Abstract
We previously identified KEPI as a morphine-regulated gene using subtractive hybridization and differential display PCR. Upon phosphorylation by protein kinase C, KEPI becomes a powerful inhibitor of protein phosphatase 1. To gain insights into KEPI functions, we created KEPI knockout (KO) mice on mixed 129S6 × C57BL/6 genetic backgrounds. KEPI maps onto mouse chromosome 10 close to the locus that contains the μ-opioid receptor (Oprm1) and provides a major quantitative trait locus for morphine effects. Analysis of single nucleotide polymorphisms in and near the Oprm1 locus identified a doubly-recombinant mouse with C57BL/6 markers within 1 Mb on either side of the KEPI deletion. This strategy minimized the amount of 129S6 DNA surrounding the transgene and documented the C57BL/6 origin of the Oprm1 gene in this founder and its offspring. Recombinant KEPIKO mice displayed a) normal analgesic responses and normal locomotion after initial morphine treatments, b) accelerated development of tolerance to analgesic effects of morphine, c) elevated activity of protein phosphatase 1 in thalamus, d) attenuated morphine reward as assessed by conditioned place preference. These data support roles for KEPI action in adaptive responses to repeated administration of morphine that include analgesic tolerance and drug reward.
Keywords: quantitative trait locus, morphine analgesia, morphine reward, morphine tolerance, protein phosphatase 1, conditioned place preference
Protein phosphorylation is implicated in many aspects of brain function. Phosphorylation of serine and threonine residues depends on the balanced activities of more than three hundred serine/threonine protein kinases (Manning et al., 2002, Caenepeel et al., 2004) and a smaller number of serine/threonine protein phosphatases. The regulation of these phosphatases is complex, however. For example, protein phosphatase 1 (PP1), a principal serine/threonine phosphatase, is regulated through its association with over 50 proteins that modulate its activity temporally and spatially. These PP1 subunits include inhibitor-1 (Ppp1r1a), DARPP-32 (Ppp1r1b), Cpi17 (Ppp1r14a), PHI-1 (Ppp1r14b), and KEPI (Ppp1r14c) (Perez and Lewis, 1992, Eto et al., 1999, McLaren et al., 2000, Yamawaki et al., 2001, Liu et al., 2002). These inhibitors are virtually all phosphoproteins themselves; their activities are regulated by kinases and phosphatases.
We identified KEPI as a morphine-regulated gene using subtractive hybridization and differential display PCR (Liu et al., 2002). After phosphorylation by protein kinase C (PKC), KEPI inhibits PP1 with an IC50 of about 10−9 M (Liu et al., 2002). KEPI maps to mouse chromosome 10 near the gene for the μ-opioid receptor (Oprm1), a principal site for morphine actions (Sora et al., 1997, Sora et al., 2001). KEPI is expressed in heart, muscle, and the central nervous system (CNS). Its multifocal, largely neuronal expression patterns in the CNS include regions associated with reward, locomotor control and nociception, such as striatum, nucleus accumbens, amygdala, thalamus, periaqueductal grey, and spinal cord (Liu et al., 2002, Gong et al., 2005).
To gain insights into KEPI functions at biochemical and behavioral levels, we created refined and characterized KEPI KO mice. While the initial animals were developed on a mixed 129S6 × C57BL/6J background, the proximity of the KEPI gene to Oprm1 lead us to consider possible effects of “hitch-hiking” genes from the 129S6 genetic background, including Oprm1 (Gerlai, 1996). Oprm1 variants provide a major contribution to a quantitative trait locus (QTL) that influences morphine antinociception and self-administration (Berrettini et al., 1994, Belknap et al., 1995, Bergeson et al., 2001). We thus identified recombinant KO animals with C57BL/6 markers in the appropriate genomic region by analyzing single nucleotide polymorphisms (SNPs) in and near the Oprm1 locus. This strategy allowed us to evaluate the contribution of KEPI to morphine-related traits without confounding influences from Oprm1 variants, and provides a generally useful approach for evaluating mice in which engineered gene variants lie near QTLs for phenotypes in which the gene of interest may be involved.
Experimental procedures
Generation of KEPI KO mice
A 129/SvEvTac genomic library in λ fixII vector (Stratagene, Cedar Creek, TX) was screened with a 32P-labeled probe amplified from the mouse 129S6 (129S6/SvEvTac) genomic DNA by PCR. The primers used for the probe amplification (J24 and J25, Table 1) targeted the first exon. Two positive clones, λ13 and λ23 were further examined by PCR with primer pairs annealing 5’ (J30 – J31, Table 1) and 3’ (J32 – J33, Table 1) from the first exon and by restriction analysis to confirm that they contained the KEPI gene. λ13 was digested with Xba I and the 3.5 kb fragment was cloned into the pBSIIKS+ vector (Stratagene). Digestion of λ23 with BamH I produced a 3.9 kb piece downstream of exon 1 which was inserted 3’ from the Xba I insert. A Spe I fragment of the Neo cassette was inserted between the two arms and the Sal I fragment of the TK gene was then inserted in the opposite orientation downstream of the BamH I arm (Figure 1a). The final construct, pJD9, was linearized with Not I and electroporated into 129.3 mouse MC1 (126S6/SvEvTac) embryonic stem (ES) cells (JHU Transgenic Core Laboratory). Primers vNEOf (in the Neo cassette, Table 1) and J39 (outside the construct, Table 1) were used for a PCR screen of 376 colonies resistant to G418 and Gancyclovir. Plasmid pJD10 derived from pJD9 by inserting a PCR amplicon (J37–J38, Table 1) into a Dra III site was used as a positive control. Embryonic stem (ES) cell clones positive by PCR were further confirmed by Southern blot after DNA digestion with Bgl II (Figure 1b). The 640-bp probe spanning the 5’ Xba I site was generated by PCR with primers J40 and J31 (Table 1). Eight of the correctly targeted ES cell clones were karyotyped and two with the correct number of chromosomes were microinjected into C57BL/6J E3.5 blastocysts at the Johns Hopkins University transgenic core facility. Heterozygous KEPI KO offspring of the resulting male chimeras were mated with each other and their progeny genotyped by PCR with primers KEPIf, KEPIr, and vNEOf (Table 1) to amplify wild type (WT) (527 bp) and KO (922 bp) alleles simultaneously (Figure 1c). To prepare DNA for genotyping, ear punches (approximately 1.5 mm diameter) were digested for 6 hours at 55 °C in 50 µl of a buffer containing 10 mM TrisHCl pH 8.5, 5 mM EDTA, 0.1 % SDS, 200 mM NaCl, and 10 mg/ml proteinase K. The resulting hydrolyzate was diluted 4 × with H2O and directly used for PCR reaction.
Table 1. DNA primers used in the study.
| Primer Name | Primer Sequence | Primer Name | Primer Sequence |
|---|---|---|---|
| J24 | CACTCCCAGCTGTCCTAGGAG | rs29358253f | CTCTAATGCCAGTGGAGCGT |
| J25 | CTAGGAAGTCGTCCGAGGTGG | rs29358253r | CACCGCAATCGGATAGCACT |
| J30 | GGAGTTCTGGCTTCTAGAAGAAC | rs29358253ex | CTCTGTACTTCTCCCTTCTC |
| J31 | CATCATGTGGTCACCACATGCC | rs29382724f | CCAGAGCAAGCGTCTCTGG |
| J32 | TCTGGCACACTGTCTTCCCTGA | rs29382724r | GAGTGGGATGCCAAATTACCT |
| J33 | CATGGCTAGCATGCCCTGGCA | rs29382724ex | TGCAATCGGGTTCTGCTAAC |
| J37 | ACTGACACTACGTGAGAAGTTTCTCCCACCTTCA | rs29382421f | CCATTGGAAATACGGTGACTC |
| J38 | TCAGAGGTAGAGTGCACTTGCC | rs29382421r | CAAAGCACATTGTACACATGAG |
| J39 | CTTCACTGGCAACAGTTATGGAG | rs29382421ex | CACACGTCTTCTAAAACGAAC |
| J40 | AAGGGAGGCAATGATTGCTGCC | rs4228112f | GATCCACAGAACGGAGAGGA |
| vNEOf | CCGCTATCAGGACATAGCGTTG | rs4228112r | GCGCATTTCGAGTCTGCTTTT |
| KEPIf | GAGTCTGCCTATGTGTATTTCCAT | rs4228112ex | CTCTATTCTTTGGTATGGATTG |
| KEPIr | CTCATAGACTATCTTGTTCCTCTC | rs29361179f | TCGTCCACCTTCAGCATCCA |
| J85 | CTCCGCAAGGATCCAGCGTCTAGG | rs29361179r | GCCCTTGACCAACTCAACAC |
| J86 | CGAAGCTCTTTCCTCCTCGCT | rs29361179ex | GTTTGTCATTACAATGAGCCA |
| rs8244279f | TTCAGCAGTAATGTAGATAAGGT | qGAPDHf | GCATGGCCTTCCGTGTTC |
| rs8244279r | GGAGACCGATGAATGTGAGC | qGAPDHr | CACCACCTTCTTGATGTCATC |
| rs8244279ex | AGTTCATATGGCTGAGCTGAA | qPKCCf | GCACCTCCTTTCAGACCACG |
| rs16821161f | TGTGGAAGGCATGGTGGAGA | qPKCCr | GGTCAGTGCTGGCGCTGCC |
| rs16821161r | ATGGATTTGAGGCACACAAACT | qPKCEf | ACTGGGTACTGCTGGAGCAG |
| rs16821161ex | GGAACCGACTTGACGTAGC | qPKCEr | GTAAGTATTGGCTCTTCCCGC |
| rs29360540f | CAGAGCCAAACATGGTAGCTC | qPP1CAf | GTGCCATGATGAGTGTGGATG |
| rs29360540r | GAGGCAGTGTCTCACGCTG | qPP1CAr | CAGGCCGCTGAACTGCCCAT |
| rs29360540ex | TGAGGCTGATATAGGATGAAG | qPP1CBf | GGCGAGTTTGACAATGCTGGT |
| rs29334299f | CATCCCATGCTCTTATTTGGG | qPP1CBr | CCCACCATACTGGTACTTAGC |
| rs29334299r | ATGTAGGCAGCCTGCCTGG | qPP1CCf | CTCCATAAGCATGATTTGGATCT |
| rs29334299ex | AGCAGAAACTCGATGGGCC | qPP1CCr | CAGAGTGACTAACTGCCTCTTT |
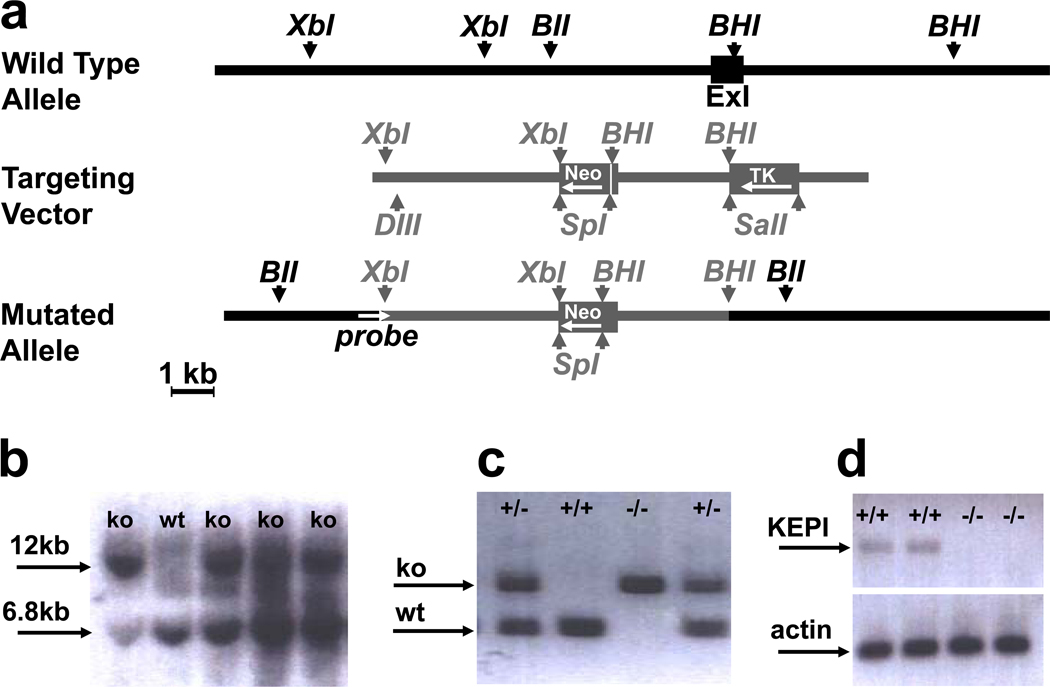
Figure 1. Generation and analysis of KEPI KO mice.
(a) Genomic organization of the KEPI gene (black) and the gene targeting construct (gray). Full boxes represent exons and arrow heads important restriction sites: XbI, Xba I; BII, Bgl II; BHI, BamH I; Sp1, Spe I; DIII, Dra III. Direction of transcription for Neo and TK is opposite to that of KEPI as indicated by the arrows. (b) Southern blot analysis of clones positive by PCR. Bgl II digestion produced a 6.8 kb fragment from the WT allele, and a 12.0 kb fragment from the mutated allele because a BGL II site was deleted. (c) PCR genotyping of KEPI mice. (d) RT-PCR analysis of brain expression of the KEPI (primers J85, J86) and actin (ACTf, ACTr) in WT and mutant mice.
SNP genotyping
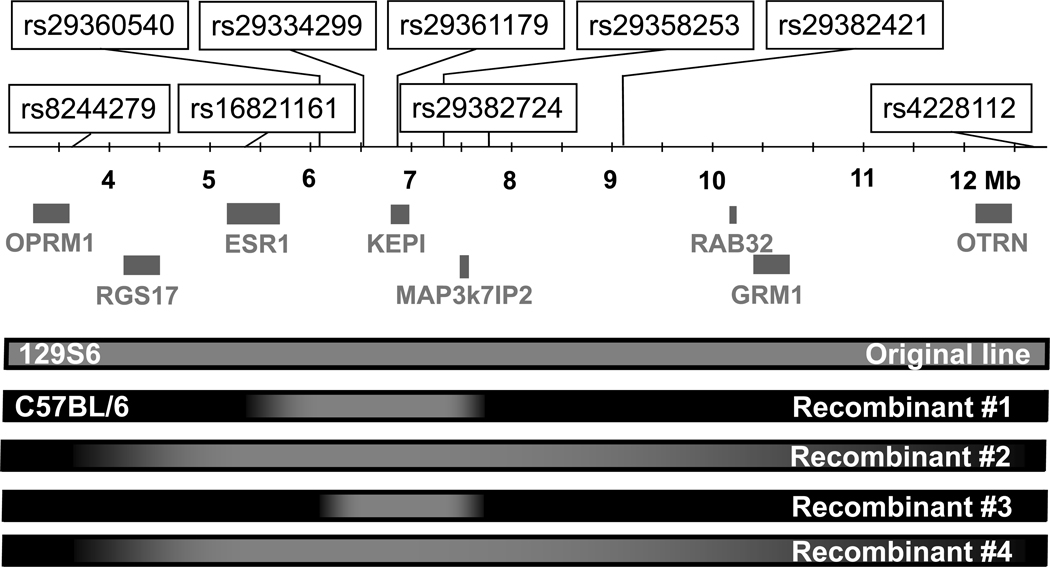
SNPs around the KEPI locus were selected from the Mouse Genome Informatics database (http://www.informatics.jax.org) and genotyped by primer extension followed by MALDI-TOF-based allele detection (Sequenom, San Diego, CA, USA) according to the manufacturer’s protocol. DNA was prepared as described above and further diluted threefold. Primers for primary PCRs and extension primer sequences are listed in Table 1. One male with cross-overs closest to KEPI (Figure 2) was mated with a C57BL/6J female and the resulting heterozygote offspring were mated to produce the “Recombinant KEPI KO Line”.
Figure 2. SNP markers on chromosome 10 genotyped in this study.
Grey boxes under the ruler represent genes near the KEPI locus that may be relevant to morphine-associated phenotypes. Bars at the bottom represent genetic background around the KEPI deletion in the original KO line and four recombinants as determined by SNP genotyping: grey, 129S6; black, C57BL/6.
RNA extraction and reverse transcription-PCR
Total brain RNA was prepared with RNeasy Lipid Tissue Kit (Qiagen, Valencia, CA). cDNA was synthesized from oligo dT primers using Superscript III (Invitrogen, Carlsbad, CA). Quantitative real time PCR was performed on ABI Prism 7900HT Sequence Detection System with primers as listed in Table 1 and SYBR Green PCR Master Mix (Applied Biosystems, Warrington, UK) according to the manufacturer’s instructions. Expression levels were normalized against GAPDH.
Preparation of brain extracts for phosphatase assay
Unless otherwise stated, all the steps were performed at 0–4 °C. Mice were euthanized by spinal cord dislocation followed by decapitation, brains rapidly dissected, frozen, and stored at −80 °C until used. The brain tissue was homogenized with a Teflon Dounce Homogenizer in 20-fold volume of ice-cold buffer containing 10 mM Tris-HCl, pH 7.5, 250 mM Sucrose, 1 mM EDTA, 1 mM EGTA, 50 µM leupeptin, 30 µM pepstatin A, and 1% β-mercaptoethanol, and centrifuged at 18,600 × g for 15 minutes. The supernatant was applied to a PD MiniTrap G-25 column (GE healthcare, Buckinghamshire, UK) equilibrated with exchange buffer (50 mM Tris-HCl, pH 7.0, 0.1 mM EDTA, and 1% β-mercaptoethanol) and eluted with the same buffer according to manufacturer’s protocol. The protein content of the eluent was measured with the Bradford Protein Assay (Bio-Rad, Hercules, CA) and diluted to 0.25 mg/ml with the exchange buffer.
Protein phosphatase activity
Serine/threonine phosphatase activity in brain extracts was assayed with [γ32P]-labeled phosphorylase a as a substrate as modified from (Cohen et al., 1988). To prepare the substrate, 100 µl of 100 mg/ml phosphorylase b (4 × recrystalized, AMP-free from Sigma, St. Louis, MO) dissolved in kinase buffer (50 mM β-glycerophosphate, 2 mM EDTA, 0.1% (v/v) β-mercaptoethanol, 50% (v/v) glycerol) was incubated at 30 °C in 310 µl phosphorylation buffer (250 mM Tris-HCl, pH8.2, 16.7 mM MgCl2, 1.67 mM ATP, 0.83 mM CaCl2, 133 mM β-glycerophosphate) with 250 μCi of [γ32P]-ATP in 80 µl H2O (3,000 Ci/mmol) and 10 µl of 20 mg/ml phosphorylase kinase (Sigma) dissolved in kinase buffer. After 1 hour, the labeled phosphorylase a was precipitated with 0.5 ml of 100% saturated (NH4)2SO4 for 30 minutes on ice and centrifuged at 10,000 × g at 4 °C. The supernatant was discarded and the pellet washed 3 times with 50% saturated (NH4)2SO4 to remove the unbound [γ32P]-ATP. The pellet was dissolved in 2.5 ml 50 mM Tris-HCl, pH 7.0, 0.1 mM EDTA, 15 mM caffeine, 0.1% β-ME and passed through a PD MiniTrap G-25 column (GE healthcare, Buckinghamshire, UK) pre-equilibrated in the same buffer. Concentration of the purified [32P]-phosphorylase b was adjusted to 24 µM. Phosphatase reactions were carried out at 30°C for 20 minutes in 80 µl total volume containing 20 µl of [32P]-phosphorylase a, 20 µl of brain extracts, and 40 µl of phosphatase buffer freshly prepared by mixing equal volumes of buffer A (0.1 mM EDTA, 1 mg/ml of BSA) and buffer B (20 mM imidazole-HCl, pH 7.63, 0.1% β-ME). The reaction was terminated by addition of 160 µl of ice-cold trichloroacetic acid and the precipitated proteins were removed by centrifugation at 12,000 × g for 5 min at 4 °C. The released [32P] in the supernatant was quantified by scintillation counting.
Behavioral testing
Mice from the original KEPI KO line were evaluated for their general health, reflexes and gross behavioral abnormalities as suggested by J. Crawley (Crawley, 1999).
Motor coordination and motor learning were measured on an accelerating rotarod over three consecutive days with one test per day as described in the literature (Lalonde et al., 1995, Crawley, 1999). The starting speed was 4 rpm and it gradually increased to 40 rpm over 5 min, which was the cut off time. Data from all three days were analyzed by repeated measures ANOVA.
Memory and learning was evaluated in the Morris Water Maze as previously described (Morris, 1984, Crawley, 1999). The apparatus consisted of a black pool (90 cm diameter) filled with room temperature water made opaque with white tempera paint. A 9 cm diameter platform was located in the center of one quadrant. For the first 6 trials the platform was visible, during all subsequent trials it was hidden 0.5 cm below the water level. Each trial lasted a maximum of 60 seconds followed by a 15 second rest period on the platform. After two trials, the mice were returned to their home cages for about 2 hours and then submitted to another 2-trial session for total of 4 trials per day. For each trial, the latency to reach the platform was recorded. After acquisition (defined by an average latency less than 10 seconds) a 60 second probe trial was conducted in which the platform was removed. The data from the probe trial were analyzed with Ethovision software (Noldus, Netherlands) to determine the time spent in each quadrant. After this probe trial, the platform was placed in a different quadrant to assess reversal learning.
Anxiety was assessed in two standard paradigms (Crawley, 1999). (1) Open Field. Mice were placed singly in an open field (42 × 42 cm) illuminated with red light, for twenty minutes. The time each animal spent in the central quadrant and the number of fecal pellets was recorded. (2) Light-Dark Test: The testing cage (18 × 36 cm) consisted of two compartments: a dark chamber (18 × 18 cm) with black walls and a small opening (5 cm) leading to a Plexiglas compartment illuminated with red light. Each animal was placed into the dark chamber. The latency to emerge was recorded by an observer and the time spent outside the dark chamber during the 10 min trial was measured with the Optovarimax system (Columbus Instruments, OH).
Depression-related behavior was assessed in the forced swim test. Mice were placed in the center of a 4 L beaker (18 cm diameter) containing 3 L of room temperature water for 15 min then towel-dried and returned to their home cages. The following day the animals were submitted to the forced swim test for 5 minutes. Their behavior was digitally recorded and later analyzed by a visual observation using the TIMER program (NIH) for 3 behavioral categories: swimming, climbing, and immobility (Hall et al., 1998).
Morphine analgesia. Morphine analgesia of both KEPI KO lines was assessed in the tail-flick and hot-plate paradigms (D'amour and Smith, 1941, Woolfe and MacDonald, 1944). First, each mouse was gently wrapped in a towel, the end of its tail was immersed 0.5 inches into a 53°C hot water bath, and the tail-flick latency was recorded. Immediately afterwards, the animal was placed onto a 55°C hot plate and the latency to jump or to lick one of the hind paws was assessed. To prevent injury, a cut-off time of 15 s was used for the tail-flick test and 30 s for the hot-plate test. Before each experiment, animals were exposed to the testing procedures for three days (3 tests at 25 min intervals per day), so that their baseline responses to saline injections stabilized. On the day of the experiment, 3 baseline measurements were performed before each animal was injected with the lowest dose of morphine followed by increasing doses to generate a cumulative dose-response curve. Analgesic responses were assessed 25 min after each dose immediately followed by injection with the next dose of morphine. This cumulative dosing regimen produced cumulative doses of 3, 10, and 30 mg/kg morphine. Analgesia was expressed as a percentage of the maximal possible effect (% MPE) calculated as follows: % MPE = 100 × (morphine latency – baseline)/(cut-off – baseline).
Morphine tolerance. In this experiment, mice from the recombinant KEPI KO line were injected twice daily with morphine and analgesia was assessed for 5 consecutive days. On the morning of the first day, after assessment of the baseline responses, the animals were injected with morphine, following a cumulative dosing regimen that produced morphine doses of 2, 4, 8, 16, and 32 mg/kg. Analgesic responses were assessed 25 min after each dose in a 51°C hot water bath and on a 53°C hot plate as described above, and the next dose of morphine was administered immediately afterwards. Over the next four days between 8 and 9 am, after assessment of baseline responses, tail withdrawal was assessed after administration of 8 mg/kg morphine, and hot plate responses after an additional injection of 8 mg/kg morphine (16 mg/kg total). The doses for each test were selected based on the ED50 values established on the first day (Table 2). The evening injections of 32 mg/kg morphine were administered 8 hours after testing.
Table 2. Behavioral phenotype of the original KEPI mice.
Number of subjects is in brackets.
| Age | Sex | +/+ | +/− | −/− | P | |||
|---|---|---|---|---|---|---|---|---|
| Motor functions | ||||||||
| latency to fall off of an accelerating rotarod (s) | 3 M | M, F | 220 (33) | 231 (21) | 221 (26) | 0.52 | ||
| 6 M | M, F | 185 (22) | 164 (24) | 176 (33) | 0.84 | |||
| 12 M | M, F | 150 (20) | 127 (19) | 168 (18) | 0.53 | |||
| Learning in Morris Water Maze | ||||||||
| Latency to platform quadrant (s) | 2–3 M | M, F | 4.4 (16) | 2.8 (15) | 2.4 (14) | 0.03* | ||
| Latency to new location (s) | 2–3 M | M, F | 2.2 (16) | 1.9 (15) | 1.2 (14) | 0.41 | ||
| Time in new/time in old | 2–3 M | M, F | 5.5 (16) | 8.9 (15) | 5.9 (14) | 0.24 | ||
| Anxiety & Stress-Induced Anxiety | ||||||||
| Dark box | - Latency to emerge (s) | baseline | 2–3 M | M, F | 59 (15) | 80 (16) | 111 (16) | 0.68 |
| stress | 2–3 M | M, F | 410 (16) | 415 (16) | 344 (12)) | 0.68 | ||
| - Time outside (s) | baseline | 2–3 M | M, F | 188 (15)) | 161 (16) | 159 (16) | 0.77 | |
| stress | 2–3 M | M, F | 115 (16) | 107 (16) | 118 (16) | 0.98 | ||
| Open field | - Time in center (s) | baseline | 2–3 M | M, F | 62 (16) | 59 (16) | 56 (16) | 0.91 |
| stress | 2–3 M | M, F | 24 (14) | 23 (15) | 27 (15) | 0.09 | ||
Morphine dependence in mice treated twice daily for eight days with increasing doses of morphine (15–75 mg/kg in 5 mg increments) was assessed by precipitating a withdrawal syndrome with naloxone (1mg/kg, s.c.) 2 hours after the last morphine treatment. Immediately after the naloxone injection, each mouse was placed into a Plexiglas chamber (18 × 18 × 18 cm) and the number of jumps recorded for 15 minutes.
Morphine reward was assessed with the conditioned place preference (CPP) paradigm using a two-compartment Plexiglas chamber as described previously (Hall et al., 2003). After a 20 min pre-test in which subjects had access to both sides of the apparatus, morphine conditioning was conducted over a 3-day period with two 30-min sessions per day. During these sessions the subjects were injected with morphine sulphate when confined to the compartment with a wire mesh floor mounted over Plexiglas, or with saline, when confined to the side with corncob bedding (mice generally demonstrated a slight preference for the corncob side over the wire side). Control subjects received saline injections on both sides. 24 hours after the last conditioning session, subjects were again given access to both sides of the apparatus for a 20 min post-test. The preference score was calculated as the difference between time spent on the drug-paired wire mesh side during the post-test and the time spent on that side during the pre-test.
Morphine locomotion was recorded during the first conditioning session and total distance (m) traveled was calculated from infrared beam breaks by the Optovarimax System.
Experiments were carried out in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals under protocols approved by the NIDA-IRP Animal Care and Use Committee.
Drugs
Morphine sulphate (NIDA IRP Drug Supply Program) was dissolved in saline and administered s.c. in a volume of 10 ml/kg.
Data analysis
The data from the behavioral tests were analyzed by linear regression (JMP software, SAS Institute) with genotype and sex as factors and age as a covariate. The additional factors of “Dose” for analysis of morphine analgesia data and “Day” for morphine tolerance data were used in repeated measures MANOVA. Data from the phosphatase assay were subjected to ANOVA with genotype and treatment as factors.
Results
Generation of KEPI KO mice
Homologous recombination was employed to disrupt exon 1 of the KEPI gene in embryonic stem cells (Figure 1a). Exon 1 contains the initiation codon and the potential PP1 docking motif; its disruption should thus inactivate the KEPI gene product. Correctly targeted ES cell clones were identified by PCR and confirmed by Southern blotting (Figure 1b). Recombinant ES cells were injected into blastocysts, which were then implanted into pseudo-pregnant females resulting in six chimeric offspring. Mating of heterozygous progeny of the chimeric mice produced WT, heterozygous KO and homozygous KO pups in ratios that were modestly but significantly different from those based on the expected Mendelian frequencies (n = 952; χ2 test p = 0.015). In particular, fewer KO pups were born than expected (χ2 test p = 0.007). Pups were genotyped by PCR with a set of 3 oligonucleotide primers that simultaneously amplified WT and KO alleles (Figure 1c). Disruption of the KEPI gene was further confirmed by RT-PCR. Amplification of cDNA from WT samples with oligonucleotides J85 (homologous to the deleted part of the gene) and J86 produced a band of the expected size, whereas the amplification of cDNA from homozygous knockouts did not (Figure 1d).
Establishing the recombinant line of KEPI KO mice
KEPI maps onto mouse chromosome 10 close to the Oprm1 gene, an area that has been identified as a major QTL influencing morphine antinociception and self-administration (Berrettini et al., 1994, Bergeson et al., 2001). Allelic differences between the 129S6 (129S6/SvEvTac) strain (source of the embryonic stem cells in which the KEPI gene was deleted) and the C57BL/6J strain (used for breeding with the KEPI chimeras) in this region are thus likely to contribute to phenotypic differences between WT and KO animals. To investigate the genetic background linked to the KEPI deletion in the initial KO mice, we analyzed 4 SNP markers in this area: rs8244279 in Oprm1, rs29361179 in KEPI, rs16821161 between Oprm1 and KEPI, and rs4228112 located 5 Mb centromeric from KEPI. Analysis of the F1 and F2 generation of KEPI KO animals confirmed that the KEPI deletion was always associated with the 129S6 genotype, while the wild-type allele always carried C57BL/6 markers (Figure 2, Original Line). Screening 120 KO animals from the F2 and F3 generations allowed us to identify four mice with recombination of 129S6 and C57BL/6 chromosomes between the KEPI and Oprm1 loci (Figure 2). Analysis of additional markers revealed that the chromosomal variant with recombination sites closest to KEPI contained less than 1 Mb of the 129S6-derived sequence at the KEPI locus and none at the Oprm1 locus (Figure 2, Recombinant #3). Breeding the carrier of this chromosome, in which the “hitch-hiking” 129S6 DNA was almost entirely eliminated, produced the “recombinant KEPI KO line”. Thus, in just three generations of breeding, we were able to minimize the risk that hitchhiking 129S6 sequences at or around Oprm1 would confound the behavioral and other phenotypic effects of KEPI deletion.
Phosphatase Activity
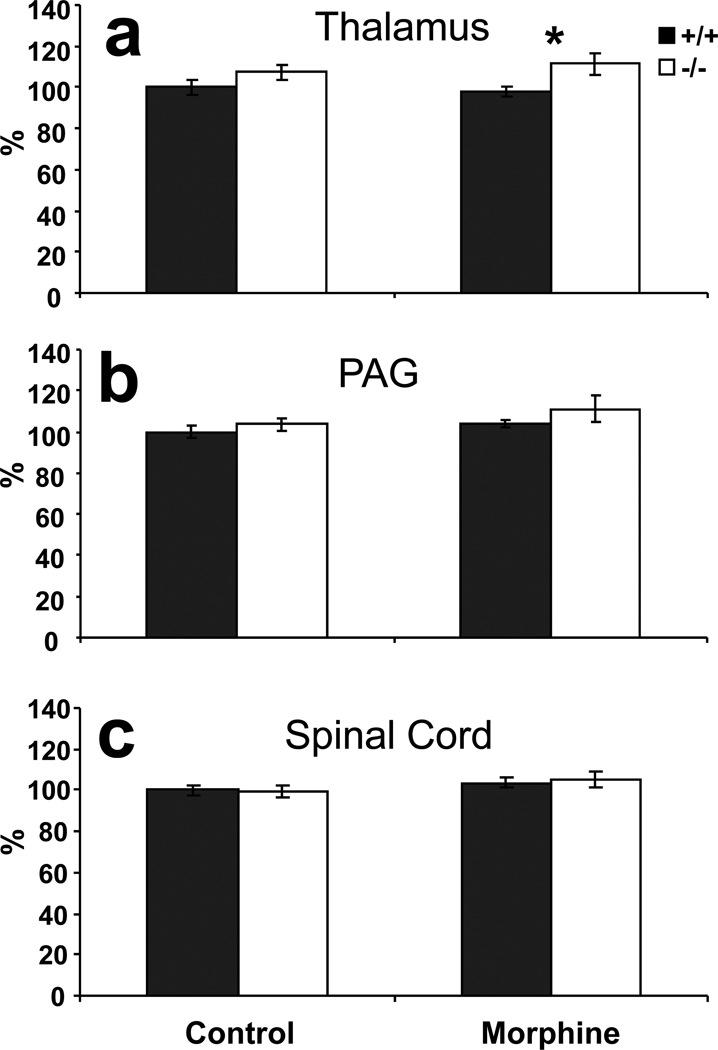
To assess the effects of the KEPI deletion on PP1 activity in areas associated with analgesia and pain transmission, we measured phosphatase activity in thalamus, periaqueductal grey (PAG) and spinal cord (Figure 3). Phosphatase 1 activity was significantly increased in thalami of KEPI KO mice (Figure 3a, ANOVA effect of genotype p = 0.03 after correction for multiple comparisons since three different brain regions were studied), with a nominally significant difference after repeated morphine treatment (t test p = 0.03). Phosphatase activity was not significantly different between the two genotypes in the PAG, although there was a modest trend towards higher activity in the KO mice (Figure 3b ANOVA nominal p = 0.16 for effect of genotype). In the spinal cord, the PP1 activities in WT and KO mice were indistinguishable from each other (Figure 3c), in agreement with behavioral data.
Figure 3. Phosphatase activity in control and morphine-treated KEPI mice.
Morphine-treated mice received morphine injections twice per day (16 mg/kg AM and 32 mg/kg PM) and were sacrificed on the third day, 1 hour after the AM injection. Control animals received saline injections under the same schedule. (a) Phosphatase 1 activity was significantly elevated in the thalamus of KEPI KO mice (ANOVA effect of genotype p = 0.03 after correction for multiple comparisons). (b) and (c) There were no differences in phosphatase activity in extracts from PAG or spinal cord. Data are expressed as means ± SEM (n = 9 per group).
Expression levels of PKC1 and PP1
To assess compensatory changes in response to elevated PP1 activity in different brain areas, expression levels of the major PKC1 and PP1 isoforms were evaluated by real time quantitative PCR. The data suggest that PKCγ, and particularly PP1β were significantly down regulated in thalamus (Figure 4a, nominal t-test p = 0.046 for PKCγ and p = 0.00002 for PP1β). The down regulation of PP1β remains highly significant even after Bonferoni correction for multiple comparisons (p = 0.0002). In PAG, where no significant differences in the phosphatase activity had been observed (Figure 3b), the expression levels of PKC1 and PP1 isoforms remained unchanged (Figure 4b).
Figure 4. Expression levels of PKC1 and PP1 isoforms in KEPI KO mice.
(a) Data from the quantitative RT-PCR show that expression level of PP1β in thalami of naive KEPI KO mice was significantly down regulated (t-test p = 0.0002 after correction for for multiple comparisons; n = 10 per group). (b) There were no differences between the genotypes in the expression of PKC1 and PP1 isoforms in KEPI mice in PAG (n=6 per group). Data are expressed as means ± SEM.
Behavioral phenotype
No gross neurological or behavioral abnormalities were observed in the original KEPI KO mice in a number of tests (Table 2). KO mice gained weight at the same rate as their WT siblings during their first year of life (MANOVA p=0.96). The animals displayed posture, gait and reflexes that were indistinguishable from those of their WT littermates. Eye blink, ear twitch or whisker reflexes elicited by light touches with cotton swabs were all similar to responses in WT mice. KEPI knockouts failed to display any unusual behavior such as wild running, freezing, circling, or stereotypy. Furthermore, the KEPI mutation had no negative consequences on locomotor coordination or learning on the accelerating rotarod, or in spatial learning assessed in the Morris Water Maze (Table 2). These results suggest that the differences observed between the KEPI knockouts and the wild-type controls reported below were not confounded by general learning or motor deficits.
Morphine analgesia
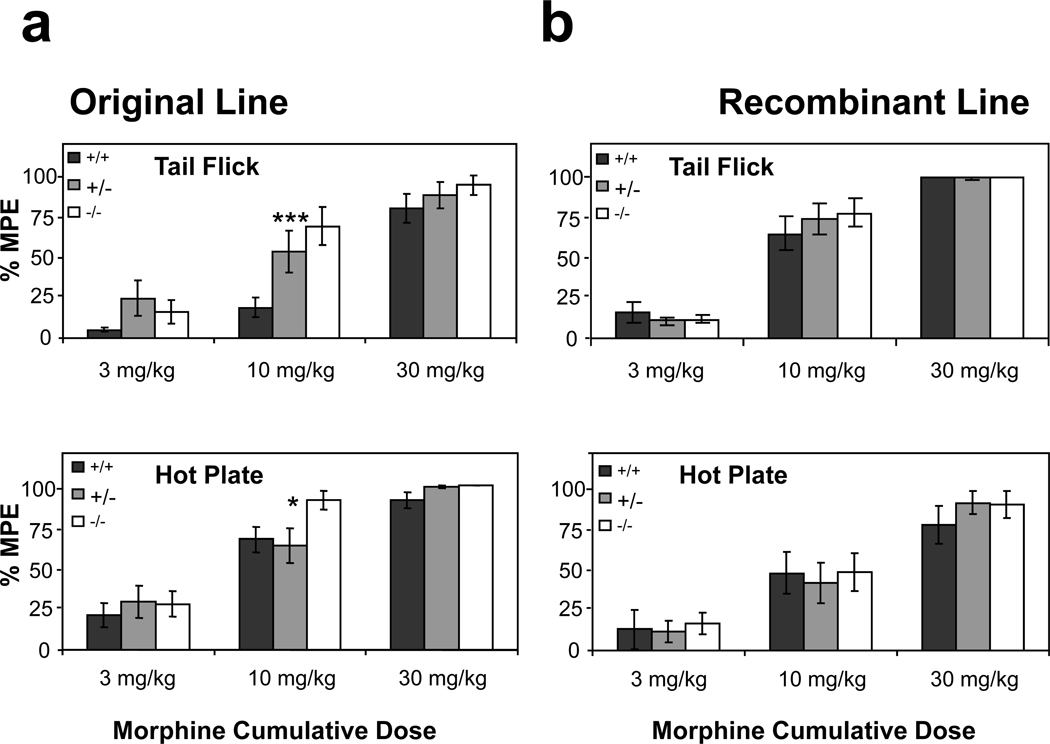
To evaluate the effect of KEPI KO and its interaction with the genetic background near the KEPI and Oprm1 loci on the phenotype of KEPI KO mice, we assessed morphine analgesia in both the “original” and the “recombinant” KEPI KO lines. The original KEPI KO mice with the Oprm1 allele from the 129S6 strain appeared to be more sensitive to morphine analgesia than their WT littermates which carried the Oprm1 allele from the C57BL/6 strain (Figure 5a). In the hot plate assay, there was a significant effect of KEPI genotype (MANOVA p<0.05). In the tail flick assay, there was a significant effect of genotype (MANOVA p<0.001), and a significant genotype × dose interaction (MANOVA p<0.005). By contrast, there was no significant difference between the two KEPI genotypes from the recombinant line (Figure 5b). These results suggest that the increased sensitivity to morphine analgesia of the original KO mice is likely to be due to allelic differences at the Oprm1 locus, and not to disruption of the KEPI gene. By the time that these data were analyzed we had discontinued production of mice from the “original” KEPI KO line.
Figure 5. Morphine analgesia in two lines of KEPI KO mice.
(a) In both, tail flick (MANOVA p < 0.001 for the effect of genotype, p < 0.005 for genotype * dose interaction) and hot plate (MANOVA p < 0.05 for the effect of genotype) assays, the original KEPI KO mice were more sensitive to morphine analgesia then their WT littermates. (b) There was no significant difference between the two genotypes from the recombinant line. MPE, maximal possible effect. Data are expressed as means ± SEM (n = 12 per group).
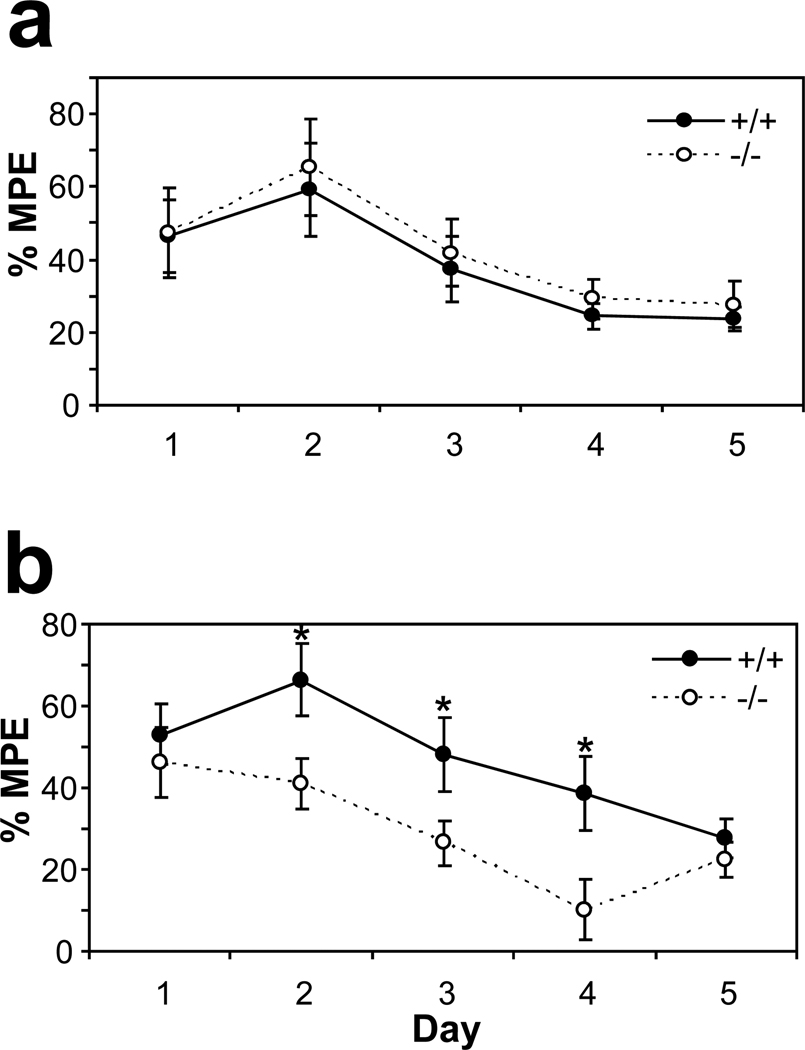
Morphine tolerance
We followed the development of tolerance to analgesic effects of morphine in recombinant KEPI KO mice using the tail flick and the hot plate paradigms (Figure 6). On the first day of testing, there were no differences between WT and KO animals in their responses to morphine, as we had already observed in the previous experiment. During the following days, antinociceptive efficacy of morphine gradually diminished in both WT and KEPI KO mice. In the tail flick assay, this measure of tolerance was essentially identical between the WT and KO animals (Figure 6a). On the hot plate, however, tolerance to morphine developed faster in the KO mice (Figure 6b; MANOVA p<0.05 for genotype; and p<0.05 for genotype × day interaction). On days 2, 3, and 4 their responses to morphine were significantly lower than the responses of their WT littermates (t-test p<0.05).
Figure 6. Development of morphine tolerance and dependence in the recombinant KEPI KO mice.
(a) In the tail flick assay, responses to 8 mg/kg morphine were identical between the WT and KO animals. (b) In the hot plate assay, tolerance to morphine developed faster in the KO mice. MPE, maximal possible effect. (MANOVA p < 0.05 for genotype; and p<0.05 for genotype * day;
n = 12per group). Data are expressed as means ± SEM.
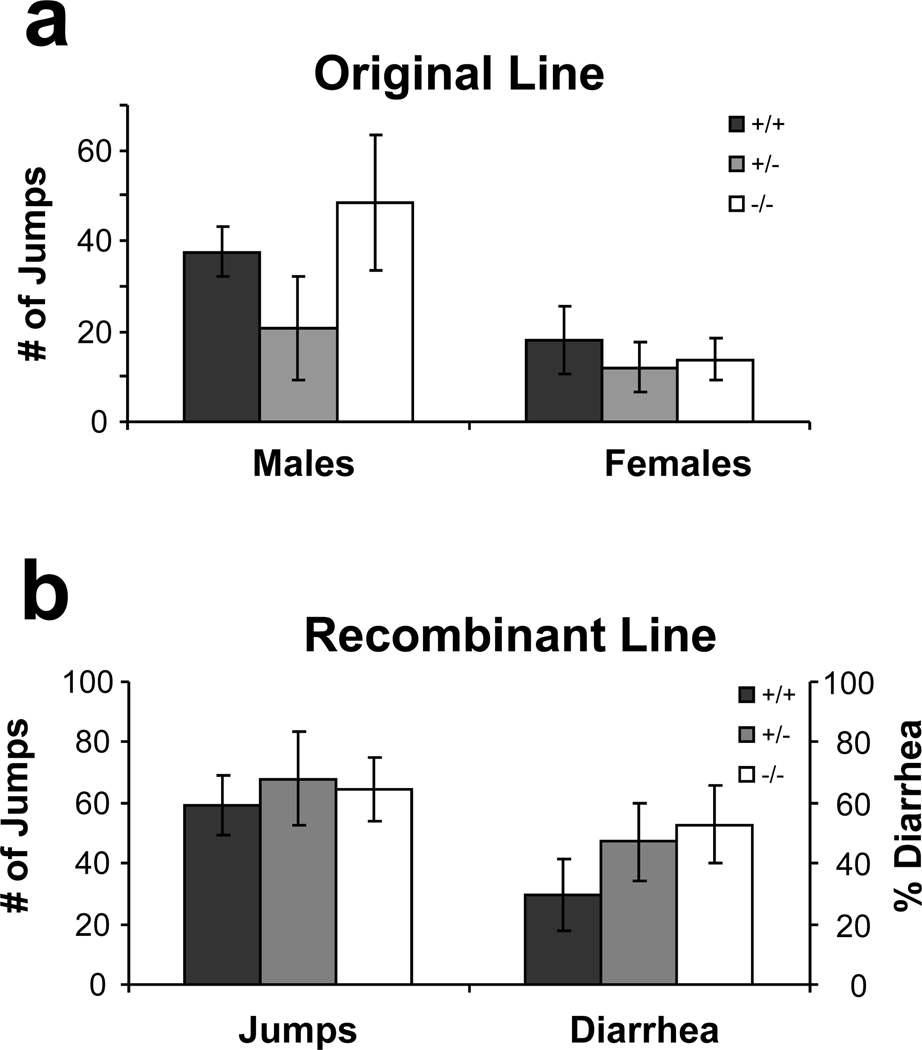
Morphine Dependence
Development of physical dependence in both the original and recombinant KEPI mice was evaluated after eight days of twice-daily morphine treatments. In neither of these two lines did deletion of KEPI have a significant effect on the quantity of naloxone-precipitated jumping (Figure 7). Females from the original line, however, exhibited significantly less jumping than the males (Figure 7a; ANOVA p = 0.0061 for sex). This difference was not detectable in the recombinant animals (Figure 7b).
Figure 7. Naloxone-precipitated withdrawal in two lines of KEPI KO mice.
(a) After 8 days of twice-daily morphine treatment, females from the original line exhibited significantly less naloxone-precipitated jumping than the males (ANOVA p<0.01; n = 6–7 per group). (b) In the recombinant line, sex had no effect on this behavior (n = 17 per group). There was no difference between the genotypes in either of the two lines. Data are expressed as means ± SEM.
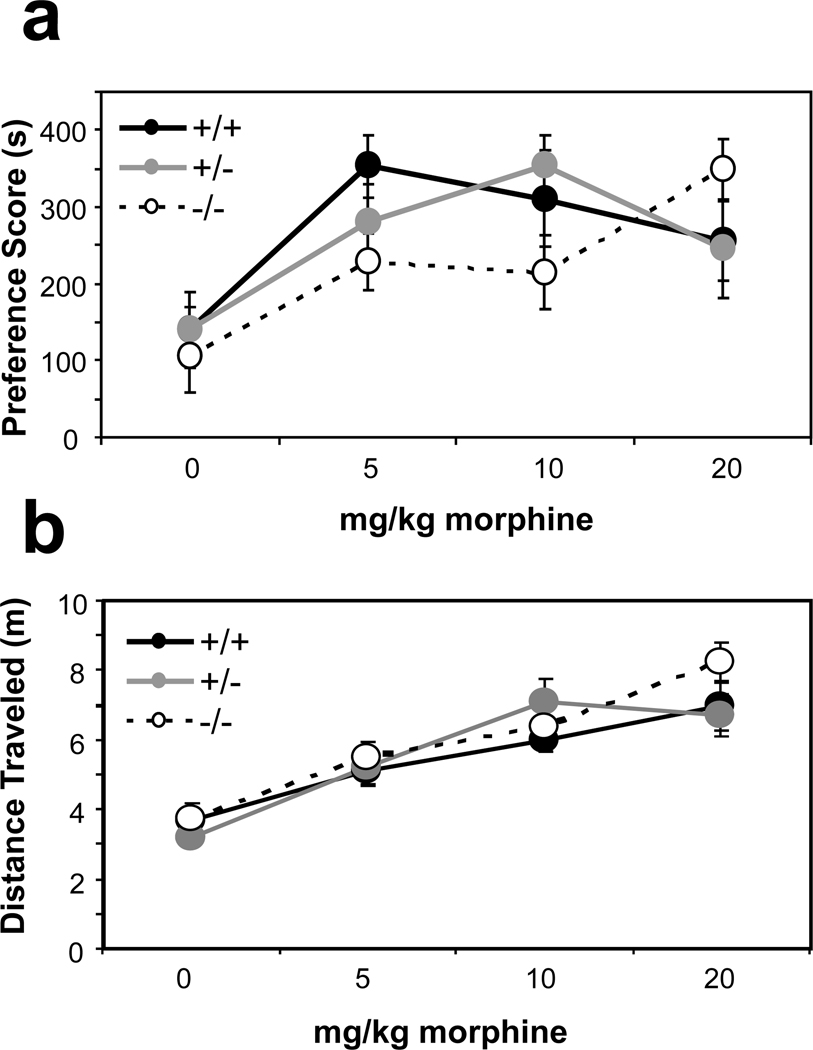
Morphine reward and locomotion
We assessed morphine reward in the recombinant KEPI line using a conditioned place preference (CPP) paradigm. When compared to the WT animals, KEPI homozygote KO mice displayed an apparent rightward shift in the dose-response curve for morphine CPP, while heterozygote KO mice showed an intermediate phenotype (Figure 8a; Linear Regression Model p = 0.001 for genotype*dose interaction, p < 0.0001 for dose, and p = 0.024 for dose*age interaction). There were no differences between genotypes in locomotion, either after saline or morphine injections as recorded during the first conditioning session (Figure 8b).
Figure 8. Morphine conditioned place preference and locomotion in the recombinant KEPI KO mice.
(a) KEPI homozygote knockouts displayed a rightward shift in the dose-response curve for morphine conditioned place preference when compared to WT animals. Heterozygote knockouts showed an intermediate phenotype (Linear Regression p = 0.001 for genotype * dose interaction). (b) There was no difference in morphine-induced locomotion as recorded during the first conditioning session. Data are expressed as means ± SEM (n = 12 per group).
Discussion
In this study, we have demonstrated that KEPI KO mice display significantly diminished responses to repeated administration of morphine in both hot plate and conditioned place preference tests. The same animals display normal analgesic responses and normal locomotion after their first exposures to the drug and the same level of naloxone-precipitated withdrawal. We also demonstrate use of genomic markers around a KO locus to rapidly identify recombinant mice that allowed us to remove “hitch-hiking genes” carried over from embryonic stem cells. Given that these hitch hiking genes might confound the KO phenotype in other cases, as well as this one, we discuss this technical feature of the study separately.
The effects of genetic background on phenotypes observed in genetically-modified mice have to be considered with care. A confounding effect of genetic background may be especially likely in areas of the genome near the transgenic insertion. This part of genome might differ between KO and WT mice, even in congenic animals produced in lines different from those that donated the embryonic stem cells. In most instances, KO mice are created when the KO DNA construct is introduced into embryonic stem cells from 129S6-mice, which are then microinjected into host blastocysts to produce chimeric offspring. To increase fertility and facilitate phenotypic analysis, particularly those related to phenotypes for which this genetic background is more favorable (such as preference for abused drugs), the chimeras are often backcrossed with C57BL/6 mice. As a result, the genetic background close to the targeted gene will reflect the 129S6 donor, whereas wild-type mice will have the corresponding region derived from C57BL/6 DNA (Hospital, 2001). Consequently, the phenotype of the KO mouse may result not from the mutated gene but from “hitchhiking” donor gene(s). Our original KEPI KO line provides an example of this phenomenon. Since KEPI lies close to a major QTL influencing morphine antinociception (Belknap et al., 1995, Bergeson et al., 2001), and the genetic markers in this area differ between the original KO and WT animals, the increased sensitivity to morphine analgesia most probably reflected the differences in morphine sensitivity between the C57BL/6 and the 129 strains (Mogil and Wilson, 1997, Kest et al., 2002, Leo et al., 2008). The fact that the differences in responses to morphine found in the original KO line were not observed in the recombinant KO line supports this assertion.
To reduce the length of the retained chromosomal segment, marker-assisted selection has long been used in the field of plant genetics (Ragot, 1995, Hospital, 2001, Hospital, 2005). In mouse models, marker assisted back-crossing has been applied to breaking QTLs into smaller intervals (Ferraro et al., 2007, Bennett et al., 2008, Doyle et al., 2008). However, to our knowledge, this approach has not been previously used to change the genetic background surrounding a gene deletion. This approach can be efficient and save generations of backcrossing. Indeed, Hospital (Hospital, 2001) calculated that the chance of obtaining double recombinants for marker pairs as close as 2 cM from the deletion site to be favorable in just 3 generations of backcrossing. When 49 individuals/generation are genotyped, there should be a 48 % chance of obtaining such double recombinants. Our results lie well within this estimate. With larger numbers of animals per generation, we obtained 4 such recombinants. One of them, the founder of the recombinant line, displayed C57BL6 markers within less than 1 Mb centromeric and telomeric to the KEPI deletion. We can thus conclude with a large degree of confidence that the phenotype of the recombinant KEPI KO mice described above reflects the direct or indirect effects of KEPI deletion, and not a genetic background difference between the knockouts and the WT controls.
Since KEPI was originally identified as a morphine regulated gene (Liu et al., 2002), we have focused on effects of the KEPI deletion on morphine-related behavioral traits. We centered our studies on the recombinant KEPI line because of apparent differences in analgesic responses resulting from differential association of C57 and 129S6 Oprm1 alleles with WT and KO animals in the original KEPI KO line (Figures 2 and 5). In contrast to the effects observed in the original KEPI knockouts, we did not observe any differences between WT and recombinant KEPI KO mice in baseline responses to noxious thermal stimuli or in baseline locomotion. However, during a course of repeated morphine injections, KO animals developed tolerance to the analgesic effects of morphine faster than the wild-type mice. Accelerated tolerance was not apparent in the tail withdrawal test, a measure of “spinal” analgesia, but only in hot plate test assessments of largely “supraspinal” nociceptive responses. Since the hot plate test is known to display “behavioral tolerance” with repeated testing, we addressed this issue by “habituating” the animals for three days to the testing procedure, using two “baseline” nociception measurements per day. Since this treatment did not lead to development of significant behavioral tolerance (Repeated Measures ANOVA p = 0.50 for day effect and p = 0.42 for day * genotype interaction; n = 9 per group) we believe that the observed latency changes are likely to be attributable to the development of analgesic tolerance to morphine. These behavioral observations are in good agreement with biochemical data that document elevated PP1 activity in thalamus but not in spinal cord (Figure 3). Elevated serine/threonine phosphatase activity, probably in response to increased kinase activity, has previously been observed after morphine treatment. Inhibition of this phosphatase activity antagonized morphine antinociception (Moncada et al., 2003, Gabra et al., 2007). This antagonism occurred at low concentrations of okadaic acid which predominantly inhibit protein phosphatase 2A. When okadaic acid concentrations are raised to levels at which PP2a and PP1 are both inhibited, this effect was reversed. PP1 thus seemed to counterbalance protein phosphatase 2A activity (Moncada et al., 2003). Selective inhibition of PP1 thus appears to enhance morphine analgesia and increased PP1 activity to suppress it. This picture agrees with our observations in KEPI KO mice, in which we detected both increased PP1 activity and decreased responsiveness to morphine after repeated morphine treatments.
While KEPI KO animals showed enhanced tolerance development after chronic morphine treatment, they did not show increased dependence. Selective effects on tolerance versus dependence have been reported in mice with a number of genetic manipulations, including p75 neurotrophin receptor deletion, mGluR5 deletion and Gβ5 knockdown (Sanchez-Blazquez et al., 2003, Bogulavsky et al., 2009, Trang et al., 2009). Tolerance and dependence, although often correlated, can thus be biologically dissociated. Even more interesting is the fact that KEPI deletion produced accelerated tolerance development only in hot plate test, which assesses mainly “supraspinal” nociceptive responses and not in the tail withdrawal test, a measure of “spinal” analgesia. A selective effect on spinal versus supraspinal tolerance development has been observed in β-arrestin-2 KO mice (Bohn et al., 2000, Bohn et al., 2002) and may stem from differential regulation of opioid signaling in different brain areas. However, β-arrestin-2 deletion, and most other reported manipulations, attenuate tolerance rather than enhance it (Table 3). Several previous reports do identify accelerated development of tolerance in genetically altered mice, however (Hendry et al., 2000, Charlton et al., 2008, Solecki et al., 2008). Of these, deletion of the guanine nucleotide binding protein αz subunit produces effects that are most similar to those produced by KEPI deletion. Akin to KEPI knockouts, mice deficient in the alpha subunit of the Gz protein, Gzα, displayed accelerated tolerance development only in the test for supraspinal analgesia (hot plate), and not in tail withdrawal test, the measure of “spinal” analgesia (Hendry et al., 2000). Thus intact KEPI and Gzα may each be necessary for minimizing the development of morphine tolerance in the supraspinal systems that contribute to morphine analgesia.
Table 3. Genes involved in regulation of morphine analgesia, tolerance, withdrawal and reward.
Effect of knockout or knockdown of these genes on morphine related behaviors.
| Gene | Analgesia | Tolerance | Dependence | Reward | Reference |
|---|---|---|---|---|---|
| GRK6 | normal | normal | normal | Normal, CPP | (Raehal et al., 2009) |
| DOPr | normal | abolished | normal | reduced, CPP | (Nitsche et al., 2002) |
| p75NTR | normal | abolished | normal | (Trang et al., 2009) | |
| GluR5 | normal | abolished | normal | normal, CPP | (Bogulavsky et al., 2009) |
| AQP4 | enhanced | attenuated | abolished | (Wu et al., 2008) | |
| (PSD)-93 | attenuated | abolished | (Liaw et al., 2008) | ||
| FosB | normal | enhanced | (Solecki et al., 2008) | ||
| Spinophilin | reduced | enhanced | enhanced | enhanced | (Charlton et al., 2008) |
| RGS14 | enhanced | attenuated | (Rodriguez-Munoz et al., 2007) | ||
| β2-AR | enhanced | abolished | reduced | (Liang et al., 2007) | |
| PKCγ | normal | attenuated | increased | (Zeitz et al., 2001) | |
| PKCε | enhanced | attenuated | enhanced | (Newton et al., 2007) | |
| AC1 | normal | normal | reduced | reduced | (Li et al., 2006) |
| AC8 | normal | attenuated | |||
| OFQ/N | normal | attenuated | normal | (Chung et al., 2006) | |
| CaMKIV | normal | attenuated | normal | (Ko et al., 2006) | |
| AC5 | attenuated | attenuated | attenuated | attenuated | (Kim et al., 2006) |
| ppENK | normal | attenuated | normal | normal, CPP | (Nitsche et al., 2002) |
| PLCβ1 | attenuated | attenuated | (Liu et al., 2006) | ||
| GPR10 | enhanced | attenuated | attenuated | enhanced | (Laurent et al., 2005) |
| Cav2.3 | enhanced | abolished | (Yokoyama et al., 2004) | ||
| α2A | normal | normal | reduced | (Ozdogan et al., 2004) | |
| Gαz | normal | enhanced | (Hendry et al., 2000) | ||
| GluRε1 | normal | attenuated | attenuated | reduced, CPP | (Miyamoto et al., 2004) |
| RGS9 | enhanced | attenuated | enhanced | enhanced | (Zachariou et al., 2003) |
| HO-2 | normal | abolished | (Liang et al., 2003) | ||
| RGS7 | enhanced | attenuated | (Garzon et al., 2003) | ||
| 12-LO | enhanced | normal | enhanced | (Walters et al., 2003) | |
| Gβ5 | enhanced | enhanced | normal | (Sanchez-Blazquez et al., 2003) | |
| NT4 | normal | attenuated | normal | (Smith et al., 2003) | |
| M5 | normal | normal | attenuated | reduced, CPP | (Basile et al., 2002) |
| PhLPL | attenuated | prolonged | normal | (Garzon et al., 2002) | |
| GluR-A | normal | attenuated | attenuated | (Vekovischeva et al., 2001) | |
| IL-6 | attenuated | enhanced | (Bianchi et al., 1999) | ||
| aCGRP | normal | normal | attenuated | Normal, SA | (Salmon et al., 2001) |
| mPKCI | enhanced | enhanced | (Guang et al., 2004) | ||
| CREB | normal | attenuated | (Maldonado et al., 1996) |
Recombinant KEPI knockouts displayed an apparent rightward shift in the dose-response relationship for morphine reward in conditioned place preference testing. The conditioned place preference paradigm provides combined information about rewarding and conditioned effects of morphine. We thus cannot distinguish among possible contributions of alterations in a) initial rewarding effects of morphine, b) tolerance or c) conditioned effects in producing the observed behavioral outcomes. The effects of KEPI deletion on conditioned place preference and on tolerance in hot plate testing appear modest, as do the measured increases in phosphatase activity. However, it is important to note that adaptive changes in the brains of these constitutive KO animals, such as the observed decrease in expression of PP1β, might modulate the effects of KEPI deletion. Other studies have documented the robustness of the phosphatase system and its resistance to large perturbations. For example, while deletion or mutation of the protein kinase A-activated PP1 inhibitor DARPP-32 displays sizable effects on psychostimulant-induced phosphorylation of Erk2, CREB, and GSK-3 (Fienberg et al., 1998, Svenningsson et al., 2003, Valjent et al., 2005), effects on cocaine reward are much more modest (Zachariou et al., 2002). Deletion of DARPP-32 or removal of its protein kinase A phosphorylation site attenuate morphine-induced hyperlocomotion but fail to influence morphine reward (Borgkvist et al., 2007). Interestingly, these observations in DARP-32 KO mice are opposite to those findings noted here in KEPI knockout mice. Such differences point clearly to the specific roles that each of these PP1 inhibitors play in the regulation of different behavioral responses to drugs of abuse.
Certain limitations are important to keep in mind when considering the present results. The modest size and the specificity of effects noted in both biochemical and behavioral experiments are consistent with the presence of diverse and numerous regulatory influences on PP1 and other phosphatases. Brain distribution data suggests that these diverse PP1 regulators are likely to play different roles in distinct brain regions. It is reasonable to postulate that different PP1 regulators differ in their ability to adapt to the lifelong absence of KEPI, which may then affect the set of effects ultimately observed in KEPI knockout mice. Although we make a comparison here between the two lines, it was not our initial intention to make such a comparison, but rather to develop a line to study KEPI in which the confounding effects of flanking genes were not present. Consequently, we eliminated the “original” line prior to analysis of all of the data from the recombinant line limiting our ability to make more detailing comparisons, including more detailed analysis of EC50 values. In addition, the cumulative dosing method used here, and elsewhere in gene knockout mice (e.g. Sora et al, 2001) to examine dose-effect relationships does not provide for very accurate estimations of EC50 values, although it does provide for meaningful comparisons between genotypes. Despite these limitations, significant knockout effects on overall dose/response relationships in the initial strain of knockout mice, in the context of the lack of such significant effects in the recombinant line, suggest that the most likely explanation for this set of data is that these effects were driven by background differences at the nearby OPRM1 locus in the original line.
Neurobiological adaptations to chronic morphine treatment are a subject of longstanding and continuing research. According to one current model, after treatment with some μ opioid receptor (MOR) agonists, MOR phosphorylation by G protein coupled receptor kinases (GRKs) leads to the receptor desensitization via binding of the regulatory protein β–arrestin, receptor internalization and eventual recycling back to the plasma membrane. A resulting protracted desensitization is postulated to contribute to tolerance development observed in vivo, as suggested by the diminished tolerance of β–arrestin-2 KO mice (Bohn et al., 2000, Bohn et al., 2002). However, morphine, unlike opioid peptides, fails to promote receptor phosphorylation, arrestin binding, endocytosis or desensitization in many circumstances (Arden et al., 1995, Keith et al., 1996, Keith et al., 1998, Whistler and von Zastrow, 1998, Zhang et al., 1998). Adaptations in downstream signaling pathways that include protein kinase C and protein kinase A-dependent systems provide alternative sites at which to seek additional contributions to morphine-induced tolerance and desensitization (Nestler and Tallman, 1988, Narita et al., 1994). Identification of substrates whose phosphorylation is altered in KEPI knockouts provides one avenue for identification of such downstream pathways. Morphine is reported to alter phosphorylation of many proteins that can play roles in MOR signaling, including MOR, GRK, G β-subunit, β-arrestin, adenylate cyclase, NMDA receptors, CREB, p38MAPK, and ERK; these and other phosphoproteins would all be interesting to study in the KEPI KO mice. (Chen and Huang, 1991, Guitart et al., 1992, Zhang et al., 1996, Gutstein et al., 1997, Chakrabarti et al., 1998, Chakrabarti et al., 2001).
In conclusion, the results from this analysis of recombinant KEPI KO mice demonstrate that KEPI contributes to adaptive responses to repeated administration of morphine including analgesic tolerance and drug reward. The data from marker-assisted backcrossing that allowed the production of the recombinant KEPI KO mice demonstrate that most of the genetic background flanking the deletion site can be replaced in just few generations of breeding and indicates that this method may be a generally useful approach for avoiding the confounding effects of “hitch-hiking” genes.
Acknowledgements
We thank Dr. Qing-Rong Liu for help with the phosphatase assay and several consultations during this work. We acknowledge valuable support of the breeding assistant Kriss Knestaut from Charles River Laboratories, help with ES cells from Teri Perona, and financial support from the National Institute on Drug Abuse Intramural Research Program (NIH/ DHHS).
List of Abbreviations
- CNS
central nervous system
- CPP
conditioned place preference
- DNA
deoxyribonucleic acid
- ES
embryonic stem
- KO
knockout
- MPE
maximal possible effect
- PAG
periaqueductal grey
- PCR
polymerase chain reaction
- PKC
protein kinase C
- PP1
protein phosphatase 1
- QTL
quantitative trait locus
- SNP
single nucleotide polymorphism
- WT
wild type
References
- Arden JR, Segredo V, Wang Z, Lameh J, Sadee W. Phosphorylation and agonist-specific intracellular trafficking of an epitope-tagged mu-opioid receptor expressed in HEK 293 cells. J Neurochem. 1995;65:1636–1645. doi: 10.1046/j.1471-4159.1995.65041636.x. [DOI] [PubMed] [Google Scholar]
- Basile AS, Fedorova I, Zapata A, Liu X, Shippenberg T, Duttaroy A, Yamada M, Wess J. Deletion of the M5 muscarinic acetylcholine receptor attenuates morphine reinforcement and withdrawal but not morphine analgesia. Proc Natl Acad Sci U S A. 2002;99:11452–11457. doi: 10.1073/pnas.162371899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belknap JK, Mogil JS, Helms ML, Richards SP, O'Toole LA, Bergeson SE, Buck KJ. Localization to chromosome 10 of a locus influencing morphine analgesia in crosses derived from C57BL/6 and DBA/2 strains. Life Sci. 1995;57:PL117–PL124. doi: 10.1016/0024-3205(95)02040-p. [DOI] [PubMed] [Google Scholar]
- Bennett B, Carosone-Link P, Beeson M, Gordon L, Phares-Zook N, Johnson TE. Genetic dissection of quantitative trait locus for ethanol sensitivity in long- and short-sleep mice. Genes Brain Behav. 2008;7:659–668. doi: 10.1111/j.1601-183X.2008.00403.x. [DOI] [PubMed] [Google Scholar]
- Bergeson SE, Helms ML, O'Toole LA, Jarvis MW, Hain HS, Mogil JS, Belknap JK. Quantitative trait loci influencing morphine antinociception in four mapping populations. Mamm Genome. 2001;12:546–553. doi: 10.1007/s003350020022. [DOI] [PubMed] [Google Scholar]
- Berrettini WH, Ferraro TN, Alexander RC, Buchberg AM, Vogel WH. Quantitative trait loci mapping of three loci controlling morphine preference using inbred mouse strains. Nat Genet. 1994;7:54–58. doi: 10.1038/ng0594-54. [DOI] [PubMed] [Google Scholar]
- Bianchi M, Maggi R, Pimpinelli F, Rubino T, Parolaro D, Poli V, Ciliberto G, Panerai AE, Sacerdote P. Presence of a reduced opioid response in interleukin-6 knock out mice. Eur J Neurosci. 1999;11:1501–1507. doi: 10.1046/j.1460-9568.1999.00563.x. [DOI] [PubMed] [Google Scholar]
- Bogulavsky JJ, Gregus AM, Kim PT, Costa AC, Rajadhyaksha AM, Inturrisi CE. Deletion of the glutamate receptor 5 subunit of kainate receptors affects the development of morphine tolerance. J Pharmacol Exp Ther. 2009;328:579–587. doi: 10.1124/jpet.108.144121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn LM, Gainetdinov RR, Lin FT, Lefkowitz RJ, Caron MG. Mu-opioid receptor desensitization by beta-arrestin-2 determines morphine tolerance but not dependence. Nature. 2000;408:720–723. doi: 10.1038/35047086. [DOI] [PubMed] [Google Scholar]
- Bohn LM, Lefkowitz RJ, Caron MG. Differential mechanisms of morphine antinociceptive tolerance revealed in (beta)arrestin-2 knock-out mice. J Neurosci. 2002;22:10494–10500. doi: 10.1523/JNEUROSCI.22-23-10494.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgkvist A, Usiello A, Greengard P, Fisone G. Activation of the cAMP/PKA/DARPP-32 signaling pathway is required for morphine psychomotor stimulation but not for morphine reward. Neuropsychopharmacology. 2007;32:1995–2003. doi: 10.1038/sj.npp.1301321. [DOI] [PubMed] [Google Scholar]
- Caenepeel S, Charydczak G, Sudarsanam S, Hunter T, Manning G. The mouse kinome: discovery and comparative genomics of all mouse protein kinases. Proc Natl Acad Sci U S A. 2004;101:11707–11712. doi: 10.1073/pnas.0306880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Oppermann M, Gintzler AR. Chronic morphine induces the concomitant phosphorylation and altered association of multiple signaling proteins: a novel mechanism for modulating cell signaling. Proc Natl Acad Sci U S A. 2001;98:4209–4214. doi: 10.1073/pnas.071031798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Wang L, Tang WJ, Gintzler AR. Chronic morphine augments adenylyl cyclase phosphorylation: relevance to altered signaling during tolerance/dependence. Mol Pharmacol. 1998;54:949–953. doi: 10.1124/mol.54.6.949. [DOI] [PubMed] [Google Scholar]
- Charlton JJ, Allen PB, Psifogeorgou K, Chakravarty S, Gomes I, Neve RL, Devi LA, Greengard P, Nestler EJ, Zachariou V. Multiple actions of spinophilin regulate mu opioid receptor function. Neuron. 2008;58:238–247. doi: 10.1016/j.neuron.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Huang LY. Sustained potentiation of NMDA receptor-mediated glutamate responses through activation of protein kinase C by a mu opioid. Neuron. 1991;7:319–326. doi: 10.1016/0896-6273(91)90270-a. [DOI] [PubMed] [Google Scholar]
- Chung S, Pohl S, Zeng J, Civelli O, Reinscheid RK. Endogenous orphanin FQ/nociceptin is involved in the development of morphine tolerance. J Pharmacol Exp Ther. 2006;318:262–267. doi: 10.1124/jpet.106.103960. [DOI] [PubMed] [Google Scholar]
- Cohen P, Alemany S, Hemmings BA, Resink TJ, Stralfors P, Tung HY. Protein phosphatase-1 and protein phosphatase-2A from rabbit skeletal muscle. Methods Enzymol. 1988;159:390–408. doi: 10.1016/0076-6879(88)59039-0. [DOI] [PubMed] [Google Scholar]
- Crawley JN. Behavioral phenotyping of transgenic and knockout mice: experimental design and evaluation of general health, sensory functions, motor abilities, and specific behavioral tests. Brain Res. 1999;835:18–26. doi: 10.1016/s0006-8993(98)01258-x. [DOI] [PubMed] [Google Scholar]
- D'amour F, Smith D. A method for determining loss of pain sensation. J Pharmacol Exp Ther. 1941;72:74–79. [Google Scholar]
- Doyle GA, Furlong PJ, Schwebel CL, Smith GG, Lohoff FW, Buono RJ, Berrettini WH, Ferraro TN. Fine Mapping of a Major QTL Influencing Morphine Preference in C57BL/6 and DBA/2 Mice Using Congenic Strains. Neuropsychopharmacology. 2008 doi: 10.1038/npp.2008.14. [DOI] [PubMed] [Google Scholar]
- Eto M, Karginov A, Brautigan DL. A novel phosphoprotein inhibitor of protein type-1 phosphatase holoenzymes. Biochemistry. 1999;38:16952–16957. doi: 10.1021/bi992030o. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Smith GG, Schwebel CL, Lohoff FW, Furlong P, Berrettini WH, Buono RJ. Quantitative trait locus for seizure susceptibility on mouse chromosome 5 confirmed with reciprocal congenic strains. Physiol Genomics. 2007;31:458–462. doi: 10.1152/physiolgenomics.00123.2007. [DOI] [PubMed] [Google Scholar]
- Fienberg AA, Hiroi N, Mermelstein PG, Song W, Snyder GL, Nishi A, Cheramy A, O'Callaghan JP, Miller DB, Cole DG, Corbett R, Haile CN, Cooper DC, Onn SP, Grace AA, Ouimet CC, White FJ, Hyman SE, Surmeier DJ, Girault J, Nestler EJ, Greengard P. DARPP-32: regulator of the efficacy of dopaminergic neurotransmission. Science. 1998;281:838–842. doi: 10.1126/science.281.5378.838. [DOI] [PubMed] [Google Scholar]
- Gabra BH, Bailey CP, Kelly E, Sanders AV, Henderson G, Smith FL, Dewey WL. Evidence for an important role of protein phosphatases in the mechanism of morphine tolerance. Brain Res. 2007;1159:86–93. doi: 10.1016/j.brainres.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon J, Lopez-Fando A, Sanchez-Blazquez P. The R7 subfamily of RGS proteins assists tachyphylaxis and acute tolerance at mu-opioid receptors. Neuropsychopharmacology. 2003;28:1983–1990. doi: 10.1038/sj.npp.1300263. [DOI] [PubMed] [Google Scholar]
- Garzon J, Rodriguez-Diaz M, Lopez-Fando A, Garcia-Espana A, Sanchez-Blazquez P. Glycosylated phosducin-like protein long regulates opioid receptor function in mouse brain. Neuropharmacology. 2002;42:813–828. doi: 10.1016/s0028-3908(02)00027-8. [DOI] [PubMed] [Google Scholar]
- Gerlai R. Gene-targeting studies of mammalian behavior: is it the mutation or the background genotype? Trends Neurosci. 1996;19:177–181. doi: 10.1016/s0166-2236(96)20020-7. [DOI] [PubMed] [Google Scholar]
- Gong JP, Liu QR, Zhang PW, Wang Y, Uhl GR. Mouse brain localization of the protein kinase C-enhanced phosphatase 1 inhibitor KEPI (kinase C-enhanced PP1 inhibitor) Neuroscience. 2005;132:713–727. doi: 10.1016/j.neuroscience.2004.11.052. [DOI] [PubMed] [Google Scholar]
- Guang W, Wang H, Su T, Weinstein IB, Wang JB. Role of mPKCI, a novel mu-opioid receptor interactive protein, in receptor desensitization, phosphorylation, and morphine-induced analgesia. Mol Pharmacol. 2004;66:1285–1292. doi: 10.1124/mol.66.5.. [DOI] [PubMed] [Google Scholar]
- Guitart X, Thompson MA, Mirante CK, Greenberg ME, Nestler EJ. Regulation of cyclic AMP response element-binding protein (CREB) phosphorylation by acute and chronic morphine in the rat locus coeruleus. J Neurochem. 1992;58:1168–1171. doi: 10.1111/j.1471-4159.1992.tb09377.x. [DOI] [PubMed] [Google Scholar]
- Gutstein HB, Rubie EA, Mansour A, Akil H, Woodgett JR. Opioid effects on mitogen-activated protein kinase signaling cascades. Anesthesiology. 1997;87:1118–1126. doi: 10.1097/00000542-199711000-00016. [DOI] [PubMed] [Google Scholar]
- Hall FS, Drgonova J, Goeb M, Uhl GR. Reduced behavioral effects of cocaine in heterozygous brain-derived neurotrophic factor (BDNF) knockout mice. Neuropsychopharmacology. 2003;28:1485–1490. doi: 10.1038/sj.npp.1300192. [DOI] [PubMed] [Google Scholar]
- Hall FS, Huang S, Fong GF, Pert A. The effects of social isolation on the forced swim test in Fawn hooded and Wistar rats. J Neurosci Methods. 1998;79:47–51. doi: 10.1016/s0165-0270(97)00155-6. [DOI] [PubMed] [Google Scholar]
- Hendry IA, Kelleher KL, Bartlett SE, Leck KJ, Reynolds AJ, Heydon K, Mellick A, Megirian D, Matthaei KI. Hypertolerance to morphine in G(z alpha)-deficient mice. Brain Res. 2000;870:10–19. doi: 10.1016/s0006-8993(00)02387-8. [DOI] [PubMed] [Google Scholar]
- Hospital F. Size of donor chromosome segments around introgressed loci and reduction of linkage drag in marker-assisted backcross programs. Genetics. 2001;158:1363–1379. doi: 10.1093/genetics/158.3.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital F. Selection in backcross programmes. Philos Trans R Soc Lond B Biol Sci. 2005;360:1503–1511. doi: 10.1098/rstb.2005.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith DE, Anton B, Murray SR, Zaki PA, Chu PC, Lissin DV, Monteillet-Agius G, Stewart PL, Evans CJ, von Zastrow M. mu-Opioid receptor internalization: opiate drugs have differential effects on a conserved endocytic mechanism in vitro and in the mammalian brain. Mol Pharmacol. 1998;53:377–384. [PubMed] [Google Scholar]
- Keith DE, Murray SR, Zaki PA, Chu PC, Lissin DV, Kang L, Evans CJ, von Zastrow M. Morphine activates opioid receptors without causing their rapid internalization. J Biol Chem. 1996;271:19021–19024. doi: 10.1074/jbc.271.32.19021. [DOI] [PubMed] [Google Scholar]
- Kest B, Hopkins E, Palmese CA, Adler M, Mogil JS. Genetic variation in morphine analgesic tolerance: a survey of 11 inbred mouse strains. Pharmacol Biochem Behav. 2002;73:821–828. doi: 10.1016/s0091-3057(02)00908-5. [DOI] [PubMed] [Google Scholar]
- Kim KS, Lee KW, Lee KW, Im JY, Yoo JY, Kim SW, Lee JK, Nestler EJ, Han PL. Adenylyl cyclase type 5 (AC5) is an essential mediator of morphine action. Proc Natl Acad Sci U S A. 2006;103:3908–3913. doi: 10.1073/pnas.0508812103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko SW, Jia Y, Xu H, Yim SJ, Jang DH, Lee YS, Zhao MG, Toyoda H, Wu LJ, Chatila T, Kaang BK, Zhuo M. Evidence for a role of CaMKIV in the development of opioid analgesic tolerance. Eur J Neurosci. 2006;23:2158–2168. doi: 10.1111/j.1460-9568.2006.04748.x. [DOI] [PubMed] [Google Scholar]
- Lalonde R, Bensoula AN, Filali M. Rotorod sensorimotor learning in cerebellar mutant mice. Neurosci Res. 1995;22:423–426. doi: 10.1016/0168-0102(95)00916-h. [DOI] [PubMed] [Google Scholar]
- Laurent P, Becker JA, Valverde O, Ledent C, de Kerchove d'Exaerde A, Schiffmann SN, Maldonado R, Vassart G, Parmentier M. The prolactin-releasing peptide antagonizes the opioid system through its receptor GPR10. Nat Neurosci. 2005;8:1735–1741. doi: 10.1038/nn1585. [DOI] [PubMed] [Google Scholar]
- Leo S, Straetemans R, D'Hooge R, Meert T. Differences in nociceptive behavioral performance between C57BL/6J, 129S6/SvEv, B6 129 F1 and NMRI mice. Behav Brain Res. 2008;190:233–242. doi: 10.1016/j.bbr.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Li S, Lee ML, Bruchas MR, Chan GC, Storm DR, Chavkin C. Calmodulin-stimulated adenylyl cyclase gene deletion affects morphine responses. Mol Pharmacol. 2006;70:1742–1749. doi: 10.1124/mol.106.025783. [DOI] [PubMed] [Google Scholar]
- Liang D, Li X, Lighthall G, Clark JD. Heme oxygenase type 2 modulates behavioral and molecular changes during chronic exposure to morphine. Neuroscience. 2003;121:999–1005. doi: 10.1016/s0306-4522(03)00483-4. [DOI] [PubMed] [Google Scholar]
- Liang DY, Shi X, Li X, Li J, Clark JD. The beta2 adrenergic receptor regulates morphine tolerance and physical dependence. Behav Brain Res. 2007;181:118–126. doi: 10.1016/j.bbr.2007.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw WJ, Zhu XG, Yaster M, Johns RA, Gauda EB, Tao YX. Distinct expression of synaptic NR2A and NR2B in the central nervous system and impaired morphine tolerance and physical dependence in mice deficient in postsynaptic density-93 protein. Mol Pain. 2008;4:45. doi: 10.1186/1744-8069-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NJ, vonGizycki H, Gintzler AR. Phospholipase Cbeta1 modulates pain sensitivity, opioid antinociception and opioid tolerance formation. Brain Res. 2006;1069:47–53. doi: 10.1016/j.brainres.2005.09.069. [DOI] [PubMed] [Google Scholar]
- Liu QR, Zhang PW, Zhen Q, Walther D, Wang XB, Uhl GR. KEPI, a PKC-dependent protein phosphatase 1 inhibitor regulated by morphine. J Biol Chem. 2002;277:13312–13320. doi: 10.1074/jbc.M107558200. [DOI] [PubMed] [Google Scholar]
- Maldonado R, Blendy JA, Tzavara E, Gass P, Roques BP, Hanoune J, Schutz G. Reduction of morphine abstinence in mice with a mutation in the gene encoding CREB. Science. 1996;273:657–659. doi: 10.1126/science.273.5275.657. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- McLaren L, Boyle S, Mason JO, Bard JB. Expression and genomic characterization of protein phosphatase inhibitor-1: a novel marker for mesothelium in the mouse. Mech Dev. 2000;96:237–241. doi: 10.1016/s0925-4773(00)00388-9. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Yamada K, Nagai T, Mori H, Mishina M, Furukawa H, Noda Y, Nabeshima T. Behavioural adaptations to addictive drugs in mice lacking the NMDA receptor epsilon1 subunit. Eur J Neurosci. 2004;19:151–158. doi: 10.1111/j.1460-9568.2004.03086.x. [DOI] [PubMed] [Google Scholar]
- Mogil JS, Wilson SG. Nociceptive and morphine antinociceptive sensitivity of 129 and C57BL/6 inbred mouse strains: implications for transgenic knock-out studies. Eur J Pain. 1997;1:293–297. doi: 10.1016/s1090-3801(97)90038-0. [DOI] [PubMed] [Google Scholar]
- Moncada A, Cendan CM, Baeyens JM, Del Pozo E. Effects of serine/threonine protein phosphatase inhibitors on morphine-induced antinociception in the tail flick test in mice. Eur J Pharmacol. 2003;465:53–60. doi: 10.1016/s0014-2999(03)01461-4. [DOI] [PubMed] [Google Scholar]
- Morris R. Developments of a water-maze procedure for studying spatial learning in the rat. J Neurosci Methods. 1984;11:47–60. doi: 10.1016/0165-0270(84)90007-4. [DOI] [PubMed] [Google Scholar]
- Narita M, Makimura M, Feng Y, Hoskins B, Ho IK. Influence of chronic morphine treatment on protein kinase C activity: comparison with butorphanol and implication for opioid tolerance. Brain Res. 1994;650:175–179. doi: 10.1016/0006-8993(94)90224-0. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Tallman JF. Chronic morphine treatment increases cyclic AMP-dependent protein kinase activity in the rat locus coeruleus. Mol Pharmacol. 1988;33:127–132. [PubMed] [Google Scholar]
- Newton PM, Kim JA, McGeehan AJ, Paredes JP, Chu K, Wallace MJ, Roberts AJ, Hodge CW, Messing RO. Increased response to morphine in mice lacking protein kinase C epsilon. Genes Brain Behav. 2007;6:329–338. doi: 10.1111/j.1601-183X.2006.00261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsche JF, Schuller AG, King MA, Zengh M, Pasternak GW, Pintar JE. Genetic dissociation of opiate tolerance and physical dependence in delta-opioid receptor-1 and preproenkephalin knock-out mice. J Neurosci. 2002;22:10906–10913. doi: 10.1523/JNEUROSCI.22-24-10906.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdogan UK, Lahdesmaki J, Hakala K, Scheinin M. The involvement of alpha 2A-adrenoceptors in morphine analgesia, tolerance and withdrawal in mice. Eur J Pharmacol. 2004;497:161–171. doi: 10.1016/j.ejphar.2004.06.051. [DOI] [PubMed] [Google Scholar]
- Perez RG, Lewis RM. Regional distribution of DARPP-32 (dopamine- and adenosine 3',5'-monophosphate-regulated phosphoprotein of Mr = 32,000) mRNA in mouse brain. J Comp Neurol. 1992;318:304–315. doi: 10.1002/cne.903180307. [DOI] [PubMed] [Google Scholar]
- Raehal KM, Schmid CL, Medvedev IO, Gainetdinov RR, Premont RT, Bohn LM. Morphine-induced physiological and behavioral responses in mice lacking G protein-coupled receptor kinase 6. Drug Alcohol Depend. 2009 doi: 10.1016/j.drugalcdep.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragot M. Les Cilloques. vol. no.72. Paris: INRA; 1995. Marker-assisted backcrossing: a practical example. Techniques et utilisations des marqueurs moleculaires. [Google Scholar]
- Rodriguez-Munoz M, de la Torre-Madrid E, Gaitan G, Sanchez-Blazquez P, Garzon J. RGS14 prevents morphine from internalizing Mu-opioid receptors in periaqueductal gray neurons. Cell Signal. 2007;19:2558–2571. doi: 10.1016/j.cellsig.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Salmon AM, Damaj MI, Marubio LM, Epping-Jordan MP, Merlo-Pich E, Changeux JP. Altered neuroadaptation in opiate dependence and neurogenic inflammatory nociception in alpha CGRP-deficient mice. Nat Neurosci. 2001;4:357–358. doi: 10.1038/86001. [DOI] [PubMed] [Google Scholar]
- Sanchez-Blazquez P, Rodriguez-Diaz M, Lopez-Fando A, Rodriguez-Munoz M, Garzon J. The GBeta5 subunit that associates with the R7 subfamily of RGS proteins regulates mu-opioid effects. Neuropharmacology. 2003;45:82–95. doi: 10.1016/s0028-3908(03)00149-7. [DOI] [PubMed] [Google Scholar]
- Smith DJ, Leil TA, Liu X. Neurotrophin-4 is required for tolerance to morphine in the mouse. Neurosci Lett. 2003;340:103–106. doi: 10.1016/s0304-3940(03)00084-3. [DOI] [PubMed] [Google Scholar]
- Solecki W, Krowka T, Kubik J, Kaczmarek L, Przewlocki R. Increased analgesic tolerance to acute morphine in fosB knock-out mice: a gender study. Pharmacol Biochem Behav. 2008;90:512–516. doi: 10.1016/j.pbb.2008.03.031. [DOI] [PubMed] [Google Scholar]
- Sora I, Elmer G, Funada M, Pieper J, Li XF, Hall FS, Uhl GR. Mu opiate receptor gene dose effects on different morphine actions: evidence for differential in vivo mu receptor reserve. Neuropsychopharmacology. 2001;25:41–54. doi: 10.1016/S0893-133X(00)00252-9. [DOI] [PubMed] [Google Scholar]
- Sora I, Takahashi N, Funada M, Ujike H, Revay RS, Donovan DM, Miner LL, Uhl GR. Opiate receptor knockout mice define mu receptor roles in endogenous nociceptive responses and morphine-induced analgesia. Proc Natl Acad Sci U S A. 1997;94:1544–1549. doi: 10.1073/pnas.94.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsson P, Tzavara ET, Carruthers R, Rachleff I, Wattler S, Nehls M, McKinzie DL, Fienberg AA, Nomikos GG, Greengard P. Diverse psychotomimetics act through a common signaling pathway. Science. 2003;302:1412–1415. doi: 10.1126/science.1089681. [DOI] [PubMed] [Google Scholar]
- Trang T, Koblic P, Kawaja M, Jhamandas K. Attenuation of opioid analgesic tolerance in p75 neurotrophin receptor null mutant mice. Neurosci Lett. 2009;451:69–73. doi: 10.1016/j.neulet.2008.12.032. [DOI] [PubMed] [Google Scholar]
- Valjent E, Pascoli V, Svenningsson P, Paul S, Enslen H, Corvol JC, Stipanovich A, Caboche J, Lombroso PJ, Nairn AC, Greengard P, Herve D, Girault JA. Regulation of a protein phosphatase cascade allows convergent dopamine and glutamate signals to activate ERK in the striatum. Proc Natl Acad Sci U S A. 2005;102:491–496. doi: 10.1073/pnas.0408305102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vekovischeva OY, Zamanillo D, Echenko O, Seppala T, Uusi-Oukari M, Honkanen A, Seeburg PH, Sprengel R, Korpi ER. Morphine-induced dependence and sensitization are altered in mice deficient in AMPA-type glutamate receptor-A subunits. J Neurosci. 2001;21:4451–4459. doi: 10.1523/JNEUROSCI.21-12-04451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters CL, Wang BC, Godfrey M, Sun D, Funk CD, Blendy JA. Augmented responses to morphine and cocaine in mice with a 12-lipoxygenase gene disruption. Psychopharmacology (Berl) 2003;170:124–131. doi: 10.1007/s00213-003-1526-7. [DOI] [PubMed] [Google Scholar]
- Whistler JL, von Zastrow M. Morphine-activated opioid receptors elude desensitization by beta-arrestin. Proc Natl Acad Sci U S A. 1998;95:9914–9919. doi: 10.1073/pnas.95.17.9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe G, MacDonald AD. The evaluation of the analgesic action of pethidine hydrochloride (demerol) J Pharmacol Exptl Therap. 1944;80:300–307. [Google Scholar]
- Wu N, Lu XQ, Yan HT, Su RB, Wang JF, Liu Y, Hu G, Li J. Aquaporin 4 deficiency modulates morphine pharmacological actions. Neurosci Lett. 2008;448:221–225. doi: 10.1016/j.neulet.2008.10.065. [DOI] [PubMed] [Google Scholar]
- Yamawaki K, Ito M, Machida H, Moriki N, Okamoto R, Isaka N, Shimpo H, Kohda A, Okumura K, Hartshorne DJ, Nakano T. Identification of human CPI-17, an inhibitory phosphoprotein for myosin phosphatase. Biochem Biophys Res Commun. 2001;285:1040–1045. doi: 10.1006/bbrc.2001.5290. [DOI] [PubMed] [Google Scholar]
- Yokoyama K, Kurihara T, Saegusa H, Zong S, Makita K, Tanabe T. Blocking the R-type (Cav23) Ca2+ channel enhanced morphine analgesia and reduced morphine tolerance. Eur J Neurosci. 2004;20:3516–3519. doi: 10.1111/j.1460-9568.2004.03810.x. [DOI] [PubMed] [Google Scholar]
- Zachariou V, Benoit-Marand M, Allen PB, Ingrassia P, Fienberg AA, Gonon F, Greengard P, Picciotto MR. Reduction of cocaine place preference in mice lacking the protein phosphatase 1 inhibitors DARPP 32 or Inhibitor 1. Biol Psychiatry. 2002;51:612–620. doi: 10.1016/s0006-3223(01)01318-x. [DOI] [PubMed] [Google Scholar]
- Zachariou V, Georgescu D, Sanchez N, Rahman Z, DiLeone R, Berton O, Neve RL, Sim-Selley LJ, Selley DE, Gold SJ, Nestler EJ. Essential role for RGS9 in opiate action. Proc Natl Acad Sci U S A. 2003;100:13656–13661. doi: 10.1073/pnas.2232594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitz KP, Malmberg AB, Gilbert H, Basbaum AI. Reduced development of tolerance to the analgesic effects of morphine and clonidine in PKC gamma mutant mice. Pain. 2001;94:245–253. doi: 10.1016/S0304-3959(01)00353-0. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ferguson SS, Barak LS, Bodduluri SR, Laporte SA, Law PY, Caron MG. Role for G protein-coupled receptor kinase in agonist-specific regulation of mu-opioid receptor responsiveness. Proc Natl Acad Sci U S A. 1998;95:7157–7162. doi: 10.1073/pnas.95.12.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yu Y, Mackin S, Weight FF, Uhl GR, Wang JB. Differential mu opiate receptor phosphorylation and desensitization induced by agonists and phorbol esters. J Biol Chem. 1996;271:11449–11454. doi: 10.1074/jbc.271.19.11449. [DOI] [PubMed] [Google Scholar]