Abstract
Early onset hereditary motor and sensory neuropathies are rare disorders encompassing congenital hypomyelinating neuropathy with disease onset in the direct post-natal period and Dejerine–Sottas neuropathy starting in infancy. The clinical spectrum, however, reaches beyond the boundaries of these two historically defined disease entities. De novo dominant mutations in PMP22, MPZ and EGR2 are known to be a typical cause of very early onset hereditary neuropathies. In addition, mutations in several other dominant and recessive genes for Charcot–Marie–Tooth disease may lead to similar phenotypes. To estimate mutation frequencies and to gain detailed insights into the genetic and phenotypic heterogeneity of early onset hereditary neuropathies, we selected a heterogeneous cohort of 77 unrelated patients who presented with symptoms of peripheral neuropathy within the first year of life. The majority of these patients were isolated in their family. We performed systematic mutation screening by means of direct sequencing of the coding regions of 11 genes: MFN2, PMP22, MPZ, EGR2, GDAP1, NEFL, FGD4, MTMR2, PRX, SBF2 and SH3TC2. In addition, screening for the Charcot–Marie–Tooth type 1A duplication on chromosome 17p11.2-12 was performed. In 35 patients (45%), mutations were identified. Mutations in MPZ, PMP22 and EGR2 were found most frequently in patients presenting with early hypotonia and breathing difficulties. The recessive genes FGD4, PRX, MTMR2, SBF2, SH3TC2 and GDAP1 were mutated in patients presenting with early foot deformities and variable delay in motor milestones after an uneventful neonatal period. Several patients displaying congenital foot deformities but an otherwise normal early development carried the Charcot–Marie–Tooth type 1A duplication. This study clearly illustrates the genetic heterogeneity underlying hereditary neuropathies with infantile onset.
Keywords: early onset hereditary neuropathies, congenital hypomyelinating neuropathy, Dejerine–Sottas neuropathy, genotype–phenotype correlations, Charcot–Marie–Tooth disease
Introduction
Hereditary motor and sensory neuropathies with onset in infancy are rare disorders that were first described by Dejerine and Sottas, 1893 in the late 19th century as a separate disease, distinct from the more commonly occurring Charcot–Marie–Tooth neuropathy (Gabreels-Festen, 2002; Plante-Bordeneuve and Said, 2002). Today, we know that these early onset hereditary neuropathies are not a single entity but rather represent a broad clinical and genetic spectrum of disorders that is still incompletely understood (Ryan and Ouvrier, 2005).
Presenting symptoms in the neonate can be severe and often include hypotonia and respiratory insufficiency (Phillips et al., 1999; Smit et al., 2008). In other patients, clinical presentation consists of early foot deformities, delay in early motor milestones, distal sensory loss and weakness with progressive gait difficulties (Plante-Bordeneuve and Said, 2002; Wilmshurst et al., 2003; Burns et al., 2009). These early and severely affected patients are often isolated in their family, obscuring the inheritance pattern; either de novo mutations in dominant genes or recessive alleles inherited from unaffected parents are at play (Gabreels-Festen, 2002; Plante-Bordeneuve and Said, 2002; Parman et al., 2004).
Two historically described forms have remained in current literature, although they are now known to represent elements of the broader disease spectrum of demyelinating neuropathies rather than distinct entities (Gabreels-Festen, 2002, 2005; Scherer, 2006). These are congenital hypomyelinating neuropathy with disease onset in the direct post-natal period and Dejerine–Sottas neuropathy, starting in infancy. Typically, de novo mutations in myelin protein zero (MPZ), peripheral myelin protein 22 (PMP22) and early growth response 2 (EGR2) are described as the cause of congenital hypomyelinating neuropathy and Dejerine–Sottas neuropathy. Clinical and genetic variability of hereditary motor and sensory neuropathy reaches beyond these boundaries however.
The currently understood genetic spectrum of early onset hereditary motor and sensory neuropathy is very heterogeneous, including both dominant and recessive mutations in multiple genes (Ryan and Ouvrier, 2005). As already mentioned, de novo and occasionally also inherited dominant mutations in EGR2, MPZ and PMP22 may occur (Plante-Bordeneuve and Said, 2002; Smit et al., 2008). Recessive mutations in myotubularin-related protein 2 (MTMR2), set-binding factor 2 (SBF2) and FYVE, RhoGEF, and PH domain-containing protein 4 (FGD4) are typically found in patients with redundant myelin loops or myelin outfoldings on sural nerve biopsies (Bolino et al., 2000; Nelis et al., 2002a; Azzedine et al., 2003; Fabrizi et al., 2009). Recessive mutations in periaxin (PRX), SH3 domain and tetratricopeptide repeat domain 2 (SH3TC2) and ganglioside-induced differentiation-associated protein 1 (GDAP1) are also associated with very early disease onset (Nelis et al., 2002b; Takashima et al., 2002; Senderek et al., 2003). Mutations in mitofusin 2 (MFN2), GDAP1, EGR2 and neurofilament light chain (NEFL) can behave both as dominant and recessive traits (Timmerman et al., 1999; Nicholson et al., 2008; Yum et al., 2009; Zimon et al., 2011). Since many patients still remain without a molecular diagnosis, the above-mentioned list of genes is by no means exhaustive.
The majority of the early onset hereditary neuropathies represent demyelinating phenotypes with sometimes profoundly slowed motor nerve conduction velocities, indicative of severe demyelination or even amyelination. Noteworthy examples are PRX and FGD4 mutation carriers (Takashima et al., 2002; Fabrizi et al., 2009) and also congenital onset forms with mutations in MPZ, PMP22 and EGR2 (Gabreels-Festen, 2002). Nerve conduction studies in MTMR2, SBF2, SH3TC2 and NEFL mutation carriers typically also show slowed nerve conduction velocity in the demyelinating range (Nelis et al., 2002a; Azzedine et al., 2003; Senderek et al., 2003; Parman et al., 2004; Yum et al., 2009). In addition to the demyelinating phenotypes, severe early onset axonal neuropathies can be due to MFN2 or GDAP1 mutations (Nelis et al., 2002b; Nicholson et al., 2008; Feely et al., 2011).
In very young children presenting with neuropathy in the absence of a remarkable familial history, acquired causes of peripheral neuropathy such as inflammatory neuropathies, toxic causes and nutritional deficiencies have to be considered. The disease history and electrophysiology may provide hints in that direction. Acquired causes, however, are proportionally less common than hereditary causes (Connolly, 2001; Wilmshurst et al., 2003). More often, peripheral neuropathies in very young children may be seen as part of a syndromic (metabolic) hereditary disorder. Although other clinical features than those of the peripheral neuropathy usually dominate the phenotype, severe demyelinating neuropathy leading to motor development delay may be observed as the initial and only clinical sign in these disorders (Wilmshurst et al., 2003; Ryan and Ouvrier, 2005; Scherer, 2006; Landrieu et al., 2011).
Detailed electrophysiological testing is essential to confirm the clinical diagnosis and further subdivide patients into demyelinating and axonal phenotypes. Performing such examinations is challenging in very young children, and normal values vary significantly in the first 5 years of life due to ongoing maturation of the PNS and limb growth. For this reason, results of nerve conduction studies must be interpreted with utmost caution (Garcia et al., 2000; Wilmshurst et al., 2003).
In general, the need for diagnostic sural nerve biopsies has drastically decreased due to the extensive knowledge of the various genetic causes of hereditary neuropathies and the increasing availability of molecular testing. However, in the specific case of early onset neuropathies, nerve biopsies may still be diagnostically meaningful in selected patients (Wilmshurst et al., 2003).
Hereditary neuropathies are most often of mixed motor and sensory type, both in adults and children. Some forms of hereditary motor neuropathy and hereditary sensory and autonomic neuropathy can start in infancy as well (Dierick et al., 2008; Rotthier et al., 2009). These subforms can be distinguished through careful analysis of clinical presentation, electrophysiology and if possible neuropathology. As an important differential diagnosis, Infantile spinal muscular atrophy with respiratory distress type 1 (SMARD1) (spinal muscular atrophy with repiratory distress type 1) due to mutations in immunoglobulin mu binding protein 2 (IGHMBP2) should be ruled out because these patients may initially be diagnosed as early onset hereditary motor sensory neuropathy due to very low motor nerve conduction velocities in the upper and lower limbs (Pitt et al., 2003). Additional assessment of at least one sensory nerve that is usually normal in these patients may help to distinguish this disorder from typical early onset hereditary motor sensory neuropathy 1.
Since the first description by Dejerine and Sottas, many detailed studies have been published on clinical, electrophysiological and neuropathological aspects of early onset hereditary neuropathies (Dejerine and Sottas, 1893). Molecular genetic studies so far have not focused extensively on systematic genetic screenings but rather on single patient reports, smaller patient series or gene identification studies. These studies offer only a partial view on the genetic variability and the correlation with the clinical phenotypes of childhood neuropathies.
Early onset neuropathies pose a particular diagnostic challenge both to the treating child neurologist confronted with often severely affected patients and to geneticists seeking to provide molecular diagnosis and appropriate genetic counselling.
In the current study, we report on the findings from the systematic screening of 11 relevant genes in a cohort of 77 unrelated patients with presumed hereditary motor and sensory neuropathy with onset in the first year of life. Several of these patients have been reported before, either in smaller case reports or as part of gene identification studies. Patients presenting with pure hereditary motor neuropathy or hereditary sensory and autonomic neuropathy have been studied previously and were therefore not included (Dierick et al., 2008; Rotthier et al., 2009). These genotype–phenotype correlations provide important insights that are of particular relevance in the context of molecular diagnostics of rare hereditary neuropathies with an early disease onset.
Patients and methods
Patient cohort
In this study, 77 unrelated index patients were included who presented with symptoms of motor and sensory neuropathy within the first year of life. In order to grasp the full spectrum of phenotypes, we did not apply exclusion criteria regarding electrophysiology, inheritance pattern, associated clinical features or neuropathology. Presenting symptoms varied widely ranging from early neonatal hypotonia with feeding and breathing difficulties over progressive delay of motor milestones to early foot deformities in the first 12 months of life. A diagnosis of demyelinating neuropathy was made for 45 patients and axonal neuropathy for 15 patients. For the remaining 17 patients, no clear distinction could be made. Detailed electrophysiology was available for 50 patients. For a total of 29 patients, neuropathological examination of a sural nerve biopsy was performed. In total, 61 patients were isolated in their family, in 10 patients a dominantly inherited phenotype was seen in the family and in six patients familial history was suggestive of a recessive trait. For 21 out of the 77 index patients consanguinity was noted; 18 of these were isolated in their family.
Patients were referred for genetic diagnosis from various European countries, the Middle East and the USA and were clinically evaluated by neuro-paediatricians with expertise in the field of rare neuromuscular disorders. Parents or legal representatives of all patients signed an informed consent form prior to enrolment. The local institutional review boards approved the study.
Mutation screening
Genomic DNA was extracted by means of standard protocols from blood samples obtained from patients, healthy unrelated controls and, if available, family members. The coding regions and exon–intron boundaries of 11 genes (MFN2, PMP22, MPZ, EGR2, GDAP1, NEFL, FGD4, MTMR2, PRX, SBF2 and SH3TC2) were polymerase chain reaction amplified using primer oligonucleotides designed with Primer3 (Rozen and Skaletsky, 2000) Primer sequences and polymerase chain reaction conditions are available on request.
Polymerase chain reaction products were purified with the Exonuclease I-Shrimp Alkaline Phosphatase enzymes (USB). Mutation screening was performed by bidirectional sequencing using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Fragments were electrophoretically separated on an ABI3730xl DNA Analyser (Applied Biosystems). Sequence analysis was performed with the SeqMan™II (DNASTAR Inc.) programme. The nucleotide numbering was based on the published online protein and messenger RNA sequence of the respective genes (www.ncbi.nlm.nih.gov). The conventions of the Human Genome Variation Society nomenclature (http://www.hgvs.org/mutnomen) were used for the description of the identified mutations. Sequence variants were confirmed by an independent polymerase chain reaction and sequencing of the original or new stock DNA samples. Segregation of the mutations was performed when DNA samples of family members were available. For newly identified mutations, a minimum of 360 control chromosomes were screened.
Copy number variation studies
The Multiplex Amplicon Quantification technique was applied (Sleegers et al., 2006) in order to investigate the presence of pathogenic copy number variations on the second allele in patients with heterozygous variations in MTMR2 and PRX. The same technique was used to screen for the CMT1A duplication/HNPP deletion in the 17p11.2-12 region.
Multiplex amplicon quantification consists of a multiplex polymerase chain reaction amplification of fluorescently labelled target and control amplicons, followed by fragment analysis on an ABI3730 DNA Analyser (Applied Biosystems). In this assay, target amplicons are located in and around the exons of the corresponding genes and eight control amplicons are located at randomly selected genomic positions outside the corresponding target regions and other known copy number variations. These amplicons were polymerase chain reaction amplified in a single reaction containing 20 ng of genomic DNA. Peak areas of the target amplicons were normalized to these of the control amplicons. Comparison of normalized peak areas between patients and reference individuals resulted in a dosage quotient for each target amplicon, calculated by the Multiplex Amplicon Quantification software package (www.multiplicon.com). Dosage quotient values <0.75 were considered indicative for a deletion.
To further confirm an identified partial duplication of PMP22, the commercially available Multiplex Ligation-dependant Probe Amplification kit (P033-CMT1) was used. The same technique was used to check for additional partial deletions of MFN2 (P143-MFN2-MPZ). Manufacturer's instructions were applied (MRC).
Results
Genetic findings
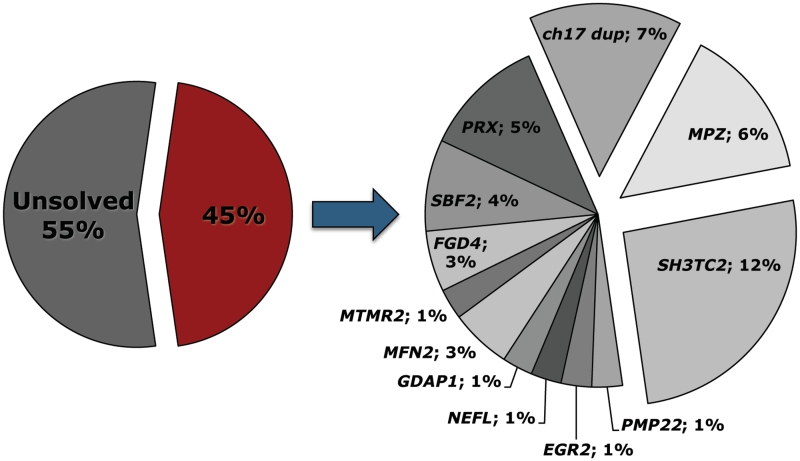
Extensive mutation screening of 11 genes in a large cohort of 77 unrelated patients with neuropathy with onset in the first year of life revealed pathogenic sequence variants in 35 patients, representing 45% of the total cohort. Sequence variants were found in all screened genes. The genetic findings are presented per gene of interest in Table 1 and mutation frequencies per gene are shown in Fig. 1.
Table 1.
Overview of genetic findings in 35 patients with hereditary neuropathy with onset in the first year of life
| Gene | Mutation | Individual | Inheritance | Segregation | Ethnicity | Consanguinity | Reference/additional remark |
|---|---|---|---|---|---|---|---|
| CMT1A | Duplication | CMT-AII2_5.1 | Autosomal dominant | + | Belgian | − | Raeymaekers et al. (1991) |
| PN-1745.1 | Isolated case | − | Belgian | − | |||
| PN-491.1 | Autosomal dominant | + | Austrian | − | |||
| PN-908.3 | Autosomal dominant | + | Spanish | − | |||
| PMP22 | Partial duplication exon 4 | CMT-127.04 | Isolated case | − | European | − | |
| EGR2 | Arg359Trp | PN-27.1 | Isolated case, de novo | + | Ashkenazy Jewish | − | Timmerman et al. (1999) |
| MFN2 | Arg104Trp | CMT-797.01 | Isolated case, de novo | + | Greek | − | |
| Arg400Pro | CMT-756.01 | Isolated case, | + | Italian/Irish | − | Parental mosaicism in asymptomatic mother | |
| MPZ | His81Arg | CMT-65.07 | Autosomal dominant | + | British | − | Sorour et al. (1997). Two additional family members born with clubfeet |
| Ala209GlufsX24 | PN-1540.1 | Isolated case, de novo | + | Belgian | − | ||
| Asp134Glu | PN-506.1 | Autosomal dominant | + | Belgian | − | Nelis et al. (1994). Two additional family members with delayed motor milestones | |
| Arg98Cys | PN-752.1 | Isolated case | − | Belgian | − | ||
| Arg98Cys | PN-966.4 | De novo | + | Austrian | − | Similarly affected identical twins | |
| NEFL | Asn98Ser | PN-1385.1 | Isolated case | − | Finnish | − | |
| PMP22 | Ser72Leu | PN-750.3 | Isolated case, de novo | + | Belgian | − | Ceuterick-de Groote et al. (2001) |
| FGD4 | Tyr587fsX14 hmz | CMT-190.01 | Isolated case | + | Italian | + | Fabrizi et al,(2009) |
| Arg224stop hmz | CMT-230.01 | Isolated case | + | Turkish | + | Stendel et al. (2007) | |
| GDAP1 | Ser194stop hmz | PN-860.3 | Autosomal recessive | + | Moroccan | + | Nelis et al. (2002) |
| MTMR2 | His416ArgfsX6 hmz | CMT-201.01 | Isolated case | − | Turkish | − | |
| PRX | Gly1258ThrfsX124 hmz | PN-1699.1 | Isolated case | − | Druze (Israel) | + | |
| Ala244ArgfsX69 hmz | PN-2175.1 | Isolated case | + | Moroccan | + | ||
| Cys715stop hmz | PN-44.1 | Autosomal recessive | + | Belgian | + | Takashima et al. (2002) | |
| Leu83CysfsX14 hmz | PN-761.3 | Isolated case | + | Maghreb | + | Takashima et al. (2002) | |
| SBF2 | Gln513Stop hmz | CMT-194.01 | Isolated case | − | Turkish | + | |
| Tyr1594Stop hmz | CMT-220.01 | Isolated case | + | Turkish | + | ||
| Leu1316PhefsX9 + Arg1433Ser | PN-1101.2 | Isolated case | + | Polish | − | ||
| SH3TC2 | Arg954Stop + Ala878Asp | CMT-191.01 | Isolated case | + | Italian | − | |
| Arg954Stop hmz | CMT-192.01 | Isolated case | + | Italian | − | ||
| Arg529Gln hmz | CMT-133.01 | Isolated case | + | Turkish | + | Senderek et al. (2003) | |
| IVS5-2A>G hmz | CMT-189.V.5 | Autosomal recessive | + | Italian | + | Senderek et al. (2003) | |
| Glu657Lys hmz | CMT-234.01 | Isolated case | + | Turkish | + | Senderek et al. (2003) | |
| Arg583AlafsX586 hmz | CMT-235.01 | Isolated case | + | Turkish | + | Senderek et al. (2003) | |
| Leu832HisfsX839 hmz | PN-1289.1 | Isolated case | − | Iranian | + | Senderek et al. (2003) | |
| Arg954stop hmz | PN-1321.1 | Isolated case | + | Belgian | − | ||
| Arg954stop hmz | PN-754.3 | Isolated case | + | Dutch | − | ||
Mutations are heterozygous unless stated otherwise, novel mutations are shown in bold, hmz, homozygous.
Figure 1.
Gene distribution among 35 patients with pathogenic mutations out of a cohort of 77 unrelated index patients.
Some patients have been reported previously as part of gene identification studies or smaller case series; the relevant references are provided in Table 1 (Raeymaekers et al., 1991; Nelis et al., 1994, 2002b; Sorour et al., 1997; Timmerman et al., 1999; Ceuterick-de Groote et al., 2001; Takashima et al., 2002; Senderek et al., 2003; Stendel et al., 2007; Fabrizi et al., 2009).
A total of five patients carried a de novo heterozygous mutation in MPZ, EGR2, PMP22 or MFN2. Four isolated patients were heterozygous for mutations in NEFL or MPZ, the CMT1A duplication or a partial PMP22 duplication; for these, however, the de novo character could not be confirmed due to lack of DNA samples from additional family members. Mutations in MPZ and the CMT1A duplication were transmitted as a dominant trait in five patients; 20 patients inherited recessive mutations in GDAP1, MTMR2, SBF2, FGD4, PRX or SH3TC2 from their unaffected parents. In one isolated patient (CMT-756.01), a novel heterozygous Arg400Pro mutation in MFN2 was identified. Low levels of the same mutant allele in the electropherogram of the unaffected mother suggest a parental mosaicism for this mutation. This mutation targets a highly conserved amino acid and is probably damaging for the protein as predicted by a high Polyphen2 score (Adzhubei et al., 2010).
Additional Multiplex Amplicon Quantification and Multiplex Ligation-dependant Probe Amplification assays in the patients with heterozygous sequence variants in PRX, MTMR2 and MFN2 did not provide evidence for partial intragenic deletions.
Clinical findings
Detailed clinical and electrophysiological findings in patients carrying pathogenic mutations are presented in Tables 2 and 3. The data from the nerve conduction studies have to be interpreted with caution since the age at which electrophysiological testing was performed varied widely among patients making the application of one standardized set of normal values impossible (Garcia et al., 2000).
Table 2.
Overview of clinical findings in 35 patients with hereditary neuropathy with onset in the first year of life
| Gene | Individual | Onset age | Symptoms at onset | Age at last examination (yrs) | Motor delay | Respiratory insufficiency | Foot deformities | Walking | Nerve pathology | Additional features |
|---|---|---|---|---|---|---|---|---|---|---|
| CMT1A dup | CMT-AII2_5.1 | Congenital | Foot deformities | 65 | Absent | Absent | Present | Never walked normally | Early scoliosis, bilateral deafness | |
| PN-1745.1 | Congenital | Club feet | 2 | Present | Absent | Present | Gait difficulties, walked at 22 m | Foot surgery at 6 m | ||
| PN-491.1 | Congenital | Foot deformities | 4 | Absent | Absent | Present | Progressive gait difficulties (4 y) | |||
| PN-908.3 | Congenital | Foot deformities | 19 | Absent | Absent | Present | Unsteady gait with steppage, unaided | Focally folded myelin, rare OBF (8 y) | Multiple foot surgery, younger brother similar phenotype, father mildly affected | |
| PMP22 exon 4 duplication | CMT-127.04 | Congenital | Hypotonia | 13 | Present | Never walked independently, wheelchair bound by age 7 y | Hypomyelination, classic and basal lamina OBF and very short myelinated internodes | Severe scoliosis requiring surgery, Parents and two sibs asymptomatic and normal nerve conduction velocities | ||
| EGR2 | PN-27.1 | Congenital | Hypotonia with breathing difficulties | 13 | Present | Present | Disturbed, walked unaided at 3 y | Severe fibre loss, demyelination and focally folded myelin | Died at 16 y due to pneumonia | |
| MFN2 | CMT-797.01 | <1 y | Developmental delay | 5 | Present | Absent | Absent | Walked without assistance >39 m, AFOs | ||
| CMT-756.01 | <1 y | Delayed motor milestones | 10 | Present | Absent | Present | Delayed walking, pronounced steppage gait | Brisk tendon reflexes, no clinical sensory loss | ||
| MPZ | CMT-65.07 | Congenital | Clubfeet | 28 | Absent | Absent | Present | Progressive walking difficulties in primary school | Sensory ataxia, no papillary accommodation reflex | |
| PN-1540.1 | Congenital | Hypotonia, breathing difficulties | 27 | Present | Present | Present | Never walked unsupported, wheelchair bound since age 6 y | Hypomyelination | Severe scoliosis requiring surgery | |
| PN-506.1 | <1 y | Delayed motor milestones | 16 | Present | Absent | Present | Walked with support at 21 m, wheelchair bound since age 15 y | Severe demyelination | ||
| PN-752.1 | <1 y | Delayed motor milestones | 38 | Present | Absent | Present | Started walking at 33 m | Demyelination | Scoliosis, nystagmus, sensory ataxia | |
| PN-966.4 | <1 y | Delayed motor milestones | 7 | Present | Absent | Absent | Started walking at 30 m, never walked normally | Proximal weakness, identical twin with similar phenotype | ||
| NEFL | PN-1385.1 | 3-4 m | Hypotonia, growth retardation | 4 | Present | Absent | Absent | Started walking at 25 m | ||
| PMP22 | PN-750.3 | Congenital | Hypotonia | 4 | Present | Absent | Present | Walks with aid at 2 y, never walked independently | Hypomyelinating neuropathy on skin biopsy | |
| FGD4 | CMT-190.01 | <1 y | Delayed motor milestones | 21 | Present | Absent | Present | Walked with aid at 17 m, unsteady gait with steppage ever since | Hypertrophic demyelinating neuropathy with focally folded myelin | Scoliosis |
| CMT-230.01 | <1 y | Delayed motor milestones | 30 | Present | Absent | Present | Delayed walking | |||
| GDAP1 | PN-860.3 | 2 m | Foot deformity | 7 | Present | Absent | Present | Started walking at 16 m with limp, progressive deterioration | Large myelinated axons Absent, regenerating clusters, rare OBF | |
| MTMR2 | CMT-201.01 | 8 m | Delayed motor milestones | 8 | Present | Absent | Present | Delayed and disturbed from the onset | Fibre density↓, focally folded myelin, OBF | |
| PRX | PN-1699.1 | Congenital | Hypotonia | 11 | Present | Absent | Present | Delayed, only walked with aid at the age of 4 y, walks unsteady at 11 y | Moderate proximal weakness, 47XXX karyotype | |
| PN-2175.1 | <1 y | Delayed motor milestones | 4 | Present | Absent | Absent | Delayed, walked with assistance at 16 m, without assistance at 30 m | |||
| PN-44.1 | <1 y | Delayed motor milestones | 50 | Present | Absent | ? | Delayed | Loss of myelinated axons, OBF, myelin outfoldings, no septate-like junctions in paranodal myelin | Hearing loss, scoliosis | |
| PN-761.3 | <1 y | Delayed motor milestones | 6 | Present | Absent | ? | Delayed, stood with support at 4 y, walked at 5 y | Severe loss large myelinated fibres, basal lamina OBF | ||
| SBF2 | CMT-194.01 | <1 y | Delayed motor milestones | 15 | Present | Absent | Present | Delayed, walked unsupported at 2, 5–3 y, frequent falls | Loss of myelinated axons, small OBF, focally folded myelin | Hypophonia |
| CMT-220.01 | <1 y | Delayed motor milestones | 8 | Present | Absent | Present | Delayed, walked unsupported at 2, 5–3 y | |||
| PN-1101.1 | <1 y | Delayed motor milestones | 10 | Present | Absent | Present | Delayed, walked with support at 2 y | Focally folded and uncompacted myelin, OBF | Anisocoria, facial weakness | |
| SH3TC2 | CMT-191.01 | <1 y | Delayed motor milestones | 10 | Present | Absent | Present | Delayed walking, unsteady gait, frequent falls, steppage, walks with AFO | Hypertrophic de-remyelinating neuropathy with focally folded myelin and basal-lamina OBF | Severe scoliosis since age 10 y, requiring surgery at age 16 y, short stature |
| CMT-192.01 | <1 y | Hypotonia | 12 | Absent | Present | Present | Started walking at 13 m but with frequent falls, wheelchair bound at 12 y | Hypertrophic de-remyelinating neuropathy with focally folded myelin and basal-lamina OBF | Severe scoliosis since age 2 y requiring surgery at age 12 y, bilateral facial weakness | |
| CMT-133.01 | <1 y | Delayed motor milestones | 8 | Present | Absent | Absent | Walked at 30 m | Branching of SC on nerve biopsy | ||
| CMT-189.V.5 | <1 y | Delayed motor milestones | 15 | Present | Present | Absent | Walked at 24 m, progressive worsening, wheelchair bound at 15 y | Pronounced scoliosis at 7 y, progressive over time with respiratory difficulties at 30 y | ||
| CMT-234.01 | <1 y | Delayed motor milestones | 25 | Present | Absent | Present | Delayed walking, walks with aid | Nystagmus | ||
| CMT-235.01 | <1 y | Delayed motor milestones | 19 | Present | Absent | Present | Walked with aid at 18 m | OBF, SC branching | ||
| PN-1289.1 | <1 y | Delayed motor milestones | 17 | Present | Present | Started walking at 24 m | Congenital nystagmus, scoliosis requiring surgery | |||
| PN-1321.1 | <1 y | Delayed motor milestones | 42 | Present | Absent | Present | Delayed walking at 26 m, progressive over time, wheelchair dependency in adulthood | Hypertrophic demyelinating neuropathy with basal lamina OBF | Pronounced scoliosis with short stature, sensorineural hearing loss | |
| PN-754.3 | <1 y | Delayed motor milestones | 10 | Present | Absent | Present | Started walking at 20 m | Demyelinating neuropathy | Pronounced scoliosis |
Foot deformities include pes cavus, pes planus, hammer toes and club feet.
AFO = ankle foot orthosis; m = months; OBF = onion bulb formation; SC = Schwann cell; y = years.
Table 3.
Overview of nerve conduction studies in 35 patients with hereditary neuropathy with onset in the first year of life
| Gene | Patient | Age | Median motor |
Ulnar motor |
Peroneal motor |
Tibial motor |
Median sensory |
Ulnar sensory |
Sural sensory |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amp | CV | Amp | CV | Amp | CV | Amp | CV | Amp | CV | Amp | CV | Amp | CV | |||
| CMT1A dup | CMT-AII2_5.1 | – | – | 17.0 | – | 20.0 | – | – | – | – | – | 23.0 | – | – | – | – |
| PN-1745.1 | 20 m | – | – | – | – | 2.0 | 18.0 | 4.6 | 20.0 | – | – | – | – | – | – | |
| PN-491.1 | 4 y | – | – | – | – | 1.5 | 17.5 | – | – | – | – | – | – | – | – | |
| PN-908.3 | 19 y | 5.5 | 19.3 | – | – | 0.05 | 12.3 | – | – | – | – | – | – | A | A | |
| PMP22 exon 4 duplication | CMT-127.04 | 13 y | A | A | A | A | A | A | A | A | A | A | A | A | – | – |
| EGR2 | PN-27.1 | 6 y | 1.5 | 8.0 | – | – | 0.4 | 8.0 | – | – | A | A | – | A | A | |
| MFN2 | CMT-797.01 | – | ↓ | – | ↓ | – | ↓ | – | ↓ | – | A | A | A | A | A | A |
| CMT756.01 | 9 y | – | – | – | – | A | A | ↓ | – | – | – | – | – | N | – | |
| MPZ | CMT-65.07 | – | – | 11.1 | – | – | – | – | – | – | A | A | – | – | – | – |
| PN-1540.1 | 20 y | 1.4 | 2.8 | A | A | A | A | A | A | A | A | – | – | A | A | |
| PN-506.1 | 4 y | 0.6 | 7.0 | 1.2 | 17.0 | |||||||||||
| PN-752.1 | 38 y | 0.2 | 5.0 | 0.7 | 7.0 | – | 5.0 | 0.02 | 6.0 | – | – | – | – | – | – | |
| PN-966.4 | – | – | 11.0 | – | 6.0 | – | – | – | – | – | – | – | – | – | – | |
| NEFL | PN-1385.1 | 5 y | – | 29.0 | – | – | – | 24.0 | – | – | A | A | A | A | A | A |
| PMP22 | PN-750.3 | 2 y | A | A | – | – | A | A | A | A | A | A | – | – | A | A |
| FGD4 | CMT-190.01 | 11 y | 1.6 | 6.2 | 2.9 | 8.4 | A | A | A | A | A | A | A | A | A | A |
| CMT-230.01 | – | – | 5.0 | – | – | – | – | – | – | A | A | – | – | – | – | |
| GDAP1 | PN-860.3 | 3 y | 1.9 | 42.0 | 1.5 | 50.0 | A | A | – | – | A | A | – | – | A | A |
| MTMR2 | CMT-201.01 | 7 y | 0.6 | 13.0 | – | – | A | A | 0.6 | 13.0 | A | A | – | – | A | A |
| PRX | PN-1699.1 | 11 y | – | – | – | – | A | A | A | A | – | – | – | – | A | A |
| PN-2175.1 | 3 y | 1.9 | 6.9 | – | – | – | – | – | – | – | – | – | – | – | – | |
| PN-44.1 | 41 y | 1.1 | 3.0 | 0.5 | 3.0 | – | – | – | – | A | A | – | – | – | – | |
| PN-761.3 | 5 y | A | A | – | – | – | – | – | – | – | 46.3 | – | – | A | A | |
| SBF2 | CMT-194.01 | 8 y | – | – | – | 26.0 | – | – | – | – | – | – | – | – | – | – |
| CMT-220.01 | 9 y | – | 16.0 | – | 22.0 | – | – | – | – | – | – | – | – | – | – | |
| PN-1101.1 | 10 y | – | – | – | 21.0 | – | 14.0 | – | – | – | – | – | – | – | – | |
| SH3TC2 | CMT-191.01 | 10 y | 9.1 | 34.5 | 6.2 | 25.0 | 1.4 | 18.4 | – | – | – | – | – | – | 0.2 | 29.4 |
| CMT-192.01 | 10 y | 0.2 | 39.6 | 4.5 | 30.0 | 0.1 | 19.5 | – | – | – | – | – | – | 7.0 | 40.6 | |
| CMT-133.01 | – | – | 27.0 | – | 37.0 | – | 24.0 | – | – | A | A | – | – | – | – | |
| CMT-189.V.5 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| CMT-234.01 | – | – | 12.0 | – | – | – | – | – | – | – | – | – | – | – | – | |
| CMT-235.01 | – | – | 26.0 | – | – | – | 21.0 | – | – | – | 34.0 | – | – | A | A | |
| PN-1289.1 | 20 y | 3.9 | 31.6 | 4.3 | 37.9 | 0.1 | 26.5 | – | – | A | A | – | – | – | – | |
| PN-1321.1 | 33 y | 0.7 | 11.9 | 1.2 | 11.7 | 0.8 | 22.6 | 0.2 | 12.7 | – | 33.0 | – | – | – | – | |
| PN-754.3 | 2 y | 1.5 | 27.0 | – | – | 1.0 | 23.0 | – | – | 15.0 | 42.0 | – | – | – | – | |
A = absent response; Age = age at examination; Amp = amplitude (motor: in millivolt; sensory: in microvolt); CV = conduction velocity (in metre per second); N = normal; – = not available; ↓ = reduced.
All patients with pathogenic mutations presented with symptoms suggestive of peripheral neuropathy within the first year of life. Presenting symptoms were nonetheless variable. A small number of patients present very soon after birth with hypotonia, occasionally associated with breathing difficulties. A second group of patients present with congenital or very early foot deformities often in combination with progressive delay in motor milestones in the first year of life, after an otherwise normal neonatal period.
Discussion
We performed a broad and systematic genetic screening in a large cohort of patients presenting with infantile onset hereditary neuropathy. Overall, a molecular diagnosis could be reached in 45% of patients by screening for mutations in a total of 11 genes and for the CMT1A duplication in the 17p11.2-12 region (Fig. 1). Although the overall mutation frequency was high, the genetic heterogeneity is extensive resulting in a diagnostic yield per gene that rarely exceeds more than a few per cent of the total cohort. Both dominant and recessive mutations are implemented in this age group with the recessive ones being slightly more prevalent in our series (20 index patients versus 15). Most common were the CMT1A duplication and mutations in MPZ, PMP22, PRX and SH3TC2, accounting together for 69% of the identified pathogenic variations.
As far as dominant mutations are concerned, these are often de novo events, as was previously described in patients with congenital hypomyelinating neuropathy or Dejerine–Sottas neuropathy (Gabreels-Festen, 2002; Plante-Bordeneuve and Said, 2002). However, we also show a few cases of inherited dominant traits (mainly for MPZ and the CMT1A duplication) that can also produce severe early onset phenotypes in some patients, in spite of the generally milder phenotype in the other family members (Raeymaekers et al., 1991; Nelis et al., 1994; Sorour et al., 1997). These young patients represent the far end of the disease spectrum associated with the various demyelinating types of classical Charcot–Marie–Tooth disease. In these instances, the familial history is useful in guiding the molecular testing, but one has to be equally alert for such mutations in isolated patients. It is important to note that more mildly affected or even asymptomatic individuals are still at risk of having offspring with a more severe onset phenotype. This was recently shown for other dominantly inherited Charcot–Marie–Tooth variants caused by mutations in TRPV4 and GDAP1 (Zimon et al., 2010, 2011). There are also reports in the literature of transmitted cases of severe Charcot–Marie–Tooth phenotypes (Smit et al., 2008). For the dominant genes, mutations in MPZ were equally frequent as patients carrying the CMT1A duplication. In the particular case of patient CMT-756.01 with the novel heterozygous Arg400Pro mutation in MFN2, a possible parental mosaicism was identified. This finding has important implications towards genetic counselling and emphasizes the importance of critical analysis of DNA samples of clinically unaffected parents.
Mutations in recessive genes are often found in isolated patients, many of whom are the product of a consanguineous union. The cohort used for this study contained a total of 21 index patients from such consanguineous families; this may have influenced the total frequency of recessive forms to some extent. It is, however, important to note that of the 20 index patients with recessive mutations, at least five stem from non-consanguineous European families (PN-1101.2, CMT-191.01, CMT-192.01, PN-1321.1 and PN-754.3). Therefore, a certain degree of suspicion for recessive forms of neuropathy is warranted in non-consanguineous populations, especially since the inheritance pattern would not be evident in small kinships.
Overall, we found SH3TC2 to be the most commonly mutated recessive gene in this cohort. SH3TC2 is associated with variable phenotypes and is known to be relatively common among the recessive Charcot–Marie–Tooth variants (Houlden et al., 2009).
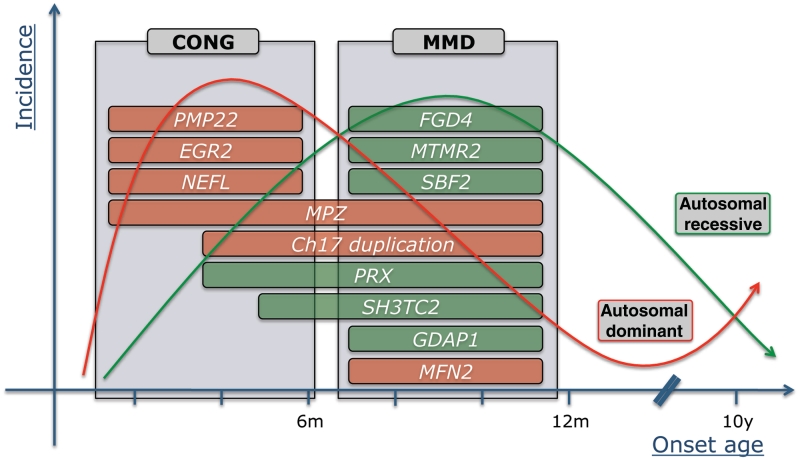
From a phenotypic point of view, it is important to note that even within this group of patients with neuropathy with a very early disease onset, there is considerable clinical variability. A few different subgroups of patients can be delineated that in turn correspond to selected subsets of the genes screened in this study (Fig. 2).
Figure 2.
Gene distribution for patients with a known pathogenic mutation in function of disease onset, inheritance pattern and phenotypic subgroup. The x-axis represents the age at onset in months and years, the y-axis displays the estimated incidence of patients with dominant (red line) and recessive (green line) subtypes of hereditary neuropathy. CONG = congenital onset phenotype characterized by neonatal hypotonia ± breathing and feeding difficulties; MMD = motor milestone delay phenotype with or without early foot deformities and progressive delay in motor milestones within the first year of life; m = month; y = year.
The first group comprises genuinely congenital onset phenotypes presenting soon after birth with symptoms such as hypotonia and breathing difficulties. In these patients, de novo mutations in MPZ, EGR2 and PMP22 are typically found (Phillips et al., 1999; Timmerman et al., 1999; Ceuterick-de Groote et al., 2001). In addition, we identified a heterozygous NEFL mutation that was previously reported to cause severe early onset phenotypes (Yoshihara et al., 2002). More surprisingly, we detected a previously unreported partial duplication of exon 4 of PMP22 in an isolated patient presenting with a congenital onset phenotype. Although the vast majority of copy number variations in the 17p11.2-12 region is the recurrent 1.4 Mb CMT1A duplication/deletion, some atypical non-recurrent copy number variations have been reported including the entire PMP22 gene, single PMP22 exons or regulatory sequences (Zhang et al., 2010). The de novo character of this variation in patient CMT-127.04 could not be proven due to lack of the parent's DNA samples; it may therefore still be a benign polymorphism. However, this PMP22 exonic duplication may well be pathogenic through such mechanisms as exon shuffling or insertional translocation (Zhang et al., 2010). No other sequence variants were found in patient CMT-127.04. To date, this partial duplication of exon 4 had not been encountered in >400 samples that were screened for diagnostic purposes with the same Multiplex Amplicon Quantification assay.
Recessive genes are far less prominent in the subgroup of the congenital cases with only two patients with mutations in PRX and SH3TC2, respectively.
The second phenotypic subgroup consists of patients who display early and progressive delay in motor development in the first year, often in combination with early foot deformities after an otherwise normal neonatal period. Recessive mutations in FGD4, PRX, MTMR2, SBF2 and GDAP1 are strongly represented in this subgroup. A few patients with dominant mutations in MPZ and MFN2 further broaden the spectrum (Fig. 2).
The third phenotypic subgroup is largely overlapping with the second and consists of patients with a CMT1A duplication presenting with early and often congenital foot deformities, a more or less normal early motor development and progressive gait difficulties later on in childhood. These patients represent the far end of the severity spectrum of classical CMT1A. This finding underscores the importance of ruling out the CMT1A duplication in any patient with a demyelinating neuropathy, even in very young children (Fig. 2).
The electrophysiological findings in our cohort show that early onset hereditary neuropathies are more frequently of a demyelinating type rather than axonal. Severe slowing of nerve conduction velocity is seen in patients with mutations in PRX and FGD4 and also in the myelin-associated genes MPZ, EGR and PMP22. A more variable range of slowing in motor nerve conduction velocity is observed in demyelinating neuropathies caused by mutations in MTMR2, SBF2 and SH3TC2. Although axonal forms are proportionally less common, early onset neuropathies cannot be restricted to demyelinating neuropathies alone. We identified three index patients with axonal neuropathies carrying mutations in GDAP1 and MFN2. Of note is that a sural nerve biopsy performed in Patient PN-860.03 (homozygous GDAP1 mutation) showed limited (secondary) demyelinating changes in addition to manifest signs of axonal neuropathy (Nelis et al., 2002b). The direct comparison of the results of the nerve conduction studies presented here is troublesome because of the broad range of ages at which the electrophysiological exams were performed and the problem of normative values in young children (Garcia et al., 2000). However, the general trends in these electrophysiological findings are clinically meaningful and hold true despite the above-mentioned drawbacks. Thorough electrophysiological testing remains the cornerstone of the diagnosis in the context of hereditary neuropathies and should, in this particular instance, be performed by electrophysiologists specialized in paediatric neuromuscular disorders (Wilmshurst et al., 2003; Pitt, 2011). To differentiate from metabolic diseases, routine screening should be performed such as biochemical markers for lysosomal storage disorders in urine, very long chain fatty acids, isoelectric focusing of serum transferine and organic acids (Landrieu et al., 2011).
The current study focuses on patients with early onset forms of hereditary motor and sensory neuropathy. Overlap with pure motor forms (hereditary motor neuropathy) has been described, however, especially in axonal forms of hereditary motor and sensory neuropathy. In that context, it is important to note that recently, MFN2 mutations were found in various patients with pure motor phenotypes (Feely et al., 2011). Therefore, it may be useful to extend future mutation screenings of MFN2 to patients with early onset forms of hereditary motor neuropathy.
In general, nerve biopsies are considered invasive procedures that are best avoided if a diagnosis can be established using other methods. In this series, in 19 of 35 patients with proven mutations, neuropathological examination of a sural nerve or skin biopsy was performed. This indicates that such a procedure may still be important in the diagnosis of hereditary neuropathies in young children for a number of reasons (Vallat et al., 2011). First, a nerve (and muscle) biopsy can be helpful in differentiating peripheral neuropathy from other causes of severe hypotonia in newborns. Secondly, patients with early onset neuropathies are often isolated in their family, making the differentiation from other rare acquired causes more difficult, especially in the absence of consanguinity among the parents. In addition, electrophysiological testing may be inconclusive with regard to discerning demyelinating from axonal forms due to technical limitations and severe denervation (Wilmshurst et al., 2003). Finally, pathology may unveil certain specific findings that can help to orient molecular testing. Myelin outfoldings and redundant myelin loops, for example, are a conspicuous feature that suggests possible mutations in the myotubularin genes MTMR2 and SBF2 (Nelis et al., 2002a; Azzedine et al., 2003). However, these findings should be interpreted cautiously, because in the current study, and in previous reports, similar changes were also observed in patients with mutations in FGD4, SH3TC2, PRX and MPZ (Takashima et al., 2002; Kochanski et al., 2004; Fabrizi et al., 2009; Houlden et al., 2009). Basal lamina onion bulbs are in turn considered a feature suggestive of SH3TC2 mutation carriers (Houlden et al., 2009).
The current study contributes to our understanding of the clinical and genetic basis of hereditary neuropathies with disease onset in the first year of life. A molecular diagnosis could be reached in 45% of patients from a heterogeneous screening cohort of 77 unrelated index patients leaving more than half of the patients without genetic diagnosis to date. This further underscores the fact that other still unknown mutations must exist in addition to yet unreported phenotypic variants associated with known or unknown disease-associated genes.
Organizing rational molecular diagnostic testing in the specific case of early onset neuropathies is cumbersome due to the low mutation frequency per gene and the broad genetic heterogeneity (Ryan and Ouvrier, 2005). Some directions can be drawn up based on the findings of this study. In patients with a congenital disease onset, mutations in dominant genes are more likely to be the cause, while mutations in recessive genes are more probable in patients with progressive delay in motor development in the first year of life, especially if parents are consanguineous. Axonal subtypes are probably restricted to a smaller subset of genes that may be tested preferentially. Other findings such as myelin outfoldings on nerve pathology may help to prioritize molecular testing to some extent. Likewise, severe and early scoliosis, although very suggestive of SH3TC2 mutations (Houlden et al., 2009), may also be seen in patients carrying mutations in other genes and can in fact be considered a feature potentially to be found in many types of severe and progressive neuropathy of early childhood.
In conclusion, reaching a correct genetic diagnosis in children with severe early onset hereditary motor and sensory neuropathy remains a major challenge with conventional screening techniques. A future solution for this diagnostic conundrum may lie in the more systematic diagnostic application of recently developed technologies for massive parallel sequencing. While operational costs of these methods plummet, capacity and overall robustness grows exponentially. These technologies allow for the simultaneous screening of larger sets of genes and eventually of patient's entire exome or even genome (Hoischen et al., 2010; Lupski et al., 2010; Montenegro et al., 2011). By doing so, mutations in one of the myriad of known genes can be identified quickly and also new genetic risk factors can be scrutinized on the same datasets. Such approaches would ultimately allow geneticists to base the entire process of molecular diagnosis in a patient on a single test. The downside of these technologies is that they will yield numerous sequence variants of unknown significance. Although further validation of sequence variants in known genes may still be relatively straightforward, the identification of pathogenic mutations in novel genes will be more problematic and will require pooling of multiple unrelated patients and the use of robust bioinformatics tools (Depristo et al., 2011).
Funding
University of Antwerp; Fund for Scientific Research (FWO-Flanders); Medical Foundation Queen Elisabeth (GSKE); ‘Association Belge contre les Maladies Neuromusculaires’ (ABMM); Interuniversity Attraction Poles P6/43 programme of the Belgian Federal Science Policy Office (BELSPO); ‘Methusalem excellence grant’ of the Flemish Government; Austrian Science Fond (FWF; P19455-B05); PhD fellowships of the FWO-Flanders (to J.B., M.Z. and K.P.); Bogazici University Research Fund (00M102, in part).
Acknowledgements
We are grateful to the patients and their families for their willingness to cooperate in this research project. We also wish to thank the Genetic Service Facility (VIB) for the sequencing support (http://www.vibgeneticservicefacility.be/).
Glossary
Abbreviations
- EGR2
early growth response 2
- FGD4
FYVE, RhoGEF, and PH domain-containing protein 4
- GDAP1
ganglioside-induced differentiation-associated protein 1
- MFN2
mitofusin 2
- MPZ
myelin protein zero
- MTMR2
myotubularin-related protein 2
- NEFL
neurofilament light chain
- PMP22
peripheral myelin protein 22
- PRX
periaxin
- SBF2
set-binding factor 2
- SH3TC2
SH3 domain and tetratricopeptide repeat domain 2.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzedine H, Bolino A, Taieb T, Birouk N, Di Duca M, Bouhouche A, et al. Mutations in MTMR13, a new pseudophosphatase homologue of MTMR2 and Sbf1, in two families with an autosomal recessive demyelinating form of Charcot-Marie-Tooth disease associated with early-onset glaucoma. Am J Hum Genet. 2003;72:1141–53. doi: 10.1086/375034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolino A, Muglia M, Conforti FL, LeGuern E, Salih MA, Georgiou DM, et al. Charcot-Marie-Tooth type 4B is caused by mutations in the gene encoding myotubularin-related protein-2. Nat Genet. 2000;25:17–9. doi: 10.1038/75542. [DOI] [PubMed] [Google Scholar]
- Burns J, Ryan MM, Ouvrier RA. Evolution of foot and ankle manifestations in children with CMT1A. Muscle Nerve. 2009;39:158–66. doi: 10.1002/mus.21140. [DOI] [PubMed] [Google Scholar]
- Ceuterick-de Groote C, De Jonghe P, Timmerman V, Van Goethem G, Lofgren A, Ceulemans B, et al. Infantile demyelinating neuropathy associated with a de novo point mutation on Ser72 in PMP22 and basal lamina onion bulbs in skin biopsy. Pathol Res Pract. 2001;197:193–8. doi: 10.1078/0344-0338-00033. [DOI] [PubMed] [Google Scholar]
- Connolly AM. Chronic inflammatory demyelinating polyneuropathy in childhood. Pediatr Neurol. 2001;24:177–82. doi: 10.1016/s0887-8994(00)00237-x. [DOI] [PubMed] [Google Scholar]
- Depristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierick I, Baets J, Irobi J, Jacobs A, De Vriendt E, Deconinck T, et al. Relative contribution of mutations in genes for autosomal dominant distal hereditary motor neuropathies: a genotype-phenotype correlation study. Brain. 2008;131:1217–27. doi: 10.1093/brain/awn029. [DOI] [PubMed] [Google Scholar]
- Dejerine J, Sottas J. Sur la nevrite interstitielle, hypertrophique et progressive de I'enfance. C R Soc Biol. 1893;45:63–96. [Google Scholar]
- Fabrizi GM, Taioli F, Cavallaro T, Ferrari S, Bertolasi L, Casarotto M, et al. Further evidence that mutations in FGD4/frabin cause Charcot-Marie-Tooth disease type 4H. Neurology. 2009;72:1160–4. doi: 10.1212/01.wnl.0000345373.58618.b6. [DOI] [PubMed] [Google Scholar]
- Feely SM, Laura M, Siskind CE, Sottile S, Davis M, Gibbons VS, et al. MFN2 mutations cause severe phenotypes in most patients with CMT2A. Neurology. 2011;76:1690–6. doi: 10.1212/WNL.0b013e31821a441e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabreels-Festen A. Dejerine-Sottas syndrome grown to maturity: overview of genetic and morphological heterogeneity and follow-up of 25 patients. J Anat. 2002;200:341–56. doi: 10.1046/j.1469-7580.2002.00043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabreels-Festen A, Thomas PK. Autosomal recessive hereditary motor and sensory neuropathies. In: Dyck PJ, Thomas PK, editors. Peripheral neuropathy. Philadelphia: Saunders; 2005. pp. 1769–90. [Google Scholar]
- Garcia A, Calleja J, Antolin FM, Berciano J. Peripheral motor and sensory nerve conduction studies in normal infants and children. Clin Neurophysiol. 2000;111:513–20. doi: 10.1016/s1388-2457(99)00279-5. [DOI] [PubMed] [Google Scholar]
- Hoischen A, Gilissen C, Arts P, Wieskamp N, van der Vliet W, Vermeer S, et al. Massively parallel sequencing of ataxia genes after array-based enrichment. Hum Mutat. 2010;31:494–9. doi: 10.1002/humu.21221. [DOI] [PubMed] [Google Scholar]
- Houlden H, Laura M, Ginsberg L, Jungbluth H, Robb SA, Blake J, et al. The phenotype of Charcot-Marie-Tooth disease type 4C due to SH3TC2 mutations and possible predisposition to an inflammatory neuropathy. Neuromuscul Disord. 2009;19:264–9. doi: 10.1016/j.nmd.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Kochanski A, Drac H, Kabzinska D, Hausmanowa-Petrusewicz I. A novel mutation, Thr65Ala, in the MPZ gene in a patient with Charcot-Marie-Tooth type 1B disease with focally folded myelin. Neuromuscul Disord. 2004;14:229–32. doi: 10.1016/j.nmd.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Landrieu P, Baets J, De Jonghe P. Hereditary motor-sensory, sensory and motor neuropathies in childhood. In: Dulac O, Lassonde M, Sarnat H, editors. Pediatric neurology handbook. Elsevier; 2011. [Google Scholar]
- Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362:1181–91. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro G, Powell E, Huang J, Speziani F, Edwards YJ, Beecham G, et al. Exome sequencing allows for rapid gene identification in a Charcot-Marie-Tooth family. Ann Neurol. 2011;69:464–70. doi: 10.1002/ana.22235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelis E, Erdem S, Tan E, Lofgren A, Ceuterick C, De Jonghe P, et al. A novel homozygous missense mutation in the myotubularin-related protein 2 gene associated with recessive Charcot-Marie-Tooth disease with irregularly folded myelin sheaths. Neuromuscul Disord. 2002a;12:869–73. doi: 10.1016/s0960-8966(02)00046-9. [DOI] [PubMed] [Google Scholar]
- Nelis E, Erdem S, Van Den Bergh PY, Belpaire-Dethiou MC, Ceuterick C, Van Gerwen V, et al. Mutations in GDAP1: autosomal recessive CMT with demyelination and axonopathy. Neurology. 2002b;59:1865–72. doi: 10.1212/01.wnl.0000036272.36047.54. [DOI] [PubMed] [Google Scholar]
- Nelis E, Timmerman V, De Jonghe P, Muylle L, Martin JJ, Van Broeckhoven C. Linkage and mutation analysis in an extended family with Charcot-Marie-Tooth disease type 1B. J Med Genet. 1994;31:811–5. doi: 10.1136/jmg.31.10.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson GA, Magdelaine C, Zhu D, Grew S, Ryan MM, Sturtz F, et al. Severe early-onset axonal neuropathy with homozygous and compound heterozygous MFN2 mutations. Neurology. 2008;70:1678–81. doi: 10.1212/01.wnl.0000311275.89032.22. [DOI] [PubMed] [Google Scholar]
- Parman Y, Battaloglu E, Baris I, Bilir B, Poyraz M, Bissar-Tadmouri N, et al. Clinicopathological and genetic study of early-onset demyelinating neuropathy. Brain. 2004;127:2540–50. doi: 10.1093/brain/awh275. [DOI] [PubMed] [Google Scholar]
- Phillips JP, Warner LE, Lupski JR, Garg BP. Congenital hypomyelinating neuropathy: two patients with long-term follow-up. Pediatr Neurol. 1999;20:226–32. doi: 10.1016/s0887-8994(98)00138-6. [DOI] [PubMed] [Google Scholar]
- Pitt M. Paediatric electromyography in the modern world: a personal view. Dev Med Child Neurol. 2011;53:120–4. doi: 10.1111/j.1469-8749.2010.03831.x. [DOI] [PubMed] [Google Scholar]
- Pitt M, Houlden H, Jacobs J, Mok Q, Harding B, Reilly M, et al. Severe infantile neuropathy with diaphragmatic weakness and its relationship to SMARD1. Brain. 2003;126:2682–92. doi: 10.1093/brain/awg278. [DOI] [PubMed] [Google Scholar]
- Plante-Bordeneuve V, Said G. Dejerine-Sottas disease and hereditary demyelinating polyneuropathy of infancy. Muscle Nerve. 2002;26:608–21. doi: 10.1002/mus.10197. [DOI] [PubMed] [Google Scholar]
- Raeymaekers P, Timmerman V, Nelis E, De Jonghe P, Hoogendijk JE, Baas F, et al. Duplication in chromosome 17p11.2 in Charcot-Marie-Tooth neuropathy type 1a (CMT 1a). The HMSN Collaborative Research Group. Neuromuscul Disord. 1991;1:93–7. doi: 10.1016/0960-8966(91)90055-w. [DOI] [PubMed] [Google Scholar]
- Rotthier A, Baets J, Vriendt ED, Jacobs A, Auer-Grumbach M, Levy N, et al. Genes for hereditary sensory and autonomic neuropathies: a genotype-phenotype correlation. Brain. 2009;132:2699–711. doi: 10.1093/brain/awp198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Ryan MM, Ouvrier R. Hereditary peripheral neuropathies of childhood. Curr Opin Neurol. 2005;18:105–10. doi: 10.1097/01.wco.0000162849.37273.af. [DOI] [PubMed] [Google Scholar]
- Scherer SS. Finding the causes of inherited neuropathies. Arch Neurol. 2006;63:812–6. doi: 10.1001/archneur.63.6.812. [DOI] [PubMed] [Google Scholar]
- Senderek J, Bergmann C, Stendel C, Kirfel J, Verpoorten N, De Jonghe P, et al. Mutations in a gene encoding a novel SH3/TPR domain protein cause autosomal recessive Charcot-Marie-Tooth type 4C neuropathy. Am J Hum Genet. 2003;73:1106–19. doi: 10.1086/379525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleegers K, Brouwers N, Gijselinck I, Theuns J, Goossens D, Wauters J, et al. APP duplication is sufficient to cause early onset Alzheimer's dementia with cerebral amyloid angiopathy. Brain. 2006;129:2977–83. doi: 10.1093/brain/awl203. [DOI] [PubMed] [Google Scholar]
- Smit LS, Roofthooft D, van Ruissen F, Baas F, van Doorn PA. Congenital hypomyelinating neuropathy, a long term follow-up study in an affected family. Neuromuscul Disord. 2008;18:59–62. doi: 10.1016/j.nmd.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Sorour E, MacMillan J, Upadhyaya M. Novel mutation of the myelin P0 gene in a CMT1B family. Hum Mutat. 1997;9:74–7. doi: 10.1002/(SICI)1098-1004(1997)9:1<74::AID-HUMU16>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Stendel C, Roos A, Deconinck T, Pereira J, Castagner F, Niemann A, et al. Peripheral nerve demyelination caused by a mutant Rho GTPase guanine nucleotide exchange factor, frabin/FGD4. Am J Hum Genet. 2007;81:158–64. doi: 10.1086/518770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima H, Boerkoel CF, De Jonghe P, Ceuterick C, Martin JJ, Voit T, et al. Periaxin mutations cause a broad spectrum of demyelinating neuropathies. Ann Neurol. 2002;51:709–15. doi: 10.1002/ana.10213. [DOI] [PubMed] [Google Scholar]
- Timmerman V, De Jonghe P, Ceuterick C, De Vriendt E, Lofgren A, Nelis E, et al. Novel missense mutation in the early growth response 2 gene associated with Dejerine-Sottas syndrome phenotype. Neurology. 1999;52:1827–32. doi: 10.1212/wnl.52.9.1827. [DOI] [PubMed] [Google Scholar]
- Vallat JM, Funalot B, Magy L. Nerve biopsy: requirements for diagnosis and clinical value. Acta Neuropathol. 2011;121:313–26. doi: 10.1007/s00401-011-0804-4. [DOI] [PubMed] [Google Scholar]
- Wilmshurst JM, Pollard JD, Nicholson G, Antony J, Ouvrier R. Peripheral neuropathies of infancy. Dev Med Child Neurol. 2003;45:408–14. doi: 10.1017/s0012162203000768. [DOI] [PubMed] [Google Scholar]
- Yoshihara T, Yamamoto M, Hattori N, Misu K, Mori K, Koike H, et al. Identification of novel sequence variants in the neurofilament-light gene in a Japanese population: analysis of Charcot-Marie-Tooth disease patients and normal individuals. J Peripher Nerv Syst. 2002;7:221–4. doi: 10.1046/j.1529-8027.2002.02028.x. [DOI] [PubMed] [Google Scholar]
- Yum SW, Zhang J, Mo K, Li J, Scherer SS. A novel recessive Nefl mutation causes a severe, early-onset axonal neuropathy. Ann Neurol. 2009;66:759–70. doi: 10.1002/ana.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Seeman P, Liu P, Weterman MA, Gonzaga-Jauregui C, Towne CF, et al. Mechanisms for nonrecurrent genomic rearrangements associated with CMT1A or HNPP: rare CNVs as a cause for missing heritability. Am J Hum Genet. 2010;86:892–903. doi: 10.1016/j.ajhg.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimon M, Baets J, Auer-Grumbach M, Berciano J, Garcia A, Lopez-Laso E, et al. Dominant mutations in the cation channel gene transient receptor potential vanilloid 4 cause an unusual spectrum of neuropathies. Brain. 2010;133:1798–809. doi: 10.1093/brain/awq109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimon M, Baets J, Fabrizi GM, Jaakkola E, Kabzińska D, Pilch J, et al. Dominant GDAP1 mutations cause predominantly mild CMT phenotypes. Neurology. 2011 doi: 10.1212/WNL.0b013e318228fc70. Jul 13. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]