Abstract
Previous work has demonstrated the important role of E2F transcription activity in the induction of S phase during the transition from quiescence to proliferation. In addition to the E2F-dependent activation of a number of genes encoding DNA replication activities such as DNA Pol α, we now show that the majority of genes encoding initiation proteins, including Cdc6 and the Mcm proteins, are activated following the stimulation of cell growth and are regulated by E2F. The transcription of a subset of these genes, which includes Cdc6, cyclin E, and cdk2, is also regulated during the cell cycle. Moreover, whereas overall E2F DNA-binding activity accumulates during the initial G1 following a growth stimulus, only E2F3-binding activity reaccumulates at subsequent G1/S transitions, coincident with the expression of the cell-cycle-regulated subset of E2F-target genes. Finally, we show that immunodepletion of E2F3 activity inhibits the induction of S phase in proliferating cells. We propose that E2F3 activity plays an important role during the cell cycle of proliferating cells, controlling the expression of genes whose products are rate limiting for initiation of DNA replication, thereby imparting a more dramatic control of entry into S phase than would otherwise be achieved by post-transcriptional control alone.
Keywords: E2F3-binding activity, E2F transcription control, S-phase induction, cell cycle
Numerous genes have been identified that control the transition of cells through the cell cycle, including those encoding proteins that are critical for the initiation of DNA replication. The elucidation of the events associated with the control of DNA replication is largely the result of the combination of genetic and biochemical analyses in budding and fission yeast. These studies have identified a series of proteins that assemble a functional origin of replication (for review, see Stillman 1996; Dutta and Bell 1997; Newlon 1997). At the core of the functional origin is the six-component origin recognition complex (ORC) that interacts with origin sequences throughout the cell cycle and is critical for DNA replication. Other work has defined a second complex of six proteins known as the Mcm proteins, that associate with the DNA-bound ORC and are also essential for replication. Finally, the cdc6 gene, initially identified in genetic screens as a critical cell cycle regulatory gene governing the G1/S transition, encodes a protein that appears to facilitate the association of the Mcm complex with ORC (Donovan et al. 1997; Tanaka et al. 1997). The additional observation that the level of the Cdc6 product varies in the cell cycle suggests that the G1/S accumulation of Cdc6 may contribute to the regulation of origin function.
Intensive efforts devoted to the analysis of mammalian cell growth control has complemented and extended the studies in yeast. Although basic aspects of the G1 regulatory events are conserved in yeast and higher eukaryotes, it is clear that the requirements of cell growth control that couple proliferation with cell differentiation have added additional complexity. This work has revealed a pathway controlling the progression of cells out of quiescence, through G1, and into S phase that involves the action of G1 cyclin-dependent kinases (cdks) to inactivate the Rb tumor suppressor and related proteins, which then leads to the accumulation of E2F transcription factor activity (for review, see Nevins 1992; Helin and Harlow 1993; Hunter and Pines 1994; Weinberg 1995; Sherr 1996). The importance of this pathway for mammalian cell growth control is indicated by the fact that disruption of the pathway, either the activation of positive-acting components such as the G1 cyclins or the inactivation of negative components such as p53, Rb, and the cdk inhibitors, can lead to the loss of cell growth control that underlies the development of virtually all forms of human cancer (Weinberg 1995; Hunter 1997).
In some respects, the role of the E2F transcription factors appears similar to the Saccharomyces cerevisiae transcriptional regulatory proteins SWI4/6/MBF and the Schizosaccharomyces pombe Cdc10 protein. In particular, E2F regulates transcription of a large number of genes that encode DNA replication activities including DNA polymerase α (Pol α) PCNA, ribonucleotide reductase, and others. Many of these genes are controlled by the SWI/MBF/Cdc10 proteins in yeast. This similarity now extends to the control of cdc6 gene transcription based on our recent work (Yan et al. 1998). Consistent with the role for the E2F transcription factors in the regulation of genes encoding DNA replication activities (Nevins 1992; Helin and Harlow 1993), E2F can induce DNA replication in otherwise quiescent cells (Johnson et al. 1993; Qin et al. 1994; Shan and Lee 1994; DeGregori et al. 1995b).
Although many of the molecular events involved in the control of mammalian cell growth have been well characterized, particularly the various signal transduction pathways that are activated when quiescent cells are stimulated to grow, the role of these activities in the control of cell cycle transitions in proliferating cell populations is much less clear. Because proliferating cells maintain a defined G1 phase, the timing of initiation of DNA replication must be subject to tight control during each cell cycle. Although much of the understanding of origin function has derived from yeast, it is clear that homologs of many, if not all, of these activities can be found in mammalian cells. Each of the ORC components is highly conserved as are the Mcm proteins. In addition, recent work leading to the isolation of clones of mammalian (Williams et al. 1997) and Xenopus Cdc6 (Coleman et al. 1996) has revealed a significant conservation in the structure of this protein.
Recent work that has revealed E2F control of several of the genes encoding initiation proteins such as Orc1 (Ohtani et al. 1996) and Cdc6 (Yan et al. 1998) suggests a central role for E2F in the control of DNA replication. In light of this, and given the fact that control of initiation of DNA replication is critical in proliferating cells as well as during the transition out of quiescence, we have investigated the role of E2F in transcription control during the cell cycle of proliferating cells.
Results
The transcription of a large number of genes encoding DNA replication activities, including Cdc6 and the Mcm proteins, is regulated by cell growth and dependent on E2F
Previous work has documented the role of E2F in the growth-regulated expression of genes encoding proteins such as DHFR, DNA Pol α, thymidine kinase, and various other genes that encode activities important for DNA replication (Nevins 1992). These genes are expressed at low or undetectable levels in quiescent, nondividing fibroblasts and are induced following growth stimulation. Recent experiments have extended this group of growth-regulated genes to those encoding proteins that mediate the initiation of replication, including Orc1 (Ohtani et al. 1996), one of six components of the ORC, and Cdc6 (Williams et al. 1997; Yan et al. 1998), a protein that is essential for the formation of a functional initiation complex. In addition to the role of Cdc6 and ORC in facilitating origin function, a number of experiments now point to the role of a second complex, involving the 6 Mcm proteins (Mcm2–Mcm7), as critical for initiation of DNA replication. In light of the fact that Orc1 and Cdc6 have been found to be tightly regulated by cell proliferation and dependent on E2F, and taking advantage of the cloning of each of the genes encoding mammalian Mcm proteins (Holthoff et al. 1996; Kiyono et al. 1996; Tsuruga et al. 1997), we have investigated the potential cell growth-dependent control of expression of these genes.
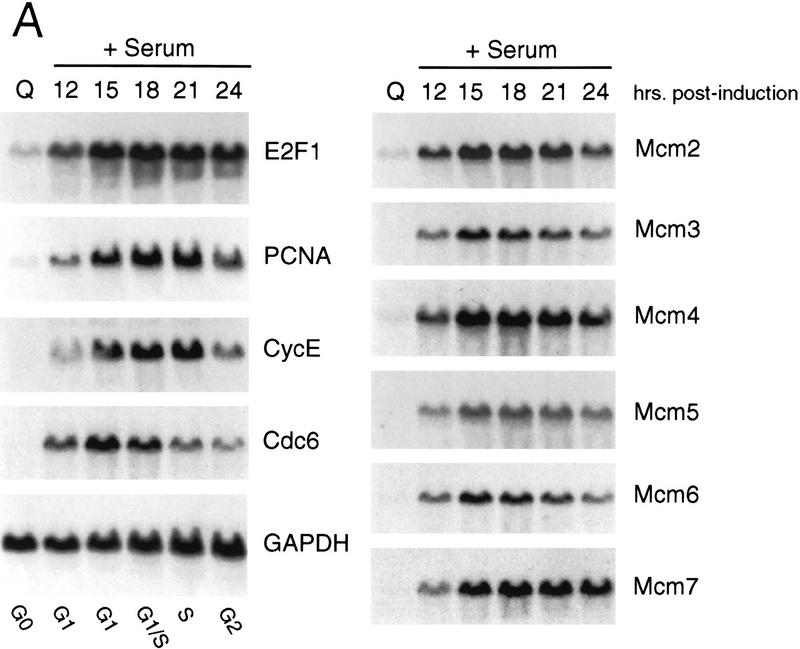
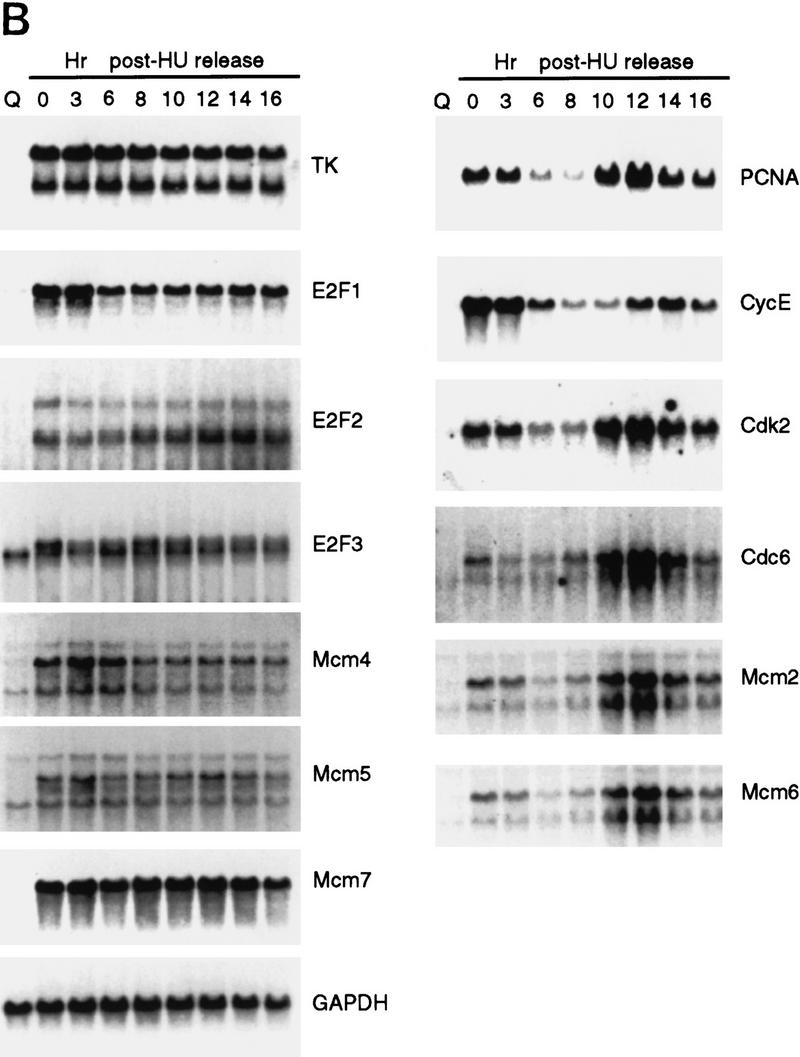
As shown in Figure 1A, analysis of RNA from quiescent cells and from growth-stimulated cells revealed a large induction of each of the mcm genes. The pattern of accumulation of the Mcm transcripts paralleled that of genes encoding E2F1 as well as PCNA, cyclin E, and Cdc6, which were shown previously to be regulated by cell growth. Only very low levels of the RNAs were detected in the quiescent cells which then increased 10- to 20-fold following growth stimulation. Clearly, a large number of genes encoding DNA replication activities are tightly regulated by cell growth. We also note distinctions in the pattern of accumulation of these transcripts as cells pass through S phase. For instance, whereas E2F1 and Mcm7 RNAs clearly remained constant following the initial accumulation, cyclin E, PCNA, and Cdc6 were consistently observed to decline as cells moved through S phase.
Figure 1.
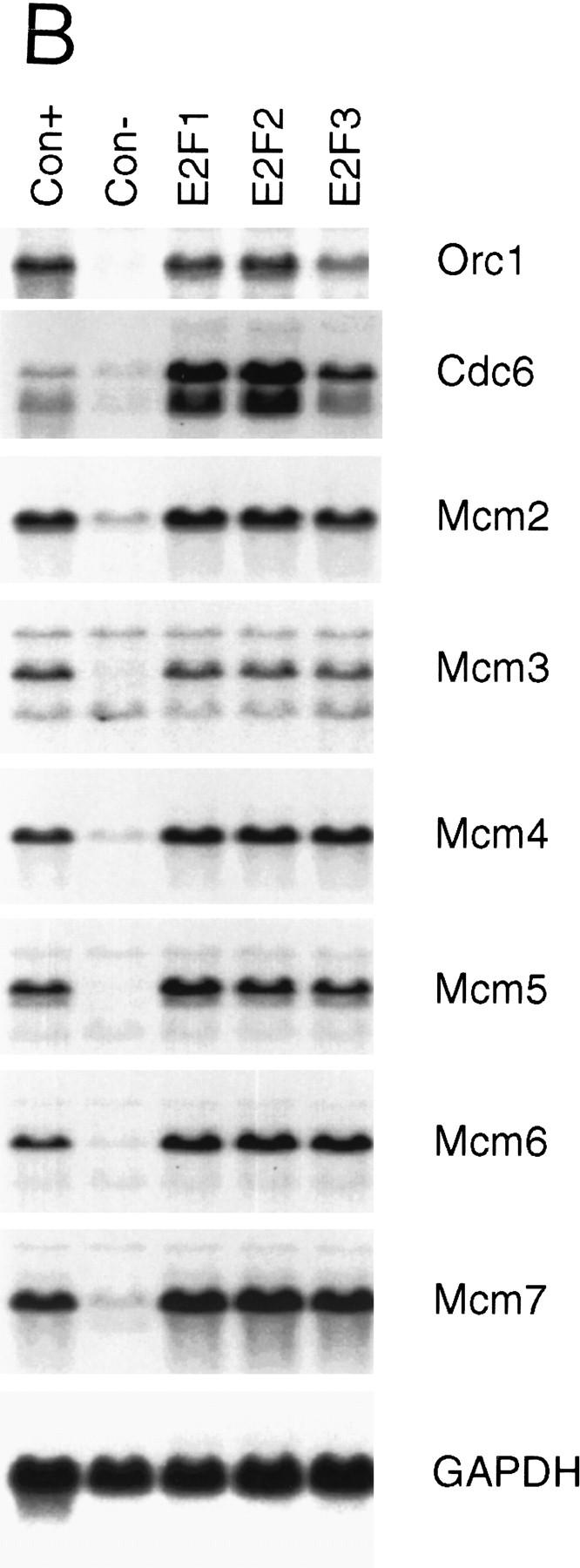
Growth regulated expression of genes encoding DNA replication proteins. (A) Quiescent Ref 52 cells (Q) were stimulated with media containing 10% serum. Cells were harvested at the indicated times, processed for Northern analysis as described in Materials and Methods, and hybridized with the indicated probes. The position of the cell with respect to cell cycle is indicated below based on FACS analysis of similarly treated samples. (B) Quiescent cells were infected with recombinant adenoviruses expressing either E2F1, E2F2, or E2F3 proteins, or with a control virus containing an empty expression cassette (m.o.i. of 100, 100, 200, and 100, respectively). Cells were harvested 18 hr postinfection and processed and analyzed as in A. A portion of the cells that were infected with the control virus were stimulated with media containing 10% serum for an additional 18 hr (Con +).
Given the role of E2F in the control of transcription of many growth-regulated genes including cdc6, and the observation that the mcm6 gene promoter contains sequences that match E2F consensus sites (Tsuruga et al. 1997), we assayed the effect of E2F overproduction on the expression of each of the mcm genes. Quiescent REF52 fibroblasts were infected with recombinant adenoviruses that express the E2F1, E2F2, or E2F3 products. RNA was prepared and then assayed for expression of each of the Mcms. As shown in Figure 1B, expression of E2F1, E2F2, or E2F3 resulted in a large induction of each of the mcm genes, similar to the induction of the cdc6 and orc1 genes, and equivalent to that achieved following serum stimulation. We thus conclude that the mcm genes are indeed regulated as a function of cell growth and that they are also subject to control by E2F, coincident with the control of many other genes encoding DNA replication activities.
A subset of E2F targets, including Cdc6, cyclin E, and Cdk2 are cell cycle regulated in proliferating cells
Mechanisms that control the initiation of DNA replication are important not only at the initial G1/S transition as cells re-enter a cell cycle from a quiescent state, but also at each subsequent G1/S as cells proliferate in the presence of growth factors. Although numerous experiments have documented the role of E2F activity in the induction of the initial S phase, following exit from quiescence, coincident with the activation of a variety of genes that encode DNA replication proteins, including Cdc6 (Williams et al. 1997), little is known of the role of E2F in transcription regulation following this initial G1/S transition, when cells continue to proliferate. Given the observation that many of the proteins that determine initiation of replication are subject to E2F control, together with the fact that initiation must be regulated during each cell cycle, we have explored the possible role of E2F-dependent transcription regulation during a cell cycle in proliferating cells.
We initially examined the cell cycle expression of a series of genes, shown previously to be targets for E2F control and that are regulated by cell growth signals (DeGregori et al. 1995a; DeGregori et al. 1997). Because cell synchrony is rapidly lost as quiescent cells re-enter the cell cycle and proceed to the next cell cycle, we have used hydroxyurea (HU) to achieve a G1/S block and thus cell cycle synchronization, as an alternate approach to examine cell cycle progression. Growth-arrested cells were stimulated to grow in the presence of HU, resulting in a cell cycle arrest at the first G1/S. The block is readily reversed by removal of the HU, generating a population of cells that move synchronously through S phase, G2/M, the next G1, and a following S phase (Fig. 2A). The use of an inhibitor such as HU to achieve cell cycle synchronization does not appear to have secondary effects, because upon removal of the drug, cells continue to proliferate with normal kinetics of transition through the cell cycle. RNA was prepared from these cultures at various times during the experiment and analyzed by Northern blotting, using a variety of probes that detect RNAs known to be E2F-regulated. As seen in the data presented in Figure 1 as well as in previous work, each of these E2F target genes was tightly regulated by cell growth, with little or no expression in quiescent cells and a large increase in cells that had accumulated at G1/S (Fig. 2B, lanes 1,2). Analysis of the expression of these genes as cells moved into and through the cell cycle following release from the HU arrest revealed two categories of genes. One group, typified by the genes encoding E2F1, E2F2, and E2F3, as well as thymidine kinase and three of the genes encoding Mcms (Mcm4, Mcm5, Mcm7), were constantly expressed, without significant fluctuations following cell cycle re-entry. Thus, these genes appear to be growth regulated but not cell cycle regulated. In contrast, a second group of genes, which includes those encoding Cdc6, PCNA, Cdk2, and cyclin E, as well as two of the Mcm proteins (Mcm2 and Mcm6), were cell cycle regulated in addition to being growth regulated. In each case, the RNA declined as the cells progressed through S phase and into G2/M and then began to rise as the cells progressed towards the next G1/S.
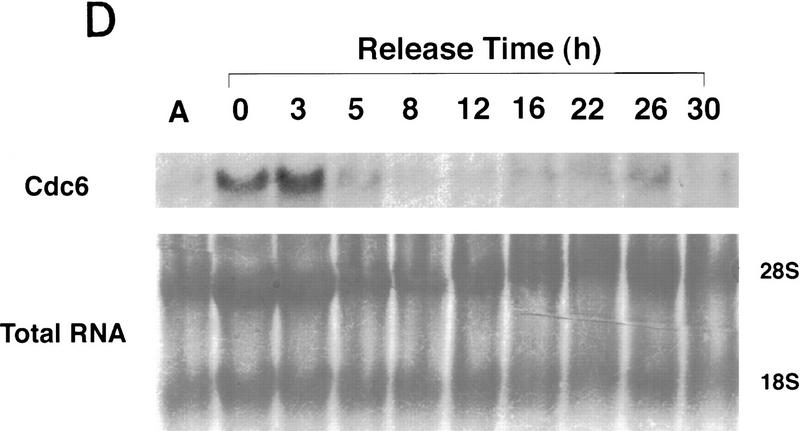
Figure 2.
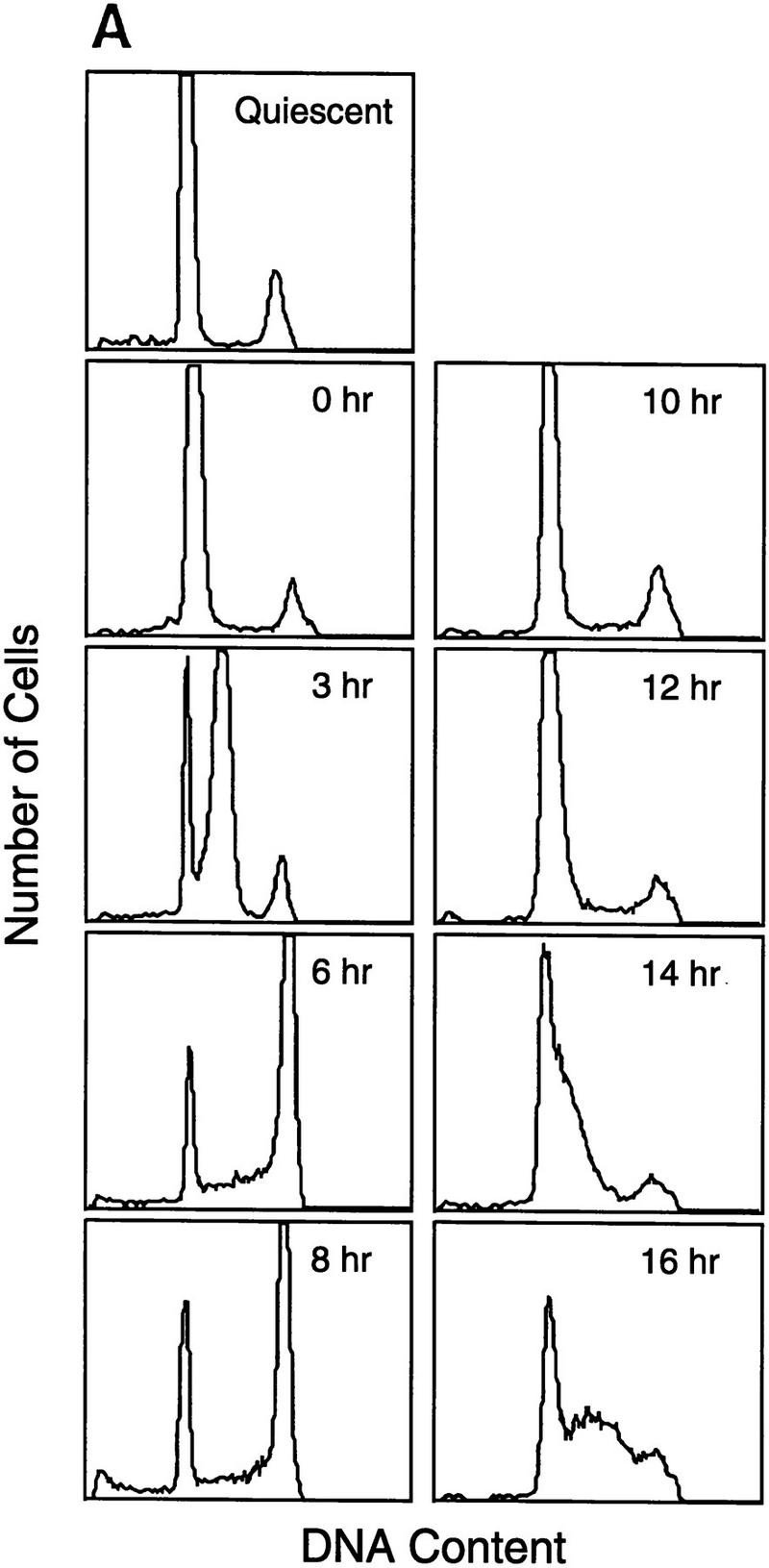
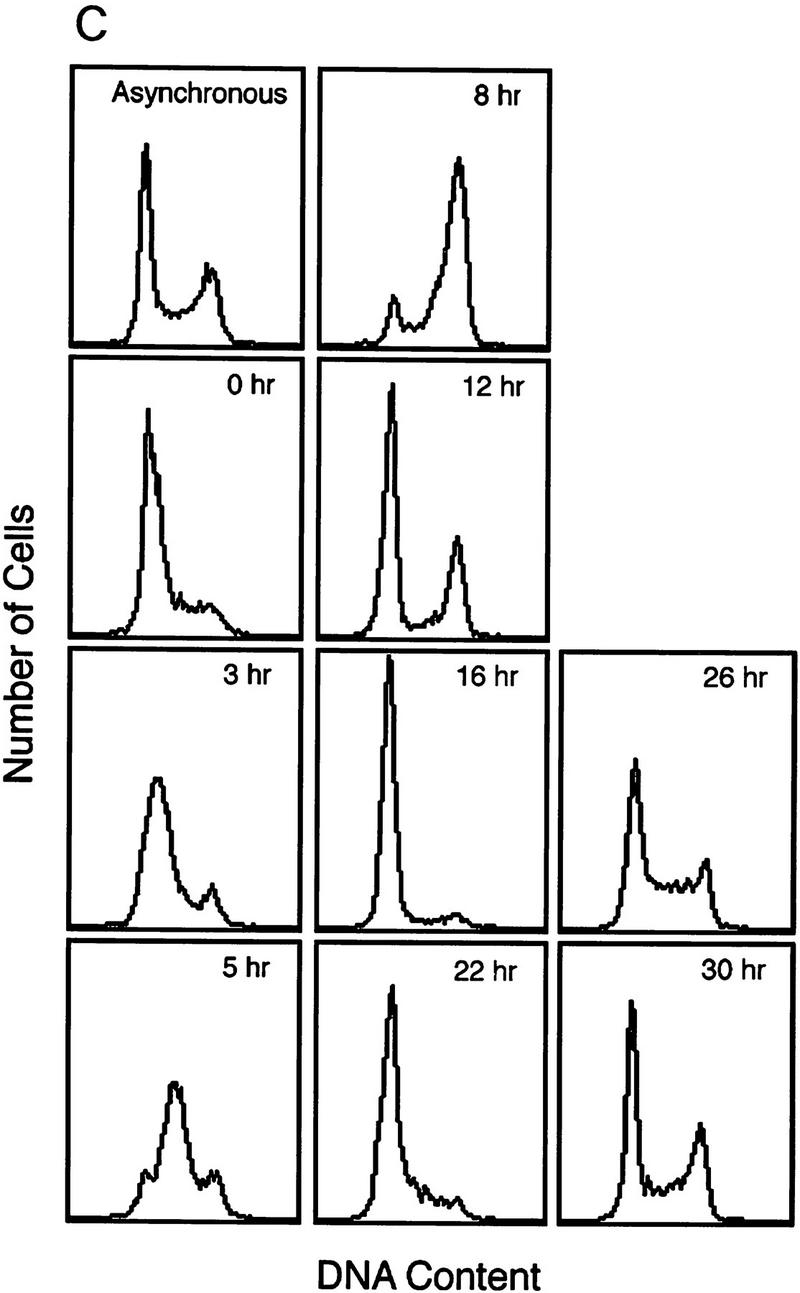
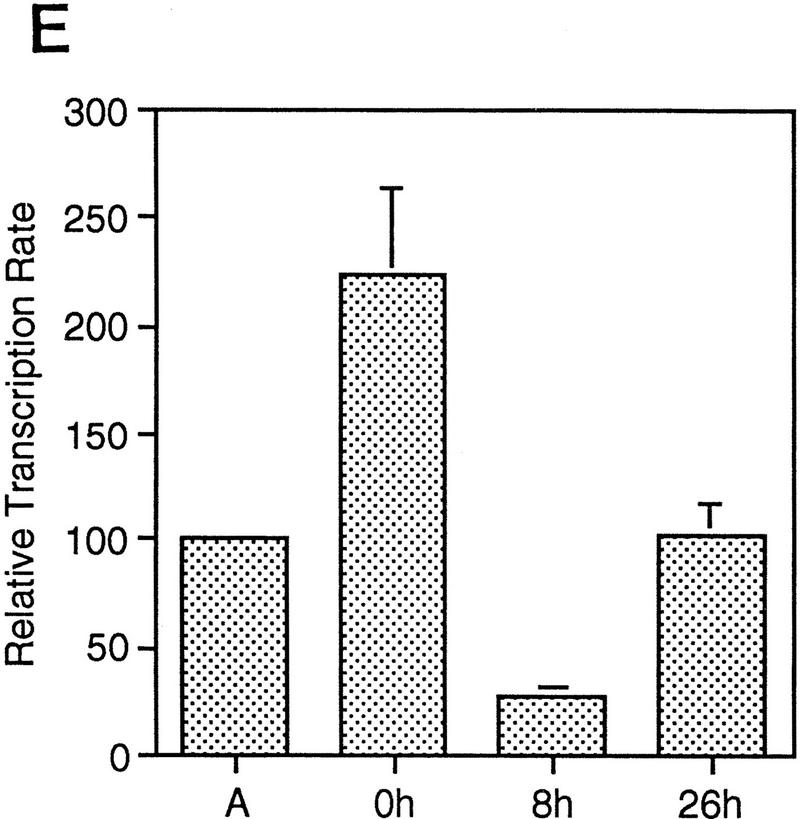
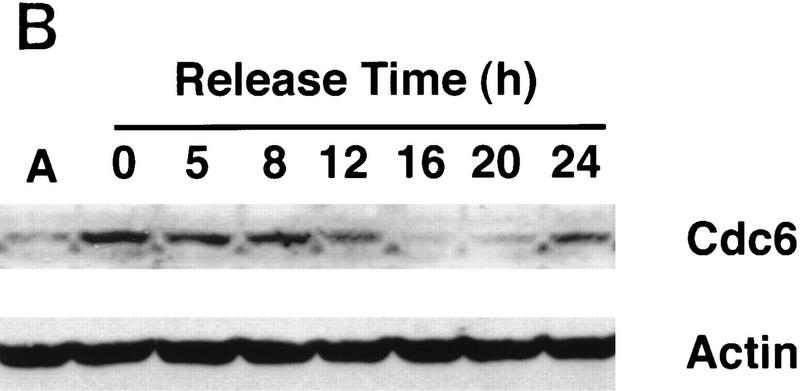
A subset of E2F targets, including Cdc6, Cyclin E, and Cdk2, are regulated during the cell cycle. (A) Quiescent REF 52 cells were stimulated with media containing 10% serum in the presence of 2 mm HU. After 21 hr, the G1/S-arrested cells were washed free of HU and then grown in medium containing 10% serum. At the indicated times following the release from the HU block, cells were harvested, stained with propidium iodide, and processed for flow cytometry as described in Materials and Methods. (B) Quiescent REF52 cells (Q) or samples harvested at the indicated times following the release from the HU block were processed for Northern analysis as described in Materials and Methods. Northern blots were hybridized with the indicated probes. Notably, two E2F3-specific RNAs are consistently detected by Northern analysis; the slower-migrating species represents the growth-regulated RNA that coincides with the accumulation of E2F3 protein (see Fig. 5A; data not shown). (C) HeLa cells were arrested at G1/S by incubation with 2 mm thymidine for 16 hr, washed, and released in media containing 10% fetal calf serum. At the indicated times, cells were harvested and prepared for FACS analysis as described in Materials and Methods. (D) At the indicated times following release from the thymidine block, HeLa cells were harvested and processed for Northern analysis. The blot was probed with a human specific Cdc6 probe. Blots were stained with methylene blue to confirm equal loading. (A) Asynchronous cells. (E) At the indicated times following the release of HeLa cells from a thymidine block, nuclei were isolated and used for nuclear run-on transcription assays as described in Materials and Methods. The Cdc6-specific transcription rate is presented relative to the transcription rate of the β-actin control. Data are presented as a mean of six independent determinations.
To further verify the cell cycle-regulated expression of E2F target genes, we have assayed the expression of these genes in HeLa cells, a human epithelial cervical carcinoma cell line. Although growth regulation is largely lost in these cells, in part because of the expression of the papillomavirus oncoproteins, one would expect basic aspects of cell cycle regulation to be maintained in any cell type, including tumor cells. Indeed, as shown in Figure 2C, Northern assays for Cdc6 reveal a very similar pattern of accumulation during the cell cycle as observed in the REF52 cells. Moreover, at least for Cdc6, the cell cycle fluctuation of RNA levels appears to be a result of transcriptional control as seen by nuclear run-on measurements in synchronized HeLa cell populations (Fig. 2D).
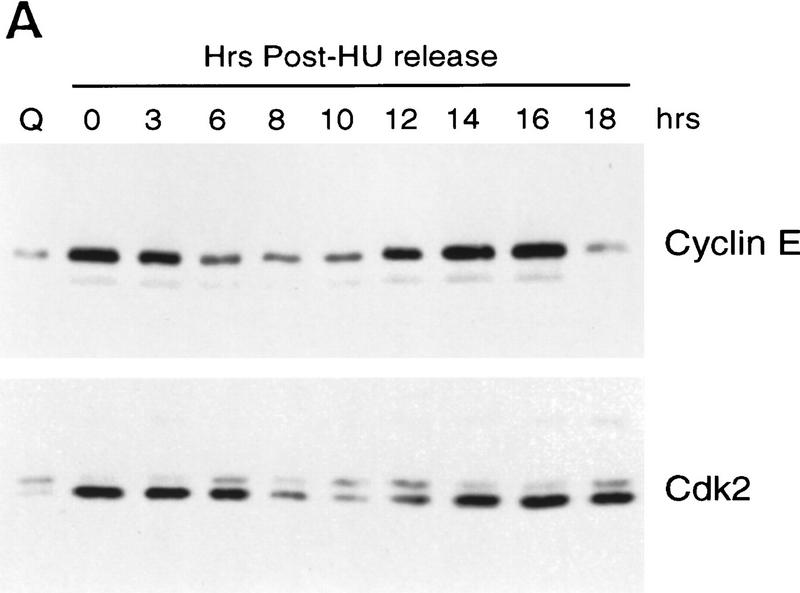
The control of cyclin E and cdk2 expression was also reflected in the accumulation of each of the proteins during the cell cycle, as measured by Western blot assays (Fig. 3A). In each case, the protein declined as cells left the G1/S arrest and passed through S phase and then reaccumulated as cells entered the next G1. We have also measured Cdc6 protein accumulation during the cell cycle although because of antibody specificities, we have not been able to assay for Cdc6 protein in Ref52 cells. However, assays of HeLa cell fractions yielded a result that paralleled that of cyclin E and cdk2 and closely matched the accumulation of Cdc6 RNA during the HeLa cell cycle (Fig. 3B).
Figure 3.
Cell cycle regulation of Cyclin E, Cdk2, and Cdc6 protein accumulation. (A) Protein lysates (10 μg per lane) derived from REF52 cells treated as in the HU arrest/release experiments decribed in Fig. 2A were subjected to 10% SDS-PAGE, Western blotted, and probed with either cyclin E- or cdk2-specific antibodies as indicated. (B) Protein samples (60 μg per lane) from HeLa cells treated as in the thymidine arrest/release experiment described in Fig. 2C were subjected to 10% SDS-PAGE, transferred to nitrocellulose, and probed with antibodies specific to human Cdc6 protein.
Cell cycle control of E2F activity
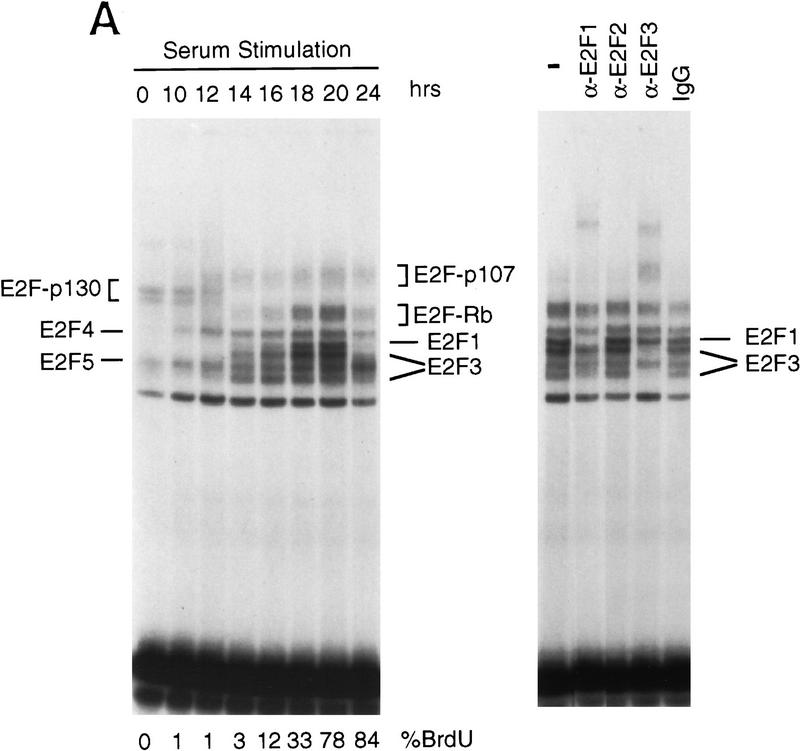
Many previous experiments have detailed the changes in E2F DNA-binding activities in relation to growth stimulation. As shown in Figure 4A, this includes the predominance of an E2F4,5–p130 complex in quiescent cells, the disappearance of this complex as cells are stimulated to grow, and the appearance of free E2F4 and E2F5 activities as well as an E2F4,5–p107 complex, which also contains cyclin A and cdk2, as cells progress into S phase. During G1, the accumulation of the regulated E2F activities (E2F1, and E2F3) was also observed in parallel with an increase in Rb–E2F complexes that likely reflects the overall accumulation of E2F activity during this time period. We were aided in these assays by the use of gel electrophoretic conditions that allowed the separation of individual E2F–DNA complexes (Ikeda et al. 1996). As such, it was possible to identify gel-shift bands representing the accumulating E2F1 and E2F3 activities, confirmed by antibody supershift (Fig. 4A, right), as well as by overexpression of individual E2F proteins (data not shown) as cells progressed through G1/S.
Figure 4.
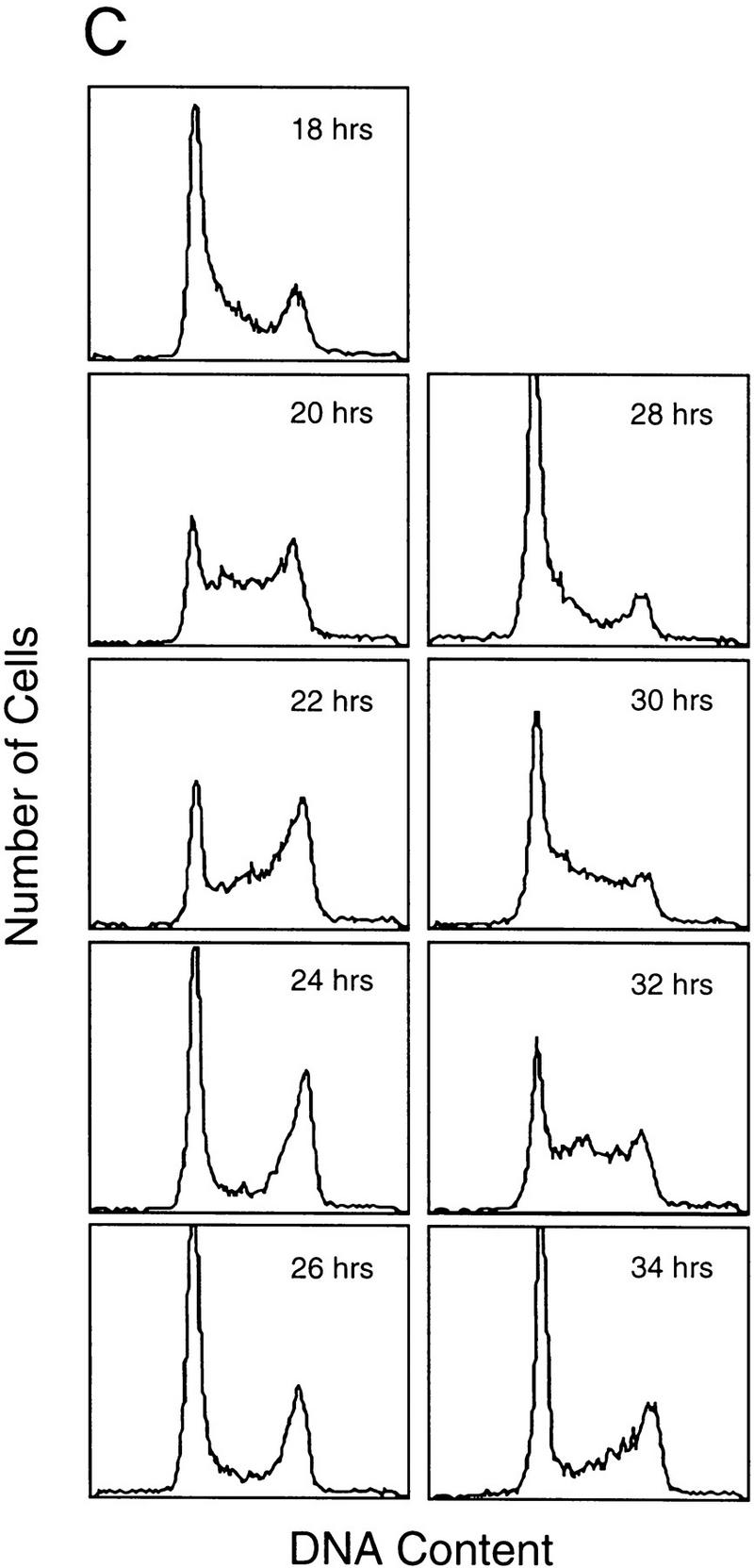
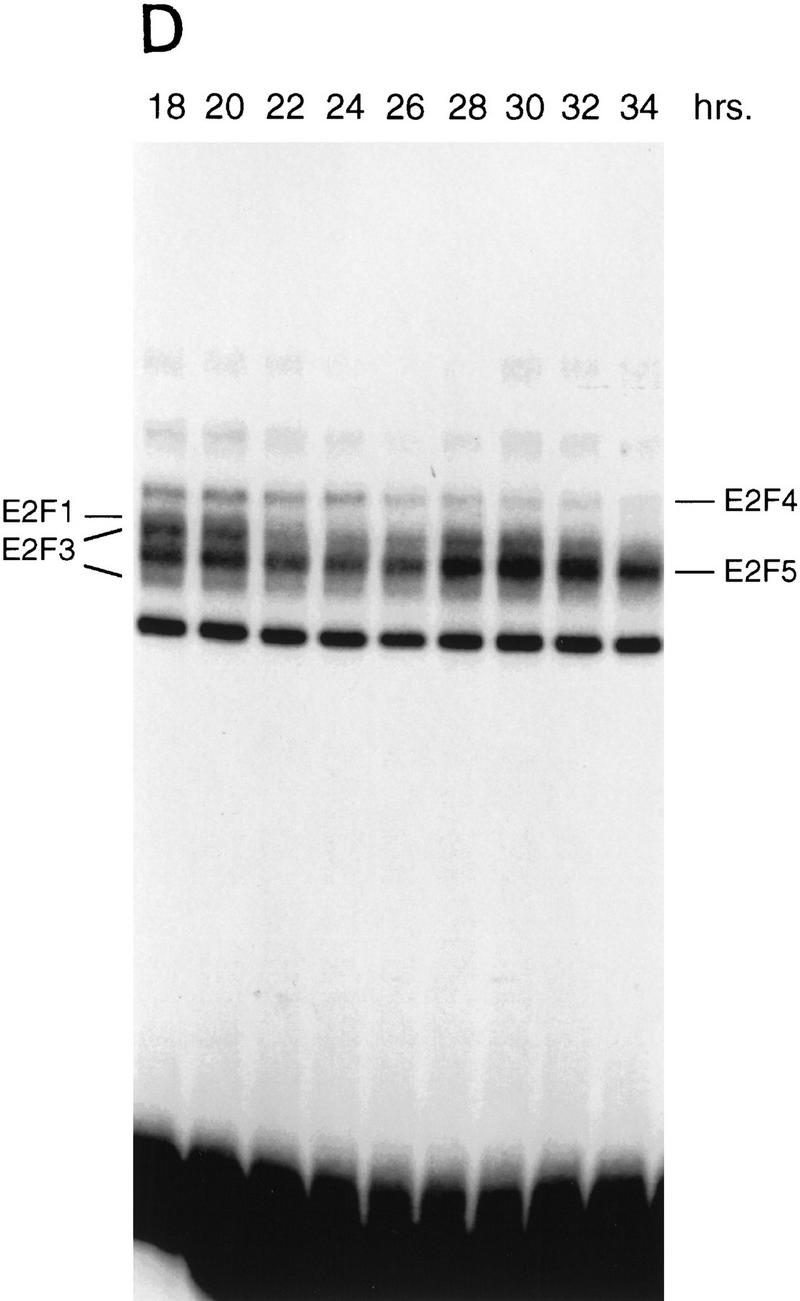
Cell cycle control of E2F activity. (A) Nuclear extracts prepared at various times following the stimulation of quiescent REF52 cells (Q) were assayed for E2F DNA-binding activity by electrophoretic mobility-shift assays (EMSA) using an E2F-specific 32P-labeled DNA probe (left). Cells similarly stimulated were incubated with BrdU (10 μm), fixed at the indicated times, subsequently immunostained with BrdU-specific antibodies, and visualized by immunofluorescent microscopy. The percentage of BrdU-positive cells at each time point is indicated below the DNA-shift gel. The nuclear extract sample from the 20-hr time point presented at left (G1/S sample) was incubated with either IgG-, E2F1-, E2F2-, or E2F3-specific antibodies prior to being subjected similarly to EMSA (right). The E2F4- and E2F5-specific bands indicated on the DNA gel-shift have been identified similarly using specific antibodies against the respective proteins (data not shown). We have been unable to identify an E2F2-specific DNA-binding activity in these assays. (B) Nuclear extracts prepared at various times following the release of cells from an HU block as described in Fig. 2A, as well as from quiescent (Q) REF52 cells, were assayed for E2F DNA-binding activity by EMSA using an E2F-specific 32P-labeled DNA probe (left). The position of the cells with respect to cell cycle is indicated below based on the FACS analysis shown in Fig. 2A. Cytoplasmic extracts prepared from the same time point samples were assayed for E2F-binding activity by EMSA using the same E2F-specific probe (right). (C) Ref52 cells were brought to quiescence by serum starvation and then stimulated to grow by addition of fresh medium with serum. Samples were taken at the indicated times and processed for FACS analysis as described in Materials and Methods. (D) Nuclear extracts prepared at various times following serum stimulation and assayed for E2F DNA-binding activity by EMSA using an E2F-specific 32P-labeled DNA probe.
The observation that several genes that have been shown to be growth regulated by E2F are also regulated during the cell cycle raises the possibility that E2F activity might be regulated in the cell cycle. To address this possibility, we analyzed nuclear extracts from the same HU-block experiment utilized for the Northern analyses (Fig. 2) and measured E2F DNA-binding activity as a function of cell-cycle progression. Assay of the E2F activity in nuclear extracts from the G1/S cells resulting from a HU arrest yielded a pattern similar to that seen in the serum-stimulated cell extracts (Fig. 4A and 4B, lane 20 hr and lane 2, respectively). As the G1/S-arrested cells were released from the block, allowing the cells to progress through the cell cycle, the accumulated E2F1 and E2F3 activities were seen to decline and then disappear within 6 hr of release, a time in which the cells had passed through S and accumulated in G2 (Fig. 2A). The specific disappearance of E2F1 and E2F3 DNA-binding activity was similarly observed in cells that have been stimulated with serum for 24 hr (Fig. 4A, 24 hr lane), a time in which most cells acquired a G2 DNA content (data not shown). In contrast, there was no change in the abundance of the E2F4 or E2F5 activities in the nuclear extracts during this period of time (Fig. 4B). In addition, whereas E2F1 and E2F3 activities were exclusively nuclear, >80% of the E2F4 and E2F5 activity was cytoplasmic, and the level of these activities did not vary during the cell cycle (Fig. 4B, right). As cells passed through G2/M and entered the next G1, there was a reaccumulation of E2F3 activity that peaked as cells entered the next S phase, coincident with the expression of the various cell cycle-regulated E2F targets. In contrast, there was little or no reaccumulation of the E2F1 activity. We have not detected E2F2 DNA-binding activity in these assays.
To ensure that the fluctuations in E2F activity were not related to the HU-induced cell synchronization, we also assayed E2F accumulation through two cell cycles following the stimulation of cell growth by serum addition. REF52 cells were brought to quiescence by serum starvation and then stimulated to re-enter the cell cycle by serum addition. Samples were taken at various times and assayed for DNA content by FACS analysis and E2F DNA-binding activity. As shown in Figure 4C, the cells maintained good synchrony as they passed from the initial cell cycle and into the second G1. Assays for E2F DNA-binding activity (Fig. 4D) revealed a pattern of accumulation that closely reflected that seen in the HU-synchronized cells. In particular, E2F1 and E2F3 activity accumulated during the initial G1/S, declined, and then E2F3 but not E2F1 reappeared in the second G1 phase. Based on these results, we conclude that the control of E2F3 accumulation is indeed linked to cell-cycle regulation and not the method of synchronization.
The decline in E2F1 and E2F3 DNA-binding activities reflects post-transcriptional regulation (Fig. 2B) and, at least for E2F1 activity, is consistent with previous work that has demonstrated an ability of cyclin A/cdk2 to bind to the amino-terminus of the E2F1 protein, phosphorylate the associated DP1 protein specifically, and result in the inactivation of the E2F1 DNA-binding activity (Krek et al. 1994; Xu et al. 1994; Krek et al. 1995; Dynlacht et al. 1997). The fact that the E2F3 protein shares the cyclin A/cdk2-binding motif suggests that this E2F activity may be regulated similarly. Indeed, the kinetics of the decline of E2F1 and E2F3 DNA-binding activity as cells pass through S phase coincides with the accumulation of cyclin A-dependent kinase activity, and the reappearance of E2F3 activity in the next G1 follows the decline in cyclin A-dependent kinase activity after cells pass through G2/M (Fig. 5B).
Figure 5.
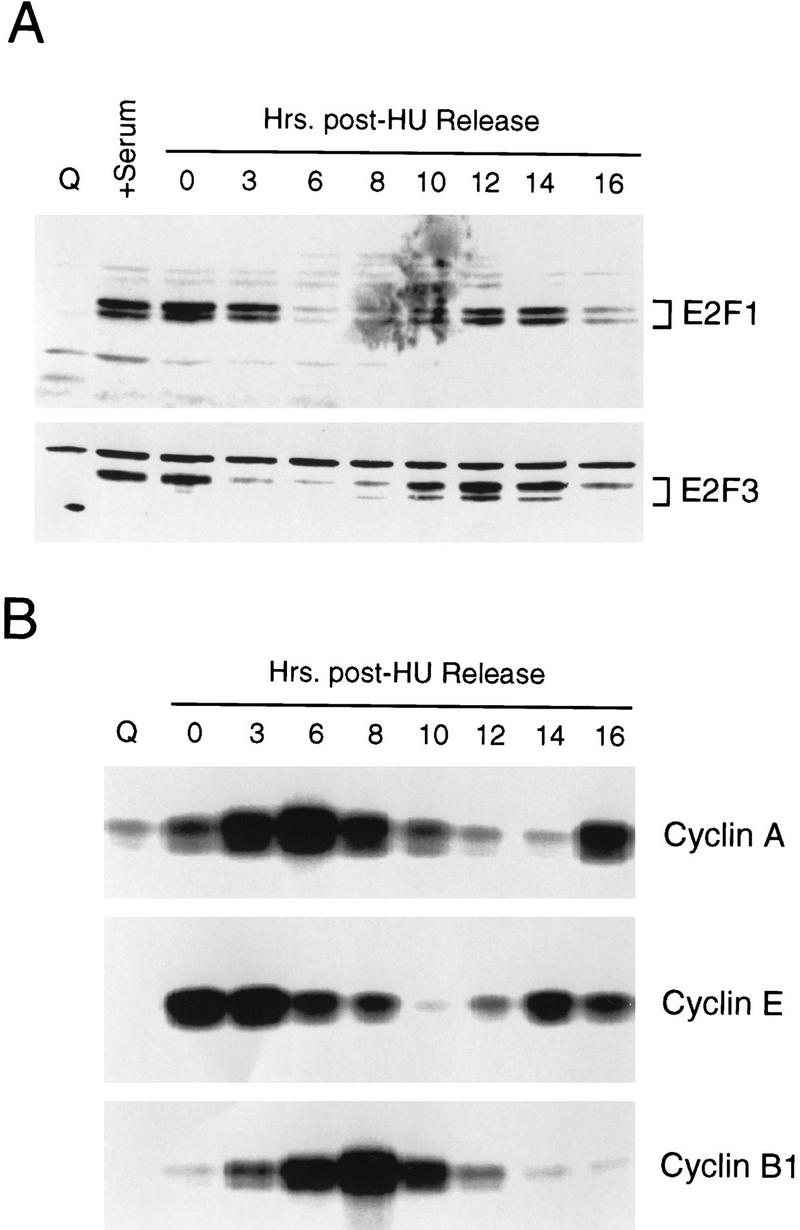
Cell cycle control of E2F protein accumulation. (A) Quiescent REF52 cells (Q), or REF52 cells synchronized at G1/S by HU treatment as described in Fig. 2A, were harvested at the indicated times following the release of the G1/S block, and processed for Western blot analysis using either E2F1- or E2F3-specific antibodies. For comparison, samples from quiescent cells (Q) or quiescent cells stimulated for 19 hr with 10% serum (+serum) were also included. (B) Aliquots from the same HU-treated samples, or from quiescent cells (Q) were processed for cyclin A-, cyclin E-, or cyclin B1-dependent kinase activity by immunoprecipitation of kinase complexes using the indicated antibodies specific for the respective cyclin activities and using histone H1 as a substrate. The kinase reactions were analyzed by 10% SDS-PAGE.
Additional work has shown that the E2F1 protein is subject to ubiquitin-dependent degradation (Hateboer et al. 1996; Hofmann et al. 1996). Analysis of endogenous E2F1 and E2F3 protein levels by Western blot assays revealed a cyclic accumulation of these proteins (Fig. 5A) that, at least for E2F3, coincides with the pattern of accumulation of DNA-binding activity. Interestingly, although there was no reaccumulation of E2F1 DNA-binding activity in the second cell cycle, the E2F1 protein did reaccumulate. Based on these results and previous experiments, we conclude that the accumulation of E2F1 and E2F3 DNA binding activity during a cell cycle may be governed by at least two events—the cyclic accumulation of cyclin A/cdk2 that affects DNA-binding activity and the control of protein stability by the ubiquitin-dependent proteasome pathway. Moreover, additional control must provide specificity in preventing the reaccumulation of E2F1 DNA-binding activity during G1/S of proliferating cells.
E2F3 is required for S phase in cycling cells
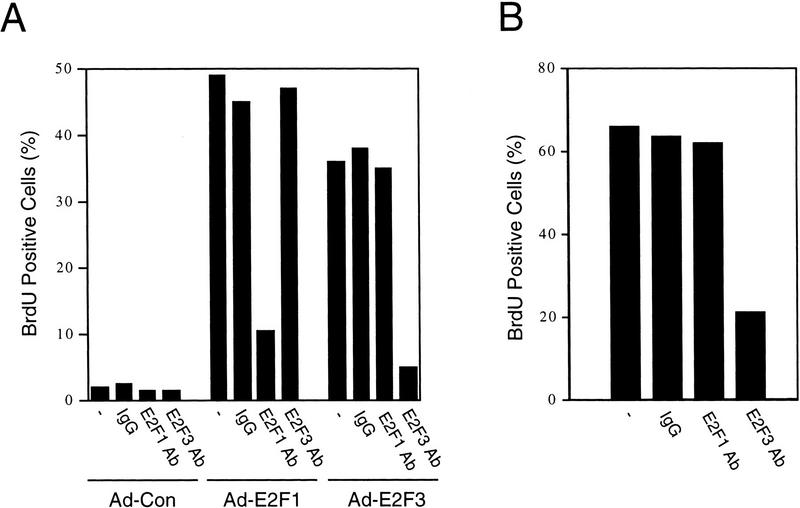
The data shown thus far demonstrate that a specific subset of E2F activity, namely E2F3, is cell cycle regulated, coincident with the regulation of several E2F targets that are critically important for initiation of replication. To provide direct evidence for a role for E2F3 in allowing S phase in cycling cells, we have made use of antibodies that specifically recognize the E2F3 protein to determine whether the injection of these antibodies into cells released from a G1/S block would prevent entry of these cells into the next S phase. Specificity of the antibodies was indicated by their ability to specifically recognize the cognate E2F activities in gel-shift assays (Fig. 4A; data not shown). As an additional test for function of the antibodies, as well as a test for specificity, we have measured the effect of antibody microinjection on the ability of either E2F1 or E2F3 to induce S phase when these proteins are expressed in quiescent REF52 cells. Our previous experiments have shown that infection of quiescent fibroblasts with recombinant adenoviruses expressing either E2F1 or E2F3 (Ad–E2F1 and Ad–E2F3, respectively) results in an induction of S phase as measured by BrdU incorporation (DeGregori et al. 1997). As shown in Fig. 6A, injection of the E2F3 antibody blocked the induction of S phase by Ad–E2F3 but did not affect Ad–E2F1-induced S phase. Conversely, injection of an E2F1-specific antibody blocked the induction of S phase by Ad–E2F1, without affecting Ad–E2F3-induced S phase. Injection of control antibodies (IgG) into fibroblasts had no effect on S-phase entry relative to uninjected cells. We thus conclude that the antibodies are capable of blocking the function of their respective substrate proteins and that there is specificity in this action.
Figure 6.
Inhibition of E2F3 activity inhibits the cell cycle induction of S phase. (A) Quiescent REF52 cells were infected with either Ad–E2F1, Ad–E2F3, or Ad–Con (m.o.i. of 100, 200, and 100, respectively). Cells were microinjected 4 hr postinfection with IgG, or with E2F1- or E2F3-specific antibodies (at an antibody concentration of 1 μg/ml, containing fluorescein-conjugated dextran as a marker for detecting injected cells). BrdU was added 12 hr later (10 μm) and cells were incubated an additional 4 hr prior to fixation and immunostaining with BrdU-specific antibodies. Microinjected cells were visualized by fluorescent microscopy and the percentage of cells staining positively for BrdU is presented above. Approximately 150–250 cells were microinjected in two separate experiments, a representative experiment is shown. (−) The quantitation of uninjected cells in the same tissue-culture plate. (B) REF52 cells were synchronized by HU treatment as described in Fig. 2A, and 5–7 hr following the release from the G1/S block, a time when cells are predominantly in G2, cells were microinjected with IgG, or either with E2F1- or E2F3-specific antibodies. BrdU was then added to the cells 14 hr after the HU release and incubated for a further 3 hr, after which cells were fixed, immunostained, and quantitated for BrdU incorporation as in A. Approximately 150–250 cells were microinjected for each condition, and a representative experiment (similar results were obtained in five independent experiments) is presented. (−) The quantitation of uninjected cells in the same tissue culture plate.
Given the ability of the E2F1 and E2F3 antibody to specifically block function of the cognate protein, we then assessed the role of these E2F proteins in regulating S-phase entry in fibroblasts that were synchronized at G1/S by HU treatment and then released into the cell cycle by removal of HU. When cells were in G2 (5–7 hr following HU release), several hundred cells were then microinjected with either the E2F1-or the E2F3-specific antibody together with fluorescein-labeled dextran as a marker. The entry of cells into the subsequent S phase was monitored by labeling with BrdU for 3 hr starting in late G1 (14 hr following HU release). Samples were then fixed and stained for incorporation of BrdU. As shown in Figure 6B, injection of the E2F3-specific antibody consistently led to a threefold decrease in the number of BrdU-positive cells as compared to the injection of the control antibody or cells that were not injected. In contrast, injection of the E2F1-specific antibody was without effect. Based on these results, we conclude that E2F3 activity, but not E2F1 activity, is indeed important for S-phase entry during a cell cycle.
Discussion
Previous work has provided strong evidence for a role of E2F activity in cell growth control. The work we describe here now defines a role for E2F in the control of transcription during the cell cycle. In particular, these experiments demonstrate that a subset of the E2F activity is regulated during the cell cycle and that this accumulation coincides with the expression of several genes, including cdc6, cyclin E, and cdk2, each previously identified as E2F-regulated genes. Most importantly, our results indicate that E2F activity is required for the efficient induction of S phase in proliferating cells and reveal a specific role for the E2F3 family member in this process.
Distinct roles for E2F family members in cell growth and cell cycle
A key point evident from the data presented here is the evidence for distinct behavior and roles for the individual E2F proteins during the cell cycle. Numerous experiments have detailed the regulation of E2F1 accumulation during G1 (10- to 20-fold induction), distinct from the relatively constant level of E2F4 or E2F5 (2- to 3-fold induction). This accumulation results at least in part from the transcriptional derepression of the E2F1 gene as cells leave a quiescent state (Johnson et al. 1994). It is also evident from the studies presented here that E2F3 behaves in a similar manner during the transition through the initial G1. Both E2F1 and E2F3 activities then decline as cells pass through S phase, likely resulting from cyclin A/cdk2-mediated phosphorylation. The dichotomy of behavior of E2F1 and E2F3 as cells continue to cycle and enter the next G1 is particularly striking since E2F3 DNA-binding activity reaccumulates in the succeeding G1, whereas E2F1 activity does not reappear after the initial G1. The underlying basis for this differential control is unclear, although it does not appear to involve differences in translational or stability control since the steady-state accumulation of E2F1 and E2F3 proteins during the cell cycle is quite similar. One possibility might relate to control by Rb, as our preliminary experiments suggest that Rb associates preferentially with E2F1. Possibly, there is sufficient Rb available once cells have entered a cell cycle to titrate the available E2F1 but, because of differential affinities, this level of Rb is insufficient to prevent the accumulation of E2F3 activity.
The differential regulation of E2F1 and E2F3 during the cell cycle, whereby E2F3 continues to cycle but E2F1 does not reappear after the accumulation during the initial G1/S, is also interesting in light of experiments that define a role for E2F1 as a signal for apoptosis. Previous experiments have shown that E2F1-mediated induction of S phase is frequently followed by apoptosis, largely dependent on p53 (Qin et al. 1994; Shan and Lee 1994; Wu and Levine 1994; Kowalik et al. 1995). Intriguingly, our recent experiments have shown that this activity is unique to the E2F1 protein, despite the fact that other E2F family members also induce S phase (DeGregori et al. 1997). Thus, the fact that E2F1 DNA-binding activity accumulates as cells are stimulated to grow and re-enter a cell cycle, but does not accumulate once cells are in a cell cycle, suggests the possibility that E2F1 plays a role as a growth checkpoint, ensuring that cell cycle re-entry has properly occurred. Once the cells are then growing in the presence of growth factors, this checkpoint might no longer be critical and only the cyclic accumulation of E2F3 activity represents the E2F requirement.
A central role for E2F in control of DNA replication activities
Based on a large volume of work directed at understanding E2F function, it is now clear that this transcriptional activity plays a key and central role in the activation of genes that encode DNA replication activities. Indeed, considering the fact that the group of E2F-regulated genes now includes those encoding deoxynucleotide enzymes (DHFR, RR, TK, TS), DNA synthetic enzymes (DNA Pol α, PCNA), and proteins that mediate the recognition and utilization of replication origins (Orc1, Cdc6, Mcms), it appears that E2F transcriptional activity may coordinate the accumulation of most if not all of the essential activities necessary for DNA replication. The results presented here also suggest that the role for E2F goes beyond the coordination of production of the activities as cells re-enter the cell cycle from a quiescent state but also involves the coordination of control of several rate-limiting activities that dictate G1/S control during the cell cycle of proliferating cells. In particular, the fact that various studies point to an essential role for Cdc6 as well as cyclin E/cdk2 in S-phase induction, together with the finding that the synthesis of these proteins does indeed oscillate during the cell cycle coincident with E2F3 activity, strongly suggests that E2F control is critical for this event.
Various lines of evidence point to roles for Cdc6, as well as cyclin E/cdk2, as rate-limiting activities for initiation of DNA replication. Likewise, our previous work has shown that the accumulation of E2F activity in otherwise quiescent cells can lead to an induction of S phase. The fact that Cdc6, cyclin E, and cdk2 are E2F targets, and that their expression is regulated during the cell cycle in parallel with E2F3 accumulation, suggests that the E2F-dependent control of Cdc6, together with cyclin E and cdk2, may well represent a rate-determining event for initiation of DNA replication. Our preliminary experiments have shown that whereas expression of Cdc6 alone is not sufficient to induce S phase, expression of Cdc6 together with that of cyclin E/cdk2 does induce S phase in otherwise quiescent cells (G. Leone, J. DeGregori, R.S. Williams, Z. Yan, and J. Nevins, unpubl.).
Importance of transcriptional and post-transcriptional control in the cell cycle
In principle, cell cycle control of Cdc6 accumulation, as well as cyclin E/cdk2 accumulation, could be achieved without transcription control. A constant level of the mRNAs, and thus constant synthesis of the proteins, coupled with protein degradation during mitosis, would result in oscillation of the activities during the cell cycle. Nevertheless, our observations suggest additional complexity whereby a tight interrelationship in the control of E2F3, cyclin E/cdk2, and cyclin A/cdk2 is evident. Based on the experiments presented here, we suggest that the expression of Cdc6, cyclin E, and cdk2, which is seen to fluctuate during the cell cycle, is regulated, at least in part, by the cyclic accumulation of E2F3 transcriptional activity. The accumulation of cyclin A/cdk2 during S phase would lead to the elimination of E2F3 (and E2F1) DNA-binding activity. This, together with the subsequent degradation of these proteins, would lead to a decline in the expression of a subset of E2F target genes. As cells progress through G2 and mitosis, degradation of cyclin E and cyclin A, leading to the decline in the associated kinase activities, would then reset the clock. Continued synthesis of E2F3 results in a reaccumulation of E2F activity in the following G1, induction of cyclin E and Cdc6 synthesis, and induction of S phase. This process continues as long as the cell is growing in the presence of growth factors. We suggest that transcriptional regulation by E2F of the key initiation activities such as Cdc6 and cyclin E/cdk2 provides an additional level of control of the accumulation of these activities, enhancing the magnitude that would result from post-transcriptional control alone.
Materials and methods
Cells and viruses
Viral stocks were created as described previously (Schwarz et al. 1995), and the virus was purified by CsCl density-gradient centrifugation as described (Nevins et al. 1997). Viral titers were determined by an indirect immunofluorescent assay specific for the viral 72-kd E2 gene product as described (DeGregori et al. 1995a) and defined as focus forming units (FFU) per ml. The construction of the recombinant viruses Ad–E2F1, Ad–E2F2, Ad–E2F3, and Ad–Con (a control virus, previously termed AdMb or Ad–CMV, lacking a cDNA insert) have been described (DeGregori et al. 1997).
REF52 cells were grown in DMEM containing 10% serum (5% fetal bovine serum and 5% calf serum). To bring cells to quiescence, cells were plated at ∼6500 cells/cm2, or at 3000 cells/cm2 for the HU-block/release experiments, and incubated overnight. The next day, the cells were washed once with DMEM and the culture medium replaced with DMEM containing 0.25% serum. Cells were incubated further for 36 hr prior to virus infection or serum stimulation.
Where indicated, cells were subsequently serum stimulated by replacement with media containing 10% serum. For HU-block/release experiments, quiescent cells were stimulated for 21 hr with 10% serum containing 2 mm HU, washed twice with DMEM, and refed with media containing 10% serum (t = 0).
For infections with recombinant adenoviruses, quiescent REF52 cells on plates were infected in DMEM with 20 mm HEPES at pH 7.2 for 75 min at 37°C at a cell-to-volume ratio of 0.5 × 106 cells/ml (0.5 ml for a 35-mm plate, 2 ml for a 100-mm plate, or 5 ml for a 150-mm plate). Following infection, four volumes of media containing 0.25% or 10% serum (indicated as +serum) was added to each plate, and the cells were incubated at 37°C (DeGregori et al. 1995a).
HeLa cells were plated at a density of 1 × 106 cells/100-mm plate (Northern and Western blotting analysis) or 3 × 106 cells/150-mm plate (nuclear run-on assays) in DMEM supplemented with 10% fetal bovine serum (complete growth medium) and grown at 37°C, 5% CO2, for 24 hr. To obtain synchronized cell populations, cells were blocked by adding 2 mm thymidine to complete growth medium for 16 hr. To release the cells from G1/S arrest, cultures were washed three times and then incubated in prewarmed complete growth medium.
Flow cytometry and BrdU incorporation assays
Cell synchrony was assessed by flow cytometry (Smith et al. 1996). BrdU incorporation was determined as described previously (DeGregori et al. 1995b).
Nuclear and cytoplasmic extract preparation
REF52 cells on tissue-culture plates were washed twice with PBS and scraped (in PBS) into microcentrifuge tubes placed in ice. Cells were pelleted and resuspended in 10 volumes of hypotonic lysis buffer (10 mm HEPES at pH 7.5, 10 mm KCl, 3 mm MgCl2, 0.05% NP-40, 1 mm EDTA, 10 mm NaF, 0.1 mm NaVO4, 1 mm PMSF, 1 mm DTT, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 10 mm β-glycerophosphate) by pipetting up and down 10 times, and incubated on ice for 30 min. Nuclei were pelleted at 500g for 5 min at 4°C, and an equal volume of 2× gel-shift lysis buffer (90 mm HEPES at pH 7.9, 0.5 m KCl, 0.15% NP-40, 0.2 mm EGTA, 20% glycerol, 10 mm NaF, 0.1 mm NaVO4, 1 mm PMSF, 1 mm DTT, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 10 mm β-glycerophosphate) was added, mixed, and stored at −70°C (cytoplasmic fraction). The nuclei were washed once with 10 volumes of hypotonic lysis buffer and repelleted. Nuclei were then lysed in 10 volumes of gel shift buffer (50 mm HEPES at pH 7.9, 250 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 0.1% NP-40, 10% glycerol, 10 mm NaF, 0.1 mm NaVO4, 1 mm PMSF, 1 mm DTT, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 10 mm β-glycerophosphate) on ice for 30 min. Lysates were then spun at full speed in a microcentrifuge for 10 min at 4°C. The supernatants were then stored at −70°C (nuclear fraction).
E2F DNA-binding assays
E2F assays were performed as described previously (Ikeda et al. 1996). Supershift analysis was carried out as previously described (Ikeda et al. 1996) using antibodies specific against E2F1 (SC-251x), E2F2 (SC-633x), E2F3 (SC-878x), or IgG as control (Santa Cruz Biotechnologies).
Kinase assays
Kinase assays were performed as described previously (DeGregori et al. 1995) using histone H1 as a substrate.
Northern analysis
Northern analysis of RNA from REF52 cells was performed as described (DeGregori et al. 1995a). For HeLa cell experiments, total RNA was purified using Tri-Pure reagent (Boehringer Mannheim) according to the manufacturer’s protocol. Thirty micrograms of total RNA was separated by agarose gel electrophoresis under denaturing conditions, transferred to nylon membranes, and probed under condition of high stringency with Cdc6 cDNA radiolabeled with 32P as described previously.
Western analysis
REF52 cell lysates (nuclear or cytoplasmic extracts) containing equal amounts of protein were boiled for 5 min in protein sample buffer and subjected to SDS-PAGE on 10% polyacrylamide gels. Proteins were transferred onto PVDF membrane as described previously (Ikeda et al. 1996), and the PVDF membrane was blocked in TBS (25 mm Tris at pH 7.4, 137 mm NaCl, 2.7 mm KCl) containing 10% skim milk for 2 hr. Blots were then incubated with primary antibodies in TBS containing 5% skim milk overnight at 4°C, and washed subsequently in TBS containing 0.1% Tween 20 for 30 min. Blots were then incubated in TBS containing 5% skim milk and secondary antibodies for 1 hr at room temperature, and then washed for 30 min. Blots were processed with Amersham’s ECL system as described by the manufacturer. Antibodies against E2F1 (SC-251), E2F3 (SC-879), cyclin E (SC-481), and cdk2 (SC-163) were from Santa Cruz Biotechnologies.
HeLa cells were lysed in buffer containing 50 mm HEPES at pH 7.6, 50 mm NaCl, 0.1% SDS, 2 mm EDTA, 2 mm EGTA, and 1% NP-40. Whole-cell lysates were mixed immediately with protein gel-loading buffer and boiled for 3 min. Sixty micrograms of total protein was separated on 10% SDS-polyacrylamide gels, transferred to nitrocellulose membranes, and probed with a polyclonal anti-Cdc6 antibody (Williams et al. 1997) or an anti-actin antibody (Boehringer Mannheim).
Nuclear run-on transcription assays
Nuclei were prepared from HeLa cells by lysis in buffer containing 10 mm Tris at pH 7.4, 10 mm NaCl, 5 mm MgCl2, and 0.5% NP-40, as described (Ferrell 1993). RNA transcripts from 1 × 107 nuclei were extended in the presence of [32P]UTP (Amersham), purified with Tri-Pure reagent (Boehringer Mannheim), and hybridized to cDNA (Cdc6 or βactin) immobilized on nylon membranes. Cdc6 transcription rate was calculated as the ratio of radioactivity bound to Cdc6 cDNA relative to that bound to β-actin cDNA, and values from cells synchronized at specific stages of the cell cycle were compared to values from asynchronously growing HeLa cells.
Microinjection
REF52 cells were plated on 35-mm2 plates, treated as indicated in the legend to Fig. 6, and HEPES (pH 7.9) was added to 30 mm prior to microinjection. Antibodies at a concentration of 1 μg/ml, containing 1% fluorescein-conjugated dextran (Pierce Chemical) were microcentrifuged for 15 min (to remove aggregates) and injected into the cytoplasm of ∼150–250 cells per plate using an Eppendorf microinjection system with femtotips needles (Eppendorf). Antibodies specific for E2F1 (SC-251L) and E2F3 (SC-879L) were from Santa Cruz Biotechnologies, and rabbit anti-mouse IgG (as control) was from Cappel Labs. Cells were then incubated at 37°C and BrdU was added as indicated in the legend to Fig. 6. Cells were washed once with PBS and fixed first with 4% paraformaldehyde (in PBS) for 15 min at room temperature and subsequently with methanol/acetone (1:1) for 10 min. Fixed cells were washed once with PBS and incubated with 2 n HCl for 20 min, washed three times with PBS, and further incubated in 1% BSA/PBS (in 1% BSA/PBS) for 10 min. Cells injected with E2F1-specific antibodies were preblocked with 100 μg/ml rabbit anti-mouse IgG in 1% BSA/PBS for 45 min, washed extensively, and then incubated with1% BSA/PBS for an additional 5 min. Cells were then incubated for 1 hr at room temperature with anti-BrdU Solution (Amersham, cat. no. RPN-202), washed three times with PBS and once with 1% BSA/PBS, and then incubated with rhodamine-conjugated goat anti-mouse IgG (1:75, Boehringer-Mannheim), washed extensively, and visualized by immunofluorescence microscopy.
Acknowledgments
We thank Mike Cook and Lynn Martinek of the Duke Comprehensive Cancer Center Flow Cytometry Facility for flow cytometric analysis, Hiroshi Kimura for the generous gift of plasmids encoding the Mcm proteins, and Kaye Culler for assistance in the preparation of the manuscript. G.L. is supported by a fellowship from the Medical Research Council of Canada and the Alberta Heritage Foundation for Medical Research. This work was supported by a grant from the National Institutes of Heralth (HL06296) to R.S.W.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL J.Nevins@duke.edu; FAX (919) 681-8973.
References
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis and G1/S regulatory genes. Mol Cell Biol. 1995a;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J, Leone G, Ohtani K, Miron A, Nevins JR. E2F1 accumulation bypasses a G1 arrest resulting from the inhibition of G1 cyclin-dependent kinase activity. Genes & Dev. 1995b;9:2873–2887. doi: 10.1101/gad.9.23.2873. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JFX. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Bell SP. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- Dynlacht BD, Moberg K, Lees JA, Harlow E, Zhu L. Specific regulation of E2F family members by cyclin-dependent kinases. Mol Cell Biol. 1997;17:3867–3875. doi: 10.1128/mcb.17.7.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell RE. RNA methodologies. San Diego, CA: Academic Press; 1993. Analysis of nuclear RNA; pp. 235–253. [Google Scholar]
- Hateboer G, Kerkhoven RM, Shvarts A, Bernards R, Beijersbergen RL. Degradation of E2F by the ubiquitin-proteasome pathway: Regulation by retinoblastoma family proteins and adenovirus transforming proteins. Genes & Dev. 1996;10:2960–2970. doi: 10.1101/gad.10.23.2960. [DOI] [PubMed] [Google Scholar]
- Helin K, Harlow E. The retinoblastoma protein as a transcriptional repressor. Trends Cell Biol. 1993;3:43–46. doi: 10.1016/0962-8924(93)90150-y. [DOI] [PubMed] [Google Scholar]
- Hofmann F, Martelli F, Livingston DM, Wang Z. The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes & Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- Holthoff HP, Hameister H, Knippers R. A novel human Mcm protein: Homology to the yeast replication protein Mis5 and chromosomal location. Genomics. 1996;37:131–134. doi: 10.1006/geno.1996.0530. [DOI] [PubMed] [Google Scholar]
- Hunter T. Oncoprotein networks. Cell. 1997;88:333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- Hunter T, Pines J. Cyclins and cancer II: Cyclin D and CDK inhibitors come of age. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- Ikeda M-A, Jakoi L, Nevins JR. A unique role for the Rb protein in controlling E2F accumulation during cell growth and differentiation. Proc Natl Acad Sci. 1996;93:3215–3220. doi: 10.1073/pnas.93.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Schwarz JK, Cress WD, Nevins JR. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- Johnson DG, Ohtani K, Nevins JR. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes & Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- Kiyono T, Fujita M, Hayashhi Y, Ishibashi M. Cloning of a cDNA encoding a human homologue of CDC47, a member of the MCM family. Biochim Biophys Acta. 1996;1307:31–34. doi: 10.1016/0167-4781(96)00057-7. [DOI] [PubMed] [Google Scholar]
- Kowalik TF, DeGregori J, Schwarz JK, Nevins JR. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek W, Ewen ME, Shirodkar S, Arany Z, Kaelin WG, Livingston DM. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- Krek W, Xu G, Livingston DM. Cyclin A-kinase regulation of E2F-1 DNA binding function underlies suppression of an S phase checkpoint. Cell. 1995;83:1149–1158. doi: 10.1016/0092-8674(95)90141-8. [DOI] [PubMed] [Google Scholar]
- Nevins JR. E2F: A link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- Nevins JR, DeGregori J, Jakoi L, Leone G. Functional analysis of E2F. Methods Enzymol. 1997;283:205–219. doi: 10.1016/s0076-6879(97)83017-0. [DOI] [PubMed] [Google Scholar]
- Newlon CS. Putting it all together: Building a prereplicative complex. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- Ohtani K, DeGregori J, Leone G, Herendeen DR, Kelly TJ, Nevins JR. Expression of the HsOrc1 gene, a human ORC homolog, is regulated by cell proliferation via the E2F transcription factor. Mol Cell Biol. 1996;16:6977–6984. doi: 10.1128/mcb.16.12.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X-Q, Livingston DM, Kaelin WG, Adams PD. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JK, Bassing CH, Kovesdi I, Datto MB, Blazing M, George S, Wang X-F, Nevins JR. Expression of the E2F1 transcription factor overcomes type β-transforming growth factor-mediated growth suppression. Proc Natl Acad Sci. 1995;92:483–487. doi: 10.1073/pnas.92.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan B, Lee W-H. Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Mol Cell Biol. 1994;14:8166–8173. doi: 10.1128/mcb.14.12.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- Smith EJ, Leone G, DeGregori J, Jakoi L, Nevins JR. The accumulation of an E2F-p130 transcriptional repressor distinguishes a G0 from a G1 cell state. Mol Cell Biol. 1996;16:6965–6976. doi: 10.1128/mcb.16.12.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Tsuruga H, Yabuta N, Hosoya S, Tamura K, Endo Y, Nojima H. HSMCM6: A new member of the human MCM/P1 family encodes a protein homologous to fission yeast Mis5. Genes Cells. 1997;2:381–399. doi: 10.1046/j.1365-2443.1997.1290327.x. [DOI] [PubMed] [Google Scholar]
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- Williams RS, Shohet RV, Stillman B. A human protein related to yeast Cdc6p. Proc Natl Acad Sci. 1997;94:142–147. doi: 10.1073/pnas.94.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Levine AJ. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Sheppard K-A, Peng C-Y, Yee AS, Piwnica-Worms H. Cyclin A/CDK2 binds directly to E2F-1 and inhibits the DNA-binding activity of E2F-1/DP-1 by phosphorylation. Mol Cell Biol. 1994;14:8420–8431. doi: 10.1128/mcb.14.12.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, DeGregori J, Shohet RV, Leone G, Stillman B, Nevins JR, Williams RS. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc Natl Acad Sci. 1998;95:3603–3608. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]