Abstract
Human hnRNP A1 is a versatile single-stranded nucleic acid-binding protein that functions in various aspects of mRNA maturation and in telomere length regulation. The crystal structure of UP1, the amino-terminal domain of human hnRNP A1 containing two RNA-recognition motifs (RRMs), bound to a 12-nucleotide single-stranded telomeric DNA has been determined at 2.1 Å resolution. The structure of the complex reveals the basis for sequence-specific recognition of the single-stranded overhangs of human telomeres by hnRNP A1. It also provides insights into the basis for high-affinity binding of hnRNP A1 to certain RNA sequences, and for nucleic acid binding and functional synergy between the RRMs. In the crystal structure, a UP1 dimer binds to two strands of DNA, and each strand contacts RRM1 of one monomer and RRM2 of the other. The two DNA strands are antiparallel, and regions of the protein flanking each RRM make important contacts with DNA. The extensive protein–protein interface seen in the crystal structure of the protein–DNA complex and the evolutionary conservation of the interface residues suggest the importance of specific protein–protein interactions for the sequence-specific recognition of single-stranded nucleic acids. Models for regular packaging of telomere 3′ overhangs and for juxtaposition of alternative 5′ splice sites are proposed.
Keywords: hnRNP A1, telomere, RNA-recognition motif, x-ray crystallography
Heterogeneous nuclear ribonucleoprotein (hnRNP) A1 is one of the most abundant and best-studied components of hnRNP complexes. Together with other hnRNP proteins, hnRNP A1 packages nascent pre-messenger RNA (pre-mRNA) for processing in the nucleus (for review, see Dreyfuss et al. 1993; McAfee et al. 1997). Although it accumulates predominantly in the nucleus, hnRNP A1 shuttles continuously between the nucleus and the cytoplasm (Piñol-Roma and Dreyfuss 1992). Nuclear localization and import-export signals within hnRNP A1 have been mapped to a 38 amino acid sequence, known as M9, located near the carboxyl terminus of the protein (for review, see Izaurralde and Adam 1998). Because hnRNP A1 associates with poly(A)+ RNA both in the nucleus and in the cytoplasm, and injection of hnRNP A1 with an intact M9 region into frog oocytes inhibits mRNA export, it is likely that hnRNP A1 is involved in transporting mature mRNA from the nucleus to the cytoplasm.
Nuclear hnRNP A1 and other closely related hnRNP A/B proteins can modulate the use of alternative 5′ splice sites in a concentration-dependent manner. hnRNP A1 activates distal 5′ splice sites and promotes alternative exon skipping both in vitro and in vivo (Fu et al. 1992; Mayeda and Krainer 1992; Mayeda et al. 1993; Cáceres et al. 1994; Yang et al. 1994). The alternative splicing activity of hnRNP A1 counteracts that of members of the serine-arginine (SR) family of splicing factors, such as SF2/ASF and SC35. The relative levels or activities of these two classes of antagonistic factors can determine the alternative splicing patterns of a variety of pre-mRNAs. hnRNP A1 facilitates duplex formation by complementary single-stranded polynucleotides (for review, see Pontius 1993); therefore, it may also contribute to spliceosome assembly by promoting annealing reactions between the RNA components of the small nuclear ribonucleoprotein (snRNP) particles and pre-mRNA. In addition, hnRNP A1 has been reported to bind to certain 3′ splice sites under splicing conditions in vitro (Swanson and Dreyfuss 1988), and to interact stably with U2 and U4 snRNPs (Buvoli et al. 1992).
Recently, hnRNP A1 was shown to be involved in telomere biogenesis (LaBranche et al. 1998); mouse cells deficient in hnRNP A1 expression have short telomeres, and stable restoration of hnRNP A1 expression in these cells results in longer telomeres. This result confirms and extends early observations that hnRNP A1 and other closely related hnRNP proteins can bind specifically to single-stranded d(TTAGGG)n human telomeric DNA repeats in vitro (McKay and Cooke 1992; Ishikawa et al. 1993; Erlitzki and Fry 1997). Eukaryotic chromosomal ends contain simple repeats of short nucleotide sequences, known as telomeric repeats. Most of the length of the telomeric repeats is double stranded, consisting of a G-rich strand (3′ end) and a complementary C-rich strand (5′ end) (for review, see Blackburn and Greider 1995; Zakian 1995). The double-stranded region is bound by specific protein factors that are important for the integrity and proper function of the chromosomal ends (for review, see König and Rhodes 1997). In all organisms investigated, the 3′ ends of chromosomes extend beyond the complementary C-rich strand. In vertebrates, the 3′ overhang consists of tandem repeats of the hexamer TTAGGG, with varying overall lengths in different species (McElligot and Wellinger 1997; Wright et al. 1997). Although the telomeric repeats can form G-quartet high-order structures in vitro, it is not clear whether such structures exist in vivo (for review, see Blackburn and Greider 1995; Wellinger and Sen 1997). The single-stranded telomere overhangs are recognized as the substrate for elongation by telomerase (Lee et al. 1993; Lingner and Cech 1996), a ribonucleoprotein enzyme.
Proteins that interact specifically with single-stranded telomeric DNA have been isolated and characterized in several organisms, including Oxytrichia nova (Price and Cech 1987), Euplotes crassus (Wang et al. 1992), Tetrahymena thermophila (Sheng et al. 1995), Stylonychia mytillis (Fang and Cech 1991), Xenopus laevis (Cardenas et al. 1993), and Saccharomyces cerevisiae (Lin and Zakian 1996; Nugent et al. 1996; Virta-Pearlman et al. 1996). The cocrystal structure of a heterodimeric telomere-binding protein from Oxytrichia nova (OnTEBP) with single-stranded DNA (ssDNA) has been determined recently (Horvath et al. 1998). The structure shows that the oligonucleotide/oligosaccharide-binding (OB) structural motifs (Murzin 1993) of OnTEBP are responsible for ssDNA binding. Recognition of telomeric ssDNA is mediated by RNA-recognition motifs (RRMs) in the case of two proteins, hnRNP A1 and Gbp1b. Gbp1b is a putative telomere-binding protein from Chlamydomonas reinhardtii (Johnston et al. 1999). An amino-terminal proteolytic fragment of hnRNP A1, known as unwinding protein 1 (UP1) (Herrick and Alberts 1976; Pandolfo et al. 1985), is sufficient to bind telomeric DNA (Ishikawa et al. 1993) and retains full activity in telomere length maintenance (LaBranche et al. 1998). Surprisingly, UP1, but not full-length hnRNP A1, was found to associate with telomerase (LaBranche et al. 1998). UP1 encompasses the amino-terminal two-thirds of hnRNP A1, including both RRMs of the protein. The RRM is an ancient and extremely common RNA-binding module, within which the RNP-2 hexamer and RNP-1 octamer submotifs are highly conserved (for review, see Birney et al. 1993). The crystal structure of the UP1 domain was determined recently (Shamoo et al. 1997; Xu et al. 1997). The structure revealed that the two RRMs are anti-parallel and held in close contact, mainly by two pairs of Arg–Asp salt bridges. The carboxy-terminal region of hnRNP A1 is particularly rich in glycine and includes several Arg–Gly–Gly (RGG) repeats that constitute an additional RNA-binding motif (Kiledjian and Dreyfuss 1992). In contrast to the activity in telomere biogenesis, maximal RNA binding and alternative splicing activities of hnRNP A1 require the carboxy-terminal glycine-rich domain, in addition to both RRMs (Cobianchi et al. 1988; Merrill et al. 1988; Kumar et al. 1990; Casas-Finet et al. 1993; Mayeda et al. 1994; Shamoo et al. 1994). This difference in domain requirements is strong evidence that the effect of hnRNP A1 on telomere length is not an indirect consequence of its alternative splicing activity.
Interestingly, purified hnRNP A1 binds tightly to short RNAs containing one or more copies of the motif UAGGGA/U (Burd and Dreyfuss 1994; Abdul-Manan and Williams 1996; Abdul-Manan et al. 1996; Mayeda et al. 1998). The deoxy form of one version of this sequence is identical to the human telomeric repeat, d(TTAGGG)n. It is likely that hnRNP A1 utilizes similar structural principles for recognition of single-stranded telomeric DNA and high-affinity RNA sequences. We report here the crystal structure of the amino-terminal UP1 domain of hnRNP A1 complexed with human telomeric ssDNA repeats, d(TTAGGG)2, at 2.1 Å resolution.
Results
The amino-terminal 196-amino acid UP1 domain of human hnRNP A1 (Fig. 1A) was cocrystallized with a 12-nucleotide ssDNA, d(TTAGGGTTAGGG), designated TR2. The crystals belong to space group P43212, and have unit-cell dimensions of a = b = 51.20 Å, and c = 171.09 Å. There is one protein and one ssDNA molecule per asymmetric unit. The structure was determined by a combination of multiple isomorphous replacement with anomalous scattering (MIRAS) and molecular replacement methods (Table 1). The refined model consists of 183 amino acids (residues 8–190), 11 nucleotides, and 144 ordered water molecules. The final model has an R-factor of 19.5% (free R-factor = 24.9%) and excellent stereochemistry. The Ramachandran plot of the main chain parameters shows 95.1% (154 residues) of the nonglycine, nonproline residues in the most favored region, and none in the disallowed regions (Laskowski et al. 1993).
Figure 1.
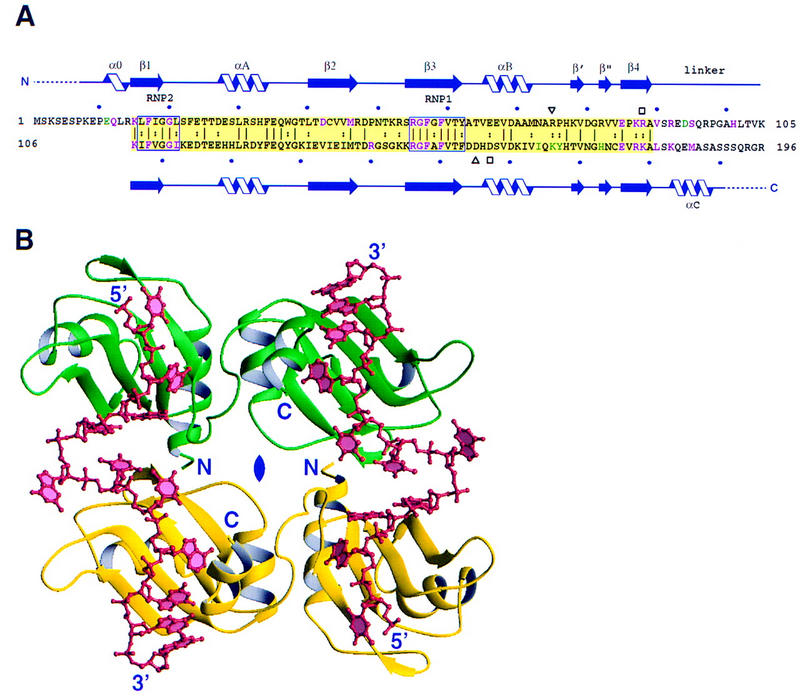
Structure of the UP1 complex with d(TTAGGG)2. (A) Amino acid sequence of the UP1 domain of human hnRNP A1. The two RRMs are shaded yellow and the conserved RNP2 and RNP1 submotifs are boxed and labeled. Sequence identity and similarity between the two RRMs are indicated by vertical lines and colons, respectively. The residue numbers are shown at the beginning and end of each line, and the bullets above and below mark 10-residue increments. The secondary structure elements are shown above and below the sequence. Dotted lines represent regions that are disordered in the structure. Two pairs of Asp–Arg residues involved in salt bridges between RRM1 and RRM2 of each monomer are indicated by a pair of open squares and a pair of open triangles, respectively. Residues shown in green are involved in protein–protein interactions between the monomers, and residues shown in magenta are involved in contacting ssDNA. (B) Overall view of the UP1–TR2 complex. The protein molecules are shown in yellow or green as a ribbon model (Carson 1991) and the DNA molecules in red as a ball-and-stick model. One asymmetric unit contains one protein molecule and one strand of DNA. The protein monomers are related by a dyad axis (labeled in blue) perpendicular to the plane of the figure. The antiparallel DNA strands have the same symmetry relationship. The DNA and protein termini are labeled in blue.
Table 1.
Statistics from the crystallographic analysis
| Native
|
HgCl2
|
||||
|---|---|---|---|---|---|
| Data sets
|
1
|
2
|
PCMBS
|
1
|
2
|
| Diffraction data | |||||
| Resolution (Å) | 2.07 | 2.80 | 2.80 | 2.80 | 2.80 |
| Observed reflections | 169730 | 35408 | 42921 | 28960 | 59342 |
| Unique reflections | 14247 | 5217 | 5854 | 5425 | 5711 |
| Completeness (%) | 96.1 | 85.1 | 95.0 | 88.8 | 94.2 |
| Average I/ς | 14.2 | 29.5 | 18.8 | 12.1 | 23.2 |
| Rmerge (%)a | 5.1 | 2.5 | 3.7 | 5.9 | 3.5 |
| Phasing | |||||
| Hg sites | 1 | 1 | 1 | ||
| Phasing powerb at 3.0 Å | |||||
| isomorphous | 1.60 | 1.79 | 1.62 | ||
| anomalous | 1.06 | 0.99 | 1.80 | ||
| Overall figure of merit | 0.67 | ||||
| RCullisc | 0.481 | 0.577 | 0.584 | ||
| RKrautd | |||||
| isomorphous | 0.100 | 0.167 | 0.184 | ||
| anomalous | 0.154 | 0.317 | 0.474 | ||
| Refinement | |||||
| Resolution range (Å) | 47.67–2.10 | ||||
| R-factore (Rfree) | 19.5% (24.9%) | ||||
| Reflections with |F| > 2ς | 11767 | ||||
| No. of protein atoms | 1465 | ||||
| No. of DNA atoms | 234 | ||||
| No. of water molecules | 144 | ||||
| Average B-factor | 29.2 Å2 | ||||
| r.m.s deviations | |||||
| bond lengths | 0.005 Å | ||||
| bond angles | 1.19° | ||||
| dihedrals | 23.2° | ||||
| improper | 0.81° | ||||
Rmerge = ∑|I − <I>|/∑<I>, where I and <I> are the measured and averaged intensities of multiple measurements of the same reflection. The summation is over all the observed reflections.
Phasing power = r.m.s. (<FH>/E), where FH is the calculated structure factor of the heavy atoms and E is the residual lack of closure.
RCullis = ∑∥FPH − FP| − FH(calc)|/∑|FPH − FP|, where FPH and FP denote observed derivative and native crystal structure factors, respectively, and FH denotes the calculated heavy atom structure factor.
RKraut = ∑∥FPH| − |FPH(calc)∥/∑|FPH| for isomorphous data, and RKraut = ∑(∥FPH+| − |FPH+(calc)∥ + ∥FPH−| − |FPH−(calc)∥)∑(|FPH+| + |FPH−|) for anomalous data.
R-factor = ∑∥FO| − |FC∥/∑ |FO|, where FOdenotes the observed structure factor amplitude and FC denotes the structure factor calculated from the model.
Description of the structure
The overall structure of the UP1–TR2 complex is shown in Figure 1B. Two strands of TR2 bind to two protein molecules. The two protein and two TR2 molecules are related by a crystallographic twofold rotation. The two ssDNA molecules are antiparallel, with each 5′ terminus located near RRM1 of one of the protein monomers, and the 3′ terminus located near RRM2 of the symmetry-related protein monomer.
Significant conformational changes of UP1 are observed on DNA binding. First, there is a ∼15° change in the relative position of the two RRMs, compared with the protein-only form (Shamoo et al. 1997; Xu et al. 1997), bringing the two β-sheet surfaces closer together (Fig. 2A). This rotation is hinged around the two conserved pairs of Asp–Arg salt-bridges (Arg-75–Asp-155 and Arg-88–Asp-157), which were found in the protein-only structure and are preserved in the structure of the UP1–TR2 complex (Fig. 2B). In spite of the large domain movement, there are only minor changes within each RRM. The root-mean-square (r.m.s.) deviations of Cα positions are 0.386 Å for RRM1 (residues 15–89) and 0.685 Å for RRM2 (residues 106–180), when each RRM is individually aligned to compare the complex and the protein-only forms. Second, the linker connecting the two RRMs becomes ordered on TR2 binding (Fig. 2A). Third, the region near the carboxyl terminus of UP1 (Lys-183–Ser-190), which was disordered in the absence of DNA, also becomes ordered in the presence of DNA and forms an α-helix (Fig. 2A). Both regions make important contacts with DNA (see below).
Figure 2.
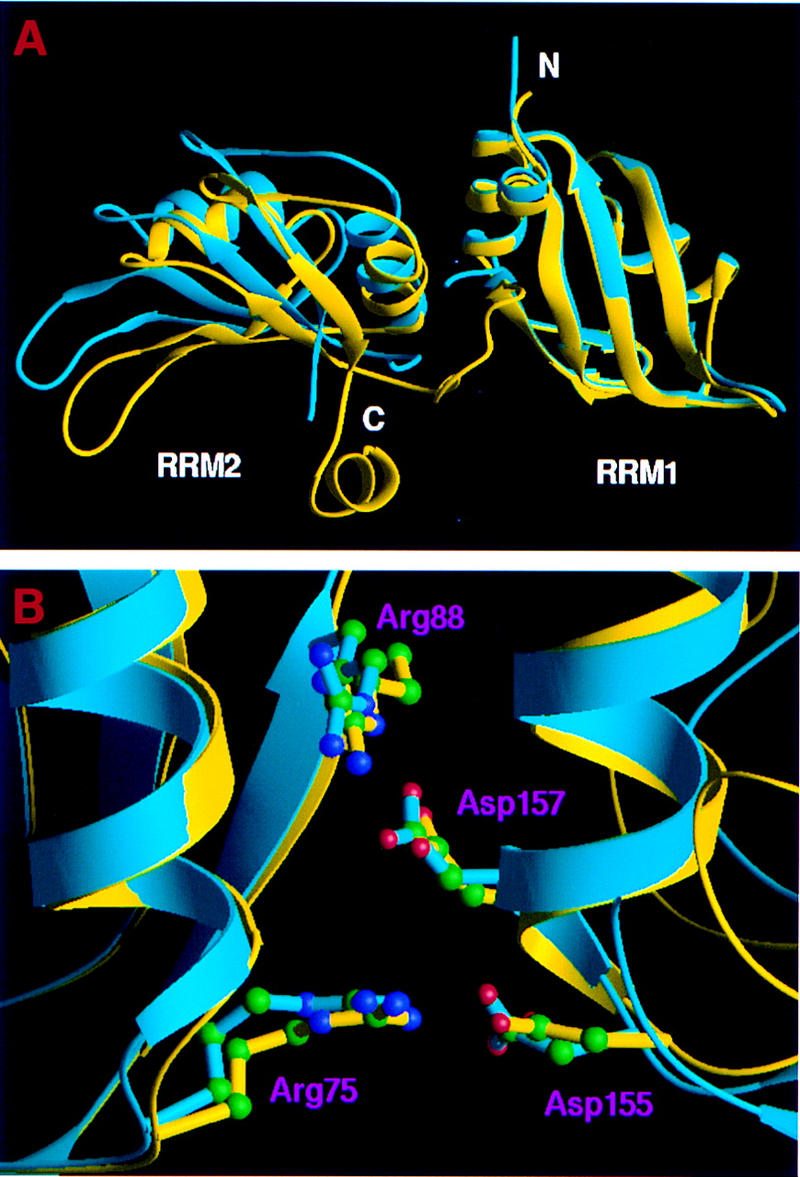
Conformational change of UP1 upon DNA binding. (A) Superposition of the Cα chains of UP1 from the UP1–TR2 complex (yellow) and from the protein-only structure (cyan) shows large domain movement attributable to DNA binding. The Cα atoms of RRM1 (residues 15–89) were used for least-squares alignment. (B) A close view of the salt bridges in both DNA-bound (yellow) and free (cyan) forms of UP1. The pairs of arginines and aspartates are shown in ball-and-stick representation. Green (C); blue (N); red (O).
The telomeric DNA repeats bind to UP1 in a genuinely single-stranded form. The two strands are antiparallel and the interstrand backbone distance ranges from ∼25–50 Å, that is, the two strands never come into contact. The path of each strand of TR2 follows two arcs, with each arc traversing one RRM module (Figs. 1B and 3A). The transition from one arc to the other occurs over a single nucleotide, Gua-6. This abrupt change of direction forces the guanine base to loop out. This base has weak electron density (Fig. 3A), presumably because it is highly mobile when exposed to solvent. The upstream arc consists of five nucleotides, Thy-2–Gua-6, and runs across the amino-terminal RRM1 of one UP1 monomer (Figs. 1B and 3B). The 5′-most nucleotide, Thy-1, cannot be modeled reliably because of weak electron density (Fig. 3). There are few interactions between the bases in this arc. The downstream arc consists of six nucleotides, Thy-7–Gua-12, and runs across the carboxy-terminal RRM2 of a symmetry-related neighboring UP1 monomer (Fig. 1B). As with the upstream arc, there are few interactions among the first 4 bases. However, the last three guanines (Gua-10–Gua-12) show base-stacking interactions (Fig. 1B). The bases of Gua-4 and Gua-10 are in the syn orientation, whereas all other ordered bases are in the anti orientation. 2′-endo sugar puckering is observed throughout.
Figure 3.
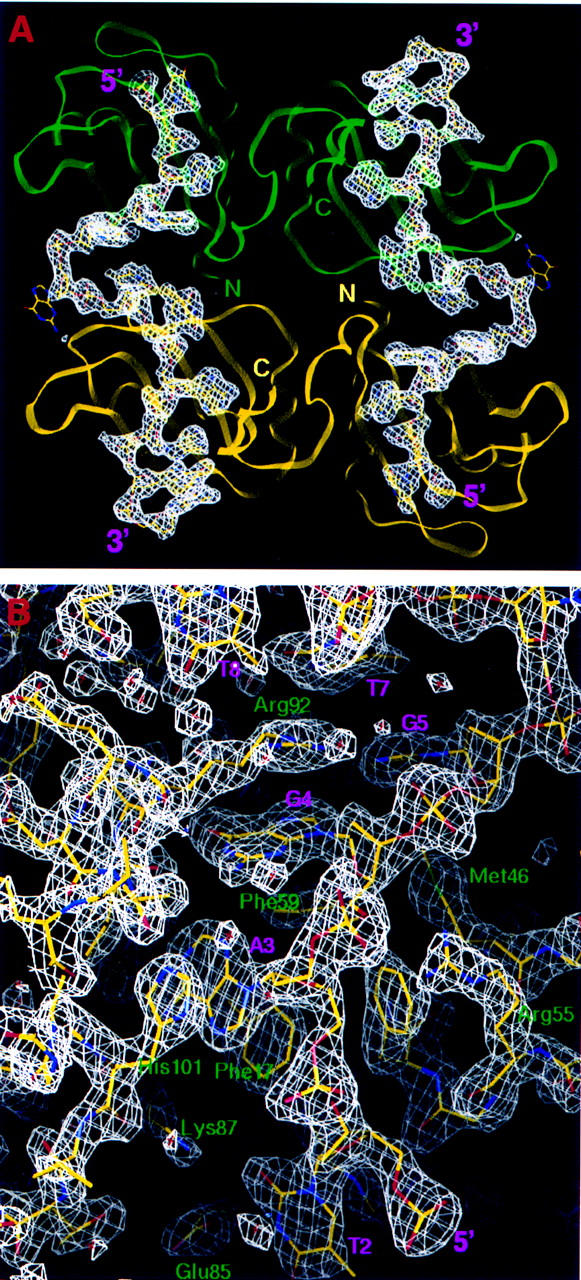
Electron density maps showing the bound telomeric DNA. (A) Omit difference map (FO − FC, φC) showing the path of the bound ssDNA. The map is contoured at 2.5ς. The DNA molecules are shown in a ball-and-stick model and the protein molecules in ribbon representation. The coloring scheme for the protein monomers is the same as in Fig. 1B. (B) Electron density surrounding the nucleic-acid-binding region in RRM1. The (2FO − FC, φC) map was contoured at 1.5ς level, and the refined protein and DNA models are represented in a ball-and-stick model.
Protein–DNA interactions
Detailed UP1–ssDNA interactions are shown in Figures 4 and 5A. Many of the contacts with ssDNA are similar for both RRMs, a feature consistent with the high degree of sequence homology between the two RRMs (Fig. 1A). Specifically, three consecutive nucleotides (T2, A3, and G4) in the first telomeric repeating unit superimpose extremely well with the corresponding nucleotides (T8, A9, and G10) in the second repeating unit (Fig. 5B). Two conserved phenylalanines (Phe-17 and Phe-59 in RRM1, and Phe-108 and Phe-150 in RRM2) located in the RNP-2 and RNP-1 submotifs of each RRM interact directly with AG dinucleotides (Ade-3–Gua-4, and Ade-9–Gua-10, respectively) by aromatic ring stacking (Fig. 5A). The same four phenylalanines have been shown to be the major sites of covalent adduct formation when hnRNP A1 is UV cross-linked to oligo(dT) (Merrill et al. 1988). A third phenylalanine, Phe-57 in RRM1 and Phe-148 in RRM2, does not contact the bases directly but, rather, interacts with the backbone of a guanine (Gua-3 and Gua-9) via van der Waals contacts. Another RNP-1 residue, Arg-55 in RRM1 and Arg-146 in RRM2, interacts with the backbone of a guanine (Gua-4 and Gua-10, respectively) via charge interaction with the phosphate and hydrogen bonding with O5′. A lysine (Lys-15 in RRM1, and Lys-106 in RRM2) immediately amino-terminal to the RNP-2 submotif forms a hydrogen bond with the N7 atom of this same guanine. This lysine contacts the next adjacent guanine via a water molecule in RRM1, but directly in RRM2. In addition, two charged residues (Glu-85 and Lys-87 in RRM1, Glu-176 and Arg-178 in RRM2) located in β4 interact with a thymine, Thy-2 in RRM1 and Thy-8 in RRM2, respectively. Arg-178 also interacts with Ade-9. Apart from the side-chain interactions, similar hydrogen bonds for main chain atoms of amino acids 88–90 and 179–181 are formed with AG dinucleotides, that is, Ade-3–Gua-4 and Ade-9–Gua-10, respectively.
Figure 4.
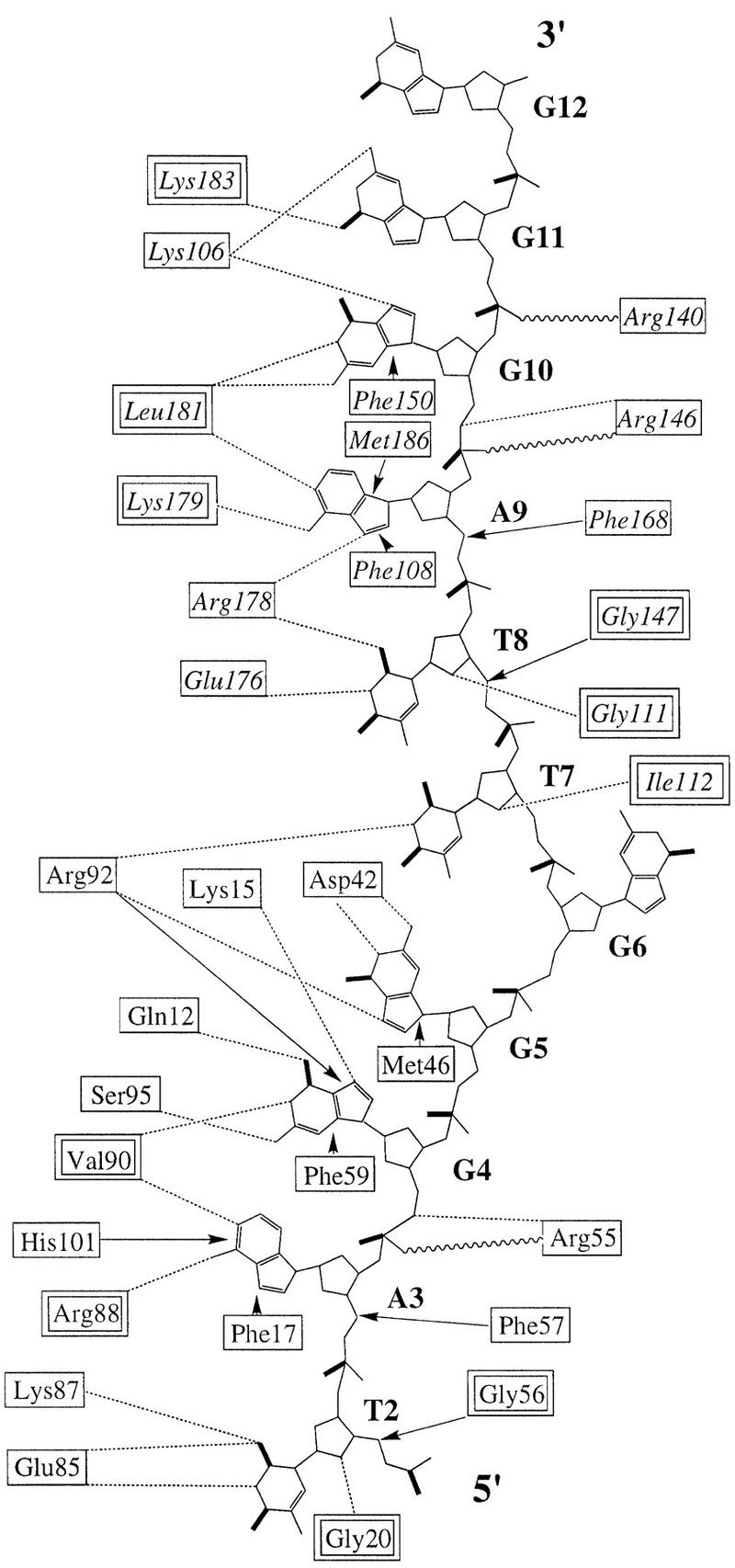
Summary of contacts between UP1 and TR2. Residues enclosed in single-line boxes indicate side-chain contacts with DNA; residues enclosed in double-line boxes contact DNA with main chain atoms. (Dotted lines) hydrogen bonds; (wavy lines) charge interactions; (arrows) base-stacking or van der Waals interactions. For the DNA molecule, a thick line represents either carbonyl groups at the base or double-bonded phosphate oxygens. RRM1 residues from one monomer are shown in roman type, and RRM2 residues from the other monomer are shown in italics.
Figure 5.
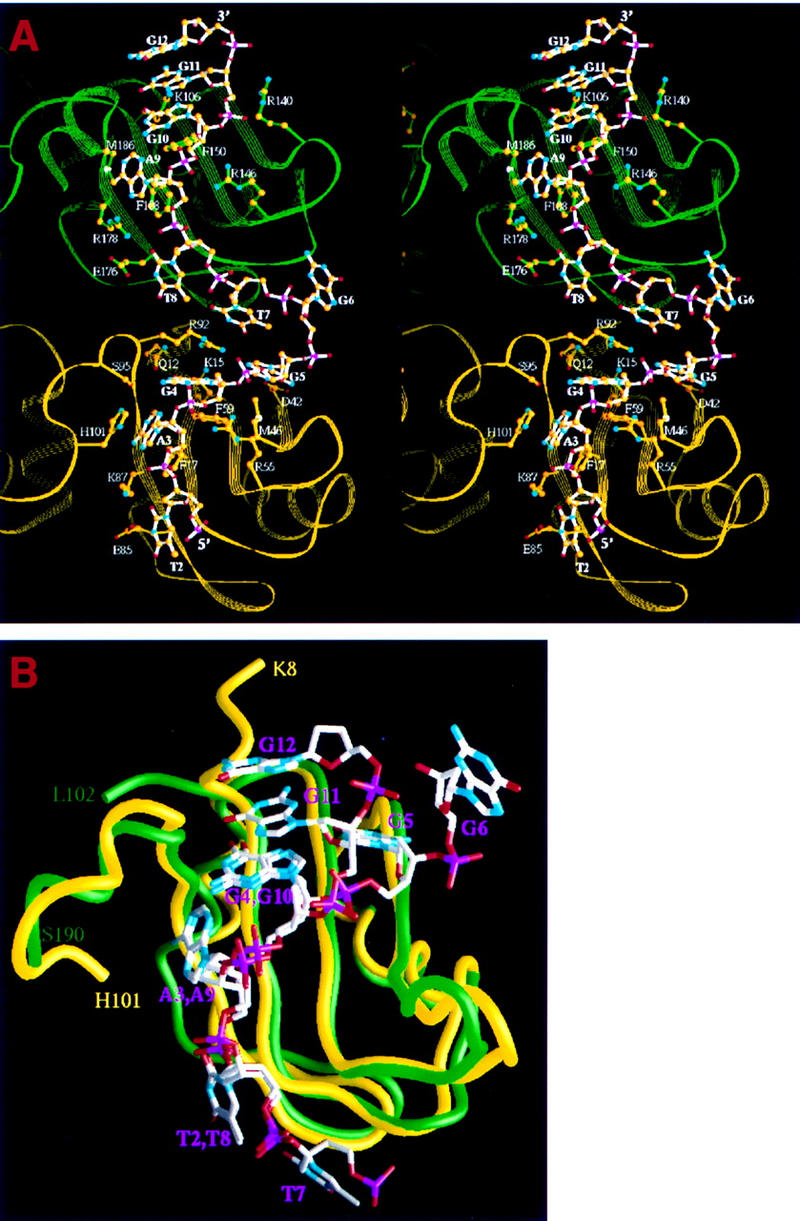
UP1 contacts with bound single-stranded telomeric DNA. (A) Stereo ribbon diagram showing side-chain contacts with DNA. Only one of the telomeric strands is shown, interacting with RRM1 of one monomer (yellow) and RRM2 of the other (green). The DNA strand and relevant protein side chains are shown in a ball-and-stick representation. The atom color scheme is gold (C); red (O); cyan (N); magenta (P). The bond color scheme is white (DNA); yellow (RRM1); green (RRM2). The identity and number of the nucleotides (in boldface) and amino acids are indicated. (B) Superimposition of first and second telomeric repeats interacting with the respective RRMs. The backbones of RRM1 (yellow) and RRM2 (green) were superimposed (r.m.s. deviation =1.46 Å). The resulting positions of the telomeric repeats show a high degree of similarity for three consecutive nucleotides (T2, A3, G4 and T8, A9, G10), and significant divergence for the following two nucleotides (G5, G6 and G11, G12).
One major difference between RRM1 and RRM2 interactions with DNA is seen in β2. In RRM1, the side chain of Asp-42 makes two hydrogen bonds with the N1 and N2 atoms of Gua-5, whereas the equivalent residue in RRM2, Val-133, does not contact DNA at all. Met-46, another residue located in β2 of RRM1, is within the range for making van der Waals contact with the base of Gua-5, whereas the RRM2 counterpart, Met-137, is further away from the DNA bases. Instead, Arg-140 in RRM2 interacts electrostatically with the backbone phosphate of Gua-11, whereas its RRM1 counterpart, Pro-49, does not play a role in DNA binding.
The inter-RRM linker segment is defined to include amino acids 90–105. Three residues in this segment make direct contacts with DNA bases. Arg-92 makes contacts with three bases, Gua-4, Gua-5, and Thy-7, and the guanidino moiety is locked between the three bases (Figs. 4 and 5A). Ser-95 makes a hydrogen bond with the N2 atom of Gua-4. The imidazole ring of His-101 stacks with the purine ring of Ade-3, which is sandwiched between His-101 and Phe-17.
We predicted that the amino-terminal 310 helix of UP1 would be involved in nucleic acid binding (Xu et al. 1997). The present structure confirmed this prediction. O1 of Gln-12 forms a hydrogen bond with O6 of Gua-4 (Figs. 4 and 5A). Glu-11 does not interact with DNA directly, but it stabilizes the conformation of Lys-15, which makes direct contact with DNA. At the carboxyl terminus, the main-chain amide group and carbonyl group of Leu-181 hydrogen bond with the guanine rings of Gua-8 and Gua-9, respectively. The segment including residues 183–190 was disordered in the protein-only structure, but becomes ordered and forms an α-helix when bound to DNA. The amide group of Lys-183 makes a hydrogen bond with Gua-11, and its side chain interacts with the DNA backbone via a water-mediated interaction. Met-186 makes a van der Waals contact with Ade-9, such that the adenine ring is sandwiched between Met-186 and Phe-108.
Protein–protein interactions
In the present structure, DNA binding is achieved through a dimer of UP1, whereas in the absence of DNA, UP1 crystallized as a monomer (Shamoo et al. 1997; Xu et al. 1997). In the DNA complex there is an extensive interface between the symmetry-related protein monomers, which forms two contiguous DNA-binding clefts (Fig. 6A). A total surface area of 1574 Å2 is shielded from the solvent by protein–protein interactions between the monomers. The dimer interface involves contacts that are distinct from the crystal lattice contacts in the protein-only structure. The intermolecular interaction is mediated mainly through six residues, of which four (Ile-164, Lys-166, Tyr-167, and His-173) are within RRM2 and two (Glu-11 and Asp-94) are outside of the two-RRM region (Fig. 1A). The intersubunit contacts include the following: hydrogen bonding between Glu-11 and His-173; a charge interaction between Asp-94 and Lys-166; hydrogen bonding between Tyr-167 and the carbonyl group of Ile-164 (Fig. 6B). Because of the twofold symmetry, each of these interactions occurs twice in identical fashion.
Figure 6.
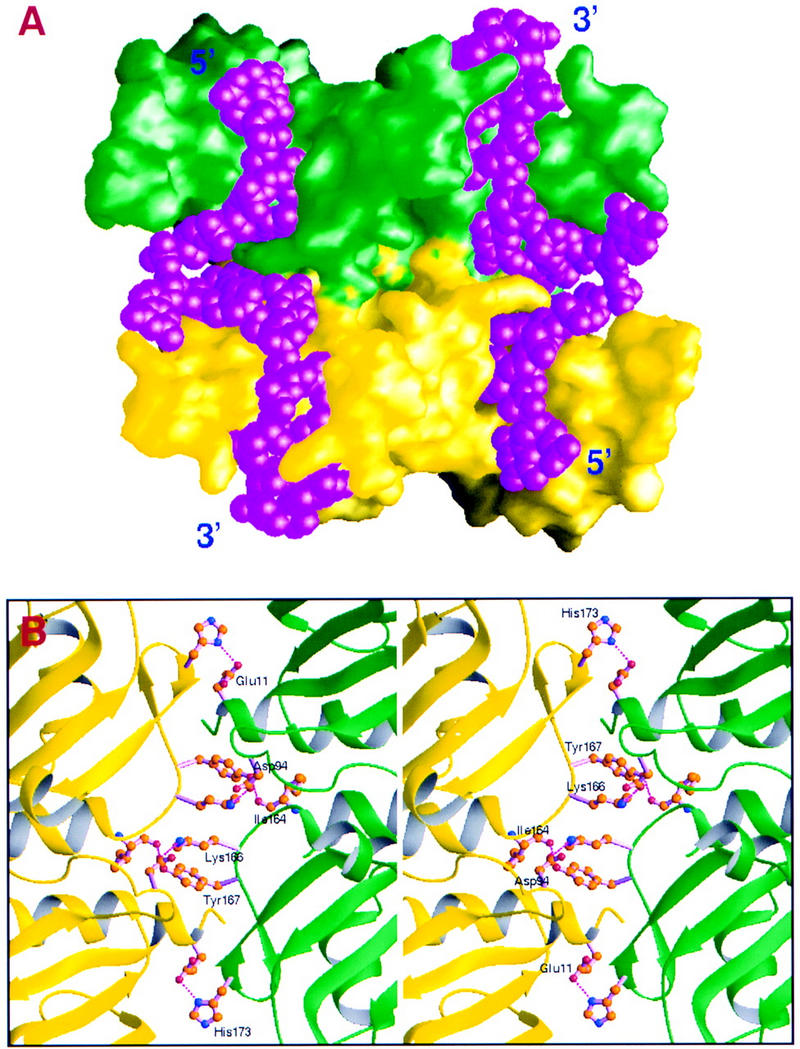
Protein–protein interactions in the UP1–TR2 dimer. (A) Molecular surfaces showing extensive contacts between the two protein monomers bound to the same two strands of DNA. The DNA fits into contiguous clefts in the two monomers. The molecular surfaces were generated with the program GRASP (Nicholls et al. 1991) with a 1.4 Å probe radius. The same color-code convention as in previous figures was used for the proteins, and the DNA molecules are shown in a CPK model colored in magenta. (B) Stereo figure showing amino acids involved in the protein–protein interface. (Magenta dotted lines) Hydrogen bonds; (red) oxygen atoms; (blue) nitrogen atoms; (brown) carbon atoms.
Discussion
Structural implications for interactions of hnRNP A1/UP1 with RNA
Biochemical studies have demonstrated that hnRNP A1 can bind specifically and with high affinity to single-stranded RNA or DNA sequences of similar sequence. Although physiological RNA targets remain to be better defined, some functionally significant RNA targets with sequences related to the high affinity motif have recently been found (Chabot et al. 1997; Li et al. 1997; del Gatto-Konczak et al. 1999; M. Caputi, A. Mayeda, A.R. Krainer, and A.M. Zahler, in prep.). The sequence specificity of hnRNP A1 appears to be similar for RNA and ssDNA, as no qualitative differences in binding were observed between these polynucleotides (Swanson and Dreyfuss 1988; Buvoli et al. 1990; Ishikawa et al. 1993; Abdul-Manan and Williams 1996). The binding similarity implies a similar underlying structural basis for sequence-specific interactions with RNA and ssDNA. We observed indistinguishable UP1 complexes by gel mobility shift with TR2 or with the RNA version of the same sequence (data not shown). These similarities enable us to infer the structural basis of RNA–hnRNP A1 interaction from the present structure of the UP1–ssDNA complex, at least in a qualitative sense. The presence of 2′ OH groups and the 3′-endo sugar puckering preferred by RNA should not interfere with the global features of the model, although small readjustments of the backbone conformation are expected. Moreover, the 5-methyl groups of the thymines are not involved in significant interactions with UP1.
In the cocrystal structure, RRM1 and RRM2 within the same protein monomer bind to two separate strands of ssDNA, which are antiparallel. Because the two RRMs within each monomer are also antiparallel, the 5′ → 3′ polarity of ssDNA with respect to the RRM orientation is the same for each RRM. Confirming our earlier conjecture (Xu et al. 1997), this nucleic acid directionality with respect to the RRM orientation is the same as that observed in the structures of an RNA–hairpin bound to the single amino-terminal RRM of U1A protein and also in the recently solved U2B′′–U2A′–RNA ternary complex (Oubridge et al. 1994; Allain et al. 1996; Price et al. 1998). Therefore, it appears that RRMs bind RNA or ssDNA molecules in a preferred 5′ → 3′ direction.
Direct interaction with 4 nucleotides (TAGG) is observed in RRM1 and with 5 nucleotides (TTAGG) in RRM2 (Fig. 4). These sequences are not only present in the high-affinity hnRNP A1-binding sequences selected from a random-sequence RNA pool by an in vitro iterative procedure (SELEX), but they can also be identified in the consensus sequences selected with hnRNP A1 lacking either RRM (Burd and Dreyfuss 1994). A 4-nucleotide sequence in an extended conformation is required to traverse the four β-strands in an RRM, following the nucleic acid path observed in the present structure and in the structures of U1A and U2B′′ and, therefore, this appears to be the minimal length required for recognition by an individual RRM. Interactions with the TAG trinucleotide (nucleotides 2–4 for RRM1 and nucleotides 8–10 for RRM2) are virtually identical and in register in both RRMs (Fig. 5B). This trinucleotide sequence is part of the consensus vertebrate 3′ splice site. In fact, binding of hnRNP A1 to 3′ splice-site sequences has been reported (Swanson and Dreyfuss 1988; Buvoli et al. 1990; Ishikawa et al. 1993). Interactions with the other nucleotides differ between the two RRMs. Asp-42 in RRM1 makes two hydrogen bonds with Gua-5 in RRM1, whereas the corresponding amino acid in RRM2, Val-133, does not interact with ssDNA at all. According to the present structure, replacing Gua-5 with an adenine would maintain one of the two hydrogen bonds, but replacing it with a pyrimidine would prevent all the hydrogen bonds with Asp-42 because of distance constraints. Thy-7 is within the range of van der Waals interaction with Gly-111 and Ile-112 in RRM2, but the corresponding Thy-1 is not ordered in the structure.
The inter-RRM linker is highly conserved in length and sequence and has been implicated in nucleic acid binding and alternative splicing function (Burd and Dreyfuss 1994; Mayeda et al. 1998). The present structure demonstrates the direct involvement of the linker segment in both protein–protein and protein–ssDNA interactions. Several residues located within this region contact ssDNA directly (Fig. 4). Most interestingly, Arg-92 makes contacts with three nucleotides, Gua-4, Gua-5, and Thy-7. The two guanines are bound by RRM1, whereas the thymine is the first nucleotide of the TTAGG sequence bound by RRM2. The remaining direct DNA contacts made by the linker are located in the RRM1 region. However, the presence of both RRMs and their spatial positioning are important for the linker to function in nucleic acid binding, because they constrain its spatial location and range of motion. The spatial arrangement of the two RRMs of hnRNP A1, which is largely determined by the two salt bridges, is such that the linker is exposed to the RNA-binding surface and is readily accessible for interaction with RNA or DNA. Conversely, the linker segment influences the spatial positioning of the two RRMs. In the cocrystal structure, the relative movement of the two RRMs appears to be primarily dictated by the covalent joining of the two RRMs by the linker. Thus, interaction with DNA on the RRM1 side pulls the linker toward RRM1, and this movement in turn pulls RRM2 closer to RRM1. This cooperative phenomenon is likely to be the origin of nucleic acid binding and functional synergy between the two RRMs (Shamoo et al. 1995, Mayeda et al. 1998 and references therein).
Additional contributions to nucleic acid binding from both the amino-terminal and carboxy-terminal regions outside of the RRMs of UP1 appear to be localized, that is, they contribute to nucleic acid binding by RRM1 and RRM2, respectively. Likewise, structural studies of the amino-terminal U1A RRM (Oubridge et al. 1994; Allain et al. 1996) and the RRM of hnRNP C (Gorlach et al. 1992), have shown that regions immediately adjacent to the classical RRM are often important in RNA binding. Interestingly, the RNA-binding role of the U1A RRM carboxy-terminal helix, which undergoes large conformational changes on RNA binding and makes contacts with bound RNA (Oubridge et al. 1994; Allain et al. 1996), appears to be analogous to that of α0, the amino-terminal 310-helix of UP1. When RRM2 of UP1 is superimposed with the U1A amino-terminal RRM, αC points in the opposite direction compared with the carboxy-terminal helix of the U1A RRM. This difference may reflect the fact that the U1A RRM binds to an RNA stem–loop, whereas hnRNP A1 binds to single-stranded nucleic acids.
Significance of UP1 dimerization
The structure reveals an interesting mode of nucleic acid binding by UP1, in which one single-stranded telomeric DNA binds to the amino-terminal RRM of one protein monomer and the carboxy-terminal RRM of another (Fig. 1B). This mode of binding may also contribute to the above-mentioned binding and functional synergy between the RRMs. The closely interacting UP1 monomers are related by a twofold crystallographic symmetry, raising the possibility that dimerization is induced by packing forces in the crystal lattice, and therefore is not physiological. The nucleic acid-free form of UP1 crystallized in a monomeric form (Shamoo et al. 1997; Xu et al. 1997), and the contact sites in the crystal lattice differ from those observed in the crystal of the UP1–TR2 complex. However, discounting the potential physiological relevance of the observed protein–protein interaction on the basis of these in vitro observations would be premature. Several lines of evidence suggest that this mode of dimerization may be important. (1) The extensive area of interface of 1574 Å2 is indicative of a specific protein–protein interaction. For example, a survey of protease–inhibitor or antibody–antigen complexes revealed that the interfaces bury a surface area of 1500 ± 250 Å2 (Janin and Chothia 1990; Janin and Rodier 1995), whereas a typical crystal-lattice contact buries <1200 Å2; (2) the residues that are critical for the dimer interface are all highly conserved in the hnRNP A/B family of proteins (not shown; see also Mayeda et al. 1998); (3) Gbp1p, a putative telomere-binding protein from C. reinhardtii that also contains two RRMs, can interact with single-stranded telomeric oligonucleotides as a monomer or as a dimer (Johnston et al. 1999). Dimeric Gbp1p shows strong preference for binding ssDNA; and (4) the conformational change, that is, the relative rotation of RRM2 with respect to RRM1 caused by nucleic acid binding is important for optimal dimer formation, because modeling the similar dimeric state with the protein-only UP1 structure gave an unfavorable protein–protein interaction (not shown). These properties are consistent with the specific association of hnRNP A1 molecules being induced by DNA or RNA binding.
It is possible that UP1 dimerization is efficiently promoted by the particular oligonucleotide sequence used, which contains two tandem telomeric repeats. Nucleic acid-induced UP1 or hnRNP A1 dimerization may account for the unusually high binding affinity of hnRNP A1 for the SELEX winner sequence, which also contains two hexamer repeats separated by two nucleotides (Burd and Dreyfuss 1994; Abdul-Manan and Williams 1996; Abdul-Manan et al. 1996). On the other hand, purified hnRNP A1 can bind to oligoribonucleotides containing only one copy of UAGGGU/A (Burd and Dreyfuss 1994; Mayeda et al. 1998). In such cases, it is not known whether the same protein dimerization and binding stoichiometry applies.
We have used a variety of physical methods to test whether UP1 can dimerize in solution in the presence of nucleic acid, but the results have been ambiguous, that is, we could neither demonstrate nor rule out that dimers can form. However, binding of UP1 to TR2 assayed by gel mobility shift required both of the telomeric repeat sequences and the wild-type forms of both RRMs, confirming the synergistic behavior of the RRMs evident in the structure (data not shown). It is possible that stable dimers cannot form under physiological conditions, and that the crystal structure and interface residue conservation reflect interactions of a more transient nature. Our recent hnRNP A1 domain-swap and domain-duplication data showed that although RNA binding is not severely affected in these variants, efficient alternative splice-site switching activity requires the presence of one copy of RRM2 preceding the carboxy-terminal glycine-rich domain, whereas the amino-terminal RRM can be either RRM1 or a copy of RRM2 (Mayeda et al. 1998). The alternative splicing activity of the different hnRNP A1 variants correlates well with the ability to model the dimerization interface, which involves residues in RRM2 but not in RRM1. This observation suggests that protein dimer formation may be required for alternative splicing activity, and hence, that the observed dimer interface in the UP1–TR2 structure may have important physiological implications. It should be possible to test this hypothesis by mutational and functional analyses.
hnRNP A1/UP1 and telomere length regulation
As hnRNP A1, the UP1 fragment, and other very closely related proteins have been found to associate with the single-stranded overhangs of vertebrate telomeric repeats (McKay and Cooke 1992; Ishikawa et al. 1993; Erlitzki and Fry 1997), and more importantly, a recent study demonstrated a functional role for hnRNP A1 and/or UP1 in telomere length regulation in mouse cells (LaBranche et al. 1998), the present structure has important implications for the structure and function of vertebrate telomeres. Vertebrate chromosomes have single-stranded 3′ overhangs of the G-rich strand, and at least one of the ends of each chromosome has overhangs longer than 45 nucleotides, and possibly as long as 275 nucleotides (McElligot and Wellinger 1997; Wright et al. 1997). Although two or more telomeric repeats can form intra- and intermolecular tetraplex structures with G-quartet motifs (for review, see Blackburn and Greider 1995; Wellinger and Sen 1997), in the present structure the telomeric sequences remain single stranded, as also seen in the structure of O. nova OnTEBP bound to telomeric ssDNA (Horvath et al. 1998). In vertebrates, the entire length of the overhangs may be complexed with hnRNP A1 or UP1 in a repetitive nucleoprotein structure that may be important for telomere-length regulation and/or to protect the single-stranded overhangs from nucleases. To this end, or perhaps as an additional function, hnRNP A1/UP1 may affect the distribution of telomeric structures between single-stranded and tetraplex forms.
The crystal structure of OnTEBP complexed with ssDNA shows that there are four OB structural motifs in the heterodimeric protein complex, three in the α subunit and one in the β subunit (Horvath et al. 1998). Three OB modules, two at the amino terminus of the α subunit and one from the β subunit, interact with ssDNA directly. The carboxy-terminal OB module in the α subunit is involved in protein–protein interaction with the β subunit. The two OB modules at the amino terminus of the α subunit are tightly associated and form a single recognition surface. The DNA is bound between the α subunit amino-terminal surface and the β subunit surface. It is interesting that both OB and RRM folds are involved in telomere recognition, because both are common single-stranded nucleic acid binding motifs. A β-sheet is involved in single-stranded nucleic acid recognition by both types of motif, although the detailed protein–DNA interactions differ significantly between the two structures. An unexpected finding from the present structure is the apparent involvement of protein–protein interactions in determining the nucleic acid binding specificity. Notably, the spatial arrangement of the two α subunit amino-terminal OB folds is similar to that of the two RRMs (from different UP1 molecules that bind to the same DNA strand) in the UP1–TR2 structure; in each case the two modules are in close contact and in parallel orientation. This similarity provides additional indirect evidence that protein–protein interaction is important for specific nucleic acid recognition by UP1. Because vertebrate telomeric 3′ overhangs are considerably longer than the dodecamers used in this study, it is possible that a repetitive nucleoprotein structure coats the telomeric ends. Several possible UP1–telomeric DNA higher order configurations are shown in Figure 7A–C.
Figure 7.
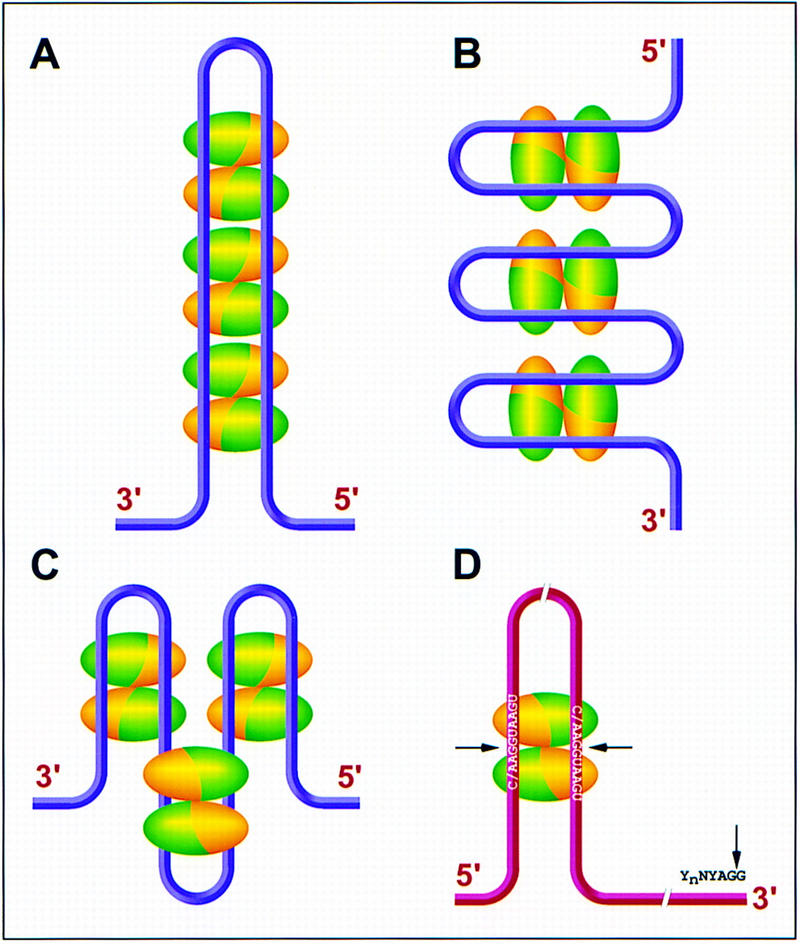
Models of possible hnRNP A1/UP1 interactions with single-stranded telomere overhangs and pre-mRNA alternative 5′ splice sites. Each ellipsoid represents one UP1 monomer; RRM1 is green and RRM2 is gold. Each pair of antiparallel ellipsoids constitutes a UP1 dimer. (A–C) Three models for UP1 interactions with telomeric DNA. The magenta strings represent the single-stranded telomeric DNA 3′ overhangs. Within each dimer, the ssDNA always follows a path from RRM1 to RRM2 in the 5′ to 3′ direction. (D) Model for UP1 interactions with an alternatively spliced pre-mRNA. The pre-mRNA is shown as a red string, with two antiparallel alternative 5′ splice sites brought into close proximity (∼25 Å) by a dimer of UP1 (or hnRNP A1), and a single 3′ splice site. (Arrows) Splice junctions.
Implications for alternative splicing regulation
hnRNP A1 is an abundant protein associated with most nascent transcripts in the cell (for review, see McAfee et al. 1997). It has global concentration-dependent effects on alternative pre-mRNA splicing that require RNA binding via both of its RRMs (Mayeda et al. 1994). The hnRNP A1 SELEX consensus hexamer sequence UAGGGA/U coincides with the telomeric repeat TTAGGG and also bears some resemblance to portions of the vertebrate consensus 5′ and 3′ splice sites (C/AAG:GUAAGU; YNYAG:G) (Burd and Dreyfuss 1994). It is not clear at present whether binding to high-affinity sites, or to one or both splice sites, is necessary for the global effects of hnRNP A1 in alternative splicing. However, binding of hnRNP A1 to the sequence UAGAGU, which resembles the above consensus, within an intron of the hnRNP A1 pre-mRNA, has been implicated in autoregulation at the level of alternative splicing (Chabot et al. 1997). Likewise, specific hnRNP A1 binding to other similar sequences, which in all cases include one or two copies of the UAG trinucleotide, is involved in splicing silencing of fibroblast growth factor receptor 2 and HIV pre-mRNAs, and in subgenomic mRNA transcription of mouse hepatitis virus RNA (Li et al. 1997; del Gatto-Konczak et al. 1999; M. Caputi, A. Mayeda, A.R. Kraimer, and A.M. Zahler, in prep.). In view of its participation in many different cellular processes, it is likely that hnRNP A1 can function both as a sequence-specific and general nucleic acid-binding protein, depending on the process.
The structural basis for the sequence specificity of UP1 is predicted to be essentially identical for RNA and for ssDNA. With the short oligonucleotides used for cocrystallization in the present study, the structure shows that each RRM within a UP1 monomer binds to a different molecule of ssDNA. However, the crystal structure is consistent with the involvement of either one or both of the hnRNP A1 RRMs interacting with a single, long nucleic acid molecule (Shamoo et al. 1997). For example, when there is only one high-affinity binding site, such as a splice site, it is possible that only one of the RRMs from a single hnRNP A1 molecule is used. When multiple binding sites are present in a single RNA molecule, each site may be bound by separate hnRNP A1 molecules and the protein molecules may associate with each other to bring distant binding sites closer together. It is also possible that these protein molecules do not interact with each other, that is, the sites are bound independently. Another scenario would be that the two binding sites interact with separate RRMs within the same protein molecule.
One or more of these scenarios may be relevant to the cellular functions of hnRNP A1. For example, the hnRNP A1-binding sites could be pre-mRNA splice sites. hnRNP A1 may then bring distant splice sites into close proximity and facilitate the proper folding of pre-mRNA for efficient splicing. In the case of alternative 5′ splice sites, which may be quite far apart on the same pre-mRNA molecule, binding to each of the RRMs within an hnRNP A1 monomer, or across a UP1 dimer, would place the two 5′ splice sites in an antiparallel orientation and within 25 Å of each other (Fig. 7D). This would present the two splice sites to the splicing machinery in a context in which they are easily distinguishable, thus explaining in part how hnRNP A1 can promote the selection of distal alternative 5′ splice sites (Mayeda and Krainer 1992). The next step would be the base pairing of the 5′ terminus of U1 snRNA to the appropriate 5′ splice site, which may be facilitated by the RNA annealing activity of the carboxy-terminal domain of hnRNP A1 (Pontius 1993). Each RRM may also bind to separate nucleic acid molecules to bring them together for efficient duplex formation (Shamoo et al. 1997). Conversely, hnRNP A1 may unwind base-paired RNA or DNA by binding to transiently single-stranded regions and thus promoting the opening up of the duplex. Whether any of the above scenarios is realized in nature clearly warrants further structure-function studies.
Materials and methods
Crystallization and data collection
Recombinant human UP1 was expressed and purified as described (Mayeda et al. 1994; Jokhan et al. 1997). Oligonucleotide TR2 was synthesized on an Applied Biosystems machine by standard phosphoramidite chemistry. The oligonucleotide was precipitated twice with ethanol from 20 mm MgCl2 solutions and analyzed for purity by urea–PAGE and UV shadowing. The UP1–TR2 complex was formed by mixing UP1 protein and TR2 oligonucleotide at a 1:1 molar ratio and incubating on ice for 1 hr. The final protein concentration was ∼12 mg/ml. The crystals were grown by the hanging-drop vapor diffusion method. The reservoir contained 0.1 m Tris (pH 8.5), 15% glycerol, and 2.0 m (NH4)2HPO4. The crystals reached a maximal size of ∼0.2 × 0.2 × 0.4 mm after 3 days. The space group for these crystals is P43212, with cell dimensions of a = b = 51.20 Å, c = 171.09 Å. A native data set, Native-1, was collected with a one-cell Brandeis CCD detector at beam-line X12C, and all other data sets, except HgCl2-1, were collected at beam-line X26C. Both beam-lines are at the National Synchrotron Light Source, Brookhaven National Laboratory. The derivative data set HgCl2-1 was collected on a Rigaku X-ray generator (focused Cu Kα) with an Raxis-II imaging plate detector. All data were collected at 100°K. Derivatives were prepared by soaking the crystals with either 2.5 mm p-chloromercuribenzene sulfonate (PCMBS) or 2.5 mm mercuric chloride (HgCl2) for two days. A native data set, Native-2, was collected from a crystal soaked for >2 months with 2.5 mm lead acetate, but no lead atom sites were detected. X-ray wavelengths used for synchrotron data sets were 1.15 Å for Native-1, 1.127 Å for PCMBS, and 1.115 Å for Native-2 and HgCl2-2. All data reduction was carried out by the HKL program suite (Otwinowski 1993).
Phasing and refinement
One mercury site was identified for each of the derivatives by isomorphous and anomalous difference Patterson maps. Interestingly, the Native-2 data set, which was obtained from crystals presoaked with lead acetate, showed a higher degree of isomorphism with the mercury derivatives, and was therefore used for phasing. Refinement of heavy-atom parameters and phase calculations were done by the PHASES suite of programs (Furey and Swaminathan 1996). The initial 3 Å MIRAS phases were improved by solvent flattening (Wang 1985). The resulting solvent-flattened electron density map clearly shows the protein and DNA molecules. The protein model was placed into the electron density by molecular replacement with the coordinates of RRM1 and RRM2 from the protein-only UP1 structure (pdb code 1up1) with the AmoRe program (Navaza 1994). The molecular replacement solution has an R-factor of 42.9% and a correlation coefficient of 41.1%, by use of the data in the resolution range of 10.0–4.0 Å. In addition to the protein model, 11 (of 12) nucleotides can be built into the solvent-flattened MIRAS electron density map. Modeling of the DNA molecule and protein model rebuilding were performed with the graphics program O (Jones et al. 1991). Model refinements were carried out with XPLOR and CNS (Brünger et al. 1998). Initially, data in the resolution range of 8.0–3.0 Å were used, and the starting crystallographic R-factor was 40.5%. Both higher and lower resolution data were then included with bulk-solvent correction in the refinement. Multiple rounds of model rebuilding and refinement were carried out before the placement of ordered water molecules and the refinement of the temperature factors. During the X-PLOR and CNS refinements, the Rfree value was monitored by use of 10% of the data. The stereochemical quality of the refined model was assessed by the PROCHECK program (Laskowski et al. 1993).
The atomic coordinates of the UP1–TR2 complex have been deposited with the Brookhaven Protein Data Bank (pdb code 2up1). The coordinates can also be obtained from R.-M. Xu at xur@cshl.org.
Acknowledgments
We thank M. Capel and R.M. Sweet for help with data collection at beamlines X26C and X12C at the National Synchrotron Light Source, Brookhaven National Laboratory. We are grateful to S. Munroe, D. Bastia, T. Messick, and D. Vaughn for comments on the manuscript. This work was supported in part by the W.M. Keck Foundation and by National Institutes of Health grant GM55874 (R.-M.X.) and National Cancer Institute program project CA13106 (A.R.K.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL xur@cshl.org; FAX (516) 3678873.
References
- Abdul-Manan N, Williams KR. hnRNP A1 binds promiscuously to oligoribonucleotides: Utilization of random and homo-oligonucleotides to discriminate sequence from base-specific binding. Nucleic Acids Res. 1996;24:4063–4070. doi: 10.1093/nar/24.20.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdul-Manan N, O’Malley SM, Williams KR. Origins of binding specificity of the A1 heterogeneous nuclear ribonucleoprotein. Biochemistry. 1996;35:3545–3554. doi: 10.1021/bi952298p. [DOI] [PubMed] [Google Scholar]
- Allain FH-T, Gubser CC, Howe PWA, Nagai K, Neuhaus D, Varani G. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formation. Nature. 1996;380:646–650. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Greider CW. Telomeres. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- Brünger AT, Adams PD, Clore GM, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: A new software system for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1, significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvoli M, Cobianchi F, Biamonti G, Riva S. Recombinant hnRNP A1 and its N-terminal domain show preferential affinity for oligodeoxyribonucleotides homologous to intron/exon acceptor sites. Nucleic Acids Res. 1990;18:6595–6600. doi: 10.1093/nar/18.22.6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvoli M, Cobianchi F, Riva S. Interaction of hnRNP A1 with snRNPs and pre-mRNAs: Evidence for a possible role of A1 RNA annealing activity in the first steps of spliceosome assembly. Nucleic Acids Res. 1992;20:5017–5025. doi: 10.1093/nar/20.19.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Stamm S, Helfman D, Krainer AR. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Nature. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- Cardenas ME, Bianchi A, de Lange T. A Xenopus egg factor with DNA-binding properties characteristic of terminus-specific telomeric proteins. Genes & Dev. 1993;7:883–894. doi: 10.1101/gad.7.5.883. [DOI] [PubMed] [Google Scholar]
- Carson M. Ribbons 2.0. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- Casas-Finet JR, Smith JD, Jr, Kumar A, Kim JG, Wilson SH, Kapel RL. Mammalian heterogeneous ribonucleoprotein A1 and its constituent domains: Nucleic acid interaction, structural stability and self-association. J Mol Biol. 1993;229:873–889. doi: 10.1006/jmbi.1993.1093. [DOI] [PubMed] [Google Scholar]
- Chabot B, Blanchette M, Lapierre I, La Branche H. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol Cell Biol. 1997;17:1776–1786. doi: 10.1128/mcb.17.4.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobianchi F, Karpel RL, Williams KR, Notario V, Wilson SH. Mammalian heterogeneous nuclear ribonucleoprotein complex protein A1: Large scale overproduction in Escherichia coli and cooperative binding to single-stranded nucleic acids. J Biol Chem. 1988;263:1063–1071. [PubMed] [Google Scholar]
- Del Gatto-Konczak F, Olive M, Gesnel M, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Piñol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Erlitzki R, Fry M. Sequence-specific binding protein of single-stranded and unimolecular quadruplex telomeric DNA from rat hepatocytes. J Biol Chem. 1997;272:15881–15890. doi: 10.1074/jbc.272.25.15881. [DOI] [PubMed] [Google Scholar]
- Fang GW, Cech TR. Molecular cloning of telomere-binding protein genes from Stylonychia mytilis. Nucleic Acids Res. 1991;19:5515–5518. doi: 10.1093/nar/19.20.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-D, Mayeda A, Maniatis T, Krainer AR. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furey W, Swaminathan S. PHASES-95: A program package for the processing and analysis of diffraction data from macromolecules. Methods Enzymol. 1996;277:590–620. doi: 10.1016/s0076-6879(97)77033-2. [DOI] [PubMed] [Google Scholar]
- Gorlach M, Wittekind M, Beckman RA, Mueller L, Dreyfuss G. Interaction of the RNA-binding domain of the hnRNP C proteins with RNA. EMBO J. 1992;11:3289–3295. doi: 10.1002/j.1460-2075.1992.tb05407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick G, Alberts B. Purification and physical characterization of nucleic acid helix-unwinding proteins from calf thymus. J Biol Chem. 1976;251:2124–2132. [PubMed] [Google Scholar]
- Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- Ishikawa F, Matunis MJ, Dreyfuss G, Cech TR. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol Cell Biol. 1993;13:4301–4310. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- Janin J, Chothia C. The structure of protein-protein recognition sites. J Biol Chem. 1990;265:16027–16030. [PubMed] [Google Scholar]
- Janin J, Rodier F. Protein-protein interaction at crystal contacts. Proteins. 1995;23:580–587. doi: 10.1002/prot.340230413. [DOI] [PubMed] [Google Scholar]
- Jokhan L, Dong A-P, Mayeda A, Krainer AR, Xu R-M. Crystallization and preliminary X-ray diffraction studies of UP1, the two-RRM domain of hnRNP A1. Acta Crystallogr. 1997;D53:615–618. doi: 10.1107/S0907444997003326. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Johnston SD, Lew JE, Berman J. Gbp1p, a protein with RNA recognition motifs, binds single-stranded telomeric DNA and changes its binding specificity upon dimerization. Mol Cell Biol. 1999;19:923–933. doi: 10.1128/mcb.19.1.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: Binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König P, Rhodes D. Recognition of telomeric DNA. Trends Biochem Sci. 1997;22:43–47. doi: 10.1016/s0968-0004(97)01008-6. [DOI] [PubMed] [Google Scholar]
- Kumar A, Casas-Finet JR, Luneau CJ, Karpel RL, Merrill BM, Williams KR, Wilson SH. Mammalian heterogeneous nuclear ribonucleoprotein A1: Nucleic acid binding properties of the COOH-terminal domain. J Biol Chem. 1990;265:17094–17100. [PubMed] [Google Scholar]
- LaBranche H, Dupuis S, Ben-David Y, Bani M-R, Wellinger RJ, Chabot B. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat Genet. 1998;19:199–202. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structure coordinates. J Appl Crystallogr. 1993;A42:140–149. [Google Scholar]
- Lee MS, Gallagher RC, Bradley J, Blackburn EH. In vivo and in vitro studies of telomeres and telomerase. Cold Spring Harb Symp Quant Biol. 1993;58:707–718. doi: 10.1101/sqb.1993.058.01.078. [DOI] [PubMed] [Google Scholar]
- Li HP, Zhang X, Duncan R, Comai L, Lai MM. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc Natl Acad Sci. 1997;94:9544–9599. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Zakian VA. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: Requirement of a primer 3′ overhang. Proc Natl Acad Sci. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Helfman DM, Krainer AR. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol Cell Biol. 1993;13:2993–3001. doi: 10.1128/mcb.13.5.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Munroe SH, Cáceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Munroe SH, Xu R-M, Krainer AR. Distinct functions of the closely related tandem RNA-recognition motifs of hnRNP A1. RNA. 1998;4:1111–1123. doi: 10.1017/s135583829898089x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAfee JG, Huang M, Soltaninassab S, Rech JE, Iyengar S, LeStourgeon WM. The packaging of pre-mRNA. In: Krainer AR, editor. Eukaryotic mRNA processing. Oxford, UK: IRL Press; 1997. pp. 68–102. [Google Scholar]
- McElligott R, Wellinger RJ. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay SJ, Cooke H. hnRNP A2/B1 binds specifically to single stranded vertebrate telomeric repeat TTAGGGn. Nucleic Acids Res. 1992;20:6461–6464. doi: 10.1093/nar/20.24.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill BM, Stone KL, Cobianchi F, Wilson SH, Williams KR. Phenylalanines that are conserved among several RNA-binding proteins form part of a nucleic acid-binding pocket in the A1 heterogeneous nuclear ribonucleoprotein. J Biol Chem. 1988;263:3307–3313. [PubMed] [Google Scholar]
- Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J. AMoRe: An automated package for molecular replacement. Acta Crystallogr. 1994;50:157–163. [Google Scholar]
- Nicholls A, Sharp K, Honig B. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. Oscillation data reduction program. In: Sawyer L, Issacs N, Bailey S, editors. Data collection and processing, Proceedings of the CCP4 study weekend. Warrington, UK: SERC Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- Oubridge C, Ito N, Evans PR, Teo C-H, Nagai K. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- Pandolfo M, Valentini O, Biamonti G, Morandi C, Riva S. Single stranded DNA binding proteins derive from hnRNP proteins by proteolysis in mammalian cells. Nucleic Acids Res. 1985;13:6577–6590. doi: 10.1093/nar/13.18.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Pontius BW. Close encounters: Why unstructured, polymeric domains can increase rates of specific macromolecular association. Trends Biochem Sci. 1993;18:181–186. doi: 10.1016/0968-0004(93)90111-y. [DOI] [PubMed] [Google Scholar]
- Price CM, Cech TR. Telomeric DNA-protein interactions of Oxytricha macronuclear DNA. Genes & Dev. 1987;1:783–793. doi: 10.1101/gad.1.8.783. [DOI] [PubMed] [Google Scholar]
- Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B′′–U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- Shamoo Y, Abdul-Manan N, Patten AM, Crawford JK, Pellegrini MC, Williams KR. Both RNA-binding domains in heterogeneous nuclear ribonucleoprotein A1 contribute toward single-stranded-RNA binding. Biochemistry. 1994;33:8272–8281. doi: 10.1021/bi00193a014. [DOI] [PubMed] [Google Scholar]
- Shamoo Y, Abdul-Manan N, Williams KR. Multiple RNA binding domains (RBDs) just don’t add up. Nucleic Acids Res. 1995;23:725–728. doi: 10.1093/nar/23.5.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamoo Y, Krueger U, Rice LM, Williams KR, Steitz TA. Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 Å resolution. Nat Struct Biol. 1997;4:215–222. doi: 10.1038/nsb0397-215. [DOI] [PubMed] [Google Scholar]
- Sheng H, Hou Z, Schierer T, Dobbs DL, Henderson E. Identification and characterization of a putative telomere end-binding protein from Tetrahymena thermophila. Mol Cell Biol. 1995;15:1144–1153. doi: 10.1128/mcb.15.3.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Dreyfuss G. RNA binding specificity of hnRNP proteins: A subset bind to the 3′ end of introns. EMBO J. 1988;7:3519–3529. doi: 10.1002/j.1460-2075.1988.tb03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virta-Pearlman V, Morris DK, Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes & Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- Wang B-C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- Wang W, Skopp R, Scofield M, Price C. Euplotes crassus has genes encoding telomere-binding proteins and telomere-binding protein homologs. Nucleic Acids Res. 1992;20:6621–6629. doi: 10.1093/nar/20.24.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RJ, Sen D. The DNA structures at the ends of eukaryotic chromosomes. Eur J Cancer. 1997;33:735–749. doi: 10.1016/S0959-8049(97)00067-1. [DOI] [PubMed] [Google Scholar]
- Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes & Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R-M, Jokhan L, Cheng X, Mayeda A, Krainer AR. Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA-recognition motifs. Structure. 1997;5:559–570. doi: 10.1016/s0969-2126(97)00211-6. [DOI] [PubMed] [Google Scholar]
- Yang X, Bani MR, Lu SJ, Rowan S, Ben-David Y, Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc Natl Acad Sci. 1994;91:6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakian VA. Telomeres: Beginning to understand the end. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]


