates could be evaluated.
Telomere protein subunits modulate telomerase activity
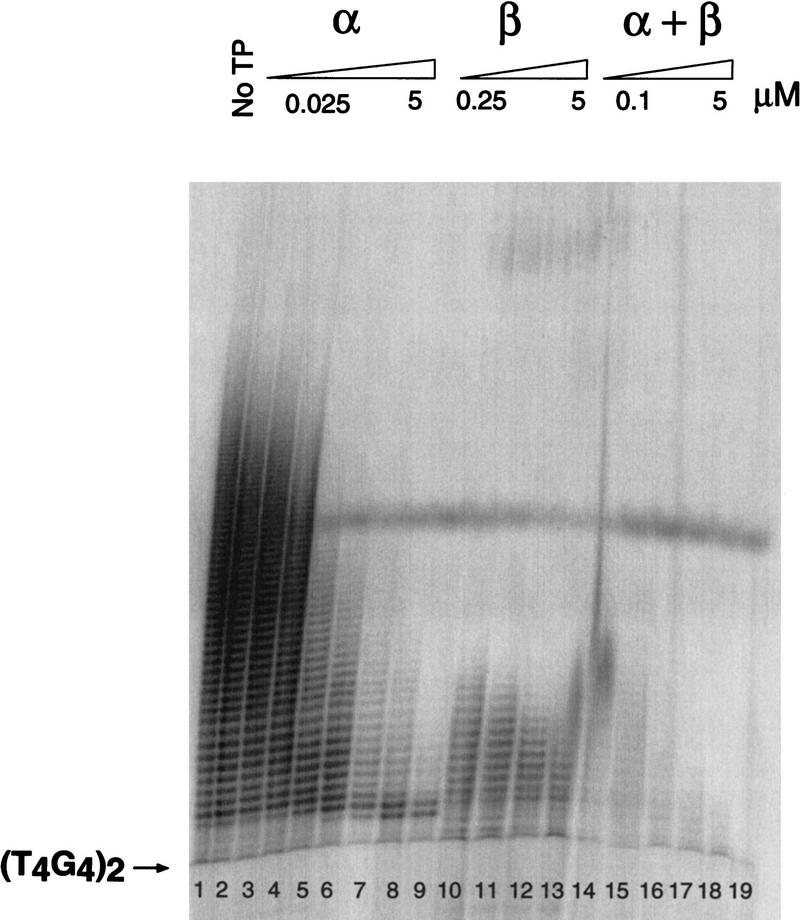
The ability of telomerase to use telomeric DNA complexed with telomere protein subunits was analyzed. O. nova telomere proteins were prebound to the DNA substrate, followed by the addition of telomerase-containing Oxytricha trifallax nuclear extract. (The justification for use of the nuclear extract from a different species of Oxytricha is given in a subsequent section.) The synthesis of telomeric repeats was monitored by denaturing gel electrophoresis and quantified relative to an internal normalization marker (Fig. 3).
Figure 3.
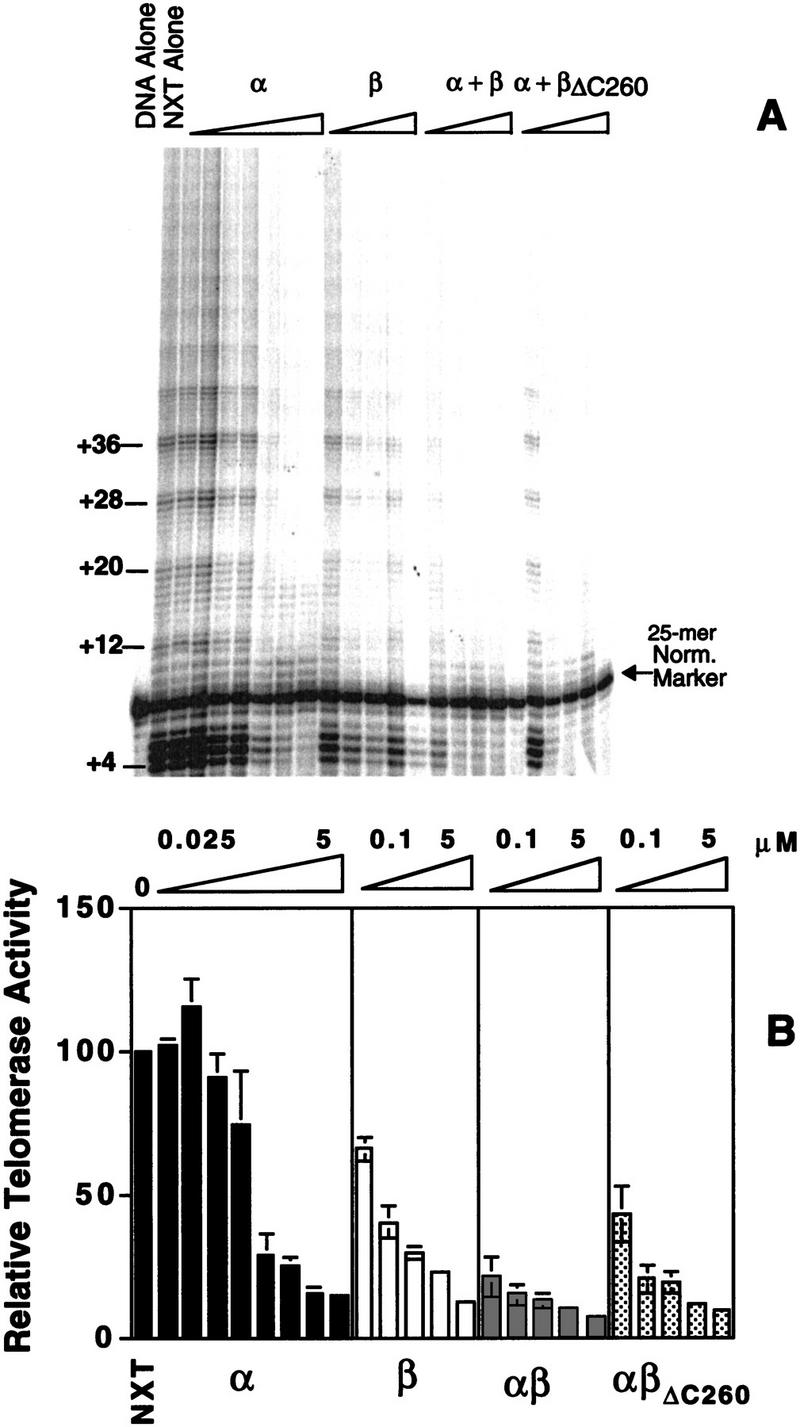
Telomere protein modulation of telomerase elongation of telomeric DNA. (A) A telomerase activity assay was performed with 5′-radiolabeled DNA substrate (T4G4)2 in the presence of the telomere protein subunits. α, β, or α + β were preincubated with DNA, followed by the addition of telomerase and nucleotides. The β subunit was present at the following concentrations: 0.025, 0.050, 0.1, 0.25, 0.5, 1, 2.5, and 5 μm. The α subunit was present at 0.1, 0.25, 0.5, 1, and 5 μm. In studies with the α + β and α + βΔC260, both subunits were present at 0.1, 0.25, 0.5, 1, and 5 μm. A 25-mer oligonucleotide was added after the reaction was stopped but before ethanol precipitation, serving as a size marker and for internal normalization during quantification. (B) The bar graph quantifies the effects of O. trifallax telomere protein on telomerase activity. βΔC260 denotes a truncated version of the β subunit (the lysine-rich carboxyl terminus is deleted). Standard deviations for three independent experiments are depicted by the error bars.
In the absence of telomere protein, telomerase added multiple repeats to the (T4G4)2 primer. Given the large excess of unlabeled primer, this indicates that the DNA substrate bound initially remains bound through additional rounds of extension (i.e., the extension is processive). Telomerase was also able to extend the DNA in the presence of up to 250 nm α subunit. In this concentration range the α · DNA complex was formed, as verified by agarose gel electrophoresis. At higher concentrations of the α subunit, where the α · DNA · α complex was the predominant species, extension was greatly diminished.
In the presence of the β subunit alone the level of telomerase activity was decreased. Telomerase activity is known to be attenuated in the presence of G-quartet structures (Zahler et al. 1991), and the β subunit carboxy-terminal domain is known to mediate the formation of these structures (Fang and Cech 1993a). Consistent with this interpretation, when a truncated β subunit (βΔC260) with the carboxy-terminal domain deleted is used, the effect of the protein on extension is minimal (see last section of Results).
The ability of telomerase to elongate telomeric DNA is attenuated and eliminated rapidly with increasing concentrations of both the α and β subunits (Fig. 3). When a ternary complex is formed in the presence of the α subunit and a βΔC260 subunit, telomerase activity is also abolished, but the reduction in activity occurs at higher protein concentrations. This result suggests that the carboxyl terminus of the β subunit may play a role in ternary complex stabilization or exclusion of telomerase.
The modulation of telomerase activity is telomere protein specific
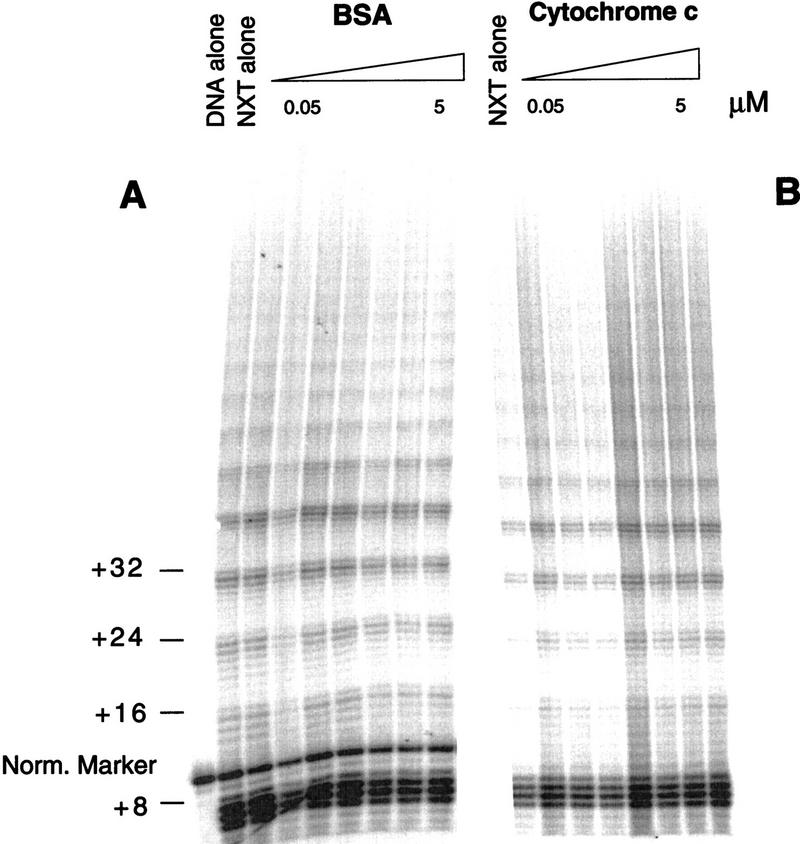
To test whether the influence on telomerase activity was specific to telomere DNA-binding protein subunits, a titration, analogous to that performed with the telomere proteins (Fig. 3), was completed with bovine serum albumin (BSA) (Fig. 4A). Even at the highest concentration (5 μm) of BSA, there was no effect on telomerase activity. Because the telomere protein subunits are highly basic, the identical experiment was performed with cytochrome c, a very basic protein that binds nonspecifically to single- and double-stranded DNA (Fig. 4B). Again, there was no effect on telomerase activity.
Figure 4.
The effects on telomerase activity are telomere protein specific. Telomerase elongation was monitored in the presence of BSA (A) and the more basic cytochrome c (B). Both were preincubated with the DNA substrate (T4G4)2 at the following concentrations: 0.5, 0.1, 0.25, 0.5, 1, 2.5, and 5 μm. In the BSA experiment, a 25-mer 5′-radiolabeled oligonucleotide was added as a size marker and internal normalization standard.
Telomere protein displaces telomerase bound to telomeric DNA
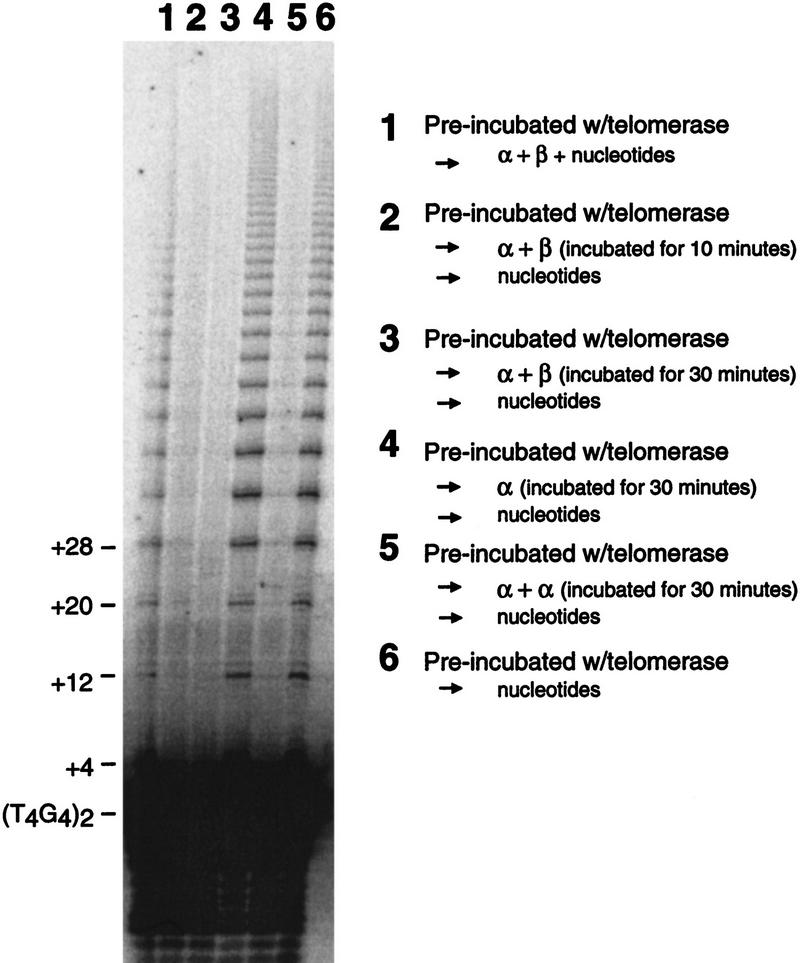
Because the telomere protein–DNA ternary complex is not elongated by telomerase, it was hypothesized that the telomere proteins may provide a mechanism of length regulation. In theory, this could be accomplished by the displacement of telomerase by the telomere protein after synthesis of a desired number of repeats. To investigate the ability of telomere protein subunits to displace telomerase in a specific fashion, telomerase was prebound to a 5′-radiolabeled DNA substrate in the absence of nucleotides. Telomere protein and nucleotides were then added to initiate the reaction. As shown in Figure 5, telomerase activity was detectable (lane 1), although efficiency of primer elongation was decreased substantially compared to extension in the absence of telomere DNA-binding protein (lane 6). When telomere protein was incubated with the telomerase–telomeric DNA complex before reaction initiation, elongation of the telomeric DNA in both the ternary complex and the α · DNA · α complex was eliminated (Fig. 5, lane 2,3,5). In contrast, the α · DNA complex permitted DNA elongation (Fig. 5, lane 4).
Figure 5.
Telomere proteins are able to displace telomerase at the telomeric terminus. Telomerase was preincubated with the 5′-end-labeled oligonucleotide substrate (T4G4)2 for 10 min before the addition of telomere-binding protein subunits and nucleotides or nucleotide alone. Lane description can be found within the figure. “Nucleotides” refers to dGTP and dTTP. The concentrations of the α + β subunit were 1 μm in lanes 1–3; the concentration of the α subunit was 100 nm in lane 4, and 2.5 μm in lane 5. The identification of the +4 extension product was based on a lighter exposure.
These results indicate that telomere protein subunits are able to displace telomerase. It is also possible that telomerase is dissociating from the telomeric DNA and allowing access by the telomere proteins. Then, if the binding proteins have a greater affinity for the telomeric DNA, they will not be displaced by telomerase. However, this cannot be the sole explanation as processive elongation (repetitive rounds of replication without dissociation) occurs in the absence of the telomere protein subunits (see Fig. 3; data not shown; for a detailed study with Euplotes aediculatus, see Hammond et al. 1997). The inhibition of ongoing processive elongation by two α subunits or α + β suggests that these proteins actively displace telomerase.
The α subunit as a length regulator
The observation that the α · DNA complex allows telomerase-mediated extension, whereas the presence of two α subunits inhibits telomerase, suggested a role for the α subunit in telomere length regulation. To investigate this possibility further, the oligonucleotide A8T4G4 was extended in the presence of increasing concentrations of the α subunit (Fig. 6). The rationale was that extension by one repeat (8 nucleotides) would enable two α subunits to be bound, at which point elongation would be terminated. Instead, at concentrations where the second α subunit is bound, it appeared that telomerase added only 4 nucleotides and then halted (Fig. 6). One possible explanation is that a second α subunit may not require a full telomeric repeat for binding (i.e., 4 nucleotides could support a second subunit; for example, 2 α subunits will bind to the 12-mer G4T4G4; J.M. Bevilacqua and S.C. Schultz, unpubl.). In any case, the α subunit provides reasonably precise length regulation in vitro, but it is not the same specificity that would be sufficient to explain the presence of the 16-nucleotide 3′ overhang in vivo.
Figure 6.
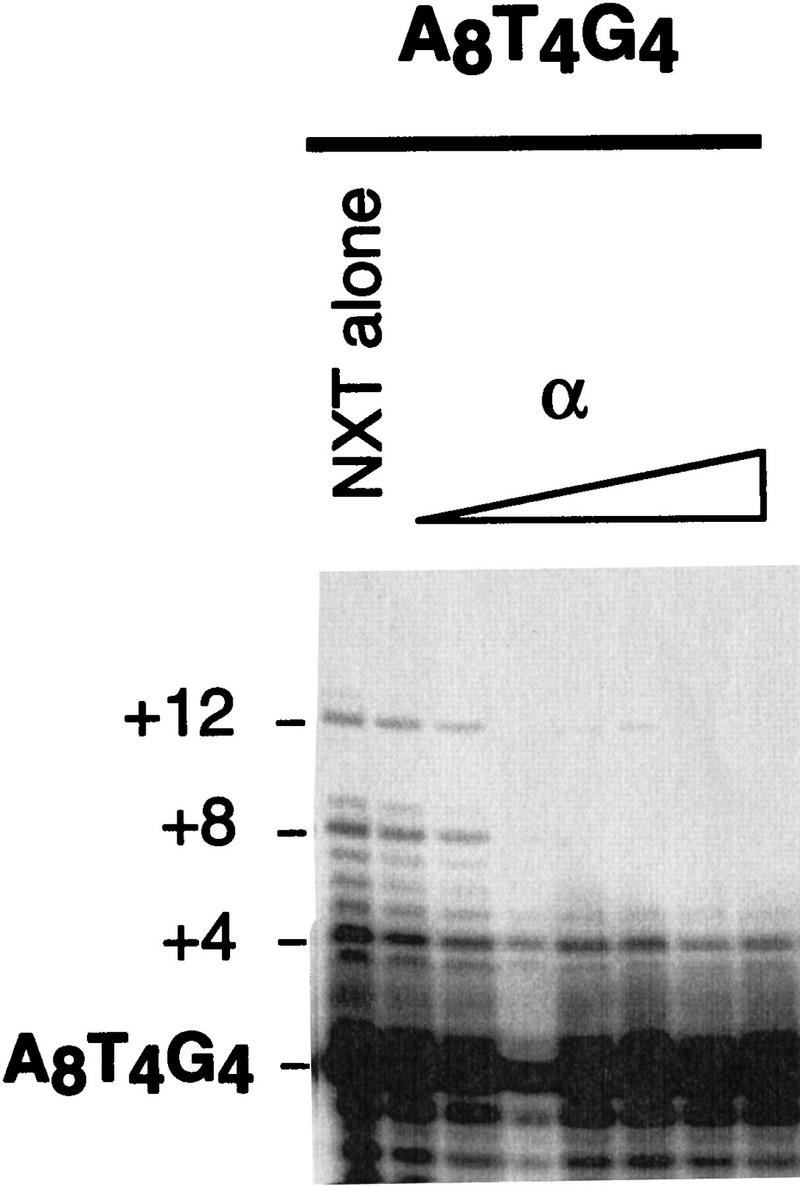
Extension of an oligonucleotide with one α binding register. 5′-Radiolabeled A8T4G4 was incubated with 0.05, 0.1, 0.25, 0.5, 1, 2.5, and 5 μm α subunit, then with nuclear extract. Lane 4 represents a poor ethanol precipitation.
Telomerase is able to extend a telomeric substrate bound by a single α subunit
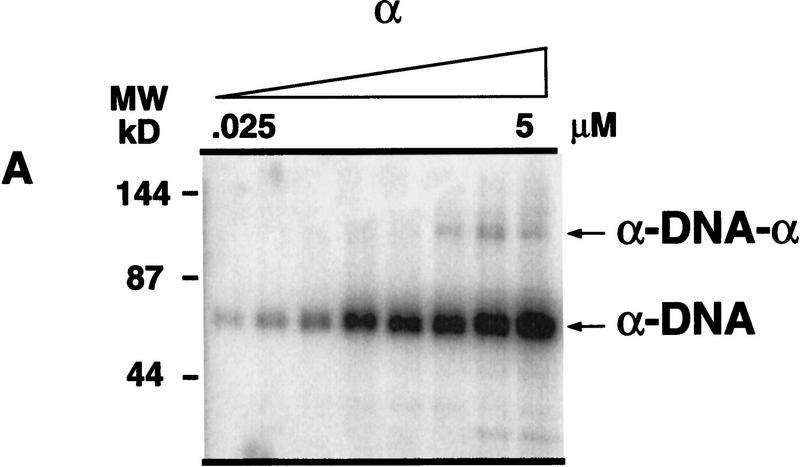
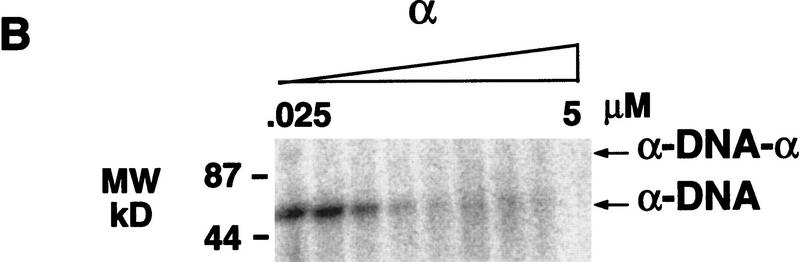
In the presence of the α subunit, at concentration ranges where the α · DNA complex was visualized by gel-shift analysis, telomerase was able to extend the telomeric DNA substrate (see Fig. 3A). However, it was unclear whether the α · DNA complex was extended, or whether the elongation was attributable to the inherent instability of the α · DNA complex. The α off-rate is very rapid (half-life < 1 min; Fang et al. 1993; data not shown), and thus, the extension of transiently “free” DNA is a possibility. To address this point, the α subunit was cross-linked to the telomeric DNA. To monitor cross-linking efficiency, the DNA substrate was 5′-radiolabeled and incubated with increasing concentrations of the α subunit. The samples were irradiated, and the formation of the α–DNA and α–DNA–α complexes (where dashes represent covalent linkage) was monitored by denaturing gel electrophoresis (Fig. 7A).
Figure 7.
Cross-linking of the α subunit to the DNA substrate (T4G4)2. (A) To monitor complex formation, the DNA was 5′-radiolabeled, incubated with the α subunit, and cross-linked. Cross-linked complexes were monitored by denaturing polyacrylamide gel electrophoresis. (B) To evaluate whether the α–DNA and α–DNA–α cross-linked complexes were extended by telomerase, increasing concentrations of the α subunit were cross-linked to nonlabeled DNA. 32P-Radiolabeled nucleotides and nuclear extract were added, and the label incorporation into the complexes was monitored by gel electrophoresis.
To determine whether telomerase is able to extend the cross-linked complexes, unlabeled DNA substrate (T4G4)2 was cross-linked to the α subunit; telomerase-containing extract and radiolabeled nucleotide were then added, and the complexes were analyzed by gel electrophoresis. Incorporation of labeled nucleotide into the α–DNA complex indicated that the complex was a substrate for telomerase, whereas there was no incorporation of nucleotide in the α–DNA–α complex (Fig. 7B). The inhibition of telomerase extension seen at higher α subunit concentrations in Figure 7B is explained by the formation of the α · DNA · α complexes, which are not substrates for telomerase. The α · DNA · α complexes are usually cross-linked only once, and therefore, run as α · DNA complexes in the denaturing gel. This explains the large abundance of the α–DNA complex in Figure 7A.
The telomere-binding protein subunits inhibit extension by another DNA polymerase
To evaluate whether the telomere protein subunits modulate telomerase activity specifically or whether binding of the subunits also affects other polymerases, the activity of calf thymus terminal deoxynucleotidyltransferase (TdT) was analyzed. Unlabeled (T4G4)2 was incubated with telomere protein subunits followed by the addition of enzyme and radiolabeled dTTP. Nucleotide addition by TdT was monitored (Fig. 8). Previously, Shippen et al. (1994) reported that the native α · β telomere protein complex inhibited DNA extension by TdT. Consistent with these results, the recombinant telomere protein subunits in the ternary complex inhibited TdT activity (Fig. 8). The α · DNA · α complex also abolished elongation; however, TdT was able to access its substrate when bound by a single α subunit (Fig. 8).
Figure 8.
Telomere proteins modulate TdT activity. Telomere protein subunits were prebound to DNA followed by the addition of TdT. (Lane 1) The activity of TdT in the absence of telomere protein (TP) subunits; (lanes 2–9) α subunit was present at 0.025, 0.05, 0.1, 0.25, 0.5, 1, 2.5, and 5 μm; (lanes 10–14) the β subunit was present at 0.1, 0.25, 0.5, 1, and 5 μm; and (lanes 15–19) the ternary complex was formed in the presence of 0.1, 0.25, 0.5, 1, and 5 μm of each subunit.
Because the telomere DNA-binding protein subunits inhibit both TdT and telomerase, these data suggest that the telomere proteins are inhibitory in a substrate-specific fashion rather than by distinct protein–protein interactions. This conclusion is further supported by the fact that the initial rate of telomerase elongation was not altered significantly when one α subunit was bound. Thus, we propose that the telomere protein subunits regulate telomerase in vitro by controlling the accessibility of the telomeric DNA substrate.
Justification for use of a heterologous system
Because of recent difficulties in extracting active telomerase from O. nova cells, telomerase-containing nuclear extract was prepared from the closely related O. trifallax. Accordingly, all experiments described thus far were performed with O. trifallax telomerase and O. nova telomere protein. Although the O. trifallax telomere proteins have not been purified, their gene sequences have been determined (DuBois and Prescott 1997). The derived amino acid sequences of the O. trifallax and O. nova telomere DNA-binding proteins are 80% identical and 90% similar, with the differing amino acids dispersed throughout the proteins. To ascertain whether the species difference was affecting the observed results, several tests were performed. The telomerase ribonucleoprotein complexes from O. nova and O. trifallax were compared by native gel analysis. The apparent size and charge of the two complexes were indistinguishable (Fig. 9A). An E. aediculatus extract included for comparison showed a higher mobility telomerase ribonucleoprotein complex. In addition, an assay was performed in a homologous system with O. nova nuclear extract isolated previously (Lingner et al. 1994). The effect of the O. nova telomere proteins on O. nova telomerase activity was analogous to that obtained in the heterologous experiments (cf. Fig. 9B and Fig. 3A). As in Figure 3, α + full-length β was inhibitory at a lower concentration than α + βΔC260. From these results, it is clear that the telomerase and telomere proteins from these two species are very similar. Hence, it is unlikely that the use of the heterologous system influenced our results.
Figure 9.
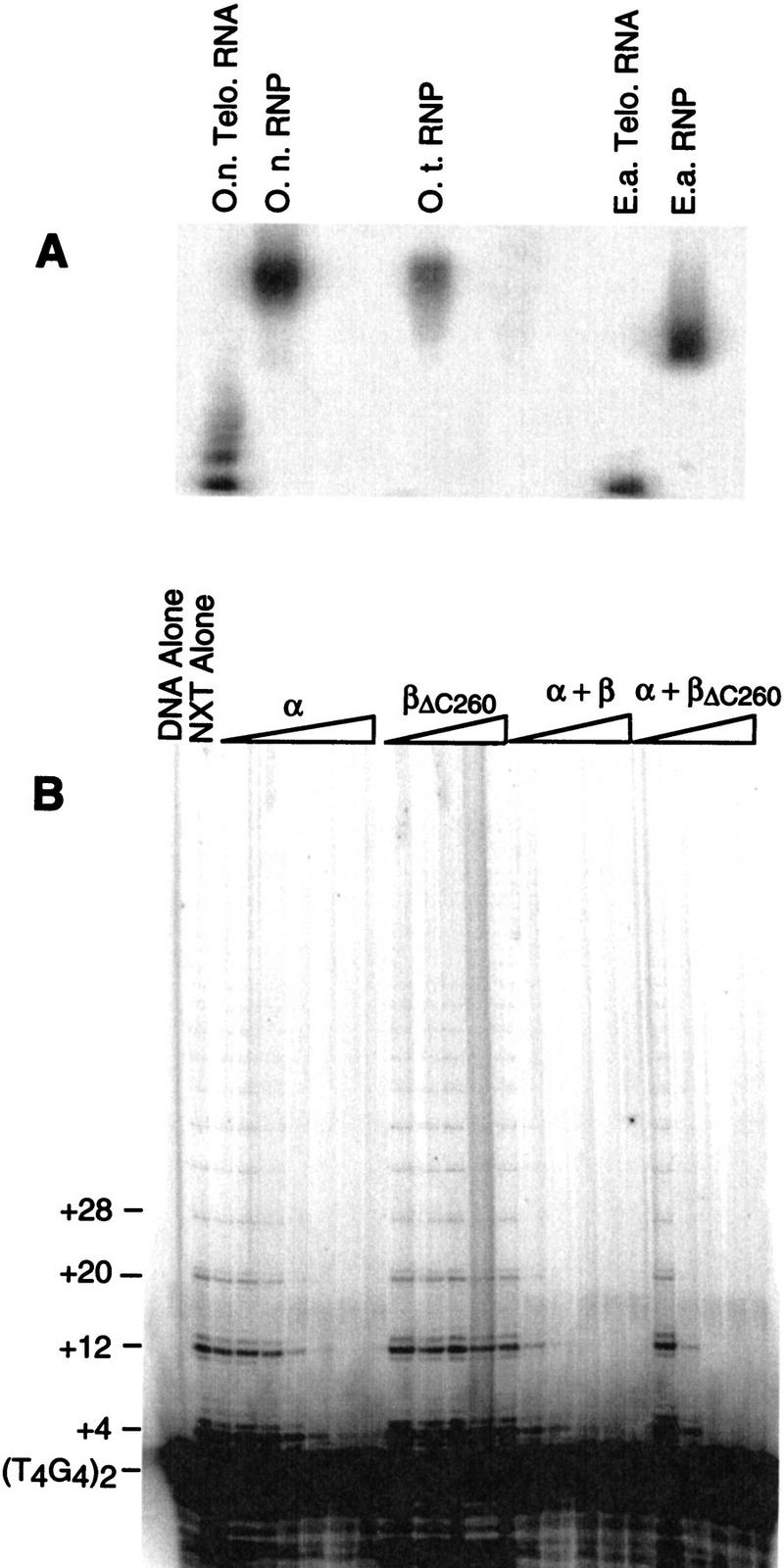
Analogous studies with a homologous system. (A) The telomerase ribonucleoprotein (RNP) complexes were compared on a native agarose gel visualized by Northern hybridization to the RNA subunits. (O.t.) Oxytricha trifallax; (O.n.) Oxytricha nova; (E.a.) Euplotes aediculatus. (B) The extension efficiency of O. nova telomerase was measured in the presence of O. nova telomere proteins. Protein concentrations are identical to those in Fig. 3.
Discussion
Although there is no evidence for a conserved sequence motif or superfamily of single-stranded DNA-binding telomere proteins, it is likely that the functional properties are conserved. These proteins are thought to be crucial in the protection of telomeric DNA, but they may also participate in other aspects of telomere function (Fang and Cech 1995a; Lin and Zakian 1996; Nugent et al. 1996), as has been found for double-stranded DNA-binding telomere proteins (Krauskopf and Blackburn 1996; Cooper et al. 1997; Marcand et al. 1997; van Steensel and de Lange 1997). One of these roles may be the regulation of telomerase. Because it now appears that a single-stranded telomeric overhang is a conserved feature of eukaryotic chromosomes (see introductory section), it may be generally true that single-stranded telomere DNA-binding proteins coordinate their efforts with telomerase for the length regulation and maintenance of the telomere. To characterize further the effects of single-stranded DNA-binding telomere proteins on telomerase activity, the present study evaluated the interaction of purified Oxytricha α and β subunits with telomerase. The results are summarized in the model depicted in Figure 10A.
Figure 10.
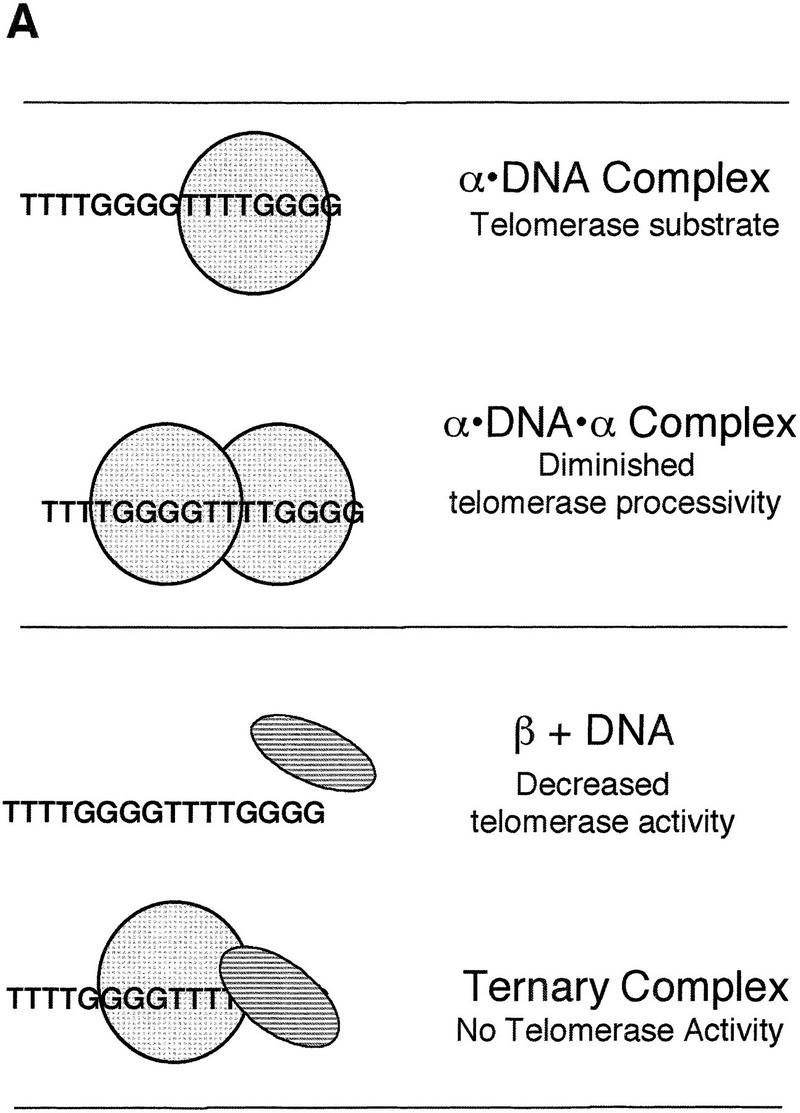
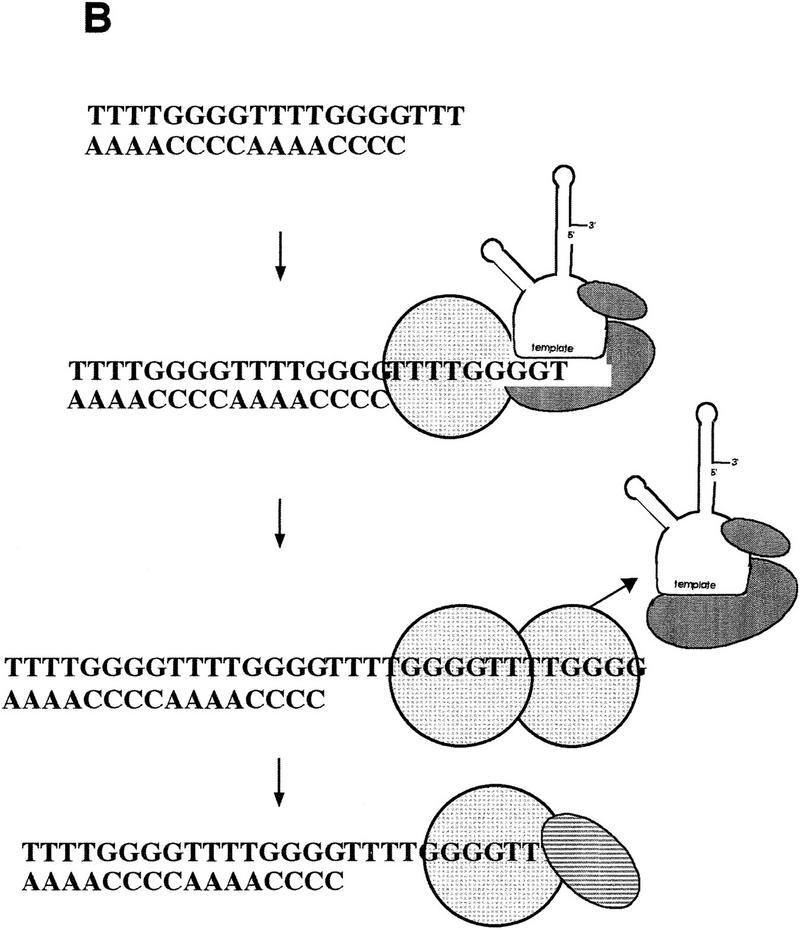
Telomere protein modulation of telomerase: a model for telomere length regulation. (A) The effects of telomere protein subunits on telomerase elongation of telomeric DNA are summarized schematically. The α subunit is light gray and hatched, whereas the β subunit is a darker gray and striped. (B) A proposed model for the in vivo telomere protein involvement in telomere length regulation. Exactly when telomere proteins bind relative to telomerase extension is speculative.
On the basis of the results presented, it appears that telomeric DNA bound to one α subunit can provide a substrate for efficient telomerase extension. This is true even if α is bound to the 3′-terminal repeat (Fig. 6). In addition, the α subunit may contribute to length regulation. After initiation and addition of a minimal binding site (∼4–8 nucleotides) by telomerase, one α subunit is bound, which still allows further extension by telomerase. Once another 8-base repeat is added (i.e., the physiological length of the telomeric overhang is achieved), a second α subunit binds, telomerase is displaced, and extension is halted. The β subunit can then displace one α subunit and a stable ternary complex is formed, protecting the telomere. This model (Fig. 10B) is supported by the following evidence: the α · DNA · α does not allow telomerase elongation of telomeric DNA and the α · DNA · α complex is able to displace telomerase. Although binding of two α subunits resulted in a defined telomere length in vitro, it produced a 12-nucleotide telomeric tract instead of the 16-nucleotide extension present in the in vivo steady state; thus, our in vitro system does not fully recapitulate telomere length regulation.
There is no direct evidence for the formation of the α · DNA · α complex in vivo. However, it seems very likely to exist as an intermediate in ternary complex formation, because (1) the α and β subunits do not form a stable heterodimer in the absence of DNA, but instead bind individually and form a heterodimer on the DNA (Fang and Cech 1993b), and (2) during in vitro reconstitution, the formation of the α · DNA · α complex precedes that of α · β · DNA (J.M. Bevilacqua and S.C.Schultz, unpubl.). Binding of the α subunit could serve as a mechanism to count the repeats in the single-stranded region of the telomere, perhaps by inducing a conformational change in the telomeric DNA. It has been proposed that telomere length in yeast is determined not so much by governing telomerase, but rather by counting the number of Rap1 proteins bound (Marcand et al. 1997), with special importance of Rap1p binding to the sequence repeat at the very end of the telomeric tract (Krauskopf and Blackburn 1996). Because of the consistent maintenance of a defined telomere length in Oxytricha, this could also be the case with the α subunit.
Shippen et al. (1994) first addressed Oxytricha telomerase · telomere protein interactions, and reported that telomerase was able to extend the α · β · DNA complex. Unlike the current report, native rather than recombinant proteins were used, and telomere protein complex integrity during extension was not defined. There are two feasible, and not mutually exclusive, explanations for the different results found in the two studies. First, the β subunit is particularly susceptible to proteolytic digestion (Gottschling and Zakian 1986; Price and Cech 1987), and thus in a native protein system, there could be insufficient β to maintain all the DNA in ternary complexes. Thus, Shippen et al. (1994) might have been observing extension of α · DNA complexes, which we also show to be substrates for telomerase. Consistent with this interpretation, Shippen et al. suggest that their telomere complexes may exist in different populations that have different accessibility to telomerase. Second, the native proteins are known to be post-translationally modified (Hicke et al. 1995), and it is possible that such modification could activate the ternary complex for extension by telomerase through a conformational change or protein dissociation. Recombinant proteins presumably lack these modifications. Thus, the effect of protein modifications on the accessibility of the telomere to telomerase remains to be investigated.
We find that the α · β · DNA complex, reconstituted from purified recombinant proteins, is not a substrate for telomerase. Most telomeres exist as an α · β · DNA complex in vivo (Price and Cech 1987; Gray et al. 1991). How, then, does telomerase gain access to its telomeric DNA substrate to permit the next round of DNA replication? It cannot wait for protein dissociation, as the ternary complex has a half-life of ∼100 hr in vitro (Fang et al. 1993), which is much longer than the Oxytricha cell cycle (6–8 hr). Cell-cycle-regulated post-translational modification of the protein (Hicke et al. 1995), degradation of one or both subunits, or even active removal by another enzyme are all conceivable solutions to this problem.
Because the α · β · DNA complex is inhibitory for DNA extension mediated by TdT as well as telomerase, it is evident that these telomere proteins are acting by binding to the substrate rather than by a specific protein–protein interaction. In addition, when a single α subunit is bound, the initial rate of telomerase extension is not affected (data not shown), which demonstrates that the subunit can bind in such a way to allow accessibility of the telomeric 3′ terminus to telomerase.
In summary, the evidence presented here suggests that single-stranded telomeric DNA-binding proteins may contribute to telomere length regulation by interacting with the chromosome end and thereby repressing telomerase.
Materials and methods
Oligonucleotides and clones
DNA oligonucleotides were synthesized and purified on denaturing polyacrylamide gels. For telomerase assays, oligonucleotides were 5′-end labeled by incubating at 37°C for 30 min in 10-μl reactions containing 20 pmoles of oligonucleotide, 10 units of T4 polynucleotide kinase (New England Biolabs), 1 μl of 10× PNK buffer (commercially supplied, New England Biolabs), 1 μl [α-32P]ATP [150 mCi/ml, 6000 Ci/mmol; New England Nuclear (NEN)]. Radiolabeled oligonucleotide was separated from unincorporated nucleotides by Sephadex–G25 chromatography. The primary oligonucleotide sequence used in these experiments was 5′-TTTTGGGGTTTTGGGG-3′, the sequence of the native Oxytricha single-stranded telomere overhang. Other oligonucleotides used are denoted in the figure legends or the text.
The Northern hybridization probes for Figure 9 were DNA fragments complementary to the telomerase RNAs for those species. Fragments were obtained by excising the sequence from a cloned plasmid (O. nova and E. aediculatus; Lingner et al. 1994), or by PCR (O. trifallax). In all cases, the fragment was radiolabeled as follows: DNA was denatured by heating at 95°C for 5 min and brief ice-water cooling for 1 min, then 5 μl of [α-32P]dCTP (3000 Ci/mmole, 40 mCi/ml; Easytide NEN) and 5 μl of High Prime (Boehringer Mannheim) were added. Labeling proceeded at 37°C for 30 min. Labeled DNA was separated from unincorporated nucleotides by Sephadex–G25 chromatography. Before addition to native gel blots, the labeled DNA probe was denatured as outlined above.
Purification of the telomere protein subunits α and β
The purification protocol was developed by J.A. Ruggles and S.C. Schultz (unpubl.). Basically, 6 liters of BL21 plysS Escherichia coli cells containing the plasmids p56a and p41a (encoding the α and β genes, respectively) were grown for ∼2 hr at 37°C (to an OD600 ≅ 0.3 or 0.4). Cultures were then shifted to 25°C and cooled for ∼1 hr. IPTG (0.5 mm) was added to induce protein expression. After a 6-hr induction, cells were harvested by centrifugation (10 min at 4000 rpm) and frozen at −20°C. Cells were lysed by thawing in 50 mm HEPES (pH 7.5), 50 mm NaCl, 0.02% NaN3, and 1 mm NaEDTA; the cell mix was sonicated for three 40-sec bursts to break remaining intact cell walls and to eliminate higher order chromosomal structure. Cell debris was removed by centrifugation (30 min at 12,000 rpm). Ammonium sulfate precipitations (35% and 70%) were performed and the final supernatant was dialyzed. The α subunit was further purified by FPLC with sequential chromatography on S-Sepharose, Q-Sepharose and a final sizing column. The β and βΔC260 subunits were further purified by FPLC using S-Sepharose and a sizing column. Samples were dialyzed, concentrated (if needed), and filtered before each column chromatography step. Protein integrity was determined by Coomassie-stained SDS-PAGE and Western blot analysis, and proteins were found to be >95% homogeneous. Protein concentration was determined by Bradford colorimetric assay (Bio-Rad).
Growth of O. trifallax
O. trifallax was grown as described previously (Swanton et al. 1980) under nonsterile conditions with Chlorogonium as the food source. Growth was conducted in glass baking dishes at room temperature.
Preparation of nuclear extract
Nuclear extracts were prepared by a modified version of a previously described protocol (Lingner et al. 1994). Cells were isolated and concentrated by multiple-step filtration using 10 μm Nytex filters, and harvested by centrifugation for 2 min at 2000 rpm at 4°C in a low speed clinical centrifuge. Cells were lysed using an overhead stirring rod in 1% NP-40 in the presence of 0.5 mg/ml spermidine phosphate, 0.5 mm PMSF, 10 mm Tris-acetate, and 10 mM MgCl2. Nuclei were isolated by centrifugation through 10% sucrose at 7500 rpm for 10 min. The nuclei were washed once to remove further cellular debris. Nuclear yield and integrity were monitored under the microscope using 5% acetocarmine. The nuclei were then lysed in 500 μl/10 grams cells of the following buffer: 50 mm Tris-acetate (pH 7.5), 10 mm MgCl2, 10% glycerol, 0.1% NP-40, 0.5 mm PMSF, and 0.4 m K-glutamate. Homogenization (25 strokes) in a 2-ml glass Dounce aided the salt lysis. The nuclear membrane and debris were pelleted by microcentrifugation at 14,000 rpm for 30 min. Supernatant was isolated, quick frozen in liquid N2, and stored at −80°C. Extracts exhibited activity for several months.
Gel-shift analysis of telomere protein complex formation
Telomere protein complex formation was monitored by a novel horizontal agarose gel system first developed by J.M. Bevilacqua and S.C. Schultz (unpubl.) and further optimized for these studies. The telomere protein subunits were preincubated with 5′-radiolabeled substrate at 4°C overnight or at 25°C for 1–2 hr in 20 mm HEPES (pH 7.5), 1 mm EDTA, 200 mm NaCl, and 2.5% NP-40. Reactions were then electrophoresed on a 1% agarose gel (0.4 cm thick, 10 cm long, 15 cm wide) buffered by 40 mm Tris-bis propane–acetate (pH 7.0), 1 mm EDTA at 4°C. The samples were loaded in the middle of the gel such that the positively charged complexes and free protein migrated in the opposite direction as the negatively charged unbound DNA substrate. Gels were electrophoresed at 90 V for ∼2 hr. Gels were dried on Hybond N+ membranes to minimize loss of DNA and analyzed using a PhosphorImager or by autoradiography.
Telomerase activity assays
In vitro telomerase activity was evaluated by monitoring the addition of nucleotides to an oligonucleotide substrate. Assays followed the basic outline described previously by Lingner et al. (1994). Two variations were used. In the first, 0.5 nm of 5′-radiolabeled oligonucleotide was incubated with telomerase-containing nuclear extract and 125 μm dGTP and dTTP, in telomerase assay buffer (1 mm MgCl2, 1 mm DTT, 50 mm K-glutamate, and 20 mm Tris-acetate). Alternatively, 25 nm nonlabeled oligonucleotide was incubated with telomerase, dGTP, and [α-32P]dTTP (400 Ci/mmole, 10 mCi/ml; Amersham) in telomerase assay buffer. For telomerase elongation, samples were incubated at 25°C for 30 min, unless otherwise indicated. When telomere protein was present in the activity assays, complexes were formed as described above, followed by the addition of telomerase. Analogous methodology was used when the effects of BSA and cytochrome c on telomerase activity were studied. Reactions were terminated by the addition of stop solution [10 mm Tris-HCl (pH 7.5), 15 mm EDTA, 0.6% SDS, and 0.1 mg/ml proteinase K] and incubated at 45°C. In standard reaction, proteinase K digestion was conducted for 30 min, but in cases where telomere protein was present, the time of incubation was increased to 2 hr. The samples were then ethanol precipitated after the addition of Na acetate (final concentration, 250 mm) and ∼20 μg of glycogen (Boehringer Mannheim), followed by a 70% ethanol wash. The DNA pellet was dried, resuspended in loading dye (90% formamide, 0.5× TBE, 0.05% bromophenol blue and xylene cyanol), and heated at 50°C for 5 min. Samples were electrophoresed on an 8% polyacrylamide/7 m urea/1× TBE sequencing gel. The gel was dried and subjected to PhosphorImager analysis. For quantification purposes, a radiolabeled oligonucleotide standard was added to reactions after termination but before ethanol precipitation to provide an internal normalization marker to combat the occasional inconsistencies prone to ethanol precipitations.
In “order of addition” experiments, telomerase was incubated with the DNA substrate for 10 min at 25°C to allow association, followed by the addition of telomere protein and nucleotides to initiate extension. Telomere protein incubations were done at 25°C for the denoted times. Once extension was initiated by the addition of nucleotides, the reactions proceeded as described previously.
TdT assay
The effect of telomere DNA-binding proteins on the extension efficiency of TdT (New England Biolabs) was investigated in a manner as similar to telomerase activity assays as possible, although the reaction buffer was optimized for TdT activity. Telomere protein complex formation in this buffer was confirmed by agarose gel electrophoresis (data not shown). Telomere protein subunits were incubated with the DNA substrate (T4G4)2 overnight at 4°C in 1× TdT buffer (New England Biolabs). TdT (5 units) and 1 μl of [α-32P]dTTP (400 Ci/mmole, 10 mCi/ml; Amersham) were added to a final volume of 40 μl, and the reaction mix was placed at 37°C for 1 hr. Reactions were terminated by the addition of 200 μl of stop solution (see above) and incubated for 2 hr at 45°C. Elongation products were analyzed by 8% polyacrylamide/7 m urea/1× TBE sequencing gel electrophoresis.
Cross-linking of the α subunit to DNA
To form a covalent α–DNA complex, the α subunit was cross-linked to its DNA substrate (Hammond et al. 1997). Various concentrations of α were preincubated with 5′-radiolabeled oligonucleotide in a final volume of 10 μl. After a 2-hr incubation, the reactions were spotted on Parafilm stretched over a metal block placed on ice. The samples were irradiated ∼12 cm from the bulbs for 20 min at 312 nm in a Stratalinker 1800 (Stratagene). The reactions were stopped by the addition of 5 μl of stop buffer [187 μm Tris-HCl (pH 6.8), 30% glycerol, 6% SDS, 0.075% bromophenol blue]. The samples were heated at 95°C for 3 min and complex formation was monitored by gel electrophoresis on 4%–20% Novex gradient gels in Tris–glycine–SDS buffer. Molecular weight markers (Bio-Rad) were included. The gels were dried and PhosphorImager analysis was performed.
In experiments in which the ability of telomerase to extend the α-DNA cross-linked complex was tested, the α subunit was cross-linked to unlabeled DNA. After cross-linking, [α-32P]dTTP (400 Ci/mmole, 10 mCi/ml; Amersham), ddGTP and telomerase were added and the samples were incubated at 25°C for 30 min. The reactions were halted by the addition of stop buffer (see above). The reactions were then centrifuged through a Centricon-30 concentrator (Amicon) to remove unincorporated nucleotides and extended unbound oligonucleotide. The eluent was analyzed by gel electrophoresis as described above. Any incorporated label at the appropriate molecular weight represents complex that was extended.
Native gel analysis of RNP complex formation
Nondenaturing 1% agarose gels were run in 75 mm Tris–glycine buffer at 4°C. Telomerase RNAs from Oxytricha and Euplotes were run as standards. The gel was denatured in 50% urea for 15 min and transferred to Hybond N+ membrane (Amersham). After transfer, the blot was cross-linked at 254 nm and incubated at 65°C in Church buffer (0.5 m NaPO4, 1 mm EDTA, 7% SDS, and 1% BSA) with radiolabeled denatured probe. The blot was hybridized overnight, washed thoroughly in 0.1% SDS/0.1× SSC, and analyzed using the PhosphorImager.
Acknowledgments
We express our sincere thanks to David Prescott for the assistance in growing Oxytricha, for all the advice, and for the generous gift of cells; Joachim Lingner for DNA, O. nova nuclear extract, and discussion; Arthur Zaug and Phil Hammond for helpful discussion; and Tracy Bryan and Jamie Sperger for critical reading of the manuscript. This research was supported in part by National Institutes of Health (NIH) grant GM28039 to T.R.C. and by NIH grant AG11636 to S.C.S. S.J.F.A. was the recipient of an American Cancer Society Postdoctoral Fellowship. J.M.B. was the recipient of an NIH postdoctoral fellowship GM17155. T.R.C. is an Investigator of the Howard Hughes Medical Institute and an American Cancer Society Professor.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL thomas.cech@colorado.edu; FAX (303) 492-6194.
References
- Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- ————— Telomerases. Annu Rev Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu C-P, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of lifespan in normal human cells by introduction of telomerase. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Brun C, Marcand S, Gilson E. Proteins that bind to double-stranded regions of telomeric DNA. Trends Cell Biol. 1997;7:317–324. doi: 10.1016/S0962-8924(97)01092-1. [DOI] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- DuBois ML, Prescott DM. Volatility of internal eliminated segments in germ line genes of hypotrichous ciliates. Mol Cell Biol. 1997;17:326–337. doi: 10.1128/mcb.17.1.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Cech TR. Characterization of a G-quartet formation reaction promoted by the β subunit of Oxytricha telomere-binding protein. Biochemistry. 1993a;32:11646–11657. doi: 10.1021/bi00094a022. [DOI] [PubMed] [Google Scholar]
- ————— Oxytricha telomere-binding protein: DNA-dependent dimerization of the α and β subunits. Proc Natl Acad Sci. 1993b;90:6056–6060. doi: 10.1073/pnas.90.13.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— . Telomere proteins. In: Blackburn E, Greider C, editors. Telomeres. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1995a. pp. 69–105. [Google Scholar]
- ————— Telomerase RNA in the replication band and spherical subnuclear organelles in hypotrichous ciliates. J Cell Biol. 1995b;130:243–253. doi: 10.1083/jcb.130.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Gray JT, Cech TR. Oxytricha telomere-binding protein: Separable DNA-binding and dimerization domains of the α-subunit. Genes & Dev. 1993;7:870–882. doi: 10.1101/gad.7.5.870. [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Zakian VA. Telomere proteins: Specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell. 1986;47:195–205. doi: 10.1016/0092-8674(86)90442-3. [DOI] [PubMed] [Google Scholar]
- Gray JT, Celander DW, Price CM, Cech TR. Cloning and expression of genes for the Oxytricha telomere-binding protein: Specific subunit interactions in the telomeric complex. Cell. 1991;67:807–814. doi: 10.1016/0092-8674(91)90075-a. [DOI] [PubMed] [Google Scholar]
- Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- Hammond PW, Lively TN, Cech TR. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol Cell Biol. 1997;17:296–308. doi: 10.1128/mcb.17.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley CB, Villeponteau B. Telomeres and telomerase in aging and cancer. Curr Opin Genet Dev. 1995;5:249–255. doi: 10.1016/0959-437x(95)80016-6. [DOI] [PubMed] [Google Scholar]
- Hicke BJ, Celander DW, MacDonald GH, Price CM, Cech TR. Two versions of the gene encoding the 41-kilodalton subunit of the telomere binding protein of Oxytricha nova. Proc Natl Acad Sci. 1990;87:1481–1485. doi: 10.1073/pnas.87.4.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke BJ, Rempel R, Maller J, Swank R, Hamaguchi J, Bradbury EM, Prescott DM, Cech TR. Phosphorylation of the Oxytricha telomere protein: Possible cell cycle regulation. Nucleic Acids Res. 1995;23:1887–1893. doi: 10.1093/nar/23.11.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher LA, Swanton MT, Donini P, Prescott DM. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc Natl Acad Sci. 1981;78:3015–3019. doi: 10.1073/pnas.78.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauskopf A, Blackburn EH. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature. 1996;383:354–357. doi: 10.1038/383354a0. [DOI] [PubMed] [Google Scholar]
- Lin JJ, Zakian VA. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner, J. and T.R. Cech. 1998. Telomerase and chromosome end maintenance. Curr. Opin. Genet. Dev. 8 (in press). [DOI] [PubMed]
- Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes & Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- Lundblad V, Szostak J. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- McElligott R, Wellinger RJ. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- Price CM, Cech TR. Telomeric DNA–protein interactions of Oxytricha macronuclear DNA. Genes & Dev. 1987;1:783–793. doi: 10.1101/gad.1.8.783. [DOI] [PubMed] [Google Scholar]
- Raghuraman MK, Cech TR. Effect of monovalent cation-induced telomeric DNA structure on the binding of Oxytricha telomeric protein. Nucleic Acids Res. 1990;18:4543–4552. doi: 10.1093/nar/18.15.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippen DE, Blackburn EH, Price CM. DNA bound by the Oxytricha telomere protein is accessible to telomerase and other DNA polymerases. Proc Natl Acad Sci. 1994;91:405–409. doi: 10.1073/pnas.91.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopp R, Wang W, Price C. rTP: A candidate telomere protein that is associated with DNA replication. Chromosoma. 1996;105:82–91. doi: 10.1007/BF02509517. [DOI] [PubMed] [Google Scholar]
- Swanton MT, Greslin AF, Prescott DM. Arrangement of coding and non-coding sequences in the DNA molecules coding for rRNAs in Oxytricha sp. Chromosoma. 1980;77:203–215. doi: 10.1007/BF00329545. [DOI] [PubMed] [Google Scholar]
- van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Wolf AJ, Zakian VA. Saccharomyces telomeres acquire single-strand TG1-2 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Ethier K, Labrecque P, Zakian VA. Evidence for a new step in telomere maintenance. Cell. 1996;85:423–433. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]
- Williamson JR. G-quartet structures in telomeric DNA. Annu Rev Biophys Biomol Struc. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes & Dev. 1997;11:2810–2821. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- Zakian VA. Structure and function of telomeres. Annu Rev Genet. 1989;23:579–604. doi: 10.1146/annurev.ge.23.120189.003051. [DOI] [PubMed] [Google Scholar]
- ————— Telomeres: Beginning to understand the end. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]
- ————— Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu Rev Genet. 1996;30:141–172. doi: 10.1146/annurev.genet.30.1.141. [DOI] [PubMed] [Google Scholar]