Abstract
Tetrahymena thermophila is the best studied of the ciliates, a diversified and successful lineage of eukaryotic protists. Mirroring the way in which many metazoans partition their germ line and soma into distinct cell types, ciliates separate germ line and soma into two distinct nuclei in a single cell. The diploid, transcriptionally silent micronucleus undergoes meiosis and fertilization during sexual reproduction and determines the genotype of the progeny; in contrast, the expressed macronucleus contains many copies of hundreds of small chromosomes, determines the cell’s phenotype, and is inherited only through vegetative reproduction. Here we demonstrate the power of HAPPY physical mapping to aid the complete assembly of T. thermophila macronuclear chromosomes from shotgun sequence scaffolds. The finished genome, one of only two ciliate genomes shotgun sequenced, will shed valuable additional light upon the biology of this extraordinary, diverse, and, from a genomics standpoint, as yet largely unexplored evolutionary branch of eukaryotes.
Keywords: Assembly closure, Chromosome breakage sequence, Germ-line nucleus, Internally eliminated sequence, Link validation, Macronuclear chromosomes, Sequence scaffolds, Somatic nucleus, Telomeres, Whole-genome-shotgun sequence
Ciliates are one of the most abundant, successful, and diverse groups of unicellular eukaryotes. They are a mainly free-living group of alveolates, which also include the dinoflagellates and the obligate parasitic apicomplexans (e.g., Plasmodium, the malaria parasite). Although the ciliates have conserved much animal biology, their last common ancestor with the animals is very near to—if not at—the root of the eukaryotic phylogenetic tree (reviewed by [1]). The ciliates have occupied every major type of aquatic environment and are important elements at the base of the animal food chain.
The best studied member of this group, Tetrahymena thermophila, has been the subject of study for some 80 years [2]. Major discoveries made using this model organism include catalytic RNA (ribozymes), telomere structure and telomerase, the cell motor dynein, and the role of histone acetylation in gene expression [3]. Conventional genetic techniques in Tetrahymena were elaborated by Nanney and his collaborators in the 1950s and powerful molecular technologies for experimental gene manipulation exist for this organism (reviewed in [3]).
Ciliates possess not one but two types of nuclei, distinct in their appearance and function [4,5]. The small micronucleus (MIC) is the germ line, which in Tetrahymena contains five pairs of chromosomes that are transcriptionally silent in vegetative cells, while the larger (somatic) macronucleus (MAC) contains about 250–300 smaller, transcriptionally active chromosomes totaling slightly over 100 Mb and determines the phenotype of the cell. MAC chromosome sizes range from 21 kb to an estimated 3.3 Mb, and all are believed to be maintained at comparable copy number, averaging ~45 copies per G1-stage MAC [6]. The only known exception is the smallest MAC chromosome, which encodes the major ribosomal RNAs and is present at approximately 9000 copies per MAC.
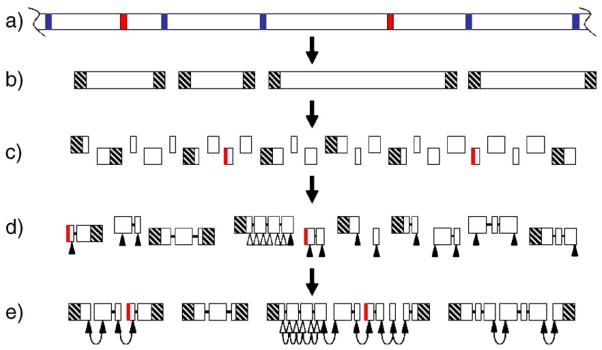
The new micronucleus and macronucleus differentiate from mitotic products of the diploid fertilization nucleus. During MAC differentiation, the germ-line-derived chromosomes are generated by programmed chromosome breakage, telomere addition, and amplification (Figs. 1a–1b). Breakage occurs at conserved chromosome breakage sequences (CBSs); about 75 bp, consisting of the CBS and some flanking sequence (breakage-eliminated sequences or BESs), are lost. Each of the resulting MAC chromosomes is capped at both ends by the addition of a readily identifiable telomere, a roughly 100- to 400-bp-long tract of tandem repeats of the hexanucleotide GGGGTT/CCCCAA. All evidence indicates that the sequences adjacent to each of the ~550 MAC telomeres are unique [7–9]. A second type of DNA rearrangement occurs during MAC differentiation: the splicing out and physical loss of “internally eliminated sequences,” or IESs [10]. Roughly 6000 distinct IESs are eliminated, representing 15% of the MIC sequence complexity. Most of the repetitive sequence in the MIC is lost from the MAC through IES removal. IESs and BESs are collectively called “MIC-limited,” while the remainder is referred to as “MAC-destined.”
Fig. 1.
The genome of Tetrahymena thermophila and strategy for the assembly of the macronuclear genome. (a) The micronuclear genome consists of large chromosomes (only part of one chromosome is shown). Copies of these chromosomes are cut at CBSs (blue), and IESs (red) are spliced out, to produce the smaller macronuclear chromosomes (b), which are capped with telomeres (hatched). (c) Shotgun sequencing of the macronuclear chromosomes yields many sequence contigs, some of which may consist of—or be prematurely truncated by the incorporation of—IESs from contaminating micronuclear DNA (red vertical lines). (d) Contigs can be linked to form scaffolds using read-pair information (heavy black lines). Sequences close to nontelomeric scaffold ends (filled triangles) plus some from within larger scaffolds (open triangles) are chosen for use as markers. (e) HAPPY mapping reveals physical links between markers, allowing the scaffolds to be grouped into superscaffolds (hoops indicate HAPPY links) and also confirming the correct assembly within some larger scaffolds.
Tetrahymena preserves a fairly complete set of ancestral animal eukaryotic biological processes and shares a high degree of functional conservation with the human genome [11]. These features, coupled with its ease of culture, experimental tractability, and membership in a hitherto unsequenced group of organisms, made it a focus of a recent genome sequencing program. The T. thermophila MAC was sequenced using a whole-genome shotgun approach at The Institute for Genome Research (TIGR; http://www.tigr.org/tdb/e2k1/ttg/). Sequence reads, sequence assemblies, predicted gene sequences, and annotations are publicly available at TIGR (ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/t_thermophila/), at the Tetrahymena Genome Database (http://www.ciliate.org/), and at NCBI (http://www.ncbi.nlm.nih.gov/). The current release of the macronuclear genome assembly (November 2003) consists of 1971 sequence scaffolds. One hundred twenty-five scaffolds, comprising about 45% of the genome, extend from telomere to telomere (E.P. Hamilton and E. Orias, unpublished observations) and thus represent complete chromosomes.
Completion of the macronuclear sequence by closing the remaining gaps is considered essential for this valuable model organism. It is estimated that over 100 macronuclear chromosomes remain to be completely assembled from the 1846 sequence scaffolds that carry telomeric sequence at neither or only one end. Gaps between sequence scaffolds are encountered in most, if not all, genome projects and consist mainly of three types. First, repeat gaps occur wherever tracts of repetitive sequence extend beyond the distances spanned by a single small-insert clone; such regions cannot be assembled unambiguously, even though data exist to fill the gap. Second, statistical gaps occur wherever the random selection of clones for sequencing happens to miss a particular region; such gaps can in principle be closed by continuing the shotgun to greater depth but, at an average shotgun coverage of ninefold, a point of diminishing returns has been reached. Third, and most intractable, are the gaps caused by cloning bias: certain sequences in any genome are refractory to cloning in the bacterial host and will simply be absent from the small-insert libraries.
How can the remainder of the MAC genome be assembled? In many genome projects, the long-range ordering of sequence scaffolds is achieved by the use of large-insert clones such as yeast or bacterial artificial chromosomes (YACs or BACs), which can span the gaps left in the shotgun assembly. Unfortunately, no one has been able to make stable sequencing libraries with inserts larger than ~7 kb from the very AT-rich (76%) Tetrahymena genome. An alternative method to span the gaps would be to try bridging open scaffold ends by PCR amplification. Because we lack prior knowledge of which scaffolds adjoin one another in the genome, this work scales as the square of the number of open ends. Thus, at least ~2.5 million PCR primer combinations would be required, and a more cost-effective method is needed.
Fortunately, this crucial linking information can be provided by HAPPY mapping. This technique, which has been applied to a number of genomes [12–14], enables the distances between sequences to be estimated, even if the intervening DNA cannot be cloned and DNA polymorphisms are not available. Genomic DNA is sheared randomly and aliquoted into samples, each containing only a fraction of a genome’s worth of fragments. These samples are then tested, by PCR, for their content of specific sequences (markers). Markers that lie close to one another in the genome (compared to the average size of the fragments) tend to remain on the same DNA fragments after shearing and hence are often found together in the same sample. The frequency of this cosegregation can be used to estimate the distances between the markers and hence to construct a map of their relative positions in the genome. The process is analogous to genetic linkage mapping, in which cosegregation of loci in meiosis reflects their proximity on the chromosomes. The work scales linearly with the number of markers to be mapped.
We report here the results of a pilot HAPPY mapping project to provide linking information to help complete the macronuclear genome of Tetrahymena. Sequences chosen near the ends of the shotgun sequence scaffolds have been used as markers and tested on a HAPPY mapping panel. Pairs of markers that are found to cosegregate strongly are inferred to adjoin one another on the same MAC chromosome. Our initial aim (Figs. 1d–1e) is to map all shotgun scaffolds of >2 kb in size (and a few smaller scaffolds that are capped at one end by telomeric sequences), linking them by HAPPY mapping to define superscaffolds representing each of the >100 remaining unassembled macronuclear chromosomes. The results of our initial survey show that the method is very effective for linking MAC sequence scaffolds. We also discuss our plans to use the completed macronuclear genome sequence as a springboard for the completion of the micronuclear chromosome assembly.
Results
Identification of interscaffold links by HAPPY mapping
Markers were designed near scaffold ends and typed against the HAPPY panel as described under Materials and methods. The distribution of positive typings among all markers peaked in the 40–45 interval (of 88 panel members), i.e., within sampling error of the targeted 50%. Markers that gave few (<20) or no positive typings with the panel were set aside, as these often are due to poor PCR amplification. (Our unpublished observations indicate that some of these scaffolds may represent IES DNA contamination; they are being investigated). Also set aside were those markers that gave an anomalously high proportion of positive typings (>65 positives of the 88 HAPPY panel aliquots); such markers generally represent sequences that are present in multiple copies in the genome.
For the 520 markers that were retained in the analysis, pairwise linkages were calculated (Materials and methods), and groups of markers linked at LOD ≥5 (odds in favor of linkage ≥105:1) were identified. A total of 40 such linkage groups, involving 47 links between previously unconnected scaffolds, were identified (Table 1). For each such group, the linkage data were inspected to deduce the correct order and, when possible, orientation of the linked scaffolds. Scaffold orientation (and, in a small number of cases, order) remains ambiguous for some of the smaller (typically <4 kb) scaffolds, particularly in cases in which the scaffold contains only a single, central marker or only one of the two markers from either end of the scaffold has thus far been mapped. An additional 7 groups of markers linked at LOD 4.5–5 were identified involving 10 links between previously unconnected scaffolds (Table 1).
Table 1.
HAPPY links obtained after mapping 520 markers
| Link a | θ | Respective scaffold sizes (bp) | Superscaffold size (bp) | Gap (kb) b | Weak links c |
|---|---|---|---|---|---|
| n-8253825-8254470+ | 0.32 | 7698, 379103 | 386801 | −0.83 | 0 |
| +8253906′-8254686+ | 0.37 | 8877, 541833 | 550710 | 0 | |
| *-(8253909)-(8254228)-* | 0.26 | 2014, 1492 | 3506 | −0.35 | 0 |
| h-8253981-8254728′-x | 0.28 | 17149, 224744 | 241893 | −0.7 | 1 |
| *-8253985-8254230′-x | 0.23 | 9556, 18120 | 27676 | −0.8 | 0 |
| *-8253992-8254601-* | 0.34 | 141270, 46535 | 187805 | 0 | |
| n-8254003′-8254247′-(8253973′+-8253829′+) d,e | 0.42, 0.41, 0.46 | 20084, 42068, 1918, 3705 | 64070 | ND, ND, −0.05 | 0, 0, 1 |
| x-[8254053]-8254685′-* | 0.4 | 3763, 42582 | 46345 | −0.7 | 1 |
| *-8254104-8254754-* | 0.28 | 298947, 392787 | 691734 | −1.148 | 1 |
| *-[8254146]-{8253925′-8254240}-(8255519)-* | 0.25, 0.41, 0.32 | 3199, 8181, 8180, 1335 | 20895 | ND, ND, +1.0 | 0, 0, 1 |
| x-8254152-8254481+ | 0.36 | 16921, 36977 | 53898 | 0 | |
| *-8254216-8254399-8254445′-* | 0.32, 0.39 | 32465, 6376, 6660 | 45501 | 0, 0 | |
| *-[8254220]-8254598′-* | 0.38 | 4918, 267927 | 272845 | −2.2 | 0 |
| *-[8254233′]-8254693+ | 0.39 | 5989, 27987 | 33976 | −0.5 | 1 |
| x-8254242-8254432-* | 0.43 | 5041, 10358 | 15399 | −0.8 | 0 |
| x-[8254246]-[8254027′]-x(8254031) | 0.39, 0.39 | 4444, 6276, 4048 | 14768 | −3.0, −3.0 | 0, 0 |
| *-[8254253]-(8255219)-* | 0.01 | 3786, 1254 | 5040 | 0 | |
| *-(8254294)-(8254829)-* | 0.41 | 1511, 1636 | 3147 | 0 | |
| *-(8254317)-(8254325)-* | 0.33 | 2189, 2708 | 4897 | −0.5 | 1 |
| *-8254355′-8253932-x | 0.33 | 9713, 20693 | 30406 | 0 | |
| +8254361-8254021+ e | 0.44 | 73 558, 21071 | 94629 | +1.351 | 0 |
| *-8254407-8254779′-x | 0.33 | 49038, 131984 | 181022 | −0.6 | 0 |
| *-8254418′-8254265+ | 0.39 | 100878, 13464 | 114342 | [−1.3] | 0 |
| +8254423-8254034+ f | 0.25 | 91433, 211760 | 303193 | −1.083 | 14 |
| *-8254425-8254500-x | 0.3 | 12302, 54835 | 67137 | +0.078 | 1 |
| *-8254426-8254443′-* | 0.29 | 20663, 9862 | 30525 | 0 | |
| +8254431-8253850′+ g | 0.29 | 715652, 425044 | 1140696 | −0.002 | 1 |
| x-8254442′-8254479+ | 0.39 | 17810, 384242 | 402052 | −3.7 | 1 |
| *-8254485-8254222+ | 0.37 | 158875, 20223 | 179098 | −1.5 | 1 |
| x-8254492-8254674′-* | 0.39 | 83344, 268780 | 352124 | 0 | |
| *-(8254531)-8254690-x | 0.42 | 2582, 46225 | 48807 | −0.5 | 1 |
| *-{(8254532)-(8254539)}-8254642-h | 0.23, 0.34 | 2710, 2382, 29839 | 34931 | −0.6, −0.75 | 1,9 |
| *-8254561-8254526-* e | 0.43 | 18747, 18366 | 37113 | −0.548 | 20 |
| +8254593-8254780′-8254599+ | 0.28, 0.39 | 533928, 67241, 80448 | 681617 | ND, −0.6 | 0, 12 |
| *-8254603-(8254409)-* | 0.27 | 126171, 2561 | 128732 | −0.6 | 1 |
| *-8254604-8253899-* | 0.37 | 20288, 65344 | 85632 | 0 | |
| n-8254605-8254824-* | 0.36 | 384764, 137162 | 521926 | +0.195 | 1 |
| *-8254628-8253928′-* | 0.32 | 16361, 20789 | 37150 | 0 | |
| *-8254643′-8254618-* | 0.35 | 8282, 3441 | 11723 | +0.3 | 7 |
| *-8254655′-8254671′-8253977-* | 0.16, 0.39 | 16966, 583021, 12455 | 612442 | −1.5, h ND | 11, 0 |
| +8254662-8254612-h | 0.23 | 56407, 22412 | 78819 | −0.9 | 1 |
| *-8254676-8254033-he,f | 0.42 | 73992, 32639 | 106631 | 0 | |
| *-8254727-8254799′-* | 0.39 | 12815, 233521 | 246336 | −1.2 | 9 |
| +8254814′-8254673+ e | 0.35 | 591214, 199572 | 790786 | −3.6 | 7 |
| *-8254819′-8254667+ f | 0.38 | 1807540, 356833 | 2164373 | −1.4 i | 6 |
| h-8254821′-8254781-* e | 0.43 | 14120, 58936 | 73056 | 0 | |
| *-8254585′-8253989 e-8254729′-* | 0.41, 0.39 | 74110, 8063, 22475 | 104648 | ND, −0.5 | 0, 0 |
| Average j | 0.34, 0.34 | −0.90 |
HAPPY mapping scaffold links. Symbols: ′, scaffold reversed; +, telomere-capped end; –, link (if in middle of a group); –* (or *-), the group ends or begins with a mapped marker that does not yet link on further; -x (or x-), the marker at that end of the group failed to type (needs to be repeated or redesigned); -n (or n-), no marker has yet been designed for this end of the group; -h (or h-), the terminal mapped marker is present at very high copy number (presumed duplicate/multicopy sequence—see text) and hence cannot be linked reliably; [ ], orientation of scaffold not confirmed (but the orientation shown is likeliest); ( ), orientation not known (e.g., for a small scaffold with only a single internal marker); { }, order of bracketed scaffolds not confirmed. Two small telomere-capped scaffolds that were linked to form a 2.4-kb superscaffold (not shown) were found to be rDNA sequences that had escaped the filtering process.
Estimated gap length (positive number) or overlap (negative number) between linked scaffolds. Bold numbers shown in square brackets have been verified by PCR only. Bold numbers without brackets were verified by sequencing a linking PCR product. The other estimates were based on weak links or overlaps confirmed by BLAST alignment of the scaffold ends with >96% identity. When both weak link and alignment data were available, the estimate is based on the latter, deemed to be more accurate. ND or blank means no data available to make an estimate.
Number of weak links supporting the HAPPY link; those shown in boldface indicate overlaps that were confirmed by BLAST alignment.
In HAPPY superscaffold 8254003, the small third and fourth scaffolds are both shown as telomere-capped because both scaffolds—largely nonoverlapping and correctly ordered as shown—were linked to the same telomere by independent pair-read information.
LOD at least 4.5 but less than 5.0. For all others LOD was at least 5.
Both scaffolds map to the same MIC chromosome.
Confirmed by genetic coassortment.
Match ends 937 bp from the 3′ end of 8254671.
Match excludes first two contigs (2416 bp) of 8254819.
For the θ column: averages for all links and for supported links (i.e., those for which gap estimates are given).
The linking of these scaffolds to form superscaffolds has so far defined 7 complete MAC chromosomes (that is, linked groups of two or more scaffolds, of which the first and last are capped by telomeres), in addition to the 125 complete chromosomes that were represented by single scaffolds in the initial shotgun assembly. The remaining links define superscaffolds that remain uncapped at one or both ends—it is anticipated that these superscaffolds will merge or link to others as markers on as-yet-unmapped scaffolds are mapped.
Validating HAPPY links
To test the reliability of the interscaffold links defined by the HAPPY data, the genome sequence was scrutinized for any independent evidence that pairs of HAPPY-linked scaffolds are indeed connected. One approach was to look for reasonable “weak links” between the two scaffolds. Usually paired-end sequence reads (the reads from each end of a sequencing library clone, also called “mate pairs” or “mates”) are assembled into the same scaffold. Mate pairs assembled by the Celera assembler at TIGR into different scaffolds, for whatever reason, are considered weak links. Some of these links were not used because there was only a single mate pair linking two scaffolds and the Celera assembler requires two links. Sometimes the links were not used because they did not support a current scaffold assembly; for instance, the mates might be too far apart in an existing scaffold (the average size of the inserts in each sequencing library being known). A link might also have been rejected because it conflicted with another, better supported link. A file, euttg.weaklinks, containing information on nearly 5000 weak links from the Tetrahymena genome project is available at the TIGR Web site at ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/t_thermophila/Assemblies_and_Sequences/Assembly2_Extra_Info/Assembly2-Nov-2003.scaffolds.euttg.weaklinks.Z. The existence of one or more weak links between scaffolds, especially when the reads are located in the correct scaffold ends with spacing compatible with the clone insert size range, can be considered good supporting evidence for the link. Of the 57 HAPPY linkage groups, 24 are supported by one or more weak links (Table 1). The estimated gap (positive numbers) or overlap (negative numbers) lengths obtained from the weak links file are shown in Table 1.
Prompted by the weak link data, which suggested that many of the links identified by HAPPY mapping represented overlaps, we searched, by BLAST alignment [15], for all overlaps between the linked ends of scaffolds. Overlaps with at least 96% sequence identity were indeed found in 31 cases (Table 1); 15 of these also had weak link support (see boldface weak links in Table 1). These data provide independent support for the validity of these HAPPY links. Altogether, about half of the links identified by HAPPY mapping (31 of 57, or 53%) are believed to correspond to overlaps with an average size of ~900 bp, as determined by scaffold sequence alignment or weak link data (Table 1).
To test directly the validity of this corroborating evidence, 12 selected HAPPY links were tested by PCR amplification. Those chosen included links identified early in the study, links between large scaffolds, links involving two scaffolds with telomere-capped ends, and links with good weak link substantiation (see Table 1). PCR primers, designed in unique regions of sequence lying nearest to the linked ends, were used to amplify PCR products using whole-genome Tetrahymena DNA as template. Nine tests gave specific products, and eight of these have been sequenced further and rigorously confirm the scaffold link (shown in boldface in Table 1). In the three cases in which PCR was attempted but no product was obtained, we have not yet ruled out problems with long-range PCR and/or primer design; thus none of these cases provide compelling evidence that the HAPPY links are incorrect. Note that the test of the link between scaffolds 8254361 and 8254021 was totally “blind” in that only HAPPY mapping indicated that these two scaffolds might be linked.
Scaffolds representing about half of the Tetrahymena genome have been genetically mapped (E. Orias and E.P. Hamilton, unpublished results). Four cases in which pairs of scaffolds joined by a HAPPY link had been genetically mapped provided opportunities to refute HAPPY links rigorously. In one case, DNA polymorphisms on the two scaffolds coassorted, thus providing strong genetic evidence [16] that in vivo the two scaffolds indeed belong to the same MAC chromosome. The probability of such a match occurring by chance alone is only ~0.004. For the other three links, coassortment data were not available, but the coupled scaffolds independently mapped to the same MIC chromosome arm. For each linked pair, the probability of a chance coincidence is higher, but still only about 0.1 each. Thus all four HAPPY links are supported by the available genetic mapping data.
In summary, no HAPPY links have been rigorously refuted and many have been supported by independent tests and mapping information. No previous linking information was available for the majority of the HAPPY links, and HAPPY mapping was indispensable for their identification. HAPPY links identified a subset of weak links, which, without the HAPPY data, would have remained overlooked in the weak links file. The Celera weak links are not a good primary source of linking information because they contain mainly accumulated accidents and problems of various types. Likewise, because many scaffolds are terminated by repeated sequence, finding matches between scaffold ends provides corroborating evidence of a HAPPY link but cannot reliably substitute for it.
The number of interscaffold linkages found by HAPPY mapping so far is consistent with theoretical predictions
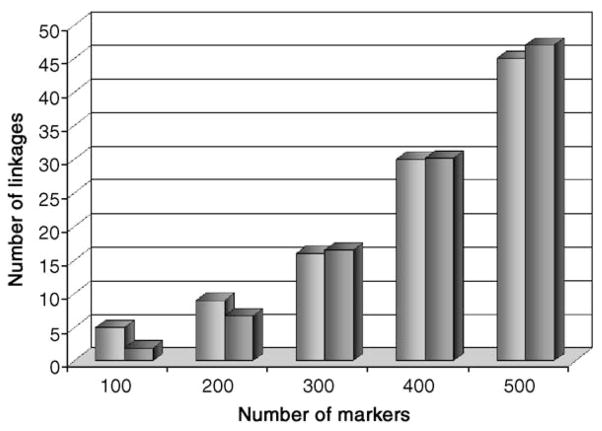
We sought to discover whether the actual number of interscaffold linkages found by the HAPPY mapping was consistent with that expected at this stage in the project and hence to infer the likely outcome of mapping the ends of all larger (>2 kb) scaffolds. We performed multiple simulations involving increasing numbers of markers tested, as described under Materials and methods, and compared these with the actual results obtained over the course of the project to date. As can be seen (Fig. 2), the actual and expected results are very similar. The number of linkages (in both the real and the simulated data) increases slowly at first; as more markers are mapped it becomes more likely that a newly mapped marker will link to its previously-mapped “twin” from the other side of the gap, and the number of linkages starts to increase more steeply.
Fig. 2.
Actual and predicted results from HAPPY mapping. For numbers of mapped markers between 100 and 520, the number of interscaffold HAPPY linkages predicted by simulation (darker gray bars; each value is the mean of 10 simulations) is compared with the number of linkages observed (lighter gray bars).
This concordance between the real and the simulated results indicates that interscaffold linkages are being robustly detected and that the typing of all uncapped scaffold termini will allow almost all of the scaffolds to be assembled into superscaffolds corresponding to complete macronuclear chromosomes. It also indicates that the present shotgun assembly represents the majority of the macronuclear sequence: if many of the interscaffold gaps were much larger than a few kilobases, we would fail to detect strong linkage between the markers on either side of the gap, and the number of linkages that we detect would fall below that expected by simulation.
Intrascaffold links indicate a substantially correct shotgun assembly
To check the quality of the shotgun assembly (on which the further assembly of the macronuclear genome rests), we designed and tested HAPPY markers from within two of the largest sequence scaffolds. Linkage between consecutive contigs within a scaffold would confirm the read-pair-based assembly of contigs into scaffolds. Fifty such markers, giving between 20 and 65 positives on the HAPPY panel, have been successfully typed to date. They span 20 gaps between consecutive contigs. In 14 of these instances, the lod score between the markers on either side of the gap is >5. In 3 instances, the lod score is low (between 1.6 and 3), but one or both of the relevant markers give a higher than average number of positives (49–60 positives) and may represent a multi- (or two-) copy sequence. In the remaining 3 instances, the lod score is low (2.0–3.7) despite the relevant markers giving normal numbers of positives on the mapping panel, but still strongly indicative of linkage (odds for linkage between 100:1 and 5000:1).
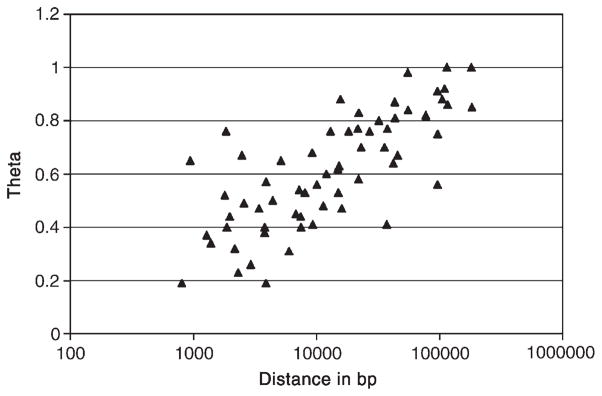
Further confirmation of the shotgun assembly comes from links seen between pairs of markers that were designed for linking scaffolds but that, fortuitously, also represent both ends of small scaffolds. When all 62 such pairs in the current dataset are examined, linkage between paired scaffold ends falls exponentially with increasing scaffold length as expected (Fig. 3), but with a number of outliers (paired ends from short scaffolds that show lower than expected, though still mostly significant, LODs). However, when we discard linkages involving markers with <30 or >50 positives (which may arise from poorly typed or MIC-derived markers or from double- or multicopy sequences, respectively), all but 1 of the remaining 37 linkages show the expected linkages. Therefore, current data strongly indicate that the current assembly is largely correct.
Fig. 3.
Plot of θ (inversely related to linkage) as a function of base pair distance for intrascaffold marker pairs. The scatter results from a combination of sampling error, fragment length heterogeneity, and imperfect typings for a few markers. Outliers are discussed in the text.
Discussion
The whole-genome shotgun sequence of the T. thermophila MAC genome presents a unique challenge, the assembly of more than 200 chromosomes. Their assembly was further hampered by the inability to make large-insert clones (YACs or BACs) to aid in the long-range linking of sequence scaffolds. This work shows that HAPPY mapping can be used to circumvent this problem. A shotgun sequence inevitably fails to represent a small (but unknown) proportion of the genome and a larger (but still unknown) proportion of genes that span the gaps within scaffolds (which separate sequence contigs) or between scaffolds. For a great many questions of biological and evolutionary interest, being able to state what is absent from a genome can be as important as what is present, and a shotgun sequence fails to provide this information. Nor does a shotgun sequence provide genome-wide positional context and long-range structure, features that are increasingly recognized as important in understanding genome function and evolution. Because of its relatively low frequency of repetitive sequence, the T. thermophila MAC genome is one of the very few eukaryotic genomes for which obtaining a complete sequence and assembly seems within reach.
To date, the observations on the rate of linkage discovery by HAPPY mapping and the confirmation of physical linkage between marker pairs imply that the underlying shotgun assembly of the T. thermophila MAC genome is robust and substantially accurate. The distances between markers among links supported by independent evidence (PCR results, scaffold end overlaps, and/or assembler-generated weak links) appear to be representative of those among all the identified links, as the average θ values measured for both sets are indistinguishable (0.34; Table 1). If the links found to date are representative of the genome, then the interscaffold gaps in the current assembly are small, averaging much less than 1 kb. An independent analysis using measured θ for all links, a regression of the data on intrascaffold θ values (Fig. 3), and the known distances between markers and scaffold ends leads to a similar conclusion (overlaps of a few hundred base pairs; not shown). Although these estimates are crude, they suggest that the current assembly of the MAC genome covers all but at most a few hundred kilobases (<1% of the assembled sequence) that are attributable to the interscaffold gaps.
All the current evidence suggests that HAPPY mapping will enable the vast majority of the current scaffolds to be assembled into superscaffolds that correspond to complete macronuclear chromosomes, leaving only small, well-defined gaps or overlaps to be closed by directed approaches. We intend to complete the mapping of not only the larger (>2 kb) scaffolds, but also the ~1200 smaller scaffolds, a task that will require the testing of an additional 1700 markers. Some MAC scaffolds may prove difficult to map (for example, very small ones that may lack any segment of unique sequence or those flanked by gaps of extreme size that approximate or exceed DNA fragment size in the mapping panel). However, our experience after testing nearly a quarter of the scaffold ends encourages us to believe that these would be very few; furthermore, they should prove tractable to more focused efforts once the majority of the MAC chromosomes have been assembled.
Our work shows that the fragment size to which the DNA was sheared (50–100 kb) provides more than adequate statistical significance for detecting linkages across interscaffold gaps and building an inventory of the scaffolds that belong to every MAC chromosome. However, it does introduce two limitations. First, the resolution is not sufficient to order and orient very small scaffolds with respect to adjacent ones. Second, the scatter in the relationship of θ vs physical distance (Fig. 3) does not allow a precise estimate of individual gap sizes, especially in our special case in which the map-making information for each linkage is often necessarily restricted to data from just two markers. These limitations are unlikely to be serious in the long term, given that most of the gaps appear to be much smaller than 1 kb. For example, a limited number of PCR amplifications (conventional or long-range) should make is possible to order and orient the small number of scaffolds in any given MAC chromosome and, at the same time, rather precisely determine gap lengths. While the preliminary results could hardly be more encouraging, only after all the markers have been tested will it become clear how well the results match our expectations.
The results obtained so far also open up the possibility of using HAPPY mapping to assemble the MIC genome. Since the MAC genome is derived from the MIC by a conceptually simple process of fragmentation and splicing (Figs. 1a and 1b), the reconstruction of the MIC genome sequence can be treated largely as problem of sequence assembly at a higher level. The MAC chromosomes (once completed) can essentially be considered as sequence contigs, to be ordered and oriented to produce a “scaffold” for each of the larger MIC chromosomes—an in silico reversal of in vivo steps (a) and (b) of Fig. 1. This can be accomplished by using HAPPY mapping to link end-to-end all of the MAC chromosomes, this time using a mapping panel made from purified MIC DNA and markers from the telomere-adjacent regions of each MAC chromosome. Given their estimated numbers, less than 600 markers should suffice for this purpose. Since the gaps between adjacent MAC chromosome DNAs are known to be even smaller than between incomplete MAC scaffolds (i.e., less than 75 bp), a mapping panel of similarly sheared purified MIC DNA should give excellent performance. The completed MIC assemblies, although lacking IESs, should be colinear with the genetic maps of MIC chromosomes and should facilitate mutant gene cloning by “forward genetics.”
Germ-line and soma differentiation in a single cell has always been a ciliate phenomenon of great general interest, not only in itself but because of the rich eukaryotic biology that accompanies this process. This includes the thousands of distinct programmed, site-specific DNA rearrangements, the structural and functional diversification of chromatin in the MIC and MAC, and recently discovered roles of MIC-derived RNAi in guiding these DNA rearrangements, centromere function, and meiotic chromosome pairing [17,18]. The complete assembly and the finished sequence of the germ-line and somatic genomes will eventually provide the first genome-wide, nucleotide-level description of the starting substrate and end product of these remarkable developmental processes.
Materials and methods
HAPPY mapping
DNA was prepared in agarose strings to minimize shearing, as follows. T. thermophila SB210 was cultured in 500 ml of growth medium (20 g/L protease peptone (Difco), 428 mg/L FeEDTA, 250 mg/L each of penicillin and streptomycin) at 22–24°C for 48 h, then supplemented with a further 500 ml of growth medium, and incubated as before for a further 4 h. Cells were pelleted (1100g, 3 min), resuspended in water at 2 × 106 cells/ml at 37°C, and mixed with an equal volume of 2% w/v low-melting-point agarose in water. The cell/agarose suspension was drawn up into glass capillaries (internal diameter ± 1 mm; 100 μl Supracaps; Brand) and allowed to set at 4°C for 2–4 min. The solidified agarose strings were then expelled from the capillaries into lysis solution (0.2% w/v SDS, 10 mM Tris–HCl, pH 8.0, 0.5 M Na2EDTA, 1 mg/ml proteinase-K) and incubated at 56°C for 28 h, the lysis solution being replaced after the first 4 h of incubation. Strings were then transferred to wash buffer (1% w/v lithium dodecyl sulfate, 10 mM Tris–HCl, pH 8.0, 1 mM EDTA) at 4°C, the buffer being changed after 4 h and again after 48 h. Samples of the DNA were checked by PFGE (8 mm of string loaded per lane) and were found to contain DNA from <100 kb to >2.2 Mb, with the larger macronuclear chromosomes visible as distinct bands (data not shown).
The HAPPY mapping panel was prepared by equilibrating 1 cm of agarose string into 2.5 ml of 0.5× PCR Buffer II (Perkin–Elmer) and incubating at 69°C with occasional gentle inversion to melt the agarose and disperse the DNA. In our experience, this procedure reliably shears the dilute DNA to an average fragment size of around 50–100 kb, appropriate for detecting linkages robustly over distances up to 10–20 kb. Ten microliters of this solution was then diluted (using wide-bore pipette tips to minimize DNA shearing) in 30 ml of HPLC-grade water, and 5-μl aliquots of this solution were dispensed into each of 88 wells of a microtiter plate; the remaining 8 wells each received 5 μl of HPLC-grade water as negative controls. This dilution was chosen to give approximately 0.7 haploid genomes per aliquot and hence, assuming a Poisson distribution of fragments in the aliquots, approximately 50% of aliquots being positive for any given single-copy sequence, this being the most informative fraction for HAPPY mapping. The correctness of the DNA concentration was confirmed by the results for the first few markers tested on the panel (see below), each of which was found in approximately half of the aliquots.
Primer-extension preamplification (PEP) was performed by supplementing each well with 0.7 μl of 10× PCR Buffer II (Perkin–Elmer), 0.7 μl of 25 mM MgCl2, 0.06 μl of 25 mM dNTPs, 0.07 μl of 1 mM N15 (fully degenerate 15-mer oligonucleotides; Operon Technologies), 0.28 μl of Taq polymerase (AmpliTaq; Perkin–Elmer, 5 U/μl), and 0.19 μl of water. Reactions were cycled with an initial step of 5 min at 93°C followed by 50 cycles of 94°C for 30 s, 37°C for 2 min, 37–55°C ramp over 3 min, and 55°C for 4 min. PEP products were diluted to 200 μl each and stored at −80°C until used as templates for marker typing (below).
For each marker to be mapped, heminested primers (forward external, forward internal and reverse) were selected using software (P.H.D., unpublished); primers were selected as far as possible to have a length of 18–22 nucleotides, to have a melting temperature (calculated as 2 × [A+T] + 4 × [G+C]) of 54–62°C, with two G or C nucleotides at the 3′ and one G or C nucleotide at the 5′ end, and to give an internal amplimer of between 70 and 120 bp (external amplimer length 90–250bp) with an A+T content not exceeding 70%.
Marker typing was performed for batches of 96 markers at a time, using protocols essentially identical to those reported previously [12,13]: a multiplex PCR was performed with 96 pairs of forward-external and reverse primers, and the products were diluted and used as templates for marker-specific (monoplex) PCRs, each using a single primer pair (forward-internal and reverse primers). Results were scored either by gel electrophoresis or by melting-curve analysis.
Typing results were recorded using custom software (P.H.D., unpublished), and pair-wise linkages (lod scores and associated θ values, indicating respectively the certainty of linkage and the distance between markers) were computed as previously described. Briefly, each possible pair of markers in the dataset is considered in turn. For that pair, the probability of obtaining the observed frequency of cosegregation is calculated, first under the assumption that θ (the probability of the DNA between the loci being broken) is 0 and then under the assumption of progressively greater values of θ up to 1 (complete breakage between loci, the loci being infinitely far apart). The value of θ giving the highest likelihood of the observed pattern of cosegregation is then inferred to be the optimal estimate of θ between those loci, reflecting the distance between them relative to the average fragment size. The lod score, reflecting the confidence in the linkage, is the logarithm of the ratio of the likelihood of obtaining the observed cosegregation pattern at the optimal θ to that at θ = 1. Groups of markers linked by lod scores of >5.0 were identified, and the linkages between these markers (and hence the arrangement of the scaffolds from which they originated) were determined by inspection.
Marker target selection
The current (November 2003) assembly of the whole-genome shotgun consists of 1971 scaffolds. Of these, 125 are telomere-capped at both ends and are inferred to represent complete macronuclear chromosomes. A further 120 scaffolds are telomere capped at one end only (and are hence presumed each to represent one end of a MAC chromosome), while 1726 are capped at neither end and are inferred to represent internal chromosome segments (E. Orias and E.P. Hamilton, unpublished observations). We term these three types of scaffolds as “doubly capped,” “singly capped,” and “uncapped,” respectively.
For mapping, we selected uncapped scaffolds that were >2 kb in size (based on the assembler’s convention that any intrascaffold sequence gaps are represented by 100 “N” nucleotides), plus a small number of smaller uncapped scaffolds, plus all singly capped scaffolds regardless of size. A PCR-based marker (see Materials and methods) was designed, wherever possible, within approximately 1–2 kb of each uncapped scaffold end; for those uncapped scaffolds that were <4 kb in size, a single marker, located approximately in the center of the scaffold, was designed, since the expected resolution of the HAPPY map would not allow these smaller scaffolds to be oriented even using markers from each end.
A further 78 markers were designed in contigs comprising two of the larger scaffolds—8254803 (2.2 Mb long, consisting of 20 contigs) and 8254798 (983 kb long, consisting of 23 contigs)—as a positive control, as well as to test the correct assembly of these scaffolds. In these cases, markers were designed in the larger contigs within 2 kb of each end or within the central part of smaller contigs (<4 kb).
Validating HAPPY links
Scaffolds linked by HAPPY mapping were checked for regions of sequence overlap using the BLAST 2 sequences program at NCBI (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi; [19]). To confirm directly HAPPY links by PCR amplification, primers were designed in unique regions of scaffold sequence nearest to the linked ends, using the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi; [20]). Whole-genome DNA was made from inbred strain B of T. thermophila as detailed in [21]. If the putative size of the PCR product (assuming a negligible gap between scaffolds) was under 2 kb, Taq PCR conditions for each pair of primers were optimized using a temperature gradient PCR machine (PCR Express; Hybaid), which varied the annealing temperature over a 15°C range, usually from 45 to 60°C. Each 25-μl PCR contained 5 μl of genomic DNA (5 ng/μl), 2.5 μl of 10× PCR buffer, 2.5 μl of 10 mM MgCl2, 4 μl of dNTPs, 1.25 μl of each primer at 4 μM, 8.4 μl of H2O, and 0.125 μl of AmpliTaq (5 U/μl; Perkin–Elmer, Cat. No. N801-0060). Cycling conditions were 5 min at 90°C, followed by 35 cycles of 1 min at 90°C, 1 min at the annealing temperature, and 2 min at 68°C, followed by a terminal extension period of 7 min at 68°C. If the putative size of the PCR product was over 2 kb, Phusion DNA polymerase (New England Biolabs) was used for long-range PCR. Each 50-μl reaction contained 2 μl of genomic DNA (100 ng/μl), 10 μl of 5× Phusion HF buffer, 1 μl of 10 mM dNTPs, 6.25 μl of each primer at 4 μM, 24 μl of H2O, and 0.5 μl of Phusion (2 U/μl). Cycling conditions were 30 s at 98°C, followed by 35 cycles of 10 s at 98°C, 1 min at 60°C, and 3 min at 72°C, followed by a terminal extension period of 5 min at 72°C. PCR products were cloned into the plasmid pCR2.1-TOPO (Invitrogen) and transformed into chemically competent TOP10 cells according to the supplier’s instructions. PCR product (purified with a Millipore Ultrafree filter) or plasmid DNA was sequenced using the Big Dye Terminator Cycle-Sequencing-Ready Reaction kit (PE Applied Biosystems). Nucleotide sequences were determined using an ABI 310 genetic analyzer.
Predicting the number of links expected as a function of the number of markers tested
We began with the simplifying assumption that the current assembly can be represented as a set of 1500 consecutive scaffolds, with 3000 scaffold ends to be linked in pairs to bridge 1499 interscaffold gaps. This assumption disregards the ±400 chromosomal termini but accurately models the number of gaps in the current assembly. We then simulated the expected results of mapping various numbers (N) of markers by choosing a subset of N of the 1500 scaffold termini at random and counting the number of instances in which the right-hand end of one scaffold and the left-hand end of the following scaffold were both present in this chosen subset: such instances should correspond to gaps closed after HAPPY mapping N markers.
Acknowledgments
We acknowledge support from NIH Grants RR-09231 from the National Center for Research Resources to E.O. and R01 GM067012-03 from the National Institute of General Medical Sciences to J.A.E. Sequence data for the T. thermophila scaffolds were obtained from The Institute for Genomic Research (http://www.tigr.org/tdb/e2k1/ttg/). We thank Steven Salzberg and Art Delcher (TIGR) for making public and calling our attention to Tetrahymena weak link files at the TIGR Web site, and to Martin Wu (TIGR) for providing a repeat-masked version of the genome.
References
- 1.Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 2.Lwoff A. Sur la nutrition des infusoires. C R Acad Sci Paris. 1923;176:928–930. [Google Scholar]
- 3.Turkewitz AP, Orias E, Kapler G. Functional genomics: the coming of age for Tetrahymena thermophila. Trends Genet. 2002;18:35–40. doi: 10.1016/s0168-9525(01)02560-4. [DOI] [PubMed] [Google Scholar]
- 4.Maupas E. La rejeunissement karyogamique chez les cilies. Arch Zool Exp Genet. 1889;7:149–517. [Google Scholar]
- 5.Sonneborn TM. Recent advances in the genetics of Paramecium and Euplotes. Adv Genet. 1947;1:263–358. doi: 10.1016/s0065-2660(08)60488-5. [DOI] [PubMed] [Google Scholar]
- 6.Doerder FP, Deak JC, Lief JH. Rate of phenotypic assortment in Tetrahymena thermophila. Dev Genet. 1992;13:126–132. doi: 10.1002/dvg.1020130206. [DOI] [PubMed] [Google Scholar]
- 7.Yao M-C. Site-specific chromosome breakage and DNA deletion in ciliates. In: Berg DE, Howe MM, editors. Am Soc Microbiol. Mobile DNA; Washington, DC: 1989. pp. 715–734. [Google Scholar]
- 8.Cassidy-Hanley D, et al. Genome-wide characterization of Tetrahymena thermophila chromosome breakage sites: II. Physical and genetic mapping. Genetics. 2005;170:1623–1631. doi: 10.1534/genetics.104.031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamilton E, et al. The highly conserved family of Tetrahymena thermophila chromosome breakage elements contains an invariant 10 base pair core. Eukaryotic Cell. 2006;5:771–780. doi: 10.1128/EC.5.4.771-780.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coyne RS, Chalker DL, Yao MC. Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring. Annu Rev Genet. 1996;30:557–578. doi: 10.1146/annurev.genet.30.1.557. [DOI] [PubMed] [Google Scholar]
- 11.Fillingham JS, et al. Analysis of expressed sequence tags (ESTs) in the ciliated protozoan Tetrahymena thermophila. J Eukaryotic Microbiol. 2002;49:99–107. doi: 10.1111/j.1550-7408.2002.tb00350.x. [DOI] [PubMed] [Google Scholar]
- 12.Bankier AT, et al. Integrated mapping, chromosomal sequencing and sequence analysis of Cryptosporidium parvum. Genomics. 2003;1:1787–1799. doi: 10.1101/gr.1555203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eichinger L, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall N, et al. Sequence of Plasmodium falciparum chromosomes 1, 3–9 and 13. Nature. 2002;419:527–531. doi: 10.1038/nature01095. [DOI] [PubMed] [Google Scholar]
- 15.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 16.Wong L, et al. Autonomously replicating macronuclear DNA pieces are the physical basis of coassortment groups in Tetrahymena thermophila. Genetics. 2000;155:1119–1125. doi: 10.1093/genetics/155.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mochizuki K, Gorovsky MA. Small RNAs in genome rearrangement in Tetrahymena. Curr Opin Genet Dev. 2004;14:181–187. doi: 10.1016/j.gde.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Mochizuki K, Gorovsky MA. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tatusova TA, Madden TL. Blast 2 sequences—A new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 20.Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press; Totowa, NJ: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 21.Hamilton EP, et al. Genome-wide characterization of Tetrahymena thermophila chromosome breakage sites. I. Cloning and identification of function sites. Genetics. 2005;170:1611–1621. doi: 10.1534/genetics.104.031401. [DOI] [PMC free article] [PubMed] [Google Scholar]