Abstract
Methylglyoxal is a highly reactive α-ketoaldehyde that is produced endogenously and present in the environment and foods. It can modify DNA and proteins to form advanced glycation end products (AGEs). Emerging evidence has shown that N2-(1-carboxyethyl)-2′-deoxyguanosine (N2-CEdG) is a major marker for AGE-linked DNA adducts. Here, we report, for the first time, the preparation of oligodeoxyribonucleotides (ODNs) containing individual diastereomers of N2-CEdG via a postoligomerization synthesis method, which provided authentic substrates for examining the replication and repair of this lesion. In addition, thermodynamic parameters derived from melting temperature data revealed that the two diastereomers of N2-CEdG destabilized significantly the double helix as represented by a 4 kcal/mol increase in Gibbs free energy for duplex formation at 25°C. Primer extension assay results demonstrated that both diastereomers of N2-CEdG could block considerably the replication synthesis mediated by the exonuclease-free Klenow fragment of Escherichia coli DNA polymerase I. Strikingly, the polymerase incorporated incorrect nucleotides, dGMP and dAMP, opposite the lesion more preferentially than the correct nucleotide, dCMP.
Introduction
Humans are exposed to methylglyoxal (MG), a highly reactive α-ketoaldehyde, from a variety of sources. General exogenous sources of MG include cigarette smoke 1, food and beverages such as soy sauce, coffee and whiskey 2,3. On the other hand, the fragmentation of triose phosphate during glycolysis, metabolism of acetone, and catabolism of aminoacetone all contribute to the endogenous formation of MG 4,5. Many factors, including aging, hyperglycemia, inflammation, oxidative stress and uremia, could enhance the accumulation of MG in vivo 6. In this respect, as high as 310 μM of methylglyoxal was detected in Chinese hamster ovary cells 7. In addition, a previous clinical study indicated that the median level of MG increased by several fold in blood samples from type I and II diabetic patients, and the concentration of MG correlates positively with the duration and development of type I diabetes 8.
As a reactive dicarbonyl compound, MG is involved in numerous pathological processes in vivo by binding irreversibly to proteins, primarily on arginine residues 9–11, DNA and other substrates to form advanced glycation end products (AGEs) 12,13. If remain unrepaired, those AGEs could eventually lead to the inhibition of enzyme activity 14, transcription activation 15 and apoptosis 16.
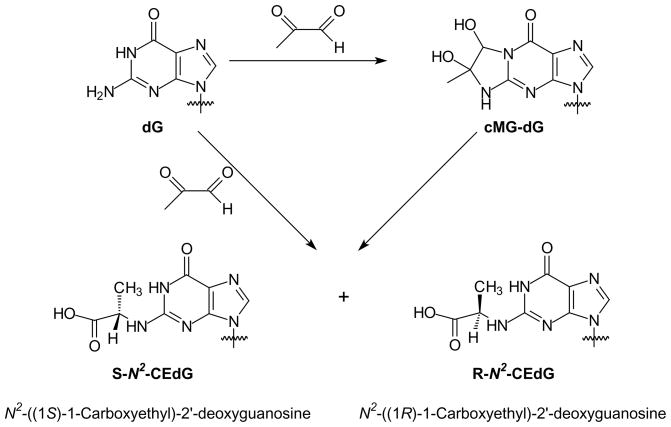
Several MG adducts of DNA have been previously reported, and guanine is the predominant modification site induced by MG when calf thymus DNA was used as substrate 13,17,18. In an earlier study, N2-(1-carboxyethyl)-9-methylguanine was formed from the incubation of 9-methylguanine with glucose or MG, thus it was postulated that N2-(1-carboxyethyl)-2′-deoxyguanosine( N2-CEdG) could be induced in DNA by MG 19. Indeed Frischmann et al. 13 showed recently that N2-CEdG was the stable adduct formed in calf thymus DNA upon prolonged exposure to MG at physiological concentration and temperature. These authors further proposed that the N2-CEdG could be produced from either direct coupling of 2′-deoxyguanosine (dG) with MG or from the conversion of 1, N2-(1,2-dihydroxy-2-methyl)ethano-2′-deoxyguanosine (cMG-dG) (Scheme 1).
Scheme 1.
Formation of N2-CEdG.
N2-CEdG was also detected in urine samples from healthy human subjects using immunoaffinity chromatography with a polyclonal antibody raised against N2-CEdG 20, and the lesion was found to occur more frequently in kidney and aorta cells of diabetic and uremic patients 21. Moreover, the lesion was found in cultured human smooth muscle cells and bovine aorta endothelium cells with the same immunoassay technique 20.
Prior studies have demonstrated that methylglyoxal-DNA adducts could induce single strand breaks and high mutation frequencies in Escherichia coli (E. coli)cells 22, and G:C→C:G and G:C→T:A transversions in supF gene in mammalian cells 23. In these previous mutagenesis studies, lesion-containing DNA substrates were prepared via the direct treatment of undamaged DNA with dihydroxyacetone or methylglyoxal 14,16,22,23. The identity and homogeneity of adducts, however, were not carefully examined. In this context, it is known that the commercially available methylglyoxal is likely to be contaminated with formaldehyde and other impurities such as pyruvate, lactate and formate 24, which may induce side reactions and complicate the interpretation of the results. We sought to understand the biological implications of N2-CEdG at the molecular level. As a first step toward that goal, herein, we developed a postoligomerization method for the site-specific and stereospecific insertion of N2-CEdG into ODNs. We also examined the effects of N2-CEdG on duplex stability and carried out in vitro replication studies on the lesion-bearing substrates with Klenow fragment.
Experimental Procedures
Materials
All chemicals, unless otherwise specified, were from Sigma-Aldrich (St. Louis, MO) or EM Science. Common reagents for solid-phase DNA synthesis were obtained from Glen Research Co. (Sterling, VA), and unmodified ODNs used in this study were purchased from Integrated DNA Technologies (Coralville, IA). [γ-32P]ATP was obtained from Amersham Biosciences Co. (Piscataway, NJ).
Mass spectrometry (MS), NMR and circular dichroism (CD)
Electrospray ionization-mass spectrometry (ESI-MS) and tandem MS (MS/MS) experiments were carried out on an LCQ Deca XP ion-trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA). A mixture of acetonitrile and water (50:50, v/v) was used as solvent for electrospray. The spray voltage was 3.0 kV, and the temperature of the heated capillary was maintained at 300°C. High-resolution mass spectra (HRMS) were acquired on an Agilent 6210 TOF LC/MS instrument (Agilent Technologies, Palo Alto, CA) equipped with an electrospray ionization (ESI) source. 1H NMR spectra were recorded at 300 MHz on a Varian Inova 300 NMR spectrometer, and 31P NMR spectra were acquired at 80 MHz on the same instrument. CD spectra were recorded on a JASCO J-815 CD spectrometer (JASCO Inc., Easton, MD).
HPLC
The purification of ODNs was performed on a Beckman HPLC System (32 Karat software version 3.0, pump module 125) with a UV detector (module 126) monitoring at 260 nm. A 4.6×250 mm Apollo C18 column (5 μm in particle size and 300 Å in pore size, Alltech Associate Inc., Deerfield, IL) was used, and a 50-mM triethylammonium acetate buffer (TEAA, pH 6.6, Solution A) and a mixture of 50-mM TEAA and acetonitrile (70/30, v/v, Solution B) were employed as mobile phases. The flow rate was 0.8 mL/min. Three different HPLC gradients were used: (I) 5 min 0–20% B, 40 min 20–40% B, 10 min 40–100% B and 5 min at 100% B; (II) 5 min 0–30% B, 40 min 30–50% B, 10 min 50–100% B and 5 min at 100% B; (III) 5 min 0–30% B, 40 min 30–60% B, 10 min 60–100% B and 5 min at 100% B. The purified ODNs were desalted on the same HPLC system with H2O as mobile phase A and acetonitrile as mobile phase B where a gradient of 25 min 0% B, 2 min 0–60% B and 25 min at 60% B was employed.
Synthesis of 5′-O-(4,4′-dimethoxytrityl)-2-fluoro-O6-(trimethylsilylethyl)-2′-deoxyinosine (5, Scheme S1)
Compound 5 was prepared following the previously published procedures (spectra shown in the Supporting Information) 25,26.
Synthesis of 5′-O-(4,4′-dimethoxytrityl)-2-fluoro-O6-(trimethylsilylethyl)-2′-deoxyinosine-3′-O-[(2-cyanoethyl)-N,N-diisopropyl-phosphoramidite] (6, Scheme S1)
To a flask, which was suspended in an ice bath and contained a solution of compound 5 (100 mg, 0.149 mmol) in dry CH2Cl2 (1.5 mL), was added N,N -diisopropylethylamine (DIEA, 52 μL, 0.298 mmol) followed by dropwise addition of 2-cyanoethyl-NN, -diisopropyl chlorophosphoramidite (50 μL, 0.224 mmol). The mixture was stirred at room temperature for 30 min under argon atmosphere. The reaction was quenched by cooling the mixture in an ice bath followed by slow addition of CH3OH (0.40 mL). The solution was quickly extracted with EtOAc (8 mL), and the organic layer was washed with saturated NaHCO3 (4 mL) and brine (4 mL) and then dried with anhydrous Na2SO4. The solvent was evaporated in vacuo to yield 6 in white foam that was used directly for ODN synthesis. 31P NMR (CDCl3): δ 149.9, 149.8. ESI-MS: m/z 873.1 [M + H]+.
Synthesis of standard S- and R-N2-CEdG
L- or D-alanine (2 mg, 24.5 μmol) was added to a mixture containing 2-fluoro-O6-(trimethylsilylethyl)-2′-deoxyinosine (2 mg, 5.8 μmol), DMSO (500 μL) and diisopropylethylamine (DIEA, 100 μL). The resulting solution was stirred at 55°C for 48 h, and the solvent was removed in vacuo. The dried residues were dissolved in 5% acetic acid and stirred at room temperature for 1 h. The S- and R-N2-CEdG were then isolated from the resulting reaction mixtures by HPLC.
S-N2-CEdG
1H NMR (300 MHz, D2O, 25°C): δ7.96 (s, 1H, H-8), 6.35 (t, 1H, H-1′, J = 6.6 Hz), 4.66 (m, 1H, H-3′), 4.29 (q, 1H, CH, J = 7.3 Hz), 4.10 (m, 1H, H-4′, J = 9.9, 4.1 Hz), 3.78 (m, 2H, H-5′ and H-5″), 2.97 (m, 1H, H-2′), 2.54 (m, 1H, H-2″), 1.47 (d, 3H, CH3, J = 7.3 Hz) (Figure S6). HRMS (ESI): [M+Na]+ calcd m/z 362.1077, found 362.1078.
R-N2-CEdG
1H NMR (300 MHz, D2O, 25°C): δ 7.92 (s, 1H, H-8), 6.33 (t, 1H, H-1′, J = 6.9 Hz), 4.66 (m, 1H, H-3′), 4.24 (q, 1H, CH, J = 7.3 Hz), 4.09 (m, 1H, H-4′, J = 6.3, 4.1 Hz), 3.85 (m, 2H, H-5′ and H-5″), 3.11 (m, 1H, H-2′), 2.44 (m, 1H, H-2″), 1.47 (d, 3H, CH3, J = 7.3 Hz) (Figure S7). HRMS (ESI): [M+Na]+ calcd m/z 362.1077, found 362.1082.
ODN synthesis
ODNs (sequences shown in Table 1) were synthesized on a Beckman Oligo 1000S DNA synthesizer (Fullerton, CA) at 1 μmol scale. The synthesized phosphoramidite building block was dissolved in anhydrous acetonitrile at a concentration of 0.07 M. Normal phosphoramidite building blocks (Glen Research Inc., Sterling, VA) of dA, dC, dG, and dT were employed, and standard ODN assembly protocol was used without any modification.
Table 1.
Sequences of Synthesized ODNs
| ODNs | sequencesa | observed m/z | theoretical m/z |
|---|---|---|---|
| ODN1/2 | 5′-GAG TAG XAT GAG-3′ | 3845.0 | 3845.5 |
| ODN3/4 | 5′-ATG GCX CAC TAT GAT CCT AG-3′ | 6188.0 | 6188.1 |
ODN1/ODN3, X = S-N2-CEG; ODN2/ODN4, X= R-N2-CEG.
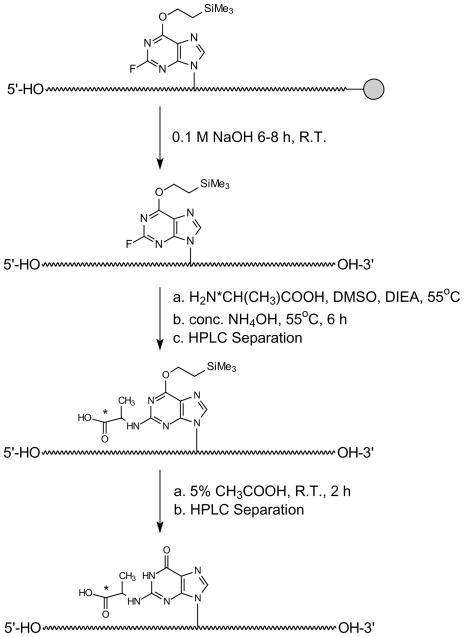
After synthesis, the products were cleaved from the controlled-pore glass (CPG) support and partially deprotected with 0.1 M NaOH (3 mL) at room temperature for 8–12 h. The supernatant was removed and the beads were washed with H2O (2×3 mL). The combined aqueous solution was neutralized with 0.1 M acetic acid to pH 7.0 and lyophilized to dryness. The dried residue was mixed with DIEA (150 μL, 0.86 mmol), L-alanine or D-alanine (4 mg, 0.045 mmol) and DMSO (500 μL). After being stirred at 55°C for 48–72 h, the reaction mixture was allowed to cool to room temperature and dried. Concentrated NH4OH (600 μL) was added and the tightly sealed vial was heated at 55°C for 6–8 h. After cooling to room temperature, the solvent was removed by using a Speed-Vac. The dried pellet was reconstituted in water and purified by using reversed-phase HPLC with gradient I for ODN1/ODN2 and gradient II for ODN3/ODN4 (Table 1) to give O6-trimethylsilylethyl (O6-tse)-protected oligomers. The fraction containing O6-tse-protected oligomer was collected, dried and the dried residue was treated with 5% acetic acid (500 μL) at room temperature for 2 h, neutralized with 1.0 M NaOH, and purified by reversed-phase HPLC to yield the fully-deprotected ODNs (Scheme 2). In this respect, gradient I was employed for the purification of ODN1/ODN2, whereas gradient III was used for the purification of ODN3/ODN4.
Scheme 2.
Postoligomerization method for the incorporation of N2-CEdG into ODNs (“*” indicates the chiral carbon).
Enzymatic digestion of synthetic ODNs
One unit of nuclease P1, 0.01 unit of calf spleen phosphodiesterase, and 2.5 μL buffer solution, which contained sodium acetate (300 mM, pH 5.0) and zinc acetate (10 mM), were added to a 20-μL solution of ODN1 or ODN2 (10 nmol). The digestion was continued at 37°C for 6 h. To the digestion mixture were then added 10 units of alkaline phosphatase, 0.05 unit of snake venom phosphodiesterase, and 5 μL of 0.5 M Tris-HCl (pH 8.9). The digestion was continued at 37°C for 6 h, and the enzymes were removed by passing through a 10-kDa cut-off Centricon membrane (Millipore, Billerica, MA). The resulting mixtures were subjected subsequently to HPLC analysis, which was carried out on an Agilent 1100 HPLC system with a UV detector monitoring at 260 nm. A 4.6×250 mm Apollo C18 column was used, and a 10-mM ammonium formate buffer (pH 6.9, Solution A) and a mixture of 10-mM ammonium formate and acetonitrile (70/30, v/v, Solution B) were employed as mobile phases. The flow rate was 0.8 mL/min where a gradient of 5 min 0–10% B followed by 40 min 10–40% B was employed.
Thermodynamic studies
UV absorbance-versus-temperature profiles were recorded on a Varian Cary 500 spectrophotometer (Varian, Inc., Palo Alto, CA). The modified ODNs and their complementary strands were dissolved in a 1.2-mL solution containing 100 mM NaCl, 10 mM sodium phosphate (pH 7.0), and 50 μM EDTA at a total ODN concentration (Ct) of 1.0, 2.0, 4.0, 8.0, or 16 μM. The absorbance was recorded in the reverse and forward directions for a temperature range of 80-10°C at the rate of 1°C/min, and the melting temperature (Tm) value was obtained by the derivative method. The thermodynamic parameters were obtained from the van’t Hoff plot 27, in which the reciprocal of Tm was plotted against ln(Ct/4):
and
where R is the universal gas constant (1.987 cal mol−1 K−1). The error limits for ΔG, ΔH, and ΔS derived from fitted parameters were calculated by using the previously described equations 28,29.
In vitro replication studies with exonuclease-free Klenow fragment of E. coli DNA polymerase I (Kf−)
The primer extension experiments were carried out under standing-start conditions. The 20mer lesion-containing template (ODN3/4) or normal template (5 nM) with dG in lieu of the N2-CEdG was annealed with a 5′-[32P]-labeled 15mer primer (10 nM), to which were then added a mixture of all four dNTPs (200 μM each) and the Klenow fragment. The reaction was carried out in a 20-μL solution containing 10 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 50 mM NaCl and 1 mM dithiothreitol (DTT) at 37°C for 60 min. In vitro replication experiments were also carried out in the presence of one type of dNTP at a time; the concentration of dNTP was 1.0 mM, 0.1 unit of Klenow fragment was added, and the reaction was continued at 37°C for 10 min.
The reaction was terminated by adding 8 μL gel-loading buffer, which contained 80% formamide, 10 mM EDTA at pH 8.0, 1 mg/mL xylene cyanol, and 1 mg/mL bromophenol. The products were resolved on 20% denaturing polyacrylamide gels containing 8 M urea. Gel images were obtained by using a Typhoon 9410 Variable Mode Imager (Amersham Biosciences Co.) and ImageQuant 5.2 software (Amersham Biosciences Co.).
Results
Synthesis of N2-CEdG-containing ODNs
Site-specific incorporation of N2-CEdG into ODNs has not been reported, though treatment of unmodified DNA with 1,3-dihydroxyacetone or methylglyoxal was previously employed for the introduction of the lesion into DNA for mutagenesis studies 14,16,22,23. However, the identity and homogeneity of the products induced by 1,3-dihydroxyacetone or methylglyoxal were not carefully assessed. Herein, we developed a postoligomerization method for the site-specific insertion of N2-CEdG into ODNs (Scheme 2). The method utilized the activity of the precursor nucleoside, 2-fluoro-2′-deoxyinosine (FdI), which was first synthesized and applied for the postoligomerization synthesis of N2-guanine-adduct-containing ODNs by Harris and coworkers 25,26. We reasoned that the nucleophilic attack by the NH2 group of alanine should also displace the fluoride in FdI and afford the desired N2-CEdG adducts. Additionally, the availability of two enantiomers of alanine should offer us the opportunity to generate separately ODNs carrying either of the two diastereomeric lesions (Schemes 1&2).
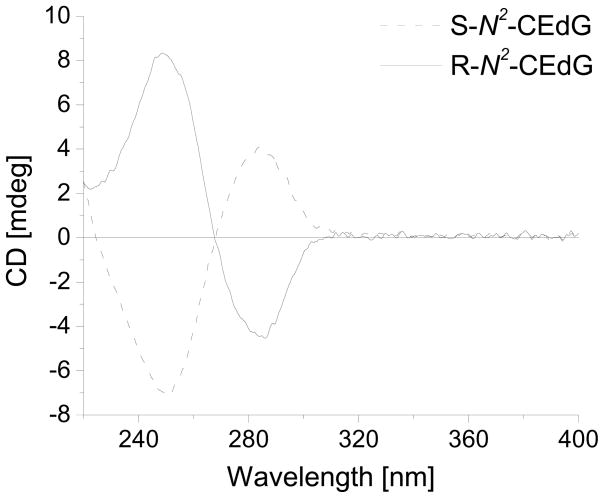
To test this idea, we first carried out the reaction at the mononucleoside level. In this respect, we treated 2-fluoro-O6-(trimethylsilylethyl)-2′-deoxyinosine with L- or D-alanine, removed the trimethylsilylethyl group in the resulting nucleoside with 5% acetic acid, isolated the modified nucleoside from the reaction mixtures by HPLC (See Experimental), and characterized the products by NMR, MS and circular dichroism spectroscopy. It turned out that the 1H-NMR spectra for the above synthetic S- and R-N2-CEdG were consistent with the previously reported data (Figures S6&S7) 30. In addition, the measured masses for these two modified nucleosides were in keeping with the calculated ones (See Experimental). Moreover, the independently prepared R- and S-N2-CEdG exhibited similar traces, yet opposite features in CD signals (Figure 1), supporting that they are indeed diastereomers.
Figure 1.
CD spectra of standard S-N2-CEdG and R-N2-CEdG (50 μM).
Having established that the synthetic route allowed for the successful preparation of the R- and S-N2-CEdG, we next synthesized the phosphoramidite building block of 2-fluoro-O6-(trimethylsilylethyl)-2′-deoxyinosine following the published procedures except that compound 5 (Scheme S1) was phosphitylated with 2-cyanoethyl-N,N-diisopropylchlorophosphoramidite instead of 2-cyanoethyl-N,N,N′,N′-tetraisopropyl phosphoramidite 25,26. By using this building block, we then synthesized several ODNs.
After being cleaved from the CPG support, the partially deprotected ODNs were treated with either L- or D-alanine to achieve stereospecific formation of S-N2-CEdG and R-N2-CEdG, respectively (Scheme 2). The temperature and time for this treatment were optimized to minimize the degradation and maximize the formation of the desired product. It turned out that the best results were obtained after ODN1/ODN2 and ODN3/ODN4 were treated with alanine at 55°C for 48 h and 72 h, respectively. In this regard, the reaction did not proceed to completion in reasonable incubation time at lower temperature, whereas slightly more side products and the spontaneous removal of O6-trimethylsilylethyl protecting group were observed at higher temperature.
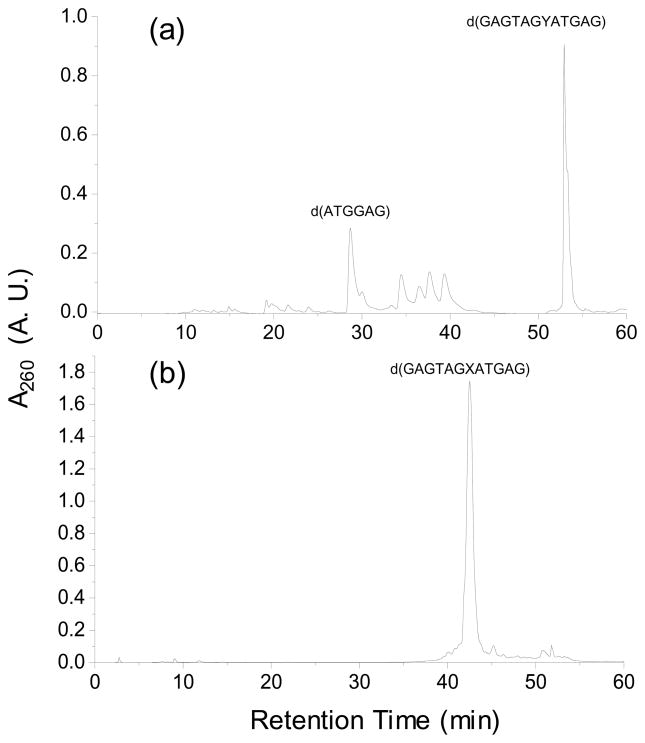
After incorporating the N2-carboxyethyl functionality into ODNs, we removed completely the protecting group on normal nucleobases by treatment with concentrated NH4OH, under which conditions the O6-trimethylsilylethyl protecting group remained intact. Because the O6-(trimethylsilylethyl)-N2-CEdG-bearing ODNs could be readily resolved by HPLC from the corresponding ODNs containing unmodified dG or FdI (Figures 2a and 3a), we separated the mixture by HPLC prior to removing the trimethylsilylethyl protecting group. After the HPLC separation and the removal of the trimethylsilylethyl group with 5% acetic acid treatment, we separated again the resulting mixture by HPLC to obtain the desired lesion-bearing ODN substrates (Figures 2b and 3b).
Figure 2.
HPLC traces for the separation of the synthesized 12mer ODNs: (a) d(GAGTAGYATGAG), Y=O6-tse-S-N2-CEdG; (b) d(GAGTAGXATGAG), X= S-N2-CEdG.
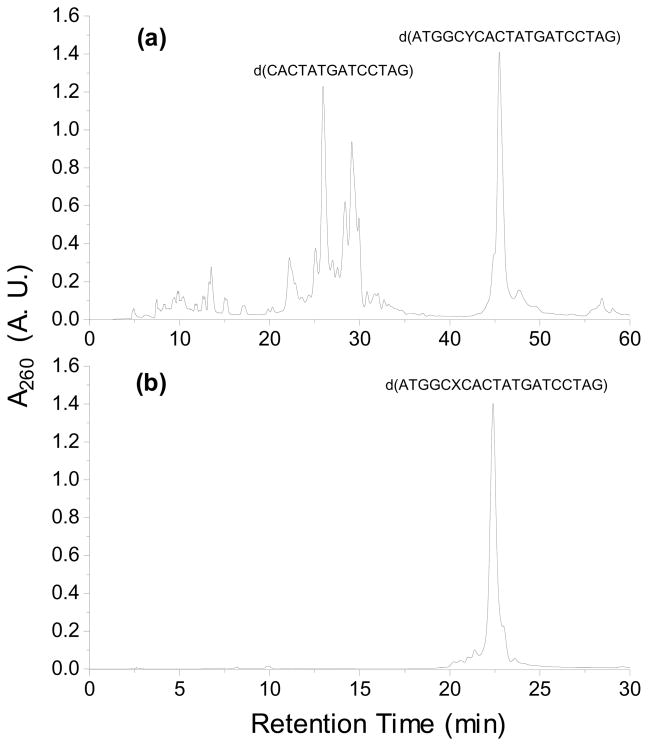
Figure 3.
HPLC traces for the separation of the synthesized 20mer ODNs: (a) d(ATGGAYCACTATGATCCTAG), Y=O6-tse-S-N2-CEdG; (b) d(ATGGAXCACTATGATCCTAG), X= S-N2-CEdG.
We next assessed the stability of lesion in ODNs by incubating the ODNs in 50 mM phosphate buffer (pH 7.0) at 37°C for up to 72 h, LC-MS/MS analysis revealed that there is no obvious degradation of the lesion (data not shown).
ESI-MS and MS/MS characterizations of ODNs containing an N2-CEdG
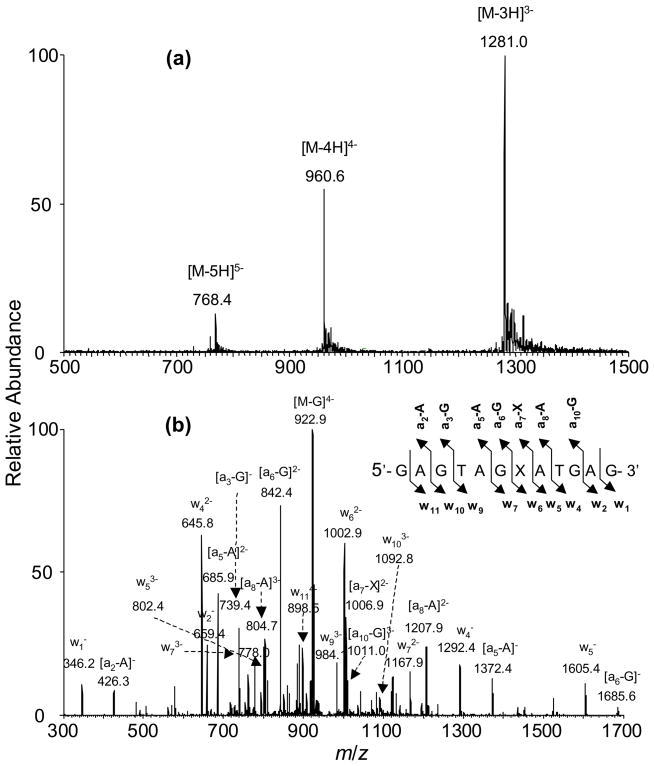
We next characterized the above ODNs by ESI-MS and MS/MS (Table 1, NFigure 4, and Figure S8). Here, we chose to use d(GAGTAGXATGAG) (ODN1, X represents S-2-CEdG) as an example to illustrate how we use ESI-MS and MS/MS to confirm the site of N2-CEdG incorporation. ESI-MS showed that the deconvoluted mass of ODN1 is 72 Da higher than the calculated mass of the unmodified d(GAGTAGGATGAG) (Table 1, NFigure 4a), which is consistent with the presence of an 2-CEdG adduct in this substrate. Moreover, high-resolution ESI-MS result revealed that the measured m/z for the monoisotopic peak of the [M – 3H]3− ion was 1280.550, which differed from the calculated m/z of 1280.558 for the corresponding peak by only 6.3 ppm (Figure S9). Moreover, the experimentally determined isotope profile was consistent with the calculated one (Figure S9).
Figure 4.
ESI-MS and MS/MS characterizations of d(GAGTAGXATGAG), X=S-N2-CEdG: (a) Negative-ion ESI-MS; (b) product-ion spectrum of the [M-4H]4− ion (m/z 960.6).
The product-ion spectrum of the [M – 4H]4− ion (m/z 960.6, Figure 4b) of the ODN showed the formation of wn ions, that is, w1−, w2−, w4−, w5−, w62−, w72−, w93−, w103− and w114− and [an - Base] ions, that is, [a2 - A]−, [a3 - G]−, [a5 - A]−, [a6 - G]−, [a7 - X]2−, [a8 - A]2− and [a10 - G]3− ions [nomenclature for fragment ions follows that reported by McLuckey et al. 31; “A”,“C”, and “G” represent adenine, cytosine, and guanine, respectively]. The measured masses for the w1, w2, w4, and w5 ions were the same as the calculated masses for the corresponding ions of the unmodified ODN, whereas the w6, w7, w9, w10 and w11 ions exhibited 72 Da higher in mass than the corresponding fragments formed from the unmodified d(GAGTAGGATGAG). These results are consistent with the presence of an N2-CEdG at the seventh position in the ODN. The above conclusion is further substantiated by the observed masses for the [an - Base] ions. In this respect, the measured masses for the [a8 - A] and [a10 - G] ions are 72 Da higher than, whereas the measured masses of the [a2 - A], [a3 - G], [a5 - A], [a6 - G] and [a7 - X] ions are the same as the calculated ones of the corresponding fragment ions for the unmodified d(GAGTAGGATGAG). The sequences of ODN3 and ODN4 were also confirmed by the similar ESI-MS and MS/MS analyses (Figure S8 shows the MS and MS/MS for ODN3).
Homogeneity and Purity of Synthetic ODNs
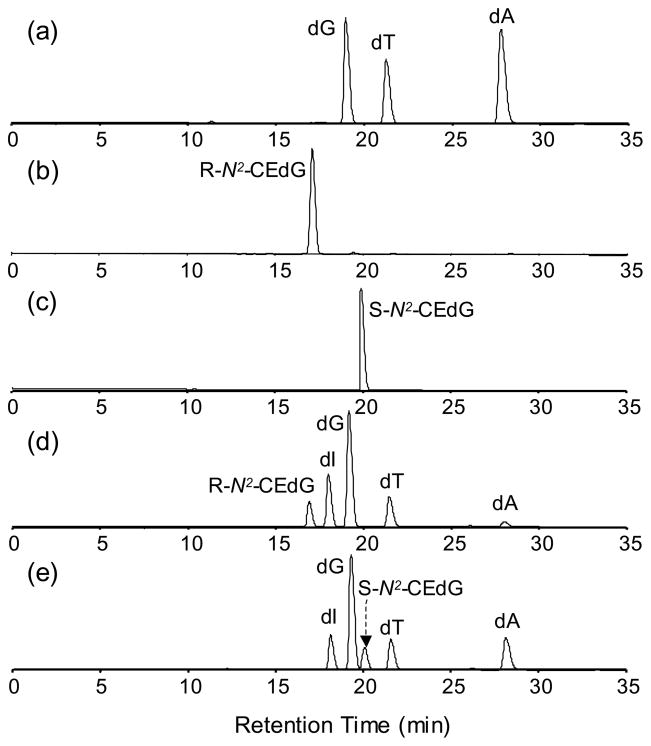
To confirm the stereospecific incorporation of S- and R-N2-CEdG into ODNs, we further digested ODNs 1&2 into mononucleosides (See Experimental) and separated the digestion mixtures by HPLC. For comparison, we also analyzed synthetic S-N2-CEdG, R-N2-CEdG as well as standard dG, dT and dA by HPLC under identical experimental conditions (Figure 5). Indeed the HPLC traces revealed the presence of R-N2-CEdG in the digestion mixture of ODN2 (the 17.1-min fraction, Figure 5b&d), whereas the S-N2-CEdG was not detectable. Likewise, S-N2-CEdG, but not R-N2-CEdG, was present in the digestion mixture of ODN1 (the 20.2-min fraction, Figure 5c&e). The above results, therefore, demonstrated unambiguously that the postoligomerization method allowed for the independent incorporation of the two diastereomers of N2-CEdG into ODNs. It is worth noting that the presence of residual adenosine deaminase in the commercial enzymes caused the deamination of some dA into 2′-deoxyinosine (dI, Figure 5d&e). The identities of the mononucleosides liberated from the enzymatic digestion were confirmed by ESI-MS measurements (Data not shown). In this context, it is important to emphasize that high-resolution ESI-MS results supported the absence of adenine deamination in the ODNs prior to the enzymatic digestion (vide supra).
Figure 5.
HPLC traces for the separation of: (a) the mixture of dG, dT and dA; (b) R-N2-CEdG; (c) S-N2-CEdG; (d) the enzymatic digestion mixture of ODN2; and (e) the enzymatic digestion mixture of ODN1. A UV detector was set at 260 nm for monitoring the effluents. The presence of residual adenosine deaminase in the commercial enzymes caused the deamination of some dA to dI.
We further integrated the peak areas for N2-CEdG, dT, and dG in the HPLC traces for the digestion mixtures of ODNs 1&2 and estimated the molar ratios of these nucleosides based on the peak area ratios with the consideration of the molar extinction coefficients of the three nucleosides. In this regard, we measured, by using a previously described 1H-NMR method 32, the extinction coefficient of N2-CEdG at 260 nm, which was 13,600 L mol−1 cm−1. It turned out that the estimated molar ratios of N2-CEdG:dT:dG were 1.0:2.0:5.1 and 1.0:2.0:5.2 for ODN1 and ODN2, respectively, which are consistent with the presence of one N2-CEdG, two dT and five dG residues in these two ODNs. Because of the deamination of dA to dI during enzymatic digestion (vide supra), the peak area for dA was not determined.
Thermodynamic properties of lesion-containing DNA duplexes
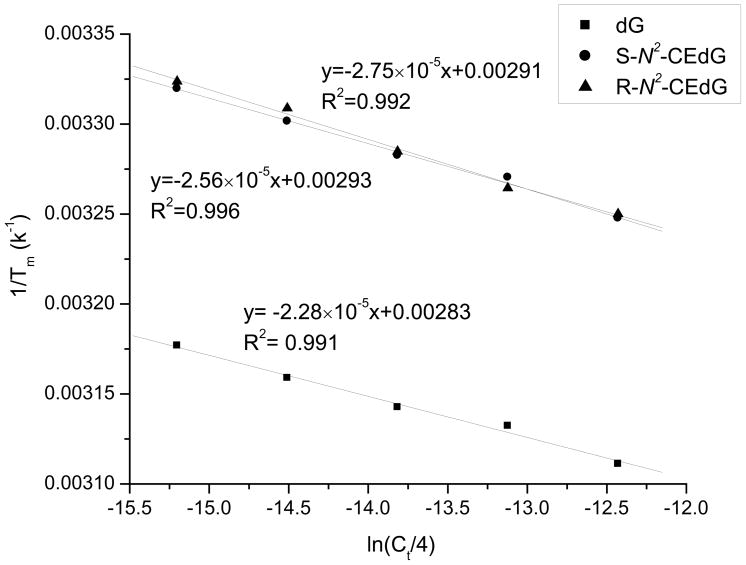
To examine how the presence of N2-CEdG affects duplex stability, we further determined the thermodynamic parameters for the formation of duplex DNA harboring an S-N2-CEdG, R-N2-CEdG, or unmodified dG by melting temperature measurements. The ODN sequences used in this study were listed in Table 1, and the ΔH and ΔS were calculated from the intercept and slope found in the van’t Hoff plot following the equations described in the Experimental Section (Figure 6). It turned out that the presence of S-N2-CEdG and R-N2-CEdG destabilized respectively the duplex by 3.8 and 4.0 kcal/mol in free energy at 25°C (Table 2). Correspondingly the presence of N2-CE-dG results in a marked decrease in melting temperature; at the total ODN concentration of 2.0 μM, the replacement of dG with an S-N2-CEdG or R-N2-CEdG leads to a drop in melting temperature from 43.6°C to 29.9°C and 29.2°C, respectively.
Figure 6.
Plots of 1/Tm vs. ln(Ct/4) for the duplex ODNs containing dG and N2-CEdG adducts. The duplex is d(GAGTAGXATGAG)/d(CTCATCCTACTC), where X represents dG(■), S-N2-CEdG(●) and R-N2-CEdG(▲).
Table 2.
Thermodynamic Parameters for Duplex Formation
| duplex | Tma (°C) | ΔH (kcal mol−1) | ΔS (cal mol−1 K−1) | ΔG25 °C (kcal mol−1) | ΔΔG25 °Cb (kcal mol−1) |
|---|---|---|---|---|---|
| 5′-CTCATCCTACTC-3′ 3′-GAGTAXGATGAG-5′ | |||||
| X = G | 43.6 ± 0.1 | −87.0 ± 4.5 | 246 ± 16 | −13.7 ± 0.6 | |
| X = S-N2-CEG | 29.9 ± 0.2 | −77.7 ± 1.0 | 228 ± 4 | −9.9 ± 0.2 | 3.8 |
| X = R-N2-CEG | 29.2 ± 0.1 | −72.2 ± 1.9 | 210 ± 6 | −9.7 ± 0.3 | 4.0 |
Ct = 2.0 μM.
ΔΔG25 °C = ΔG25 °C (lesion-containing duplex) - ΔG25 °C (unmodified duplex)
In vitro replication studies with Klenow fragment
We next examined the bypass of N2-CEdG with Klenow fragment by carrying out in vitro primer extension assays. To this end, we assessed the ability for the Klenow fragment, which has been used extensively as a model replicative polymerase, to extend a 5′-[32P]-labeled 15-mer primer annealed with a 20-mer, N2-CEdG-bearing template in the presence of all four dNTPs. The primer extension results unveiled that the synthesis stops mostly after the incorporation of one nucleotide opposite N2-CEdG for the lesion-bearing substrates, though a small percentage of bypass product could also be detected. However, the full-length products from the replication of the undamaged template can be readily obtained (Figure 7a). In this regard, the percentages of the full-length products formed from the extension of the normal, S- and R-N2-CEdG-bearing substrates in the presence of 0.3 unit of Kf− were estimated to be 80%, 22%, and 5%, respectively (Figure 7a).
Figure 7.
In vitro replication studies of N2-CEdG-bearing and control undamaged substrates with Klenow fragment of E. coli polymerase I (X represents S-N2-CEdG, R-N2-CEdG or unmodified dG). The concentrations of the primer and template were 5 and 10 nM, respectively, and the reaction volume was 20 μL. (a) The primer extension was carried out in the presence of all four dNTPs at a concentration of 200 μM each, and the amounts Kf− were as indicated in the figure. The reactions were continued at 37°C for 60 min. (b) The reaction was performed in the presence of one type of dNTP at a time ([dNTP] =1.0 mM) at 37°C for 10 min, and 0.1 unit of Kf− was used.
When the above primer/template complex was incubated with Kf– in the presence of one type of nucleotide at a time, we found that dGMP and dAMP were incorporated opposite the lesion with much greater efficiency than the correct nucleotide, dCMP. Moreover, the dAMP insertion was more favored while the template strand bears an S-N2-CEdG than while it contains an R-N2-CEdG (Figure 7b). In this context, we quantified the percentages of nucleotide incorporation by integrating the band intensities for the unextended primer and the extended products. It turned out that the percentages for the incorporation of dAMP, dGMP, dCMP, and dTMP opposite S-N2-CEdG were 50±10%, 72±4%, 11±3%, and 8±1%, respectively. The corresponding percentages for the incorporation of these nucleotides opposite R-N2-CEdG were 37±8%, 85±3%, 9±1%, and 9±2%, whereas the percentages for the insertion of corresponding nucleotides opposite dG in the control substrate were 30±1%, 73±2%, 88±1%, and 61±0.2% (These results were based on three or more independent measurements).
Discussion
Recently, a number of studies have shown that methylglyoxal is mutagenic in vivo 22,23,33. Under physiological conditions, the major MG adduct of DNA was shown to be N2-CEdG 13. Obtaining ODNs bearing a site-specifically incorporated N2-CEdG constitutes a crucial step towards examining the biological implications of the lesion at the molecular level. In this study, we developed a facile postoligomerization method for the site-specific incorporation of individual diastereomers of N2-CEdG into ODNs, which provided an opportunity to investigate the stereochemical effect on the replication and repair of N2-CEdG. We demonstrated that the method facilitated the preparation of 12- and 20mer ODNs in reasonably good yield. After HPLC separation and desalting, we were able to obtain 70–100 nmol of the lesion-bearing ODNs from a 1-μmol scale synthesis. Thus, the synthetic strategy facilitates the availability of sufficient ODN substrates for future NMR structural studies. In addition, these lesion-bearing substrates can be readily employed to construct, by enzymatic ligation, longer lesion-bearing ODNs for in-vitro DNA repair studies 34. Likewise, the lesion-housing substrates can be ligated into plasmid DNA for in-vivo replication and repair studies 35.
Thermodynamic parameters for duplex formation, as derived from melting temperature measurements, revealed that the introduction of an N2-CEdG into duplex DNA led to an increase in ΔG at 25°C by 3.8 kcal/mol and 4.0 kcal/mol for the S and R-diastereomers, respectively (Table 2). The increase in ΔG for duplex formation is in the similar range as those reported for some other single-nucleobase lesions. For instance, 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG) and 5,6-dihydroxy-5,6-dihydrothymidine (thymidine glycol) caused 3.4 36 and 4.7 kcal/mol 37 increase in free energy for duplex formation. In this context, it is of note that the presence of a carboxyethyl functionality on the N2 position of guanine is expected to alter the Watson-Crick base pairing property of this nucleobase thereby destabilizing duplex DNA. Although the oxidation of dG to a 8-oxodG does not alter the hydrogen bonding properties of the nucleobase per se, the N-glycosidic linkage in 8-oxodG is known to equilibrate between the syn and anti conformations, with the former being energetically more favorable 38. The presence of 8-oxodG in syn conformation renders this nucleobase to base pair preferentially with dA 38. Similarly, the formation of thymidine glycol does not change the hydrogen bonding portion of the nucleobase itself; however, NMR structure studies indicated that thymidine glycol induces a large, localized structural change in duplex DNA with the modified nucleobase being extrahelical 39. Therefore, the similar extent of destabilization to DNA double helix induced by N2-CEdG, 8-oxodG and thymidine glycol suggest that the presence of an N2-CEdG in duplex DNA may also cause significant change to local DNA structure.
The destabilization to duplex DNA induced by lesions may provide insights into recognition of the lesions by repair enzyme 40. In this respect, nucleotide excision repair (NER) has been proposed to be involved in both the repair and the fixation of methylglyoxal-induced mutations 41.
The N2 position of guanine, located in the minor groove of duplex DNA, is a major site for modification induced by various carcinogens 42. Previous studies showed that N2-methylguanine could stall slightly the Klenow fragment-mediated primer extension reaction one base prior to the lesion, but DNA synthesis past the lesion was readily completed with small amount of misincorporation of dTMP 43. On the other hand, N2-ethylguanine could retard DNA synthesis but preferentially miscodes with dGMP when Klenow fragment was employed 44.
Our primer extension experiments with Klenow fragment demonstrated that N2-CEdG blocked markedly the Klenow fragment-mediated DNA synthesis past the lesion site. In addition, the polymerase incorporated dGMP and dAMP opposite the lesion much more efficiently than the correct nucleotide, dCMP. This result suggests that the presence of the bulky N2-carboxyethyl group in the minor groove may render the hydrogen bonding property of the modified guanine not to be recognized by the Klenow fragment during nucleotide insertion. Under such circumstances purine nucleotides can conceivably be incorporated more efficiently than pyrimidine nucleotides because the former nucleotides exhibit more favorable stacking interaction with the nucleotide at the 3′ terminus of the primer than the latter nucleotides 45. In addition, the result of the in-vitro replication study is consistent with previous studies showing that MG induced G:C→ C:G and G:C→T:A transversions in supF gene in mammalian cells 23. Moreover, the insertion of dAMP opposite the S diastereomer of N2-CEdG occurs more readily than that opposite the R diastereomer, highlighting the effect of difference in stereochemistry on the selectivity of nucleotide incorporation.
Future steady-state kinetic measurements on nucleotide incorporation and in vivo mutagenesis studies with the structurely defined lesion-carrying substrates as described in this paper will further illuminate the genotoxic effects of MG at the molecular level.
Supplementary Material
Acknowledgments
The authors want to thank the National Institutes of Health for supporting this research (Grants No. R01 CA96906 and R01 CA101864).
Footnotes
Supporting Information Available
NMR spectra of synthetic compounds, MS and MS/MS of 20mer ODNs. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Moreetesta P, Saintjalm Y. J Chromatogr. 1981;217:197–208. [Google Scholar]
- 2.Nagao M, Fujita Y, Wakabayashi K, Nukaya H, Kosuge T, Sugimura T. Environ Health Perspect. 1986;67:89–91. doi: 10.1289/ehp.866789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagao M, Fujita Y, Sugimura T, Kosuge T. IARC Sci Publ. 1986:283–291. [PubMed] [Google Scholar]
- 4.Kalapos MP. Toxicol Lett. 1999;110:145–175. doi: 10.1016/s0378-4274(99)00160-5. [DOI] [PubMed] [Google Scholar]
- 5.Phillips SA, Thornalley PJ. Eur J Biochem. 1993;212:101–105. doi: 10.1111/j.1432-1033.1993.tb17638.x. [DOI] [PubMed] [Google Scholar]
- 6.Ramasamy R, Yan SF, Schmidt AM. Cell. 2006;124:258–260. doi: 10.1016/j.cell.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Chaplen FWR, Fahl WE, Cameron DC. Proc Natl Acad Sci USA. 1998;95:5533–5538. doi: 10.1073/pnas.95.10.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLellan AC, Thornalley PJ, Benn J, Sonksen PH. Clin Sci. 1994;87:21–29. doi: 10.1042/cs0870021. [DOI] [PubMed] [Google Scholar]
- 9.Ahmed N, Thornalley PJ. Biochem J. 2002;364:15–24. doi: 10.1042/bj3640015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Y, Wang YS. Biochemistry. 2006;45:15654–15660. doi: 10.1021/bi061410o. [DOI] [PubMed] [Google Scholar]
- 11.Oya T, Hattori N, Mizuno Y, Miyata S, Maeda S, Osawa T, Uchida K. J Biol Chem. 1999;274:18492–18502. doi: 10.1074/jbc.274.26.18492. [DOI] [PubMed] [Google Scholar]
- 12.Lo TWC, Westwood ME, McLellan AC, Selwood T, Thornalley PJ. J Biol Chem. 1994;269:32299–32305. [PubMed] [Google Scholar]
- 13.Frischmann M, Bidmon C, Angerer J, Pischetsrieder M. Chem Res Toxicol. 2005;18:1586–1592. doi: 10.1021/tx0501278. [DOI] [PubMed] [Google Scholar]
- 14.Murata-Kamiya N, Kamiya H. Nucleic Acids Res. 2001;29:3433–3438. doi: 10.1093/nar/29.16.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maeta K, Izawa S, Okazaki S, Kuge S, Inoue Y. Mol Cell Biol. 2004;24:8753–8764. doi: 10.1128/MCB.24.19.8753-8764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukunaga M, Miyata S, Liu BF, Miyazaki H, Hirota Y, Higo S, Hamada Y, Ueyama S, Kasuga M. Biochem Biophys Res Commun. 2004;320:689–695. doi: 10.1016/j.bbrc.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 17.Shapiro R, Cohen BI, Shiuey SJ, Maurer H. Biochemistry. 1969;8:238. doi: 10.1021/bi00829a034. [DOI] [PubMed] [Google Scholar]
- 18.Krymkiew N. FEBS Lett. 1973;29:51–54. [Google Scholar]
- 19.Papoulis A, Alabed Y, Bucala R. Biochemistry. 1995;34:648–655. doi: 10.1021/bi00002a032. [DOI] [PubMed] [Google Scholar]
- 20.Schneider M, Georgescu A, Bidmon C, Tutsch M, Fleischmann EH, Popov D, Pischetsrieder M. Mol Nutr Food Res. 2006;50:424–429. doi: 10.1002/mnfr.200500217. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Nakamura S, Miyazaki S, Morita T, Suzuki M, Pischetsrieder M, Niwa T. Kidney Intl. 2006;69:388–392. doi: 10.1038/sj.ki.5000064. [DOI] [PubMed] [Google Scholar]
- 22.Pischetsrieder M, Seidel W, Munch G, Schinzel R. Biochem Biophys Res Commun. 1999;264:544–549. doi: 10.1006/bbrc.1999.1528. [DOI] [PubMed] [Google Scholar]
- 23.Murata-Kamiya N, Kamiya H, Kaji H, Kasai H. Mutat Res. 2000;468:173–182. doi: 10.1016/s1383-5718(00)00044-9. [DOI] [PubMed] [Google Scholar]
- 24.Pourmotabbed T, Creighton DJ. J Biol Chem. 1986;261:14240–14244. [PubMed] [Google Scholar]
- 25.Harris CM, Zhou L, Strand EA, Harris TM. J Am Chem Soc. 1991;113:4328–4329. [Google Scholar]
- 26.DeCorte BL, Tsarouhtsis D, Kuchimanchi S, Cooper MD, Horton P, Harris CM, Harris TM. Chem Res Toxicol. 1996;9:630–637. doi: 10.1021/tx9501795. [DOI] [PubMed] [Google Scholar]
- 27.Breslauer KJ. Methods Enzymol. 1995;259:221–242. doi: 10.1016/0076-6879(95)59046-3. [DOI] [PubMed] [Google Scholar]
- 28.Persmark M, Guengerich FP. Biochemistry. 1994;33:8662–8672. doi: 10.1021/bi00195a006. [DOI] [PubMed] [Google Scholar]
- 29.Santalucia J, Kierzek R, Turner DH. J Am Chem Soc. 1991;113:4313–4322. [Google Scholar]
- 30.Ochs S, Severin T. Liebigs Ann Chem. 1994;8:851–853. [Google Scholar]
- 31.McLuckey SA, Vanberkel GJ, Glish GL. J Am Soc Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 32.Hong H, Wang Y. J Am Chem Soc. 2005;127:13969–13977. doi: 10.1021/ja0531677. [DOI] [PubMed] [Google Scholar]
- 33.Seidel W, Pischetsrieder M. Cell Mol Biol. 1998;44:1165–1170. [PubMed] [Google Scholar]
- 34.Gu C, Zhang Q, Yang Z, Wang Y, Zou Y, Wang Y. Biochemistry. 2006;45:10739–10746. doi: 10.1021/bi060423z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delaney JC, Essigmann JM. Methods Enzymol. 2006;408:1–15. doi: 10.1016/S0076-6879(06)08001-3. [DOI] [PubMed] [Google Scholar]
- 36.Plum GE, Grollman AP, Johnson F, Breslauer KJ. Biochemistry. 1995;34:16148–16160. doi: 10.1021/bi00049a030. [DOI] [PubMed] [Google Scholar]
- 37.Iwai S. Chem Eur J. 2001;7:4343–4351. doi: 10.1002/1521-3765(20011015)7:20<4343::aid-chem4343>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 38.Grollman AP, Moriya M. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 39.Kao JY, Goljer I, Phan TA, Bolton PH. J Biol Chem. 1993;268:17787–17793. [PubMed] [Google Scholar]
- 40.Pilch DS, Plum GE, Breslauer KJ. Curr Opin Struct Biol. 1995;5:334–342. doi: 10.1016/0959-440x(95)80095-6. [DOI] [PubMed] [Google Scholar]
- 41.Murata-Kamiya N, Kaji H, Kasai H. Mutat Res. 1999;442:19–28. doi: 10.1016/s1383-5718(99)00054-6. [DOI] [PubMed] [Google Scholar]
- 42.Guengerich FP. Chem Rev. 2006;106:420–452. doi: 10.1021/cr0404693. [DOI] [PubMed] [Google Scholar]
- 43.Yasui M, Matsui S, Ihara M, Laxmi YRS, Shibutani S, Matsuda T. Nucleic Acids Res. 2001;29:1994–2001. doi: 10.1093/nar/29.9.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terashima I, Matsuda T, Fang TW, Suzuki N, Kobayashi J, Kohda K, Shibutani S. Biochemistry. 2001;40:4106–4114. doi: 10.1021/bi002719p. [DOI] [PubMed] [Google Scholar]
- 45.Saenger W. Principles of Nucleic Acid Structure. Springer-Verlag New York Inc; New York: 1984. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.