Summary
Elevated catecholamines in the heart evoke transcriptional activation of the Myocyte Enhancer Factor (MEF) pathway to induce a cellular response known as pathological myocardial hypertrophy. We have discovered that the A-Kinase Anchoring Protein AKAP-Lbc is up-regulated in hypertrophic cardiomyocytes. It coordinates activation and movement of signaling proteins that initiate MEF2-mediated transcriptional reprogramming events. Live-cell imaging, fluorescent kinase activity reporters and RNA interference techniques show that AKAP-Lbc couples activation of protein kinase D (PKD) with the phosphorylation-dependent nuclear export of the class II histone deacetylase HDAC5. These studies uncover a role for AKAP-Lbc in which increased expression of the anchoring protein selectively amplifies a signaling pathway that drives cardiac myocytes towards a pathophysiological outcome.
Introduction
Heart disease is a complex disorder that is a leading cause of death worldwide. Unfavorable environmental factors and genetic profiles contribute to the etiology of several cardiac disorders. These factors can adversely modify heart muscle contraction by interfering with the fine-tuning of cardiomyocyte signal transduction cascades (Bassel-Duby and Olson, 2006). Activation of β-adrenergic receptors during periods of cardiac stress initially improves cardiac output by increasing heart rate and contractility. However chronic mobilization of the same signaling pathway ultimately harms the heart (Perrino and Rockman, 2007). This is compounded by increased risk factors such as arterial hypertension and valvular heart diseases that place additional biomechanical stress on the heart, inducing a cellular response known as pathological cardiac hypertrophy. Hallmarks of this syndrome include increased cardiomyocyte size and a greater organization of the sarcomere (Frey and Olson, 2003). At the molecular level, hypertrophic signals, such as elevated adrenergic activity, evoke transcriptional activation of the Myocyte Enhancer Factor (MEF) (Bassel-Duby and Olson, 2006). This induces a reprogramming of cardiac gene expression that drives cardiomyocytes toward a development paradigm known as the “fetal gene response” (Molkentin and Dorn, 2001).
A body of work suggests that signaling from the plasma membrane to the transcription factor MEF2 is altered during pathological cardiac hypertrophy (Black and Olson, 1998). MEF2 activity is controlled through its direct association with histone deacetylases (HDACs). These enzymes deacetylate nucleosomal histones, thus promoting chromatin condensation and transcriptional repression (Kouzarides, 2007). The class II histone deacetylases (HDACs 4, 5, 7 & 9) are preferentially expressed in muscle cells (Grozinger and Schreiber, 2000). Phosphorylation of these enzymes by calmodulin dependent (CaM) kinases or the related protein kinase D releases them from association with MEF2 to permit their export from nucleus (Backs et al., 2006; Bers and Guo, 2005; Sucharov et al., 2006; Vega et al., 2004; Wu et al., 2006). The ensuing de-repression of MEF2 activity favors transcription of cardiac genes that promote the fetal gene response causing a hypertrophic phenotype (Chang et al., 2005). Although each component in this pathway is well defined, much less is known about their integration into a dynamic cell-signaling cascade.
Cardiac A-Kinase Anchoring Proteins (AKAPs) play an active role in this process by incorporating protein kinases, protein phosphatases, guanine nucleotide exchange factors and phosphodiesterases (PDEs) into signaling complexes that respond to catecholamine evoked changes in the production of second messengers (Chen et al., 2007; Dodge-Kafka et al., 2005; Fink et al., 2001; Lygren et al., 2007). In cardiomyocytes AKAP-Lbc functions as a scaffolding protein for PKA and PKC to mediate activation of a third enzyme, protein kinase D (PKD1) (Carnegie et al., 2004). In this report we show that AKAP-Lbc is upregulated in response to hypertrophic stimuli and functions to enhance the efficiency of signaling through a PKD/HDAC5/MEF2 pathway that elicits the fetal gene response.
Results
AKAP-Lbc expression is elevated in hypertrophic NRVM
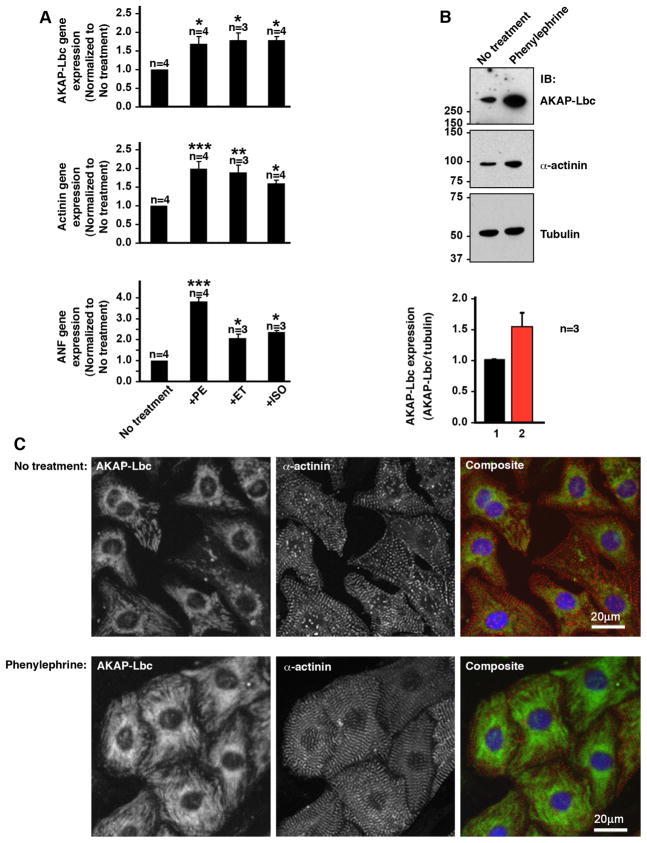
Initial experiments focused on establishing whether hypertrophic stimuli alter AKAP-Lbc expression. Neonatal rat ventricular myocytes (NRVM) were exposed to the α1 adrenergic agonist phenylephrine (PE, 10 μM) for 48 hours to evoke a cellular model of cardiac hypertrophy (Clerk and Sugden, 1999). Quantitative RT-PCR showed that PE induced AKAP-Lbc mRNA levels 1.7 ± 0.2 fold (n=4) over untreated cells (Fig 1A, top panel, column 2). Data were normalized against the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene. Comparable increases in AKAP-Lbc mRNA levels were measured in NRVM treated with other hypertrophic agents such as endothelin (ET, 100 nM) [1.8 ± 0.2, n=3] and isoproterenol (ISO, 10 μM) [1.8 ± 0.1, n=4] (Fig 1A, top panel, columns 3 & 4). Control experiments confirmed that each agonist up-regulated two hypertrophic marker genes, atrial natriuretic factor (ANF) and α-actinin (Fig 1A mid & bottom panels, columns 2–4).
Figure 1. AKAP-Lbc expression is elevated in hypertrophic rat neonatal cardiomyocytes.
A) RT-PCR analysis of gene expression from RNA prepared from rat neonatal cardiomyocytes (NRVM) treated with hypertrophic agonists. Cells were treated with agonists for 48hrs prior to lysis with TriZol and preparation of RNA. All gene expression data was normalized to GAPDH gene expression; AKAP-Lbc (top panel); α-actinin (middle panel) and ANF (bottom panel). Lane 1: no treatment. Lane 2: PE; phenylephrine (working concentration: 10 μM). Lane 3: ET; endothelin-1 (working concentration: 100 nM). Lane 4: ISO; isoproterenol (working concentration: 10 μM).
Data are expressed as mean ± SEM. Differences in quantitative variables were examined by one-way analysis of variance (ANOVA). A P value < 0.05 was considered significant (*), a P value < 0.01 was considered very significant (**) and a P value < 0.001 was considered extremely significant (***). All analyses were performed using InStat.
B) Immunoblot detection of AKAP-Lbc (top), α-actinin (middle) and tubulin (bottom) in control and PE-treated NRVM. Cells were treated with PE for 48 hrs prior to lysis. Quantitation of the average amounts of AKAP-Lbc was performed over three independent experiments. NIH Image J software was used to calculate the relative intensity of each AKAP-Lbc signal normalized to a tubulin loading control.
C) Immunocytochemical detection of AKAP-Lbc in control and PE treated NRVM. Cells were treated with PE for 48hrs prior to fixation, permeabilization and immunostaining. Immunodetection of AKAP-Lbc (green; left panels), immunostaining for the sarcomeric marker protein α-actinin (red; middle panels) and composite images with the nuclear stain DAPI (blue; right panels) are presented. Images were collected on a BioRad MRC 1024 confocal microscope using identical settings for control and hypertrophic NRVM.
Parallel studies monitored changes in AKAP-Lbc protein levels following PE treatment of NRVM (Fig 1B). AKAP-Lbc was elevated 1.5 ± 0.2 fold (n=3) over untreated controls. (Fig 1B, top panel, lane 2). Increased expression of α-actinin indicated hypertrophy (Fig 1B, mid panel, lane 2) and tubulin immunoblotting demonstrated equivalent protein loading (Fig 1B, bottom panel). Cellular changes in AKAP-Lbc were further evaluated by immunocytochemistry. Affinity purified antibodies were used to detect the anchoring protein (green) in control NRVM (Fig 1C). Cells were also stained with the sarcomeric marker α-actinin (red) and DAPI (blue) to denote the nucleus (Fig 1C, top panels). PE treatment increased cell size and enhanced the AKAP-Lbc staining concentrated in perinuclear regions when compared to untreated NRVM (Fig 1C). Taken together these data illustrate that AKAP-Lbc is elevated in hypertrophic cardiomyocytes. These results substantiate gene array data showing that this anchoring protein (also called AKAP13) is elevated in the hearts of mice with heart failure (van Oort et al., 2006).
Gene silencing of AKAP-Lbc in NRVM
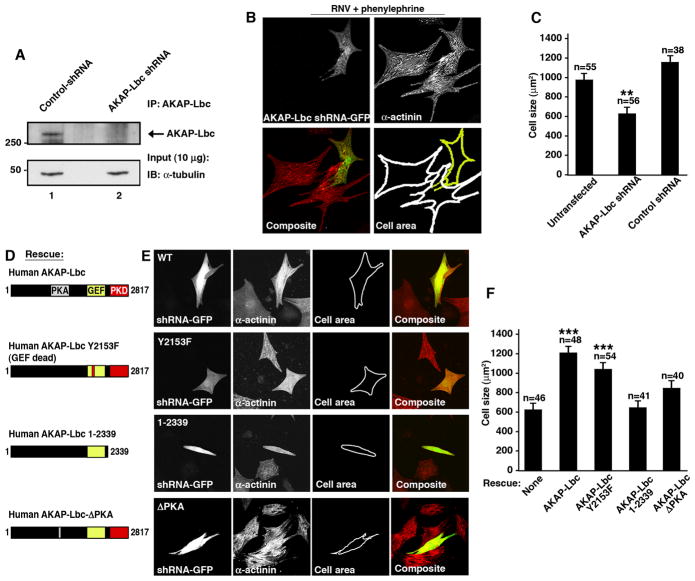
RNA interference (RNAi) was used to test whether reduced expression of the anchoring protein perturbed aspects of the hypertrophic response. Lentiviral vectors were used to maximize the infection of NRVM with shRNA-GFP constructs (Appert-Collin et al., 2007). Gene silencing was confirmed upon immunoblot analysis of AKAP-Lbc immune complexes (Fig 2A, top panel, lane 2). Detection of tubulin was used as a loading control (Fig 2A, bottom panel). Cells expressing the AKAP-Lbc shRNA constructs were identified by GFP fluorescence (Fig 2B, top left panel). Immunostaining for α-actinin identified all NRVM (Fig 2B, top right panel). A composite image is presented (Fig 2B, bottom left panel). Phenylephrine effects were assessed by two independent methods: measuring changes in cell size (Kodama et al., 2000), (Fig 2B, bottom right panel) and immunocytochemical detection of ANF (Supplementary data S1). Following treatment with PE, the cells transfected with control shRNA were 1.8 ± 0.2 fold larger (n=38) than NRVM expressing the rat AKAP-Lbc-shRNA/GFP construct (Fig 2C, columns 2 & 3). Likewise, significant ANF expression was induced by PE treatment of control shRNA cells, whereas induction of ANF was barely detected in cells expressing the AKAP-Lbc shRNA construct (Supplementary data S1). Thus, gene silencing of AKAP-Lbc blunts PE-induced hypertrophic responses in NRVM.
Figure 2. Effects of RNAi knockdown of AKAP-Lbc in NRVM.
A) Immunoblot showing RNAi knockdown of AKAP-Lbc in NRVM. Endogenous AKAP-Lbc was immunoprecipitated from NRVM lysates after 72hrs exposure to shRNA. Proteins were separated by SDS-PAGE and detected by Western blot. AKAP-Lbc (top panel), tubulin loading control (lower panel).
B) Detection of AKAP-Lbc shRNA/GFP and measurement of cell size in hypertrophic NRVM. NRVM expressing AKAP-Lbc shRNA/GFP were detected by green fluorescence (top left panel). NRVM were fixed and stained for α-actinin (red, top right panel) 72hrs after transfection of shRNA and incubation with PE (for 48hrs). A composite image is presented (bottom, left panel). The outlines for control NRVM (white) and cells infected with the AKAP-Lbc shRNA-GFP (yellow) are indicated (bottom, right panel).
C) The cell size of untransfected NRVM, cells expressing AKAP-Lbc shRNA/GFP, or a control shRNA was measured using NIH Image J software. NRVM were treated with PE for 48 hrs prior to analysis. The amalgamated data are presented (number of cells measured in each group is indicated).
D) Schematic diagram of human AKAP-Lbc constructs used to rescue the anchoring protein shRNA phenotype: hAKAP-Lbc, hAKAP-Lbc-Y2153F (a GEF-dead mutant), hAKAP-Lbc-ΔPKA (a mutant unable to bind PKA) and AKAP-Lbc 1-2339 (a C-terminal truncation mutant that is unable to bind PKD; Supplementary data S3).
E) Measurement of cell size in hypertrophic cells expressing AKAP-Lbc shRNA/GFP with human AKAP-Lbc rescue constructs. NRVM were fixed and stained for α-actinin (red) 72hrs after transfection of shRNA and incubation with PE. NRVM expressing AKAP-Lbc shRNA/GFP were detected by green fluorescence. Co-expression of rescue constructs was verified by anti-FLAG immunostaining (Supplementary data S2c).
F) Measurement of cell size following PE treatment. Control NRVM (column 1), NRVM expressing hAKAP-Lbc (column 2), hAKAP-Lbc-Y2153F (column 3), AKAP-Lbc 1-2339 (column 4) and AKAP-Lbc-ΔPKA (column 5).
Additional support for this notion was provided by experiments that combined RNAi silencing of the endogenous anchoring protein and rescue with human AKAP-Lbc forms that are refractory to the rat-specific shRNA (Fig 2D–F). Rescue with full-length human AKAP-Lbc ortholog induced a 1.9 ± 0.1 fold increase (n=48) in average cell size compared to NRVM where the endogenous rat anchoring protein was knocked down (Fig 2D–F). AKAP-Lbc is a chimeric molecule that functions as a kinase anchoring protein and a guanine nucleotide exchange factor (GEF) for the small molecular weight GTPase Rho (Diviani et al., 2001). Both enzyme classes have been implicated in the onset of cardiac hypertrophy (Clerk and Sugden, 2000). Therefore, gene silencing/rescue experiments were performed with modified AKAP-Lbc forms to delineate the contribution of each signaling pathway in the PE response. When NRVM were rescued with the AKAP-Lbc Y2153F mutant that lacks Rho GEF activity, we observed an agonist-induced increase in cell size (1.7 ± 0.1 fold, n=54, Fig 2D–F) and elevated ANF levels (Supplementary data S2). A smaller agonist-induced increase in cell size (1.2 ± 0.2 fold, n=40, Fig 2D–F) was observed when NRVM were rescued with the AKAP-Lbc-ΔPKA mutant that cannot anchor PKA. Thus anchoring of PKA may also contribute to this process. However a more significant observation was that phenylephrine induced increases in NRVM size and ANF expression were blocked when rescue experiments were performed with a truncated AKAP-Lbc form (AKAP-Lbc 1-2339) that does not bind PKD1 (Fig 2D–F & Supplementary data S2 & S3). These latter results imply that the PKD scaffolding function of AKAP-Lbc is the principle factor in transduction of the hypertrophic response. Additional mapping experiments identified multiple PKD1 binding sites in the c-terminal quarter of AKAP-Lbc (Supplementary data S4).
AKAP-Lbc facilitates nuclear export of HDAC5
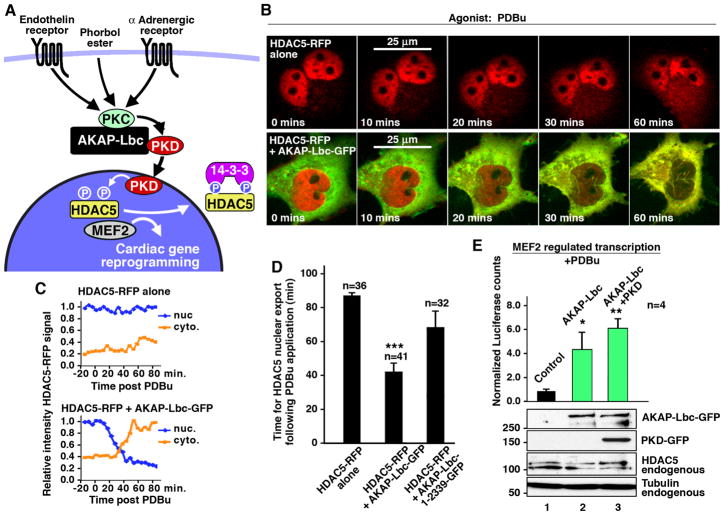
A variety of kinase signaling pathways induce cardiac gene reprogramming during hypertrophy. A major pathway involves phosphorylation of class II HDACs (HDACs 4, 5, 7 & 9) by the calmodulin dependent kinases CaMKII or CaMKIV, or by PKD (McKinsey, 2007). Phosphorylation of HDACs promotes their nuclear export, resulting in de-repression of MEF2 activity and concomitant activation of target genes to propagate the hypertrophic response (Fig 3A). Recent investigations suggest that CaM kinases selectively target HDAC4 whereas PKD1 preferentially regulates HDAC5 action (Backs et al., 2006; Wu et al., 2006). Therefore we postulated that AKAP-Lbc may facilitate the activation of the PKD1 isoform may enhance nuclear export of HDAC5.
Figure 3. Imaging AKAP-Lbc mediated nuclear export of HDAC5 and regulation of transcription in Cos7 cells.
A) Schematic diagram depicting the regulation of MEF2 transcription in response to hypertrophic agonists. We postulate that AKAP-Lbc plays a central role in this pathway, facilitating activation of PKD, causing phosphorylation and subsequent nuclear export of HDAC5, thereby leading to de-repression of MEF2 transcription and cardiac gene reprogramming events.
B) Live-cell imaging techniques were used to monitor the cellular location of HDAC5 in Cos7 cells treated with phorbol ester (PDBu). PDBu was applied to Cos7 cells at 0 mins and images were collected at designated times (indicated in each frame). Representative images from cells expressing HDAC5-RFP (red; top panels) and cells co-expressing HDAC5-RFP with AKAP-Lbc-GFP (green; bottom panels).
C) Quantitation of the relative nuclear and cytoplasmic intensity of HDAC5-RFP signal carried out using NIH Image J software.
D) Amalgamated data indicating the time of HDAC5 nuclear export in Cos7 cells following application of PDBu. Data are expressed as mean ± SEM. Differences in quantitative variables were examined by ANOVA using the software InStat.
E) Analysis of MEF2-regulated transcriptional activity in response to PDBu in Cos7 cells co-expressing AKAP-Lbc (lane 2) and PKD + AKAP-Lbc (lane 3). A Western blot of lysates shows even loading across samples (tubulin loading control), equal ectopic expression of AKAP-Lbc in lanes 2 and 3 and PKD in lane 4.
Initial support for this model was provided by live-cell imaging experiments in Cos7 cells. We monitored the movement of fluorescently tagged HDAC5 in response to phorbol ester (PDBu, 50 nM), a pharmacological activator of PKD (Waldron and Rozengurt, 2003). NIH Image-J software was used to measure the relative intensity of fluorescent signals in defined regions of the nucleus and cytoplasm of each cell. Measurements were conducted at 5 minute intervals for the duration of the experiments (Fig 3B–D). There was no discernable movement of HDAC5-red fluorescent protein (RFP) in control cells for up to 81.3 ± 7 min (n=36) after the application of PDBu. Yet the mean time for HDAC5-RFP nuclear export in the presence of AKAP-Lbc-GFP was 42 ± 5 min (n= 41 cells) after the application of PDBu (Fig 3B–D and Supplementary Movies 3B). Control experiments were performed with the AKAP-Lbc 1-2339-GFP fragment that is unable to interact with PKD1. The mean time for HDAC5-RFP nuclear export was 67 ± 9 min (n= 32 cells) after the application of PDBu and suggested that AKAP-Lbc influences PKD1 to enhance nuclear export of the histone deacetylase (Fig 3D).
Further studies were performed to assess the contribution of the GEF domain in AKAP-Lbc and CaM kinases in this process. The rate of HDAC5-RFP nuclear export either in the presence of AKAP-Lbc Y2153F (Supplementary data S5) or in cells pretreated with the CaM kinase inhibitor KN93 (Supplementary data S6) was similar to cells expressing AKAP-Lbc-GFP. This suggests that AKAP-Lbc-mediated nuclear export of HDAC5 proceeds via a mechanism that involves activation of PKD1 and that this can occur in a manner that is independent of Rho activation and CaM Kinase activity.
Nuclear export of class II HDACs triggers a fetal gene response in cardiomyocytes through stimulation of a MEF2 transcriptional activation pathway. Therefore we used a luciferase reporter (provided by Dr. Eric Olson, University of Texas-Southwestern) to examine MEF2-mediated transcription. Cos7 cells express low levels of AKAP-Lbc, PKD and the histone deacetylase thereby providing an ideal model cell line to study the role of this pathway in transcriptional activation using the MEF2 reporter. PDBu-dependent stimulation of MEF2 transcription was increased 4.9 ± 1.7 fold (n=4) in the presence of AKAP-Lbc and was further enhanced to 6.9 ± 1.4 fold (n=4) upon overexpression of PKD with the anchoring protein (Fig 3E). Controls did not detect any PDBu-dependent stimulation of transcriptional activity driven by a β-galactosidase reporter. These findings strengthen our view that AKAP-Lbc mediated activation of PKD can stimulate MEF2 transcription.
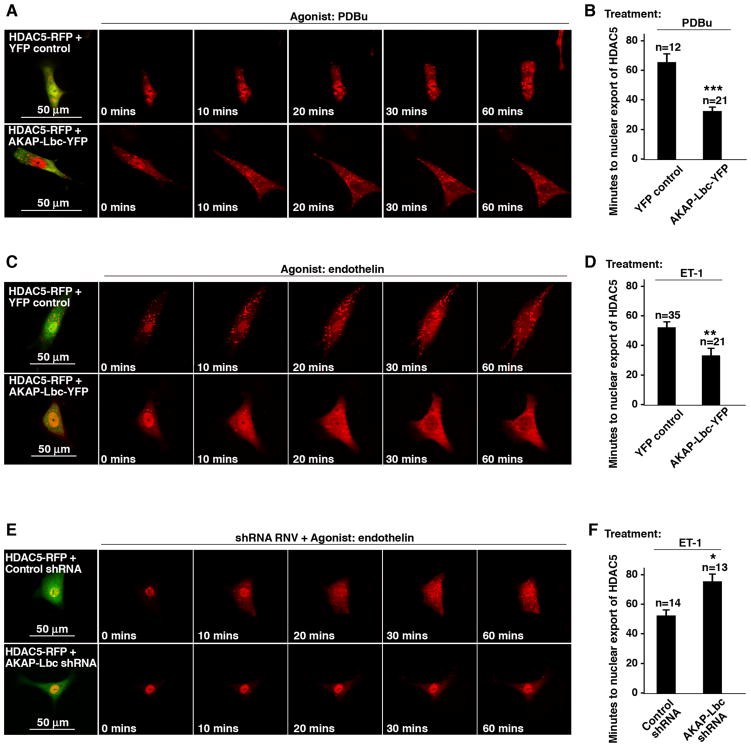
The remainder of our analyses were conducted in primary cultures of NRVM to probe the spatiotemporal dynamics of this signaling pathway in a more appropriate cellular context. Nuclear export of HDAC5-RFP was complete within 66 ± 6 min (n=12 cells) after application of PDBu in control NRVM (Fig 4A; top panels & Fig 4B, column 1). In cells expressing AKAP-Lbc-YFP a more rapid PDBu-dependent nuclear export of HDAC5-RFP was observed. Movement of HDAC5 from the nucleus was complete within 33 ± 3 min (n= 21 cells) (Fig 4A & B and Supplementary Movies 4A).
Figure 4. AKAP-Lbc mediated HDAC5 nuclear export in NRVM.
A) Time course of HDAC5-RFP (red) nuclear export in response to PDBu (top panels) and nuclear export of HDAC5-RFP when co-expressed with AKAP-Lbc-YFP (bottom panels). PDBu was applied to cells at 0 mins. AKAP-Lbc-YFP and control cells were identified by YFP fluorescence (first panels).
B) Amalgamated data indicating the time of HDAC5 nuclear export following application of PDBu in NRVM expressing HDAC5-RFP and YFP (control cells, column 1) or co-expressing HDAC5-RFP with AKAP-Lbc-YFP (column 2). Data are expressed as mean ± SEM. Differences in quantitative variables were examined by ANOVA, or an unpaired two-tailed t test. All analyses were performed using InStat.
C) Time course of HDAC5-RFP (red) nuclear export in NRVM in response to endothelin-1 (ET) (top panels) and nuclear export of HDAC5-RFP when co-expressed with AKAP-Lbc-YFP (bottom panels). ET-1 was applied to cells at 0 mins.
D) Amalgamated data indicating the time of HDAC5 nuclear export following application of ET-1 in NRVM expressing HDAC5-RFP and YFP (control cells, lane 1) or co-expressing HDAC5-RFP with AKAP-Lbc-YFP (column 2). Data are expressed as mean ± SEM. Differences in quantitative variables were examined by ANOVA, or an unpaired two-tailed t test using Instat.
E) The time course of ET-1 nuclear export HDAC5-RFP (red) in NRVM co-transfected with control shRNA (top panels) or NRVM co-transfected with AKAP-Lbc-shRNA (green; bottom panels). ET-1 was applied to cells at 0 mins.
F) Amalgamated data indicating the time of HDAC5 nuclear export following application of ET-1 in NRVM expressing HDAC5-RFP and control shRNA (column 1) or co-expressing HDAC5-RFP with AKAP-Lbc-shRNA (column 2). Data are expressed as mean ± SEM. Differences in quantitative variables were examined by ANOVA, or an unpaired two-tailed t test using Instat.
Similar data was obtained upon stimulation with endothelin-1 (ET-1, 100 nM), a potent vasoconstrictor that is implicated in the onset of myocardial inflammation and hypertrophy (Sugden and Clerk, 2005; Teerlink, 2005). Endothelin-dependent movement of HDAC5-RFP took 53.3 ± 4 min (n=35 cells) in control cells, but was reduced to 34 ± 5 min (n= 21 cells) upon overexpression of the anchoring protein (Fig 4C & D and Supplementary Movies 4C). Finally, we measured the rate of ET-1 dependent HDAC5-RFP nuclear export in cells in which the endogenous rat AKAP-Lbc expression was silenced by RNAi. AKAP-Lbc shRNA and control shRNA cells were identified by the co-expression of GFP as a marker (Fig 4E, first panels). Endothelin-dependent nuclear export of HDAC5-RFP was significantly delayed upon depletion of the anchoring protein [75 ± 5 min, n=13] as compared to shRNA controls [53 ± 8 min, n=14] (Fig. 4E & F and Supplementary Movies 4E). These data reinforce our contention that AKAP-Lbc contributes to the enhancement of HDAC5 nuclear export in cardiomyocytes.
In situ analysis of PKD activity
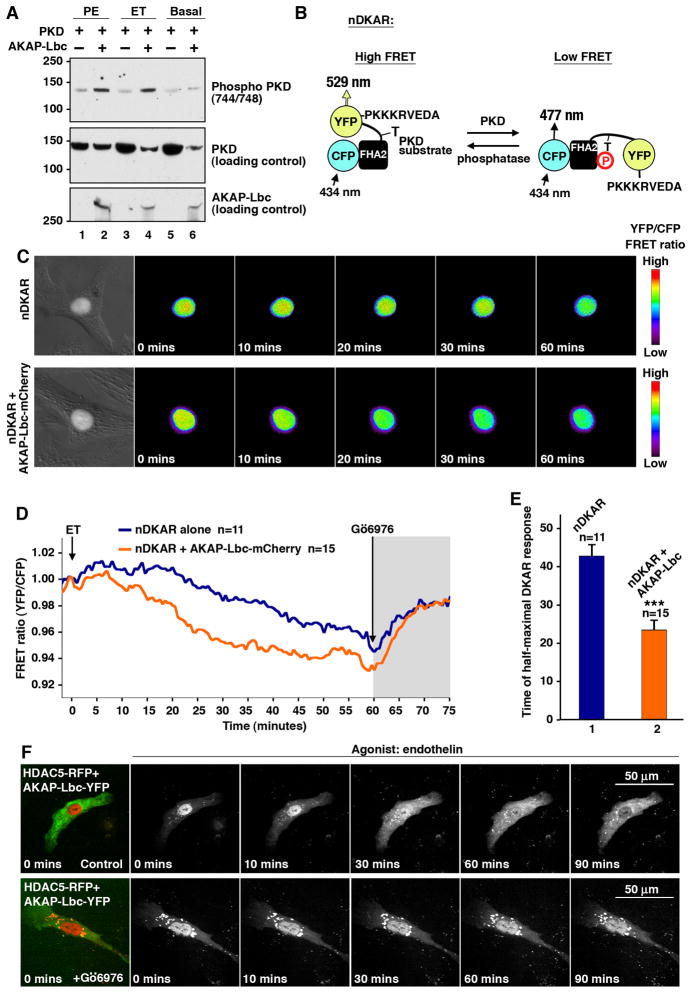
Our working hypothesis is that AKAP-Lbc mediated activation of PKD potentiates the hypertrophic response. Accordingly, overexpression of AKAP-Lbc enhanced activation of the kinase as assessed by immunoblot using anti-phospho-PKD Ser 744/748 antibodies when phenylephrine, endothelin or PDBu was used as the agonist (Fig 5A & Supplementary data S7). The time course of these events was further evaluated using a genetically encoded fluorescence based D-Kinase Activity Reporter (DKAR) (Kunkel et al., 2007). PKD phosphorylation of a consensus substrate sequence (LSRQLTAAVSE) results in a decrease in Fluorescence Resonance Energy Transfer (FRET) as measured by dynamic changes in the YFP/CFP ratio (Fig. 5B).
Figure 5. In situ analysis of PKD activity.
A) Activation of PKD by AKAP-Lbc. HEK293 cells expressing PKD alone or co-expressing PKD with AKAP-Lbc were untreated or stimulated with PE, or ET-1 prior to lysis. Proteins were separated by SDS-PAGE. PKD activation was assessed by immunoblot using anti-phospho-Ser-744/748 PKD antibodies (top panel). Loading controls for total PKD (middle panel) and AKAP-Lbc (bottom panel) are presented.
B) Diagram showing the mechanism of action for the nuclear D Kinase Activity Reporter (nDKAR). A decrease in Fluorescence Resonance Energy Transfer (FRET) equates to an increase in PKD activity.
C) Pseudo-color images following the time course changes in the nDKAR FRET ratio (YFP/CFP) in NRVM upon stimulation with ET-1. The first panels are brightfield images of the cells, showing the location of nDKAR. Top panels show a typical response of nDKAR after application of ET-1. The bottom panels show a typical response of nDKAR in NRVM co-expressing AKAP-Lbc.
D) Amalgamated traces of nDKAR FRET ratio in response to ET-1 in NRVM expressing nDKAR alone (blue line; n=11 cells) or co-expressing nDKAR and AKAP-Lbc (orange line; n=15 cells). ET-1 was added at 0 mins. The selective inhibitor Go 6976 was added after 60 mins to block PKD activity.
E) The time of half-maximal nDKAR response was measured and averaged for cells expressing nDKAR alone (column 1) or cells co-expressing nDKAR and AKAP-Lbc (column 2). Data are expressed as mean ± SEM. Differences in quantitative variables were examined by ANOVA, using the software InStat.
F) Top panels: time course showing nuclear export of HDAC5-RFP when co-expressed with AKAP-Lbc-YFP in NRVM in response to endothelin-1 (ET). Bottom panels: nuclear export of HDAC5-RFP when co-expressed with AKAP-Lbc-YFP in cells treated with the inhibitor Go 6976 prior to ET-1 application (at 0 mins).
A nuclear localization signal (PKKKRKVEDA) was engineered into the parent DKAR construct. This allowed us to selectively monitor PKD activity in the nucleus where HDAC5 is a likely substrate. ET-1 was applied to NRVM at time zero and fluorescent images were collected at 30 sec intervals for 90 minutes (Fig 5C–E). Increased PKD activity was scored as a decrease in the YFP/CFP ratio in a defined area of the nucleus. Half-maximal responses were reached within 44 ± 3 min (n=11) following application of ET-1 in NRVM expressing nDKAR alone (Fig 5C upper panels, Fig 5D blue line & Fig 5E column 1). However, the rate and magnitude of the DKAR response was increased in NRVM expressing AKAP-Lbc-mCherry showing a half maximal activation within 25 ± 3 min (n=15, Fig 5C lower panels, Fig 5D orange line & Fig 5E column 2). Control experiments confirmed that the nDKAR FRET ratio was reversed upon application of Gö6976 (500 nM), the most selective, commercially available PKD inhibitor (Fig 5D). Hence AKAP-Lbc acts as a catalyst to favor PKD activation and apparently increases the pool of active enzyme in the nucleus. Both factors drive NRVM toward the nuclear exclusion of HDAC5. Additional imaging experiments confirmed that pre-treatment of NRVM with Gö6976 blocks agonist-induced nuclear export of HDAC5-RFP when compared to controls (Fig 5F).
PKA and PKD catalyze the phosphorylation dependent recruitment of 14-3-3 and nuclear export of HDAC5
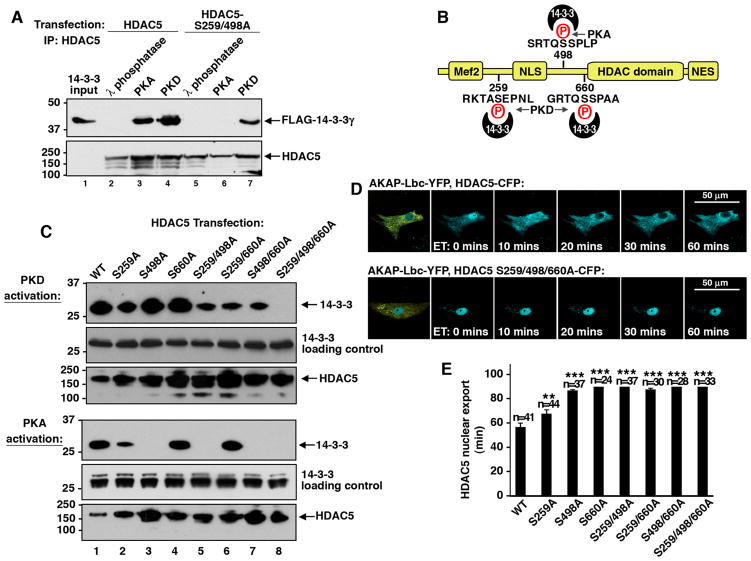
A pair of consensus 14-3-3 binding sites have been identified that encompass serine residues 259 and 498 on HDAC5 (McKinsey et al., 2000). Sequence analysis of these regions showed that serine 259 lies within the context of a PKD substrate motif whereas the amino acid residues surrounding serine 498 form a PKA phosphorylation site. Radiolabelling of HDAC5 with purified kinases and peptide array analysis was used to further define the relevant phosphorylation sites on the histone deacetylase (Supplementary data S8). In vitro binding studies confirmed that phosphorylation of HDAC5 by either PKA or PKD1 permitted the recruitment of 14-3-3 (Fig 6A, lanes 1–4). However, 14-3-3 still bound to an HDAC5 S259/498A double mutant upon phosphorylation by PKD1 (Fig 6A, lanes 5–7). This inferred that additional 14-3-3 binding sites must reside on HDAC5. Upon further inspection another consensus 14-3-3 binding motif was identified that included a possible PKD substrate site at serine 660 (Fig 6B). Cell based studies with a family of HDAC5 point mutants confirmed this hypothesis (Fig 6C bottom panel, lane 8). PKD1 dependent recruitment of 14-3-3 was abolished only when all three sites were inactivated by the introduction of a non-phosphorylatable alanine residue (Fig 6C top panel, lane 8). This differed from the PKA dependent recruitment of 14-3-3 that was lost in HDAC5 mutants lacking Ser 498 (Fig 6C, bottom panels, lanes 3, 5, 7 & 8). These observations were consistent with live-cell imaging experiments conducted with CFP tagged versions of the HDAC5. Any HDAC5 point mutant that lacked either Ser 498 or Ser 660 was not exported from the nucleus when compared to the wild-type control (Fig 6D and Supplementary Movies 6D). In contrast mutation of serine 259 to alanine minimally reduced the rate of HDAC nuclear export from 57 ± 3 min (n=41) to 68 ± 3 min (n=44) when compared to the wild type control (Fig 6E, column 2). Thus serine residues 498 and 660 may represent the principal phosphorylation sites on HDAC5 that are required for the 14-3-3 dependent nuclear export. Furthermore, two AKAP-Lbc associated kinases PKA and PKD1 may cooperate to promote movement of HDAC5 into the cytoplasm.
Figure 6. Analysis of HDAC5 phosphorylation and induction of 14-3-3-binding.
A) Phosphorylation of HDAC5 by PKA or PKD induces 14-3-3 binding. Epitope tagged-HDAC5 (lanes 2–4) or an HDAC5-S259/498A double mutant (lanes 5–7) were immunoprecipitated from HEK293 cells and incubated with cell lysates expressing FLAG-14-3-3γ (top panel). 14-3-3 binding was assessed under conditions where HDAC5 was either de-phosphorylated by incubation with λ-phosphatase (lanes 2 & 5) or phosphorylated by PKA (lanes 3 & 6) or PKD (lanes 4 & 7) in vitro.
B) Diagram of phosphorylation sites that regulate 14-3-3 binding on HDAC5. Consensus 14-3-3 binding sites are denoted. Kinase phosphorylation sites are indicated in red.
C) Mapping the essential 14-3-3 binding sites on HDAC5. A family of seven HDAC5 mutants where serines 259, 498 and 660 were systematically replaced with alanine were screened for interaction with 14-3-3 in HEK293 cells. Experiments were performed using PDBu to activate PKD (top group) or with forskolin/IBMX to activate PKA (bottom group). The presence of 14-3-3 in HDAC5 immune complexes was assessed by immunoblot (top panels). Loading controls for 14-3-3 (middle panels) and HDAC5 (bottom panels) are presented.
D) Cellular analysis of 14-3-3 binding sites on HDAC5 nuclear export. The movement of each HDAC5 14-3-3 binding mutant was analyzed in NRVM. Representative images of a time course depicting ET-1-dependent response of HDAC-CFP (top panels) and the HDAC5-CFP S259S, S498A, S660A triple mutant (bottom panels).
E) Graph of amalgamated data from experiments measuring the rate of nuclear export (mins) of each HDAC5 mutant. Statistical significance and number of cells measured is indicated above each column.
Discussion
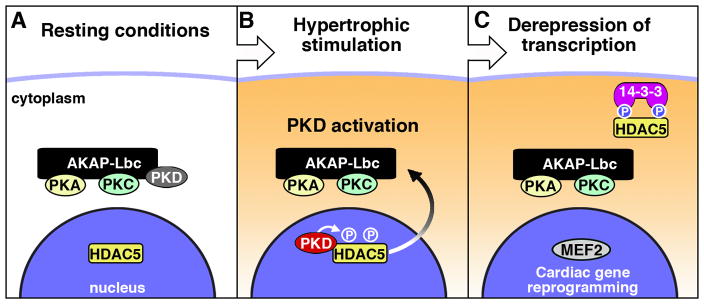
We propose that AKAP-Lbc is a previously unrecognized factor that selectively directs catecholamine signals to the transcriptional machinery to potentiate the hypertrophic response. The anchoring protein is both up-regulated under pathophysiological conditions and organizes a signaling pathway that contributes to the onset of myocardial hypertrophy. Although most of our study was conducted in neonatal rat cardiac myocytes, further support for this concept was provided by analysis of human heart tissue samples obtained post mortem from two individuals exhibiting hypertrophic myopathy; a 46 year old male, with a left ventricular ejection fraction (LVEF) of 42%, and a 62 year old female, LVEF 12%. A 2 ± 0.5 fold increase in AKAP-Lbc mRNA was measured over normal age-matched patient controls (Supplementary data S9). While induction of cardiomyopathy is a multifaceted process, we contend that AKAP-Lbc mobilizes a subset of effectors to enhance progression of this syndrome. Our data support a model where AKAP-Lbc facilitates activation of protein kinase D, which in turn phosphorylates the histone deacetylase HDAC5 to promote its nuclear export (Fig 7A & 7B). Finally, the concomitant reduction in nuclear histone deacetylase activity favors MEF2 transcription and the onset of cardiac hypertrophy (Fig 7C).
Figure 7.
Model showing the role of AKAP-Lbc in the regulation of cardiac hypertrophy, through the PKD-HDAC5-MEF2 signaling pathway.
In response to signals from α1-adrenergic and endothelin receptors AKAP-Lbc facilitates activation of anchored kinases that mobilize the transcriptional machinery. Signals downstream of the α1-AR can also stimulate the small molecular weight GTPase Rho during hypertrophy (Maruyama et al., 2002). Additionally, several Rho effectors were identified in a genetic screen for modulators of class II HDAC phosphorylation (Chang et al., 2005). These included Rho-GEF1/p115-GEF, also known as “Lbc’s second cousin” (Lsc; mouse ortholog) on the basis of its homology to the anchoring protein. Since AKAP-Lbc also possesses intrinsic Rho guanine nucleotide exchange factor (GEF) activity (Diviani et al., 2001) it was important to delineate whether hypertrophic agonists exert their effects through activation of anchored kinases or via the stimulation of Rho. Two findings point toward kinase scaffolding as the predominant mechanism. First, AKAP-Lbc RNAi/rescue experiments showed that PKD binding is necessary for PE induced hypertrophy, while disruption of the AKAP-Lbc GEF activity had minimal effect (as assessed by cell size or induction of ANF; Fig 2E & F, Supplementary data S2). Second, live-cell imaging showed that disruption of the GEF activity in AKAP-Lbc had no effect on the rate of agonist-dependent HDAC5-RFP nuclear export when compared with the wild type anchoring protein (Supplementary data S5).
While activation of PKD appears to be the primary function of AKAP-Lbc, it should be noted that the anchoring protein may independently contribute to Rho activation through a Gα12 pathway. It has recently been reported that overexpression of dominant interfering mutants of Gα12 reduced phenylephrine-induced changes in cardiomyocyte size (Appert-Collin et al., 2007). Therefore AKAP-Lbc may provide a locus for crosstalk between the PKD and Rho signaling pathways in a context dependent manner. In the immune system Rho is required for PKD’s effects on pre-T cell differentiation at the membrane but is not necessary for PKD activation in the cytoplasm of mature T cells (Mullin et al., 2006). Indeed, AKAP-Lbc may function as a pleiotropic effector by integrating PKD and Rho action in certain subcellular locations while segregating these signaling pathways in other regions of the cell.
Live-cell imaging with FRET based kinase activity reporters and fluorescently tagged proteins offer different perspectives on the same cellular process. This has enabled us to correlate changes in nuclear PKD activity with HDAC nuclear export. We now have a clearer picture of how individual steps in this hypertrophic signaling pathway are coupled. Overexpression of AKAP-Lbc increases the rate and strength of PKD activation (Fig 5D) and accelerates the nuclear export of HDAC5 (Fig 4A–D). On the contrary a reduction in AKAP-Lbc expression (by shRNA) virtually abolishes the movement of the histone deacetylase. Technical limitations precluded the use of DKAR in cells expressing the AKAP-Lbc shRNA with a GFP marker. Regardless, these experiments provide some of the most compelling evidence to date on how AKAPs organize discrete molecular events that relay signals from one cellular compartment to another.
HDACs are primed for nuclear export upon the phosphorylation-dependent recruitment of 14-3-3 proteins (Grozinger and Schreiber, 2000; Kao et al., 2001; McKinsey et al., 2001). Our data add to the understanding of this concept in two ways. First of all, we show that PKA phosphorylation of serine 498 on HDAC5 facilitates 14-3-3 binding. Since PKA and PKD are components of the AKAP-Lbc scaffold it would appear that HDAC5 phosphorylation may represent a point of convergence for the phospholipid and cAMP signaling pathways. However, PKA phosphorylation alone is not sufficient to induce nuclear export as treatment with isoproterenol to elevate intracellular cAMP has no effect on HDAC5 translocation in NRVM (J.S & G.K.C personal communication).
Secondly, we have characterized an additional and physiologically relevant 14-3-3 interaction site at serine 660 of the histone deacetylase. This site acts in conjunction with the previously recognised sites at serines 259 and 498. Phosphorylation of two of these sites is required for nuclear export as mutation of either Serine 498 or Serine 660 blocks HDAC5 movement. In contrast, mutation of Ser 259 reduces 14-3-3 binding but has a negligible effect on HDAC5 movement (Fig 6B & E). These observations tally with current models for 14-3-3 action where the dimeric protein sequentially binds a pair of phosphoserine motifs on its binding partner to elicit a biological response (Yaffe, 2002). The initial binding event occurs at a “gatekeeper site” which in the case of HDAC5 could be either Ser 498 or Ser 660. Taken together these findings further substantiate the view that PKD is the principle HDAC5 kinase.
As the concept of kinase anchoring has gained acceptance an underlying question is how AKAPs influence physiologically important phosphorylation events. One emerging theme is that aberrant signaling through AKAP complexes contributes to certain heart diseases (Diviani, 2007; McConnachie et al., 2006). For example, Yotiao anchors PKA and the phosphatase PP1 with the KCNQ1 potassium channel subunit to shape the duration of cardiac action potentials in response to β-adrenergic agonists (Chen et al., 2005; Marx et al., 2002; Westphal et al., 1999). Inherited mutations in Yotiao or the KCNQ1 channel subunit have been linked to Long QT Syndrome, a rare genetic disease characterized by stress induced cardiac arrhythmias and sudden death (Chen et al., 2007; Kass and Moss, 2003). Likewise, the cardiac muscle-selective anchoring protein, mAKAP, assembles a cAMP signaling module consisting of PKA, PDE4D3, and Epac-1 (Dodge et al., 2001). A larger mAKAP-ryanodine receptor (RyR) complex controls excitation-contraction coupling at the sarcoplasmic reticulum (Marx et al., 2001). Pathophysiological changes in the enzyme composition of this mAKAP-RyR complex that alter the phosphorylation state of the channel have been linked to the onset of certain forms exercise induced arrhythmias in mice (Lehnart et al., 2005). We now implicate AKAP-Lbc as a co-factor that amplifies PKD signaling to HDAC5 to initiate a hypertrophic phenotype. Myocardial hypertrophy is extremely common, affecting 14–18% of the general adult population (Benjamin and Levy, 1999; World Health Organization). Therefore it will be important to explore the role of the AKAP-Lbc/PKD/HDAC5 signaling pathway in whole animal models, to establish whether AKAP-Lbc is a valid biomarker for hypertrophic cardiomyopathy and to determine which genes are initiated upon up-regulation of the anchoring protein.
Experimental Procedures
All Western blotting, RII overlays, immunoprecipitations and phosphorylation experiments were performed as described previously (Westphal et al., 1999).
Cell culture, immunocytochemistry and live cell imaging
Cos7 cells were cultured on glass coverslips. All transfections were carried out using Effectene reagent (Qiagen, Valencia, CA). Neonatal rat cardiomyocytes were cultured as described in (Pare et al, 2005). After 1 day in culture, cells were treated with vehicle or agonist for a further 48hrs at 37°C, prior to lysis and evaluation of AKAP-Lbc expression by Western blot or RT-PCR.
Preparation of primary NRVM and confocal microscopy experiments, NRVM were as described in (Dodge-Kafka et al., 2005). Cells were fixed in 3.7% paraformaldehyde in PBS followed by staining for α-actinin or ANF. Secondary antibodies used were from Jackson Immunoresearch. To determine the effect of shRNA expression on cellular hypertrophy, myocytes were co-transfected with rat AKAP-Lbc shRNA-GFP or control (human AKAP-Lbc) shRNA plasmid and pEYFP (Clontech). For rescue experiments FLAG-human AKAP-Lbc constructs were used. Anti-actinin was used to stain sarcomeric Z-disks to distinguish myocytes from contaminating fibroblasts. Images were acquired using a BioRad MRC1024 confocal microscope.
For all live cell imaging experiments NRVM were cultured on glass coverslips coated with 1% gelatin (Sigma). For ectopic expression of protein, NRVM were electroporated using a modified Amaxa Nucleofector protocol. After incubation at 37 °C for 18–48 h post electroporation, cells were washed twice with Hank’s balanced salt solution, mounted in a Ludin chamber (Life Imaging Services) and imaged using a Leica AS MDW workstation. Images were acquired as described in (Dodge-Kafka et al., 2005). Cells were treated with ET-1 (100 nM working concentration) for 60 mins prior to treatment with the PKC and PKD inhibitor Go 6976 (500 nM) to validate that we were measuring changes in PKD activity rather than other basophilic kinases. Nuclear phosphatase activity was inhibited by pretreatment with calyculin A (10 nM). Additional experiments were performed using cells that were pretreated with the CaMK inhibitor KN93 (5 μM working concentration for 30 mins) to ensure that nDKAR was not detecting CaMK activity in NRVM.
Statistical analyses
All data are expressed as mean ± standard error of the mean. Differences in quantitative variables were examined by one-way analysis of variance (ANOVA), or an unpaired two- tailed t test. A P value < 0.05 was considered significant (*), a P value < 0.01 was considered very significant (**) and a P value < 0.001 was considered extremely significant (***). All analyses were performed using InStat.
Note: Experimental details for RT-PCR, expression constructs used, in vitro HDAC5 phosphorylation, 14-3-3 binding assays and MEF2-luciferase assays are included in the Supplementary data.
Supplementary Material
Acknowledgments
This work was supported by the Fondation Leducq and the NIH (J.D.S. HL088366, A.C.N. DK54441). We thank Eric N. Olson for providing the MEF2 luciferase reporter and HDAC5-GFP constructs, Qinghong Zhang & Richard Goodman for help with luciferase assays, Lisette Maddison & Wenbiao Chen for assistance with RT-PCR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Appert-Collin A, Cotecchia S, Nenniger-Tosato M, Pedrazzini T, Diviani D. The A-kinase anchoring protein (AKAP)-Lbc-signaling complex mediates alpha1 adrenergic receptor-induced cardiomyocyte hypertrophy. Proc Natl Acad Sci U S A. 2007;104:10140–10145. doi: 10.1073/pnas.0701099104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backs J, Song K, Bezprozvannaya S, Chang S, Olson EN. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J Clin Invest. 2006;116:1853–1864. doi: 10.1172/JCI27438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- Benjamin EJ, Levy D. Why is left ventricular hypertrophy so predictive of morbidity and mortality? Am J Med Sci. 1999;317(3):168–175. doi: 10.1097/00000441-199903000-00006. [DOI] [PubMed] [Google Scholar]
- Bers DM, Guo T. Calcium signaling in cardiac ventricular myocytes. Ann N Y Acad Sci. 2005;1047:86–98. doi: 10.1196/annals.1341.008. [DOI] [PubMed] [Google Scholar]
- Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- Carnegie GK, Smith FD, McConnachie G, Langeberg LK, Scott JD. AKAP-Lbc nucleates a protein kinase D activation scaffold. Mol Cell. 2004;15:889–899. doi: 10.1016/j.molcel.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Chang S, Bezprozvannaya S, Li S, Olson EN. An expression screen reveals modulators of class II histone deacetylase phosphorylation. Proc Natl Acad Sci U S A. 2005;102:8120–8125. doi: 10.1073/pnas.0503275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Kurokawa J, Kass RS. Phosphorylation of the A-kinase-anchoring protein Yotiao contributes to protein kinase A regulation of a heart potassium channel. J Biol Chem. 2005;280:31347–31352. doi: 10.1074/jbc.M505191200. [DOI] [PubMed] [Google Scholar]
- Chen L, Marquardt ML, Tester DJ, Sampson KJ, Ackerman MJ, Kass RS. Mutation of an A-kinase-anchoring protein causes long-QT syndrome. Proc Natl Acad Sci U S A. 2007;104:20990–20995. doi: 10.1073/pnas.0710527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerk A, Sugden PH. Activation of protein kinase cascades in the heart by hypertrophic G protein-coupled receptor agonists. Am J Cardiol. 1999;83:64H–69H. doi: 10.1016/s0002-9149(99)00261-1. [DOI] [PubMed] [Google Scholar]
- Clerk A, Sugden PH. Small guanine nucleotide-binding proteins and myocardial hypertrophy. Circ Res. 2000;86:1019–1023. doi: 10.1161/01.res.86.10.1019. [DOI] [PubMed] [Google Scholar]
- Diviani D. Modulation of cardiac function by A-kinase anchoring proteins. Curr Opin Pharmacol. 2007 doi: 10.1016/j.coph.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Diviani D, Soderling J, Scott JD. AKAP-Lbc anchors protein kinase A and nucleates Galpha 12-selective Rho-mediated stress fiber formation. J Biol Chem. 2001;276:44247–44257. doi: 10.1074/jbc.M106629200. [DOI] [PubMed] [Google Scholar]
- Dodge KL, Khouangsathiene S, Kapiloff MS, Mouton R, Hill EV, Houslay MD, Langeberg LK, Scott JD. mAKAP assembles a protein kinase A/PDE4 phosphodiesterase cAMP signaling module. EMBO J. 2001;20:1921–1930. doi: 10.1093/emboj/20.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge-Kafka KL, Soughayer J, Pare GC, Carlisle Michel JJ, Langeberg LK, Kapiloff MS, Scott JD. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature. 2005;437:574–578. doi: 10.1038/nature03966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink MA, Zakhary DR, Mackey JA, Desnoyer RW, Apperson-Hansen C, Damron DS, Bond M. AKAP-mediated targeting of protein kinase a regulates contractility in cardiac myocytes. Circ Res. 2001;88:291–297. doi: 10.1161/01.res.88.3.291. [DOI] [PubMed] [Google Scholar]
- Frey N, Olson EN. Cardiac hypertrophy: the good, the bad, and the ugly. Annu Rev Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc Natl Acad Sci U S A. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Verdel A, Tsai CC, Simon C, Juguilon H, Khochbin S. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J Biol Chem. 2001;276:47496–47507. doi: 10.1074/jbc.M107631200. [DOI] [PubMed] [Google Scholar]
- Kass RS, Moss AJ. Long QT syndrome: novel insights into the mechanisms of cardiac arrhythmias. J Clin Invest. 2003;112:810–815. doi: 10.1172/JCI19844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama H, Fukuda K, Pan J, Sano M, Takahashi T, Kato T, Makino S, Manabe T, Murata M, Ogawa S. Significance of ERK cascade compared with JAK/STAT and PI3-K pathway in gp130-mediated cardiac hypertrophy. Am J Physiol Heart Circ Physiol. 2000;279:H1635–1644. doi: 10.1152/ajpheart.2000.279.4.H1635. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kunkel MT, Toker A, Tsien RY, Newton AC. Calcium-dependent regulation of protein kinase D revealed by a genetically encoded kinase activity reporter. J Biol Chem. 2007;282:6733–6742. doi: 10.1074/jbc.M608086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnart SE, Wehrens XH, Reiken S, Warrier S, Belevych AE, Harvey RD, Richter W, Jin SL, Conti M, Marks AR. Phosphodiesterase 4D deficiency in the ryanodine-receptor complex promotes heart failure and arrhythmias. Cell. 2005;123:25–35. doi: 10.1016/j.cell.2005.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygren B, Carlson CR, Santamaria K, Lissandron V, McSorley T, Litzenberg J, Lorenz D, Wiesner B, Rosenthal W, Zaccolo M, et al. AKAP complex regulates Ca2+ re-uptake into heart sarcoplasmic reticulum. EMBO Rep. 2007;8:1061–1067. doi: 10.1038/sj.embor.7401081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama Y, Nishida M, Sugimoto Y, Tanabe S, Turner JH, Kozasa T, Wada T, Nagao T, Kurose H. Galpha(12/13) mediates alpha(1)-adrenergic receptor-induced cardiac hypertrophy. Circ Res. 2002;91:961–969. doi: 10.1161/01.res.0000043282.39776.7c. [DOI] [PubMed] [Google Scholar]
- Marx SO, Kurokawa J, Reiken S, Motoike H, D’Armiento J, Marks AR, Kass RS. Requirement of a macromolecular signaling complex for beta adrenergic receptor modulation of the KCNQ1-KCNE1 potassium channel. Science. 2002;295:496–499. doi: 10.1126/science.1066843. [DOI] [PubMed] [Google Scholar]
- Marx SO, Reiken S, Hisamatsu Y, Gaburjakova M, Gaburjakova J, Yang YM, Rosemblit N, Marks AR. Phosphorylation-dependent regulation of ryanodine receptors: a novel role for leucine/isoleucine zippers. J Cell Biol. 2001;153:699–708. doi: 10.1083/jcb.153.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnachie G, Langeberg LK, Scott JD. AKAP signaling complexes: getting to the heart of the matter. Trends Mol Med. 2006;12:317–323. doi: 10.1016/j.molmed.2006.05.008. [DOI] [PubMed] [Google Scholar]
- McKinsey TA. Derepression of pathological cardiac genes by members of the CaM kinase superfamily. Cardiovasc Res. 2007;73:667–677. doi: 10.1016/j.cardiores.2006.11.036. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci U S A. 2000;97:14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol Cell Biol. 2001;21:6312–6321. doi: 10.1128/MCB.21.18.6312-6321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Dorn IG., 2nd Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu Rev Physiol. 2001;63:391–426. doi: 10.1146/annurev.physiol.63.1.391. [DOI] [PubMed] [Google Scholar]
- Mullin MJ, Lightfoot K, Marklund U, Cantrell DA. Differential requirement for RhoA GTPase depending on the cellular localization of protein kinase D. J Biol Chem. 2006;281:25089–25096. doi: 10.1074/jbc.M603591200. [DOI] [PubMed] [Google Scholar]
- Perrino C, Rockman HA. Reversal of cardiac remodeling by modulation of adrenergic receptors: a new frontier in heart failure. Curr Opin Cardiol. 2007;22:443–449. doi: 10.1097/HCO.0b013e3282294d72. [DOI] [PubMed] [Google Scholar]
- Sucharov CC, Langer S, Bristow M, Leinwand L. Shuttling of HDAC5 in H9C2 cells regulates YY1 function through CaMKIV/PKD and PP2A. Am J Physiol Cell Physiol. 2006;291:C1029–1037. doi: 10.1152/ajpcell.00059.2006. [DOI] [PubMed] [Google Scholar]
- Sugden PH, Clerk A. Endothelin signalling in the cardiac myocyte and its pathophysiological relevance. Curr Vasc Pharmacol. 2005;3:343–351. doi: 10.2174/157016105774329390. [DOI] [PubMed] [Google Scholar]
- Teerlink JR. Endothelins: pathophysiology and treatment implications in chronic heart failure. Curr Heart Fail Rep. 2005;2:191–197. doi: 10.1007/BF02696649. [DOI] [PubMed] [Google Scholar]
- van Oort RJ, van Rooij E, Bourajjaj M, Schimmel J, Jansen MA, van der Nagel R, Doevendans PA, Schneider MD, van Echteld CJ, De Windt LJ. MEF2 activates a genetic program promoting chamber dilation and contractile dysfunction in calcineurin-induced heart failure. Circulation. 2006;114:298–308. doi: 10.1161/CIRCULATIONAHA.105.608968. [DOI] [PubMed] [Google Scholar]
- Vega RB, Harrison BC, Meadows E, Roberts CR, Papst PJ, Olson EN, McKinsey TA. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol. 2004;24:8374–8385. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron RT, Rozengurt E. Protein kinase C phosphorylates protein kinase D activation loop Ser744 and Ser748 and releases autoinhibition by the pleckstrin homology domain. J Biol Chem. 2003;278:154–163. doi: 10.1074/jbc.M208075200. [DOI] [PubMed] [Google Scholar]
- Westphal RS, Tavalin SJ, Lin JW, Alto NM, Fraser ID, Langeberg LK, Sheng M, Scott JD. Regulation of NMDA receptors by an associated phosphatase-kinase signaling complex. Science. 1999;285:93–96. doi: 10.1126/science.285.5424.93. [DOI] [PubMed] [Google Scholar]
- Wu X, Zhang T, Bossuyt J, Li X, McKinsey TA, Dedman JR, Olson EN, Chen J, Brown JH, Bers DM. Local InsP3-dependent perinuclear Ca2+ signaling in cardiac myocyte excitation-transcription coupling. J Clin Invest. 2006;116:675–682. doi: 10.1172/JCI27374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB. How do 14-3-3 proteins work?-- Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–57. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.