This study presents the crystal structure of the trifunctional THI20 from S. cerevisiae, an enzyme with an N-terminal HMP kinase (ThiD-like) domain and a C-terminal thiaminase II (TenA-like) domain. In addition, two distinct structural classes of TenA have been identified by comparison to other known structures.
Keywords: thiamin salvage, thiamin metabolism, TenA, ThiD, thiaminases, HMP kinases
Abstract
In a recently characterized thiamin-salvage pathway, thiamin-degradation products are hydrolyzed by thiaminase II, yielding 4-amino-5-hydroxymethyl-2-methylpyrimidine (HMP). This compound is an intermediate in thiamin biosynthesis that, once phosphorylated by an HMP kinase, can be used to synthesize thiamin monophosphate. Here, the crystal structure of Saccharomyces cerevisiae THI20, a trifunctional enzyme containing an N-terminal HMP kinase/HMP-P kinase (ThiD-like) domain and a C-terminal thiaminase II (TenA-like) domain, is presented. Comparison to structures of the monofunctional enzymes reveals that while the ThiD-like dimer observed in THI20 resembles other ThiD structures, the TenA-like domain, which is tetrameric in all previously reported structures, forms a dimer. Similarly, the active site of the ThiD-like domain of THI20 is highly similar to other known ThiD enzymes, while the TenA-like active site shows unique features compared with previously structurally characterized TenAs. In addition, a survey of known TenA structures revealed two structural classes, both of which have distinct conserved features. The TenA domain of THI20 possesses some features of both classes, consistent with its ability to hydrolyze both thiamin and the thiamin-degradation product 2-methyl-4-amino-5-aminomethylpyrimidine.
1. Introduction
Thiamin (vitamin B1) is an essential cofactor in all organisms and is necessary for the combustion of carbohydrates and the biosynthesis of amino acids (Begley & Ealick, 2010 ▶; Jordan, 2010 ▶). Unlike microorganisms and plants, humans cannot synthesize thiamin and must rely upon dietary sources of this important nutrient. The biosynthesis of thiamin pyrophosphate (ThDP), the active form of the cofactor, has been extensively studied in prokaryotes and nearly all of the required individual enzymes have been structurally and biochemically characterized (Jurgenson et al., 2009 ▶). While less is known about the eukaryotic pathway, the pace of research has recently accelerated and important details are beginning to emerge (Jurgenson et al., 2009 ▶; Nosaka, 2006 ▶; Paul et al., 2010 ▶).
In Bacillus subtilis, thiamin monophosphate (ThMP) biosynthesis requires nine gene products. Six gene products (ThiS, ThiF, ThiO, ThiG, TenI and YrvO) are required to produce 2-carboxy-4-methyl-5-(β-hydroxyethyl)thiazole phosphate (Thz-P) from 1-deoxyxyulose 5-phosphate, cysteine and glycine. Gram-negative bacteria utilize tyrosine instead of glycine and require the radical SAM enzyme ThiH instead of ThiO. ThiC catalyzes the complex rearrangement of 5-aminoimidazole ribonucleotide to form 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate (HMP-P), and ThiD phosphorylates HMP-P to form HMP pyrophosphate (HMP-PP). Thz-P and HMP-PP are coupled by ThiE to form ThMP. A final phosphorylation by ThiL results in ThDP.
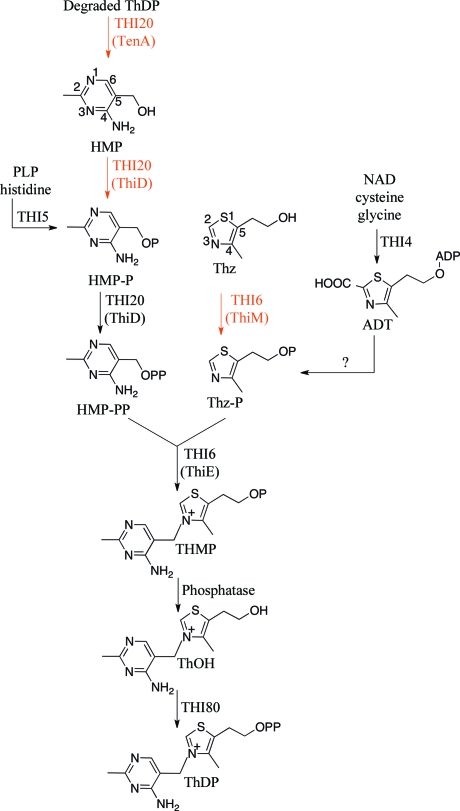
In Saccharomyces cerevisiae, only four gene products are required for ThMP biosynthesis (Fig. 1 ▶). THI4 catalyzes the conversion of NAD, glycine and cysteine to adenylated Thz-P (ADT). THI5 catalyzes the conversion of histidine and pyridoxal 5′-phosphate (PLP) to HMP-P, which is phosphorylated by the N-terminal domain of THI20 to form HMP-PP. The N-terminal domain of THI6 catalyzes the coupling reaction to form ThMP. Two additional enzymes, a phosphatase and thiamin pyrophosphokinase (THI80), are required to convert ThMP to ThDP.
Figure 1.
The role of THI20 in thiamin biosynthesis and salvage in S. cerevisiae. The salvage pathway is indicated by red arrows and enzyme names. The yeast enzymes are designated by all-upper-case letters and the corresponding bacterial enzymes are shown in parentheses. The conversion of ADT to Thz-P during biosynthesis is not understood and may require an additional enzyme.
The S. cerevisiae pathway is remarkable for three reasons: (i) the biosynthesis of thiamin requires the cannibalism of two cofactors, NAD and PLP, to form one molecule of ThDP; (ii) two biosynthetic enzymes, THI6 and THI20, also possess thiamin-salvage activities; and (iii) a single enzyme (THI4) can assemble the thiazole heterocycle, in contrast to the bacterial requirement for six gene products. The N-terminal domain of THI6 is homologous to ThiE and catalyzes the coupling of Thz-P and HMP-PP (biosynthesis), while the C-terminal domain is homologous to ThiM and catalyzes the phosphorylation of the thiazole alcohol to Thz-P (salvage). The N-terminal domain of THI20 is homologous to ThiD and catalyzes the phosphorylation of HMP-P to HMP-PP (salvage and biosynthesis) and the phosphorylation of HMP to HMP-P (salvage), while the C-terminal domain is homologous to TenA, which catalyzes the production of HMP from thiamin-degradation products (salvage).
TenA is widely distributed, but only a few TenAs have been biochemically characterized. In B. subtilis TenA was shown to be a thiaminase II which catalyzes the hydroloysis of thiamin to give HMP and thiazole alcohol (Toms et al., 2005 ▶). In B. halodurans TenA is involved in a novel salvage pathway (Jenkins et al., 2007 ▶). In this pathway, 2-methyl-4-formylamino-5-aminomethylpyrimidine, a product of thiamin degradation, is transported into the cell and following deamination is hydrolyzed by TenA to produce HMP. Phosphorylation of HMP by ThiD yields HMP-PP, which is coupled to Thz-P by thiamin phosphate synthase. Helicobacter pylori TenA is also thought to catalyze a salvage reaction; however, the identity of the pyrimidine substrate has not yet been established (Barison et al., 2009 ▶). In yeast, the N-terminal domain of THI20 has been shown to catalyze the hydrolysis of both thiamin (Haas et al., 2005 ▶) and 2-methyl-4-formylamino-5-aminomethylpyrimidine (Onozuka et al., 2008 ▶). Thiaminase I is less widely distributed but also catalyzes the degradation of thiamin (Fujita, 1954 ▶). B. subtilis thiaminase I (Campobasso et al., 2000 ▶) and B. subtilis thiaminase II (Toms et al., 2005 ▶) have very different folds; however, both utilize an active-site cysteine for initial cleavage of thiamin, resulting in a covalently bound intermediate. Thiaminase I accepts a wide variety of nucleophiles to complete the reaction, while thiaminase II is specific for water. The physiological function of thiaminase I is still unknown.
In this work, we present the X-ray crystal structure of S. cerevisiae THI20. THI20 consists of separate dimeric ThiD-like and dimeric TenA-like domains connected by two relatively flexible loop regions. The ThiD-like dimeric domain is structurally homologous to ThiDs from bacteria; however, all reported bacterial TenA structures are tetrameric. Examination of the active sites reveals that while the ThiD-like domain of THI20 is quite similar to other members of this family, the TenA-like active site shares properties with two different classes of TenA. One of these classes possesses a conserved cysteine residue that is known to participate in catalysis, while the other class of TenAs lack this cysteine but possess a pair of conserved glutamate residues in the active site. The structure of THI20 and the accompanying analysis provide insights into the architecture and function of this trifunctional enzyme and its possible relationship to other TenA-family members.
2. Materials and methods
2.1. Protein expression and purification
THI20 was cloned into a pET-28-based vector (Novagen) with a C-terminal six-His tag using standard molecular biology techniques. Escherichia coli BL21 (DE3) cells were transformed with this vector and grown at 310 K in LB broth to an OD600 of 0.4, at which point the temperature was reduced to 288 K. After 30 min, expression was induced by the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside. The cells were grown overnight, pelleted by centrifugation at 5000g and stored at 253 K. For purification, the cell pellet was thawed and then lysed by sonication in a buffer consisting of 300 mM NaCl, 10 mM imidazole and 50 mM Tris–HCl pH 7.6. The lysate was centrifuged at 40 000g and the resulting extract was loaded onto pre-equilibrated Ni–NTA resin (Qiagen). The resin was washed with ten column volumes of buffer consisting of 300 mM NaCl, 20 mM imidazole, 10% glycerol and 50 mM Tris pH 7.6 before being eluted with 300 mM NaCl, 250 mM imidazole and 50 mM Tris pH 7.6. The eluted protein was further purified by size-exclusion chromatography on an ÄKTAexplorer FPLC with a HiLoad 26/60 Superdex prep-grade G200 column running 10 mM Tris buffer pH 7.6 with 30 mM NaCl. The protein was found to be greater than 90% pure by SDS–PAGE after this chromatography step. The protein was concentrated to 10 mg ml−1 using a centrifugal concentrator and aliquots were flash-frozen and stored at 193 K.
2.2. Crystallization, data collection, data processing and structure determination
Crystals of THI20 were grown using the hanging-drop vapor-diffusion method at 291 K and appeared after approximately one week in 10–14% PEG 8000, 0.2 M calcium acetate, 0.1 M imidazole pH 7.5. Seeding the drops prior to sealing the wells yielded higher quality crystals that grew in 2–3 d. The crystals were dipped into a solution consisting of 15% PEG 8000, 0.2 M calcium acetate, 20% glycerol and 0.1 M imidazole pH 7.5 and were flash-frozen in liquid nitrogen. Data collection was performed at 100 K using a wavelength of 0.979 Å on the Northeastern Collaborative Access Team beamline 24-ID-C at the Advanced Photon Source, Argonne National Laboratory. Data were indexed, integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶). Data-collection statistics are shown in Table 1 ▶.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Resolution (Å) | 50.0–2.68 (2.75–2.68) |
| Wavelength (Å) | 0.9791 |
| Beamline | 24-ID-C, APS |
| Space group | P212121 |
| Unit-cell parameters | |
| a (Å) | 59.7 |
| b (Å) | 140.1 |
| c (Å) | 143.3 |
| No. of reflections | 87487 |
| Unique reflections | 31670 |
| Average I/σ(I) | 12.2 (1.5) |
| Multiplicity | 2.8 (1.5) |
| Completeness (%) | 92.8 (70.0) |
| Rmerge† (%) | 9.4 (32.9) |
| Data refinement | |
| No. of protein atoms | 7795 |
| No. of ligand atoms | 5 |
| No. of water atoms | 56 |
| Working-set reflections | 30019 |
| Test-set reflections | 1606 |
| R factor‡ | 23.8 |
| Rfree§ | 27.8 |
| R.m.s.d. bonds (Å) | 0.006 |
| R.m.s.d. angles (°) | 0.91 |
| Mean B factor (Å2) | 53 |
| Ramachandran plot | |
| Most favored (%) | 92.7 |
| Additionally allowed (%) | 7.1 |
| Generously allowed (%) | 0.1 |
| Disallowed (%) | 0.1¶ |
R
merge = 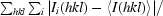
 , where 〈I(hkl)〉 is the mean intensity of the i reflections with intensities I
i(hkl) and common indices hkl.
, where 〈I(hkl)〉 is the mean intensity of the i reflections with intensities I
i(hkl) and common indices hkl.
R factor = 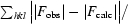
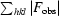 , where F
obs and F
calc are observed and calculated structure factors, respectively.
, where F
obs and F
calc are observed and calculated structure factors, respectively.
For R free, the sum is extended over a subset of reflections (5%) that were excluded from all stages of refinement.
Disallowed angles occurred at a Gly-Gly sequence (residues 200–201).
The structure of THI20 was solved using molecular replacement with the B. subtilis TenA structure (PDB entry 1yaf; Toms et al., 2005 ▶) and the Salmonella typhimurium ThiD structure (PDB code 1jxh; Cheng et al., 2002 ▶) as search models for the TenA and ThiD domains, respectively. The model was refined through successive rounds of manual model building using Coot (Emsley & Cowtan, 2004 ▶) and restrained refinement using REFMAC5 (Murshudov et al., 2011 ▶). Water molecules were added only after the model converged and were followed by two additional rounds of refinement. Data-refinement statistics are provided in Table 1 ▶.
2.3. Figure preparation
Sequence alignments and phylogenetic trees were computed using ClustalW (Thompson et al., 1994 ▶). All figures were prepared using PyMOL (DeLano, 2002 ▶) and ChemBioDraw (CambridgeSoft).
3. Results and discussion
3.1. Overall structure of THI20
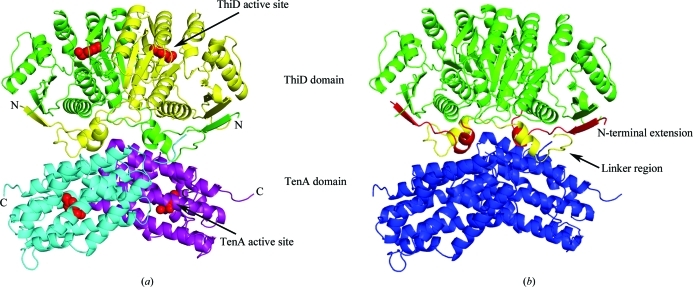
THI20 is a trifunctional enzyme that is composed of a ThiD-like N-terminal domain that catalyzes the phosphorylation of HMP to HMP-P and of HMP-P to HMP-PP and a TenA-like C-terminal domain with thiaminase activity. The structure revealed an overall dimeric organization in which N-terminal domains and C-terminal domains form ThiD-like and TenA-like local dimers (Fig. 2 ▶ a). A relatively flexible linker region composed of a loop and a short helix joins the two domains (Fig. 2 ▶ b). This linker region and a flexible N-terminal extension occupy the interface between the ThiD-like and TenA-like dimers (Fig. 2 ▶ b). The N-terminal extension is composed of a single β-strand that packs against and extends the length of the internal β-sheet of the ThiD-like domain and a short α-helix. It is the short helices that are present at the N-terminus and inter-domain linker region of THI20 that are the major points of interaction between the ThiD-like domain and the all-α-helical TenA-like domain.
Figure 2.
Stucture of S. cerevisiae THI20. (a) A ribbon diagram illustrating the structure of the THI20 dimer with each domain colored separately (ThiD-like domains colored yellow and green and TenA-like domains colored cyan and purple). The locations of the active sites are shown with a space-filling model of HMP and are indicated by arrows. (b) A model of THI20 showing the regions with no sequence similarity to other ThiD or TenA proteins. The linker region joining the ThiD-like domains (green) and the TenA-like domains (blue) is shown in yellow, while the N-terminal extension is shown in red.
3.2. Organization of the ThiD-like domain
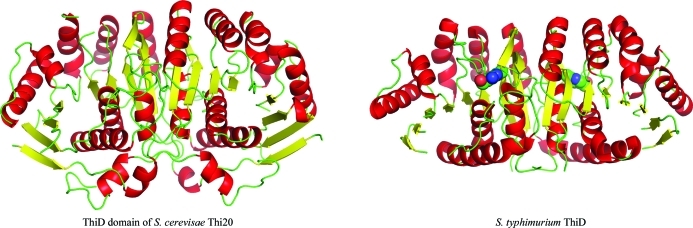
The ThiD-like domain of S. cerevisiae is an αβα sandwich with a nine-stranded β-sheet flanked by three α-helices on one side and five on the other (Fig. 3 ▶). ThiD belongs to the ribokinase superfamily and is structurally similar to several kinases within the superfamily. ThiD has been shown to form a tight dimer that is conserved in all known structures of ThiD (e.g. PDB entries 1jxh and 1ub0; Cheng et al., 2002; B. Bagautdinov, S. Kuramitsu, S. Yokoyama, M. Miyano & T. H. Tahirov, unpublished work). Superposition of the THI20 structure with S. typhimurium ThiD (Cheng et al., 2002 ▶) illustrates that this dimer organization is also conserved in THI20 (Fig. 3 ▶). The main difference between the THI20 ThiD-like domain and S. typhimurium ThiD is the presence of two additional loop regions at the N- and C-termini of the ThiD-like domain of THI20, both of which lie at the interface between the ThiD-like and TenA-like domains (Fig. 2 ▶ b).
Figure 3.
Comparison of the S. cerevisiae THI20 N-terminal domain with S. typhimurium ThiD (PDB entry 1jxh). Both structures are shown as ribbon diagrams with helices colored red, β-strands colored yellow and loop regions colored green. The active site in the model of S. typhimurium ThiD is indicated by a space-filling model of HMP (C atoms colored green, O atoms colored red and N atoms colored blue).
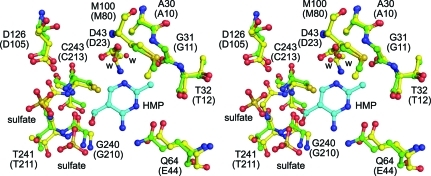
Like the other structurally characterized orthologs, the THI20 ThiD-like domain contains two active sites separated by approximately 20–25 Å. A comparison of the architecture of the active sites of the THI20 ThiD-like domain and S. typhimurium ThiD shows a high degree of similarity (Fig. 4 ▶). Despite a sequence identity of only 29%, all of the active-site residues that have been demonstrated to be necessary for substrate binding and catalysis are conserved (Cheng et al., 2002 ▶). A particularly important structural feature, the GTGC motif that makes up the anion hole, is completely conserved (residues 240–243) in the ThiD-like domain of THI20.
Figure 4.
The active site of the ThiD-like domain. A superposition of the active-site residues of the ThiD-like domain of THI20 (green C atoms) with the active site of S. typhimurium ThiD (yellow C atoms) is shown. In both cases O atoms are shown in red and N atoms are shown in blue. The HMP molecule (cyan C atoms) is from the S. typhimurium ThiD model. Water molecules are represented by red spheres and are labeled ‘w’. The residue numbers provided correspond to the THI20 sequence and the S. typhimurium ThiD sequence (in parentheses).
3.3. Organization of the TenA-like domain
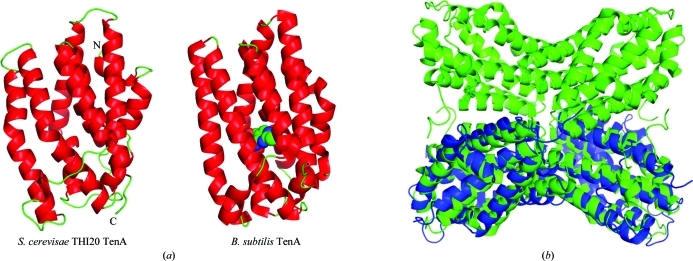
The C-terminal α-helical domain of THI20 is composed of nine mostly parallel α-helices (Fig. 5 ▶ a). This domain is identified by sequence similarity as belonging to the TenA superfamily, which is widely distributed and was originally identified as a transcriptional activator (Pang et al., 1991 ▶). More recently, however, TenA has been shown to possess thiaminase activity and is believed to participate in thiamin salvage (Jenkins et al., 2007 ▶; Toms et al., 2005 ▶). While the available structures of TenA show a tetramer as the active biological unit (Benach et al., 2005 ▶; Toms et al., 2005 ▶), the fusion of the ThiD-like and TenA-like domains in THI20 precludes the possibility of a TenA-like tetramer. Superposition of the THI20 TenA-like domain with the TenA tetramer from B. subtilis, however, reveals that the TenA-like dimer observed in THI20 is structurally homologous to half of the tetramer observed for other TenAs (Fig. 5 ▶ b).
Figure 5.
Comparison of the overall structure of the THI20 TenA-like domain to B. subtilis TenA. (a) The THI20 TenA-like protomer is shown alongside the protomer of B. subtilis TenA (PDB entry 1yak). In both cases helices are colored red and loop regions are colored green. The active site of B. subtilis TenA is indicated by HMP, which is shown as a space-filling model with green C atoms, red O atoms and blue N atoms. (b) A superposition of the THI20 TenA-like dimer (blue) with the B. subtilis TenA tetramer (green).
A large number of structures annotated as TenA have been deposited in the PDB, mostly owing to the efforts of structural genomics groups. Despite the high degree of overall structural similarity among TenA-family members, some interesting differences are present in the active sites of this enzyme. Thiaminase II enzymes are specific for water as the nucleophile in the cleavage of thiamin. The proposed mechanism identifies a conserved cysteine residue as the protein nucleophile that initiates the reaction. Alignment of several TenA sequences, however, shows that this cysteine is not conserved in all organisms (Supplementary Figs. S1 and S21). Furthermore, comparison of the sequences and active sites of available TenA structures revealed two different classes.
3.4. Differences in TenA classes
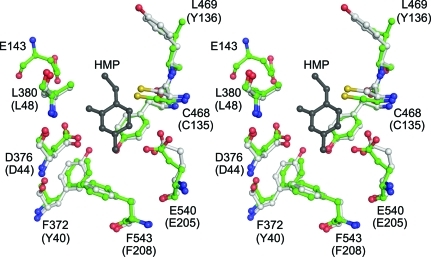
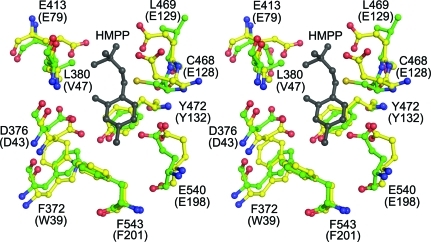
While there is a high degree of overall structural similarity between the two classes of TenA, there are distinct differences in the active sites. One of the TenA classes, which is similar to B. subtilis TenA, has a conserved cysteine that is believed to act as the nucleophile in the thiaminase reaction (Fig. 6 ▶). The other class of TenA lacks this cysteine residue but instead possesses a pair of structurally conserved glutamate residues in the active site (Fig. 7 ▶). These two glutamate residues are expected to interact primarily with the substituent at the 5-position of the pyrimidine ring. While the active sites of the members of the class of TenAs with the conserved cysteine are relatively large and open, the two conserved glutamate residues in the members of the second class of TenA fill the active site and make a much tighter fit for the ligand.
Figure 6.
The active site of TenA with the conserved cysteine. A stereodiagram is shown of the superposition of the THI20 TenA-like active site (green C atoms) with that of B. subtilis TenA (white C atoms). The residue numbers given correspond to the THI20 sequence and the B. subtilis TenA sequence (in parentheses). The HMP molecule (gray) is from the B. subtilis TenA structure. In both cases, O atoms are colored red, N atoms are colored blue and S atoms are colored yellow.
Figure 7.
The active site of TenA with conserved glutamate residues. A stereodiagram is shown of the superposition of the THI20 TenA-like active site (green C atoms) with that of P. furiosus TenA (PDB entry 1rtw; yellow C atoms). The residue numbers given correspond to the THI20 sequence and the P. furiosus TenA sequence (in parentheses). The HMP-P molecule (gray) is from the P. furiosus TenA structure. In both cases O atoms are colored red, N atoms are colored blue and S atoms are colored yellow.
Despite the differences between the two classes in this region of the active site, the residues that interact with the pyrimidine moiety appear to be highly structurally conserved. Several phenylalanine and tyrosine residues (Phe372, Phe543, Tyr472 and Tyr379 from THI20) pack against the ring and additional contacts are provided by structurally conserved aspartate (Asp376) and glutamate (Glu540) residues. It is clear from this structural comparison that both classes of TenA evolved to accommodate the pyrimidine ring but may differ in their ability to accommodate substituents at the 5-position. This suggests that the TenAs with a conserved cysteine could accept larger ligands such as thiamin, while the other class may be limited to smaller ligands that lack the thiazole moiety (e.g. 2-methyl-4-amino-5-aminomethylpyrimidine, the base-degraded form of thiamin). Despite a relatively high degree of sequence similarity within the two classes, there does not appear to be any general evolutionary trend toward one class; both classes of TenA appear in archaea, bacteria and eukaryotes (Supplementary Fig. S31). It is possible that parallel divergent evolution of these two types of TenA has been driven by differing environmental or nutritional pressures.
Although the TenA-like domain of THI20 contains the conserved cysteine residue (Cys468) and shares a similar active-site architecture with B. subtilis TenA (Fig. 6 ▶), it also possesses a glutamate residue that is not present in the B. subtilis TenA active site. This residue (Glu413) aligns well with one of the two conserved glutamate residues (Glu79 from P. furiosis) that are present in the other class of TenA proteins (Fig. 7 ▶). Recent biochemical studies on S. cerevisiae THI20 demonstrated that in addition to thiaminase II activity this enzyme can hydrolyze thiamin and the thiamin analogs pyrithiamin and oxythiamin, producing HMP (Onozuka et al., 2008 ▶). In addition, this enzyme has been shown to accept aminomethylpyrimidine as a substrate. The ability of the TenA domain of THI20 to accommodate both large and small ligands is consistent with its similarity to both classes of TenA. Further biochemical experiments are needed to determine the range of substrates that can be hydrolyzed by THI20.
Prior to the discovery that THI20 catalyzes a salvage reaction, it was unknown why a synthetic activity (HMP-P kinase) and a degradative activity (thiaminase) were fused into a single gene product. Based on the structure of THI20 and the properties of the monofunctional enzymes ThiD and TenA, it is unlikely that improving the solubility or channeling of intermediates drove the evolution of this trifunctional enzyme. There are no obvious ligand channels present in THI20 and all of the substrates and intermediates in the salvage reaction are stable in aqueous environments. In addition, both the ThiD-like and TenA-like domains are known to be stable and active in their monofunctional forms, indicating that the fusion is not likely to be a result of increased stability. In light of the recent biochemical evidence, it is likely that the evolutionary pressure towards efficient salvage of exogenous thiamin analogs led to this fusion (Jenkins et al., 2007 ▶; Onozuka et al., 2008 ▶). The marriage of the thiaminase activity of the TenA domain and the HMP(P) kinase activity of the ThiD domain leads to a single trifunctional enzyme that can recycle thiamin-degradation products (such as 2-methyl-4-formylamino-5-aminomethylpyrimidine). While these types of genetic fusions are not common in thiamin metabolism, S. cerevisiae does possess another multi-functional enzyme involved in the biosynthesis of thiamin. This enzyme, THI6, has also recently been structurally characterized (Paul et al., 2010 ▶).
Two THI20 homologs, THI21 and THI22, are also found in S. cerevisiae. THI21 and THI22 share 86% and 76% sequence identity to THI20, respectively. While THI20 and THI21 have been shown to be functionally redundant, THI22 is unable to synthesize HMP-PP from HMP or HMP-P (Kawasaki et al., 2005 ▶; Llorente et al., 1999 ▶). Considering that there are only minor differences between the sequence of THI20 and those of THI21/THI22, it is unclear why these proteins are present in S. cerevisiae. Given their similarity to THI20, it is unlikely that they catalyze a completely different reaction or even have different substrate specificities. A possible explanation for the loss of HMP(P) kinase ability in THI22 is the difference at residue 241 in the completely conserved GTGC motif that makes up the anion hole (Supplementary Fig. S41). The Thr241Ala mutation that occurs in THI22 may impair the ability of this protein to bind phosphate and thus catalyze the phosphorylation of HMP or HMP-P (Fig. 2b).
4. Conclusions
Structural and biochemical studies demonstrate that THI20 is a homodimer with an N-terminal ThiD-like dimeric domain and a C-terminal TenA-like dimeric domain. ThiD and TenA were thought to have disparate functions (thiamin biosynthetic and degradative) and their fusion into a single gene product seemed surprising; however, the roles of ThiD and TenA in thiamin salvage offer a plausible explanation in which the TenA-like domain converts thiamin-degradation products to HMP and the ThiD-like domain converts HMP to HMP-P. The fusion of the two domains links their expression and ensures efficient thiamin salvage. In addition, a comparison of the TenA domain of THI20 and other known TenAs revealed that there are different classes of TenA and that the THI20 TenA-like domain shares properties of both, suggesting its ability to salvage a wide variety of possible thiamin-degradation products. Further biochemical studies are needed to fully understand the role of TenA in the salvage pathways of various organisms.
Supplementary Material
PDB reference: THI20, 3rm5
Supplementary material file. DOI: 10.1107/S0907444911024814/be5176sup1.pdf
Acknowledgments
This work was supported by the Robert A. Welch Foundation (A-0034 to TPB) and National Institutes of Health grants DK44083 to TPB and DK67081 to SEE. The collection of data on the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source is supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the US Department of Energy, Office of Basic Energy Sciences under contract No. DE-AC02-06CH11357. We would like to acknowledge Cynthia Kinsland of the Protein Production Facility in the Department of Chemistry and Chemical Biology for help with molecular biology and Leslie Kinsland for help with manuscript preparation. JBF acknowledges the Tri-Institutional Training Program in Chemical Biology for financial support.
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: BE5176). Services for accessing this material are described at the back of the journal.
References
- Barison, N., Cendron, L., Trento, A., Angelini, A. & Zanotti, G. (2009). FEBS J. 276, 6227–6235. [DOI] [PubMed]
- Begley, T. P. & Ealick, S. E. (2010). Comprehensive Natural Products II, edited by M. Lew & L. Hung-Wen, pp. 547–559. Oxford: Elsevier.
- Benach, J., Edstrom, W. C., Lee, I., Das, K., Cooper, B., Xiao, R., Liu, J., Rost, B., Acton, T. B., Montelione, G. T. & Hunt, J. F. (2005). Acta Cryst. D61, 589–598. [DOI] [PubMed]
- Campobasso, N., Mathews, I. I., Begley, T. P. & Ealick, S. E. (2000). Biochemistry, 39, 7868–7877. [DOI] [PubMed]
- Cheng, G., Bennett, E. M., Begley, T. P. & Ealick, S. E. (2002). Structure, 10, 225–235. [DOI] [PubMed]
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fujita, A. (1954). Adv. Enzymol. Relat. Subj. Biochem. 15, 389–421. [DOI] [PubMed]
- Haas, A. L., Laun, N. P. & Begley, T. P. (2005). Bioorg. Chem. 33, 338–344. [DOI] [PubMed]
- Jenkins, A. H., Schyns, G., Potot, S., Sun, G. & Begley, T. P. (2007). Nature Chem. Biol. 3, 492–497. [DOI] [PubMed]
- Jordan, F. (2010). Comprehensive Natural Products II, edited by M. Lew & L. Hung-Wen, pp. 561–598. Oxford: Elsevier.
- Jurgenson, C. T., Begley, T. P. & Ealick, S. E. (2009). Annu. Rev. Biochem. 78, 569–603. [DOI] [PMC free article] [PubMed]
- Kawasaki, Y., Onozuka, M., Mizote, T. & Nosaka, K. (2005). Curr. Genet. 47, 156–162. [DOI] [PubMed]
- Llorente, B., Fairhead, C. & Dujon, B. (1999). Mol. Microbiol. 32, 1140–1152. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nosaka, K. (2006). Appl. Microbiol. Biotechnol. 72, 30–40. [DOI] [PubMed]
- Onozuka, M., Konno, H., Kawasaki, Y., Akaji, K. & Nosaka, K. (2008). FEMS Yeast Res. 8, 266–275. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pang, A. S., Nathoo, S. & Wong, S.-L. (1991). J. Bacteriol. 173, 46–54. [DOI] [PMC free article] [PubMed]
- Paul, D., Chatterjee, A., Begley, T. P. & Ealick, S. E. (2010). Biochemistry, 49, 9922–9934. [DOI] [PMC free article] [PubMed]
- Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed]
- Toms, A. V., Haas, A. L., Park, J.-H., Begley, T. P. & Ealick, S. E. (2005). Biochemistry, 44, 2319–2329. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: THI20, 3rm5
Supplementary material file. DOI: 10.1107/S0907444911024814/be5176sup1.pdf