Abstract
Background
S-RNase-based self-incompatibility (SI) occurs in the Solanaceae, Rosaceae and Plantaginaceae. In all three families, compatibility is controlled by a polymorphic S-locus encoding at least two genes. S-RNases determine the specificity of pollen rejection in the pistil, and S-locus F-box proteins fulfill this function in pollen. S-RNases are thought to function as S-specific cytotoxins as well as recognition proteins. Thus, incompatibility results from the cytotoxic activity of S-RNase, while compatible pollen tubes evade S-RNase cytotoxicity.
Scope
The S-specificity determinants are known, but many questions remain. In this review, the genetics of SI are introduced and the characteristics of S-RNases and pollen F-box proteins are briefly described. A variety of modifier genes also required for SI are also reviewed. Mutations affecting compatibility in pollen are especially important for defining models of compatibility and incompatibility. In Solanaceae, pollen-side mutations causing breakdown in SI have been attributed to the heteroallelic pollen effect, but a mutation in Solanum chacoense may be an exception. This has been interpreted to mean that pollen incompatibility is the default condition unless the S-locus F-box protein confers resistance to S-RNase. In Prunus, however, S-locus F-box protein gene mutations clearly cause compatibility.
Conclusions
Two alternative mechanisms have been proposed to explain compatibility and incompatibility: compatibility is explained either as a result of either degradation of non-self S-RNase or by its compartmentalization so that it does not have access to the pollen tube cytoplasm. These models are not necessarily mutually exclusive, but each makes different predictions about whether pollen compatibility or incompatibility is the default. As more factors required for SI are identified and characterized, it will be possible to determine the role each process plays in S-RNase-based SI.
Keywords: S-RNase, self-incompatibility, Solanaceae, Rosaceae, Nicotiana, Prunus
GENETICS OF SELF-INCOMPATIBILITY (SI)
Angiosperms display diverse reproductive strategies that have very different consequences for generating and maintaining genetic diversity. Plants that reproduce clonally are at one end of a spectrum and show little or no variability. Sexual reproduction generates new combinations of genes and alleles by crossing over, meiosis and gamete fusion. Clearly, so long as the cross is not so wide that fitness is reduced (as in interspecific crosses), crosses between less-related individuals favour diversity. Thus, although many plants reproduce by selfing, many others have evolved special mechanisms to enhance outcrossing. SI systems are genetically controlled mechanisms that inhibit fertilization by self-pollen or pollen from closely related plants (de Nettancourt, 2001). Outcrossing is enhanced in SI species by effectively dividing the population into compatibility groups, or mating types, where within-group crosses are sterile but crosses between groups are fertile (Darwin, 1877).
SI appears to have evolved independently in different angiosperm lineages. Three distinct mechanisms have been studied at the molecular level. SI plants in the Brassicaceae display a system in which proteins deposited by the tapetum onto the pollen coat interact with receptors in the stigmatic papillae to control whether pollen is accepted (Takayama and Isogai, 2005). In SI Papaver, low molecular-weight proteins secreted onto the stigma surface cause self-pollen to cease growth immediately and eventually die (Wheeler et al., 2010). SI species in the Solanaceae, Rosaceae and Plantaginaceae employ ribonucleases for recognition and rejection of self-pollen. Further mechanisms will likely be discovered through studies of SI in other lineages. However, at this point, systems employing ribonucleases appear to be the most phylogentically widespread (Igic and Kohn, 2001).
SI fascinated botanists in the early part of the twentieth century when the foundations of genetics were being forged. In reviewing early work, East and Park (1917) highlight instances where the presence of SI, or ‘self-sterility,’ showed signs of Mendelian inheritance. They observed important regularities in crosses among SI Nicotiana species, but the correct genetic model for cross compatibility was only described later by East and Mangelsdorf (1925). After determining the inheritance of compatibility groups, these authors concluded that compatibility is controlled by a single locus, the S-locus, and that compatibility in individual crosses depends on whether the S-allele, or ‘allelomorph’, in the pollen is also present in the pistil. As they described it, a plant would only provide stimulus for growth of pollen with an S-genotype not found in the pistil. This is now called gametophytic SI. Although these studies used Nicotiana species as the experimental system, other SI species in the Solanaceae, Rosaceae and Plantaginaceae display gametophytic control as well. Papaver rhoeas also shows gametophytic control of compatibility; although, the underlying mechanism is now known to be different (Wheeler et al., 2009, 2010). SI in the Brassicaceae is referred to as sporophytic SI since compatibility determinants are produced in the tapetum, a sporophytic tissue, and deposited on the pollen coat (Takayama and Isogai, 2005).
The fact that compatibility is controlled by an interaction between the pollen and the pistil is foundational to all modern studies of SI. It is now known that separate, but closely linked, genes determine S-specificity in the pistil and pollen. Thus, the current convention is to use the term S-haplotype to describe S-locus variants. Framed in this way, in gametophytic SI pollen is rejected when there is a match between the single S-haplotype in the haploid pollen and either of the two S-haplotypes in the diploid pistil. Although early workers appreciated that this genetic interaction is mediated by the ‘constituents’ of the pollen and pistil (Darwin, 1877), the relationship between these constituents and the genetic interaction revealed by progeny analysis was unknown.
S-SPECIFICITY DETERMINANTS IN S-RNASE-BASED SI
Linking the genetics of SI with the physiology of compatibility requires identification of the genes and gene products that determine S-specific pollen rejection. Candidate genes must meet three criteria: (1) linkage to the S-locus; (2) sequence variability in allelic genes from different S-haplotypes; and (3) expression in the pistil or pollen as appropriate. Since SI S-specific pollen rejection is the defining feature of SI, candidate genes must be tested in a genetic experiment that directly addresses specificity.
Pistil determinant
Analysis of pistil extracts allowed identification of proteins meeting the three basic criteria for S-specificity determinants. Using isoelectric focusing, Bredimeijer and Blaas (1981) showed that abundant polypeptides expressed in the pistil cosegregate with S-haplotype in Nicotiana alata. Anderson et al. (1986) obtained N-terminal sequence information and cloned the protein coded by the N. alata S2-haplotype. This breakthrough allowed many important facts about the structure and expression of the protein to be established. The S2-protein contains a secretion signal, and the gene is expressed in the stigma, the style transmitting tract, and the epidermis of the placenta (Anderson et al., 1986; Cornish et al., 1988). Proteins from different S-haplotypes are distinct in their chromatographic behaviour and glycosylation patterns (Jahnen et al., 1989; Woodward et al., 1989). Comparing sequences from different S-haplotypes revealed conserved and variable domains (Ioerger et al., 1991). These characteristics fit the important requirements of protein that functions in pollen recognition: it is secreted into the extracellular matrix (ECM) that forms the path from the stigma to the ovary and a distinct protein is expressed from each functional S-haplotype. These results are supported by identification of similar proteins in Petunia and Solanum (Broothaerts et al., 1989, 1991; Clark et al., 1990; Xu et al., 1990; Singh et al., 1991; Saba-El-Leil et al., 1994) as well as other solanaceous genera. Similar proteins have also been identified in Pyrus, Malus and Prunus (Broothaerts et al., 1995; Boskovic and Tobutt, 1996; Sassa et al., 1996) in the Rosaceae and in Antirrhinum in the Plantaginaceae (Xue et al., 1996). Interestingly, Prunus S-RNase genes contain two introns, while Maloideae and Solanaceae S-RNases contain only one (Igic and Kohn, 2001).
The ribonuclease activity of pistil S-proteins provided clues to the mechanism of self-pollen rejection. Studies of the active site residues in RNase T2 from Aspergillus oryzae revealed similarity to the N. alata S2-glycoprotein (Kawata et al., 1988; McClure et al., 1989). Direct enzyme assays showed that each S-protein from N. alata copurifies with a major ribonuclease activity in pistil extracts, and the proteins are now referred to as S-RNases (McClure et al., 1989). Ribonuclease activity suggested a potential link between the genetic function of the S-locus in pollen recognition and a plausible physiological mechanism for pollen rejection. Experiments following the fate of 32P-labelled pollen RNA in compatible and incompatible pollination tested this hypothesis. The results showed that pollen RNA is stable in compatible pollinations and degraded in incompatible pollinations (McClure et al., 1990). Further experiments showed that, as expected, S-RNase effectively inhibits translation. Moreover, experiments using 3H-labelled S-RNase showed that the protein enters pollen tubes intact and, thus, retains its potentially cytotoxic enzyme activity (Gray et al., 1991). Finally, Huang et al. (1994) showed that S-RNase ribonuclease activity is required for pollen rejection. Together, these results form the basis for the cytotoxic model for SI in the Solanaceae, Rosaceae and Plantaginaceae. In this model, S-RNases have dual functions, acting as recognition proteins as well as directly inhibiting growth of incompatible pollen.
The recognition function of S-RNase was confirmed using plant transformation and analysis of self-compatible mutants. Murfett et al. (1994) and Lee et al. (1994) showed that transforming an S-RNase gene into a new background causes a gain-of-function change that allows rejection of pollen expressing the corresponding S-genotype. Likewise, suppressing expression of a specific S-RNase causes loss of the ability to reject a specific pollen S-genotype (Lee et al., 1994; Murfett et al., 1994). Sassa et al. (1997) showed that S-RNase also determines S-specificity in the pistil in Pyrus serotina. These genetic results clearly demonstrate that S-RNases are the determinants of S-specificity in the pistil. Although structural differences exist between S-RNase genes in Pyrus and those in other taxa, the evidence suggests that the genes are derived from a common ancestor and that S-RNase-based SI may have emerged in the common ancestor to these diverse lineages (Igic and Kohn, 2001).
The most detailed sequence analysis of solanaceous sequences identified five conserved regions, C1 to C5, that account for about 40 of the residues in a typical S-RNase (Ioerger et al., 1991). Regions C2 and C3 contain histidine residues implicated in catalysis, and the others contribute to the hydrophobic core of the molecule (Ishimizu et al., 1995; Kawata et al., 1988). Although all other regions are variable, Ioerger et al. (1991) identified two areas with especially high sequence variability, HVa and HVb. Similar approaches were used to identify a single ‘hypervariable’ region in S-RNases from the Rosaceae (Ishimizu et al., 1998). The role of these variable regions and other S-RNase sequences in pollen recognition was tested in domain-swap experiments in transformable solanaceous species. Working in Nicotiana, Zurek et al. (1997) showed that all regions contribute to S-specific recognition and that swapping any region destroyed recognition. Kao and McCubbin (1996) swapped two regions between petunia S-RNase genes and also found that both were required for recognition. In contrast, Matton et al. (1997) found that swapping just four residues between very closely related S-RNase proteins in potato could switch the S-specificity of the target protein. Thus, while a small number of residues may distinguish a particular pair of S-RNases, it is not possible to conclude that HVa and HVb are sufficient to determine S-specificity. Indeed, a pair of S-RNase sequences has been identified in Prunus that are identical in the regions of the molecule usually described as the most variable (Zisovich et al., 2004).
Pollen determinant
Sequencing genomic regions surrounding S-RNase genes identified the pollen determinant of S-specificity. Since all genes located near S-RNase fulfill the criterion of linkage to the S-locus, the challenge was to identify the correct gene against a background that may include many genes showing sequence variation. Lai et al. (2002) identified an F-box protein gene (SLF, S-locus F-box) just 9 kb downstream of S-RNase in Antirrhinum. However, this gene, AhSLF2, was not immediately judged to be a good pollen S candidate because it was not as polymorphic as expected. Similar analyses of Prunus species with compact genomes also revealed F-box protein genes (Entani et al., 2003; Ushijima et al., 2003). In Prunus dulcis, allelic SLF genes have sufficient sequence variation that probes have shown S-specific hybridization patterns on genomic DNA. This reflects much greater sequence variability than seen in Antirrhinum (Lai et al., 2002) and is reminiscent of results with S-RNase genes (Anderson et al., 1989; Ioerger et al., 1991). The P. dulcis SLF genes are also expressed in pollen and, thus, appeared to be excellent candidates for S-specificity-determining genes: they are linked to S-RNase, show appropriate sequence variability, and are expressed in pollen (Ushijima et al., 2003). Similar results were reported in P. mume (Entani et al., 2003).
Transformation experiments offer the most straightforward approach to test the function of pollen S genes. Simple gain-of-function tests of the role of SLF, however, were deemed unlikely to succeed, and a more complex strategy was devised. In the solanaceous species where transformation experiments are practical, evidence suggested that the only pollen-side mutations that cause self-compatibility (SC) are due to the so-called heteroallelic pollen (HAP) effect. In these species, converting an SI diploid (e.g. S1S2) to a tetraploid causes SC (de Nettancourt, 1977), and the defect occurs only in the pollen; thus, S1S1S2S2 pistils reject S1- and S2-pollen normally, but diploid pollen is not rejected. Moreover, of the three types of diploid pollen produced by a tetraploid (i.e. S1S1, S1S2 and S2S2), SI breakdown only occurs in the HAP case, S1S2. However, noteworthy differences have been described between the behaviour of tetraploids in the Solanaceae compared with the Rosaceae, and there even appears to be variability within the latter family. Hauck et al. (2006) showed that SI only breaks down in tetraploid P. cerasus when at least two defective S-haplotypes are present. Thus, the HAP effect does not explain SC in this species. However, the HAP effect does seem to be effective in P. pseudocerasus and Malus (Lewis and Modlibowska, 1942; Huang et al., 2008; Sassa et al., 2009).
An influential series of modern mutagenesis experiments in N. alata nevertheless provided strong support for the idea that breakdown of SI in pollen occurs when two different pollen S genes are expressed together, at least in the Solanaceae. Golz et al. (1999, 2000) examined radiation-induced pollen-part mutants (PPMs) and concluded that all the mutants could be accounted for by duplications or translocations of the pollen S gene, effectively creating HAP (Golz et al., 1999, 2001). The absence of other classes of mutants was interpreted to mean that pollen S functions to provide resistance to S-RNase. The implication for transgenic tests of candidate pollen S genes is that, since pollen S function is essential, knock-outs are lethal and the HAP effect is the best way to test function.
Pollen S candidate SLF genes were identified by genomic sequencing in Petunia inflata, a species where transformation experiments could be conducted (McCubbin et al., 2000a, b; Wang et al., 2003, 2004). In a classic experiment, Sijacic et al. (2004) transformed the PiSLF2 gene into SI S1S1 and S2S3 plants. As expected from the HAP effect, expression of the transgene caused SC. In the transformed S1S1 plants, only pollen expressing PiSLF2 shows breakdown in SI; consistent with the HAP effect, S1-pollen without the transgene behaves normally. Thus, the effect of the PiSLF2 transgene is clearly gametophytic. However, S-specificity per se, can only be tested by comparing effects on both self- and non-self S-haplotypes. This complication is inherent in the HAP effect. Thus, S-specificity was demonstrated in S2S3 plants transformed with the PiSLF2 gene (Sijacic et al., 2004). Here, breakdown of SI is only seen when the transgene is expressed in S3-pollen. Since the PiSLF2 transgene did not interfere with SI in S2-pollen, the effect is S-specific. Qiao et al. (2004a) transformed the Antirrhinum AhSLF2 gene into SI Petunia hybrida and also reported breakdown of SI in pollen, but this experimental design does not directly address S-specificity since only a single S-haplotype can be tested.
As noted, studies of S-linked genes in Prunus provided strong evidence for a role of F-box protein genes in SI. However, it is now clear that important differences exist between the ways these genes, often referred to as SFB (S-locus F-box) genes, behave in the Rosaceae and Solanaceae (Tao and Iezzoni, 2010). For example, the level of sequence polymorphism between S-linked F-box genes (SLF or SFB) from different S-haplotypes is much lower in the Solanaceae and Plantaginaceae than in the Rosaceae. The significance of this is not clear, but the difference is striking (Wheeler and Newbigin, 2007). In another example, results in Malus and Pyrus unexpectedly identified multiple copies of pollen-expressed S-haplotype-specific F-box genes at the S-locus (SFBB, S-locus F-box brothers; Sassa et al., 2007).
MUTATIONS AFFECTING POLLEN COMPATIBILITY
Pollen-side breakdown of SI in the Rosaceae also differs from breakdown in the Solanaceae. SC mutants have attracted the attention of fruit tree breeders for many years, and their collections are a valuable resource for SI studies. Self-compatibe PPMs have been identified and analysed in P. avium (Ushijima et al., 2004; Sonneveld et al., 2005; Marchese et al., 2007), P. armeniaca (Vilanova et al., 2006), P. cerasus (Hauck et al., 2006), P. mume (Ushijima et al., 2004) and P. persica (Tao et al., 2007). In most cases, genomic analyses of Prunus PPMs reveal insertions or deletions in the SFB coding region, providing additional evidence that SFB functions as the pollen S-gene (Yamane and Tao, 2009). This is a sharp contrast to the situation just described in N. alata, where mutant studies suggest that loss of pollen S gene function is lethal (Golz et al., 2001). The contrast is further highlighted by the discovery that a PPM in an SC P. avium accession is caused by an SFB gene deletion in the S3′-haplotype (Sonneveld et al., 2005). Together, these data clearly show that mutation or deletion of Prunus SFB genes leads to SC. These genes are, therefore, not essential for pollen tube growth per se.
The apparent differences between PPM behaviour in Prunus and in N. alata are difficult to reconcile. However, these differences speak directly to the role of the interaction between the genetically defined pollen and pistil S-specificity determinants. What is the default condition: compatibility or incompatibility? Does interaction of the pollen and pistil S-specificity determinants confer resistance to S-RNase, or does it initiate rejection? In N. alata, the absence of mutations other than those that can be explained by the HAP effect is interpreted as evidence that interaction between S-RNase and SLF confers resistance to the cytotoxic effects of S-RNase in compatible pollinations (Golz et al., 2001). In this very reasonable view, pollen S acts as a genetic inhibitor of the action of S-RNase; the default, in the absence of pollen S, is rejection. The Prunus PPM results, however, support the opposite interpretation: mutation or deletion of pollen S leads to SC, and compatibility is the default condition.
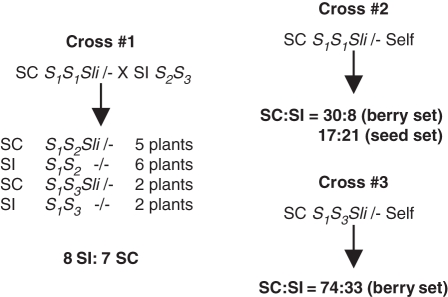
The behaviour of a PPM in Solanum chacoense offers a potentially different view from the one supported by the N. alata results and suggests that compatibility may also be the default condition for pollen in the Solanaceae. The S. chacoense S-locus inhibitor (Sli) gene has been characterized in numerous inter- and intra-specific crosses and has been followed through several generations (Hosaka and Hanneman, 1998a, b; Phumichai et al., 2005; Phumichai and Hosaka, 2006). Hosaka and Hanneman (1998a, b) interpret the behaviour of Sli as a single dominant factor that displays sporophytic inhibition of SI. Figure 1 illustrates the intriguing behaviour of Sli. In cross #1, SI S. phureja (S2S3) pollinates the inbred (i.e. seventh selfed generation) SC S. chacoense source of Sli (S1S1). In this compatible cross, the progeny segregate about 1 : 1 for SC, indicating that the defect is heterozygous. Importantly, SC is not due to a defective S-locus; both S1S2 and S1S3 SI progeny are obtained. Also, reciprocal sib-crosses (e.g. SI S1S2 × SC S1S2 and vice versa, not shown) indicate that pistil function is normal; thus, Sli only affects pollen function. The novelty of the Sli defect is evident from crosses #2 and #3, showing results from selfing the SC S1S1 parent in cross #1 and one of the SC S1S3 progeny from cross #1. Progeny from these self-pollinations also segregate for SC and SI. The existence of SI progeny shows that fully functional S-haplotypes are transmitted through pollen even when the Sli factor is not. Thus, Sli appears to act as a sporophytic factor that suppresses pollen function in a gametophytic SI system.
Fig. 1.
Genetic behaviour of S-locus inhibitor (Sli) from Solanum chacoense. Cross #1, a cross between the inbred S. chacoense S1S1 Sli source plant and an SI S. phureja plant designated S2S3. The S. chacoense parent is heterozygous for Sli and behaves as a dominant factor that inhibits SI in half the progeny. SC is unlikely to be due to a defective S-locus since half the progeny are SI. Crosses #2 and #3, selfing the S. chacoense parent or one of the SC progeny of cross #1 also results in SI progeny. Thus, functional S-haplotypes are transmitted through the pollen even when Sli is not.
This surprising behaviour of Sli contrasts dramatically with the behaviour of pollen SI defects induced by radiation or transgenesis (Golz et al., 1999, 2001; Sijacic et al., 2004). As just described, these are explained in terms of the HAP effect: pollen expressing two different pollen S-alleles is not rejected and plants that display this effect are, therefore, SC. Crucially, the HAP effect is strictly gametophytic (Golz et al., 2000). SC caused by the HAP effect can be due to changes at the S-locus or elsewhere. For instance, an Sc-haplotype with a translocation closely linked to the S-locus such that pollen S genes from two haplotypes, S1 and S2, segregate together would cause SC (Golz et al., 2001). Since the effect is expressed gametophytically, selfing an SxSc plant would yield only SC progeny (i.e. SxSc and ScSc but not SxSx). An unlinked pollen S gene would have a similar gametophytic effect. For example, when the HAP effect is used to induce SC by transforming Petunia inflata with the pollen S PiSLF2 gene, all self progeny are SC because pollen not expressing the transgene is rejected (Sijacic et al., 2004). Thus, SC of Sli-containing plants cannot be easily explained by a duplication or translocation of a pollen S gene.
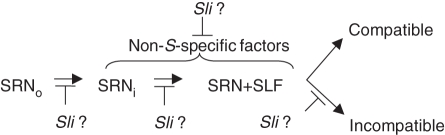
The behaviour of Sli has potentially important implications for understanding the mechanism of SI. Incompatibility is the default condition in one model (Hua et al., 2008), while compatibility is the default condition in the other (McClure, 2008). The behaviour of Sli may favour the latter because the mutant causes SC. Furthermore, the difference in PPM behaviour between Solanaceae and Rosaceae, along with differences in S-RNase gene structure and other observations, has been interpreted as evidence that S-RNase-based SI in the two families is fundamentally different (Tao and Iezzoni, 2010). Sli could, in principle, inhibit SI at several stages (Fig. 2): S-RNase uptake, the interaction of S-RNase and SLF, a later event necessary for pollen rejection, or through the action on a non-S-specific factor such. Information about the molecular nature of Sli is clearly needed to distinguish between these possibilities. Nevertheless, Sli represents an under-appreciated type of mutation that allows pollen to overcome the cytotoxic activity of S-RNase independent of its pollen S-genotype. Further studies of Sli should, therefore, reveal new insights into the mechanism of SI.
Fig. 2.
Possible functions of Sli. In S-RNase-based SI, S-RNase is secreted into the pistil extracellular matrix (SRNo). It is taken up into the pollen tube (SRNi), where it interacts with SLF (SRN + SLF) to determine whether a pollination is compatible or incompatible. Sli may prevent S-RNase uptake, its interaction with SLF, or a downstream step needed for incompatibility. It may also prevent the action of non-S-RNase factors needed for pollen rejection. It is not, however, required for pollen tube growth per se and does not appear to interfere with the compatible pathway.
NON-S-SPECIFIC FACTORS
Although the simple genetics of S-RNase-based SI demonstrate that a single locus determines the specificity of pollen rejection, other genes are also required. Candidates for these modifier genes have been identified through genetic studies of breakdown of pollen- and pistil-side functions in SI as well as through biochemical studies. The functions of modifier genes can be grouped into genes directly required for expression of S-specificity determinants (S-RNase, SLF), genes required for SI but not for pollination per se, and genes required for SI and other pollen–pistil interactions (McClure et al., 2000). Modifier genes with a unique function in SI are of special interest, as they offer insights not available through studies of S-specificity determinants alone.
Pollen factors
Biochemical and genetic studies have identified several potential modifier genes expressed in pollen. Although further studies are needed, the Sli factor already discussed has the characteristics of a pollen gene required for SI but not pollination per se. Vilanova et al. (2006) described a PPM in SC Prunus armeniaca ‘Canino’ that behaves somewhat similarly although it acts gametophytically: the gene is unlinked to the S-locus and does not affect expression of S-locus genes or cause sterility. It is noteworthy that both mutations result in SC.
Sims and Ordanic (2001) used a yeast two-hybrid (Y2H) approach to identify PhSBP1, a RING domain protein from P. hybrida pollen that binds to S-RNase. Proteins similar to SBP1 have been identified in S. chacoense, P. inflata and N. alata (O'Brien et al., 2004; Hua and Kao, 2006; Lee et al., 2008). SBP1 is expressed in a variety of tissues. In addition to binding S-RNase, SBP1 binds other proteins in Y2H assays, including the C-terminal domain of pistil arabinogalactan proteins (AGPs) such as the 120-kDa glycoprotein (120K), SLF, and certain transcription factors (Hua and Kao, 2006; Ben-Naim et al., 2007; Lee et al., 2008). Hua and Kao (2006) proposed that SBP1 forms a complex with SLF that allows degradation of non-self S-RNase in pollen and thus provides resistance to its cytotoxic effect. Although its broad expression pattern and variety of binding partners suggest that SBP1 functions in a process more general than SI per se, this does not exclude a role in degradation of S-RNase.
A number of additional putative pollen modifier genes encode proteins that form complexes with SLF. These SLF-binding proteins have been reviewed elsewhere (Hua et al., 2008; Zhang et al., 2009). Briefly, most F-box proteins are thought to function as adaptors that allow specific client proteins destined for degradation to enter one of several types of E3 ubiquitin ligase complexes. In some models for S-RNase-based SI, an SCF-like (Skp1-Cullin-F-box) E3 complex containing SLF provides for ubiquitylation of non-self S-RNase leading to its degradation, while self S-RNases fail to bind productively and thus escape degradation (Hua et al., 2008; Zhang et al., 2009). A canonical SCF E3 ubiquitin ligase complex includes a Skp1-like protein that links a cullin 1 scaffold protein to an F-box protein. Huang et al. (2006) used Y2H to identify AhSSK1, a Skp1-like protein from Antirrhinum that binds to SLF. They also found evidence that AhSSK1, in turn, binds a cullin 1-like protein and suggested that the ternary complex functions in SI. Although the F-box domain is generally thought to confer interaction with Skp1-like proteins, Hua and Kao (2006) reported that the three Skp1-like proteins expressed in P. inflata pollen do not bind SLF. Instead, as just described, these authors presented evidence that SLF binds the RING domain protein SBP1 and further suggested that it functions in a complex with a specific cullin 1 (Hua and Kao, 2008; Hua and Kao, 2006). Thus, several SLF-containing protein complexes are possible. Tests of whether the genes encoding these proteins truly behave as SI modifier genes have not been reported. However, if SLF provides for ubiquitylation and degradation of non-self S-RNase as a necessary step to overcome the cytotoxicity of S-RNase, then preventing expression of genes encoding other proteins in an SLF-containing complex would be lethal. On the other hand, the behaviour of Sli and PPMs in the Rosaceae suggest that suppressing pollen S function can lead to compatibility. Thus, direct functional tests to determine whether putative SLF-complex members are truly SI modifier genes should be considered.
Pistil factors
S-RNase is an abundant component of the pistil ECM. Other factors are also required for SI, albeit for functions other than determining S-specificity. Modifier genes encoding putative pistil SI factors have been identified through biochemical and genetic studies.
120K
S-RNase binding studies identified some of these factors and revealed possible connections between SI and other pollen-pistil interactions. Four S-RNase-binding proteins were identified in pistil extracts using immobilized S-RNase (Cruz-García et al., 2003, 2005). These include a small protein similar to chemocyanin, a pistil protein from Lilium longiflorum that displays pollen tube chemotactic activity in vitro (Kim et al., 2003). Three other S-RNase-binding proteins from N. alata are abundant transmitting tract AGPs that share a homologous cysteine-rich C-terminal domain: 120K, the pistil extensin-like protein III (PELPIII) and the transmitting tract-specific glycoprotein (TTS) (Cheung et al., 1993; Schultz et al., 1997; de Graaf et al., 2003). Given the extraordinary concentration of these proteins in the ECM, AGPs are likely to form complexes with S-RNase in planta (Cruz-Garcia et al., 2003). The functional significance of such putative S-RNase complexes is uncertain. It is noteworthy that each of the S-RNase binding proteins has been reported to interact with pollen tubes (Lind et al., 1996; Wu et al., 1995; Kim et al., 2003; de Graaf et al., 2004).
The function of 120K was tested using RNAi. Careful selection of the RNAi target region allowed production of plants with no detectable 120K protein yet near normal levels of TTS and PELP III proteins (Hancock et al., 2005). These plants fail to show S-specific pollen rejection, suggesting a direct role for 120K. Neither pollen tube growth nor uptake of S-RNase into pollen tubes is greatly affected in 120K-silenced plants (Hancock et al., 2005; Goldraij et al., 2006). Thus, 120K is not essential for these processes. Interestingly, 120K is known to enter pollen tubes, but this has not been shown for the other S-RNase-binding AGPs (Wu et al., 1995; Lind et al., 1996; de Graaf et al., 2004; Wolf et al., 2004).
Immunolocalization studies in N. alata-compatible pollen tubes using antibodies to both S-RNase and 120K showed that 120K associates with the margin of large pollen-tube vacuoles far from the tube tip (Goldraij et al., 2006) and that S-RNase is present in the lumen of these vacuoles. In N. alata, pollen rejection occurs only after a delay of several hours. Experiments comparing compatible and incompatible pollinations prior to rejection showed similar compartmentalization patterns for 120K and S-RNase. However, at later times (i.e. 36 h), the vacuolar localization of S-RNase and 120K in incompatible pollen tubes breaks down (Goldraij et al., 2006). S-RNase compartmentalization could provide a mechanism for pollen tubes to evade rejection, and this concept is the basis for one model of S-RNase-based SI (McClure, 2008).
Little is known about how S-RNase, 120K and other pistil proteins are taken up and sorted in pollen tubes. Y2H or pull-down experiments to test for interactions between pollen and pistil proteins could contribute to these potentially important processes. Lee et al. (2009) used the C-terminal domain of 120K to identify a pollen protein, NaPCCP (Pollen C2 domain-containing protein from N. alata), that binds both 120K and phosphatidyl inositol-3-phosphate. Biochemical experiments, live imaging and immunolocalization experiments showed that NaPCCP associates with the pollen tube plasma membrane and internal compartments (Lee et al., 2009). Since phosphatidyl inositol-3-phosphate is implicated in endocytosis and endomembrane transport, NaPCCP may contribute to sorting pistil proteins in pollen. However, there is no evidence that this function is restricted to proteins implicated in SI.
NaStEP
Comparisons between the pollen rejection activity associated with S-RNase transgenes in N. alata and SC N. plumbaginifolia provide clear evidence for pistil modifier genes required for SI. Murfett et al. (1996) showed that expressing S-RNase in N. plumbaginifolia does not cause S-specific pollen rejection. However, S-specific pollen rejection is fully restored by crossing in modifier genes from SC N. alata ‘Breakthrough’. Diverse cDNA cloning strategies identified candidate modifier gene transcripts (McClure et al., 1999; Juarez-Diaz et al., 2006; Busot et al., 2008). A cDNA clone corresponding to a stigma-expressed protein designated NaStEP shows a very strong differential signal (Busot et al., 2008). The encoded NaStEP protein is acidic and homologous to Kunitz-type proteinase inhibitors. It is abundant in papillar cells on the stigma surface and accumulates after pollination. Low-level expression is also detectable in the style. NaStEP protein shows an intriguing change in localization in the stigma after pollination. A putative vacuolar targeting signal is present near the N-terminus; in unpollinated stigmas, NaStEP is present in papillar cell vacuoles. However, after pollination, NaStEP is released into the stigmatic exudate (Busot et al., 2008), apparently through perforations in the papillar cell wall. Antibodies to NaStEP show cross-reacting pistil proteins in SI species, including N. alata, N. forgetiana and N. bonariensis, but not in SC N. tabacum, N. plumbaginifolia, N. benthamiana, N. longiflora and N. glauca (Busot et al., 2008). Studies are currently underway to test whether NaStEP is required for SI and whether it is taken up by pollen tubes. Although it is not known whether NaStEP is an active proteinase inhibitor, an intriguing possibility is that it may modulate breakdown of pistil proteins inside the pollen tube.
NaTrxh
A cDNA-AFLP screen identified other pistil-expressed genes with higher expression in N. alata compared with N. plumbaginifolia (Juarez-Diaz et al., 2006). One differentially expressed gene encodes NaTrxh, a protein belonging to thioredoxin h subgroup II (Fig. 3; Juarez-Diaz et al., 2006). Participation of thioredoxin h proteins in pollen rejection has been demonstrated in Brassica, where down-regulation of THL1 and THL2 leads to a partial breakdown of SI (Haffani et al., 2004). Other experiments suggest that THL1 prevents autophosphorylation of the S-receptor kinase in the absence of a ligand (Cabrillac et al., 2001). The NaTrxh protein contains an N-terminal extension that may be important for targeting or activity but does not posses a canonical secretion signal. Immunolocalization experiments, nevertheless, showed that NaTrxh is secreted into the transmitting tract ECM where it colocalizes with S-RNases and NaTTS. Moreover, the NaTrxh sequence is sufficient to confer secretion of green fluorescent protein into the extracellular space (Juarez-Diaz et al., 2006). In vitro reduction assays show that NaTrxh reduces S-RNase in crude style extracts, suggesting that it is a potential extracellular substrate (Juarez-Diaz et al., 2006). Although a functional test of NaTrxh in pollen rejection has not yet been performed, the results suggest it may function as a modifier gene in the SI response. It will be interesting to determine whether reduction of S-RNase by NaTrxh alters its three-dimensional structure or trafficking in pollen tubes. For example, NaTrxh may disrupt S-RNase–AGP complexes in pollen tube vacuoles or alter their structures and thus affect the destination in the pollen endomembrane system.
Fig. 3.
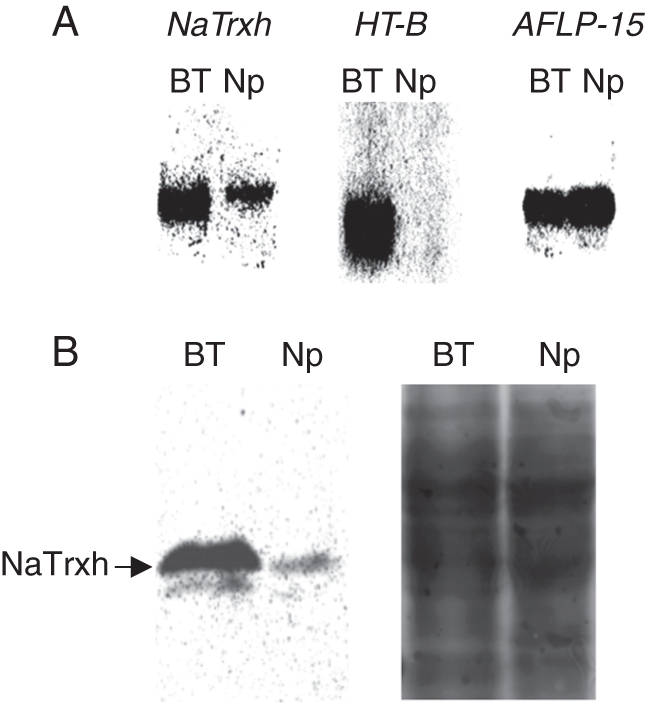
Expression of NaTrxh. (A) RNA level expression: pistil RNA (10 µg) from N. alata ‘Breakthrough’ (BT) or N. plumbaginifolia hybridized with 32P-labelled probes for NaTrxh, HT-B or a non-differential clone AFLP-15. (B) Protein blot analysis: left, total pistil protein (15 µg) probed with anti-NaTrxh; right, a similar gel stained with Coomassie Blue.
HT-B
HT-B was the first modifier gene cloned. Like NaStEP, it was identified as a differential cDNA expressed in N. alata but not in N. plumbaginifolia (McClure et al., 1999). Antisense experiments in Nicotiana showed that suppressing HT-B expression in the pistil prevents S-specific pollen rejection (McClure et al., 1999). In these experiments, an anti-HT-B antibody was used to detect expression in antisense-suppressed plants. Plants with reduced expression show partial breakdown in S-specific pollen rejection, and plants with no detectable HT-B protein allow many otherwise incompatible pollen tubes to penetrate to the base of the style. An HT-B gene was recently identified in SI P. inflata, and an RNAi construct tested its function in SI. Plants with extremely low HT-B expression show partial breakdown in SI. This observation supports a role in SI and also suggests that even small amounts of HT-B protein are sufficient (Puerta et al., 2009). Plants in the genus Solanum (including relatives of potato and tomato) express two very similar genes, HT-A and HT-B. O'Brien et al. (2002) used RNAi to test the function of these genes in S. chacoense. Plants with strongly suppressed HT-B expression show partial breakdown in SI. Since these plants retain near normal levels of HT-A mRNA, the authors concluded that HT-B is implicated in SI but that HT-A is not. Kondo et al. (2002), who examined HT-A and HT-B genes in cultivated tomato and several of its wild relatives, noted that HT-B genes are disrupted or contain stop codons in SC species. However, a recent study of SI S. habrochaites showed that both SI and SC accessions have stop codons in HT-B. Protein-level studies similarly failed to detect HT-B protein in all accessions of S. habrochaites (Covey et al., 2010). Since all indications are that even extremely low levels of HT-B protein are sufficient, it is worth reexamining the possibility that HT-A can function in S-specific pollen rejection. It is also possible, as Puerta et al. (2009) suggested, that HT-B has an indirect role in SI and strengthens the response. Such an effect could vary between different S-haplotypes.
Although HT-B's exact function is not known, it is clear that it is not required for S-RNase uptake. Goldraij et al. (2006) observed normal S-RNase uptake and compartmentalization in HT-B-suppressed plants that are incapable of S-specific pollen rejection. Thus, S-RNase uptake is not sufficient to cause pollen rejection, and S-RNase sequestered in the vacuole also is not necessarily detrimental to pollen-tube growth, at least in the absence of HT-B protein. Therefore, it is very intriguing that HT-B protein appears to be degraded after compatible pollination. Immunolocalization experiments in Nicotiana show little or no anti-HT-B reactive protein in compatible pollen tubes but substantial amounts in incompatible pollen tubes (Goldraij et al., 2006). This observation is consistent with HT-B protein being taken up and degraded in pollen tubes. Moreover, analysis of style extracts showed that, after compatible pollination, HT-B levels drop to 3–25 % of unpollinated controls. HT-B levels also drop after incompatible pollination but only by about 2-fold (Goldraij et al., 2006).
It is now clear that several HT-like proteins are expressed in the pistil. Kondo and McClure (2008) noted similarities between HT-proteins and certain glycine-rich proteins, including nodulin-24. Interestingly, the putative secretion signals are among the most conserved features of the HT/NOD-24 family. It is followed by a variable core sequence of about 50 amino acid residues. All family members contain one or more cysteine motifs, including CXXCXC, CXXXCC or CXXCC. HT-A and HT-B proteins are distinguishable by a characteristic stretch of about 20 asparagine and aspartic acid residues (ND domain) that is flanked by the cysteine motifs CXXCXC and CXXXCC. Sassa and Hirano (2006) identified the PiHTL-A and PiHTl-B transcripts in P. inflata as products of alternative splicing of a single gene. The encoded proteins contain a CXXCXCCXXXCXXXC motif but lack an ND domain. PiHTL-A and PiHTl-B do not appear to be required for SI. Kondo and McClure (2008) identified a family of small HT-family proteins in Nicotiana designated HT-M. Like the P. inflata HTL-A and -B proteins, HT-M proteins lack an ND domain and contain a single cysteine motif. Like HT-B, HT-M protein levels decrease after pollination. Unlike HT-B, the response to compatible and incompatible pollen is similar.
MODELS
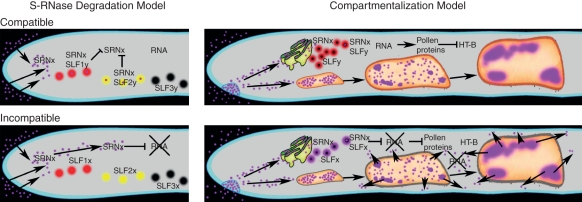
Figure 4 shows two models used to explain S-RNase-based SI: the S-RNase degradation model (left) and the compartmentalization model (right). In both, interaction between S-RNase and SLF determines whether pollination is compatible or incompatible, as shown in Fig. 2. Both models entail degradation of pollen RNA after incompatible pollination (RNA, crossed out), which leads to a general inhibition of pollen protein synthesis needed for continued growth. The latter is explicitly shown only in the compartmentalization model, which emphasizes homeostatic mechanisms that maintain the integrity of the endomembrane system and facilitate degradation of HT-B from the pistil. The explanations for how compatible pollen evades S-RNase cytotoxicity are different in the two models.
Fig. 4.
Models for S-specific pollen rejection. Pollen tubes are shown in the pistil ECM containing a single S-RNase (SRNx, purple); although, in a typical S-heterozygote two S-RNases would be present. Compatible (top, Sy-pollen tube in a pistil expressing Sx-RNase) and incompatible (bottom, Sx-pollen tube) pollinations are shown. Left: an S-RNase degradation model that implicates multiple SLF proteins (SLF1, red; SLF2, yellow; SLF3, black regardless of whether they are derived from the Sx- or Sy-haplotype) collaborating to cause S-RNase degradation. These models do not specify a route from the ECM to the pollen cytoplasm where the S-RNase-SLF interaction occurs. Right: the compartmentalization model shows most S-RNase taken up by endocytosis and trafficking by default to progressively larger vacuoles in more mature regions of the pollen tube. S-RNase must exit the endomembrane system to interact with SLF; a single SLF (red, SLFx; blue SLFy) is shown. Both models show degradation of pollen RNA (cross) in incompatible pollen tubes, a process that do not occur in compatible pollen tubes (no cross). In S-RNase degradation models, compatibility is attributed to wholesale degradation of S-RNase. The compartmentalization model, in contrast, emphasizes S-RNase isolation from the cytoplasm.
In S-RNase degradation models, interaction between S-RNase and SLF leads to wholesale destruction of S-RNase, thereby providing resistance to its cytotoxic effect (Hua et al., 2008; Zhang et al., 2009). In the version of the S-RNase-degradation model shown in Fig. 4, multiple SLF proteins (SLF1, red; SLF2, yellow; SLF3, black) collaborate to provide resistance to Sx-RNase (Kubo et al., 2010). Different S-haplotypes express a unique array of allelic SLF genes (e.g. SLF1x and SLF1y). Individual SLF proteins may or may not bind a given S-RNase. In the hypothetical compatible pollination shown (top, left), only SLF1y and SLF2y bind Sx-RNase, preventing RNA degradation. In contrast, in an incompatible pollination, none of the SLF proteins productively bind self S-RNase (SLF1x, SLF2x and SLF3x are shown without bound Sx-RNase; bottom, left), pollen RNA is degraded, and pollen tube growth is inhibited. Thus, the collaborative S-RNase degradation model rationalizes the finding that multiple SLF genes are linked to a given S-RNase gene as well as results showing variable binding between individual S-RNase and SLF proteins (Kubo et al., 2010). These features, however, make definitive tests difficult. Since the number of SLF genes in a given S-haplotype is unknown and since S-RNase binding for a given SLF protein cannot be predicted, almost any binding result or transgenic test of S-specificity can be accommodated.
Ubiquitylation and subsequent proteasomal degradation of non-self S-RNase is taken to be the fundamental process responsible for compatibility in S-RNase degradation mechanisms. Thus, the identification of SLF-containing complexes provides support for this model (Qiao et al., 2004a, b; Hua and Kao, 2006; Huang et al., 2006; Hua et al., 2007). S-specific interactions between S-RNase and SLF have been reported in vitro, but degradation of S-RNase in pollen extracts is not S-specific (Hua and Kao, 2006). Although non-self S-RNase degradation is a plausible mechanism, a direct connection between S-RNase/SLF interactions and S-RNase degradation has not been established.
The compartmentalization model (Fig. 4, right) explains resistance to non-self S-RNase by its sequestration from the cytoplasm in compatible pollen tubes. Immunolocalization experiments show large amounts of S-RNase sequestered in vacuoles of compatible pollen tubes and compartmentalization breakdown in incompatible pollen tubes (Goldraij et al., 2006). The compartmentalization model emphasizes the homeostatic effects of normal pollen gene expression (pollen proteins, Fig. 4) in relation to maintenance of the endomembrane system and elimination of HT-B protein. As previously discussed, degradation of HT-B occurs in compatible pollen tubes (Fig. 4; top, right) and is assumed to rely on normal pollen gene expression. Much S-RNase traffics to vacuoles, presumably, through another default process. Nevertheless, since SLF is a cytoplasmic protein, some S-RNase must exit the luminal compartment, possibly after retrograde transport to the endoplasmic reticulum, as has been demonstrated for other cytotoxins (McClure, 2006). In the compartmentalization model, the non-self S-RNase/SLF interaction does not cause wholesale degradation of S-RNase, which is largely inaccessible. This feature of the model rationalizes the immunolocalization experiments showing large amounts of S-RNase in compatible pollen tubes (Goldraij et al., 2006), but it does not address the question of the direct function of SLF-containing ubiquitin ligase complexes. For convenience, Fig. 4 shows alternate complexes for both compatible and incompatible interactions (i.e. SRNx/SLFy, top; SRNx/SLFx, bottom), but the model places no specific restraint on the biochemical nature of these interactions. The model only predicts that, whatever the function of the SLF-complex, a non-self S-RNase does not cause RNA degradation, a normal spectrum of pollen proteins are produced, and growth proceeds. Incompatibility, however, is seen as an active process. One speculation is that self S-RNase activates the SLF-complex to target a pollen protein whose function is to degrade HT-B, which promotes the ability of S-RNase to cause pollen rejection. Regardless of the actual target of the SLF complex, it is clear that in an incompatible pollination HT-B is stabilized, pollen RNA is degraded, the endomembrane system loses its integrity and ever more S-RNase is released. Thus, incompatibility is seen as self-reinforcing.
The compartmentalization and S-RNase degradation models are not mutually exclusive. For example, the presence of large amounts of S-RNase in pollen tube vacuoles does not exclude degradation of smaller amounts in the cytoplasm, and vice versa. However, if non-self S-RNase degradation is the dominant mechanism for S-RNase resistance, then breakdown of SI in pollen should lead to rejection. Mutant studies in N. alata, the HAP effect, and the behaviour of SLF transgenes in P. inflata are interpreted as consistent with this prediction (Golz et al., 2001; Hua et al., 2008). On the other hand, SBF mutations in several Prunus species clearly result in pollen compatibility, not incompatibility (Ushijima et al., 2004; Sonneveld et al., 2005; Vilanova et al., 2006; Marchese et al., 2007; Tao et al., 2007). Tao and Izzeoni (2010) suggest this latter observation is due to a fundamental difference between the mechanism of S-RNase-based SI in the Solanaceae and Rosaceae. Differences between the models may exist, but the major similarities (i.e. S-RNase determining S-specificity in the pistil and F-box proteins fulfilling this function in pollen) are more striking than the differences. The behaviour of Sli in Solanum may be important as well. Although no information is available about its identity or molecular mechanism, the genetic data are not consistent with gametophytic action. Therefore, Sli cannot be attributed to the well-documented HAP effect, which is strictly gametophtic. Yet, similar to the pollen-side mutations in Prunus, when Sli causes breakdown of SI in pollen, compatibility, not incompatibility, is the result. These pollen-side defects in Prunus and in Solanum are consistent with pollen rejection resulting from an active process in pollen: in the absence of recognition, pollen is compatible. Of note, this is also the situation in Papaver SI (Wheeler et al., 2010). Although the molecular mechanism in Papaver is completely different, SI signalling clearly induces a series of active processes in pollen that result in incompatibility. In this regard, it is also important that pistil-modifier genes are required for S-RNase to function in S-RNase-based SI. Some modifier genes have been identified, and the activities of additional candidates are being tested. Nevertheless, none of those tested so far affect S-RNase uptake (Goldraij et al., 2006). In the absence of ‘activation’ by modifier genes, S-RNase remains harmlessly sequestered in pollen tube vacuoles.
PROSPECTS
More questions than answers about S-RNase-based SI remain. Progress is needed at all levels. At the biochemical level, it would be helpful to establish clearly the target of the SLF complex. Is S-RNase a target or a modifier of its activity toward other proteins? At the genetic level, it would be helpful to identify all of the genes required for SI to function on both the pollen and pistil sides. Sli is an especially intriguing factor acting in pollen. Many additional factors are likely to contribute to SI in both the pollen and the pistil. Finally, at the cell biological level, it would be helpful to characterize the S-RNase trafficking pathway from the ECM to the vacuole and understand how it gains access to the cytoplasm.
ACKNOWLEDGEMENTS
The authors thank Melody Kroll for editorial assistance. This work was supported by grant (IOB 0614962) from the US National Science Foundation to B.McC.; grant (IN205009) from DGAPA and grant (81968) from CONACYT to F.C.-G.
LITERATURE CITED
- Anderson MA, Cornish EC, Mau S-L, et al. Cloning of cDNA for a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata. Nature. 1986;321:38–44. [Google Scholar]
- Anderson M, McFadden G, Bernatzky R, et al. Sequence variability of three alleles of the self-incompatibility gene of Nicotiana alata. The Plant Cell. 1989;1:483–491. doi: 10.1105/tpc.1.5.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Naim O, Eshed R, Parnis A, et al. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. The Plant Journal. 2007;46:462–476. doi: 10.1111/j.1365-313X.2006.02706.x. [DOI] [PubMed] [Google Scholar]
- Boskovic RI, Tobutt KR. Correlation of stylar ribonuclease zymograms with incompatibility alleles in sour cherry. Euphytica. 1996;90:245–250. [Google Scholar]
- Bredemeijer GMM, Blaas J. S-Specific proteins in styles of self-incompatible Nicotiana alata. Theoretical and Applied Genetics. 1981;59:185–190. doi: 10.1007/BF00264974. [DOI] [PubMed] [Google Scholar]
- Broothaerts WJ, van Laere A, Witters R, et al. Purification and N-terminal sequencing of style glycoproteins associated with self-incompatibility in Petunia hybridia. Plant Molecular Biology. 1989;14:93–102. doi: 10.1007/BF00015658. [DOI] [PubMed] [Google Scholar]
- Broothaerts W, Vanvinckeyroye P, Decock B, Van Damme J, Vendrig JC. Petunia hybridia S-proteins: ribonuclease activity and the role of their glycan side chains in self-incompatibility. Sexual Plant Reproduction. 1991;4:258–266. [Google Scholar]
- Broothaerts W, Janssens GA, Proost P, Broekaert WF. cDNA cloning and molecular analysis of two self-incompatibility alleles from apple. Plant Molecular Biology. 1995;27:499–511. doi: 10.1007/BF00019317. [DOI] [PubMed] [Google Scholar]
- Busot GY, McClure B, Ibarra-Sánchez CP, Jiménez-Durán K, Vázquez-Santana S, Cruz-García F. Pollination in Nicotiana alata stimulates synthesis and transfer to the stigmatic surface of NaStEP, a vacuolar Kunitz proteinase inhibitor homologue. Journal of Experimental Botany. 2008;59:3187–3201. doi: 10.1093/jxb/ern175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrillac D, Cock JM, Dumas C, Gaude T. The S-locus receptor kinase is inhibited by thioredoxins and activated by pollen coat proteins. Nature. 2001;410:220–223. doi: 10.1038/35065626. [DOI] [PubMed] [Google Scholar]
- Cheung AY, May B, Kawata E, Gu Q, Wu H-M. Characterization of cDNAs for stylar transmitting tissue-specific proline-rich proteins in tobacco. The Plant Journal. 1993;3:151–160. [PubMed] [Google Scholar]
- Clark KR, Okuley JJ, Collins PD, Sims TL. Sequence variability and developmental expression of S-alleles in self-incompatible and pseudo-self-compatible Petunia. The Plant Cell. 1990;2:815–826. doi: 10.1105/tpc.2.8.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish EC, Anderson MA, Clarke AE. Molecular aspects of fertilization in flowering plants. Annual Review of Cell Biology. 1988;4:209–228. doi: 10.1146/annurev.cb.04.110188.001233. [DOI] [PubMed] [Google Scholar]
- Covey PA, Kondo K, Welch L, et al. Multiple features that distinguish unilateral incongruity and self-incompatibility in the tomato clade. The Plant Journal. 2010;64:367–378. doi: 10.1111/j.1365-313X.2010.04340.x. [DOI] [PubMed] [Google Scholar]
- Cruz-García F, Hancock CN, McClure B. S-RNase complexes and pollen rejection. Journal of Experimental Botany. 2003;53:123–130. doi: 10.1093/jxb/erg045. [DOI] [PubMed] [Google Scholar]
- Cruz-García F, Hancock CN, Kim D, McClure B. Stylar glycoproteins bind to S-RNase in vitro. The Plant Journal. 2005;42:295–304. doi: 10.1111/j.1365-313X.2005.02375.x. [DOI] [PubMed] [Google Scholar]
- Darwin CR. The different forms of flowers on plants of the same species. 1st edn. London: John Murray; 1877. [Google Scholar]
- East EM, Mangelsdorf AJ. A new interpretation of the hereditary behaivor of self-sterile plants. Proceedings of the National Academy of Sciences of the USA. 1925;11:166–171. doi: 10.1073/pnas.11.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East EM, Park JB. Studies on self-sterility. I. The behaviour of self-sterile plants. Genetics. 1917;2:505–609. doi: 10.1093/genetics/2.6.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entani T, Iwano M, Shiba H, Che FS, Isogai A, Takayama S. Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen-expressed F-box gene with allelic diversity. Genes and Cells. 2003;8:203–213. doi: 10.1046/j.1365-2443.2003.00626.x. [DOI] [PubMed] [Google Scholar]
- Goldraij A, Kondo K, Lee CB, et al. Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature. 2006;439:805–810. doi: 10.1038/nature04491. [DOI] [PubMed] [Google Scholar]
- Golz JF, Su V, Clarke AE, Newbigin E. A molecular description of mutations affecting the pollen component of the Nicotiana alata S locus. Genetics. 1999;152:1123–1135. doi: 10.1093/genetics/152.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz JF, Clarke AE, Newbigin E. Mutational approaches to the study of self-incompatibility: revisiting the pollen-part mutants. Annals of Botany. 2000;85(Suppl. A):95–103. [Google Scholar]
- Golz JF, Oh H-Y, Su V, Kusaba M, Newbigin E. Genetic analysis of Nicotiana pollen-part mutants is consistent with the presence of an S-ribonuclease inhibitor at the S-locus. Proceedings of the National Academy of Sciences of the USA. 2001;98:15372–15376. doi: 10.1073/pnas.261571598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf BHJ, Knuiman BA, Derksen J, Mariani C. Characterization and localization of the transmitting tissue-specific PELPIII proteins of Nicotiana tabacum. Journal of Experimental Botany. 2003;54:55–63. doi: 10.1093/jxb/erg002. [DOI] [PubMed] [Google Scholar]
- de Graaf BHJ, Knuiman BA, van der Weerden GM, Feron R, Derksen J, Mariani C. The PELPIII glycoproteins in Solanaceae: stylar expression and transfer to pollen tube walls. Sexual Plant Reproduction. 2004;16:245–252. [Google Scholar]
- Gray JE, McClure BA, Bönig I, Anderson MA, Clarke AE. Action of the style product of the self-incompatibility gene of Nicotiana alata (S-RNase) on in vitro-grown pollen tubes. The Plant Cell. 1991;3:271–283. doi: 10.1105/tpc.3.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffani YZ, Gaude T, Cock JM, Goring D. Antisense suppression of thioredoxin h mRNA in Brassica napus cv. Westar pistils causes a low level of constitutive pollen rejection response. Plant Molecular Biology. 2004;55:619–630. doi: 10.1007/s11103-004-1126-x. [DOI] [PubMed] [Google Scholar]
- Hancock CN, Kent L, McClure BA. The 120kDa glycoprotein is required for S-specific pollen rejection in Nicotiana. The Plant Journal. 2005;43:716–723. doi: 10.1111/j.1365-313X.2005.02490.x. [DOI] [PubMed] [Google Scholar]
- Hauck NR, Yamane H, Tao R, Iezzoni AF. Accumulation of nonfunctional S-haplotypes results in the breakdown of gametophytic self-incompatibility in tetraploid Prunus. Genetics. 2006;172:1191–1198. doi: 10.1534/genetics.105.049395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka K, Hanneman RE. Genetics of self-compatibility in a self-compatible wild diploid potato species Solanum chacoense. 1. Detection of an S-locus inhibitor (Sli) gene. Euphytica. 1998a;99:191–197. [Google Scholar]
- Hosaka K, Hanneman RE. Genetics of self-compatibility in a self-compatible wild diploid potato species Solanum chacoense. 2. Localization of an S-locus inhibitor (Sli) gene on the potato genome using DNA markers. Euphytica. 1998b;103:265–271. [Google Scholar]
- Hua ZH, Kao T-H. Identification and characterization of components of a putitative Petunia S-locus F-box containing E3 ligase complex involved in S-RNase-based self-incompatibility. The Plant Cell. 2006;18:2531–2553. doi: 10.1105/tpc.106.041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Kao T-H. Identification of major lysine residues of S3-RNase of Petunia inflata involved in ubiquitin-26S proteasome-mediated degradation in vitro. The Plant Journal. 2008;54:1094–1104. doi: 10.1111/j.1365-313X.2008.03487.x. [DOI] [PubMed] [Google Scholar]
- Hua Z, Meng XY, Kao T-H. Comparison of Petunia inflata S-locus F-box protein (PiSLF) with PiSLF-like proteins reveals it unique function in S-RNase-based self-incompatibility. The Plant Cell. 2007;19:3593–3609. doi: 10.1105/tpc.107.055426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua ZH, Fields A, Kao T-H. Biochemical models for S-RNase-based self-incompatibility. Molecular Plant. 2008;1:575–585. doi: 10.1093/mp/ssn032. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhao L, Yang Q, Xue Y. AhSSK1, a novel SKP1-like protein that interacts with the S-locus F-box protein SLF. The Plant Journal. 2006;46:780–793. doi: 10.1111/j.1365-313X.2006.02735.x. [DOI] [PubMed] [Google Scholar]
- Huang S, Lee H-S, Karunanandaa B, Kao T-H. Ribonuclease activity of Petunia inflata S proteins is essential for rejection of self-pollen. The Plant Cell. 1994;6:1021–1028. doi: 10.1105/tpc.6.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S-X, Wu H-Q, Li Y-R, et al. Competitive interaction between two functional S-haplotypes confers self-compatibility on tetraploid Chinese cherry (Prunu pseudocerasus Lindl cv. Nanjing Chuisi) Plant Cell Reports. 2008;27:1075–1085. doi: 10.1007/s00299-008-0528-7. [DOI] [PubMed] [Google Scholar]
- Igic B, Kohn JR. Evolutionary relationships among self-incompatibility RNases. Proceedings of the National Academy of Sciences of the USA. 2001;98:13167–13171. doi: 10.1073/pnas.231386798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger TR, Gohlke JR, Xu B, Kao T-H. Primary structural features of the self-incompatibility protein in Solanaceae. Sexual Plant Reproduction. 1991;4:81–87. [Google Scholar]
- Ishimizu T, Endo T, Yamaguchi-Kabata Y, Nakamura KT, Sakiyama F, Norioka S. Identification of regions in which positive selection may operate in S-RNase of Rosaceae: implication for S-allele-specific recognition sites in S-RNase. FEBS Letters. 1998;440:337–342. doi: 10.1016/s0014-5793(98)01470-7. [DOI] [PubMed] [Google Scholar]
- Ishimizu T, Miyagi M, Norioka S, Liu Y-H, Clarke AE, Sakiyama F. Identification of histidine-31 and cysteine-95 in the active site of self-incompatibility associated S6-RNase in Nicotiana alata. Journal of Biochemistry. 1995;118:1007–1013. doi: 10.1093/jb/118.5.1007. [DOI] [PubMed] [Google Scholar]
- Jahnen W, Batterham MP, Clarke AE, Moritz RL, Simpson RJ. Identification, isolation, and N-terminal sequencing of style glycoproteins associated with self-incompatibility in Nicotiana alata. The Plant Cell. 1989;1:493–499. doi: 10.1105/tpc.1.5.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez-Diaz JA, McClure B, Vazquez-Santana S, et al. A novel thioredoxin h is secreted in Nicotiana alata and reduces S-RNases. Journal of Biological Chemistry. 2006;281:3418–3424. doi: 10.1074/jbc.M511687200. [DOI] [PubMed] [Google Scholar]
- Kao T-H, McCubbin AG. How flowering plants discriminate between self and non-self pollen to prevent inbreeding. Proceedings of the National Academy of Sciences of the USA. 1996;93:12059–12065. doi: 10.1073/pnas.93.22.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata Y, Sakiyama F, Tamaoki H. Amino acid sequence of ribonuclease T2 from Aspergillus oryzae. European Journal of Biochemistry. 1988;176:683–697. doi: 10.1111/j.1432-1033.1988.tb14331.x. [DOI] [PubMed] [Google Scholar]
- Kim S, Mollet J-C, Dong J, Zhang K, Park S-Y, Lord EM. Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proceedings of the National Academy of Sciences of the USA. 2003;100:16125–16130. doi: 10.1073/pnas.2533800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, McClure B. New microsome-associated HT-family proteins from Nicotiana respond to pollination and define an HT/NOD-24 protein family. Molecular Plant. 2008;1:634–644. doi: 10.1093/mp/ssn018. [DOI] [PubMed] [Google Scholar]
- Kondo K, Yamamoto M, Itahashi R, et al. Insights into the evolution of self-compatibility in Lycopersicon from a study of stylar factors. The Plant Journal. 2002;30:143–153. doi: 10.1046/j.1365-313x.2002.01275.x. [DOI] [PubMed] [Google Scholar]
- Kubo K-I, Entani T, Takara A, et al. Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science. 2010;330:796–799. doi: 10.1126/science.1195243. [DOI] [PubMed] [Google Scholar]
- Lai Z, Ma W, Han B, et al. An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Molecular Biology. 2002;50:29–42. doi: 10.1023/a:1016050018779. [DOI] [PubMed] [Google Scholar]
- Lee CB, Swatek KN, McClure B. Pollen proteins bind to the C-terminal domain of Nicotiana alata pistil arabinogalactan proteins. Journal of Biological Chemistry. 2008;283:26965–26973. doi: 10.1074/jbc.M804410200. [DOI] [PubMed] [Google Scholar]
- Lee CB, Kim S, McClure B. A pollen protein, NaPCCP, that binds pistil arabinogalactan proteins also binds phosphatidylinositol 3-phosphate and associates with the pollen tube endomembrane system. Plant Physiology. 2009;149:791–802. doi: 10.1104/pp.108.127936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-S, Huang S, Kao T-H. S proteins control rejection of incompatible pollen in Petunia inflata. Nature. 1994;367:560–563. doi: 10.1038/367560a0. [DOI] [PubMed] [Google Scholar]
- Lewis D, Modlibowska I. Genetical studies in pears. IV. Pollen tube growth and incompatibility. Journal of Genetics. 1942;43:211–222. [Google Scholar]
- Lind JL, Bönig I, Clarke AE, Anderson MA. A style-specific 120 kDa glycoprotein enters pollen tubes of Nicotiana alata in vivo. Sexual Plant Reproduction. 1996;9:75–86. [Google Scholar]
- McClure B. New views of S-RNase-based self-incompatibility. Current Opinion in Plant Biology. 2006;9:639–646. doi: 10.1016/j.pbi.2006.09.004. [DOI] [PubMed] [Google Scholar]
- McClure B. Comparing models for S-RNase-based self-incompatibility. In: Franklin-Tong V, editor. Self-incompatibility in flowering plants: evolution, diversity, and mechanisms. Berlin: Springer; 2008. pp. 217–236. [Google Scholar]
- McClure BA, Haring V, Ebert PR, et al. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature. 1989;342:955–957. doi: 10.1038/342955a0. [DOI] [PubMed] [Google Scholar]
- McClure BA, Gray JE, Anderson MA, Clarke AE. Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature. 1990;347:757–760. [Google Scholar]
- McClure BA, Mou B, Canevascini S, Bernatzky R. A small asparagine-rich protein required for S-allele-specific pollen rejection in Nicotiana. Proceedings of the National Academy of Sciences of the USA. 1999;96:13548–13553. doi: 10.1073/pnas.96.23.13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Cruz-Garcia F, Beecher BS, Sulaman W. Factors affecting inter- and intra-specific pollen rejection in Nicotiana. Annals of Botany. 2000;85:113–123. [Google Scholar]
- McCubbin AG, Wang X, Kao T-H. Identification of self-incompatibility (S-) locus linked pollen cDNA markers in Petunia inflata. Genome. 2000a;43:619–627. [PubMed] [Google Scholar]
- McCubbin AG, Zuniga C, Kao T-H. Construction of a binary bacterial artificial chromosome library of Petunia inflata and the isolation of large genomic fragments linked to the self-incompatibility (S-) locus. Genome. 2000b;43:820–826. [PubMed] [Google Scholar]
- Marchese A, Boskovic RI, Caruso T, Raimondo A, Cutuli M, Tobutt KR. A new self-incompatibility haplotype in sweet cherry ‘Kronio’, S5' attributable to a pollen-part mutation in the SFB gene. Journal of Experimental Botany. 2007;58:4347–4356. doi: 10.1093/jxb/erm322. [DOI] [PubMed] [Google Scholar]
- Matton DP, Maes O, Laublin G, et al. Hypervariable domains of self-incompatibility RNases mediate allele-specific pollen recognition. The Plant Cell. 1997;9:1757–1766. doi: 10.1105/tpc.9.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfett J, Atherton TL, Mou B, Gasser CS, McClure BA. S-RNase expressed in transgenic Nicotiana causes S-allele-specific pollen rejection. Nature. 1994;367:563–566. doi: 10.1038/367563a0. [DOI] [PubMed] [Google Scholar]
- Murfett JM, Strabala TJ, Zurek DM, Mou B, Beecher B, McClure BA. S RNase and interspecific pollen rejection in the genus Nicotiana: multiple pollen rejection pathways contribute to unilateral incompatibility between self-incompatible and self-compatible species. The Plant Cell. 1996;8:943–958. doi: 10.1105/tpc.8.6.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nettancourt D. Incompatibility in angiosperms. Berlin: Springer-Verlag; 1977. [Google Scholar]
- de Nettancourt D. Incompatibility and incongruity in wild and cultivated plants. 2nd edn. Berlin: Springer-Verlag; 2001. [Google Scholar]
- O'Brien M, Kapfer C, Major G, et al. Molecular analysis of the stylar-expressed Solanum chacoense asparagine-rich protein family related to the HT modifier of gametophytic self-incompatibility in Nicotiana. The Plant Journal. 2002;32:1–12. doi: 10.1046/j.1365-313x.2002.01486.x. [DOI] [PubMed] [Google Scholar]
- O'Brien M, Major G, Chanta S-C, Matton DP. Isolation of S-RNase binding proteins from Solanum chacoense: identification of an SBP1 (RING finger protein) orthologue. Sexual Plant Reproduction. 2004;17:81–87. [Google Scholar]
- Phumichai C, Hosaka K. Cryptic improvement for fertility by continuous selfing of diploid potatoes using Sli gene. Euphytica. 2006;149:251–258. [Google Scholar]
- Phumichai C, Mori H, Kobayashi A, Kamijima O, Hosaka K. Toward the development of highly homozygous diploid potato lines using the self-compatibility controlling Sli gene. Genome. 2005;48:977–984. doi: 10.1139/g05-066. [DOI] [PubMed] [Google Scholar]
- Puerta AR, Ushijima K, Koba T, Sassa H. Identification and functional analysis of pistil self-incompatibility factor HT-B of Petunia. Journal of Experimental Botany. 2009;60:1309–1318. doi: 10.1093/jxb/erp005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Wang F, Zhao L, et al. The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility. The Plant Cell. 2004a;16:2307–2322. doi: 10.1105/tpc.104.024919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Wang H, Zhao L, et al. The F-box protein AhSLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. The Plant Cell. 2004b;16:582–595. doi: 10.1105/tpc.017673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba-El-Leil M, Rivard S, Morse D, Cappadocia M. The S11 and S13 self-incompatibility alleles in Solanum chacoense Britt. are remarkably similar. Plant Molecular Biology. 1994;24:571–583. doi: 10.1007/BF00023555. [DOI] [PubMed] [Google Scholar]
- Sassa H, Nishio T, Kowyama Y, Hirano H, Koba T, Ikehashi H. Self-incompatibility (S) alleles alleles of the Rosaceae encode members of a distinct class of the T2/S-ribonuclease superfamily. Molecular and General Genetics. 1996;250:547–557. doi: 10.1007/BF02174443. [DOI] [PubMed] [Google Scholar]
- Sassa H, Hirano H, Nishio T, Koba T. Style-specific self-compatible mutation caused by deletion of the S-RNase gene in Japanese pear (Pyrus serotina) The Plant Journal. 1997;12:223–227. [Google Scholar]
- Sassa H, Hirano H. Identification of a new class of pistil-specific proteins of Petunia inflata that is structurally similar to, but functionally distinct from, the self-incompatibility factor HT. Molecular Genetics and Genomics. 2006;275:97–104. doi: 10.1007/s00438-005-0067-7. [DOI] [PubMed] [Google Scholar]
- Sassa H, Kakui H, Miyamoto M, et al. S locus F-box brothers: multiple and pollen-specific F-box genes with S haplotype-specific polymorphisms in apple and Japanese pear. Genetics. 2007;175:1869–1881. doi: 10.1534/genetics.106.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa H, Kakui H, Minamikawa M. Pollen-expressed F-box gene family and mechanism of S-RNase-based gametophytic self-incompatibility (GSI) in Rosaceae. Sexual Plant Reproduction. 2009;23:39–43. doi: 10.1007/s00497-009-0111-6. [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Hauser K, Lind JL, et al. Molecular characterization of a cDNA sequence encoding the backbone of a style-specific 120 kDa glycoprotein which has features of both extensins and arabinogalactan proteins. Plant Molecular Biology. 1997;35:833–845. doi: 10.1023/a:1005816520060. [DOI] [PubMed] [Google Scholar]
- Sijacic P, Wang X, Skirpan AL, et al. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature. 2004;429:302–305. doi: 10.1038/nature02523. [DOI] [PubMed] [Google Scholar]
- Sims TL, Ordanic M. Identification of a S-ribonuclease binding protein in Petunia hybrida. Plant Molecular Biology. 2001;47:771–783. doi: 10.1023/a:1013639528858. [DOI] [PubMed] [Google Scholar]
- Singh A, Ai Y, Kao T-H. Characterization of ribonuclease activity of three S-allele-associated proteins of Petunia inflata. Plant Physiology. 1991;96:61–68. doi: 10.1104/pp.96.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneveld T, Tobutt KR, Vaughan SP, Robbins TP. Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. The Plant Cell. 2005;17:37–51. doi: 10.1105/tpc.104.026963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Isogai A. Self-incompatibility in plants. Annual Review of Plant Biology. 2005;56:467–489. doi: 10.1146/annurev.arplant.56.032604.144249. [DOI] [PubMed] [Google Scholar]
- Tao R, Iezzoni AF. The S-RNase-based gametophytic self-incompatibility system in Prunus exhibits distinct genetic and molecular features. Scientia Horticulturae. 2010;124:423–433. [Google Scholar]
- Tao R, Watari A, Hanada T, et al. Self-compatible peach (Prunus persica) has mutant versions of the S-haplotypes found in self-incompatible species. Plant Molecular Biology. 2007;63:109–123. doi: 10.1007/s11103-006-9076-0. [DOI] [PubMed] [Google Scholar]
- Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H. Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. The Plant Cell. 2003;15:771–781. [Google Scholar]
- Ushijima K, Yamane H, Watari A, et al. The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. The Plant Journal. 2004;39:573–586. doi: 10.1111/j.1365-313X.2004.02154.x. [DOI] [PubMed] [Google Scholar]
- Vilanova S, Badenes ML, Burgos L, Martinez-Calvo J, Llacer G, Romero C. Self-compatibility of two apricot selections is associated with pollen-part mutations of different nature. Plant Physiology. 2006;142:629–641. doi: 10.1104/pp.106.083865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang X, McCubbin AG, Kao T-H. Genetic mapping and molecular characterization of the self-incompatibility (S) locus in Petunia inflata. Plant Molecular Biology. 2003;53:565–580. doi: 10.1023/B:PLAN.0000019068.00034.09. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tsukamoto T, Yi K-W, et al. Chromosome walking in the Petunia inflata self-incompatibility (S-) locus and gene identification in an 881-kb contig containing S2-RNase. Plant Molecular Biology. 2004;54:727–742. doi: 10.1023/B:PLAN.0000040901.98982.82. [DOI] [PubMed] [Google Scholar]
- Wheeler D, Newbigin E. Expression of 10 S-class SLF-like genes in Nicotiana alata pollen and its implications for understanding the pollen factor of the S-locus. Genetics. 2007;177:2171–2180. doi: 10.1534/genetics.107.076885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler MJ, de Graaf BHJ, Hadjiosif N, et al. Identification of the pollen self-incompatibility determinant in Papaver rhoeas. Nature. 2009;459:992–995. doi: 10.1038/nature08027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler MJ, Vatovec S, Franklin-Tong V. The pollen S-determinant in Papaver: comparisons with known plant receptors and protein ligand partners. Journal of Experimental Botany. 2010;61:2015–2025. doi: 10.1093/jxb/erp383. [DOI] [PubMed] [Google Scholar]
- Wolf DH, Hilt W, Sommer T. Death gives birth to life: the essential role of the ubiquitin-proteasome system in biology. Biochimica et Biophysica Acta. 2004;1695:1–2. doi: 10.1016/j.bbamcr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Woodward JR, Bacic A, Jahnen W, Clarke A. N-Linked glycan chains on S-allele-associated glycoproteins from Nicotiana alata. The Plant Cell. 1989;1:511–514. doi: 10.1105/tpc.1.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H-M, Wang H, Cheung AY. A pollen tube growth stimulatory glycoprotein is deglycosylated by pollen tubes and displays a glycosylation gradient in the flower. Cell. 1995;83:395–403. doi: 10.1016/0092-8674(95)90428-x. [DOI] [PubMed] [Google Scholar]
- Xu B, Mu J, Nevins DL, Grun P, Kao T-H. Cloning and sequencing of cDNAs encoding two self-incompatibility associated proteins in Solanum chacoense. Molecular and General Genetics. 1990;224:341–346. doi: 10.1007/BF00262427. [DOI] [PubMed] [Google Scholar]
- Xue Y, Carpenter R, Dickinson HG, Coen ES. Origin of allelic diversity in Antirrhinum S locus RNases. The Plant Cell. 1996;8:805–814. doi: 10.1105/tpc.8.5.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane H, Tao R. Molecular basis of self-(in)compatibility and current status of S-genotyping in Rosaceous fruit trees. Journal of the Japanese Society of Horticultural Science. 2009;78:137–157. [Google Scholar]
- Zhang Y, Zhao Z, Xue Y. Roles of proteolysis in plant self-incompatibility. Annual Review of Plant Biology. 2009;60:21–42. doi: 10.1146/annurev.arplant.043008.092108. [DOI] [PubMed] [Google Scholar]
- Zisovich AH, Stern RA, Sapir G, Shafir S, Goldway M. The RHV region of S-RNase in the European pear (Pyrus communis) is not required for the determination of specific pollen rejection. Sexual Plant Reproduction. 2004;17:151–156. [Google Scholar]
- Zurek DM, Mou B, Beecher B, McClure B. Exchanging sequence domains between S-RNases from Nicotiana alata disrupts pollen recognition. The Plant Journal. 1997;11:797–808. doi: 10.1046/j.1365-313x.1997.11040797.x. [DOI] [PubMed] [Google Scholar]