Abstract
Dyneins are microtubule-based motor proteins that power ciliary beating, transport intracellular cargos, and help to construct the mitotic spindle. Evolved from ring-shaped hexameric AAA-family adenosine triphosphatases (ATPases), dynein’s large size and complexity have posed challenges for understanding its structure and mechanism. Here, we present a 6 angstrom crystal structure of a functional dimer of two ~300-kilodalton motor domains of yeast cytoplasmic dynein. The structure reveals an unusual asymmetric arrangement of ATPase domains in the ring-shaped motor domain, the manner in which the mechanical element interacts with the ATPase ring, and an unexpected interaction between two coiled coils that create a base for the microtubule binding domain. The arrangement of these elements provides clues as to how adenosine triphosphate–driven conformational changes might be transmitted across the motor domain.
The cytoskeletal motor proteins consist of the myosin family, which moves along actin filaments, and the kinesin and dynein families, which move along microtubules. These motors use a common principle to generate movement in which they bind to their track, undergo a force-producing conformational change, release from the track, and then return to their original conformation. These structural changes are coupled to chemical transitions in the motor’s adenosine triphosphatase (ATPase) cycle [adenosine triphosphate (ATP) binding, hydrolysis, and product release].
The force-generating cycles of kinesins and myosins are understood in considerable mechanistic detail. Even though they interact with different cytoskeletal polymers, kinesins and myosins share a protein fold, reflecting their common evolutionary origin (1). Dyneins, by contrast, are unrelated to kinesins/myosins and instead have evolved from the AAA family of ATPases (2). The AAA ATPases (ATPases associated with diverse cellular activities), which are present in both prokaryotes and eukaryotes, participate in diverse functions, including protein unfolding for proteolysis, disassembly of stable protein complexes, and helicase activities (3). The majority of AAA ATPases self-assemble into hexameric rings that carry out the functional activities of these enzymes (4, 5). Dynein is one of two AAA ATPases that has six distinct AAA domains concatenated within a single polypeptide chain [the other being Rea1, an ATPase involved in ribosome biogenesis (6)]. Electron microscopy (EM) studies of dynein have shown that these AAA domains fold into a ring-shaped structure similar to other AAA ATPases (7–9).
Dyneins can be divided into three major families: axonemal dyneins, which power the beating of cilia and flagella; intraflagellar transport (IFT) dyneins, which transport proteins in the axoneme; and cytoplasmic dyneins, which perform most of the minus-end–directed transport of cargos (e.g., membranes, mRNAs, nuclei, and viruses) along microtubules in eukaryotic cells (10, 11). The N-terminal third of the >500 kD dynein heavy chain (Fig. 1A), which differs considerably between each dynein subfamily, contains elements that confer oligomerization (IFT and cytoplasmic dyneins are dimers, and axonemal dyneins can be monomers, dimers, or trimers) and binding sites for accessory chains and cargos. The remainder of the heavy chain contains the motor domain, which has a conserved sequence across the dynein family. The N-terminal region of the motor domain is termed the “linker” and has been proposed to serve as a mechanical element because its position shifts in different nucleotide states (7, 8, 12). The linker is followed by four AAA domains (AAA1 to 4) that contain functional nucleotide binding sites. ATP hydrolysis at AAA1 is essential for motility, whereas the role of nucleotide binding/hydrolysis at AAA2 to 4 is less clear but appears to influence motility (13–15). AAA5 and AAA6 have lost their conserved nucleotide binding sites and may serve a structural role (2). Unlike kinesin and myosin, where the polymer and nucleotide binding sites are in close proximity (<4 nm), dynein binds to microtubules through a small globular domain at the end of a 15-nm-long anti-parallel coiled-coil stalk that emerges between AAA4 and AAA5 (16–18) (Fig. 1A). Because microtubule binding stimulates ATP turnover at AAA1 and the nucleotide state of AAA1 affects microtubule binding affinity, allosteric changes must be communicated over long distances and across multiple domains within the motor.
Fig. 1.
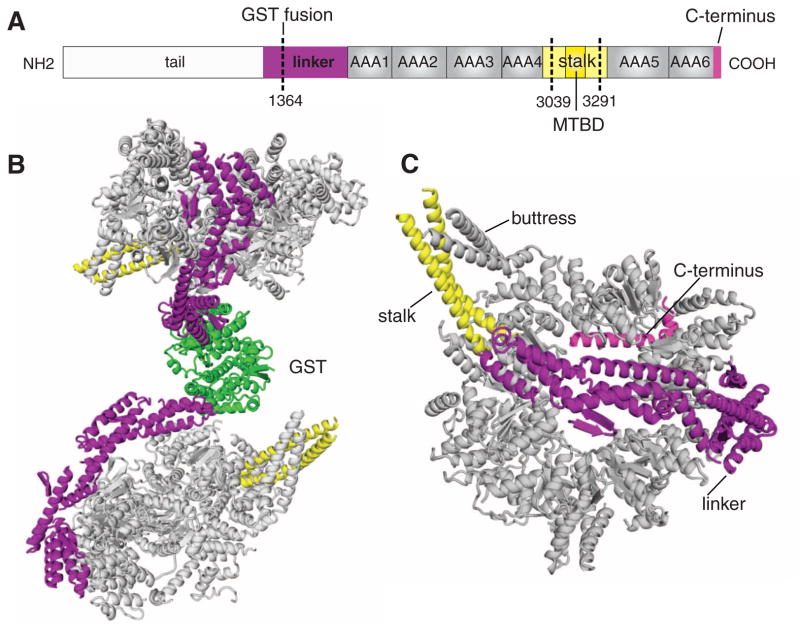
The cytoplasmic dynein motor domain crystal structure. (A) Schematic illustrating the domains of the dimeric yeast cytoplasmic dynein heavy chain. For crystallization, the linker was fused to GST, and the MTBD and part of the stalk (amino acids 3039 to 3291) were removed and replaced with a short peptide (20). (B) The complete GST-cytoplasmic dynein dimer; GST is in green. (C) View of the motor domain from the linker face. The linker, stalk, and C terminus are color-coded as in (A).
In comparison with kinesin and myosin, dynein’s mechanism of motility is poorly understood, in part because of the lack of high-resolution structural information. Here, we present the crystal structure of the dynein motor domain in the context of a 610-kD dimer of two motor domains.
Structure determination
We expressed recombinant Saccharomyces cerevisiae cytoplasmic dynein in yeast (19) using fed-batch fermentation (20). To identify a compact and mono-disperse protein that might crystallize, we varied the N-terminal truncation of the linker and removed different lengths of the coiled-coil stalk. A truncated motor domain fused to glutathione S-transferase (GST) (Fig. 1A) produced small crystals in a screen, which were subsequently optimized (20). With the exception of the removal of the microtubule binding domain (MTBD), this GST fusion corresponds to a previously characterized dimeric motor (GST-Dyn1314kD) that moves processively and at similar velocities to the wild-type yeast dynein holoenzyme (19). The unit cell (174 by 119 by 203 Å) of the diffracting crystals contained one dynein dimer and had a P21 space group (see table S1 for crystallographic information and statistics). To obtain phases, poly-tungsten clusters (W12) were incorporated into the crystals, and phases were determined to 8 Å by two-wavelength anomalous diffraction and extended to 6 Å by solvent flattening (20). The experimental phase information produced an interpretable electron density map with clear densities for helices and β sheets (fig. S1); the electron densities of the two dynein motor domains are nearly identical to one another, with minor differences in the clarity of a subset of loops connecting secondary structural elements. At this resolution, side chains cannot be resolved; hence, a polyalanine polypeptide chain was built into the electron density map. Due to the conserved structure of AAA domains, the majority of secondary structural elements predicted by sequence could be assigned in the model (uncertainties in assignment and connectivity exist for the linker and C-terminal domains, which do not have good homology models) [see supporting online material (20) for details of model building and caveats]. Because secondary structure predictions and our electron density map are both imprecise, registry errors of several amino acids will be common throughout our model. However, the primary focus of this study is to characterize the domain organization of the dynein motor.
Structural overview of a dynein dimer
The two dynein motor domains, which are dimerized by GST, are related by a pseudo–two-fold symmetry, such that the linker-face of the AAA rings appose each other and the stalk domains point in opposite directions (Fig. 1B). The presence of the linker domain makes it unlikely that the two AAA rings can directly stack on each other, as has been observed in other AAA proteins (21). The motor domain is a ring-shaped structure with a central hole; viewed from the top, the ring is somewhat asymmetric, with one side curved and the other more flattened (Fig. 1, B and C). The overall appearance is similar to negatively stained EM images of an inner arm axonemal dynein from Chlamydomonas (7) and a cytoplasmic dynein motor domain from Dictyostelium (8) (fig. S2). The linker arches over the center of the AAA ring (we refer to this side of the ring as the “linker face”) (Fig. 1, B and C). In previous EM images, it was thought that the linker might wrap around the top of the ring (7), but the crystal structure shows that the two ends of the linker contact the ring and the central portion bows well above the central hole like the handle of a basket (Fig. 1B and fig. S1B). The two contact points of the linker, adjacent to the base of the stalk and across the ring by AAA1, are consistent with the EM images of dynein in the nucleotide-free (apo) state (8) (fig. S2). Two coiled coils emerge from the ring, one of which corresponds to the “stalk” that extends to the MTBD. The second coiled coil (which we term the “buttress”) is predicted in the sequence (22). This uncharacterized coiled coil might correspond to the side of a triangular structure that has been visualized in a subset of the EM images (fig. S2B) (7). The C-terminal helices lie on the side of the ring opposite from the linker (Fig. 1C) in agreement with EM studies (fig. S2); we refer to this face of the ring as the “C-terminal face.”
Structure and assignment of the AAA domains
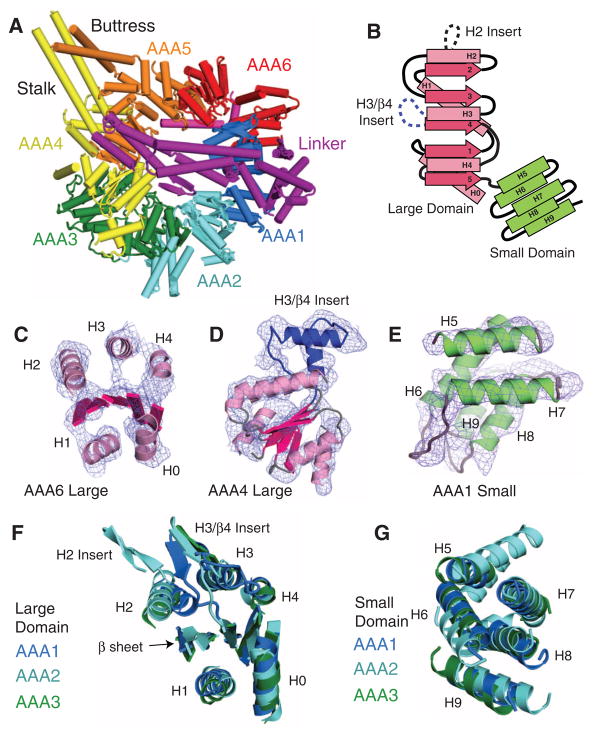
The six AAA domains can be easily identified in the electron density map (overview in Fig. 2A). AAA proteins possess an α/β large domain, which contains the nucleotide binding Walker A (P-loop) and B motifs, and a helical bundle small domain that extends from the C terminus of the large domain (Fig. 2B). The large domain (AAAL) is characterized by a parallel five-strand β sheet surrounded by two α helices (H0 and H1) on one face and three helices (H2, H3, and H4) on the opposite face, although different subclasses of AAA proteins contain unique insertions to this core structure (4). In our map of both motor domains, the AAA domains show a nearly identical pattern of electron density (fig. S3A), with five helices (H0 to H4) in similar positions to other AAA proteins (Fig. 2C and fig. S3, B and C). H0 and H1 in the six AAA domains are on the exterior of the ring, as in other hexameric AAA proteins. Although individual β strands are difficult to identify, a diffuse electron density in the position and orientation expected for the AAA β sheet is observed (Fig. 2C and fig. S1). When viewed from the side, the large domains form the top layer of the linker face of the ring.
Fig. 2.
The dynein AAA domains. (A) The six individual AAA domains are highlighted in colors. The linker spanning over the center of the ring is magenta. (B) Topology of the dynein large and small domains. (C) The electron density map [experimental (Fo) map contoured at 1 σ] and model build of the secondary structure elements (helices and central β sheet) of a dynein AAA large domain (AAA6L). (D) Experimental electron density map and model showing a unique helical insert (blue) in AAA4L. Insert sequences are shown in fig. S5. (E) Experimental electron density map and model of a dynein small domain (AAA1s). Omit maps of AAA1 and AAA2 small domains are shown in fig. S1. (F and G) Comparison of the large and small domains of AAA1 to 3. Galleries for all large and small domains can be found in figs. S3 and S4, respectively.
The six dynein AAA small domains (AAAs), which form the C-terminal face of the ring, all show a similar pattern of five helices (H5 to H9) (Fig. 2E and fig S4A), whose connectivity can be established by well-ordered loops. The general placement of the small domains in the ring and connectivity of H5 to H8 are similar to NSF (N-ethylmaleimide–sensitive fusion protein) (fig. S4C) and several other AAA proteins. Dynein has evolved a fifth helix (H9) at the bottom of the small domain, which helps to connect the small domain of one AAA domain to the large domain of the neighboring AAA in the poly-peptide chain. In contrast to an earlier suggestion (4), the extra helix in dynein is at the C terminus rather than the N terminus of the small domain; thus, the small-domain architecture is not similar to the BchI subclass of AAA proteins. AAA5s and AAA6s contain additional helices beyond H9, although their connectivity is not well established in our model. The stalk and buttress coiled coils are extensions of small-domain helices (fig. S4A).
Several pieces of information enabled us to assign AAA1 to 6 within the electron density map. First, we could use information from previous EM studies. The apo form of dynein seen by EM is strikingly similar to our crystal structure (fig. S2, A and B); the N terminus of the linker and the stalk base create fiducial markers that allow the x-ray and EM structures to be aligned with one another. The order of the AAA domains around the ring has been determined by EM after tagging particular domains with engineered fluorescent protein markers (8). When thus aligned, the order of the AAA domains is clockwise around the ring when viewed from the linker face, with AAA1 positioned close to the linker C terminus based on sequence (Fig. 2A). In the model presented in this EM study, however, the linker was assigned to the opposite face of the AAA ring, possibly because of insufficient three-dimensional discrimination.
Further evidence for assigning the individual AAA domains came from the inspection of the electron density maps for characteristic insertions to the canonical large domain AAA core structure. AAA1, AAA2, AAA3, and AAA5 have β-hairpin insertions between H3 and β4, a feature that is found in a subset of other AAA proteins (4) (Fig. 2, B and D, and fig. S5, A and B). AAA2 has an additional loop insertion within H2 (fig. S5A); a similar combination of H2 and H3 inserts is found in NtrC (23) (fig. S3B) and BchI (24) but few other AAA proteins. AAA4 also has an insertion of a pair of helices that extend from H3 and β4. Furthermore, AAA2, 4, and 5 have an extra N-terminal helix that packs against H0 and H1, as observed in certain other AAA proteins such as ClpX (fig. S3C). All of these features in the dynein sequence (fig. S5B) could be observed in the electron density maps (fig. S3) and helped to make our domain assignments.
The AAA small domains also have unique characteristics that could be matched to the electron density maps. Our AAA domain assignment places the stalk and buttress coiled coils within the small domains of AAA4 and AAA5, respectively, in agreement with their position in the sequence (fig. S5B). In addition, AAAs H5 to H8 are predicted to be longer than in other AAA domains, which is seen in the electron density map (fig. S4A) and agrees with an alignment of our crystal structure to the EM of Chlamydomonas axonemal dynein (7) (fig. S2B). When viewed from the linker face, the small domains are located clockwise with respect to each large domain, as is true of many other AAA ATPases such as ClpX (25) and HslU (26). Of the six AAA domains, AAA1 and 3 small and large domains are the most similar to one another in structure, as is true of their sequences as well.
To further validate our assignment, we obtained information on methionine positions using selenomethionine (SeMet). SeMet was incorporated into our yeast-expressed dynein to 64% occupancy, using a modification of previously described methods (20, 27). Although our 7.5 Å SeMet data was not sufficient to calculate phases, we were able to combine it with our experimentally determined phases to generate an anomalous difference Fourier density map in which we could locate approximate positions of 64 of the 126 methionines (peaks at >3 σ) in the GST-dynein dimer (table S1 and fig. S6). Although only four sites (out of 18 total methionines) were observed in the poorly ordered GST molecule, their locations corresponded to known methionine positions (fig. S6). Within the dynein motor domain, 42 sites were located in similar positions in each motor domain (21 in each monomer), whereas the remaining 18 sites were only observed in one of the two motor domains. Some of these peaks corresponded to predicted Met locations based on our model (fig. S6B). However, due to the low-resolution SeMet map, it was not possible to use this information to establish a registry of the polypeptide chain. Thus, although the secondary structure assignments in our model (fig. S5B) are likely to be mostly correct, the precise registry of the residues in our model will not be accurate (20). However, by counting the number of SeMet peaks in each of the large or small AAA domains, we could determine whether these numbers were consistent with our assignment of the six AAA domains. In the assignment shown in Fig. 2A, the number of SeMet peaks was equal to or less than that expected from the dynein sequence (fig. S6C). On the other hand, if the AAA assignment was shifted by one in either the clockwise or counterclockwise direction, then mismatches occurred, in which more SeMet peaks occurred in several domains than would be predicted from the sequence (fig. S6C). Thus, the SeMet data, the positions of large and small domain insertions, and agreement with previous EM mapping all support the AAA assignment shown in Fig. 2A.
Distinct conformations of the AAA domains produce an asymmetry of ring
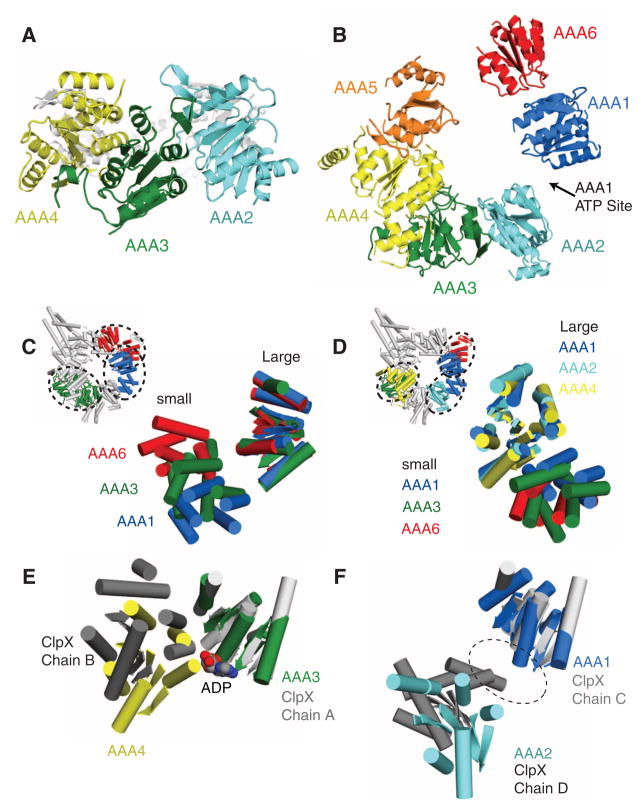
The arrangement of AAA domains around the motor domain ring is more asymmetric than any AAA hexamer structure solved to date. Viewed from the side, the AAA large domains occupy different planes within the ring (Fig. 3A). Viewed from the linker face, larger gaps are evident between the large domains of AAA1 and AAA2 and between AAA5 and AAA6 (Fig. 3B). Two major factors determine the positions of the AAA domains. First, as noted for other AAA proteins (25, 26), a flexible linker between β5 of the large domain and H5 of the small domain allows for rigid body rotations between these domains (Fig. 3C and fig. S7). Second, the small domains all pack rigidly and in a similar orientation against the large domains of the neighboring AAA (e.g., AAA1s against AAA2L) (Fig. 3D and fig. S7), as was noted for ClpX (25). Thus, AAA(n)small-AAA(n+1)large may behave as rigid units that can move relative to one another by rotations about the peptide linker that connects the large and small subdomains within a AAA domain.
Fig. 3.
Asymmetry of the dynein ring. (A) Side view of AAA ring showing the different planes occupied by the AAA1 to 3 large domains. (B) View of the linker face of the AAA ring (as in Fig. 2A), showing just the large domains. Note the large gaps between AAA1 and 2 and between AAA5 and 6. ATP is expected to bind between AAA1 and 2. (C) Comparison of the positions of the small domains relative to the large domain for AAA1, 3, and 6 (large domains aligned to AAA1); small domains can adopt a variety of different orientations due to a flexible linker joining β5 (large) to H5 (small). (D) Packing of the small domains against the neighboring AAA large domain. (Inset) AAA1s against AAA2L, AAA3s against AAA4L, and AAA6s against AAA1L. AAA large domains were aligned to AAA1. The orientations of the small domains to the neighboring large domain are similar to one another. (E) Comparison of the positions of adjacent large domains in dynein and ClpX (PDB code 3HWS). In the nucleotide-bound ClpX monomers in the hexamer (monomer chain A shown here) and AAA3 of dynein (aligned with ClpX), the adjacent large domain is closely apposed in a closed conformation. (F) In the nucleotide-free monomers of the same ClpX hexamer (monomer chain C) and AAA1, the adjacent large domain is more widely separated in an open conformation, due to the rotation of the intervening small domain. Comparison of the positions of adjacent large domains around the dynein AAA ring is found in fig. S7.
Hexameric AAA enzymes bind ATP between adjacent AAA subunits, with the clockwise neighbor [viewed from the linker face (Fig. 2A)] contributing residues that promote nucleotide hydrolysis (4, 5). In other AAA proteins, the close apposition of large domains enables nucleotide binding and hydrolysis (28, 29), whereas a greater separation is associated with a nucleotide-free or apo state (25, 30). In dynein, mutagenesis studies suggest that AAA3 can bind and hydrolyze ATP (13). Although we cannot ascertain whether nucleotide is bound to the AAA domains at our current resolution, AAA3 and AAA4 are positioned in a similar closed conformation to the interface of the nucleotide-bound subunits in the ClpX hexameric ring (Fig. 3E) (25). On the other hand, AAA1, the main hydrolytic site of dynein, is separated by a large distance from AAA2, which is more similar to the interface of the nucleotide-free subunits in ClpX (Fig. 3F) (25). The conformation of AAA1 and AAA2, combined with our crystallization in a nucleotide-free buffer and the observed position of the linker domain, makes it likely that AAA1 is in an apo state. It seems highly probable that ATP binding will close the gap between AAA1 and AAA2 and help to trigger other conformational changes in the dynein ring.
The linker domain
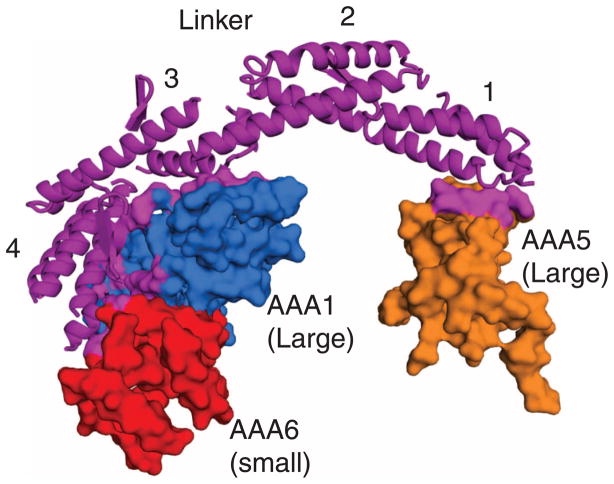
The arching linker domain is composed of four predominantly helical subdomains (Fig. 4). The C terminus subdomain 4 is composed predominantly of a four-helix bundle that interacts with the large and small domains of AAA1, as well as part of the small domain of AAA6. This extensive interface is suggestive of a stable interaction. The N-terminal subdomain 1 has a pattern of α helices that is reminiscent of the repeating unit in spectrin (31). It contacts H2 and the H3-β4 β-hairpin insert of the AAA5 large domain, which differs from previous suggestions that the linker N terminus interacts with either AAA2 (12) or AAA4 (8). The limited subdomain 1–ring contact raises the possibility that the N terminus of the linker might dissociate from AAA5 during the ATPase cycle. Subdomains 2 and 3 meet through long helices, which have a small region of overlap at their ends. The lack of extensive interactions between subdomains 2 and 3 and between subdomains 3 and 4 raises the possibility that one of these regions could act as a hinge to allow rigid body motions of subdomains 1 and 2 in other nucleotide states.
Fig. 4.
The linker domain and its interaction with the AAA ring. The linker and its four helical subdomains (subdomains 1 and 4 are N- and C-terminal, respectively) are indicated, and the contact sites (<8 Å) with the AAA domains are shown in color on the space-filling model.
The stalk and buttress
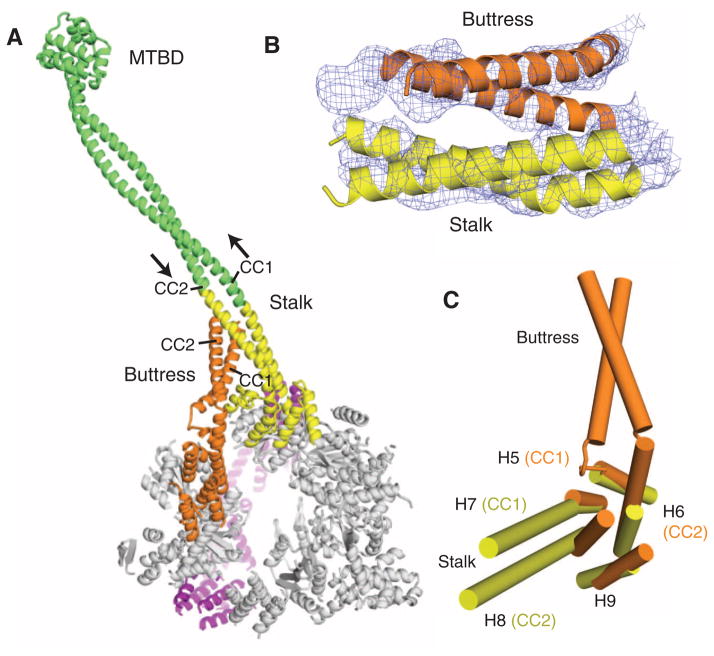
In addition to the stalk coiled coil, our crystal structure revealed a second antiparallel coiled coil that emerges from the AAA5 small domain (Fig. 5, A and B) and corresponds to a region of strong coiled-coil prediction (22). It likely corresponds to a bridging structure noted by EM (fig. S2B) that was then assumed to be part of the stalk rejoining the AAA ring (8). Strikingly, the AAA5 coiled coil extends toward and makes contact with the stalk (Fig. 5B). Based on the position of overlap and length of the helices in the stalk, we postulate that a highly conserved tryptophan in CC2 and a glycine in CC1 of the stalk (18) are positioned in the region of overlap with the AAA5 coiled coil. Based on its potential role in supporting the stalk, we have named the AAA5 coiled coil the “buttress.” This term need not imply a completely static support, and indeed the buttress may influence the conformation of the stalk during dynein’s ATPase cycle.
Fig. 5.
The stalk and buttress coiled coils. (A) The motor domain, highlighting the stalk and buttress, viewed from the C-terminal face. The stalk from this crystal structure is highlighted in yellow, and the green extension is a continuation of the stalk modeled with an antiparallel coiled coil of the proper length. The MTBD and distal coiled coil is from a previously solved crystal structure (PDB code 3ERR). (B) The experimental electron density map (1 σ contour) and model showing the likely interaction of the distal part of the buttress with the stalk. (C) The small domains of AAA4 and AAA5 show that H7 and H8 extend into the stalk coiled coil, and H5 and H6 extend into the buttress coiled coil.
The stalk and buttress coiled coils are extensions of helices in the small domains of AAA4 and 5, respectively, somewhat analogous to the insertion of an antiparallel coiled coil in the small domains of ClpB and Hsp104 (32). The two helices (CC1 and CC2) of the stalk coiled coil are extensions of the AAA4 helices H7 and H8, respectively, whereas the buttress coiled coil is an extension of AAA5 helices H5 and H6 (Fig. 5C). The stalk and the buttress both possess a kink close to their junctions with the core small domain (Fig. 5C), which causes their direction to deviate from that of the small-domain helices and allows the two coiled coils to interact.
The interaction between the stalk and the buttress provide new ideas for how the MTBD at the end of the stalk (Fig. 5A) might be regulated by the AAA ring. Previous work has suggested that the two helices (CC1 and CC2) in the stalk coiled coil might slide relative to one another, undergoing a half-heptad shift in different nucleotide states; this sliding would propagate ~10 nm to the MTBD and influence its polymer binding affinity (18, 33, 34). Our previous model suggested that CC1 might be pulled from its base, relative to a stationary CC2 (18). However, because CC1 and CC2 merge into the well-packed helices of the AAA4 small domain, it is unlikely that either helix can move at its base. The presence of the buttress, however, suggests another possibility. Through its contact with the stalk, the buttress might relay rigid body motions between AAA domains (e.g., between AAA4 and 5) into shear motions between the helices of the stalk coiled coil.
Allosteric communication
For dynein to function as a motor protein, nucleotide transitions at AAA1 (the main ATP hydrolytic site) must be conveyed to the linker and distant MTBD. The unexpected large gap between AAA1 and AAA2 provides a clue as to how such long-range allosteric communication might take place. In AAA proteins, ATP binds at the interface between adjacent AAA subunits, with each subunit contributing to nucleotide binding and hydrolysis (3–5). For nucleotide to bind and be hydrolyzed at AAA1, the AAA1 and AAA2 large domains must move toward one another (Fig. 6A), producing a closed conformation similar to that of AAA3-AAA4 and active nucleotide hydrolyzing sites of other AAA proteins (Fig. 3, E and F). Nucleotide binding would provide the energy that drives the closure of AAA1-AAA2, similar to the closing of the pocket that has been observed between the apo and nucleotide-bound states in the F1-ATPase (35, 36).
Fig. 6.
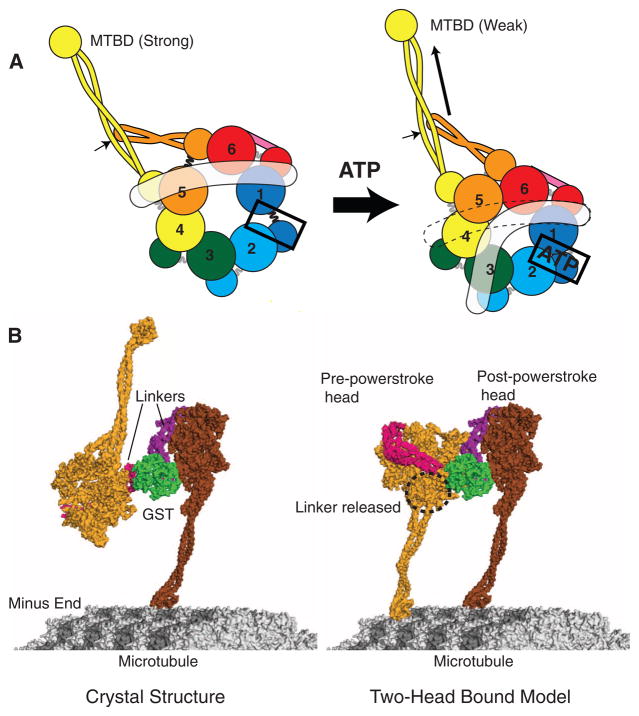
Models for dynein conformational changes. (A) A schematic model showing how ATP binding to AAA1 might propagate a conformation change to the MTBD and linker (in white). (Left) The apo state with gaps between AAA1 and 2 and between AAA5 and 6, as based on our crystal structure (Fig. 3B). (Right) The proposed consequence of ATP binding to AAA1: closure of the AAA1-2 gap, which in turn pulls upon and moves AAA2, 3, and 4. The relative motions that might ensue between AAA4 and 5 cause the detachment of the linker from AAA5. The indicated position of the linker in the ATP state is based on EM studies of Roberts et al. (8), although it may be mobile and not have a defined docked state on the ring. AAA4 and 5 movement may create a shear between the stalk (yellow coiled coil) and buttress (orange coiled coil); if the stalk preferentially interacts with the buttress through one of its helices, this could shift the registry of the stalk helices (arrow) that propagates to the MTBD to change its affinity (18, 34). (B) (Left) The dynein crystal structure with the distal stalk and MTBD, modeled as in Fig. 5A. One of the MTBDs is docked onto the microtubule as determined previously (18). (Right) One possible model in which the second head (light orange) has its MTBD docked onto the microtubule. To allow this head and its MTBD to bind to the microtubule, its linker was undocked from AAA5 and rotated about subdomain 3, and a slight bend was applied to the stalk of the front (left) head.
Closure of the AAA1–AAA2 gap by nucleotide binding could trigger a conformational change that propagates around the ring to reach the AAA4s/AAA5L unit. We speculate that this could be achieved by a domino effect, in which the closure of the AAA1-AAA2 gap induces the motions of adjacent rigid units (AAA2s/AAA3L, AAA3s/AAA4L, and AAA4s/AAA5L). Any movement in this last unit (AAA4s/AAA5L) will have the combined effect of applying tension to the binding interface between the stalk and the buttress, as well as between the linker and the AAA5 large domain. The nucleotide state of AAA2, 3, and 4 (not known in our structure) might subtly change the positions of these domains and thus regulate the transmission of the conformational change between AAA1 and AAA4s/AAA5L.
Important consequences of ATP-induced rigid body movements of AAA domains are a likely conformational change in the stalk and the detachment of the linker from its contact with AAA5 large, which has been observed by EM (8). The fate of the linker after its detachment remains unknown. The H2 and H3/β4 inserts that lie on top of the large domains in AAA2, AAA3, and AAA4 (Fig. 3F) might provide secondary docking sites for the linker. Indeed, comparable inserts provide binding sites for the AAA helicase RuvB with RuvA (37) and the AAA transcriptional activator PspF with the σ54 subunit of RNA polymerase (23). Alternatively, once detached from AAA5, the linker might be mobile, as has been suggested by its positional variation seen by EM (8). Similar to the conformational change in the neck linker of kinesin (1), a transition of the dynein linker from a detached or weakly bound “pre-powerstroke” state to a strongly, AAA5-docked “post-powerstroke” state (8) may constitute part of the mechanism for producing unidirectional motion.
Interaction of the dynein dimer with micro-tubules
Our structure of a GST-dimerized dynein provides insights and raises new questions concerning how cytoplasmic dynein binds to and moves along a microtubule. To move processively, myosin V, myosin VI, and kinesin bind with both motor domains (heads) to their tracks, at least for some portion of their motility cycles (38, 39). To enable simultaneous microtubule binding of both MTBDs, the two dynein motor domains must rearrange substantially from the conformation found in our crystal structure (Fig. 6B). Generating a two-head-bound microtubule intermediate cannot be achieved simply by a rotation around the GST-linker boundary. Moreover, the possible ways to position the two heads on the microtubule are constrained by a “short leash” between the GST and the motor domains (Fig. 1B). However, we could dock the second head to the microtubule and create a two-head-bound intermediate by rotating the motor ring, detaching the linker, and altering the stalk (Fig. 6B, right). In this tentative model, the MTBDs are bound to neighboring protofilaments of the microtubule, with one MTBD positioned 8 nm in front of the other (Fig. 6B). The front head with the detached linker is in a pre-powerstroke state, and the rear head with the docked linker is in a post-powerstroke state (7, 8), which is similar to the conformation of the mechanical elements of kinesin and myosin in their two-head-bound intermediate state (38, 39).
The two-head-bound model shown in Fig. 6B raises questions of how the rear MTBD advances past the front MTBD as dynein steps along the microtubule (19). Redocking of the linker in the front head would pull on the rear head, causing it to move forward after it dissociates from the microtubule. It is possible that the MTBD of the rear head also swings forward, through a rotation of the ring or a change in the stalk-ring angle, so that it can more easily rebind to a new tubulin binding site toward the micro-tubule minus end. Information on the relative positions of the ring, stalk/MTBD, and linker in different nucleotide states will help to resolve the sequence of conformational changes that occurs as cytoplasmic dynein steps along the microtubule.
Supplementary Material
Acknowledgments
We thank N. Zhang (UCSF), J. Holton (Advanced Light Source), H. Schmidt (MRC), E. Gleave (MRC), and M. Sirajuddin (UCSF) for their assistance and helpful discussions. This work was supported by the NIH (R.D.V.), Jane Coffin Child Foundation (A.P.C.), Lymphoma and Leukemia Foundation (A.P.C.), American Heart Association (C.C.), and Genentech Graduate Student Fellowship (C.C.). The PDB accession number for this dynein dimer structure is 3QMZ.
Footnotes
References and Notes
- 1.Vale RD, Milligan RM. Science. 2000;288:88. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 2.Neuwald AF, Aravind L, Spouge JL, Koonin EV. Genome Res. 1999;9:27. [PubMed] [Google Scholar]
- 3.Tucker PA, Sallai L. Curr Opin Struct Biol. 2007;17:641. doi: 10.1016/j.sbi.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Erzberger JP, Berger JM. Annu Rev Biophys Biomol Struct. 2006;35:93. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 5.Enemark EJ, Joshua-Tor L. Curr Opin Struct Biol. 2008;18:243. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ulbrich C, et al. Cell. 2009;138:911. doi: 10.1016/j.cell.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 7.Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Nature. 2003;421:715. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 8.Roberts AJ, et al. Cell. 2009;136:485. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samso M, Koonce MP. J Mol Biol. 2004;340:1059. doi: 10.1016/j.jmb.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 10.Wickstead B, Gull K. Traffic. 2007;8:1708. doi: 10.1111/j.1600-0854.2007.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hook P, Vallee RB. J Cell Sci. 2006;119:4369. doi: 10.1242/jcs.03176. [DOI] [PubMed] [Google Scholar]
- 12.Kon T, Mogami T, Ohkura R, Nishiura M, Sutoh K. Nat Struct Mol Biol. 2005;12:513. doi: 10.1038/nsmb930. [DOI] [PubMed] [Google Scholar]
- 13.Kon T, Nishiura M, Ohkura R, Toyoshima YY, Sutoh K. Biochemistry. 2004;43:11266. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- 14.Reck-Peterson SL, Vale RD. Proc Natl Acad Sci U S A. 2004;101:1491. doi: 10.1073/pnas.2637011100. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Cho C, Reck-Peterson SL, Vale RD. J Biol Chem. 2008;283:25839. doi: 10.1074/jbc.M802951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gee MA, Heuser JE, Vallee RB. Nature. 1997;390:636. doi: 10.1038/37663. [DOI] [PubMed] [Google Scholar]
- 17.Koonce MP. J Biol Chem. 1997;272:19714. doi: 10.1074/jbc.272.32.19714. [DOI] [PubMed] [Google Scholar]
- 18.Carter AP, et al. Science. 2008;322:1691. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reck-Peterson SL, et al. Cell. 2006;126:335. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Materials and methods are available as supporting material on Science Online.
- 21.Davies JM, Brunger AT, Weis WI. Structure. 2008;16:715. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 22.Serohijos AW, Chen Y, Ding F, Elston TC, Dokholyan NV. Proc Natl Acad Sci U S A. 2006;103:18540. doi: 10.1073/pnas.0602867103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rappas M, et al. Science. 2005;307:1972. doi: 10.1126/science.1105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fodje MN, et al. J Mol Biol. 2001;311:111. doi: 10.1006/jmbi.2001.4834. [DOI] [PubMed] [Google Scholar]
- 25.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Cell. 2009;139:744. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bochtler M, et al. Nature. 2000;403:800. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- 27.Bushnell DA, Cramer P, Kornberg RD. Structure. 2001;9:R11. doi: 10.1016/s0969-2126(00)00554-2. [DOI] [PubMed] [Google Scholar]
- 28.Enemark EJ, Joshua-Tor L. Nature. 2006;442:270. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 29.Thomsen ND, Berger JM. Cell. 2009;139:523. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. Cell. 2004;119:47. doi: 10.1016/j.cell.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 31.Grum VL, Li D, MacDonald RI, Mondragon A. Cell. 1999;98:523. doi: 10.1016/s0092-8674(00)81980-7. [DOI] [PubMed] [Google Scholar]
- 32.Doyle SM, Wickner S. Trends Biochem Sci. 2009;34:40. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 33.Gibbons IR, et al. J Biol Chem. 2005;280:23960. doi: 10.1074/jbc.M501636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kon T, et al. Nat Struct Mol Biol. 2009;16:325. doi: 10.1038/nsmb.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abrahams JP, Leslie AGW, Lutter R, Walker JE. Nature. 1994;370:621. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Oster G. Nature. 1998;396:279. doi: 10.1038/24409. [DOI] [PubMed] [Google Scholar]
- 37.Yamada K, et al. Mol Cell. 2002;10:671. doi: 10.1016/s1097-2765(02)00641-x. [DOI] [PubMed] [Google Scholar]
- 38.Gennerich A, Vale RD. Curr Opin Cell Biol. 2009;21:59. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kodera N, Yamamoto D, Ishikawa R, Ando T. Nature. 2010;468:72. doi: 10.1038/nature09450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.