Abstract
α-Ketoglutarate dehydrogenase (KGDH), a key regulatory enzyme within the Krebs cycle, is sensitive to mitochondrial redox status. Treatment of mitochondria with H2O2 results in reversible inhibition of KGDH due to glutathionylation of the cofactor, lipoic acid. Upon consumption of H2O2, glutathione is removed by glutaredoxin restoring KGDH activity. Glutathionylation appears to be enzymatically catalysed or require a unique microenvironment. This may represent an antioxidant response, diminishing the flow of electrons to the respiratory chain and protecting sulphydryl residues from oxidative damage. KGDH is, however, also susceptible to oxidative damage. 4-Hydroxy-2-nonenal (HNE), a lipid peroxidation product, reacts with lipoic acid resulting in enzyme inactivation. Evidence indicates that HNE modified lipoic acid is cleaved from KGDH, potentially the first step of a repair process. KGDH is therefore a likely redox sensor, reversibly altering metabolism to reduce oxidative damage and, under severe oxidative stress, acting as a sentinel of mitochondrial viability.
Keywords: α-Ketoglutarate dehydrogenase, glutathionylation, redox signalling, mitochondria, free radicals
Introduction
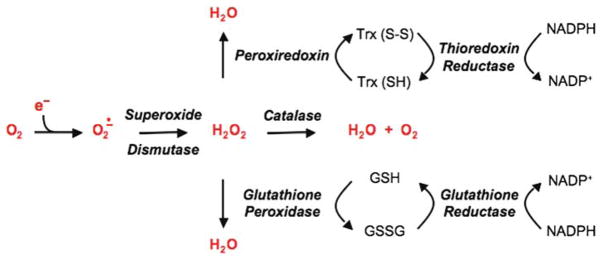
It is widely theorized that declines in physiological and cellular function observed with ageing and a variety of degenerative diseases are due, at least in part, to free radical derived damage to critical cellular components. Indirect evidence has been gleaned from studies showing positive correlations between age- and disease-related increases in certain measures of oxidative stress and the accumulation of oxidized forms of protein, DNA and/or lipids. The traditional view is that prolonged and/or elevated production of free radical species, such as occurs with age and/or disease, overwhelms the large variety of antioxidant enzymes (Scheme 1) and small molecular weight scavengers present within cells, thereby enabling reactive free radical species to interact with and damage biomolecules. This view has been challenged by the failure of traditional antioxidant interventions to consistently minimize various functional manifestations of ageing and disease. In addition, emerging evidence indicates that free radical species are important elements of certain signal transduction pathways [1–6] and can, under certain conditions such as ischemic pre-conditioning [7–14], exert beneficial effects. Moreover, mitochondria, a major intracellular source of pro-oxidants [15,16], are reversibly responsive to changes in redox status [17,18]. These findings indicate that cells possess previously unanticipated mechanisms for rapidly sensing and responding to changes in redox status and underscore the need for reconsideration of the response to oxidative stress and molecular factors that underlie free radical damage.
Scheme 1.
Multiple enzymatic processes exist for the removal of superoxide anion and hydrogen peroxide.
Evidence is presented that the mitochondrial Krebs cycle enzyme α-ketoglutarate dehydrogenase (KGDH) is a component of the mitochondrial antioxidant system and a key sensor of redox status, altering function to induce critical changes in mitochondrial and cellular metabolism to prevent oxidative damage. KGDH is uniquely sensitive to oxidative stress, capable of undergoing fully reversible free radical mediated inhibition [17,19,20] or, under certain conditions, oxidative inactivation [21–24]. KGDH represents a key regulatory site within the Krebs cycle, controlling the supply of reducing equivalents generated by the Krebs cycle and, as such, electron transport and ATP production [23,25,26]. Given that KGDH is also sensitive to free radical mediated alterations in activity, redox regulation of KGDH would provide a means for controlling energy production in response to changes in mitochondrial and cellular oxidative stress. Nevertheless, KGDH has been reported to decline in activity during a number of neurological and cardiovascular disorders associated with oxidative stress [27–33]. The purpose of this review is to highlight structural and functional features of KGDH that render the enzyme uniquely responsive to changes in mitochondrial redox status. In this context, key findings that underscore the potential antioxidant function(s) of KGDH and direct future studies addressing the physiological and pathophysiological significance of redox-dependent regulation and oxidative inactivation of KGDH are presented and discussed.
Structure and catalytic mechanism of α-ketoglutarate dehydrogenase
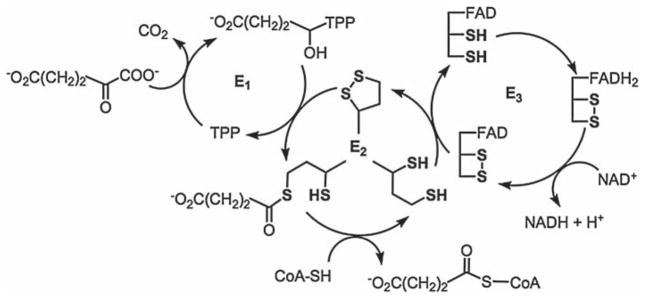
α-Ketoglutarate dehydrogenase (KGDH) is a large multi-sub-unit enzyme complex composed of multiple copies of three sub-units: E1 (α-ketoacid decarboxylase), E2 (dihydrolipoyl transacetylase) and E3 (dihydrolipoamide dehydrogenase) (Scheme 2). A variety of co-factors are utilized by KGDH to catalyse the multistep reaction that converts α-ketoglutarate to succinyl-CoA, CO2 and NADH. These co-factors are: (1) Thiamine pyrophosphate (TPP) tightly bound to the E1 sub-unit; (2) Lipoic acid covalently bound to the E2 sub-unit; (3) CoASH, which interacts with the succinyl group on lipoic acid; (4) FAD+, which is tightly bound to the E3 sub-unit; and (5) NAD+, which is reduced by the FADH2 on E3 to form NADH. The reaction mechanism for KGDH is as follows: α-Ketoglutarate is covalently bound to TPP on the E1 sub-unit. The binding process initiates the oxidative decarboxylation of α-ketoglutarate to form CO2 and a covalently bound succinyl group. The succinyl group is then transferred to oxidized lipoic acid on the E2 sub-unit. CoASH reacts with the succinyl group to form succinyl-CoA and reduced lipoic acid. Lipoic acid subsequently reduces vicinal sulphydryl groups on the E3 sub-unit. The reducing equivalents are then transferred to the tightly bound FAD+. Finally, FADH2 reduces NAD+ to NADH, completing a catalytic cycle [34–36]. It is noteworthy that enzyme catalysis involves cycling of the oxidation states of various sulphydryl groups, functionalities that readily react with (pro-)oxidants and electrophilic byproducts of lipid peroxidation [37]. Thus, the chemical features of KGDH predict a strong likelihood that the enzyme would be sensitive to changes in mitochondrial redox status.
Scheme 2.
α-Ketoglutarate dehydrogenase catalytic cycle. E1 sub-unit: α-Ketoacid decarboxylase; E2 sub-unit: Dihydrolipoyl transacetylase; and E3 sub-unit: Dihydrolipoamide dehydrogenase.
Reversible redox-dependent regulation of KGDH
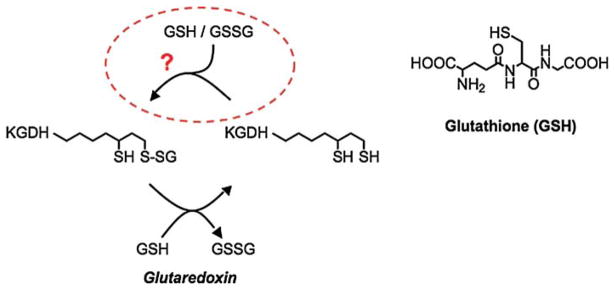
Investigations of the effects of oxidative stress on mitochondrial function revealed the capacity to reversibly alter respiratory activity. Upon treatment of isolated mitochondria with H2O2, electron transport and oxidative phosphorylation activity were diminished [17]. However, activity was fully recovered upon consumption of the H2O2, indicative of redox regulation [17]. Further investigation revealed that H2O2-induced declines in the rates of electron transport and oxidative phosphorylation were associated with reversible inhibition of the Krebs cycle enzymes KGDH [17] and aconitase [38,39]. Because KGDH represents a rate-limiting enzyme for the production of NADH and NADH production, not consumption, limits the rate of electron transport and ATP synthesis [23,25,26], inhibition of KGDH was deemed responsible for H2O2-induced loss in the rate of oxidative phosphorylation [17]. Inhibition of KGDH occurred as a result of formation of a mixed disulphide between a sulphydryl group on KGDH and glutathione (glutathionylation) [20]. Upon consumption of H2O2, glutathione is removed by the enzyme glutaredoxin resulting in restoration of KGDH activity. Glutathionylation of KGDH occurred on the covalently bound cofactor, lipoic acid (Scheme 3) [19]. In support of this, glutathionylated lipoic acid sulphydryl groups were protected from modification by sulphydryl reactive compounds such as N-ethylmaleimide or 2-alkenals [19]. The reversible nature of H2O2-induced inhibition and glutathionylation of KGDH provokes intriguing questions as to: (1) What are the functional consequences of redox-dependent regulation of KGDH and (2) What are the molecular mechanisms that govern the glutathionylation of KGDH?
Scheme 3.
α-Ketoglutarate dehydrogenase is regulated through the reversible glutathionylation of the enzyme’s cofactor lipoic acid. Evidence suggests that glutathionylation of KGDH represents an antioxidant response and is either enzyme catalysed or requires a unique microenvironment.
Functional consequences of redox regulation of KGDH
A strong possibility is that glutathionylation of KGDH may represent an antioxidant response. KGDH plays a pivotal role in controlling flux through the Krebs cycle and, thus, the supply of reducing equivalents in the form of NADH for electron transport and ATP production [23,25,26]. When pro-oxidant production increases, such as may occur when electron transport is impaired, inhibition of KGDH would be expected to limit the supply of reducing equivalents generated by the Krebs cycle. This would ultimately reduce free radical production by a compromised electron transport chain, a possibility currently under investigation in our laboratory. In addition, glutathionylation of KGDH would protect critical sulphydryl groups from damage until the oxidative stress is relieved. This would be important because each lipoic acid bound to the E2 sub-unit contains two sulphydryls and the E3 sub-unit possesses a pair of vicinal sulphydryls, each of which is required for enzyme activity [34–36]. Glutathionylation of lipoic acid protects the sulphydryls from adduction by lipid peroxidation products [19] and would lock the sulphydryls on E3 in the disulphide form reducing their reactivity with free radicals and related products. Fine-tuning of the antioxidant function of KGDH may also exist. In support of this possibility, the maximal degree of H2O2-induced KGDH inhibition does not exceed a defined threshold (50% relative to control), irrespective of H2O2 concentration [17,20]. Previous findings indicate that a sub-population of KGDH appears to directly interact with complex I and channel the reductive potential of NADH directly to the electron transport chain [40–43]. Selective glutathionylation and inhibition of complex I associated KGDH would preserve the activity of free KGDH maintaining biosynthetic processes, while slowing electron transport to reduce superoxide production.
If glutathionylation of KGDH were purely an anti-oxidant response, it might be predicted to occur in mitochondria from all tissues. We have found that this is not the case. In cardiac mitochondria treated with H2O2, reversible inhibition of KGDH by glutathionylation is observed, however skeletal muscle mitochondria exhibit no deficits in KGDH activity under similar conditions. This may reflect inherent differences in mitochondria isolated from different tissues or a lack of in-depth understanding of the molecular events that govern the glutathionylation of KGDH. Nevertheless, additional ramifications of KGDH glutathionylation and the role of pro-oxidants in other processes that depend on KGDH activity must also be considered. While KGDH is generally studied with regard to the production of NADH and, therefore, in the synthesis of ATP, the enzyme also represents a key site for the utilization of amino acids for energy production, produces succinyl-CoA for heme biosynthesis, and the enzyme’s substrate α-ketoglutarate plays a role in other important processes such as HIF-1α degradation. In addition, specific inhibition of KGDH has been shown to result in cytochrome c release from the mitochondria and cell death [44]. Therefore, in addition to playing a role in the mitochondrial antioxidant response, reversible inhibition of KGDH by redox-dependent glutathionylation may provide a means to modulate energy production during processes such as development, cancer growth, hibernation and other events that require a rapid transition from energy production to biosynthesis.
There is also evidence to support alternative functions for KGDH that may underlie observed glutathionylation of the enzyme. Thioredoxin 2 can protect KGDH from self-inactivation [45–47] and has been implicated in other roles involving KGDH. Thioredoxin 2 reduces peroxiredoxin, allowing for peroxiredoxin to undergo another round of H2O2 reduction. Thioredoxin reductase then regenerates reduced thioredoxin. In vitro evidence indicates that oxidized thioredoxin can also be reduced by free lipoic acid which in turn is reduced by KGDH to complete the catalytic cycle and regenerate thioredoxin activity [48]. There is also evidence to suggest that lipoic acid on the E2 sub-unit can participate in reduction of peroxidases and serve in other potential antioxidant roles [49,50]. In addition, the E3 sub-unit in the bacteria Mycobacterium tuberculosis has been shown to exhibit thioredoxin reductase activity [51]. Moreover, E3 has high structural and catalytic homology to glutathione reductase [52–54]. The potential therefore exists that in response to oxidative stress, KGDH shifts to a reductase role as a means of aiding the removal of pro-oxidants. This would have the added benefit of reducing electron flow and thus O2·− production. Finally, E3 has recently been reported to act as a protease upon dissociation from the multi-enzyme complex [55]. How glutathionylation of lipoic acid may be involved in these processes is not yet clear. Glutathionylation may cause structural alterations allowing for the switch in function or, alternatively, may reflect an enzyme intermediate required for peroxidase/reductase function.
Mechanism of KGDH glutathionylation
The mechanism by which KGDH undergoes glutathionylation has not yet been defined. Glutathione exists primarily in the reduced form intracellularly. However, under conditions of oxidative stress, the ratio of reduced-to-oxidized glutathione (GSH/GSSG) can decrease dramatically [20,56,57]. Under these conditions, GSSG is believed to react with low pKa cysteine residues on protein resulting in the formation of a mixed disulphide (glutathionylation). Alternatively, cysteine sulphydryl residues can undergo limited oxidation, effectively priming them for glutathionylation by GSH [58–60]. It is noteworthy that glutathionylation and inhibition of KGDH cannot be readily reconstituted using purified protein or solubilized mitochondria treated with GSSG in combination with various substrates, co-factors and oxidants. H2O2-induced inhibition requires actively respiring mitochondria [17,20]. In addition, H2O2-induced glutathionylation of KGDH appears to be tissue-specific, occurring in mitochondria isolated from cardiac but not skeletal muscle. These observations suggest the possibility that glutathionylation of KGDH represents a novel enzymatically-driven process or requires a unique microenvironment that is not readily reconstituted using purified enzyme or solubilized mitochondria. It will be important to identify the mechanism of KGDH glutathionylation. This information is critical to understanding how the process is regulated, when it is likely to occur and the physiological ramifications of redox-dependent regulation of KGDH.
Redox-dependent self-inactivation of KGDH
In light of the potential impact of KGDH glutathionylation on a number of mitochondrial and cellular processes, it is intriguing that KGDH is capable of producing superoxide anion and, subsequently, undergoing self-inactivation [46,61–64]. This occurs under conditions when NADH concentrations are elevated or low levels of NAD+ are available as electron acceptors. Superoxide anion production at the FAD component of E3 causes generation of a thiyl radical on the reduced lipoic acid bound to E2. Reduction of the thiyl radical in the presence of α-ketoglutarate and CoA results in the formation of a carbon-centred radical within the active site of E1 and, thus, inhibition of the enzyme [46,62]. This form of KGDH self-inactivation can be prevented in the presence of thioredoxin [45–47]. It is believed that thioredoxin interacts specifically with KGDH, neutralizing the thiyl radical on lipoic acid and preventing the formation of the carbon-centred radical on E1. The formation of this radical may play a role in priming lipoic acid for glutathionylation.
Oxidative inactivation of KGDH
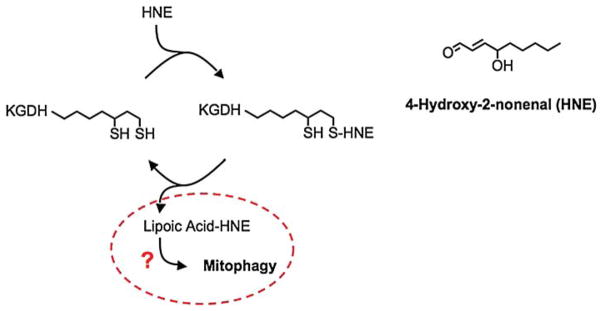
Paradoxically, the chemical properties of lipoic acid that enable glutathionylation of KGDH render the enzyme susceptible to oxidative inactivation. The cytotoxic lipid peroxidation product 4-hydroxy-2-nonenal (HNE) is formed under conditions of severe oxidative stress [37]. Treatment of cardiac mitochondria with HNE diminishes the rate of oxidative phosphorylation [22]. This is the result of selective inactivation of KGDH by virtue of the high reactivity of HNE with E2-bound lipoic acid [21]. The susceptibility of KGDH to oxidative inactivation is reflected by the fact that the enzyme has been reported to decline in activity during a large variety of neurological and cardiovascular disorders associated with oxidative stress [27–33]. This raises the question that if the enzyme can be readily protected from oxidative damage and reduce oxidative stress by virtue of glutathionylation, why then does it become oxidatively inactivated? The answer may lie in the magnitude and duration of the oxidative stress and reveal a terminal antioxidant sensing mechanism. Prolonged or excessive production of reactive oxygen-derived free radicals and pro-oxidants would be indicative of irreparable damage. Recent evidence indicates that HNE-induced inactivation of KGDH is followed by cleavage of the modified lipoic acid residue (Scheme 4). Mitochondria possess an enzyme capable of hydrolysing the lipoamide bond as well as the machinery to synthesize lipoic acid and covalently attach the co-factor to the E2 sub-unit of the enzyme [65–72]. Cleavage of the lipoic acid–HNE adduct may therefore represent the initial step in enzyme repair followed by insertion of native lipoic acid and restoration of KGDH activity, an intriguing possibility under investigation in our laboratory. However, high levels of HNE modification to the enzyme are likely to occur only when mitochondria have undergone significant alterations in structure and function. We are currently assessing whether extensive externalization of lipoic acid–HNE on the outer mitochondrial membrane may represent a signal for the recognition of irreparably damaged mitochondria and the initiation of mitophagy. In addition, selective inhibition of KGDH has been shown to result in release of cytochrome c and cell death [44], as previously discussed. The relative level of KGDH modification by the lipid peroxidation product HNE could therefore serve as a sensor of mitochondrial viability.
Scheme 4.
α-Ketoglutarate dehydrogenase is sensitive to oxidative inactivation through reaction of lipoic acid with the lipid peroxidation product 4-hydroxy-2-nonenal (HNE). The potential for removal of the modified lipoic acid and reinsertion of native lipoic acid exists and would represent enzyme repair. Could the presence of the cleaved hydrophobic lipoic acid-HNE adduct on the mitochondrial membrane serve as a signal for mitophagy?
Summary
Increases in tissue and cellular free radical production and the subsequent occurrence of free radical mediated modification(s) to various biomolecules have traditionally been viewed as deleterious to mitochondrial, cellular and physiological function. In this context, oxidative inhibition of specific enzymes has been considered primarily as a form of oxidative damage that, under normal conditions, can be countered by the integrated actions of antioxidant enzymes and small molecular weight free radical scavengers that remove well-known free radical and pro-oxidant species such as superoxide anion and H2O2. Nevertheless, it is becoming increasingly evident that free radical species are components of a series of normal cellular processes and can exert reversible and potentially beneficial effects. The Krebs cycle enzyme KGDH, an enzyme that controls the reductive potential (NADH/NAD+) of the mitochondria, can be regulated by subtle alterations in mitochondrial redox status [17,19,20] but is also susceptible to free radical damage [21–24]. In addition, at high ratios of NADH/NAD+, KGDH is a source of oxygen-derived free radical species [46,61–64]. This enzyme therefore occupies a central position at the cross-roads of the redox environment, capable of altering enzyme function and/or mitochondrial metabolism to reduce oxidative damage while being a source of free radical production and a target of oxidative inactivation.
In summary, the unique perspective gained through analyses of mitochondria exposed to oxidative stress reveals that the antioxidant response is complex, dynamic and involves processes and enzymes not previously considered in this context. The antioxidant response is not confined simply to the removal of free radical and pro-oxidant species, but also likely involves regulated alterations in metabolism to both reduce free radical production and protect critical mitochondrial components from oxidative damage. Further characterization of molecular mechanisms by which multiple mitochondrial systems co-ordinately respond to oxidative stress and induce specific molecular and functional adaptations is necessary to define perturbations in these processes that contribute to oxidative damage and loss of cellular viability during ageing and the progression of numerous degenerative diseases associated with oxidative stress.
Footnotes
Declaration of Interest
This work was supported by Grant Number R01AG016339 from the National Institute on Aging. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Aging or the National Institutes of Health.
References
- 1.Droge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 2.Ghezzi P, Bonetto V, Fratelli M. Thiol-disulfide balance: from the concept of oxidative stress to that of redox regulation. Antioxid Redox Signal. 2005;7:964–972. doi: 10.1089/ars.2005.7.964. [DOI] [PubMed] [Google Scholar]
- 3.Rhee SG. Redox signaling: hydrogen peroxide as intracellular messenger. Exp Mol Med. 1999;31:53–59. doi: 10.1038/emm.1999.9. [DOI] [PubMed] [Google Scholar]
- 4.Shelton MD, Chock PB, Mieyal JJ. Glutaredoxin: role in reversible protein s-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid Redox Signal. 2005;7:348–366. doi: 10.1089/ars.2005.7.348. [DOI] [PubMed] [Google Scholar]
- 5.Forman HJ, Maiorino M, Ursini F. Signaling functions of reactive oxygen species. Biochemistry. 49:835–842. doi: 10.1021/bi9020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yap LP, Garcia JV, Han D, Cadenas E. The energy-redox axis in aging and age-related neurodegeneration. Adv Drug Deliv Rev. 2009;61:1283–1298. doi: 10.1016/j.addr.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambrosio G, Tritto I, Chiariello M. The role of oxygen free radicals in preconditioning. J Mol Cell Cardiol. 1995;27:1035–1039. doi: 10.1016/0022-2828(95)90072-1. [DOI] [PubMed] [Google Scholar]
- 8.Baines CP, Goto M, Downey JM. Oxygen radicals released during ischemic preconditioning contribute to cardioprotection in the rabbit myocardium. J Mol Cell Cardiol. 1997;29:207–216. doi: 10.1006/jmcc.1996.0265. [DOI] [PubMed] [Google Scholar]
- 9.da Silva MM, Sartori A, Belisle E, Kowaltowski AJ. Ischemic preconditioning inhibits mitochondrial respiration, increases H2O2 release, and enhances K+ transport. Am J Physiol Heart Circ Physiol. 2003;285:154–162. doi: 10.1152/ajpheart.00955.2002. [DOI] [PubMed] [Google Scholar]
- 10.Halestrap AP, Clarke SJ, Khaliulin I. The role of mitochondria in protection of the heart by preconditioning. Biochim Biophys Acta. 2007;1767:1007–1031. doi: 10.1016/j.bbabio.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kevin LG, Camara AK, Riess ML, Novalija E, Stowe DF. Ischemic preconditioning alters real-time measure of O2 radicals in intact hearts with ischemia and reperfusion. Am J Physiol Heart Circ Physiol. 2003;284:566–574. doi: 10.1152/ajpheart.00711.2002. [DOI] [PubMed] [Google Scholar]
- 12.Novalija E, Hogg N, Kevin LG, Camara AK, Stowe DF. Ischemic preconditioning: triggering role of nitric oxide-derived oxidants in isolated hearts. J Cardiovasc Pharmacol. 2003;42:593–600. doi: 10.1097/00005344-200311000-00003. [DOI] [PubMed] [Google Scholar]
- 13.Tritto I, D’Andrea D, Eramo N, Scognamiglio A, De Simone C, Violante A, Esposito A, Chiariello M, Ambrosio G. Oxygen radicals can induce preconditioning in rabbit hearts. Circ Res. 1997;80:743–748. doi: 10.1161/01.res.80.5.743. [DOI] [PubMed] [Google Scholar]
- 14.Yaguchi Y, Satoh H, Wakahara N, Katoh H, Uehara A, Terada H, Fujise Y, Hayashi H. Protective effects of hydrogen peroxide against ischemia/reperfusion injury in perfused rat hearts. Circ J. 2003;67:253–258. doi: 10.1253/circj.67.253. [DOI] [PubMed] [Google Scholar]
- 15.Cadenas E, Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med. 2000;29:222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 16.Chance B, Williams GR. The respiratory chain and oxidaive phosphorylation. Adv Enzymol Relat Subj Biochem. 1956;17:65–134. doi: 10.1002/9780470122624.ch2. [DOI] [PubMed] [Google Scholar]
- 17.Nulton-Persson AC, Szweda LI. Modulation of mitochondrial function by hydrogen peroxide. J Biol Chem. 2001;276:23357–23361. doi: 10.1074/jbc.M100320200. [DOI] [PubMed] [Google Scholar]
- 18.Rokutan K, Kawai K, Asada K. Inactivation of 2-oxoglutarate dehydrogenase in rat liver mitochondria by its substrate and t-butyl hydroperoxide. J Biochem. 1987;101:415–422. doi: 10.1093/oxfordjournals.jbchem.a121926. [DOI] [PubMed] [Google Scholar]
- 19.Applegate MA, Humphries KM, Szweda LI. Reversible inhibition of alpha-ketoglutarate dehydrogenase by hydrogen peroxide: glutathionylation and protection of lipoic acid. Biochemistry. 2008;47:473–478. doi: 10.1021/bi7017464. [DOI] [PubMed] [Google Scholar]
- 20.Nulton-Persson AC, Starke DW, Mieyal JJ, Szweda LI. Reversible inactivation of alpha-ketoglutarate dehydrogenase in response to alterations in the mitochondrial glutathione status. Biochemistry. 2003;42:4235–4242. doi: 10.1021/bi027370f. [DOI] [PubMed] [Google Scholar]
- 21.Humphries KM, Szweda LI. Selective inactivation of alpha-ketoglutarate dehydrogenase and pyruvate dehydrogenase: reaction of lipoic acid with 4-hydroxy-2-nonenal. Biochemistry. 1998;37:15835–15841. doi: 10.1021/bi981512h. [DOI] [PubMed] [Google Scholar]
- 22.Humphries KM, Yoo Y, Szweda LI. Inhibition of NADH-linked mitochondrial respiration by 4-hydroxy-2-nonenal. Biochemistry. 1998;37:552–557. doi: 10.1021/bi971958i. [DOI] [PubMed] [Google Scholar]
- 23.Tretter L, Adam-Vizi V. Inhibition of Krebs cycle enzymes by hydrogen peroxide: a key role of [alpha]-ketoglutarate dehydrogenase in limiting NADH production under oxidative stress. J Neurosci. 2000;20:8972–8979. doi: 10.1523/JNEUROSCI.20-24-08972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korotchkina LG, Yang H, Tirosh O, Packer L, Patel MS. Protection by thiols of the mitochondrial complexes from 4-hydroxy-2-nonenal. Free Radic Biol Med. 2001;30:992–999. doi: 10.1016/s0891-5849(01)00491-9. [DOI] [PubMed] [Google Scholar]
- 25.Cooney GJ, Taegtmeyer H, Newsholme EA. Tricarboxylic acid cycle flux and enzyme activities in the isolated working rat heart. Biochem J. 1981;200:701–703. doi: 10.1042/bj2000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreno-Sanchez R, Hogue BA, Hansford RG. Influence of NAD-linked dehydrogenase activity on flux through oxidative phosphorylation. Biochem J. 1990;268:421–428. doi: 10.1042/bj2680421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibson GE, Park LC, Sheu KF, Blass JP, Calingasan NY. The alpha-ketoglutarate dehydrogenase complex in neurodegeneration. Neurochem Int. 2000;36:97–112. doi: 10.1016/s0197-0186(99)00114-x. [DOI] [PubMed] [Google Scholar]
- 28.Lucas DT, Szweda LI. Declines in mitochondrial respiration during cardiac reperfusion: age-dependent inactivation of alpha-ketoglutarate dehydrogenase. Proc Natl Acad Sci USA. 1999;96:6689–6693. doi: 10.1073/pnas.96.12.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundberg KC, Szweda LI. Preconditioning prevents loss in mitochondrial function and release of cytochrome c during prolonged cardiac ischemia/reperfusion. Arch Biochem Biophys. 2006;453:130–134. doi: 10.1016/j.abb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Sadek HA, Humphries KM, Szweda PA, Szweda LI. Selective inactivation of redox-sensitive mitochondrial enzymes during cardiac reperfusion. Arch Biochem Biophys. 2002;406:222–228. doi: 10.1016/s0003-9861(02)00446-0. [DOI] [PubMed] [Google Scholar]
- 31.Beal MF. Aging, energy, and oxidative stress in neurodegenerative diseases. Ann Neurol. 1995;38:357–366. doi: 10.1002/ana.410380304. [DOI] [PubMed] [Google Scholar]
- 32.Kish SJ. Brain energy metabolizing enzymes in Alzheimer’s disease: alpha-ketoglutarate dehydrogenase complex and cytochrome oxidase. Ann N Y Acad Sci. 1997;826:218–228. doi: 10.1111/j.1749-6632.1997.tb48473.x. [DOI] [PubMed] [Google Scholar]
- 33.Schapira AH. Mitochondrial involvement in Parkinson’s disease, Huntington’s disease, hereditary spastic paraplegia and Friedreich’s ataxia. Biochim Biophys Acta. 1999;1410:159–170. doi: 10.1016/s0005-2728(98)00164-9. [DOI] [PubMed] [Google Scholar]
- 34.Reed LJ. Multienzyme complexes. Acc Chem Res. 1974;7:40–46. [Google Scholar]
- 35.Waskiewicz DE, Hammes GG. Elementary steps in the reaction mechanism of the alpha-ketoglutarate dehydrogenase multienzyme complex from Escherichia coli: kinetics of succinylation and desuccinylation. Biochemistry. 1984;23:3136–3143. doi: 10.1021/bi00309a005. [DOI] [PubMed] [Google Scholar]
- 36.Perham RN. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional protein. Biochemistry. 1991;30:8501–8512. doi: 10.1021/bi00099a001. [DOI] [PubMed] [Google Scholar]
- 37.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 38.Bulteau AL, Ikeda-Saito M, Szweda LI. Redox-dependent modulation of aconitase activity in intact mitochondria. Biochemistry. 2003;42:14846–14855. doi: 10.1021/bi0353979. [DOI] [PubMed] [Google Scholar]
- 39.Bulteau AL, O’Neill HA, Kennedy MC, Ikeda-Saito M, Isaya G, Szweda LI. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science. 2004;305:242–245. doi: 10.1126/science.1098991. [DOI] [PubMed] [Google Scholar]
- 40.Fukushima T, Decker RV, Anderson WM, Spivey HO. Substrate channeling of NADH and binding of dehydrogenases to complex I. J Biol Chem. 1989;264:16483–16488. [PubMed] [Google Scholar]
- 41.Maas E, Bisswanger H. Localization of the alpha-oxoacid dehydrogenase multienzyme complexes within the mitochondrion. FEBS Lett. 1990;277:189–190. doi: 10.1016/0014-5793(90)80840-f. [DOI] [PubMed] [Google Scholar]
- 42.Porpaczy Z, Sumegi B, Alkonyi I. Interaction between NAD-dependent isocitrate dehydrogenase, alpha-ketoglutarate dehydrogenase complex, and NADH:ubiquinone oxidoreductase. J Biol Chem. 1987;262:9509–9514. [PubMed] [Google Scholar]
- 43.Sumegi B, Srere PA. Complex I binds several mitochondrial NAD-coupled dehydrogenases. J Biol Chem. 1984;259:15040–15045. [PubMed] [Google Scholar]
- 44.Huang HM, Ou HC, Xu H, Chen HL, Fowler C, Gibson GE. Inhibition of alpha-ketoglutarate dehydrogenase complex promotes cytochrome c release from mitochondria, caspase-3 activation, and necrotic cell death. J Neurosci Res. 2003;74:309–317. doi: 10.1002/jnr.10756. [DOI] [PubMed] [Google Scholar]
- 45.Bunik V, Raddatz G, Lemaire S, Meyer Y, Jacquot JP, Bisswanger H. Interaction of thioredoxins with target proteins: role of particular structural elements and electrostatic properties of thioredoxins in their interplay with 2-oxoacid dehydrogenase complexes. Protein Sci. 1999;8:65–74. doi: 10.1110/ps.8.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bunik VI. 2-Oxo acid dehydrogenase complexes in redox regulation. Eur J Biochem. 2003;270:1036–1042. doi: 10.1046/j.1432-1033.2003.03470.x. [DOI] [PubMed] [Google Scholar]
- 47.Bunik V, Follmann H, Bisswanger H. Activation of mitochondrial 2-oxoacid dehydrogenases by thioredoxin. Biol Chem. 1997;378:1125–1130. doi: 10.1515/bchm.1997.378.10.1125. [DOI] [PubMed] [Google Scholar]
- 48.Bunik V, Follmann H. Thioredoxin reduction dependent on alpha-ketoacid oxidation by alpha-ketoacid dehydrogenase complexes. FEBS Lett. 1993;336:197–200. doi: 10.1016/0014-5793(93)80801-z. [DOI] [PubMed] [Google Scholar]
- 49.Cussiol JR, Alegria TG, Szweda LI, Netto LE. Ohr (organic hydroperoxide resistance protein) possesses a previously undescribed activity, lipoyl-dependent peroxidase. J Biol Chem. 2010;285:21943–21950. doi: 10.1074/jbc.M110.117283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi Q, Chen HL, Xu H, Gibson GE. Reduction in the E2k subunit of the alpha-ketoglutarate dehydrogenase complex has effects independent of complex activity. J Biol Chem. 2005;280:10888–10896. doi: 10.1074/jbc.M409064200. [DOI] [PubMed] [Google Scholar]
- 51.Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002;295:1073–1077. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- 52.Carothers DJ, Pons G, Patel MS. Dihydrolipoamide dehydrogenase: functional similarities and divergent evolution of the pyridine nucleotide-disulfide oxidoreductases. Arch Biochem Biophys. 1989;268:409–425. doi: 10.1016/0003-9861(89)90309-3. [DOI] [PubMed] [Google Scholar]
- 53.Sandalova T, Zhong L, Lindqvist Y, Holmgren A, Schneider G. Three-dimensional structure of a mammalian thioredoxin reductase: implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc Natl Acad Sci USA. 2001;98:9533–9538. doi: 10.1073/pnas.171178698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams CH, Jr, Arscott LD, Schulz GE. Amino acid sequence homology between pig heart lipoamide dehydrogenase and human erythrocyte glutathione reductase. Proc Natl Acad Sci USA. 1982;79:2199–2201. doi: 10.1073/pnas.79.7.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Babady NE, Pang YP, Elpeleg O, Isaya G. Cryptic proteolytic activity of dihydrolipoamide dehydrogenase. Proc Natl Acad Sci USA. 2007;104:6158–6163. doi: 10.1073/pnas.0610618104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilbert HF. Thiol/disulfide exchange equilibria and disulfide bond stability. Method Enzymol. 1995;251:8–28. doi: 10.1016/0076-6879(95)51107-5. [DOI] [PubMed] [Google Scholar]
- 57.Klatt P, Lamas S. Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. Eur J Biochem. 2000;267:4928–4944. doi: 10.1046/j.1432-1327.2000.01601.x. [DOI] [PubMed] [Google Scholar]
- 58.Dalle-Donne I, Rossi R, Giustarini D, Colombo R, Milzani A. S-glutathionylation in protein redox regulation. Free Radic Biol Med. 2007;43:883–898. doi: 10.1016/j.freeradbiomed.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 59.Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr Opin Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 60.Hurd TR, Filipovska A, Costa NJ, Dahm CC, Murphy MP. Disulphide formation on mitochondrial protein thiols. Biochem Soc Trans. 2005;33:1390–1393. doi: 10.1042/BST0331390. [DOI] [PubMed] [Google Scholar]
- 61.Ambrus A, Tretter L, Adam-Vizi V. Inhibition of the alpha-ketoglutarate dehydrogenase-mediated reactive oxygen species generation by lipoic acid. J Neurochem. 2009;109 (Suppl 1):222–229. doi: 10.1111/j.1471-4159.2009.05942.x. [DOI] [PubMed] [Google Scholar]
- 62.Bunik VI, Sievers C. Inactivation of the 2-oxo acid dehydrogenase complexes upon generation of intrinsic radical species. Eur J Biochem. 2002;269:5004–5015. doi: 10.1046/j.1432-1033.2002.03204.x. [DOI] [PubMed] [Google Scholar]
- 63.Starkov AA, Fiskum G, Chinopoulos C, Lorenzo BJ, Browne SE, Patel MS, Beal MF. Mitochondrial alpha-ketoglutarate dehydrogenase complex generates reactive oxygen species. J Neurosci. 2004;24:7779–7788. doi: 10.1523/JNEUROSCI.1899-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tretter L, Adam-Vizi V. Generation of reactive oxygen species in the reaction catalyzed by alpha-ketoglutarate dehydrogenase. J Neurosci. 2004;24:7771–7778. doi: 10.1523/JNEUROSCI.1842-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suzuki K, Reed LJ. Lipoamidase. J Biol Chem. 1963;238:4021–4025. [PubMed] [Google Scholar]
- 66.Oizumi J, Hayakawa K. Lipoamidase is a multiple hydrolase. Biochem J. 1990;271:45–49. doi: 10.1042/bj2710045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feng D, Witkowski A, Smith S. Down-regulation of mitochondrial acyl carrier protein in mammalian cells compromises protein lipoylation and respiratory complex I and results in cell death. J Biol Chem. 2009;284:11436–11445. doi: 10.1074/jbc.M806991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cicchillo RM, Iwig DF, Jones AD, Nesbitt NM, Baleanu-Gogonea C, Souder MG, Tu L, Booker SJ. Lipoyl synthase requires two equivalents of S-adenosyl-L-methionine to synthesize one equivalent of lipoic acid. Biochemistry. 2004;43:6378–6386. doi: 10.1021/bi049528x. [DOI] [PubMed] [Google Scholar]
- 69.Miller JR, Busby RW, Jordan SW, Cheek J, Henshaw TF, Ashley GW, Broderick JB, Cronan JE, Jr, Marletta MA. Escherichia coli LipA is a lipoyl synthase: in vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoylacyl carrier protein. Biochemistry. 2000;39:15166–15178. doi: 10.1021/bi002060n. [DOI] [PubMed] [Google Scholar]
- 70.Morikawa T, Yasuno R, Wada H. Do mammalian cells synthesize lipoic acid? Identification of a mouse cDNA encoding a lipoic acid synthase located in mitochondria. FEBS Lett. 2001;498:16–21. doi: 10.1016/s0014-5793(01)02469-3. [DOI] [PubMed] [Google Scholar]
- 71.Wada H, Shintani D, Ohlrogge J. Why do mitochondria synthesize fatty acids? Evidence for involvement in lipoic acid production. Proc Natl Acad Sci USA. 1997;94:1591–1596. doi: 10.1073/pnas.94.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao X, Miller JR, Jiang Y, Marletta MA, Cronan JE. Assembly of the covalent linkage between lipoic acid and its cognate enzymes. Chem Biol. 2003;10:1293–1302. doi: 10.1016/j.chembiol.2003.11.016. [DOI] [PubMed] [Google Scholar]