Abstract
We previously identified a complex of 3′ → 5′ exoribonucleases, designated the exosome, that is expected to play a major role in diverse RNA processing and degradation pathways. Further biochemical and genetic analyses have revealed six novel components of the complex. Therefore, the complex contains 11 components, 10 of which are predicted to be 3′ → 5′ exoribonucleases on the basis of sequence homology. Human homologs were identified for 9 of the 11 yeast exosome components, three of which complement mutations in the respective yeast genes. Two of the newly identified exosome components are homologous to known components of the PM–Scl particle, a multisubunit complex recognized by autoimmune sera of patients suffering from polymyositis–scleroderma overlap syndrome. We demonstrate that the homolog of the Rrp4p exosome subunit is also a component of the PM–Scl complex, thereby providing compelling evidence that the yeast exosome and human PM–Scl complexes are functionally equivalent. The two complexes are similar in size, and biochemical fractionation and indirect immunofluorescence experiments show that, in both yeast and humans, nuclear and cytoplasmic forms of the complex exist that differ only by the presence of the Rrp6p/PM–Scl100 subunit exclusively in the nuclear complex.
Keywords: Exoribonucleases, exosome, polymyositis-scleroderma, RRP4
The RRP4 gene was identified initially in the yeast Saccharomyces cerevisiae, via a mutation that resulted in defective pre-rRNA processing (Mitchell et al. 1996). Biochemical analyses revealed that Rrp4p is a component of a protein complex that was designated the exosome (Mitchell et al. 1997). Initial characterization identified five components of the exosome; Rrp4p, Rrp41p (Ski6p), Rrp42p, Rrp43p, and Rrp44p (Dis3p). Of these, recombinant Rrp4p, Rrp41p, and Rrp44p were each demonstrated to have 3′ → 5′ exonuclease activity in vitro (Mitchell et al. 1997). The in vitro activities shown by the recombinant proteins were not, however, identical. Rrp4p is a distributive, hydrolytic enzyme, Rrp44p is a processive, hydrolytic enzyme, and Rrp41p is a processive, phosphorolytic enzyme. Consistent with this activity, Rrp44p is homologous to Escherichia coli RNase R (vacB), a member of the RNase II family of processive, hydrolytic exonucleases (Cheng et al. 1998), whereas Rrp41p is homologous to E. coli RNase PH, a phosphorolytic exonuclease (Mian 1997; Mitchell et al. 1997). Rrp42p and Rrp43p are also homologous to RNase PH (Mian 1997; Mitchell et al. 1997), and, therefore, the five initial members of the complex were all known or strongly predicted to be 3′ → 5′ exonucleases. It was, however, notable that the purified exosome complex exhibited only a distributive, hydrolytic activity in vitro; no processive or phosphorolytic activities were observed (Mitchell et al. 1996, 1997). This observation suggested that a reason for the assembly of multiple activities into one complex might be to allow their coordinate repression in the absence of activation by specific cofactors.
In all eukaryotes, the mature 5.8S, 18S, 25S/28S rRNAs are generated from a single large pre-rRNA by post-transcriptional processing. The five components of the exosome that were identified initially were all shown to be required for the 3′ processing of the 7S pre-rRNA to the mature 5.8S rRNA; genetic depletion of each gave a very similar processing defect, which closely resembled that seen in the original rrp4-1 mutation (Mitchell et al. 1996, 1997). Subsequent analyses revealed that the exosome functions not only as an RNA processing complex but is also required for specific RNA turnover pathways. The degradation of the excised spacer fragment extending from the 5′ end of the 35S primary transcript to cleavage site A0 within the 5′ external transcribed spacer (5′ ETS) region is defective in the rrp4-1 strain and in strains depleted of Rrp4p, Rrp41p, Rrp42p, Rrp43p, or Rrp44p (de la Cruz et al. 1998). A wider role for the exosome in RNA metabolism was revealed by analyses that showed that Rrp4p and Rrp41p (Ski6p) both function in the 3′ → 5′ pathway of mRNA degradation (Anderson and Parker 1998). From these observations, the exosome complex, or related complexes, were predicted to be present in both the nucleolus and the cytoplasm.
Expression of the human homolog of Rrp4p, hRrp4p, in yeast was shown to complement a rrp4-1 mutation and glycerol gradient centrifugation indicated that hRrp4p was present in HeLa cell lysates in a complex of similar size to the yeast exosome (Mitchell et al. 1997). These data suggested that a complex homologous to the exosome was present in human cells.
A large number of human autoimmune diseases have been identified. Some of these, notably scleroderma, are associated with the development of antibodies directed against nucleolar epitopes (for review, see Reimer 1990). In a relatively rare autoimmune disease, polymyositis–scleroderma overlap syndrome (Reimer et al. 1986), patients frequently develop antibodies directed against a 100-kD protein, PM–Scl100 (Bluthner and Bautz 1992; Ge et al. 1992). Less frequently another protein, PM–Scl75 (Alderuccio et al. 1991), is also targeted. These two proteins are components of a large complex, designated the PM–Scl complex, that was estimated to have between 11 (Reimer et al. 1986) and 16 (Gelpi et al. 1990) components. Interestingly, PM–Scl100 is homologous to the E. coli 3′ →5′ exoribonuclease, RNase D (Briggs et al. 1998), whereas PM–Scl75 shows homology to RNase PH (Mian 1997).
Here, we report the identification of six new components of the yeast exosome and characterize distinct nuclear and cytoplasmic forms of this complex. Two of the newly defined exosome subunits are homologous to the human PM–Scl100 and PM–Scl75 autoantigens, and these proteins are associated with the human homolog of another exosome component. Moreover, like the yeast exosome, related human complexes are localized in nucleus and cytoplasm. Together, these data provide strong evidence that the PM–Scl complex is directly homologous to the yeast exosome.
Results
Identification of new components of the exosome complex
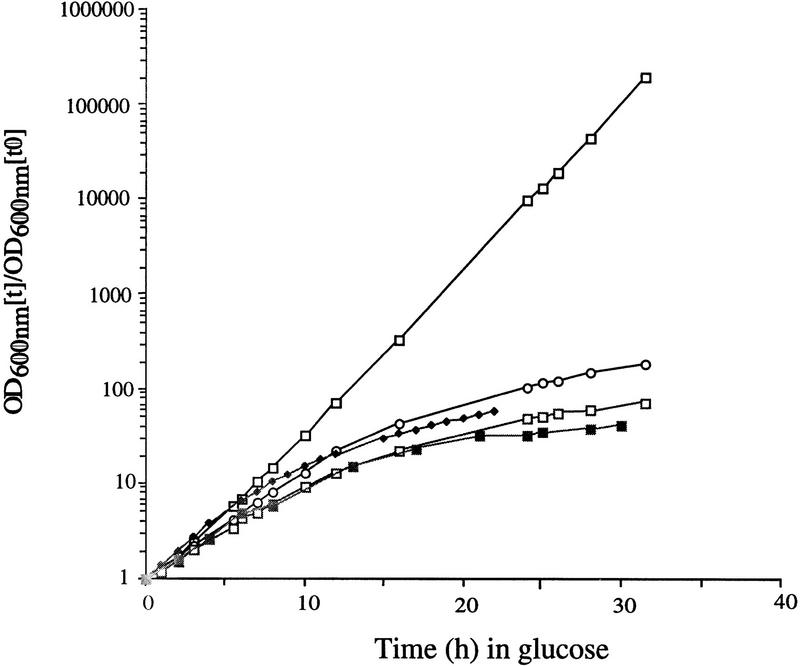
The initial characterization of components that coprecipitated with protein A-tagged Rrp4p (ProtA–Rrp4p) identified four proteins (Mitchell et al. 1997). Three of these proteins, Rrp41p, Rrp42p, and Rrp43p, were homologous to E. coli RNase PH. However, the yeast genome contains three other putative open reading frames (ORFs) with homology to RNase PH; YDR280w (RRP45), YGR095c (RRP46; Mian 1997), and YGR158c (MTR3). The RRP45 and RRP46 ORFs were each precisely deleted in diploid strains of yeast (see Materials and Methods). On sporulation of each diploid, only two viable spores were recovered per tetrad and in each case the viable spores carried the wild-type allele. We conclude that RRP45 and RRP46 are both essential, at least for spore viability. Conditional alleles were constructed by placing RRP45 and RRP46 under the control of a repressible GAL10 promoter (see Materials and Methods). In each case, the strains formed only microcolonies on solid medium containing 2% glucose (data not shown) and ceased growth following transfer from liquid RSG (raffinose/sucrose/galactose) medium to liquid glucose medium (Fig. 1). We conclude that Rrp45p and Rrp46p are essential for viability.
Figure 1.
The newly identified components of the exosome complex are required for viability. Growth curves of GAL-regulated constructs following transfer to glucose medium. Strains were pregrown in permissive, RSG medium and transferred to repressive, glucose medium for the times indicated. Strains were maintained in exponential growth by dilution with prewarmed medium. Cell densities measured by OD600 are shown corrected for dilution. (⋄) Wild type; (○) GAL::rrp45; (□) GAL::rrp46; (♦) GAL:csl4; (█) GAL::rrp40.
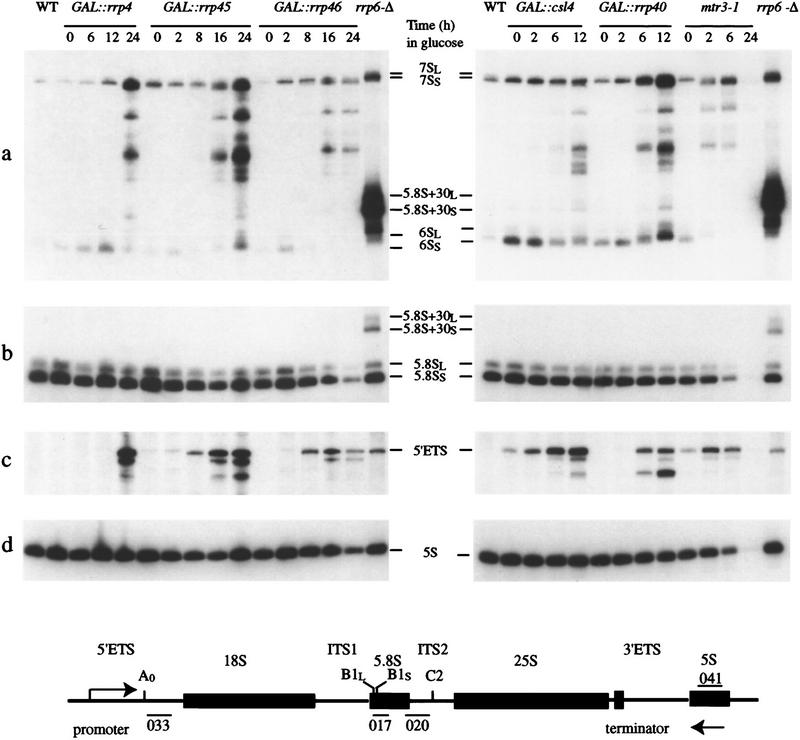
The strains depleted of Rrp45p or Rrp46p showed an accumulation of 3′ extended forms of the 5.8S rRNA that extended in a ladder to the size of the 7S pre-rRNA but not beyond (Fig. 2). This phenotype is essentially identical to that seen in strains depleted for Rrp4p (Fig. 2a) or the four other components of the exosome identified previously (Mitchell et al. 1997). Mtr3p is essential for viability (Kadowaki et al. 1995), and a strain carrying a temperature-sensitive lethal mtr3-1 allele (generously provided by A.M. Tartakoff, Case Western Reserve University, Cleveland, OH) was analyzed. This strain also accumulated 3′ extended forms of the 5.8S rRNA after transfer to the nonpermissive temperature (37°C; Fig. 2a). The mtr3-1 strain rapidly ceases growth following transfer to 37°C, and little pre-rRNA was recovered at the 24 hr time point, presumably because of the very low growth rate. In addition, the strains depleted of Rrp4p, Rrp45p, or Rrp46p or carrying mtr3-1 each accumulated the excised 5′ ETS region of the pre-rRNA, extending from the 5′ end of the primary transcript to cleavage site A0 (Fig. 2c; 5′ ETS), as well as degradation intermediates (see also de la Cruz et al. 1998). We conclude that the RNase PH homologs Rrp45p, Rrp46p, and Mtr3p are each required for the function of the exosome complex.
Figure 2.
The newly identified components of the exosome complex are required for pre-rRNA processing. Northern analysis of processing of the 5.8S and degradation of the 5′ETS region of the pre-rRNA in exosome mutants. RNA was extracted from strains carrying GAL-regulated constructs following transfer from permissive, RSG medium to repressive, glucose medium for the times indicated, or from the mtr3-1 strain following transfer from 25°C to 37°C for the times indicated. RNA was separated on an 6% polyacrylamide gel and hybridized with: (a) oligonucleotide 020 (complementary to the 5.8S/ITS2 boundary), (b) oligonucleotide 017 (hybridizing to the mature 5.8S rRNA), (c) oligonucleotide 033 (hybridizing to the 5′ETS around position +278). (d) oligonucleotide 041 (hybridizing to the 5S rRNA). The position of migration of the pre-rRNA species is indicated. The species labeled 5′ ETS extends from the transcription start site to site A0 (+610). Also shown is a cartoon of the rDNA (not to scale) with the mature rRNA regions as rectangles and the transcribed spacers as lines. The 18S, 5.8S, and 25S rRNAs are cotranscribed, separated by the internal transcribed spacers (ITS1 and ITS2) and flanked by the external transcribed spacers (5′ETS and 3′ETS). The 5S rRNA is independently transcribed in the opposite direction. The mature 5.8S rRNA is synthesized from the 7S pre-rRNA, which is 3′ extended to site C2 in ITS2. The 5′ end of the 5.8S rRNA is generated by processing at sites at B1L and B1S, which lie about 8 nucleotides apart, generating 5.8SL and 5.8SS, respectively. Because this event precedes 3′ processing, the 7S, 6S, and 5.8S + 30 pre-rRNAs all show 5′ heterogeneity, generating, e.g., 6SL and 6SS.
These observations prompted us to re-examine the biochemical purification of the exosome complex. A whole-cell extract from a strain expressing ProtA–Rrp4p under the control of the endogenous RRP4 promoter from a low-copy-number CEN plasmid (Mitchell et al. 1997) was passed over an IgG–Sepharose column, and proteins were eluted from the bound IgG–ProtA–Rrp4p complex by use of a gradient of Mg2+ (Görlich et al. 1996). Proteins were separated by SDS-PAGE, and bands were excised and subjected to sequencing analysis by mass spectroscopy (see Kuster and Mann 1998; Shevchenko et al. 1996). Most bands were identified by high mass accuracy peptide mass mapping as described by Jensen et al. (1996). Several of the bands contained more than one gene product that were, however, identified without recourse to mass spectrometric peptide sequencing. In these cases, an iterative approach was used. First all tryptic peptide masses were searched against a comprehensive protein database, identifying one yeast protein. The peptide masses remaining after detailed comparison of the spectrum against the found sequence (second pass search), were again searched in the database, yielding another yeast protein. In some cases MALDI peptide mapping did not unequivocally identify the components in a band. In these cases, nanoelectrospray on a novel quadrupole Time of Flight instrument was performed (Shevchenko et al. 1997a; Wilm et al. 1996). Two broad peaks of eluted proteins were observed; Rrp44p eluted at around 500 mm MgCl2 (Fig. 3A, lanes 4–6) whereas Rrp41p, Rrp42p, Rrp43p, Rrp45p, Rrp46p, and Mtr3p coeluted at around 1.6–1.8 m MgCl2 (Fig. 3A, lanes 16–18). Two other proteins observed in the 1.6–1.8 m MgCl2 fractions were identified as Rrp6p (YOR001w) and Rrp40p (YOL142w). The coelution of these components supports their presence in a single complex. ProtA–Rrp4p was eluted only in the acid wash of the column (Fig. 3A, lane HAc).
Figure 3.
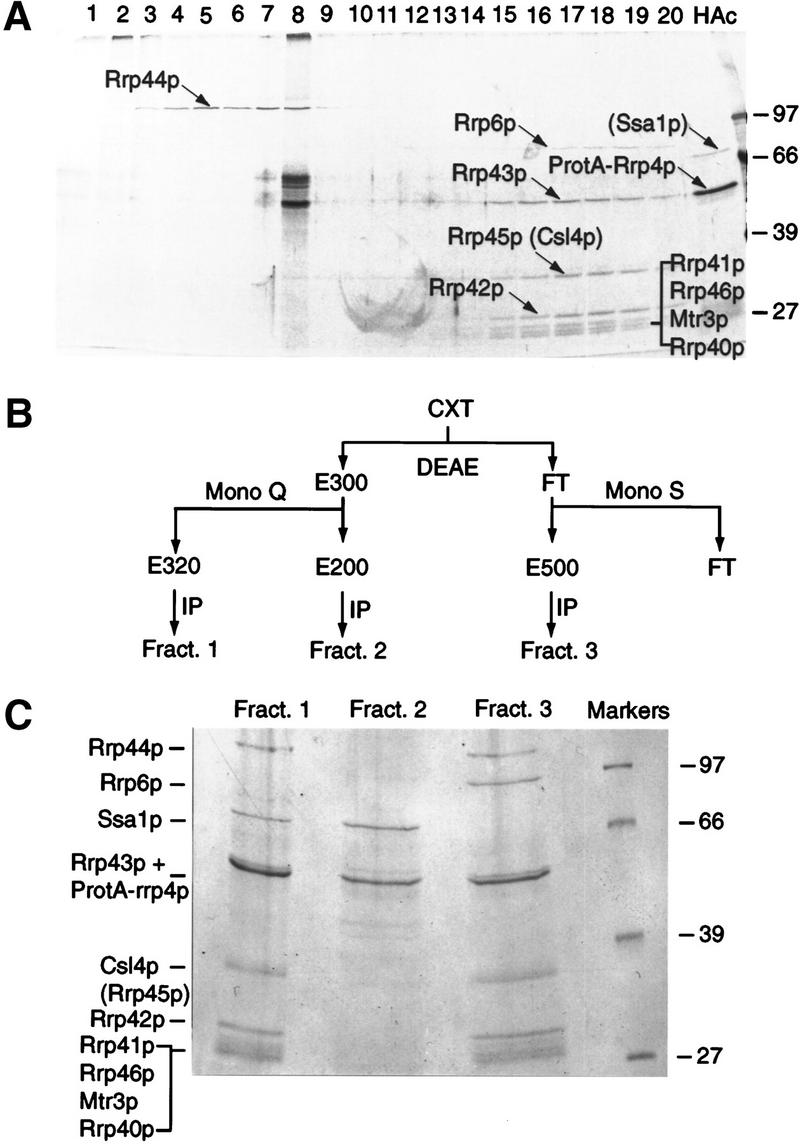
Fractionation of the exosome complex and identification of new components. (A) Proteins associated with IgG–Sepharose via binding to ProtA–Rrp4p were eluted using a gradient of MgCl2 and analyzed by SDS-PAGE. (Lanes 1–20) Material eluted with a 100 mm step gradient of MgCl2 concentration from 100 mm (lane 1) to 2 m (lane 20). (HAc) Proteins eluted by the acid wash. Proteins are visualized by silver staining. The strong bands specifically seen in lane 8 were not observed in other experiments. (B) Purification scheme. A whole-cell extract (CXT) was batch-bound to DEAE–Sepharose FF. Bound material was eluted (E300) with TMN buffer containing 300 mm NaCl/10% glycerol (TMN-300). The eluate, in TMN-100, was passed through a Mono Q column and bound material was eluted stepwise with TMN-150, TMN-200 (E200), TMN-320 (E320), and TMN-500. Material that failed to bind to DEAE–Sepharose FF (FT) was passed through a Mono S column and bound material was eluted with TMN-500 (E500). Each sample was immunoprecipitated on IgG–Sepharose. (C) Proteins present in fractions 1, 2, and 3, obtained as outlined in B, were separated by SDS-PAGE. Approximately twofold more of the material recovered in fractions 2 and 3 was loaded onto the gel, as compared with fraction 1. Proteins positively identified by mass spectroscopy are indicated. Species in brackets were not identified in the preparations shown but are predicted to be present from other analyses. Molecular weight markers are also shown. Proteins are visualized by Coomassie staining.
Following immunoprecipitation of ProtA–Rrp4p, all of the components were recovered with apparent stoichiometry, with the exception of Rrp6p, which was estimated from Coomassie staining to be approximately fivefold less abundant than the other components (data not shown). Because Rrp6p was eluted only at 1.6–1.8 m MgCl2, along with most other exosome components, it seemed unlikely that this low abundance was due to a weaker association with the exosome complex. Therefore, Rrp6p might be associated with only a subfraction of the exosome complex. To test this model, a whole-cell extract from the ProtA–Rrp4p strain was fractionated by column chromatography (see Fig. 3B). Three fractions containing ProtA–Rrp4p were recovered (Fig. 3C). The most abundant complex was recovered in fraction 1; this complex probably corresponds to the major complex purified previously by glycerol gradient centrifugation and immunoprecipitation (Mitchell et al. 1997). In addition to the previously characterized components of the exosome, fraction 1 contained Rrp40p, Rrp46p, and Mtr3p. Other protein bands in fraction 1 were identified as Csl4p (YNL232w) and the cytoplasmic Hsp70-like protein Ssa1p (YAL005c), but Rrp6p was not present. Fraction 3 contained the same exosome components as fraction 1, but lacked Ssa1p and contained Rrp6p. Rrp43p, which comigrates with ProtA–Rrp4p and the IgG heavy chain (Mitchell et al. 1997), was identified in fraction 3 but not in fraction 1 (Fig. 3C). Csl4p and Rrp45p also appear to comigrate in SDS-PAGE; from the band marked Csl4p + Rrp45p, only Rrp45p was identified from the preparation shown in Figure 3A, whereas only Csl4p was identified from the preparations shown in Figure 3C. It is, however, very likely that Rrp43p and Rrp45p are components of both complexes (see also below). Consistent with the recovery of Rrp6p in the total immunoprecipitate (Fig. 3A), approximately threefold less ProtA–Rrp4p was recovered in fraction 3 than in fraction 1 (twofold less of the material recovered in fraction 1 was loaded onto the gel in Fig. 3C than of the material in fractions 2 and 3). Fraction 2 comprises only ProtA–Rrp4p with Ssa1p, and was approximately fourfold less abundant than fraction 1. Consistent with glycerol gradient centrifugation (Mitchell et al. 1997), no free ProtA–Rrp4p was recovered. The ProtA–Rrp4p–Ssa1p complex was detected in variable yield on glycerol gradients (typically 5%–10% of total ProtA–Rrp4p; Mitchell et al. 1997) and may be due to dissociation of ProtA–Rrp4p from the complex during purification, possibly related to the presence of the protein A tag.
CSL4 was identified previously in a screen for synthetic lethality with the chromatin protein Cep1p and is essential for viability (Baker et al. 1998). Conditional alleles of CSL4 and RRP40 were constructed by placing their expression under the control of a GAL10 promoter (see Materials and Methods). In each case, the strains formed only microcolonies on solid medium containing 2% glucose (data not shown). Following transfer from liquid RSG medium to liquid glucose medium (Fig. 1) the strains ceased growth and 3′ extended forms of the 5.8S rRNA accumulated (Fig. 2a), showing a pattern of intermediates similar to other exosome mutants. Depletion of Rrp40p or Csl4p also led to the accumulation of the 5′ ETS pre-rRNA spacer fragment (Fig. 2c). Therefore, genetic depletion of any of the 10 essential components identified by copurification results in very similar defects in the processing of the 5.8S rRNA, showing that they form a single complex.
RRP6 is not essential for viability (Briggs et al. 1998), and a strain carrying a precise deletion of RRP6 was constructed (see Materials and Methods). This strain was impaired in growth at all temperatures and was nonviable at 37°C (temperature-sensitive lethal; data not shown). The rrp6-Δ strain was defective in the 3′ processing of the 5.8S rRNA, but differed from the other components of the exosome insofar as it accumulated a discrete species, 5.8S + 30, that was 3′ extended by ∼30 nucleotides (Fig. 2a; Briggs et al. 1998). The rrp6-Δ strain also accumulated the 5′ ETS region of the pre-rRNA (Fig. 2c). We conclude that the exosome includes at least 11 components, all of which are required for normal 3′ processing of the 5.8S rRNA and degradation of the 5′ ETS region. Ten of these are essential for viability, whereas the absence of Rrp6p results in temperature-sensitive lethality (see Table 1).
Table 1.
Components of the exosome
| Protein
|
Gene
|
Phenotype
|
E. coli homolog
|
Mammalian homolog
|
Comments
|
|---|---|---|---|---|---|
| Rrp4p | YHR069c | essential | S1 RNA BD | hRrp4p 43% (52%) | hRrp4p complements rrp4-1 |
| Rrp40p | YOL142w | essential | S1 RNA BD | hRrp40p 35% (48%) | homologous to Rrp4p |
| Rrp41p/Ski6p | YGR195w | essential | RNase PH | hRrp41p 35% (55%) | |
| Rrp42p | YDL111c | essential | RNase PH | hRrp42p 25% (51%) | |
| Rrp43p | YCR035c | essential | RNase PH | ||
| Rrp45p | YDR280w | essential | RNase PH | PM-Scl75 38% (64%) | human KIAA0116 and OIP2 also homologous |
| Rrp46p | YGR095c | essential | RNase PH | hRrp46p 35% (48%) | |
| Mtr3p | YGR158c | essential | RNase PH | ||
| Rrp44p/Dis3p | YOL021c | essential | RNase R | hDis3p | hDis3p complements |
| (RNase II family) | 45% | dis3-81 | |||
| Cs14p | YNL232w | essential | S1 RNA BD | hCs14p 48% (56%) | hCs14p complements csl4-1 |
| Rrp6p | YOR001w | ts lethal | RNase D | PM-Scl100 32% (52%) | component only of nuclear complex |
Rrp4p, Rrp40p, and Cs14p are not clearly homologous to known exonucleases from E. coli but are predicted to include regions homologous to the S1 RNA-binding domain (S1 RNA BD). For the human homologs numbers represent the percentage identity (similarity). In the case of Cs14p (Baker et al. 1998), Rrp40p, Rrp41p, Rrp42p, and Rrp46p, the numbers are based on consensus cDNAs assembled from ESTs and may not be fully accurate. (See text for references.)
It is unclear whether Ssa1p is a genuine component of the complex or associates with the exosome as a consequence of the protein A tag present on Rrp4p. In either event, because Ssa1p is predominantly cytoplasmic (Chirico et al. 1988; Deshaies et al. 1988), one obvious possibility was that fractions 1 and 3 contained cytoplasmic and nuclear forms of the exosome, respectively. To test this possibility, a ProtA–Rrp6p fusion was constructed (see Materials and Methods). The ProtA–Rrp6p construct complemented the temperature-sensitive lethal growth phenotype of the rrp6-Δ mutation, largely suppressed the accumulation of the 5.8S + 30 species in this strain, and cosedimented with ProtA–Rrp4p through glycerol gradients (data not shown). Therefore, we conclude that the protein A epitope does not grossly impair the ability of Rrp6p to associate with the exosome or to function in the cell.
Immunolocalization of the ProtA–Rrp6p and ProtA–Rrp4p (Mitchell et al. 1996) fusion proteins was compared to the nucleolar marker ProtA–Nop1p (Grandi et al. 1993) and staining of the nucleoplasm with DAPI. ProtA–Rrp6p gave a nuclear signal, with nucleolar enrichment and a punctate nucleoplasmic staining. ProtA–Rrp4p was also observed in the nucleoplasm and nucleolus, but was additionally detected in the cytoplasm (Fig. 4). Notably, a GFP–Rrp43p fusion protein has recently been reported to be localized to both the nucleus and cytoplasm (Zanchin and Goldfarb 1999).
Figure 4.
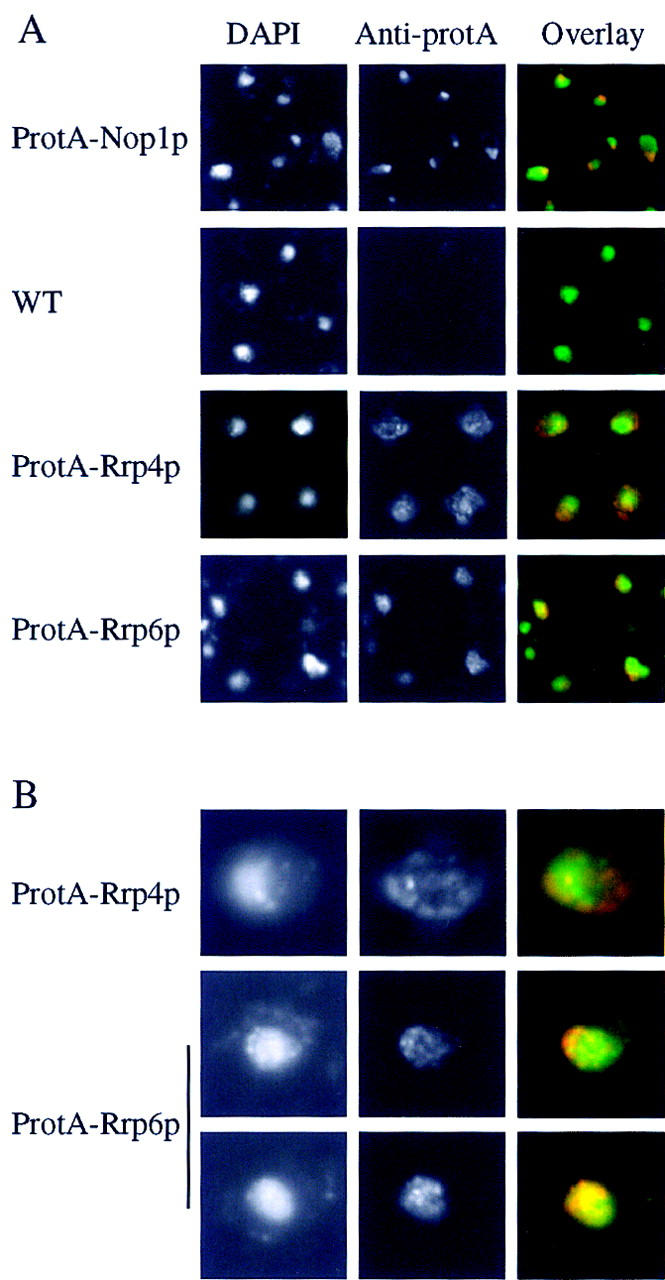
Rrp4p and Rrp6p differ in their nuclear-cytoplasmic distribution. (A) Strains expressing ProtA–Rrp4p, ProtA–Rrp6p, or ProtA–Nop1p were examined by indirect immunofluorescence using an anti-protein A antibody coupled to Texas Red. Also shown is the position of the DNA, visualized by DAPI staining. The combined image is pseudocolored with DAPI in green and Texas Red in red. For each tagged strain an otherwise isogenic wild-type control strain was also analyzed. The wild-type strain shown (P51) is isogenic with the ProtA–Rrp4p strain (see Table 2). (B) Higher resolution images are shown for the ProtA–Rrp4p and ProtA–Rrp6p to show the punctate staining pattern.
We conclude that two major forms of the exosome can be purified that contain at least 10 common components, Rrp4p, Rrp40–Rrp46p, Mtr3p, and Csl4p, all of which are essential for viability and are required for exosome function. Rrp6p is present only in a subfraction of the complex that is confined to the nucleus.
Characterization of the human PM–Scl complex
Rrp6p shows substantial homology to the human protein PM–Scl100 (Briggs et al. 1998), whereas Rrp45p is homologous to PM–Scl75 (Mian 1997), both of which are targets of autoimmune antibodies in patients suffering from polymyositis–scleroderma overlap syndrome (Alderuccio et al. 1991; Bluthner and Bautz 1992; Ge et al. 1992). Moreover, human orthologs have been identified for the Rrp4p, Rrp44p and Csl4p components of the exosome (Mitchell et al. 1997; Baker et al. 1998; Shiomi et al. 1998). Strikingly, expression of each of these cDNAs can suppress the phenotypes of mutations in the corresponding yeast genes, demonstrating their functional conservation (Mitchell et al. 1997; Baker et al. 1998; Shiomi et al. 1998). Translational searches of the human EST banks (see Materials and Methods) allowed virtual cDNAs to be assembled for hRrp40p, hRrp41p, hRrp42p, and hRrp46p; in each case, the putative human protein showed high homology to the yeast protein (Table 1). In addition, two other genes, KIAA0116 and OIP2, were found to be homologous to Rrp45p, although less so than PM–Scl75. For hRrp40p and hRrp46p, apparent products of alternative splicing were evident when the EST sequences were assembled into contigs (data not shown). In some cases, these alternative forms may have led to an overestimation of the number of discrete protein species in the PM–Scl complex.
Previous analyses showed that hRrp4p is present in a large complex (Mitchell et al. 1997). To determine whether the human homologs of other exosome components are present in the same complex, HeLa cell nuclear and cytoplasmic extracts (generously provided by Juan Valcárcel, EMBL, or prepared as described in Materials and Methods) were fractionated by glycerol gradient centrifugation. Fractions were analyzed by Western blotting with human autoimmune serum (generously provided by Walter van Venrooij, University of Nijmegen, The Netherlands) or antibodies raised against recombinant hRrp4p (Mitchell et al. 1997; Fig. 5). In the nuclear extract, PM–Scl75 and an uncharacterized protein of ∼25 kD that is also a target of the autoimmune serum (PM–Scl25) cosedimented with hRrp4p, with a peak in fractions 13 and 14. PM–Scl100 also showed substantial cosedimentation (Fig. 5A). The band at 45 kD is likely to be the species previously reported to cross-react with anti-PM–Scl75 antibodies (Alderuccio et al. 1991). PM–Scl100 was not detected in the cytoplasmic extract, but PM–Scl75 and hRrp4p cosedimented (Fig. 5B), as did PM–Scl25 (data not shown), with a peak in fractions 13 and 14.
Figure 5.
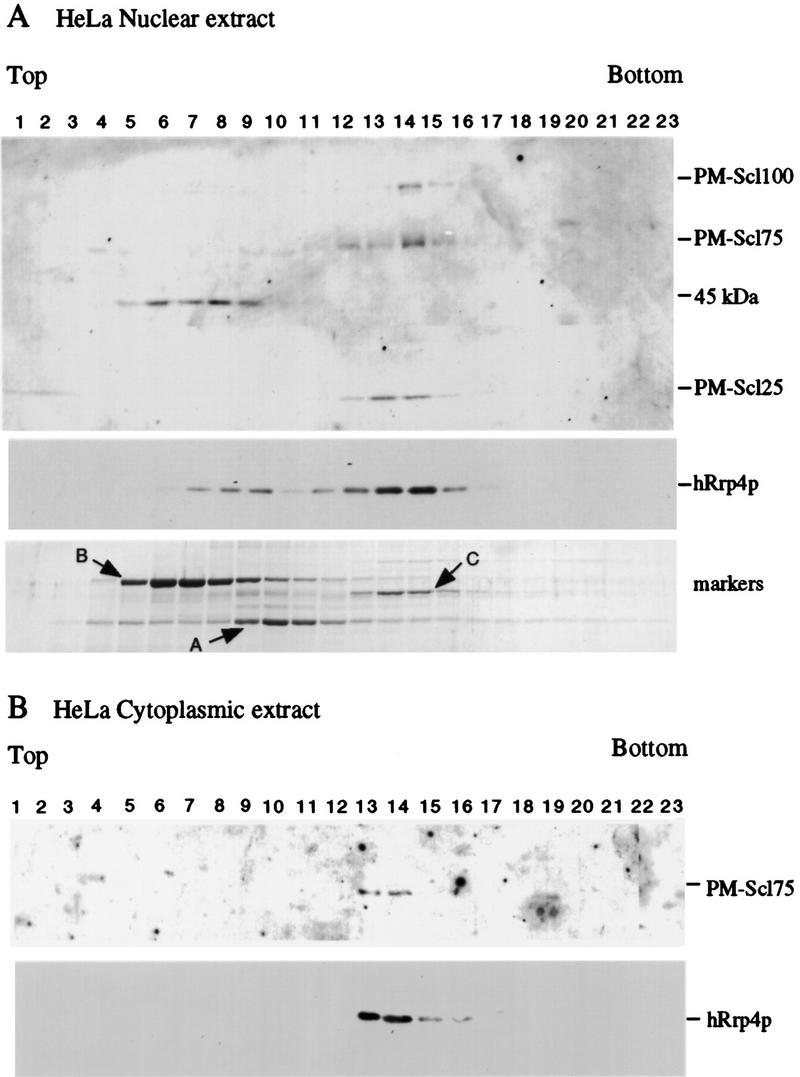
Cosedimentation of hRrp4p and the PM–Scl complex. (A) HeLa cell nuclear extract. (B) HeLa cell cytoplasmic extract. Cell extracts were fractionated by glycerol gradient centrifugation. Samples were analyzed by Western blotting decorated with human autoimmune antisera reactive against PM–Scl100, PM–Scl75, and a previously uncharacterized human protein (PM–Scl25) or with rabbit antiserum raised against recombinant hRrp4p. The serum also cross-reacts with an unrelated 45-kD protein. Also shown is the sedimentation of molecular weight markers on a gradient run in parallel with the nuclear extract. Markers: (A) alcohol deyhdrogenase from yeast (7.4S); (B) bovine serum albumin (4.3S); (C) bovine catalase (11.3S; Siegel and Monty 1966).
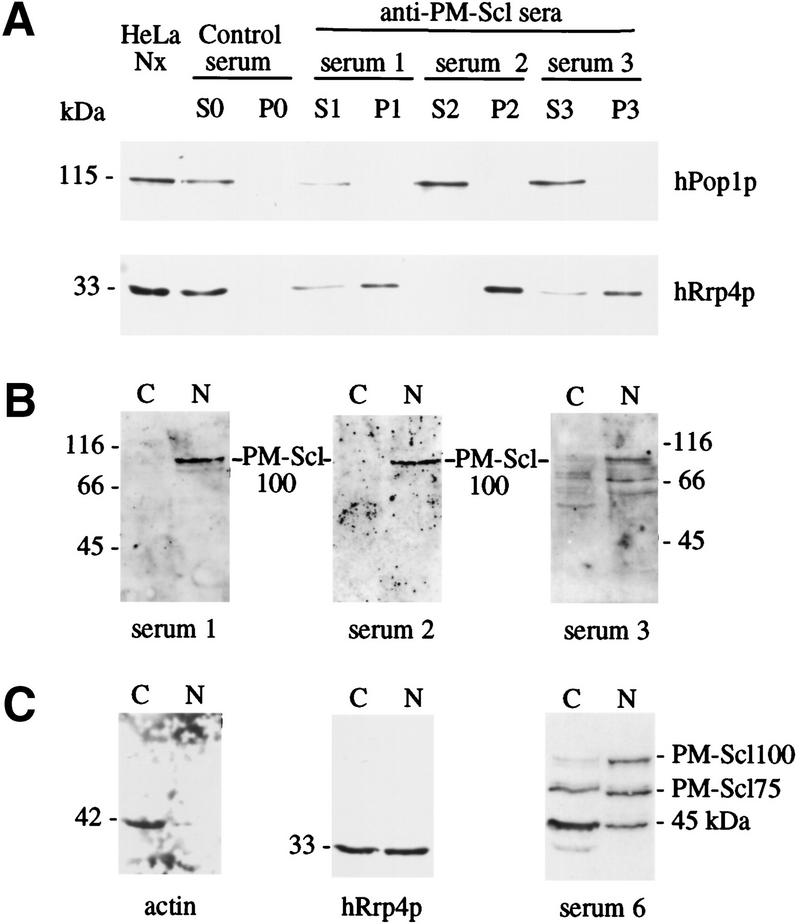
To confirm the association between PM–Scl100 and hRrp4p in the HeLa cell nuclear extract, immunoprecipitation was performed (Fig. 6). Three different autoimmune sera (sera 1–3) were used, each of which reacted specifically with PM–Scl100 on Western blots of the total nuclear extract (Fig. 6B). Following immunoprecipitation, hRrp4p was recovered in the immune precipitate (P) with each PM–Scl100 serum (Fig. 6A), but was not coprecipitated with a control human serum. In contrast, another human nucleolar protein, hPop1p, a component of RNase mitochondrial RNA processing (MRP) (Lygerou et al. 1996), was recovered exclusively in the immune supernatant (S; Fig. 6A). We conclude that hRrp4p is associated physically with PM–Scl100 in a human nuclear extract. The efficiency of precipitation of PM–Scl100 and PM–Scl75 could not be assessed in this experiment, because the secondary, anti-human antibody reacted very strongly with the human antibodies present in the immunoprecipitate.
Figure 6.
Characterization of the PM–Scl complex. (A) Three different human autoimmune sera with specificity for PM–Scl100 were used for immunoprecipitation from a HeLa nuclear extract. The total HeLa nuclear extract, supernatant (S), and pellet (P) fraction are shown. Each lane represents an equivalent quantity of lysate. Western blots were decorated with rabbit sera raised against recombinant hRrp4p or hPop1p, a component of the RNase MRP complex. (B) Western blots of total HeLa nuclear (N) and cytoplasmic (C) extracts decorated with the anti-PM–Scl100 sera used for immunoprecipitation, demonstrating the specificity of the sera. (C) Western blots of total HeLa nuclear (N) and cytoplasmic (C) extracts decorated with rabbit sera raised against recombinant hRrp4p or human actin, or with a human autoimmune serum reactive against both PM–Scl100 and PM–Scl75. Cell equivalent volumes of the nuclear and cytoplasmic fractions were loaded.
The subcellular localization of the PM–Scl complex was assessed by nuclear–cytoplasmic fractionation. Western blotting (Fig. 6C) showed that PM–Scl75 and hRrp4p were partitioned between the nuclear and cytoplasmic fractions. In contrast, PM–Scl100 was detected exclusively in the nuclear fraction (Fig. 6B,C). Rabbit antibodies directed against actin (Sigma A2066) decorated a band exclusively in the cytoplasmic fraction (Fig. 6C). Approximately equal quantities of PM–Scl75 and hRrp4p were recovered in the cytoplasmic and the nuclear fractions. Only very low amounts of PM–Scl100, PM–Scl75, and hRrp4p were detected in the residual nuclear pellet (data not shown).
We conclude that there are at least two forms of the human PM–Scl complex: a nuclear complex that includes PM–Scl100 and a cytoplasmic complex that lacks PM–Scl100. These are very likely to be directly equivalent to the nuclear and cytoplasmic forms of the yeast exosome, that similarly differ by the presence of Rrp6p, the yeast homolog of PM–Scl100, only in the nuclear complex.
Discussion
Here, we report the identification of 11 components of the nuclear exosome complex (Table 1). Remarkably, six of the components are homologous to E. coli RNase PH; Rrp41p, Rrp42p, Rrp43p, Rrp45p, Rrp46p, and Mtr3p. Of the remaining exosome components, Rrp6p is homologous to E. coli RNase D (Briggs et al. 1998), and Rrp44p is homologous to E. coli RNase R/vacB (Mitchell et al. 1997), an RNase II family member (Cheng et al. 1998). Rrp40p shows homology to Rrp4p, which was shown previously to be a 3′ → 5′ exonuclease in vitro (Mitchell et al. 1997), and, therefore, Rrp40p is also predicted to be an exonuclease. The only component of the exosome complex that does not show homology to a known exonuclease is Csl4p. It is, however, notable that both yeast Csl4p and human hCsl4p include sequences homologous to the S1 RNA-binding domain (Bycroft et al. 1997; S. Mian, pers. comm.), strongly indicating that it too interacts directly with RNA substrates. Rrp4p and Rrp40p are also predicted to contain S1 RNA-binding domains (S. Mian, pers. comm.).
We previously identified the human homolog of Rrp4p and showed that expression of the hRRP4 cDNA in yeast could suppress the temperature-sensitive lethality of the rrp4-1 allele (Mitchell et al. 1997). Subsequently, the cDNA encoding the human homolog of Rrp44p/Dis3p has been shown to partially complement a temperature-sensitive lethal dis3 allele (Shiomi et al. 1998), and the cDNA encoding hCsl4p has been shown to complement the synthetic-lethal phenotype of a csl4-1, cep1-Δ double mutant strain (Baker et al. 1998). Sequence comparisons indicate that human homologs exist for 9 of the components of the yeast exosome complex (see Table 1). Notably, Rrp6p is homologous to PM–Scl100 (Briggs et al. 1998) whereas Rrp45p is homologous to PM–Scl75. Both of these proteins are the targets of autoimmune antibodies in human patients suffering from polymyositis–scleroderma overlap syndrome (Alderuccio et al. 1991; Ge et al. 1992). The PM–Scl complex was reported to contain between 11 (Reimer et al. 1986) and 16 (Gelpi et al. 1990) proteins, as judged by SDS-PAGE analysis of immunoprecipitated proteins. We have shown by coprecipitation that hRrp4p is associated with PM–Scl100 in HeLa cell nuclear extracts, and hRrp4p cosedimented with PM–Scl75 and PM–Scl25 in both nuclear and cytoplasmic extracts. Homologs of at least three components of the exosome are present in the PM–Scl complex, providing strong evidence that these complexes are directly homologous.
Six human homologs of RNase PH were identified. These do not, however, have a 1:1 relationship with the six RNase PH homologs in the exosome. No clear human homologs were identified for Rrp43p or Mtr3p. Searches of the EST banks with these proteins identified ESTs related to KIAA0116 and OIP2; these sequences are, however, more homologous to Rrp45p than to the other yeast PH homologs (although less so than PM–Scl75). A probable interpretation is that yeast and humans have the same number of RNase PH homologs, but that some drift has occurred with duplicates of the RRP45/PM–Scl75 gene replacing other species.
Mutations in individual components of the yeast exosome inhibited both nucleolar pre-rRNA processing and cytoplasmic mRNA turnover (Anderson and Parker 1998), indicating that related complexes are present in the nucleus and the cytoplasm. Moreover, a mutation in Mtr3p leads to nuclear accumulation of poly(A)+ RNA (Kadowaki et al. 1995), as does a mutation in Dob1p/Mtr4p (de la Cruz et al. 1998; Liang et al. 1996), a putative RNA helicase required in addition to the exosome for 5.8S rRNA 3′ end maturation and degradation of the 5′ ETS fragment (de la Cruz et al. 1998). These observations suggest that the exosome may also play some role in nucleoplasmic RNA turnover or processing. Consistent with this hypothesis, GFP–Rrp43p (Zanchin and Goldfarb 1999) and ProtA–Rrp4p were detected in the nucleolus, nucleoplasm, and cytoplasm. In contrast, ProtA–Rrp6p was found to be exclusively nuclear, with a nucleolar enrichment. Two complexes could also be separated biochemically; these include 10 common components and differ in the presence of either Ssa1p, a cytoplasmic Hsp70-like protein (Chirico et al. 1988; Deshaies et al. 1988), or Rrp6p. The form lacking Rrp6p is presumed to be the cytoplasmic exosome complex, a proposal supported by the presence of Ssa1p. Approximately threefold more of this complex was recovered than the putative nuclear exosome that includes Rrp6p. Human PM–Scl100 was also restricted to the nucleus, while PM–Scl75, PM–Scl25, and hRrp4p partition between the nucleus and cytoplasm. The reported nucleolar enrichment of the human PM–Scl complex is probably a consequence of the immunodominance of PM–Scl100 in autoimmune sera (Ge et al. 1992; Gelpi et al. 1990). In fact, approximately equal amounts of the human nuclear and cytoplasmic complexes were recovered following subcellular fractionation.
We conclude that there are two forms of the exosome/PM–Scl complex in the nucleus and the cytoplasm that can be distinguished by the presence of Rrp6p/PM–Scl100 specifically in the nuclear form.
Rrp6p is not essential for viability, in contrast to the other 10 components of the exosome complex, although rrp6-Δ strains are severely impaired in growth and are temperature sensitive. Therefore, the exosome is therefore predicted to retain at least partial function in the absence of Rrp6p, a view supported by the observation that the major form of the complex lacks this protein. Conversely, all of the PM–Scl100 present in Hela cell lysates appeared to be associated with the PM–Scl complex, suggesting that Rrp6p/PM–Scl100 may not function independently of the complex in vivo.
In E. coli, the homologs of the exosome components are not present in a related complex. However, the degradosome complex includes another 3′ → 5′ exonuclease, PNPase, together with the endonuclease and exonuclease RNase E and the putative RNA helicase RhlB (Carpousis et al. 1994; Py et al. 1996; Mackie 1998; Vanzo et al. 1998). It appears that throughout evolution, major activities involved in RNA processing and degradation have been assembled into large complexes, possibly to allow their coordinate regulation. The composition of these complexes are, however, very different in bacteria and eukaryotes.
Materials and methods
Strains and media
Except where stated, strains were grown in liquid or on solid minimal medium containing 0.67% yeast nitrogen base (DIFCO) and 2% glucose with appropriate supplements. For depletion, strains carrying GAL-regulated constructs were pregrown in RSG (2% peptone, 1% yeast extract, 2% raffinose, 2% sucrose, 2% galactose) and transferred to YPD (2% peptone, 1% yeast extract, 2% glucose).
Yeast strains used and constructed in this study are listed in Table 2. Gene disruptions of RRP45 and RRP46 were generated by a PCR strategy in the diploid strain BMA38 (Baudin et al. 1993) resulting in the replacement of the complete ORF by an auxotrophic marker (see Table 2). Successful disruption was confirmed by Southern hybridization. Chromosomal DNA from the RRP45/rrp45::TRP1 and RRP46/rrp46::HIS3 strains was digested by EcoRI–HindIII or KpnI–EcoRI, respectively, and hybridized with a probe derived from the PCR products that were used for transformation. Twelve tetrads from the RRP45/rrp45::TRP1 strain and eight tetrads from RRP46/rrp46::HIS3 strain were dissected on YPD plates and incubated for 6 days at 23°C. Each showed 2:2 segregation for spore viability. All viable spores were auxotrophic for tryptophan or histidine, respectively, indicating that the disrupted alleles were lethal. The nonessential RRP6 gene was disrupted in the haploid strain YBD38 (see Table 2) by use of the Kluyveromyces lactis TRP marker, obtained by PCR amplification from plasmid pBS1408 (generously provided by Bertrand Séraphin, EMBL, Heidelberg, Germany).
Table 2.
Strains used in this study
| Strain
|
Genotype
|
Reference/Note
|
|---|---|---|
| BMA38 | MATa/α ade2-1/ade2-1 his3-Δ200/his3-Δ200 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 can1-100/can1-100 | Baudin et al. (1993) |
| YCA10 | as BMA38 but RRP45/RRP45∷TRP1 | this study |
| YCA11 | as BMA38 but RRP46/RRP46∷HIS3 | this study |
| YCA12 | MATa ade2-1 his3-Δ200 leu2-3,112 trp1-1 ura3-1 can1-100 RRP6∷Kl TRP1 | this study |
| YTK100 | MATa mtr3-1 ura3-52 | Kadowaki et al. (1995) |
| YDL401 | MATa his3Δ200 leu2Δ1 trp1 ura3-52 gal2 galΔ108 | Lafontaine and Tollervey (1996) |
| YCA20 | as YDL401 but GAL10∷RRP45 | this study |
| YCA21 | as YDL401 but GAL10∷RRP46 | this study |
| P79 | as YDL401 but GAL10∷protA–RRP4 | Mitchell et al. (1997) |
| P147 | as YDL401 but GAL10∷RRP40 | this study |
| P170 | as YDL401 but GAL10∷CSL4 | this study |
| GAL∷DOB1 | MATα ura3-1 ade2-1 his3-11,15 leu2-3,112 trp1-1 dob1∷HIS3MX6 + [pAS24–DOB1] | de la Cruz et al. (1998) |
| P49 | MATα ade2-1 his3-11 leu2-3 trp1-1 ura3-52 can1-100 rrp4Δ∷HIS3 + [pRS416/protA–RRP4] | Mitchell et al. (1996) |
| P51 | MATα ade2-1 his3-11 leu2-3 trp1-1 ura3-52 can1-100 rrp4Δ∷HIS3 + [pRS415/RRP4] | Mitchell et al. (1996) |
| YCA40 | MATa ade2-1 his3-Δ200 leu2-3,112, trp1-1 ura3-1 can1-100 RRP6∷Kl TRP1 + [pRS416/protA–RRP6] | this study |
| ProtA–Nop1 | MATα ade leu trp lys ura3 nop1∷URA3 + [pUN100–protA–NOP1] | Jansen et al. (1993) |
The oligonucleotides used to construct and test the gene disruptions were 5′RRP45::TRP1 (807); 3′RRP45::TRP (808); 5′ RRP46::HIS3 (809); 3′RRP46::HIS3 (810); 5′RRP6::Kl TRP (811); 3′RRP6::Kl TRP (812). Test oligonucleotides were 3′RRP45 (813); 3′RRP6 (815); Sc TRP (816); HIS (817); Kl TRP (818) (full sequences are available from the authors).
Conditional mutants under the control of the inducible GAL10 promoter were generated for the RRP40, RRP45, RRP46, and CSL4 genes by a one-step PCR strategy in the YDL401 strain (Lafontaine and Tollervey 1996). Transformants were selected for His+ prototrophy and screened by PCR.
The oligonucleotides used to construct the conditional mutants were 5′GAL-RRP45 (819); 3′GAL-RRP45 (820); 5′GAL-RRP46 (821); 3′GAL-RRP46 (822); 3′GAL-ProtA–RRP46 (823); 5′GAl-RRP40 (824); 3′GAl-RRP40 (825); 5′GAL-CSL4 (826); 3′GAL-CSL4 (827). The amplification of RRP6::TRP was done with oligonucleotides 5′RRP6 (834) and 3′RRP6 (835) (full sequences are available from the authors).
Construction of the ProtA–Rrp6p fusion
To construct the ProtA–RRP6 fusion gene, the RRP4 ORF was excised from plasmid pPM46 (Mitchell et al. 1997) by restriction cleavage at sites EcoRI and HindIII and replaced by the RRP6 ORF amplified by PCR from wild-type genomic DNA and flanked by the same restriction sites. The resulting plasmid was transformed into the haploid RRP6::TRP strain and shown to complement fully the RNA processing and growth phenotypes of the deleted strain. The oligonucleotides used for the PCR were 5′PRS (836) and 3′PRS (837).
Fractionation of ProtA–Rrp4p complexes
Lysate from 5-liter YPD cultures of strain P49 was prepared in TMN buffer [10 mm Tris-HCl (pH 7.6), 5 mm MgCl2, 0.1% NP-40] containing 150 mm NaCl, 1 mm PMSF, and 10% glycerol, as described (Mitchell et al. 1996). ProtA–Rrp4p complexes were purified by immunoprecipitation with IgG–Sepharose, either from clarified lysates or after fractionation by low-pressure column chromatography. Purification procedures were carried out at 4°C in buffers containing 0.5 mm PMSF and fractions were screened for the presence of ProtA–Rrp4p by Western blot analyses, using peroxidase-antiperoxidase rabbit antibody (Sigma).
Cleared lysate was applied directly to a 100-μl IgG–Sepharose 6 FF column (Pharmacia) and washed with 100 ml of TMN-150, bound material was eluted with a 0.1–2 m MgCl2 gradient (Görlich et al. 1996) in TMN-150 buffer (20 fractions of 150 μl at increments of 100 mm MgCl2). Aliquots of 5 μl were resolved by SDS-PAGE and visualized by silver staining. Fractions containing the proteins of interest were precipitated with 9 vol of isopropanol, pooled, and analyzed on 10% polyacrylamide gels containing SDS.
For fractionation, cleared lysate (30 ml) diluted to 100 mm NaCl was batch-bound to DEAE–Sepharose FF (Pharmacia). Bound material was washed three times with 30 ml of TMN buffer containing 100 mm NaCl (TMN-100), eluted with 5 × 30 ml TMN-300/10% glycerol (E300) and then frozen at −80°C. The pooled eluates were diluted to 100 mm NaCl and passed through a 10-ml Q-Sepharose FF column (Mono Q; Pharmacia). Bound material was eluted stepwise with 50 ml of TMN-150, TMN-200 (E200), TMN-320 (E320), and TMN-500. Material that failed to bind to DEAE–Sepharose FF (FT) was passed through a 10-ml SP–Sepharose FF column (Mono S; Pharmacia). After washing with 50 ml of TMN-300, bound material was eluted with 50 ml of TMN-500 (E500). Eluates from the Mono Q and Mono S columns were diluted to 150 mm NaCl and passed through small (100 μl) IgG–Sepharose 6 FF columns. Bound material was washed with 100 ml of TMN-150, and retained proteins were eluted with 1 ml of 0.5 m acetic acid. The eluates were concentrated by centrifugation under vacuum and analyzed by SDS-PAGE and nanospray mass spectrometry, as above.
Mass spectrometric analysis
Proteins bands were excised from the gel, digested in the gel, and analyzed according to the strategy described elsewhere (Shevchenko et al. 1996). High mass accuracy MALDI peptide mapping (Jensen et al. 1996) was performed on a Bruker Reflex III mass spectrometer (Bruker Daltonics, Bremen, Germany). To resolve protein mixtures an iterative approach (Jensen et al. 1997) was used. In case of uncertainty identifications were confirmed by nanoelectrospray tandem mass spectrometry on a pilot QqTOF instrument (SCIEX, Toronto, Canada; Shevchenko et al. 1997b). PeptideSearch software, developed in house, was used for protein database searching.
Glycerol gradient analysis of a HeLa cell extracts
HeLa cell lysates were prepared according to standard procedures (Dignam et al. 1983; Lee et al. 1988). Nuclear and cytoplasmic extracts were centrifuged through 12-ml glycerol density gradients as described previously (Mitchell et al. 1997). Gradient fractions were analyzed by Western blotting analysis with rabbit anti-hRrp4p serum or sera of patients suffering from polymyositis–scleroderma overlap syndrome [kindly provided by Dr. W. van Venrooij and obtained from the University Hospital (St. Radboud) of Nijmegen].
Immunofluorescence
Cells were grown in selective medium to mid-exponential phase, fixed by incubation in 4% (vol/vol) formaldehyde for 1 hr at room temperature, and spheroplasted. Then immunofluorescence was then performed as described previously (Bergès et al. 1994; Grandi et al. 1993). Protein A fusions were detected with a rabbit anti-protein A antibody (Sigma) and a secondary goat anti-rabbit antibody coupled to Texas Red (Dianova) at a 1:100 and 1:200 dilution, respectively. To stain nuclear DNA, DAPI was included in the mounting medium (Vectashield, Vector Laboratories).
Immunoprecipitation of the PM–Scl complex with patient sera
Patient sera directed specifically against PM–Scl100 (kindly provided by Dr. W. van Venrooij) were used for the immunoprecipitation experiments. HeLa cell lysates were prepared as described above. A 50% solution of protein A–Sepharose beads (10μl; Pharmacia) was washed three times in IPP 500 [500 mm NaCl, 10 mm Tris-HCl (pH 8), 0.1% NP-40, 0.5 mm PMSF] and incubated for 1 hr at room temperature with 5 μl of human autoimmune sera. Beads were washed three times with IPP500, transferred in 10 μl of IPP150 ([50 mm NaCl, 10 mm Tris-HCl (pH 8), 0.1% NP-40, 0.5 mm PMSF] and then added to 10 μl of HeLa cell nuclear extract. After incubation for 2 hr at 4°C, the supernatant was recovered and beads were washed four times with IPP150. Bound proteins were eluted from the beads by a 5 min boiling in protein gel loading buffer. Total, supernatant, and pellet proteins were analyzed by SDS-PAGE and Western blotting analysis with anti-hRrp4p serum or affinity-purified anti-hPop1 antibodies (Lygerou et al. 1996; Mitchell et al. 1997).
RNA analysis
RNA isolation and Northern blot hybridization were performed as described previously (Beltrame and Tollervey 1992; Tollervey 1987). Oligonucleotides used for rRNA and pre-rRNA analysis were 5′-TGAGAAGGAAATGACGCT (oligonucleotide 020), 5′-GCGTTGTTCATCGATGC (oligonucleotide 017), 5′-CGCTGCTCACAATGG (oligonucleotide 033), and 5′-CTACTCGGTCAGGCTC (oligonucleotide 014).
Database searches
The human EST banks were searched using the EFEAME p2n program for translational frame-shifting, on the Bioaccelerator of the European Molecular Biology Laboratory (http://www.embl-heidelberg.de). Contigs were assembled from the retrieved ESTs by use of the Gene JockeyII program. Homology was calculated by use of using the Bestfit program [Wisconsin Package Version 9.1, Genetics Computer Group (GCG), Madison, WI.].
The ESTs used for the alignments were hRRP40: HS103148, AA916866, AA715297, AA909843, AA829746, AA760696, AA748308, AA747303, HSA01383, HS479237, HS417169, HS1213865, HS1191331, HS1186630, AA937191, AA741488, HSA57832, HSA01383, HS620247, HS617138, AA736510, HS1300540, HS1273716, HS1269362, HS1229711, HS1198690, HS1191331, and HS1174014; hRRP41: HS0229, HSZZ84720, HS462881, HS1210855, HS060127, and HSAA29848; hRRP42: AA654791, HS599371, HSZZ85135, HS20834, AA581010, HS414162, HS979316, and HSZZ84357; hRRP46: HS078341, HS84856, HS1255212, HS1226957, HSZZ41259, HS1256223, HS1225454, HS1249336, and HS1172072.
Acknowledgments
We thank Walter van Venrooij for providing the PM–Scl sera, Juan Balcácel for providing HeLa cell nuclear and cytoplasmic extract, Alan Tartakoff for the mtr3-1 strain and Bertrand Seŕaphin for pBS1408. We thank Roy Parker for pointing out the homology between Rrp41p and Mtr3p and Saira Mian for pointing out the homology between Rrp4p and Rrp40p, as well as the putative S1 DNA binding domains in Rrp4p, Rrp40p, and Csl4p. This work was supported by the Wellcome Trust.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL d.tollervey@ed.ac.uk; FAX 131 650 7040.
References
- Alderuccio F, Chan EK, Tan EM. Molecular characterization of an autoantigen of PM–Scl in the polymyositis/scleroderma overlap syndrome: A unique and complete human cDNA encoding an apparent 75-kD acidic protein of the nucleolar complex. J Exp Med. 1991;173:941–952. doi: 10.1084/jem.173.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JSJ, Parker RP. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RE, Harris K, Zhang K. Mutations synthetically lethal with cep1 target S. cerevisiae kinetochore components. Genetics. 1998;149:73–85. doi: 10.1093/genetics/149.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergès T, Petfalski E, Tollervey D, Hurt EC. Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J. 1994;13:3136–3148. doi: 10.1002/j.1460-2075.1994.tb06612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluthner M, Bautz FA. Cloning and characterization of the cDNA coding for a polymyositis-scleroderma overlap syndrome-related nucleolar 100-kD protein. J Exp Med. 1992;176:973–980. doi: 10.1084/jem.176.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs MW, Burkard KT, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- Bycroft M, Hubbard TJ, Proctor M, Freund SM, Murzin AG. The solution structure of the S1 RNA binding domain: A member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–342. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM. Copurification of E. coli RNAase E and PNPase: Evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- Cheng ZF, Zuo Y, Li Z, Rudd KE, Deutscher MP. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J Biol Chem. 1998;273:14077–14080. doi: 10.1074/jbc.273.23.14077. [DOI] [PubMed] [Google Scholar]
- Chirico WJ, Waters MG, Blobel G. 70K heat shock related proteins stimulate protein translocation into microsomes. Nature. 1988;332:805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Koch BD, Werner-Washburne M, Craig EA, Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988;332:800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- Ge Q, Frank MB, O’Brien C, Targoff IN. Cloning of a complementary DNA coding for the 100-kD antigenic protein of the PM-Scl autoantigen. J Clin Invest. 1992;90:559–570. doi: 10.1172/JCI115895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelpi C, Alguero A, Angeles Martinez M, Vidal S, Juarez C, Rodriguez-Sanchez JL. Identification of protein components reactive with anti-PM/Scl autoantibodies. Clin Exp Immunol. 1990;81:59–64. doi: 10.1111/j.1365-2249.1990.tb05291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Kraft R, Kostka S, Vogel F, Hartmann E, Laskey RA, Mattaj IW, Izaurraide E. Importin provides a link between nuclear protein import and U snRNA export. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- Grandi P, Doyl V, Hurt EC. Purification of NSP1 reveals complex formation with ‘GLFG’ nucleporins and a novel nuclear pore protein NIC96. EMBO J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Tollervey D, Hurt EC. A U3 snoRNP protein with homology to splicing factor PRP4 and Gb domains is required for ribosomal RNA processing. EMBO J. 1993;12:2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen ON, Podtelejnikov A, Mann M. Delayed extraction improves specificity in database searches by matrix-assisted laser desorption/ionization peptide maps. Rapid Commun Mass Spectrom. 1996;10:1371–1378. doi: 10.1002/(SICI)1097-0231(199608)10:11<1371::AID-RCM682>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- ————— Identification of the components of simple protein mixtures by high-accuracy peptide mass mapping and database searching. Anal Chem. 1997;69:4741–4750. doi: 10.1021/ac970896z. [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Schneiter R, Hitomi M, Tartakoff AM. Mutations in nucleolar proteins lead to nucleolar accumulation of polyA+ RNA in Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1103–1110. doi: 10.1091/mbc.6.9.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuster B, Mann M. Identifying proteins and post-translational modifications by mass spectrometry. Curr Opin Struct Biol. 1998;8:393–400. doi: 10.1016/s0959-440x(98)80075-4. [DOI] [PubMed] [Google Scholar]
- Lafontaine D, Tollervey D. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 1996;24:3469–3472. doi: 10.1093/nar/24.17.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KA, Bindereif A, Green MR. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal Tech. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- Liang S, Hitomi M, Hu YH, Liu Y, Tartakoff AM. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol Cell Biol. 1996;16:5139–5146. doi: 10.1128/mcb.16.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z, Pluk H, van Venrooij WJ, Séraphin B. hPop1: An autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. EMBO J. 1996;15:5936–5948. [PMC free article] [PubMed] [Google Scholar]
- Mackie GA. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- Mian IS. Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res. 1997;25:3187–3195. doi: 10.1093/nar/25.16.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Tollervey D. The 3′-end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes & Dev. 1996;10:502–513. doi: 10.1101/gad.10.4.502. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonuclease activities. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- Reimer G. Autoantibodies against nuclear, nucleolar, and mitochondrial antigens in systemic sclerosis (scleroderma) Rheum Dis Clin North Am. 1990;16:169–183. [PubMed] [Google Scholar]
- Reimer G, Scheer U, Peters JM, Tan EM. Immunolocalization and partial characterization of a nucleolar autoantigen (PM-Scl) associated with polymyositis/scleroderma overlap syndromes. J Immunol. 1986;137:3802–3808. [PubMed] [Google Scholar]
- Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: Large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Chernushevich I, Ens W, Standing KG, Thomson B, Wilm M, Mann M. Rapid ‘de novo’ peptide sequencing by a combination of nanoelectrospray, isotopic labeling and a quadrupole/time-of-flight mass spectrometer. Rapid Commun Mass Spectrom. 1997a;11:1015–1024. doi: 10.1002/(SICI)1097-0231(19970615)11:9<1015::AID-RCM958>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Mann M. Peptide sequencing by mass spectrometry for homology searches and cloning of genes. J Protein Chem. 1997b;16:481–490. doi: 10.1023/a:1026361427575. [DOI] [PubMed] [Google Scholar]
- Shiomi T, Fukushima K, Suzuki N, Nakashima N, Noguchi E, Nishimoto T. Human Dis3p, which binds to either GTP- or GDP-Ran, complements Saccharomyces cerevisiae dis3. J Biochem. 1998;123:883–890. doi: 10.1093/oxfordjournals.jbchem.a022020. [DOI] [PubMed] [Google Scholar]
- Siegel LM, Monty KJ. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, Krisch HM, Carpousis AJ. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes & Dev. 1998;12:2770–2781. doi: 10.1101/gad.12.17.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nanoelectrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- Zanchin NI, Goldfarb DS. Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis, and the exosome subunit Rrp43p. Mol Cell Biol. 1999;19:1518–1525. doi: 10.1128/mcb.19.2.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]