Abstract
The need for an HSV-2 vaccine is great considering the increasing prevalence of HSV-2 despite the widespread use of antiviral drugs. Human clinical trials of HSV-2 vaccines that elicit neutralizing antibodies have proven to be only partially effective suggesting that induction of effective T cell responses to HSV-2 is also a critical component to an efficacious vaccine. A sensitive and specific assay to measure HSV-specific T cell responses is a necessary part of vaccine development and thus we undertook the development of an interferon-γ (IFN-γ) ELISPOT assay to measure T cell responses to HSV-2.
Methods
PBMC from HSV-seronegative (HSVneg) (n=35), HSV-1-seropositive (HSV-1+/2−) (n=20) and HSV-2-seropositive (HSV-2+) subjects (n=26) were screened by IFN-γ ELISPOT for T cell responses using 34 peptide pools representing 16 HSV-2 proteins including mostly virion and immediate-early (IE) proteins.
Results
Overall, 85% of HSV-2+ subjects had a positive response to the HSV-2 peptide pools and on average, HSV-2+ subjects responded to 3 peptide pools (range 1–10). The most frequent responses were to gD-2, UL39, UL46, ICP0, UL49, gB-2, and ICP4. In contrast, only 2 of 35 (6%) HSVneg subjects had detectable T cell responses and in both cases, responses were of low magnitude relative to responses in HSV-2+ subjects and were directed at a single peptide pool. The response rate to the HSV-2 peptide pools in HSV-1+/2− subjects was 40% suggesting that the HSV-2 peptide pools contain a significant number of type-common T cell epitopes. The IFN-γ ELISPOT assay detected CD4 and CD8 T cells directed at HSV-2 peptides as confirmed by intracellular cytokine staining and flow cytometry.
Conclusion
We have developed a quantitative IFN-γ ELISPOT assay that detects both CD4 and CD8 T cells to HSV-2 peptides. This assay does not require large quantities of PBMC to generate dendritic cells for T cell stimulation, making it an ideal assay for monitoring the immunogenicity of candidate HSV-2 vaccines designed to elicit T cell responses to HSV-2 specific epitopes.
Keywords: human, herpes, T cells
Introduction
Herpes simplex virus type 2 (HSV-2) is the causative agent of genital herpes, one of the most prevalent sexually transmitted diseases worldwide. The US seroprevalence rate of HSV-2 is estimated at 17% in 14–49 year olds (1) but in some populations of HIV-infected subjects and female sex workers, the seroprevalance rate reaches 95% (reviewed in (2)). The high seroprevalence rates of HSV-2 in developed and developing countries, the importance of subclinical shedding on HSV-2 transmission especially in subjects unaware that they have genital herpes (3, 4), the increased risk that HSV-2 plays on HIV-1 acquisition (5–7), and the severity of neonatal herpes (7) underscore the need for an effective prophylactic and therapeutic vaccine.
Animal and human studies suggest that a robust cellular immune response in addition to neutralizing antibodies is critical in protection against HSV-2 infection and thus, current vaccine strategies are directed at eliciting both neutralizing antibody and cellular immunity against HSV-2. In order to detect vaccine-induced T cell responses in PBMC, a sensitive, specific and quantitative assay will be required. Assays previously employed to detect T cell responses elicited by candidate HSV-2 vaccines in humans include lymphoproliferation (8, 9) and standard 51Cr-release assays (9). Both assays require cell expansion over 6–10 days which limits quantitative analysis of vaccine-induced responses, especially in HSV-2 seropositive subjects who possess positive baseline T cell responses to HSV-2. More recently, an IFN-γ ELISPOT was employed to detect CD8+ T cell responses to a vaccine comprised of a monovalent heat shock protein non-covalently associated with a gD-2 peptide (10). While this assay was quantitative and did not require cell expansion, large numbers of PBMC were required to select CD8+ T cells and to generate dendritic cells (DC) that were used as antigen presenting cells.
In the present study, we evaluated an IFN-γ ELISPOT assay designed to detect and quantitate T cell responses in PBMC directed at HSV-2 peptides obtained from naturally infected HSV-2 seropositive subjects compared to HSV-1 seropositive subjects and subjects seronegative for HSV-1 and HSV-2. As PBMC from individual vaccinees will be limiting in future vaccine trials, the IFN-γ ELISPOT assay was designed to stimulate unfractionated PBMC for 24 hours with pools of overlapping HSV-2 peptides without the addition of in vitro matured DC. We describe the development of this assay to measure HSV-2 peptide-specific T cell responses that should enable more detailed characterization of vaccine-induced T cell responses and assist in determining whether T cell immunity contributes to vaccine efficacy.
Materials and Methods
Subjects, HSV serology and cells
Healthy HSV-1 and HSV-2 seronegative (HSVneg), HSV-1 seropositive/HSV-2 seronegative (HSV-1+/2−) or HSV-2 seropositive (HSV-2+) subjects were enrolled into IRB-approved protocols at the University of Washington Virology Research Clinic (VRC), Seattle, WA. All subjects provided written informed consent. HSV Western blot to detect antibodies to HSV-1 and HSV-2 was performed as previously described (11, 12). Abs to CMV indicating prior CMV infection were assessed using a commercial enzyme immunoassay kit for detection of total Abs to CMV using manufacturer’s recommendations (Abbott CMV Total AB EIA, Abbott Laboratories, Abbott Park, IL) (13). Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll-Hypaque and cryopreserved within 8 hours of venipuncture as previously described (14).
Synthetic peptides and peptide pools
Open reading frames (ORFs) for peptide synthesis (n=16) were selected from among the known ORFs of HSV-2 based on studies of the prevalence of CD8+ T cell responses in subjects with genital herpes (15–17) (Table 1). ORFs were mostly virion proteins (capsid, tegument, glycoprotein) or immediate-early (IE) proteins. Amino acid sequences were derived from the HSV-2 strain HG52 genome (GenBank accession no. NC-001798). Peptides were 15 amino acids long and overlapping by 11 amino acids and synthesized in crude form by either CBI/Mimotopes (San Diego, CA) (gD-2, ICP0, ICP4, ICP22, ICP27, UL39) or New England Peptide LLC (Gardner, MA) (UL19, UL25, UL35, UL46, UL47, UL49, UL11, UL27, UL29, US5) and lyophilized. Each peptide was dissolved in 10 mg/ml in sterile endotoxin-free DMSO (Sigma) and stored at 4°C. The mass of each peptide was approximately 4–5 mg. Control peptide pools included the CEF pool (CBI/Mimotopes) comprising immunodominant CD8+ T cell epitopes within CMV, EBV and influenza (14, 17, 18) and the CMV pp65 peptide mix containing 138 15 mers overlapping by 11 amino acids (Becton Dickinson, San Jose, CA).
TABLE 1.
HSV-2 peptide pools used in the IFN-γ ELISPOT assay
| No. | HSV-2 ORF | ORF classa | Common name | ORF Length (aa) | # peptidesb | # pools | # peptides/pool |
|---|---|---|---|---|---|---|---|
| 1 | US6 | gp | gD-2 | 393 | 96 | 1 | 96 |
| 2 | RL2 | IE | ICP0 | 824 | 198 | 2 | 98,100 |
| 3 | US1 | IE | ICP22 | 413 | 101 | 2 | 50,51 |
| 4 | UL54 | IE | ICP27 | 512 | 126 | 2 | 63,63 |
| 5 | RS1 | IE | ICP4 | 1,318 | 330 | 4 | 82,82,83,83 |
| 6 | UL39 | other | ICP6 | 1,142 | 283 | 3 | 94,94,95 |
| 7 | UL19 | capsid | VP5/ICP5 | 1,375 | 341 | 4 | 85,85,85,86 |
| 8 | UL25 | capsid | 585 | 144 | 2 | 72,72 | |
| 9 | UL35 | capsid | VP26 | 112 | 26 | 1 | 26 |
| 10 | UL46 | tegument | VP11/12 | 722 | 178 | 2 | 89,89 |
| 11 | UL47 | tegument | VP13/14 | 696 | 172 | 2 | 86,86 |
| 12 | UL49 | tegument | VP22 | 300 | 74 | 1 | 74 |
| 13 | UL11 | tegument | 96 | 22 | 1 | 22 | |
| 14 | UL27 | gp | gB | 904 | 224 | 3 | 74,87,63 |
| 15 | UL29 | other | ICP8 | 1,196 | 297 | 3 | 99,99,99 |
| 16 | US5 | gp | gJ | 92 | 21 | 1 | 21 |
| Totals | 2,633 | 34 | range: 21–100 | ||||
Open reading frames (ORF) for study encoded glycoproteins (gp), immediate early (IE), capsid, tegument or other proteins.
15 mers overlapping by 11 amino acids (aa).
Peptide pools (library and array) were prepared for each ORF by grouping peptides linearly across an ORF and were used to screen PBMC. A total of 2,633 peptides were synthesized and pooled into pools composed of peptides ranging from 21–100 peptides/pool (median 85 peptides/pool) (Table 1). Array pools, used to deconvolute library pools that were positive in the PBMC screen, were prepared by arranging the peptides within a single library pool in a row-and-column format and pooling the peptides in each column or row.
IFN-γ ELISPOT
The IFN-γ ELISPOT assay was performed as previously described (17). PBMC were thawed in R10 [RPMI 1640 supplemented with 10% FBS, 2 mM l-glutamine, 25 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) buffer, 50 μg/ml streptomycin, 50 U/ml penicillin containing 50 U/ml benzonase (Novagen, Madison, WI)], and then washed and rested overnight in R10 at 37 °C, 5% CO2 before assay. Cell viability was assessed by trypan blue exclusion upon PBMC thawing and after resting overnight and viability was routinely greater than 95%. Using the IFN-γ ELISPOT kit (Mabtech, Cincinnati, OH) according to the manufacturer’s instructions, PBMC were plated at 2×105 cells/well. Peptides were added to the wells at a final concentration of 1 μg/ml for overnight stimulation. Wells containing R10 alone or R10 and DMSO served as negative controls, and those containing 1 μg/ml phytohemagglutinin-purified (PHA) (Murex, distributed by Remel, Lenexa, KS) served as positive controls. Negative controls were tested in 4 replicate wells, whereas HSV-2, CMV and CEF peptide pools and PHA were tested in duplicate wells. Spots were counted and analyzed using the automated Bioreader® (Biosys GmbH, Germany). Responses were considered positive if (1) the spot forming cells (SFC)/well were 4 times greater than the mean SFC in the 4 DMSO wells and (2) the SFC/well was ≥ 11 (17, 19). Results are expressed as SFC per 106 PBMC.
Intracellular cytokine staining and flow cytometry
To confirm peptide identity and to determine the phenotype of HSV-2 peptide-specific T cells, intracellular cytokine staining (ICS) and flow cytometry was performed using a 6-color ICS panel in a 96-well plate format modified from (20) as previously described (17). Briefly, PBMC were thawed and rested overnight in R10 media followed by stimulation with DMSO (negative control), SEB (positive control), HSV-2 peptide pools (1 μg/ml), individual HSV-2 15-mers (1 μg/ml), CEF or CMV peptide pools. During the 6-hr incubation at 37°C, Brefeldin A (10 μg/ml, Sigma, St. Louis, MO) and the co-stimulatory antibodies CD28 and CD49d (each at 1 μg/ml, BD Biosciences) were included. Antibodies CD4-FITC, CD8 PerCP-Cy5.5, IFN-γ APC, and IL-2 PE were purchased from BD Biosciences, CD3 ECD was purchased from Beckman-Coulter (Marseille, France) and the LIVE/DEAD Fixable Violet Dead Cell Stain was purchased from Invitrogen/Molecular Probes (Eugene, OR). Samples were collected from 96-well plates using High Throughput Sample (HTS, BD) device for analysis by the LSRII and all FACS analyses were performed using FlowJo® software (Treestar, Inc., OR).
Statistical analysis
Positive IFN-γ ELISPOT responses were determined from SFC/well using a cutoff of at least 11 spots and a 4-fold higher number of SFC versus the negative control DMSO (17, 19). With approximately 25 participants of each HSV serostatus, peptide-specific positivity rates within each serostatus have a standard deviation of 20%. This number of subjects also provides 90% power to detect an 8-fold increase in the positivity rate between HSVneg and HSV-2+ subjects, provided at least 50% of HSV-2+ subjects respond to at least one HSV-2 peptide pool. A variance components analysis was employed to assess inter-assay variability.
We evaluated the proportion of responses positive to each control peptide pool (CMV and CEF) and HSV-2 peptide pools by HSV serostatus. We first summed the number of hits per person, per antigen type (HSV-2, CMV, and CEF), and then performed Poisson regression to ascertain whether the rate of positivity was different by serostatus. We corrected for over dispersion (extra Poisson variability) using a scale parameter.
Results
Study Participants
A total of 81 healthy subjects were enrolled in this study; 35 subjects were HSVneg by Western blot analysis, 20 subjects were HSV-1+/2− and 26 were HSV-2+. Of the 26 HSV-2+ subjects, 12 (46%) were co-infected with HSV-1. Median ages for each group were 32 yrs for HSVneg, 29 yrs for HSV-1+/2− subjects and 47 yrs for HSV-2+ subjects. HSVneg were 51% male, HSV-1+/2− subjects were predominantly women (70%) and HSV-2+ subjects were predominantly men (69%). Of the 81 subjects enrolled, 59 (73%) were Caucasian and all subjects were HIV negative.
IFN-γ ELISPOT responses to HSV and control peptide pools
Overall, 22 of 26 (85%) of the HSV-2+ subjects, 8 of 20 (40%) of the HSV-1+/2− subjects and 2 of 35 (6%) of the HSVneg subjects had a positive IFN-γ ELISPOT response to at least one of the 34 HSV-2 peptide pools (Table 2). HSV-1+/2− subjects had a 17.5-fold increased rate of positivity to HSV-2 peptide pools (95%CI 2.2–140.2, p=.007) while HSV-2+ subjects had a 54.5-fold increased rate of positivity (95%CI 7.3–406.4, p<.001) relative to HSVneg subjects. HSV-2+ subjects had a significantly higher rate of positivity compared to HSV-1+/2− subjects (RR=3.12, 95%CI 1.55–6.28, p=.0015) (Table 2) suggesting that type-specific responses to these HSV-2 proteins may be more frequent than type-common responses. Coinfection of HSV-2+ subjects with HSV-1 had no effect on the rate of positivity to HSV-2 peptide pools: there was no significant difference in the overall rate of positivity to any HSV-2 peptide pool in HSV-2+ who were co-infected with HSV-1 (n=12) or HSV-1 seronegative (n=14) (RR=1.07 for HSV-1−/2+ versus HSV-1+/2+, 95%CI 0.57–2.01, p=.83). PHA responses were positive in all PBMC samples regardless of HSV serostatus demonstrating universal PBMC viability and general T cell responsiveness.
TABLE 2.
Response rates to HSV, CEF and CMV in subjects based on HSV serostatusa
| HSV serostatus | Total Subjects (n) | Subjects with positive responses to: | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HSV-2 | CEF | CMV | PHA | ||||||
| n | % | n | % | n | % | n | % | ||
| HSVneg | 35 | 2 | 6% | 22 | 63% | 7 | 20% | 35 | 100% |
| HSV-1+/2− | 20 | 8 | 40% | 10 | 60% | 12 | 60% | 20 | 100% |
| HSV-2+ | 26 | 22 | 85% | 18 | 69% | 19 | 73% | 26 | 100% |
PBMC from HSVneg, HSV-1+/2− and HSV-2+ subjects were screened by IFN-γ ELISPOT for responses to HSV-2 (34 overlapping peptide pools), CEF and CMV peptide pools and PHA. Responses were considered positive if the SFC in the experimental well (average of 2 wells) was 4 times the median SFC in the DMSO control wells and ≥55 SFC/106 PBMC.
IFN-γ ELISPOT positivity to the CMV peptide pool was consistent with CMV serostatus: subjects with positive IFN-γ ELISPOT responses to the pp65 CMV peptide pool were CMV seropositive and subjects with negative IFN-γ ELISPOT responses to the CMV peptide pool were CMV seronegative.
In contrast to HSV-2 response rates, there were no significant differences in response rates to CEF between HSVneg subjects (63% with positive responses) compared to HSV-1+/2− (60% with positive responses) (RR=0.95, 95%CI 0.62–1.47, p=.832) or HSV-2+ subjects (69% with positive responses) (RR=1.10, 95%CI 0.75–1.61, p=.618) (Table 2). However, for CMV, HSV-1+/2− and HSV-2+ subjects had higher response rates than HSVneg subjects (60% for HSV-1+/2− and 73% for HSV-2+ compared to 20% in HSVneg) (Table 2); ELISPOT positivity to CMV was only detected in subjects seropositive for CMV. HSV-1+/2− and HSV-2+ subjects had a 3.00-fold (95%CI 1.50–5.99, p=.002) and 3.65-fold (95%CI 1.92–6.95, p<.001) increased rate of positivity, respectively, relative to HSVneg subjects. No significant differences were detected in CMV response rates between HSV-1+/2− and HSV-2+ subjects (RR=1.22, 95%CI 0.714–2.08, p=.471) (Table 2). These data suggest that subjects infected with HSV-1 or HSV-2 were more likely to be seropositive to CMV and thus have acquired immunity to CMV than subjects who were HSVneg.
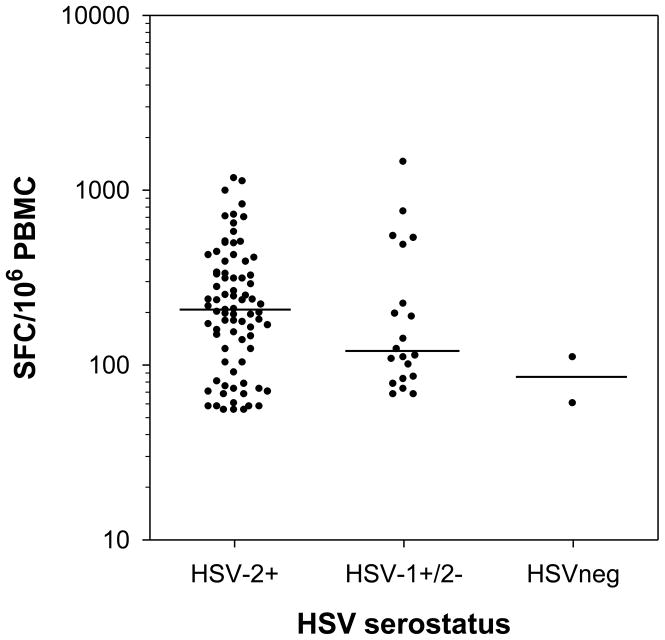
The magnitude of the IFN-γ ELISPOT response to individual HSV-2 peptide pools varied considerably between individuals. The median magnitude of positive HSV-2 peptide-specific responses in HSV-2+ subjects was 208 SFC/106 PBMC (range 55 to 1,165 SFC/106 PBMC) compared to 118 SFC/106 PBMC in HSV-1+/2− subjects (range 68 to 1,440 SFC/106 PBMC) (Figure 1). Only 2 positive HSV peptide specific responses were measured in 2 HSVneg subjects (60 and 110 SFC/106 PBMC) and the magnitudes of these responses were within the lowest range of the positive responses in the HSV-2+ and HSV-1+/2− subjects (Figure 1). These data suggest that the magnitude of HSV-2 peptide-specific responses appear to be influenced by HSV serostatus. In contrast, HSV serostatus had no effect on the magnitude of responses to CMV or CEF which ranged from 80 to 2,675 SFC/106 PBMC and 215 to 2,500 SFC/106 PBMC, respectively. The median SFC/106 PBMC for PHA was 2,160 (range 695 to 3,500) while the median SFC/106 PBMC for DMSO was 38 (range 5 to 190).
Figure 1. Magnitude of positive IFN-γ ELISPOT responses to individual HSV-2 peptide pools in HSV-2+, HSV-1+/2− and HSVneg subjects.
The magnitudes of all positive IFN-γELISPOT responses to any of the 34 HSV-2 peptide pools are displayed in each group of subjects (n=2 for HSVneg, n=8 for HSV-1+/2− and n=22 for HSV-2+ subjects) separated by HSV serostatus. The median magnitude of responses to HSV-2 peptide pools in each group is shown by the bar.
To assess the inter-assay variability in the IFN-γ ELISPOT assay, PBMC obtained from single blood draws from 5 different HSV-2+ subjects were screened for positivity to the CMV, CEF and all 34 HSV-2 peptide pools on 3 separate assay days. All participants had no response to 17 of the possible 34 HSV-2 peptide pools for all 3 repeats. Those data were dropped to avoid inflating the agreement level. Thus, for this analysis, only those HSV-2 peptide pools demonstrating any positivity in any of the 5 HSV-2+ subjects (17 HSV-2 peptide pools total) were included and considered collectively. While there was 100% agreement in assessing IFN-γ ELISPOT responses to CMV and CEF as positive or negative on all 3 days, there was 86% agreement in assessing responses to the HSV-2 peptide pools on all 3 assay days; this is likely due to the relatively higher numbers of SFC/106 PBMC measured to CEF (median 405 range 215–2,135) and CMV (median 380 range 100–1,770) compared to individual HSV-2 peptide pools (median 155 range 60–803) in these 5 HSV-2+ subjects. If the 17 pools to which there were consistently negative responses were considered in the analysis along with those pools with positive responses, the agreement was 93%. The proportion of variability in the IFN-γ ELISPOT assay due to run date was 8% (CV=93%) for CMV and CEF and 20% (CV=63%) for the HSV-2 peptide pools.
Antigen specificity of IFN-γ ELISPOT responses
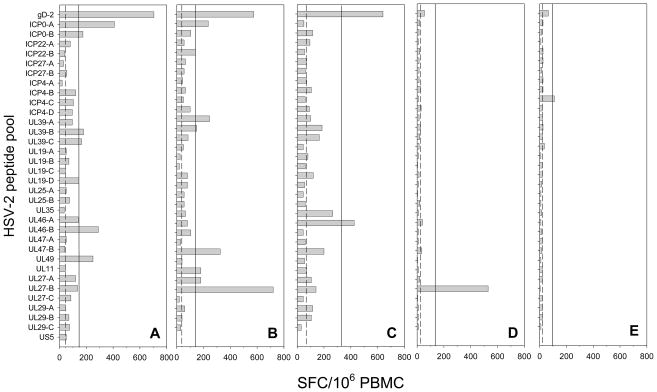
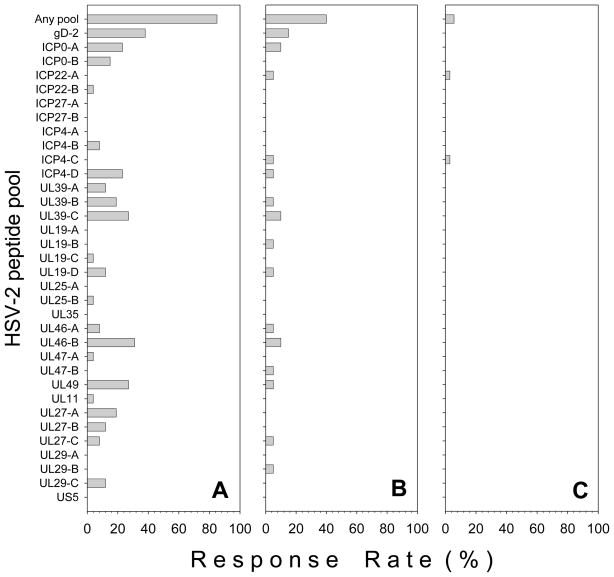
Figure 2 displays IFN-γ ELISPOT responses to the HSV-2 peptide pools used to screen PBMC from individual representative HSV-2+ (Figure 2A–C), HSV-1+/2− (Figure 2D) and HSVneg (Figure 2E) subjects. On a per pool basis, gD-2 was the most frequently recognized peptide pool in HSV-2+ subjects with a 38% response rate (Figure 3A). Frequent responses to UL46 pool B (UL46-B) (31%), UL39 pool C (UL39-C) (27%), UL49 (27%), ICP0 pool A (ICP0-A) (23%) and ICP4 pool D (ICP4-D) (23%) were also detected in HSV-2+ subjects. If the positive responses in the sub-pools were combined together to determine the frequency of responses to each of the 16 HSV-2 proteins, the highest response rates were to gD-2, UL39 and UL46 which were each recognized in 38% of all HSV-2+ subjects followed by ICP0 (31%), UL49 (27%) and UL27 and ICP4 (both 23%). No responses were detected to ICP27, UL35, and US5 in HSV-2+ subjects (Figure 3A). There were 7 HSV-2+ subjects who had a positive response in more than one sub-pool from a single HSV-2 protein; 5 HSV-2+ subjects had a response in 2–3 UL39 sub-pools, 3 subjects had a response in 2–3 UL27 sub-pools and 2 subjects had positive responses in each of the 2 ICP0 sub-pools.
Figure 2. IFN-γ ELISPOT responses in HSV-2+, HSV-1+/2− and HSVneg subjects.
PBMC were screened by IFN-γ ELISPOT using 34 pools of overlapping peptides representing 16 HSV-2 ORFs. Results are from 3 HSV-2 seropositive subjects (A–C), an HSV-1+/2− subject (D) and an HSVneg subject (E). The dashed line represents the median SFC/106 PBMC from the DMSO control wells; the solid line represents the threshold for positivity (4 times the mean of the DMSO control wells and ≥55 SFC/106 PBMC). IFN-γ ELISPOT responses to PHA control wells were 1,258 SFC/106 PBMC (HSV-2+ Subject in A), 2,553 SFC/106 PBMC (HSV-2+ Subject in B), 2,329 SFC/106 PBMC (HSV-2+ Subject in C), 1,145 SFC/106 PBMC (HSV-1+/2−Subject in D) and 2,811 SFC/106 PBMC (HSVneg Subject in E).
Figure 3. Response rates to individual HSV-2 peptide pools in HSV-2+, HSV-1+/2− and HSVneg subjects.
PBMC from HSV-2+ (n=26), HSV-1+/2− (n=20), and HSVneg subjects (n=35) were screened using 34 pools of overlapping peptides representing 16 HSV-2 ORFs. The percentage of HSV-2+ (A), HSV-1+/2− (B) and HSVneg subjects (C) who possessed positive responses to any HSV-2 peptide pool (Any pool) or to individual HSV-2 peptide pools is displayed.
The most common peptide pools recognized in HSV-1+/2− subjects were gD-2 (15%), ICP0 (10%), UL39 (10%), and UL46 (10%) (Figure 3B). Only 2 positive responses, one to ICP22 and one to ICP4 (Figure 2E; Figure 3C) were detected in 2 different HSVneg subjects.
Breadth and cumulative quantitative magnitude of IFN-γ ELISPOT responses
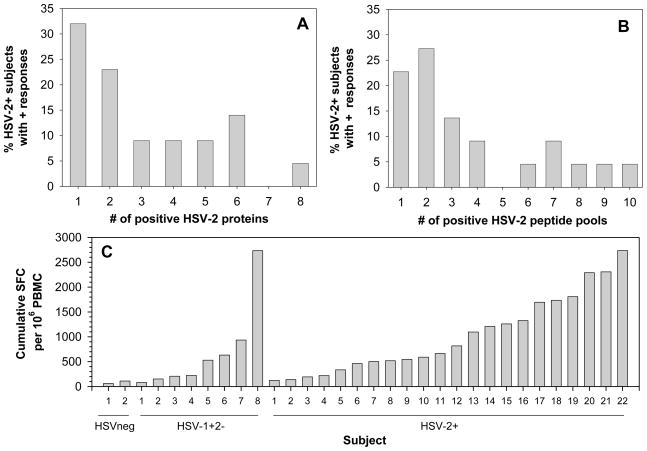
Of the 22 HSV-2+ subjects with positive IFN-γ ELISPOT responses, responses were detected to a median of 3 individual HSV-2 peptide pools (range 1–10) (Figure 4A) and to a median of 2 individual HSV-2 proteins (range 1–8) (Figure 4B). This, however, underestimates the number of epitopes recognized since multiple sub-pools were recognized in 7 HSV-2+ individuals (see below). Of the 8 HSV-1+/2− subjects with positive IFN-γ ELISPOT responses, responses were detected to a median of 2 individual HSV-2 peptides pools (range 1–7) (data not shown). As mentioned earlier, of the 2 HSVneg subjects with positive IFN-γ ELISPOT responses responses in each subject were directed at a single HSV-2 peptide pool.
Figure 4. Breadth and magnitude of HSV-2-specific responses in HSV-2+ subjects.
(A–B) Breadth of the T cell response to HSV-2. The percentage of HSV-2+ subjects who possessed 1 to 8 positive IFNγ ELISPOT responses to the 16 HSV-2 proteins (A) or 1 to 10 positive IFN-γ ELISPOT responses to the 34 HSV-2 peptide pools (B) was determined from all HSV-2+ subjects who possessed HSV-2 peptide-specific responses (n=22). (C) Magnitude of the cumulative T cell response to HSV-2. Shown are cumulative quantitative T cell responses for all subjects (n=2 for HSVneg, n=8 for HSV-1+/2− and n=22 for HSV-2+ subjects) with positive IFN-γ ELISPOT responses to the 34 HSV-2 peptide pools covering 16 HSV-2 proteins.
Figure 4C displays the cumulative quantitative T cell response in all subjects who possessed positive IFN-γ ELISPOT responses to the 34 HSV-2 peptide pools including 2 HSVneg subjects, 8 HSV-1+/2− subjects and 22 HSV-2+ subjects. Cumulative quantitative T cell responses to HSV-2 peptide pools varied by serostatus; median cumulative quantitative T cell responses were 85 (range 60–110 SFC/106 PBMC), 376 (range 83–2,732 SFC/106 PBMC) and 740 (range 123–2,735 SFC/106 PBMC) in HSVneg, HSV-1+/2− and HSV-2+ subjects, respectively (Figure 4C).
Phenotype of HSV-2 peptide-specific T cell responses
In 20 of the HSV-2+ subjects, sufficient PBMC were available to deconvolute individual HSV-2 peptide pools that were positive in the initial IFN-γ ELISPOT screen to determine the epitope(s) recognized by the responding HSV-specific T cells. Table 3 displays the 41 HSV-2 peptides that were deduced by deconvolution to be epitopes recognized by HSV-specific T cells in the 20 HSV-2+ subjects tested. In many cases, two sequential overlapping peptides were positive and in these cases, we list the 11 amino acids shared between the two peptides containing the T cell epitope. In several subjects, multiple array pools were positive and as such, a single epitope could not be deduced suggesting the presence of multiple epitopes within the ORF. This was most commonly observed in subjects with IFN-γ ELISPOT responses to gD-2 (n=8) as well as to ICP0 (n=2) and UL39 (n=2) (Table 3). The presence of multiple T cell epitopes within gD-2 is consistent with data presented in an elegant study by Kim et al. that demonstrated a high density of CD4 T cell epitopes within gD-2 that were recognized broadly across multiple HLA-DR types (21).
TABLE 3.
HSV-2 peptides recognized by HSV-2+ subjectsa
| HSV-2+ Subject | HSV-2 ORF | Peptide | Sequenceb | CD4 or CD8c |
|---|---|---|---|---|
| 2 | UL46 | UL46397–407 | KSWFGAALAPD | ndd |
|
| ||||
| 5 | UL46 | UL46625–635 | VRVYENICLRR | CD4 |
| gD-2 | multiplee | CD4f | ||
|
| ||||
| 7 | ICP0 | ICP0123–137 | VCAVCTDEIAPPLRC | nd |
| ICP0 | ICP0135–149 | LRCQSFPCLHPFCIP | nd | |
| ICP4 | ICP41009–1023 | LGNRLCGPATAAWAG | CD8 | |
| ICP4 | ICP41169–1183 | MSPREYRRAVLPALD | CD4/CD8 | |
| UL11 | UL111–15 | MGLAFSGARPCCCRH | CD4 | |
| gB-2 | gB-2513–527 | HVNDMLGRIAVAWCE | CD4 | |
|
| ||||
| 8 | ICP4 | ICP4533–547 | VAMSRRYDRAQKGFL | CD4 |
|
| ||||
| 10 | gD-2 | multiple | nd | |
|
| ||||
| 11 | UL19 | UL191237–1251 | PFAATANPWASQRFS | CD4 |
|
| ||||
| 12 | UL46 | UL46621–635 | EIPWVRVYENICLRR | CD4 |
| gD-2 | multiple | CD4f | ||
|
| ||||
| 13 | gD-2 | multiple | CD4f | |
| ICP0 | multiple | nd | ||
| UL39 | UL39957–971 | SMMKHGLRNSQFIAL | nd | |
| UL29 | UL29921–936 | SRRPPSVQAAAAWAPQ | nd | |
|
| ||||
| 14 | UL39 | UL39957–971 | SMMKHGLRNSQFIAL | nd |
| UL29 | UL29921–936 | SRRPPSVQAAAAWAPQ | nd | |
|
| ||||
| 15 | gB-2 | gB-2140–152 | IAVVFKENIAPY | CD4 |
| UL39 | multiple | nd | ||
|
| ||||
| 16 | UL49 | UL49249–263 | KNLLQRANELVNPDA | CD4 |
| ICP0 | multiple | nd | ||
| UL39 | multiple | nd | ||
| gB-2 | gB-2276–291 | YPYDEFVLATGDFVY | CD4 | |
| gB-2 | gB-2849–859 | YMALVSAMERT | CD4 | |
| gD-2 | multiple | nd | ||
|
| ||||
| 17 | UL46 | UL4669–83 | GADRSVRLAARHHNT | nd |
| UL39 | multiple | nd | ||
| gB-2 | gB-2277–287 | YPYDEFVLATG | nd | |
|
| ||||
| 18 | gD-2 | multiple | CD4f | |
| UL46 | UL46249–263 | RLGPADRRFVALSGS | nd | |
| UL46 | UL46621–635 | EIPWVRVYENICLRR | nd | |
|
| ||||
| 19 | ICP4 | ICP41013–1023 | LCGPATAAWAG | CD8 |
| gD-2 | multiple | nd | ||
| UL46 | UL46670–681 | SPPGATRAPDPG | nd | |
| ICP0 | ICP0638–651 | SASGAGERRETSLGP | nd | |
| ICP0 | ICP0650–664 | LGPRAAAPRGPRKCA | nd | |
|
| ||||
| 20 | UL19 | UL191235–1249 | PFAATANPWASQRFS | CD4 |
|
| ||||
| 21 | gD-2 | gD-2273–287 | YTSTLLPPELSDTTN | CD4 |
|
| ||||
| 22 | UL46 | UL46621–635 | EIPWVRVYENICLRR | CD4 |
| gD-2 | multiple | nd | ||
|
| ||||
| 23 | UL25 | UL25405–419 | DRLDNRLQLGMLIPG | CD8 |
|
| ||||
| 24 | ICP0 | ICP0127–137 | CTDEIAPPLRC | CD8 |
| ICP0 | ICP0638–651 | SASGAGERRETSLGP | nd | |
| ICP0 | ICP0650–664 | LGPRAAAPRGPRKCA | nd | |
| ICP4 | ICP4429–443 | QYALITRLLYTPDAE | nd | |
| ICP4 | ICP4441–455 | DAEAMGWLQNPRVAP | CD4 | |
| UL39 | UL391013–1027 | KELERTFGGKRLLDA | nd | |
|
| ||||
| 25 | ICP4 | ICP41169–1183 | MSPREYRRAVLPALD | CD4 |
| UL19 | UL19828–838 | GVRFDRVYATL | CD8 | |
| UL19 | UL19789–803 | ARAADAADDRPHRPA | CD4 | |
| UL27 | gB-2137–152 | YTEGIAVVFKENIAPY | CD4/CD8 | |
Individual HSV-2 peptides recognized by T cells from HSV-2+ subjects were determined by deconvoluting the HSV-2 peptide pools.
When sequential overlapping 15 mers within an ORF were positive, we list the 11 amino acid sequence shared between the 2 peptides.
Phenotype of responding T cells was determined by ICS and flow cytometry.
Not determined.
In some cases multiple array pools were positive indicating responses to multiple epitopes within an ORF and thus a single epitope could not be deduced.
The phenotype of responding T cells was performed using the gD-2 peptide pool to stimulated PBMC as opposed to using individual gD-2 peptides.
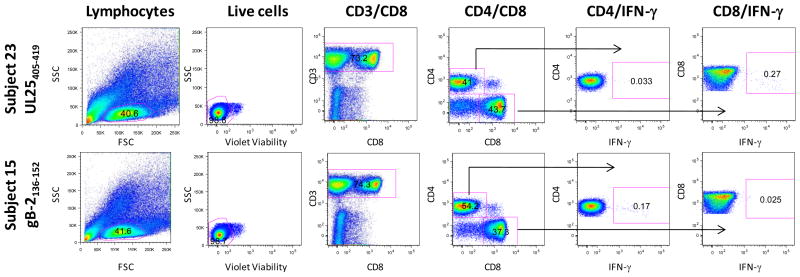
In some cases, sufficient PBMC were available to determine the phenotype of the responding T cells by ICS and flow cytometry using the T cell epitopes deduced after peptide pool deconvolution to stimulate PBMC. Figure 5 displays representative CD4 and CD8 T cell epitopes recognized by T cells from 2 different HSV-2+ subjects. The UL25405–419 peptide was recognized by CD8 T cells from HSV-2+ Subjects #23 (Figure 5; top row of graphs) and the gB-2136–152 peptide was recognized by CD4 T cells from HSV-2+ Subject #15 (Figure 5; bottom row of graphs). Background responses to DMSO control samples were 0.013% and 0.08% for CD4+/IFN-γ+ and CD8+/IFN-γ+ subsets, respectively, in PBMC from Subject #23 stimulated with the UL25405–419 peptide and 0.014 and 0.028% for CD4+/IFN-γ+ and CD8+/IFN-γ+ subsets, respectively, in PBMC from Subject #15 stimulated with the gB-2136–152 peptide. The phenotypes of the T cells responding to HSV-2-specific peptides from 14 different subjects are displayed in Table 3; 16 of the peptides were recognized by CD4+ T cells, 5 were recognized by CD8+ T cells and 2 peptides were recognized by CD4+ and CD8+ T cells. Not all peptides could be confirmed by ICS likely due to the low frequency of some HSV-2 peptide-specific CD4 and CD8 T cell responses.
Figure 5. Phenotype of HSV-2 peptide-specific T cells by ICS and flow cytometry.
To confirm the identity of the HSV-2 peptides and to determine the phenotype of the responding T cells, PBMC from all subjects with positive IFN-γ ELISPOT responses were incubated with the deduced 15-mer and the CD4 or CD8 phenotype of the T cells was determined by ICS and flow cytometry. Representative plots for a CD8 (top row of graphs) and a CD4 (bottom row of graphs) epitope are displayed from 2 HSV-2+ subjects. PBMC from HSV-2+ Subject #23 were incubated with the peptide UL25405–419 and PBMC from HSV-2+ Subject #15 were incubated with the peptide gB-2136–152 (bottom row). The gating hierarchy moves from left to right starting with lymphocytes [side scatter (SSC) vs forward scatter (FSC)], live cells (side scatter vs violet viability), CD3 T cells (CD3 vs CD8), CD4 and CD8 T cells. The gates used to identify IFN-γ producing CD4 or CD8 T cells are depicted by the arrows.
Additionally, we assessed the phenotype of T cells directed at gD-2 in 4 HSV-2+ subjects with multiple array pools positive using ICS and flow cytometry and PBMC stimulated with the gD-2 peptide pool. The phenotype of gD-2 specific T cells from HSV-2+ Subject #5 (0.63% CD4+/IFN-γ+, 0.01% CD8+/IFN-γ+), Subject #12 (0.11% CD4+/IFN-γ+, 0.01% CD8+/IFN-γ+), Subject #13 (0.18% CD4+/IFN-γ+, 0.03% CD8+/IFN-γ+) and Subject #18 (0.16% CD4+/IFN-γ+, 0.0% CD8+/IFN-γ+) were exclusively of the CD4 T cell subset. We have included the phenotype of the T cells responding to the gD-2 peptide pool from these 4 HSV-2+ subjects in Table 3.
Discussion
This study describes a sensitive, specific and quantitative IFN-γ ELISPOT assay to detect T cell responses to HSV-2 peptides. Using peptide pools representing 16 HSV-2 proteins, 85% of HSV-2 seropositive subjects had positive HSV-2 peptide-specific IFN-γ responses in unfractionated PBMC. The most frequent responses were to HSV-2 glycoproteins, IE and tegument proteins including gD-2, UL39, UL46, ICP0, UL49, gB-2 and ICP4. In many cases, the phenotype of the responding T cells was determined by ICS and flow cytometry which demonstrated that positive HSV-2 peptide-specific T cell responses detected in the IFN-γ ELISPOT contained both CD4 and CD8 T cell responses. Only 40% of HSV-1+/2− subjects responded to at least 1 HSV-2 peptide pool suggesting that type-specific responses to these HSV-2 proteins may be more frequent than type-common responses.
We previously reported on the diversity of the CD8 T cell response to HSV-2 among 37 HSV-2 seropositive subjects using an IFN-γ ELISPOT assay and peptide pools representing 48 ORFs of HSV-2 (15). Because the IFN-γ ELISPOT protocol required large numbers of cells in order to both (1) purify CD8+ T cells from PBMC and (2) to stimulate these CD8s with peptide-pulsed autologous DC using peptide pools representing 48 ORFs of HSV-2, subjects were leukapheresed, a procedure that is not feasible in clinical vaccine trials. As such, we modified the protocol in the current study and used whole unfractionated PBMC stimulated with HSV-2 peptide pools from the most immunoprevalent HSV-2 ORFs (n=16) without the requirement of in vitro matured DC. This method detected HSV-2 peptide-specific responses in 85% of HSV-2 seropositive subjects, a high response rate considering that only 20% of the HSV-2 proteome (16 out of 80 total ORFs) was used to screen PBMC. Importantly, this assay required only 16 million total PBMC, a cell number easily obtained from study subjects in the context of a vaccine trial where multiple blood draws are obtained to longitudinally measure vaccine immunogenicity. A similar pattern of antigen recognition in HSV-2 seropositive subjects was observed in this study compared to our previous study (15) suggesting that direct peptide stimulation in the absence of exogenously added DC did not lower the sensitivity of detecting HSV-2 peptide-specific responses. The observation that 15% of HSV-2+ subjects had no positive IFN-γ ELISPOT responses suggests that T cell responses in these subjects were directed at ORFs other than the 16 we selected and/or that T cells directed at these 16 ORFs were below the level of detection in the ELISPOT assay.
We have designed this ELISPOT assay to detect both CD4 and CD8 epitopes from ex vivo stimulated PBMC in the context of a candidate HSV-2 vaccine trial that will measure vaccine-induced T cell responses longitudinally pre-vaccination and multiple time points post-vaccination when total PBMC are limiting. The most efficient way to conserve PBMC is to employ the ELISPOT assay to screen for positive HSV-2 peptide pools and subsequently, use the HSV-2 peptide pools that are positive in the ELISPOT assay to stimulate PBMC in an ICS/flow cytometry assay to define the phenotype of the responding T cells. If PBMC are not limiting, the ICS assay may be preferable because it provides both phenotype and quantitation but requires 2.5 fold more PBMC than the ELISPOT assay (17). If a vaccine is designed to exclusively elicit CD4 or CD8 T cells, the sensitivity of the ELISPOT assay would be increased if the non-responding T cell subset is depleted prior to peptide stimulation although this would require significantly greater numbers of PBMC from each blood draw that may not be achievable based on the IRB standards for maximal blood draw from study subjects. Thus, the choice of T cell assays to assess the immunogenicity of a candidate HSV-2 vaccine depends upon the combination of (1) the T cell subset(s) to be analyzed, (2) the total number of blood draws to be assayed, and, (3) the total amount of blood available at each blood draw to be assayed during the vaccine trial.
It was notable that 40% of HSV-1+/2− subjects possessed T cells recognizing HSV-2 peptides due to the significant sequence homology between these 2 alpha-herpesviruses. Numerous studies have documented type-common T cell epitopes between HSV-1 and HSV-2 (21–24). These observations suggest that the appropriate negative control population for therapeutic vaccine studies is HSVneg subjects as opposed to HSV-1+/2− subjects who would likely have frequent cross-reacting T cell responses in PBMC obtained from baseline blood draws. Considering that HSV-2+ subjects had an 85% response rate to the HSV-2 peptides compared to the 40% response rate in HSV-1+/2− subjects, a significant number of epitopes must be HSV-2 type-specific. This hypothesis is further supported by the observation that more HSV-2+ subjects have mid-level T cell responses (in terms of magnitude; Figure 1) than HSV-1+/2− subjects, a phenomenon possibly due to the fact that while HSV-2+ subjects possess T cells that can respond to type-common and type-specific T cell epitopes contained within the HSV-2 peptide pools, T cells from HSV-1+/2− subjects can only recognize type-common epitopes. It is possible that HSV-1+/2− subjects who lack T cells directed at epitopes that cross-react with HSV-2 are more susceptible to HSV-2 infection. As such, therapeutic vaccines designed to prevent HSV-2 reactivation may not prevent HSV-1 reactivation unless sufficient type-common epitopes are contained in the candidate vaccine.
Of note were the significantly higher rates of CMV seropositivity and concomitant IFN-γ ELISPOT response rates to CMV in HSV-infected subjects versus HSVneg subjects. HSV-1+/2− and HSV-2+ subjects had CMV response rates of 60% and 73%, respectively, compared to a 20% response rate to CMV in HSVneg subjects. These data suggest that subjects infected with HSV-1 or HSV-2 were more likely to be seropositive to CMV and thus have acquired immunity to CMV than subjects who were HSVneg. These data are supported by a study by Zajocova et al. showing an association between seroprevalence to CMV and HSV-1 suggesting common factors related to exposure or susceptibility (25).
Interestingly, 2 subjects seronegative for HSV-1 and HSV-2 possessed positive IFN-γ ELISPOT responses to a single HSV-2 peptide pool; one subject responded to ICP22 while and one subject responded to ICP4. While these responses may be false positives since they were in the lowest range of the positive responses, it is possible that they are bona fide HSV-specific T cell responses. These subjects may be similar to those we have termed Immune Seronegative (IS) or those subjects who (1) are seronegative to HSV-1 and HSV-2, (2) have frequent exposure to HSV-2 from infected sexual partners, (3) have no clinical or virological evidence of HSV infection and/or disease, and (4) possess durable HSV-specific CD4 and CD8 T cell responses (17, 26). These subjects may share similar immunologic characteristics with those HIV-seronegative persons who appear to be resistant to HIV-1 even in the face of multiple high-risk exposures (27–29). The presence of T cell responses in some HSVneg subjects underscores the importance of quantitating pre-vaccination T cell responses in HSVneg vaccinees to determine whether T cells were induced by vaccination or induced by exposure to HSV-2 from infected partners.
In conclusion, we have designed a sensitive, specific and quantitative IFN-γ ELISPOT assay that can detect CD4 and CD8 T cell responses directed at HSV-2 peptides using unfractionated PBMC in the absence of exogenously added autologous DC making it suitable for the analysis of cellular immune responses elicited by candidate HSV-2 vaccines. Analyzing the T cell responses induced by HSV-2 vaccines will determine the contribution of T cell responses to vaccine efficacy.
Acknowledgments
We thank Steven De Rosa and Helen Horton for expert advice on IFN-γ ELISPOT and ICS and flow cytometry. We thank Michael Remington for subject recruitment and for obtaining blood from study participants. We thank Kerry Laing for supplying peptides to UL19, UL25, UL35, UL46, UL47, UL49, UL11, UL27, UL29 and US5. For excellent data management, we thank Stacy Selke and Linda Drolette. We thank Selin Caka for subject scheduling.
Role of the Funding Source
This work was supported by the National Institutes of Health grants AI-049394, AI-030731, AI-042428, AI-083418 and AI-071113. Research support was provided by Pfizer (to CMP) and GlaxoSmithKline (AW).
Footnotes
Contributors
CMP contributed to the conception and design of the study, analysis and interpretation of data, and drafted the article. ASM contributed to the design of the study, analysis and interpretation of the data and revision of the manuscript. DEM and LZ contributed to the acquisition and analysis of the data. AW contributed to the acquisition of data and revision of the manuscript. LC contributed to the conception and design of the study and revision of the manuscript. All authors have approved the final version of the manuscript for submission.
Conflict of Interest
CMP has received funding from Pfizer. LC and CMP are co-inventors on patents owned by the University of Washington concerning HSV-2 vaccines that are unrelated to the subject of this submission. AW has received grant support from GlaxoSmithKline and consulting fees from AiCuris. ASM has received consulting fees from Immune Design Corp.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Xu F, Sternberg MR, Kottiri BJ, McQuillan GM, Lee FK, Nahmias AJ, Berman SM, Markowitz LE. Trends in herpes simplex virus type 1 and type 2 seroprevalence in the United States. JAMA. 2006;296:964–973. doi: 10.1001/jama.296.8.964. [DOI] [PubMed] [Google Scholar]
- 2.Gupta R, Warren T, Wald A. Genital herpes. Lancet. 2007;370:2127–2137. doi: 10.1016/S0140-6736(07)61908-4. [DOI] [PubMed] [Google Scholar]
- 3.Fleming DT, McQuillan GM, Johnson RE, Nahmias AJ, Aral SO, Lee FK, Louis MES. Herpes simplex virus type 2 in the United States, 1976 to 1994. NEJM. 1997;337:1105–1111. doi: 10.1056/NEJM199710163371601. [DOI] [PubMed] [Google Scholar]
- 4.Leone P, Fleming DT, Gilsenan AW, Li L, Justus S. Seroprevalence of herpes simplex virus-2 in suburban primary care offices in the United States. Sex Transm Dis. 2004;31:311–316. doi: 10.1097/01.olq.0000123651.84697.d6. [DOI] [PubMed] [Google Scholar]
- 5.Freeman EE, Weiss HA, Glynn JR, Cross PL, Whitworth JA, Hayes RJ. Herpes simplex virus 2 infection increases HIV acquisition in men and women: systematic review and meta-analysis of longitudinal studies. AIDS. 2006;20:73–83. doi: 10.1097/01.aids.0000198081.09337.a7. [DOI] [PubMed] [Google Scholar]
- 6.Corey L, Wald A, Celum CL, Quinn TC. The effects of herpes simplex virus-2 on HIV-1 acquisition and transmission: a review of two overlapping epidemics. J Acquir Immune Defic Syndr. 2004;35:435–445. doi: 10.1097/00126334-200404150-00001. [DOI] [PubMed] [Google Scholar]
- 7.Kimberlin DW. Neonatal herpes simplex infection. Clin Microbiol Rev. 2004;17:1–13. doi: 10.1128/CMR.17.1.1-13.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanberry L, Spruance S, Cunningham A, Bernstein D, Mindel A, Sacks S, Tyring S, Aoki F, Slaoui M, Denis M, Vancepapeliere P, Dubin G GHVES Group. Glycoprotein-D-adjuvant vaccine to prevent genital herpes. NEJM. 2002;347:1703–1705. doi: 10.1056/NEJMoa011915. [DOI] [PubMed] [Google Scholar]
- 9.Cattamanchi A, Posavad CM, Wald A, Baine Y, Moses J, Higgins TJ, Ginsberg R, Ciccarelli R, Corey L, Koelle DM. Phase I study of a DNA vaccine for herpes simplex virus type 2 (HSV-2) in healthy HSV-2 seronegative adults by a needle-free injection system. Clin Vaccine Immunol. 2008;15:1638–1643. doi: 10.1128/CVI.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koelle DM, Magaret A, McClurkan CL, Remington ML, Warren T, Teofilovici F, Wald A. Phase I dose-escalation study of a monovalent heat shock protein 70-herpes simplex virus type 2 (HSV-2) peptide-based vaccine designed to prime or boost CD8 T-cell responses in HSV-naive and HSV-2-infected subjects. Clin Vaccine Immunol. 2008;15:773–782. doi: 10.1128/CVI.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrow RA, Friedrich D. Inaccuracy of certain commercial enzyme immunoassays in diagnosing genital infections with herpes simplex virus types 1 or 2. Am J Clin Pathol. 2003;120:839–844. doi: 10.1309/AKWH-F62Y-YJ05-4HA6. [DOI] [PubMed] [Google Scholar]
- 12.Morrow RA, Friedrich D, Meier A, Corey L. Use of “biokit HSV-2 Rapid Assay” to improve the positive predictive value of Focus HerpeSelect HSV-2 ELISA. BMC Infect Dis. 2005;5:84. doi: 10.1186/1471-2334-5-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Limaye AP, Kirby KA, Rubenfeld GD, Leisenring WM, Bulger EM, Neff MJ, Gibran NS, Huang ML, Santo Hayes TK, Corey L, Boeckh M. Cytomegalovirus reactivation in critically ill immunocompetent patients. JAMA. 2008;300:413–422. doi: 10.1001/jama.300.4.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bull M, Lee D, Stucky J, Chiu YL, Rubin A, Horton H, McElrath MJ. Defining blood processing parameters for optimal detection of cryopreserved antigen-specific responses for HIV vaccine trials. J Immunol Methods. 2007;322:57–69. doi: 10.1016/j.jim.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosken N, McGowan P, Meier A, Koelle DM, Sleath P, Wagener F, Elliott M, Grabstein K, Posavad C, Corey L. Diversity of the CD8+ T-cell response to herpes simplex virus type 2 proteins among persons with genital herpes. J Virol. 2006;80:5509–5515. doi: 10.1128/JVI.02659-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun RP, Payne LG, Dong L. Characterization of the IFN-gamma T-cell responses to immediate early antigens in humans with genital herpes. Virol J. 2006;3:54. doi: 10.1186/1743-422X-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Posavad CM, Remington M, Mueller DE, Zhao L, Magaret AS, Wald A, Corey L. Detailed characterization of T cell responses to herpes simplex virus-2 in immune seronegative persons. J Immunol. 2010;184:3250–3259. doi: 10.4049/jimmunol.0900722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Currier JR, Kuta EG, Turk E, Earhart LB, Loomis-Price L, Janetzki S, Ferrari G, Birx DL, Cox JH. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. J Immunol Methods. 2002;260:157–172. doi: 10.1016/s0022-1759(01)00535-x. [DOI] [PubMed] [Google Scholar]
- 19.Moodie Z, Huang Y, Gu L, Hural J, Self SG. Statistical positivity criteria for the analysis of ELISpot assay data in HIV-1 vaccine trials. J Immunol Methods. 2006;315:121–132. doi: 10.1016/j.jim.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 20.Horton H, Thomas EP, Stucky JA, Frank I, Moodie Z, Huang Y, Chiu YL, McElrath MJ, De Rosa SC. Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J Immunol Methods. 2007;323:39–54. doi: 10.1016/j.jim.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim M, Taylor J, Sidney J, Mikloska Z, Bodsworth N, Lagios K, Dunckley H, Byth-Wilson K, Denis M, Finlayson R, Khanna R, Sette A, Cunningham AL. Immunodominant epitopes in herpes simplex virus type 2 glycoprotein D are recognized by CD4 lymphocytes from both HSV-1 and HSV-2 seropositive subjects. J Immunol. 2008;181:6604–6615. doi: 10.4049/jimmunol.181.9.6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koelle DM, Abbo H, Peck A, Ziegweid K, Corey L. Direct recovery of herpes simplex virus (HSV)-specific T lymphocyte clones from recurrent genital HSV-2 lesions. J Inf Dis. 1994;169:956–961. doi: 10.1093/infdis/169.5.956. [DOI] [PubMed] [Google Scholar]
- 23.Posavad CM, Koelle DM, Corey L. High frequency of CD8+ cytotoxic T-lymphocyte precursors specific for herpes simplex viruses in persons with genital herpes. J Virol. 1996;70:8165–8168. doi: 10.1128/jvi.70.11.8165-8168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tigges MA, Koelle D, Hartog K, Sekulovich RE, Corey L, Burke RL. Human CD8+ herpes simplex virus-specific cytotoxic T-lymphocyte clones recognize diverse virion protein antigens. J Virol. 1992;66:1622–1634. doi: 10.1128/jvi.66.3.1622-1634.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zajacova A, Dowd JB, Aiello AE. Socioeconomic and Race/Ethnic Patterns in Persistent Infection Burden Among U.S. Adults. J Gerontol: Med Sci. 2009;64A:272–279. doi: 10.1093/gerona/gln012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Posavad CM, Wald A, Hosken N, Huang ML, Koelle DM, Ashley RL, Corey L. T cell immunity to herpes simplex viruses in seronegative subjects: silent infection or acquired immunity? J Immunol. 2003;170:4380–4388. doi: 10.4049/jimmunol.170.8.4380. [DOI] [PubMed] [Google Scholar]
- 27.Clerici M, Giorgi JV, Chou CC, Gudeman VK, Zack JA, Gupta P, Ho HN, Nishanian PG, Berzofsky JA, Shearer GM. Cell-mediated immune response to human immunodeficiency virus (HIV) type 1 in seronegative homosexual men with recent sexual exposure to HIV-1. J Inf Dis. 1992;165:1012–1019. doi: 10.1093/infdis/165.6.1012. [DOI] [PubMed] [Google Scholar]
- 28.Mazzoli S, Trabattoni D, Caputo SL, Piconi S, Ble C, Meacci F, Ruzzante S, Salvi A, Semplici F, Longhi R, Fusi ML, Tofani N, Biasin M, Villa ML, Mazzotta F, Clerici M. HIV-specific mucosal and cellular immunity in HIV-seronegative partners of HIV-seropositive individuals. Nat Med. 1997;3:1250–1257. doi: 10.1038/nm1197-1250. [DOI] [PubMed] [Google Scholar]
- 29.Rowland-Jones SL, Nixon DF, Aldhous MC, Gotch F, Ariyoshi K, Hallam N, Kroll JS, Froebel K, McMichael A. HIV-specific cytotoxic T cell activity in an HIV-exposed but uninfected infant. Lancet. 1993;341:860–861. doi: 10.1016/0140-6736(93)93063-7. [DOI] [PubMed] [Google Scholar]