Abstract
We present a microfluidic approach to characterizing temperature-dependent biomolecular interactions. Solvated L-arginine vasopressin (AVP) and its immobilized RNA aptamer (spiegelmer) were allowed to achieve equilibrium binding in a microchip at a series of selected temperatures. Unbound AVP were collected and analyzed with matrix-assisted laser desorption∕ionization mass spectrometry (MALDI-MS), yielding melting curves that reveal highly temperature-dependent zones in which affinity binding (36–45 °C) or dissociation (25–33 °C and 50–65 °C) occurs. Additionally, temperature-dependent binding isotherms were constructed; from these, thermodynamic quantities involved in binding were extracted. The results illustrated a strong change in heat capacity of interaction for this system, suggesting a considerable thermodynamic influence controlling vasopressin-spiegelmer interaction.
INTRODUCTION
Molecular binding is fundamental in biological systems and can be particularly sensitive to temperature.1 As temperature may change physiologically due to thermoregulation, understanding the mechanisms governing temperature-dependent biomolecular binding is important for developing potential applications in biosensing,2 drug development,3 and therapeutics.4 For instance, a protein can experience the fluctuating thermal stimulation in vivo, which can alter its native structure and interaction with a drug molecule. Gaining insight into the nature of this stimulation can facilitate the determination of the efficacy of the drug in question. Therapeutically, knowledge of the temperature-dependent relationship between ligand and receptor interactions may facilitate techniques to identify receptor dysfunction that leads to failures in cellular regulation as possible disease pathways. Moreover, temperature-dependent measurements can provide insight into fundamental thermodynamic properties, such as Gibbs free energy, enthalpy, and entropy, of binding systems.
Temperature-dependant receptor-ligand binding is commonly studied with scintillation,5 fluorescence probes,4 and differential calorimetry.6 Scintillation and fluorescence techniques require molecular probes, which are time-consuming to develop, could interfere with binding, and may potentially be temperature-dependant themselves. Differential calorimetry directly measures the thermal power evolved in biomolecular binding events, but is typically limited to large volumes of relatively high-concentration samples. We present an alternative, label-free approach that employs a microfluidic tool7 to characterize temperature-dependant receptor-analyte interactions. The approach involves introducing ligand molecules to surface-immobilized receptor molecules at a series of temperatures. Binding or nonbinding ligand molecules, as appropriate, are collected via microflow control and subsequently detected by matrix-assisted laser desorption∕ionization mass spectrometry (MALDI-MS). This provides an innovative tool for studying temperature-dependent binding, which eliminates the use of fluorescent or radioactive labels and allows for drastically reduced sample consumption. Although MALDI-MS incurs significant upfront investment, it is still widely used over labeled methods due to its excellent detection limits and simplicity in data interpretation. Additionally, the surface-based nature of the approach is highly relevant to biosensing platforms that use surface-immobilized receptors for analyte detection. As an example, we have characterized the temperature-dependent affinity binding of ligand molecules with aptamers, which are single-stranded oligonucleotide receptors isolated synthetically from a large randomized DNA or RNA library.8 In particular, we consider the affinity system consisting of levorotary arginine vasopressin (AVP), an antidiuretic hormonal peptide, and its nuclease resistant RNA derived aptamer termed spiegelmer (sequence: 5′-GGG-GUA-GGG-CUU-GGA-UGG-GUA-GUA-CAC-PEG18-GUG-UGC-GUG-GU-3′).9 Unlike our prior work with aptamer binding systems involving the detection of molecules for biosensing purposes, this investigation focuses on the underlying temperature-dependent behavior of the AVP-spiegelmer system, which was previously unknown. The significance lies in the fact that this approach offers an efficient, robust, and label-free method to investigating such phenomena and is relatively advantageous compared to standard techniques available.
PRINCIPLE AND METHODS
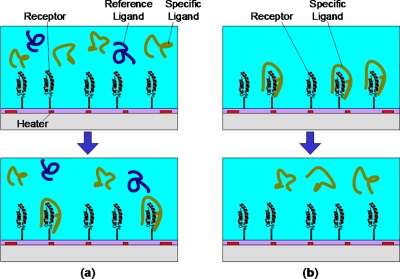
To determine the thermal binding profile, i.e., the amount of ligand bound as a function of temperature, the receptor is initially immobilized onto microfluidic solid surfaces (Figure 1a). At selected temperatures, a sample mixture with specific and non-binding ligands (used as a reference standard to facilitate assessment of binding) is introduced to surface functionalized receptor molecules. After sufficient incubation, a portion of specific ligand molecules will bind with the receptor while the reference ligands remain in solution. Specific ligands that happen to not bind and the reference ligands are subsequently transferred from the microfluidic surface and quantified with MALDI-MS. Resulting mass peaks of the specific and reference ligands are acquired. Taking the mass peak ratio of the specific and reference ligand and plotting against temperature, we acquire the binding profile. This provides a general illustration of the binding efficacy as a function of temperature for the receptor-ligand system.
Figure 1.
Principle of microfluidic characterization of temperature dependent biomolecular binding. (a) Top: A sample of specific and nonspecific reference ligands is introduced to a receptor functionalized solid surface. Bottom: After incubation at a selected temperature (controlled by integrated heaters on the surface), a certain amount of specific ligands bind to the receptor leaving unbound ligands and the reference ligands in solution. (b) Top: Similarly, a sample of specific ligands previously bound to the receptor surface can be released; bottom: following modification of the surface temperature above or below a binding temperature.
Separately, temperature-dependent receptor-ligand equilibrium binding behavior can be elucidated. This is performed by introducing binding ligands to the microfluidic surface in a range of concentrations at a prescribed temperature. Ligands are collected by regulating the surface temperature to release bound molecules into a purified solution, which is then analyzed by MALDI-MS. To reduce the experimental complexity, no reference standard is used. Instead, the AVP signal (Χ) is used for quantification,10 which is defined as the mass spectral peak height over the noise peak-to-peak amplitude (in arbitrary units). At each temperature, experimental data plotting Χ vs. ligand concentration (in Molar concentration), designated as L, can be obtained. The following single-site equilibrium binding model1 is then fitted to extract the equilibrium dissociation constant (KD):
| (1) |
where Χ∞ is the saturation AVP signal (i.e., the value of X as L→0). Then, the following relationship can be used to obtain the Gibbs free energy (ΔG) of dissociation:1
| (2) |
where R and T are the universal gas constant (8.314 J · mol−1 · K−1) and absolute temperature, respectively.
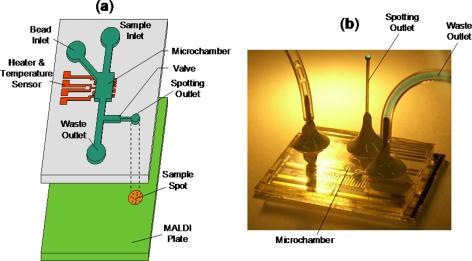
The measurements of temperature-dependent biomolecular binding behavior are conducted using a microfluidic device whose design and fabrication have been described elsewhere.7 Briefly, the device consists of a microchamber containing receptor functionalized microbeads, a microheater, and a temperature sensor. A sample spotting outlet is connected to the microchamber through a passive surface tension-based microflow gate and coupled to a MALDI analysis plate (Figure 2).7 A ligand sample (2 μl) is introduced into the microchamber to bind with surface-bound receptors at selected temperatures maintained via closed-loop control using the microheater and temperature sensor. The samples are allowed to incubate inside the microchamber for approximately 5 min. Purified water is used to then transfer the sample, via the microflow gate, from the microchamber to the spotting outlet, where it is deposited onto the MALDI plate. This step washes the microchamber and prepares it for the following sample. MALDI-MS is finally performed by placing the sample-laden MALDI plate in a mass spectrometer. The microchip is fabricated by standard soft lithography techniques, involving polydimethylsiloxane (PDMS) on glass.7 The above procedure is general for all experiments performed in this work.
Figure 2.
(a) Schematic of integrated microdevice coupled to MALDI analysis plate. (b) Photo of a typical microdevice demonstrating fluid routing through the spotting outlet.
RESULTS AND DISCUSSION
Biomolecular binding at varying temperatures
To demonstrate our approach, we examined the affinity binding of AVP with its specific spiegelmer at varying temperatures. In the investigation, angiotensin (AGT), a nonspecific polypeptide, was included as a standard in AVP solution (at equal concentration to AVP). The AVP mass peak normalized by the AGT mass peak had a value between 0, indicating the tightest possible binding, and 1, indicating non-binding conditions (Figure 1a).
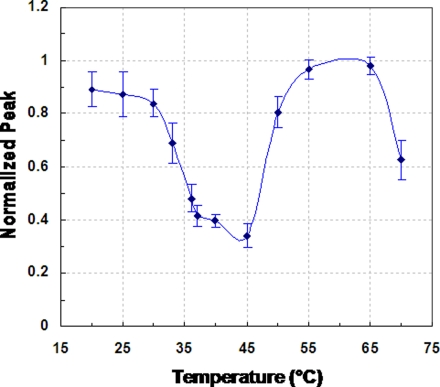
The normalized AVP mass peak as a function of temperature, obtained at an AVP concentration of 100 nM, is shown in Figure 3. (Measurements at other AVP concentrations yielded consistent results.) The error bars in the figure represented the standard deviation calculated from the measurements of three separate experimental trials. These errors reflected contributions from several sources including changes in microbead packing between experiments, and fluctuations in the ionizing electric field. As can be seen, the normalized peak exhibited a minimum value of approximately 0.34 at 45 °C. This indicates relatively strong binding between AVP and spiegelmer in this temperature range, which is consistent with the fact that the spiegelmer was synthetically isolated at physiological temperature (37 °C).9 Beyond 45 °C and up to about 65 °C, a high normalized AVP peak approaching 1 was obtained. This indicates that the AVP-spiegelmer interaction is very weak at these temperatures, resulting in almost complete disruption of binding. Similarly weak binding between AVP and the spiegelmer was also observed at temperatures lower than the physiological range (i.e., approximately from 20 to 30 °C), as reflected by normalized AVP peaks greater than 0.8. These phenomena indicate that the affinity binding strengths can become diminished when the AVP-spiegelmer system is subjected to a temperature that deviates from the temperature at which the synthetic spiegelmer isolation was performed. The deviation can be quite modest and can involve an increase as well as a decrease in temperature. As the temperature changes further, however, the diminishment of affinity can be reversed. This is demonstrated by the relatively low normalized AVP peak, indicating strong binding, near 70 °C. This could be attributed to the formation of an alternative conformational structure of the spiegelmer, leading to its binding with AVP via an affinity epitope distinct from that involved at physiological temperatures (below).
Figure 3.
Temperature dependence of the equilibrium binding between AVP and an affinity spiegelmer. Error bars acquired for each temperature represented the standard deviation calculated from three measurements.
This complex temperature-dependent binding profile was unknown previously. It demonstrates the strong and delicate effects of temperature on AVP-spiegelmer affinity binding. Of fundamental importance, this profile may also find interesting practical applications. For instance, it could be exploited to allow thermally controlled molecular release and device regeneration in biosensing, drug delivery, and biomolecular purification.7 This result also demonstrates the significance of our label-free microfluidic approach, as it would be difficult to attain with conventional approaches.
Assessment of temperature-dependent binding parameters
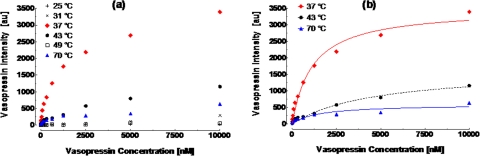
Equilibrium AVP-spiegelmer binding at a given temperature was then obtained as a function of AVP concentration. The AVP signal, X, was used to characterize the binding strength, where a higher value of X indicates stronger interaction. At a given temperature between 25 and 70 °C (Figure 4), we experimented with samples in a concentration range from 4.9 nM to 10 μM. Experimental procedure followed that which was presented in Sec. 2 (above). Consistent with the temperature-dependent binding profile above (Figure 3), strong binding was observed at or near physiological temperature (37 and 43 °C) as well as at 70 °C (Figure 4a), with the dependence of X on concentration well represented by a Langmuir-like relationship.1 Conversely, weak binding was seen at the other temperatures, which is signified by low and erratic X values. Equilibrium binding fits were performed, using GraphPad Prism© nonlinear regression software, for experimental data at 37, 43, and 70 °C only, as these temperatures exhibited measurable equilibrium binding response (Figure 4b). By fitting Eq. 1 to the experimental data in Figure 4b, we can see that the vasopressin signal monotonically increased with concentration, exhibiting a hyperbolic response expected for single-site binding,1 as predicted by Eq. 1. The fit conformed well to the experimental data, with calculated R2 values of 0.98 for both 37 and 43 °C and 0.83 for 70 °C. Consequently, monovalent equilibrium binding theory indeed applies to this system, where the interaction was reversible and bimolecular association and dissociation occur. Moreover, monovalent equilibrium binding implies that the overall affinity of interaction can be described by the equilibrium dissociation constant (KD) from Eq. 1. For instance, systems possessing low KD values generally exhibit tightly binding interaction between the receptor and the ligand, while the opposite holds true for high KD values.
Figure 4.
(a) Temperature dependent equilibrium binding of spiegelmer and AVP. (b) Equilibrium binding fit to Eq. 1 for strong binding temperature data sets (i.e., 37, 43, and 70 °C) using a nonlinear least-squares regression.
The KD, thus, obtained from fitting Eq. 1 to experimental data shows a strong dependence on temperature. At 43 and 70 °C, KD is determined to be 3.91 and 2.15 μM, respectively, which correspond to relatively strong molecular interaction between AVP and spiegelmer. At 37 °C, the dissociation constant exhibits a minimum at 1.14 μM, representing the maximum binding strength condition. This value of KD at 37 °C is about 100 times that reported from solution-based measurements,9 which could be attributed to the effects of the spiegelmer being tethered to the surface.11, 12 For example, inefficient receptor orientation and steric hindrance might have affected the affinity of the interaction. Based on knowledge of the equilibrium dissociation constant, the Gibbs free energy (ΔG) can be calculated from Eq. 2. From 37 to 43 °C, ΔG decreases from 35.3 to 32.7 kJ · mol−1, which reflects a decrease in thermodynamic stability with increasing temperature within this range. This is expected as tightly forming complexes are stable and generally more thermodynamically favorable.13 These results suggest that local folding events occur in the aptamer secondary structure to form a well-suited binding pocket for AVP, otherwise known as induced-fitting.14 Interestingly, the Gibbs free energy increases to 37.2 kJ · mol−1 at 70 °C, which implies an additional thermodynamically stable complex configuration. Although the higher ΔG at 70 °C would suggest a more favorable binding condition than at 37 °C, we recognize that this is only due to increased kinetic energy (i.e., elevated temperature contribution) input rather than actually tighter binding interaction.
Due to the tendency for typical molecular binding interactions to dissociate at such elevated temperatures, a strong AVP-spiegelmer binding at 70 °C was not expected initially (see Figures 34). At 70 °C, KD and ΔG were similar, respectively,2.15 μM and 37.2 kJ · mol−1, to their values at lower temperatures, such as 37 °C (KD and ΔG: 1.14 μM and 35.3 kJ · mol−1). This indicates a second conformational structure where dissociation is unfavorable, given the positive ΔG. At such elevated temperatures, we conjecture that binding likely involves thermodynamically driven ligand-dependent rearrangement of the spiegelmer’s three dimensional structure, as well as increased physical contact between AVP and spiegelmer due to higher kinetic energy. The later phenomenon likely contributes only a secondary role in AVP-spiegelmer binding at 70 °C, since dissociation still occurs at other elevated temperature setpoints. Resolution of this phenomenon calls for detailed structural studies (e.g., using circular dichroism experiments) and is a potential topic for future research.
CONCLUSION
To conclude, we characterized temperature-dependent biomolecular binding utilizing a microfabricated chip with biomolecule functionalized surfaces coupled to MALDI-MS. As a proof-of-concept, we observed the thermally dependent binding properties of vasopressin with its spiegelmer. A melting curve was obtained, revealing temperature zones of either strong (36–45 °C) or weak interaction (25–33 °C and 50–65 °C). Of interest was measured binding at 70 °C, which possibly is due to formation of a secondary binding structure in the spiegelmer sequence. Moreover, calculated values for ΔG suggested an induced-fit model possibly governs this interaction. These results demonstrate that our method can be used as a powerful tool for label-free characterization of temperature-dependent binding, which is important for practical applications such as biosensors, drug development, and biomolecular purification.
ACKNOWLEDGMENTS
We gratefully acknowledge financial support from the National Science Foundation (CBET-0693274, CBET 0854030, and EIA-0324845), the National Institutes of Health (RR025816-02 and CA147925-01), and the Alternatives Research and Development Foundation.
References
- Lauffenburger D. A. and Linderman J. J., Receptors: Models for Binding, Trafficking, and Signaling, Reprint, illustrated edition (Oxford University Press, New York, 1996). [Google Scholar]
- Wang J., Nucleic Acids Res. 28, 3011 (2000). 10.1093/nar/28.16.3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohlke H. and Klebe G., Angew. Chem., Int. Ed. 41, 2645 (2002). [DOI] [PubMed] [Google Scholar]
- Sinha S. S., Mitra R. K., and Pal S. K., J. Phys. Chem. B 112, 4884 (2008). 10.1021/jp709809b [DOI] [PubMed] [Google Scholar]
- Cooper M. A., Anal. Bioanal. Chem. 377, 834 (2003). 10.1007/s00216-003-2111-y [DOI] [PubMed] [Google Scholar]
- Pico G. A., Int. J. Biol. Macromol. 20, 63 (1997). 10.1016/S0141-8130(96)01153-1 [DOI] [PubMed] [Google Scholar]
- Nguyen T., Qiu C., Pei R., Stojanovic M., Ju J., and Lin Q., J. Microelectromech. Syst. 18, 1198 (2009). 10.1109/JMEMS.2009.2034392 [DOI] [Google Scholar]
- Mairal T., Ozalp V. C., Sanchez P. L., Mir M., Katakis I., and O’Sullivan C. K., Anal. Bioanal. Chem. 390, 989 (2008). 10.1007/s00216-007-1346-4 [DOI] [PubMed] [Google Scholar]
- Purschke W. G., Eulberg D., Buchner K., Vonhoff S., and Klussmann S., Proc. Natl. Acad. Sci. U.S.A. 103, 5173 (2006). 10.1073/pnas.0509663103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkovic M., Schiller J., Muller J., Muller M., Arnold K., and Arnhold J., Analyst 126, 1042 (2001). 10.1039/b101921j [DOI] [PubMed] [Google Scholar]
- Glaser R. W., Anal. Biochem. 213, 152 (1993). 10.1006/abio.1993.1399 [DOI] [PubMed] [Google Scholar]
- Nygren H., Werthen M., and Stenberg M., J. Immunol. Methods 101, 63 (1987). 10.1016/0022-1759(87)90217-1 [DOI] [PubMed] [Google Scholar]
- Lin P. H., Yen S. L., Lin M. S., Chang Y., Louis S. R., Higuchi A., and Chen W. Y., J. Phys. Chem. B 112, 6665 (2008). 10.1021/jp8000866 [DOI] [PubMed] [Google Scholar]
- Gilbert S. D., Montange R. K., Stoddard C. D., and Batey R. T., Cold Spring Harbor Symp. Quant. Biol. 71, 259 (2006). 10.1101/sqb.2006.71.015 [DOI] [PubMed] [Google Scholar]