Abstract
Epstein-Barr virus nuclear antigen 2 (EBNA2) regulation of transcription through the cell transcription factor RBPJ is essential for resting B-lymphocyte (RBL) conversion to immortal lymphoblast cell lines (LCLs). ChIP-seq of EBNA2 and RBPJ sites in LCL DNA found EBNA2 at 5,151 and RBPJ at 10,529 sites. EBNA2 sites were enriched for RBPJ (78%), early B-cell factor (EBF, 39%), RUNX (43%), ETS (39%), NFκB (22%), and PU.1 (22%) motifs. These motif associations were confirmed by LCL RBPJ ChIP-seq finding 72% RBPJ occupancy and Encyclopedia Of DNA Elements LCL ChIP-seq finding EBF, NFκB RELA, and PU.1 at 54%, 31%, and 17% of EBNA2 sites. EBNA2 and RBPJ were predominantly at intergene and intron sites and only 14% at promoter sites. K-means clustering of EBNA2 site transcription factors identified RELA-ETS, EBF-RUNX, EBF, ETS, RBPJ, and repressive RUNX clusters, which ranked from highest to lowest in H3K4me1 signals and nucleosome depletion, indicative of active chromatin. Surprisingly, although quantitatively less, the same genome sites in RBLs exhibited similar high-level H3K4me1 signals and nucleosome depletion. The EBV genome also had an LMP1 promoter EBF site, which proved critical for EBNA2 activation. LCL HiC data mapped intergenic EBNA2 sites to EBNA2 up-regulated genes. FISH and chromatin conformation capture linked EBNA2/RBPJ enhancers 428 kb 5′ of MYC to MYC. These data indicate that EBNA2 evolved to target RBL H3K4me1 modified, nucleosome-depleted, nonpromoter sites to drive B-lymphocyte proliferation in primary human infection. The primed RBL program likely supports antigen-induced proliferation.
Keywords: leukemia, lymphoma, Notch, development
Epstein Barr virus (EBV) infection is highly prevalent in all human populations. EBV is also an important cause of endemic Burkitt Lymphoma (1), Hodgkin Lymphoma (2), and lymphoproliferative disorders in immune-suppressed (3) and HIV-infected people (4). In early primary human infection, EBV infects peripheral resting B lymphocytes (RBLs) and expresses six nuclear (Epstein-Barr virus nuclear antigen, EBNA) and two integral membrane (LMP) proteins. EBNAs and LMPs convert RBLs into proliferating lymphoblastoid cells (LCLs), which are malignant in the absence of effective T-cell responses. Because EBNAs and LMPs include >4,000 amino acids, T-cell immune responses normally eliminate most EBV-infected cells. Lymphocytes with down-regulated EBV protein expression persist in tonsils and lymph nodes and are reservoirs for reactivated EBV infection (4). EBV conversion of RBLs into LCLs in vitro (5) is a relevant and experimentally useful model for EBV proliferative effects in B-lymphocytes.
In converting RBLs to LCLs, EBNA2 and EBNALP are expressed first. EBNA2 up-regulates EBV and cell gene expression, including EBV LMP1 and cell MYC, CD23, and CD21 (6–9). EBNA2 associates with DNA through RBPJ, which also mediates Notch binding to DNA (10, 11). The B-lymphocyte and macrophage lineage ETS protein, PU.1, is also important in EBNA2 activation of the EBV LMP1 promoter (12, 13). The EBNA2 C-terminal acidic domain recruits basal and activating transcription factors (TFs), including p300/CBP and PCAF Histone acetyl transferases (14–16). EBNA2 transactivation of MYC causes continuous B-cell proliferation. EBNA2 transactivation of EBV LMP1 results in NFκB and MAP kinase activations, which up-regulate prosurvival BCL2 family gene expression (17, 18). Overall, EBV conversion of RBLs to LCLs mimics antigen-induced RBL clonal expansion in lymph node germinal centers, where antigen binding to surface Ig (sIg) induces MYC-mediated proliferation (19) and T cell-expressed CD40 ligand activates B-lymphocyte CD40 receptors to up-regulate NFκB, MAP kinases, and antiapoptotic BCL2 family protein expression (20).
A major problem in understanding the mechanisms through which EBNA2 and RBPJ regulate gene expression is the finding that an RBPJ binding motif (G)TGGGAA(A) near a promoter is a poor predictor of EBNA2 dependence (21). EBNA2 and RBPJ ChIP deep-sequencing (ChIP-seq) and bioinformatic analyses (22, 23) were therefore undertaken to identify and characterize EBNA2 and RBPJ binding sites, genome-wide, in LCLs.
Results
EBNA2 and RBPJ Binding Sites in LCLs.
Independent, duplicate large-scale, EBNA2 and RBPJ ChIPs were deep-sequenced and 107 EBNA2 or RBPJ DNA reads were mapped to the human genome (hg.18) with 36% to 54% efficiency. After normalization for input DNA, QUEST (24) identified 5,151 EBNA2 and 10,529 RBPJ sites, with bidirectional sequence reads and a false-discovery rate (FDR) < 0.01 (Fig. S1A). EBNA2 and RBPJ, sites were similarly distributed relative to annotated genes. Only 14% of EBNA2 and 13% of RBPJ sites were at promoters, defined as ±2 kb of a transcription start site (TSS). Instead, EBNA2 and RBPJ were 86% to 87% at more distal sites, of which 42% to 43% were intergene sites, 8% to 9% UTR, 34% to 35% introns, and 1% exons (Fig. S1A). Overall, 72% (3,710) of EBNA2 sites were within 100 b of a significant RBPJ site (hereafter, EBNA2/RBPJ site) (Fig. S1B). The 1,441 EBNA2 sites lacking significant RBPJ signal are hereafter EBNA2-only sites and the 6,819 RBPJ sites lacking EBNA2 are RBPJ-only sites. The finding of 1,441 EBNA2 sites without significant RBPJ signal may be a result of RBPJ being occluded from antibody recognition, less stable RBPJ DNA association, or EBNA2 tethering to DNA by another transcription factor.
RBPJ at EBNA2/RBPJ Sites Is More Highly DNA-Associated.
On average, at nonpromoter, putative enhancer EBNA2/RBPJ sites, RBPJ had 34 DNA reads per kilobase per million aligned reads (hereafter, signals) versus 10 signals per RBPJ-only site; the difference was highly significant (P < 10−10) (Fig. S2). Similarly, albeit to a lesser extent, at promoter sites, EBNA2/RBPJ sites had 24 signals versus 11 signals for RBPJ-only sites (P < 10−10) (Fig. S2). Overall, EBNA2/RBPJ had more RBPJ signals than RBPJ-only sites genome-wide, consistent with the previous finding that EBNA2 significantly increased RBPJ association with a limited number of enhancer or promoter DNA sites (25).
EBNA2 Sites Are Enriched for RBPJ, ETS, EBF, RUNX, PU.1, and NFκB Motifs.
Analysis of a 500-bp window around EBNA2 sites for Transfac database motif enrichment identified significant (P < 10−100) enrichment not only for RBPJ (78%), but also for ETS (39%), EBF (39%), RUNX (43%), PU.1 (22%), and NFκB (22%) motifs, over a control set of sequences with similar GC content. These data are consistent with EBF, RELA, and PU.1 having significant affects on EBNA2 or RBPJ binding and transcription regulation. (Fig. 1A and Table S1).
Fig. 1.
EBNA2 binding site enriched TF motifs, K-means TF clusters, and associated H3K4me1 signals in LCLs and at the same genome sites in RBLs. (A) EBNA2 binding site (±250 bp) enriched TF motifs are ranked by enrichment over random sequences controlling for GC content from top to bottom. All had corrected P values <10−50. (B) EBNA2 binding sites (5,151) were K-means clustered based on the presence of RBPJ, EBF, PU.1, and NFκB subunit RELA binding by LCL ChIPseq and Homer-imputed ETS and RUNX motifs. EBNA2 site clusters are named by the principal TF components. (C) LCL EBNA2 site clusters have distinct H3K4me1 signal distributions (Left). H3K4me1 distributions at the same sites in RBLs (Right). The numbers in the upper left corners of the panels are the total signals (reads per kilobase per million mapped reads) under each curve.
To evaluate the extent to which EBNA2-associated TF site predictions correlate with TF occupancy, we used our RBPJ and ENCODE (Encyclopedia Of DNA Elements) ChIP-seq GM12878 LCL EBF, NFκB subunit RELA, and PU.1 data (26) to evaluate RBPJ, EBF, PU.1, and RELA occupancy at EBNA2 sites. EBF mapped to 27,552, RelA to 24,796, and PU.1 to 24,343 LCL sites. EBNA2 sites were 54% within 100 bp of an EBF site, 31% of a RELA site, and 17% of a PU.1 site (Fig.1B). Overall, the EBNA2-associated TF motif analysis correlated surprisingly well with TF presence by ChIP-Seq. For EBNA2 sites, RBPJ was predicted at 78% and detected at 72%. EBF was predicted at 39% and detected at 54%. PU.1 was predicted 22% and detected 17%. NFκB RELA was predicted at 22% and was detected at 31%. Thus, EBNA2 and RBPJ accessed sites that were or had been highly accessible to the EBNA2 associated TFs.
EBNA2/RBPJ, EBNA2-only, and RBPJ-only sites were associated with different TF complexes. EBF was strongly enriched at EBNA2/RBPJ sites (60%) compared with EBNA2-only sites (36%) or RBPJ-only sites (37%) (P < 10−10). In contrast, ETS motifs were enriched at EBNA2-only sites (45%) compared with EBNA2/RBPJ or RBPJ-only sites (35%) (P < 10−10). P300 sites strongly correlated with EBNA2/RBPJ and EBNA2-only sites, but were significantly less at RBPJ-only sites (P < 10−10) (Table S2). The 262 EBNA2-only sites with the smallest RBPJ signals were even more enriched for ETS motifs (57%), consistent with the possibility that ETS or an ETS-associated factor may tether EBNA2 to these sites.
EBNA2 Cofactors Correlate with Distinct Chromatin Footprints.
K-means clustering of the 5,151 EBNA2 sites for associated factors identified six clusters with distinct cofactor combinations of RBPJ, EBF, RELA, PU.1, and imputed ETS and RUNX (Fig. 1B). EBNA2 site clusters were named by their principal cofactors. RBPJ was a major component of all clusters. One cluster was comprised of EBNA2 sites in which RBPJ was the only dominant component and is referred to as RBPJ. RELA-ETS, EBF-RUNX, EBF, ETS, RBPJ, and RUNX clusters correlated with most (Fig. 1B, Top) to least (Fig. 1B, Bottom) ENCODE LCL H3K4me1 signals within ±2 kb of EBNA2 sites (Fig. 1C, Left). Differences in H3K4me1 signals between RELA-ETS and EBF-RUNX, EBF and ETS, ETS and RBPJ, and RBPJ and RUNX were each highly statistically significant (P < 10−6 Mann-Whitney U test). Importantly, H3K4me1-enriched clusters, characteristic of RELA-ETS, EBF-RUNX, EBF, and ETS, had fewer H3K4me1 signals at the EBNA2 site, indicative of EBNA2 localization at nucleosome-depleted, open chromatin sites (Fig. 1C, Left) (27).
Significant p300 signals, indicative of active transcription, also correlated with H3K4me1 signals, as noted (28, 29). EBNA2 clusters with RelA-ETS, EBF-RUNX, EBF, ETS, RBPJ, and RUNX had 41%, 37%, 30%, 15%, 6%, and 5% of sites with significant p300 signals (Table S3). These correlative data further support the hypothesis that RELA-ETS, EBF-RUNX, EBF, ETS, RBPJ, and RUNX clusters rank from highest to lowest in transcription activation.
The higher H3K4me1 and p300 association of EBNA2 with EBF-RUNX versus EBF alone (Fig. 1C, Left, upper blue and green curves) and the very low H3K4me1 effect of RUNX alone (Fig. 1C, Left, lower black curve) correlate an activating-RUNX effect with EBF and a repressive-RUNX effect unassociated with EBF, at other sites. Opposing RUNX isoforms have been described in LCLs, where RUNX3 repression of RUNX1 is important for continued growth (30).
As a complement to clustering, which detected an EBF-associated activating-RUNX effect and a repressive-RUNX effect, an additive linear model was used to correlate H3K4me1 signal levels at EBNA2 sites with the presence of EBF, RELA, PU.1, or imputed ETS or RUNX as single factors. EBF increased H3K4me1 signals 1.68-fold, RELA 1.22-fold, PU.1 1.21-fold, and imputed ETS 1.08-fold. Each effect was highly significant (P < 0.0001) (Table S4). RUNX as a single factor had no significant overall effect, consistent with the positive RUNX effect with EBF being similar in magnitude to the negative RUNX effect without EBF.
Ontogeny of H3K4me1 Modifications.
To understand how EBV infection alters RBL transcription, H3K4me1 modifications at EBNA2 sites in LCLs were compared with H3K4me1 modifications at the same sites in RBLs. Roadmap Epigenomics Mapping Center (31) ChIP-seq H3K4me1 signals in CD19(+) RBLs (Fig. 1C, Right) were compared with H3K4me1 signals at EBNA2 sites in LCLs (Fig. 1C, Left). Although H3K4me1 signals were less elevated in RBLs, they mirrored the symmetrical elevation with central depletion seen in LCLs and followed the LCL cofactor cluster site hierarchy. For example, the EBNA2, RELA, and imputed ETS cluster in LCLs had 44 H3K4me1 signals, whereas the same sites in RBLs had 39 H3K4me1 signals (Fig. 1C Left vs. Right). Importantly, H3K4me1 signal attenuation at the EBNA2 binding sites, evidence of nucleosome depletion, was also evident at the same sites in RBLs, indicating that EBNA2/EBPJ targeted nucleosome-depeleted sites. These data strongly support a model in which EBNA2 targets H3K4me1-modified and nucleosome-depleted RBL RELA-ETS, EBF-RUNX, EBF, and ETS sites for transcription up-regulation.
Differences Between EBNA2 Promoter and Putative-Enhancer Effects.
In contrast to LCL enhancer sites where RELA-ETS, EBF-RUNX, and EBF were associated with higher H3K4me1 signals (P < 0.01) versus ETS, RBPJ, and repressive RUNX (Fig. 1C, Left), and as a single-factor EBF had the largest up-regulatory effect (Table S4), at promoter sites, EBNA2 and RELA-ETS, ETS, and RBPJ clusters were associated with higher H3K4me3 signals and RUNX, EBF-RUNX, and RBPJ clusters were associated with higher H3K27me3 signals (Fig.S3 A and B, and Table S3). Furthermore, RELA-ETS and ETS complexes were more frequently at promoter sites, 29% and 18%, than other TF complexes, which were 7% to 13% at promoter sites. Moreover, these complexes at promoters were associated not only with higher H3K4me3 and lower H3K27me3 signals, but were also associated with higher P300 and highest PolII signals. These data indicate that ETS has the largest transcriptional up-regulatory effects at promoter sites.
EBF Is Critical for EBNA2 Activation of the EBV LMP1 Promoter.
EBF is essential for B-lymphocyte development and regulates gene expression from pro-B through mature B-cell stages (32). EBF may therefore be a “pioneering” factor that regulates EBNA2 access to chromatin or effects on gene expression (33), given the 54% colocalization of EBNA2 with EBF in LCLs, the embedding of RBPJ TGGGA(A) core motifs in a significant fraction of EBF motifs, and the similarity of H3K4me1 at EBNA2 sites in LCLs with the same sites in RBLs (Fig. 1C).
The hypothesis that EBF is a pioneer B-lymphocyte transcription factor that opens chromatin for EBNA2 and RBPJ binding was further evaluated by scanning EBNA2 ChIP-seq RBPJ binding sites through a 500-bp window around the EBNA2/RBPJ site to identify the 8-bp DNA sequence that has the strongest in vitro affinity for RBPJ, based on a RBPJ protein binding matrix (34). At 436 EBNA2-bound sites, high-affinity RBPJ binding sites were in an EBF motif. These data are consistent with a model in which these sites evolved to support EBF and RBPJ access, perhaps at different points in B-cell development, cell cycle, or tissue localization.
The hypothesis that EBF can be critical for EBNA2-mediated transcription was evaluated in the context of the EBV LMP1 promoter, which is EBNA2-, RBPJ-, and PU.1-dependent (12, 13, 35). Because the LMP1 promoter RBPJ, PU.1, and EBF sites are nonoverlapping (Fig. 2A), independent null EBF, RBPJ, or PU.1 mutations could be readily evaluated. In BJAB lymphoma cells, EBNA2 activated the (−335 to +40) LMP1 promoter eightfold over control expression vector. A single point mutation in the PU.1 site reduced EBNA2 activation to background (Fig. 2B). Mutation of the EBF motif abolished EBF binding and reduced EBNA2 activation from eight- to twofold (Fig. 2 B and C), similar to the RBPJ binding-site mutation effect (12). These data indicate that EBF is important for LMP1 promoter activation.
Fig. 2.
EBF is important for EBNA2 activation of the EBV LMP1 promoter. (A) Schematic of the LMP1 promoter EBNA2 response element (−227/−137), with RBPJ, EBF, and PU.1 binding sites underlined in the continuous sequence. Null EBF or PU.1 point mutations are indicated below the continuous sequence. (B) EBNA2 activated the LMP1 promoter-luciferase reporter eightfold, the EBF mutant site twofold, and the PU.1 mutant site onefold in BJAB cells. Luciferase activities were normalized to cotransfected EBNA2-independent β-galactosidase activity. EBNA2 was expressed at similar levels in all experiments as shown in the EBNA2 immune blot at the bottom of B. (C) In vitro translated EBF bound to a 32P labeled oligonucleotide that included the EBF binding site. Binding was completed by a 500-fold excess cold wild-type oligonucleotide (w), but not by 500-fold cold mutant nucleotide (m). Protein-DNA complexes were separated by PAGE and visualized by phosphoimager.
EBNA2 Up-Regulated Genes Are Enriched for EBNA2 Binding >2 kbp from the TSS.
EBNA2 was predominantly (86%) at nonpromoter sites (Fig. S1). Nonpromoter EBNA2 sites correlated with higher LCL RNA expression levels in four gene sets (Fig. S4A). A conditional EBNA2-regulated 81 gene set had 2.4 EBNA2 sites per gene distributed within 2 to 152 kb of the TSS (Fig. S4A) (21). A second gene set of RNAs that are >5 up-regulated during RBL conversion to LCLs had 1.2 EBNA2 sites per gene distributed within 2 to 152 kb of the TSS (9). Genes two- to fivefold up-regulated during RBL conversion to LCLs had 1.1 EBNA2 sites evenly split within 2 to 152 kb of the TSS. The control gene set, composed of all other genes, had 0.8 EBNA2 sites that were also evenly distributed within 2 to 152 kb of the TSS. These data indicate that EBNA2 binding to distal enhancer sites correlates with up-regulated LCL gene expression (P < 0.01).
Multiple EBNA2 Sites Have Long-Range Interactions with Promoters of the 81 EBNA2-Regulated Gene Set.
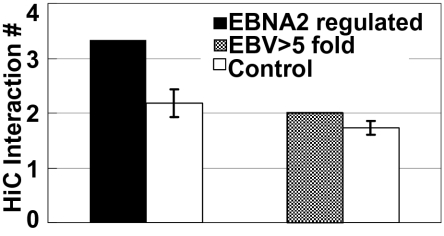
Chromatin conformation capture (3C) followed by deep-sequencing (HiC) from GM06990 LCLs revealed paired-end sequences representative of both long-range genome interaction components (36). Many LCL HiC sequences mapped with one end within ±2 kb of a nonpromoter EBNA2 site and the other end within −1 to +5 kb of the TSS of 56 EBNA2-regulated genes. Excluding EBNA2 sites <5 kb from the TSS, 3.3 different long-range interactions were detected per EBNA2-regulated gene. The EBNA2-regulated gene set was significantly enriched (corrected P < 10−4) over a control gene set, with similar expression level and distances from EBNA2 sites to the TSS, but not conditionally EBNA2-affected (Fig. 3). Overall, 61% of the interactions were within the same chromosome and 39% were interchromosomal. For intrachromosome interactions, the median distance from EBNA2 enhancer to an affected gene was 330 kb, with 90% >150 kb. These imputed EBNA2 enhancer sites overlapped 66% with an EBF site and 30% had significant p300 signals, both significantly enriched over the 54% of EBNA2 sites that have an EBF site (corrected P < 0.01) and the 22% of EBNA2 sites with significant p300 signals (corrected P < 0.05). The >fivefold up-regulated LCL RNA dataset was also enriched for interactions, with ∼2 different HiC interactions per gene (corrected P < 0.05) (Fig. 3). Interactions of EBNA2 enhancer sites with the EBNA2 regulated or >fivefold up-regulated gene sets peaked within ±2 kb of the EBNA2 binding site. EBNA2 binding-site interactions were progressively less frequent within neighboring 4-kb windows (Fig. S4B). As an additional control, the same procedure was applied to compute HiC interactions between RBPJ sites without EBNA2 and EBNA2-regulated genes. The number of detected interactions was significantly fewer than interactions with EBNA2 sites (corrected P < 0.01) (Fig. S4C). These data indicate that EBNA2 enhancers are significantly enriched for distal target interactions with EBNA2 regulated or fivefold up-regulated genes.
Fig. 3.
Nonpromoter EBNA2 binding sites are enriched for long-range interactions with EBNA2 regulated or LCL >fivefold up-regulated. HiC detected 3.2 long-range interactions between intergenic EBNA2 binding sites and EBNA2 conditionally regulated genes versus control genes (corrected P < 0.0001) and two long-range interactions between EBV >fivefold up-regulated genes (corrected P < 0.05). Control genes had similar RNA levels and distance to EBNA2 binding sites as the 81 conditionally regulated or >fivefold up-regulated genes.
EBNA2 Up-Regulates MYC Gene Expression Through Long-Range Enhancer and Promoter Looping.
Although strong PolII and H3K9ac signals were evident at the MYC promoter, consistent with known EBNA2 up-regulated MYC expression, EBNA2 and RBPJ LCL ChIP-seq failed to identify significant EBNA2 or RBPJ signals at or within 150 kb of the MYC TSS. Indeed, the nearest, above-background EBNA2 and RBPJ signals were at −168 to −186 kb of the MYC TSS (Fig. 4A and Fig. S5). These sites were associated with significant NFκB RELA, moderate-level H3K9ac and p300, and high-level H3K4me1 signals, consistent with enhancer sites. However, the strongest EBNA2 and RBPJ signals were from 11 significant EBNA2/RBPJ sites at −428 to −556 kb of the MYC TSS. These sites had high EBF, RELA, H3K4me1, and H3K9ac signals, which also coincided with high PolII and p300 signals, indicative of active enhancers (Fig. 4A and Fig. S5). Importantly, EBF and NFκB RELA are the principal EBNA2 enhancer associated up-regulatory TFs. EBNA2 induced-LMP1 constitutively and strongly activates NFκB RELA.
Fig. 4.
EBNA2 activates MYC via long-range enhancer and promoter interactions. (A) CTCF, EBNA2, RBPJ, EBF, RELA, H3K9ac, H3K4me1, PolII, and p300 signals and MYC promoter (arrow) are shown. Normalized signals are at the left end of each track. The asterisk indicates unidirectional EBNA2 reads. The three small dots at −168 kb to −186 kb indicate three significant EBNA2/RBPJ peaks. Major EBNA2/RBPJ peaks are −428 kb to −556 kb. For enlarged view, see Fig. S5B. (B) FISH for IB4 LCL and RBLs. Fixed LCLs or RBLs were hybridized with fluorescent labeled BACmid probes for DNA 428 to 556 kb upstream of MYC, (green) and 150 kb centered around MYC (orange). Cell DNA is stained blue with DAPI. Mycp indicates myc promoter and myce indicates myc enhancer. (C) One-hundred LCLs or RBLs were scored for colocalization of EBNA2 up-stream sites and MYC. Blue indicates the percentage of cells with one allele colocalized and one allele separate. (D) Chromatin conformation capture qPCR assay using conditional EBNA2 LCLs grown under permissive or nonpermissive conditions for 4 d. Cells were cross-linked with formaldehyde, DNA digested with Csp6I, diluted, ligated, and quantitated by Taqman qPCR. Fold differences were determined using the ΔΔCt method with EBNA2-expressing cells set at 1.
Surprisingly, RBL H3K4me1 signals at the principal −428 and −556 MYC regulatory sites were very similar to LCL H3K4me1 (Fig S5A), consistent with the supposition that RBLs are poised for MYC transcription activation and are then coordinately activated by EBNA2/RBPJ, with EBF and RELA (Fig. S5A). Some −428- to −556-kb EBNA2/RBPJ sites also coincided with or were near CTCF signals (Fig S5B). Strong CTCF signals were also evident at +0.2 kb, −1.9 kb, and −10 kb of the MYC TSS. CTCF may biochemically link the −428- and −556-kb sites to the MYC TSS.
To initially investigate the interaction of the −428- and −556-kb sites with MYC, FISH assays with green fluorescent Bacmid probe centered around the 428- to 556-kb enhancer site and an orange fluorescent Bacmid probe centered around MYC were used to study allele colocalization in LCLs versus RBLs. Overall, 94 of 100 LCLs and 92 of 100 RBLs had both alleles discernible. Of these, 43 LCLs and 41 RBLs had one colocalized and one separated allele. In contrast, 49 LCLs and only 1 RBL had two colocalized alleles, whereas 2 LCLs and 50 RBLs had two separated alleles (P < 0.001) (Fig. 4 B and C). In summary, LCLs significantly differed from RBLs in LCL biallelic EBNA2 enhancer site association with MYC.
Conditional EBNA2-expressing LCLs were also used to investigate the effect of EBNA2 inactivation on EBNA2 enhancer site association with MYC. After 4 d in medium permissive (+) or nonpermissive (−) for EBNA2 expression, 96 EBNA2 (+) and 69 EBNA2 (−) LCLs were evaluated. EBNA2 (+) LCLs significantly differed from EBNA2 (−) LCLs in having 62% versus 36% single allele colocalization and in having 34% versus 63% biallele separation. These data support the hypothesis that the 428- to 556-kb upstream EBNA2 enhancer site association with MYC is EBNA2-dependent.
Chromosome conformation capture (37) was used to further investigate the proximity of the −428 kb upstream EBNA2 enhancer site to MYC. After generating the 3C library, PCR was used to detect the −428-kb EBNA2 enhancer site ligated to the MYC promoter or first intron. Sequencing of the PCR product revealed the −428-kb EBNA2 enhancer linked through a Csp61 site to the MYC promoter and first intron. LCLs conditional for EBNA2 expression were used to further investigate conditional EBNA2 effects on the −428-kb EBNA2 enhancer proximity to the MYC first intron. The 3C libraries were made from EBNA2(+) and EBNA2(−) conditional LCLs (37). Taqman qPCR and Taqman probe for the putative EBNA2 enhancer and MYC intron Csp6I DNA fragment ligation junction were used to assess product abundance. EBNA2 (+) LCLs were 7.8-fold enriched for the −428-kb EBNA2 enhancer and MYC intron Csp6I ligation product over EBNA2 (−) LCLs (P < 0.01) (Fig. 4D). These data further support an EBNA2-dependent role for the −428-kb enhancer in MYC regulation.
Discussion
Previous studies of the molecular genetics and pathogenesis of EBV-induced B-cell growth support a model that EBNA2 and LMP1 usurp RBL growth and survival pathways that are normally regulated by RBL B-cell receptor antigen activation and T-cell help (4). Antigen signaling through sIg induces MYC-driven B-lymphocyte proliferation, and B-cell MHC-class II antigen presentation to germinal center CD4+ T cells induces T cell CD40-ligand expression. CD40 ligand in turn induces B-cell NFκB activation and antiapoptotic BCL2 family protein expression, which prevents MYC proliferation-induced cell death (20, 38, 39). EBNA2/RBPJ activation of MYC and of LMP1 provide similar high level MYC and NFκB activation, enabling LCL proliferation and survival.
The investigations described here elucidate the basic mechanisms through which EBNA2, genome-wide, exploits and enhances the RBL transcription program to cause continuous B-cell proliferation. EBNA2 targeted 5,151 RBL open chromatin sites, mostly through RBPJ and EBF. EBNA2, RBPJ, and EBF sites were predominantly distal intergene or intron enhancers. Only 14% were at promoter sites. EBNA2 associated with six different RBPJ, EBF, ETS, RUNX, NFκB RELA, or PU.1 complexes. Specific TF complex compositions determined much of the EBNA2 effect on nucleosome depletion, H3K4me1 signal, and other transcription-associated effects. The key EBF, PU.1, ETS, NFκB RELA, and activating RUNX1 roles in determining EBNA2 and RBPJ B-cell transcription effects is consistent with their pioneering roles in developing and mature B-lymphocyte gene expression. Component analyses detected activating and repressive RUNX effects. RUNX1 is the likely activator because EBF, PU.1, ETS, and RUNX1 are frequently associated with active B-cell enhancers (20, 33, 40, 41).
The most surprising finding is the extent to which EBV, and particularly EBNA2, have evolved to exploit the RBL transcription regulatory framework. Indeed, uninfected RBLs had very similar nucleosome depletion and H3K4me1 chromatin modification at sites subsequently targeted by EBNA2, including sites up-stream of MYC. The RBL sites were poised to activate gene expression in response to antigen stimulation and T-cell help. RBL nucleosome depletion was enabled by and efficiently enabled cell TF access. EBNA2 and RBPJ exploit these accessible sites, with EBNA2 further increasing nucleosome depletion, H3K4me1 signals, and target gene expression. These EBNA2 effects were clearly evident at the MYC locus, where EBNA2 moderately increased activating chromatin signals and markedly up-regulated myc RNA and protein.
Not only has EBNA2 evolved to activate transcription through RBPJ and EBF, but the EBV genome also evolved to require EBF for EBNA2 regulation of the LMP1 promoter. EBF developmentally activates PAX5 and is frequently linked to PAX5 in transcription up-regulatory effects. PAX5 was previously shown to be important for initial transcription from the Wp EBNA promoter in LCLs (42).
We were surprised to note that RBPJ and EBF share core DNA binding sequences and that EBNA2-associated EBF sites are enriched for high affinity RBPJ sites, consistent with site evolution in response to both EBF and RBPJ, at different points in development, cell cycle, hormonal, or tissue-specific effects. Although the structure of EBF dimers on DNA appears to preclude RBPJ association with the embedded EBF motif, RBPJ occupancy with an EBF monomer might be possible (43). Subsequent studies need to investigate the molecular basis for RBPJ and EBF sequential or concomitant occupation of these sites.
Notch also uses RBPJ to regulate MYC and cell-survival gene expression in T-cell progenitors and may have similar roles in thymic T-cell and B-cell marginal zone development (44). Hyperactivated Notch signaling can result in T-cell lymphocytic leukemia. In contrast to EBNA2/RBPJ, which is most frequently distant from affected genes, Notch/RBPJ predominantly binds near affected-gene promoter sites. Similar to EBNA2 targeting of EBF and embedded RBPJ sites in B-cells, Notch frequently associates with ZNF143 and with RBPJ sites embedded in ZNF143 sites in T-cell lymphocytic leukemia cells. However, Notch, RBPJ, and ZNF143 sites were much less associated with activating chromatin marks than EBNA2, RBPJ, and EBF sites, potentially consistent with a role for ZNF143 in distracting or modulating Notch effects. Although EBNA2/RBPJ localized with EBF and RELA to sites 100, 428, and 556 kb up-stream of MYC in LCLs, the strongest Notch/RBPJ site in T-cell lymphocytic leukemia cells was even further up-stream of MYC (45). Of interest in terms of the fundamental mechanism for EBNA2/RBPJ or Notch/RBPJ association with MYC, the 428- and 556-kb MYC promoter sites have nearby CTCF binding sites that could enable chromatin looping and upstream site association with the MYC promoter (46). Long-range enhancer MYC regulation is a characteristic of human colon, prostate, and breast cancer (47).
Interestingly, 31% of LCL EBNA2 sites had significant RELA binding, including the MYC enhancer sites. The role of LMP1 up-regulated RELA in coregulating MYC expression with EBNA2 requires further study. LMP1 activation of canonical and noncanonical NFκB is critical for LCL survival (48, 49). The data presented here indicate that EBNA2 and NFκB RELA have many other targets in common. EBNA2 and LMP1 gene coregulation creates opportunities to investigate the extent to which these transcriptional effects are EBNA2- and LMP1-dependent. Inhibitors of either pathway may have dominant or synthetic lethal effects on LCL growth or survival.
Materials and Methods
ChIP Assays and Sequencing.
IB4 LCL cells were cross-linked with formaldehyde and sonicated. Rabbit EBNA2 or RBPJ antibodies were used to immune precipitate EBNA2 or RBPJ with associated DNA. DNA was purified using a PCR purification column (Qiagen). ChIP-seq DNA libraries were prepared and sequenced, using Genome Analyzer II (Illumina). Reads were mapped to human genome hg18 (National Center for Biotechnology Information, build 36), allowing two mismatches for the first 32 bp.
ChIP-seq Peak Calling, Motif Searching, and K-Means Clustering.
Chip-Seq data and corresponding input control data were processed for Peak significance using QUEST (24) and criteria of >threefold enrichment over input and FDR < 0.01. Binding signals were normalized to reads per kilobase per million mapped reads. HOMER (http://biowhat.ucsd.edu/homer/chipseq/index.html) was used to scan a ± 250-bp region around each binding peak for TF motifs. TF enrichment was assessed using random genome sequences with similar DNA base composition as controls. K-means clustering of all TF binding sites was used to identify clusters.
Fluorescent in Situ Hybridization.
LCL and RBL cells were fixed and hybridized with SpectrumGreen (Abbott Molecular)-labeled BACmid RP11-25204 for the MYC enhancer region and Vysis Bacmid LSI MYC (8q24.12–q24.13) Spectrum Orange Probe (Abbott Molecular) for MYC. Nuclei were stained with DAPI.
Chromosome Conformation Capture and qPCR.
The 3C assays were as previously described (37). Purified library DNA was quantitated by Taqman qPCR, with fold-changes calculated using the ΔΔCt method.
Detailed materials and methods are available SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Richard Myers and Encyclopedia of DNA Elements (ENCODE) for EBF, PU.1, P300, RELA, PolII LCL ChIP-seq data; Drs. Ben Gewurz, Federica DePalma, Mark Fogg, Ellen Cahir-McFarland, Daniel Portal, and Micheal Calderwood for discussion; and Andrew Gagne and Pedro Teixeira for data conversion. This work was supported in part by National Institutes of Health Grants R01CA047006, R01CA131354, R01CA085180, and 1P50 HG004233, ENCODE Grant U54 HG004570, and Roadmap Epigenomics Mapping Center U01 ES017155 (to B.E.B.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. GSE29498).
See Commentary on page 14715.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108892108/-/DCSupplemental.
References
- 1.Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt's lymphoma. Lancet. 1964;1:702–703. doi: 10.1016/s0140-6736(64)91524-7. [DOI] [PubMed] [Google Scholar]
- 2.Johansson B, Klein G, Henle W, Henle G. Epstein-Barr virus (EBV)-associated antibody patterns in malignant lymphoma and leukemia. I. Hodgkin's disease. Int J Cancer. 1970;6:450–462. doi: 10.1002/ijc.2910060316. [DOI] [PubMed] [Google Scholar]
- 3.Ho M, et al. Epstein-Barr virus infections and DNA hybridization studies in posttransplantation lymphoma and lymphoproliferative lesions: The role of primary infection. J Infect Dis. 1985;152:876–886. doi: 10.1093/infdis/152.5.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rickinson A, Kieff E. Epstein-Barr Virus. In: Howley P, Knipe D, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2655–2700. [Google Scholar]
- 5.Henle W, Diehl V, Kohn G, Zur Hausen H, Henle G. Herpes-type virus and chromosome marker in normal leukocytes after growth with irradiated Burkitt cells. Science. 1967;157:1064–1065. doi: 10.1126/science.157.3792.1064. [DOI] [PubMed] [Google Scholar]
- 6.Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181:595–608. doi: 10.1016/0042-6822(91)90893-g. [DOI] [PubMed] [Google Scholar]
- 7.Wang F, Kikutani H, Tsang SF, Kishimoto T, Kieff E. Epstein-Barr virus nuclear protein 2 transactivates a cis-acting CD23 DNA element. J Virol. 1991;65:4101–4106. doi: 10.1128/jvi.65.8.4101-4106.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaiser C, et al. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J Virol. 1999;73:4481–4484. doi: 10.1128/jvi.73.5.4481-4484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikitin PA, et al. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe. 2010;8:510–522. doi: 10.1016/j.chom.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grossman SR, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henkel T, Ling PD, Hayward SD, Peterson MG. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265(5168):92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 12.Johannsen E, et al. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by J kappa and PU.1. J Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laux G, Adam B, Strobl LJ, Moreau-Gachelin F. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-J kappa interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 1994;13:5624–5632. doi: 10.1002/j.1460-2075.1994.tb06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen JI. A region of herpes simplex virus VP16 can substitute for a transforming domain of Epstein-Barr virus nuclear protein 2. Proc Natl Acad Sci USA. 1992;89:8030–8034. doi: 10.1073/pnas.89.17.8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong X, Wang F, Thut CJ, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J Virol. 1995;69:585–588. doi: 10.1128/jvi.69.1.585-588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Grossman SR, Kieff E. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc Natl Acad Sci USA. 2000;97:430–435. doi: 10.1073/pnas.97.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson S, et al. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell. 1991;65:1107–1115. doi: 10.1016/0092-8674(91)90007-l. [DOI] [PubMed] [Google Scholar]
- 18.Kieff E, Rickinson A. Epstein-Barr Virus and Its Replication. In: Howley P, Knipe D, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2603–2654. [Google Scholar]
- 19.Murn J, et al. A Myc-regulated transcriptional network controls B-cell fate in response to BCR triggering. BMC Genomics. 2009;10:323. doi: 10.1186/1471-2164-10-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elgueta R, de Vries VC, Noelle RJ. The immortality of humoral immunity. Immunol Rev. 2010;236:139–150. doi: 10.1111/j.1600-065X.2010.00924.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhao B, et al. RNAs induced by Epstein-Barr virus nuclear antigen 2 in lymphoblastoid cell lines. Proc Natl Acad Sci USA. 2006;103:1900–1905. doi: 10.1073/pnas.0510612103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 23.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valouev A, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Portal D, et al. EBV nuclear antigen EBNALP dismisses transcription repressors NCoR and RBPJ from enhancers and EBNA2 increases NCoR-deficient RBPJ DNA binding. Proc Natl Acad Sci USA. 2011;108:7808–7813. doi: 10.1073/pnas.1104991108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasowski M, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type–specific gene expression. Nature. 2009;459(7243):108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brady G, Whiteman HJ, Spender LC, Farrell PJ. Downregulation of RUNX1 by RUNX3 requires the RUNX3 VWRPY sequence and is essential for Epstein-Barr virus-driven B-cell proliferation. J Virol. 2009;83:6909–6916. doi: 10.1128/JVI.00216-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pongubala JM, et al. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- 33.Maier H, et al. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat Immunol. 2004;5:1069–1077. doi: 10.1038/ni1119. [DOI] [PubMed] [Google Scholar]
- 34.Del Bianco C, et al. Notch and MAML-1 complexation do not detectably alter the DNA binding specificity of the transcription factor CSL. PLoS ONE. 2010;5:e15034. doi: 10.1371/journal.pone.0015034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laux G, et al. Identification and characterization of an Epstein-Barr virus nuclear antigen 2-responsive cis element in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J Virol. 1994;68:6947–6958. doi: 10.1128/jvi.68.11.6947-6958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pomerantz MM, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelengaris S, Khan M, Evan G. c-MYC: More than just a matter of life and death. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 39.McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-specific memory B cell development. Annu Rev Immunol. 2005;23:487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
- 40.Lin YC, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Treiber T, et al. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity. 2010;32:714–725. doi: 10.1016/j.immuni.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 42.Tierney R, Kirby H, Nagra J, Rickinson A, Bell A. The Epstein-Barr virus promoter initiating B-cell transformation is activated by RFX proteins and the B-cell-specific activator protein BSAP/Pax5. J Virol. 2000;74:10458–10467. doi: 10.1128/jvi.74.22.10458-10467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Treiber N, Treiber T, Zocher G, Grosschedl R. Structure of an Ebf1:DNA complex reveals unusual DNA recognition and structural homology with Rel proteins. Genes Dev. 2010;24:2270–2275. doi: 10.1101/gad.1976610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanigaki K, et al. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat Immunol. 2002;3:443–450. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- 45.Wang H, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T lymphoblastic leukemia cells. Proc Natl Acad Sci USA. 2011;108:14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klenova EM, et al. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13:7612–7624. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmadiyeh N, et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci USA. 2010;107:9742–9746. doi: 10.1073/pnas.0910668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faumont N, et al. c-Myc and Rel/NF-kappaB are the two master transcriptional systems activated in the latency III program of Epstein-Barr virus-immortalized B cells. J Virol. 2009;83:5014–5027. doi: 10.1128/JVI.02264-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cahir-McFarland ED, Davidson DM, Schauer SL, Duong J, Kieff E. NF-kappa B inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc Natl Acad Sci USA. 2000;97:6055–6060. doi: 10.1073/pnas.100119497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.