Abstract
The phytohormones, brassinosteroids (BRs), play important roles in regulating cell elongation and cell size, and BR-related mutants in Arabidopsis display significant dwarf phenotypes. Cellulose is a biopolymer which has a major contribution to cell wall formation during cell expansion and elongation. However, whether BRs regulate cellulose synthesis, and if so, what the underlying mechanism of cell elongation induced by BRs is, is unknown. The content of cellulose and the expression levels of the cellulose synthase genes (CESAs) was measured in BR-related mutants and their wild-type counterpart. The chromatin immunoprecipitation (CHIP) experiments and genetic analysis were used to demonstrate that BRs regulate CESA genes. It was found here that the BR-deficient or BR-perceptional mutants contain less cellulose than the wild type. The expression of CESA genes, especially those related to primary cell wall synthesis, was reduced in det2-1 and bri1-301, and was only inducible by BRs in the BR-deficient mutant det2-1. CHIP experiments show that the BR-activated transcription factor BES1 can associate with upstream elements of most CESA genes particularly those related with the primary cell wall. Furthermore, over-expression of the BR receptor BRI1 in CESA1, 3, and 6 mutants can only partially rescue the dwarf phenotypes. Our findings provide potential insights into the mechanism that BRs regulate cellulose synthesis to accomplish the cell elongation process in plant development.
Keywords: Arabidopsis, brassinosteroids, cell elongation, cellulose, cellulose synthase, transcription factor
Introduction
Cellulose is the most abundant polysaccharide on earth, determines the orientation of cell expansion, and provides supporting material for plant cells (Pauly and Keegstra, 2008). The cellulose-formed microfibrils, consisting of a linear chain of several hundred to over 10 000 β-1, 4-linked glucan, are synthesized by a plasma membrane-localized cellulose synthase complex (CSC). In vascular plants, multiple cellulose synthase catalytic subunits are required for cellulose synthesis (Taylor et al., 2000). The complex extrudes up to 36 individual cellulose chains that are bound together to form the cellulose microfibril (Wightman and Turner, 2010).
The CESA gene superfamily, which encodes the catalytic subunits of cellulose synthase, has been identified in hundreds of seed plant species and characterized in Arabidopsis. The Arabidopsis genome contains ten CESA genes that include two groups with known function and some members with uncertain function (Richmond and Somerville, 2000, 2001). One group, including CESA1, CESA3, and CESA6, is preferentially expressed in expanding tissues (Desprez et al., 2002; Doblin et al., 2002; Robert et al., 2004). Mutants of cesA1, cesA3, and cesA6 are dramatically dwarfed or seedling-lethal. The other group includes CESA4, CESA7, and CESA8, and mutants of cesA4, cesA7, and cesA8 lack the characteristic secondary thickening in xylem (Scheible et al., 2001; Taylor et al., 2000, 2003). Although, the functions of the remaining CESA2, CESA5, CESA9, and CESA10 genes are poorly understood, recent studies have suggested that some of them may be functionally redundant and may compensate for the function of other CESAs under different physiological conditions (Desprez et al., 2007; Persson et al., 2007).
Plants have evolved complicated regulatory mechanisms, including the expression of CESA genes, modification of CESA protein, and intracellular trafficking and deposition of CSC subunits in the plasma membrane (Hématy and Höfte, 2006) to control cellulose biosynthesis and assembling in cell walls. Previous genetic and molecular studies have revealed that a transcription factor cascade, including NAC and MYB families, controls secondary cell wall thickening in fibres, vessels, and anthers (Mitsuda et al., 2005, 2007; Yang et al., 2007; Taylor, 2008; Zhong et al., 2008; Zhou et al., 2009). However, only transcription factor NST1/SND1 is known to be involved in cellulose synthesis by positively regulating the expressions of CESA7 and CESA8 (Zhong et al., 2007). In addition, it was recently reported that the signalling molecule nitric oxide can promote cellulose synthesis, leading to the increase in cellulose content in primary cell walls in tomato roots (Aragunde et al., 2008). Because cellulosic polysaccharides are the major part of the above-ground biomass (except grains) for many plant species, and more than 30–40% of dry matter in plants is cellulose (Pauly and Keegstra, 2008; Vogel, 2008), it is reasonable to speculate that these related signals and the supply of cellulose are likely to play an important role in biomass accumulation in plants.
It is well known that many plant hormones, such as brassinosteroids (BRs), auxins, gibberellins, and cytokinins play pivotal roles in regulating plant growth and height via promoting cell elongation and/or cell division (Gonzalez et al., 2009). During cell rapid elongation, new cell wall polymers need a large amount of cellulose deposition (Caderas et al., 2000; Refrégier et al., 2004). In some economically important crops, plant height is a major factor in determining above-ground biomass productivity (Alexopouloua et al., 2008; Yuan et al., 2008). However, it is unknown whether cellulose synthesis contributes to hormone-regulated plant growth and height.
BRs are one class of plant-specific steroid hormones that are involved in many aspects of plant growth and development (Li and Jin, 2006; Divi and Krishna, 2009), particularly in cell elongation. The biosynthetic pathway of BRs, which includes several key genes, such as CPD, DWF4, and DET2, has been established in Arabidopsis (Fujioka, 1999; Sakurai, 1999). More recently, many major components of hte BR signalling pathway have also been identified. BRs are perceived by a receptor-like kinase BRI1 (Li and Chory, 1997; Wang et al., 2001). The BR signal can activate a preformed homodimer of BRI1 (Wang et al., 2005), and induce the dissociation of a negative regulator BKI1 from the plasma membrane (Wang and Chory, 2006). Upon BRI1 activation, BSKs may be phosphorylated to inactivate a GSK3-like protein kinase BIN2 via an unkown mechanism (Tang et al., 2008). BIN2 kinase can phosphorylate and inhibit the class of plant-specific transcription factors, BES1/BZR1, which can directly bind to E-box (CANNTG) and BRRE (CGTGT/CG) elements in the promoter regions of many target genes to regulate their expression (Yin et al., 2002; He et al., 2005).
Previous studies implied that BRs may affect cell wall polymer formation, and the dwarf phenotype of the BR mutants is mainly caused by the reduced cell size, not by cell number (Kauschmann et al., 1996). In cotton, it was reported that BRs are required for fibre initiation as well as elongation of cultured cotton ovules (Sun et al., 2005; Luo et al., 2007). In Arabidopsis, BR-deficient or BR-perceptional mutants display dramatically dwarfed phenotypes (Li et al., 1996; Szekeres et al., 1996). In rice, reduction of OsDwarf2/OsDwarf1, which encodes a C-6 oxidase required for BR biosynthesis, caused a reduced elongation of the second internode and seed length (Hong et al., 2005; Tanabe et al., 2005). In maize, the dwf1 mutant is severely stunted and its encoded protein has 86% similarity with the rice DWF1 (Tao et al., 2004). By contrast, an increased BR level or activity can enhance plant size, biomass accumulation, and seed yield (Salas-Fernandez et al., 2009). However, the molecular mechanisms by which cellulose synthesis co-ordinates with the enlarged cell size caused by BR signalling are poorly understood.
In this study, biomass accumulation and cellulose content were measured in the BR-related mutants and a BRI1 over-expression line at different developmental stages, and it was found that BRs positively regulate biomass accumulation and cellulose content. The expression levels of the CESA genes were then measured by quantitative RT-PCR (qRT-PCR) and a GUS reporter driven by the CESA promoters, and it was discovered that BRs promote the expression of most CESA genes in the short term and in the long term. Further chromatin immunoprecipitation (ChIP) analysis demonstrated that BR-activated transcriptional factor BES1 can bind to the promoter regions of nine CESA genes in vivo. Using transgenic approaches, it was also found that over-expression of BRI1 in some CESA mutants cannot completely rescue their dwarf phenotype. Our results support the suggestion that BRs promote the expression of most CESA genes, which may play an essential role in regulating biomass accumulation in Arabidopsis.
Materials and methods
Plant materials, growth conditions, and hypocotyl elongation assay
A. thaliana ecotype Columbia (Col-0) was the wild type. The CESA mutants rsw1-1 (CS848759), ixr1-1 (SALK_019756), and prc1-1 (SALK_004587), and the T-DNA insertion mutants, ct-2 (SALK_091570), ct-5 (SALK_023353), and CT-9 (SALK_049129) were obtained from the ABRC (Arabidopsis Biological Resource Center). Homozygous insertion lines were verified by PCR and RT-PCR. Plants were grown on 1/2 MS plates or soil under long day (16/8 h light/dark) at 23 °C. For the hypocotyl elongation assay, seeds were planted on 1/2 MS plates, kept at 4 °C for 2 d, and then grown in the dark for 4 d or in the light for 7 d. Thirty to forty seedlings were measured for each genotype. For epiBL treatment, the 11-d-old light-grown seedlings were treated with 5 μM epiBL or DMSO (as a control) for 2 h.
Biomass and cellulose measurements
Thirty to forty aerial seedlings in different developmental stages were collected and dried at 60 °C overnight. Then the dry weight was recorded. The dry stem was ground into a fine powder in liquid nitrogen. The powder was treated as described by Updegraff (1969). Cellulose was quantified colorimetrically using the anthrone-sulphuric acid method (Laurentin and Edwards, 2003).
Plasmid construction
The 2000 bp region of each CESA gene was amplified with Col-0 genomic DNA and was cloned into the pCAMBIA1300 vector. To make BRI1 over-expression plants, BRI1 was fused with GFP into the vector of pCAMBIA2300, the resulting construct was transformed into Col-0.
CESA gene expression pattern analysis
Histochemical staining of plants expressing pCESA::GUS reporters was performed as described by Jefferson (1987). Digital images were taken with a Leica MZ FLIII stereomicroscope (Leica Microsystems, Germany).
Gene expression analysis
Total RNA was extracted from young seedlings using an RNeasy mini kit (Tiangen, http://www.tiangen.com). For RT-PCR, 2 μg of total RNA was reverse-transcribed with Super-Script II reverse transcriptase (TAKARA, http://www.takara.com.cn) as described by the manufacturer. Equal amounts of cDNA were used for PCR with 30–35 cycles. For quantitative real-time PCR, SYBR master mix (Invitrogen) and a Bio-Rad iCycler quantitative PCR system were used as described by the manufacturer. A U-box gene (at5g15400) was used to normalize the data (5′-TGCGCTGCCAGATAATACACTATT-3′ and 5′-TGCTGCCCAACATCAGGTT-3′).
ChIP assay
ChIP experiments were performed as described in the UPSTATE chip kit (http://www.millipore.com/catalogue/item/17-295) with 11-d-old Col-0 seedlings. The BES1 antibody was used to precipitate chromatin, and the GFP antibody was used as a control. Equal amounts of starting plant material and ChIP products were used for the quantitative real-time PCR reaction. Primers from 5S rRNA (used as an internal control) and CESAs were used to detect the corresponding CESA promoters in the ChIP products. The ChIP assays were repeated at least three times, and typical results were presented. The means and standard deviations were calculated from three biological repeats.
Results
BR signal influences the above-ground biomass accumulation in Arabidopsis
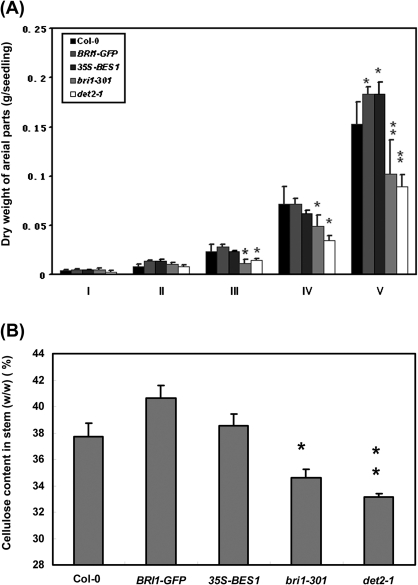
BRs are a major class of growth-promoting hormones. The BR-deficient or perceptional mutants are smaller than the wild type, while over-expression of BR-biosynthetic genes or the BR receptor BRI1 led to bigger plants (Li et al., 1996; Szekeres et al., 1996). Furthermore, the application of BRs to the BR-deficient mutant det2-1 significantly induced hypocotyl elongation (see Supplementary Fig. S1 at JXB online) (Li et al., 1996; Szekeres et al., 1996; Yin et al., 2005). To test the function of BRs in Arabidopsis further, the dry weight of the aerial parts of several genetic materials, including a weak allele of bri1, bri1-301, the biosynthetic mutant of det2-1, a BRI1-GFP over-expression line, and a 35S-BES1-GFP over-expression line, and the wild type Col-0, was measured at five different developmental stages (Stage I: having nine rosette leaves, the stage with vigorous vegetative growth; Stage II: initiation of bolting; Stage III: having one primary inflorescence with four nodes; Stage IV: having three side shoots on the primary inflorescence stalk; Stage V: having two or more secondary inflorescences, the end of plant growth) (Fig. 1A). Although the dry weight per seedling was similar between these mutants and the wild type at Stages I and II, it was found that, after bolting, there was a significant difference in the total dry weight among these materials. From Stage IV, the dry weights of det2-1 and bri1-301 were significantly less than that of the BRI1-GFP and 35S-BES1-GFP lines. At Stage V, the dry weight per seedling of det2-1 and bri1-301 was only 0.1 g, while it was 0.18 g in the BRI1-GFP line. These results indicated that BR signalling is related to dry weight above ground.
Fig. 1.
BRs regulate biomass accumulation in aerial parts of Arabidopsis. (A) Biomass accumulation in aerial parts of BRs mutants at different developmental stages: I, having nine rosette leaves, the stage with vigorous vegetative growth; II, initiation of bolting; III, having one primary inflorescence with four nodes; IV, having three side shoots on the primary inflorescence stalk; V, having two or more secondary inflorescences, the end of plant growth. (B) Cellulose content in primary inflorescence stems of BR-related mutants at stages V. Data represent means (±SE) of four independent experiments. The asterisks indicate significant levels of *P <0.05 and **P <0.01, respectively.
Because cellulose usually takes a significant portion of the total biomass, it was also tested whether the cellulose content is correlated with the amount of total biomass among these materials. The cellulose content was measured in the stems at the final developmental stage and it was found that the cellulose content in det2-1 and bri1-301 was 8% and 12% lower than that that in wild type (P <0.05), respectively. Consistent with the biomass accumulation, the amount of cellulose in the BRI1-GFP line is 7% higher than that in the wild type (Fig. 1B), and in the 35S-BES1-GFP line, the cellulose content is 3% higher than that in the wild type. These results suggest that BRs may promote cellulose biosynthesis, which contributes 30–40% of the total biomass accumulation (Pauly and Keegstra, 2008; Vogel, 2008).
BRs induce the expression of many CESA genes
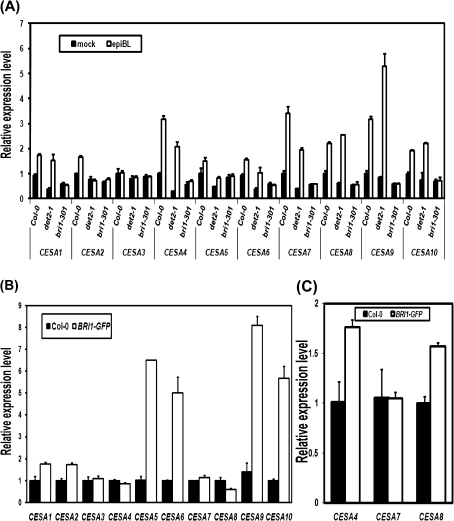
To test whether the elevated cellulose content by BRs is caused by an elevated expression of CESA genes, the expression levels of the CESA genes were measured using qRT-PCR in the BR-related mutants and the wild type. Without a 2, 4-epi-brassinolide (epiBL) treatment, the expression levels of ten CESA genes in det2-1 were reduced to about 50% of that in the wild type (Fig. 2A). After the application of epiBL to 9-d-old light-grown seedlings for 2 h, with the exception of CESA2 and CESA3 whose transcript levels were just slightly induced, the expression of the other CESA genes, including CESA1, CESA4, CESA5, CESA6, CESA7, CESA8, CESA9, and CESA10, were largely induced to approximately 1–5-fold higher than in the wild type and det2-1 (Fig. 2A). While in the BR-insensitive mutant, bri1-301, the transcript levels of these CESA genes had no significant difference before and after epiBL treatment, indicating that BRs can rapidly induce the expression of CESAs through BR signalling.
Fig. 2.
The exogenously applied epiBL induces expression of CESA genes in Arabidopsis. These CESA genes include CESA1, at4g32410; CESA2, at4g39350; CESA3, at5g05170; CESA4, at5g44030; CESA5, at5g09870; CESA6, at5g64740; CESA7, at5g17420; CESA8, at4g18780; CESA9, at2g21770; and CESA10, at2g25540. (A) Relative expression levels of CESA genes in BR-deficient and BR-insensitive mutants. Quantitative real-time RT-PCR was conduced with total RNA from 9-d-old light-grown shoots with or without the treatment of 5 μM epiBL for 2 h. (B) Relative expression levels of CESA genes in the BRI1-GFP line. (C) Expression levels of CESA4, 7, and 8 in the first internode of primary inflorescence stems of the BRI1-GFP line. Quantitative real-time RT-PCR was performed using total RNAs from 9-d-old light-grown seedlings.
The expression of these CESA genes was also tested in seedlings of the BRI1-GFP over-expression line. Compared with the wild type, the expression levels of CESA1, CESA2, CESA5, CESA6, CESA9, and CESA10 in the BRI1-GFP line were much higher than in the wild type (Fig. 2B). While the expression of CESA3, CESA4, CESA7, and CESA8 was not altered in the young seedlings of the BRI1-GFP line (Fig. 2B). Because CESA 4, 7, and 8 mainly synthesize cellulose used for the secondary cell wall, the expression levels of these genes was then measured in primary inflorescence stems at Stage V, which should contain more secondary cell wall, and it was found that, except for CESA7, the expression of CESA4 and CESA8 was significantly enhanced in the BRI1-GFP line (Fig. 2C). In summary, these results suggest that BR signalling enhances CESA gene expression for both primary and secondary growth.
To understand the biological function, expression pattern, and regulation of these CESA genes further, transgenic lines harbouring a beta-glucuronidase (GUS) reporter gene driven by the promoters of ten CESA genes in Arabidopsis were made. It was found that CESA1, CESA2, and CESA6, which showed similar expression patterns, were mainly expressed in the elongation zone of roots, in the veins of cotyledons and buds, and were weakly expressed in the leaves of light-grown seedlings (Fig. 3A, B, F). Because CESA1 and CESA6 are mainly involved in primary growth, this result indicates that CESA2 may also participate in primary growth. The expression levels of CESA3 and CESA5 were hardly detected without BR treatment (Fig. 3C, E), and their expression was only detected in the root or shoot tips following BR treatment (Fig. 3J, L). CESA9 was mainly expressed in the vasculature of cotyledons and shoot tips (Fig. 3G, N). Although CESA4 was expressed in the veins of cotyledons (Fig. 3D), CESA7 and CESA8 were only expressed in the vascular tissue of flowers, stamens, and stems at the later developmental stages, and their expression was barely detected in young seedlings, further demonstrating their important role in secondary growth (see Supplementary Fig. S2 at JXB online). However, CESA10 expression was not detected in young seedlings. These transgenic lines were also used to test whether CESA genes can be regulated by BRs. When these reporter lines were grown on medium containing 10 nm epiBL, compared with the untreated seedlings, the GUS-staining of all these transgenic lines was enhanced (Fig. 3H–N), which was consistent with our qRT-PCR analysis (see Supplementary Fig. S3 at JXB online). The expression of the CESA4 gene in long-term epiBL treatment is slightly different from the pattern of GUS-staining (see Supplementary Fig. S3 at JXB online; Fig. 3D), so it is likely that the CESA4 promoter (1.9 kb) is not long enough to drive the GUS reporter.
Fig. 3.
The GUS staining in transgenic seedlings with CESA promoter-driven GUS reporters is enhanced on the medium containing epiBL. The expression pattern and regulation by BRs of CESA genes are shown by GUS-staining of CESA promoter::GUS reporters in 5-d-old light-grown seedlings on the 1/2 MS medium (A–G), or on the 1/2 MS medium containing 10 nm epiBL (H–N).
BR-activated transcription factor BES1 can associate with upstream elements of most CESA genes in vivo
To investigate whether the enhanced expression of these CESA genes by BR signalling is directly through the BR-activated transcription factor BES1, a ChIP assay was then used to test whether BES1 can bind to the promoter regions of these CESAs. The CANNTG E-box is a primary binding element of BES1 (Yin et al., 2005), so RSTA (http://rsat.ulb.ac.be/rsat/) was used to predict how many potential binding sites of BES1 are present in the approximately 2000 bp region of each CESA promoter (see Supplementary Fig. S4A at JXB online). ChIP experiments were conducted with the anti-BES1 antibody and a control antibody, anti-GFP, and the promoter region of ACT2 (at3g18780), which does not contain any predicted BES1 binding sites, was used as a negative control. The positive control was at3g23770 (Ye et al., 2010). qPCR was used to detect the enrichment of ChIP products in the promoter region containing E-boxes (Fig. 4; see Supplementary Fig. S4B–K at JXB online). It was found that BES1 can specifically pull-down DNA fragments from the promoter regions of nine CESAs, but not CESA7. One or more binding sites were found in promoter region of CESA genes, with an enrichment of 1.5–5-fold (Fig. 4; see Supplementary Fig. S4B–K at JXB online). Taken together, it was concluded that the transcriptional factor BES1 can regulate the expression of most CESA genes by binding to their promoters, mainly through the E-box elements.
Fig. 4.
BES1 can associate with the promoter regions of most CESA genes. The BES1 antibody and the GFP antibody (as a negative control) were used to immunoprecipitate chromatin prepared from 14-d-old light-grown Col-0 seedlings. Quantitative real-time PCR was performed using primers from the indicated positions of CESA promoters. The fold changes were calculated based on the change for anti-BES1 relative to anti-GFP, after normalization to 5S rRNA, an internal control. (A) CESA1; (B) CESA2; (C) CESA3; (D) CESA4; (E) CESA5; (F) CESA6; (G) CESA8; (H) CESA9; (I) CESA10; (J) the positive control at3g23770.
CESAs act downstream of BR signalling in plant development
To determine whether BR signalling functions at upstream of CESAs to regulate cellulose synthesis, genetic analysis was conducted with some available CESA mutants: rsw1-1, a point mutation of CESA1; ixr1-1, a mutation of CESA3; prc1-1, a null mutant of CESA6; and ixr2-1, a weak allele of CESA6 were obtained. The light-grown seedlings of these CESA mutants were slightly smaller than those of the wild type under normal growing conditions (Fig. 5A, D, G, J). Double mutants of bri1-301 were then generated with some of these CESA mutants, including bri1-301 rsw1-1, bri1-301 ixr1-1, and bri1-301 prc1-1. All of these double mutants showed severely dwarf phenotypes (Fig. 5B, E, H, K), indicating that the CESA mutation can enhance the dwarf phenotype of bri1-301. Interestingly, although over-expression of the BR receptor BRI1 in these CESA mutant backgrounds can make plants larger and enhance leaf petiole elongation, they were still smaller than BRI1-OX (Fig. 5C, F, I, L). These results suggested that the enhanced BR signalling cannot completely rescue the dwarf phenotype of the CESA mutants, and a co-ordinated expression of many CESA genes downstream of BR signalling is required for plant development.
Fig. 5.
The CESA mutant enhances the dwarf phenotype of bri1-301, and over-expression of BRI1 in CESA mutants cannot completely rescue their dwarf phenotype. (A) Col-0; (B) the BR receptor mutant bri1-301; (C) the BRI1-GFP over-expression line; (D) rsw1-1, a CESA1 mutant (Ala549Val); (E) rsw1-1 bri1-301 double mutant; (F) the over-expression line of BRI1-GFP in rsw1-1; (G) ixr1-1, a CESA3 mutant (Gly998Asp); (H) ixr1-1 bri1-301 double mutant; (I) the over-expression line of BRI1-GFP in ixr1-1; (J) prc1-1, a CESA6 mutant (Q720STOP); (K) prc1-1 bri1-301 double mutant; (L) the over-expression line of BRI1-GFP in prc1-1. (This figure is available in colour at JXB online.)
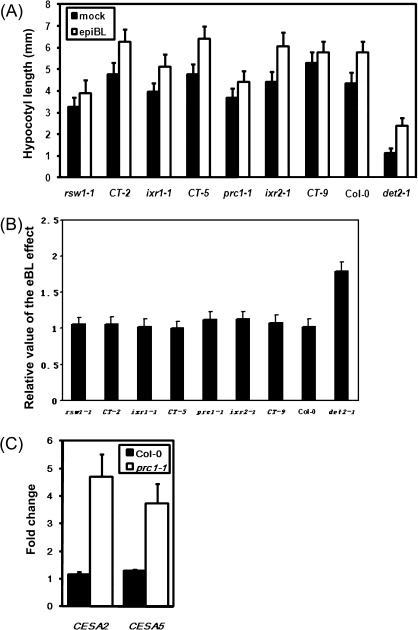
To test the ability of CESA mutants to respond to the applied BRs, the hypocotyl lengths of the wild type, det2-1, rsw1-1, ixr1-1, prc1-1, ixr2-1, and three T-DNA knockout mutants, ct-2 (CESA2), ct-5 (CESA5), and ct-9 (CESA9) (Persson et al., 2007; Desprez et al., 2007) were measured with or without an epiBL treatment. Although the weak alleles of rsw1-1, ixr1-1, and ixr2-1 can respond to BRs in the light or in the dark as indicated by a longer hypocotyl after the epiBL treatment, their hypocotyl was still shorter than that of the wild type (Fig. 6A, B), suggesting that CESA1, 3, and 6 are required for hypocotyl elongation promoted by epiBL. Moreover, the applied epiBL can largely promote hypocotyl elongation of ixr2-1 under both light and dark conditions, but it can only slightly promote the hypocotyl elongation of its non-allele prc1-1 in the light and cannot promote the hypocotyl elongation of prc1-1 in the dark (Fig. 6A, B; see Supplementary Fig. S5 at JXB online), indicating that prc1-1 was less insensitive to BRs both in light and dark. Because CESA6 is mainly involved in cellulose synthesis in the primary cell wall (Richmond and Somerville, 2000, 2001), it was suggested that CESA6 probably plays a key role in regulating BR-induced hypocotyl elongation and the knockout mutants ct-2, ct-5 and ct-9 were shown to have similar phenotypes to the wild type Col-0, suggesting that they are not essential in mediating BR-promoted cell elongation, or they are functionally redundant with other CESAs.
Fig. 6.
CESA mutants show an altered BR response. (A) Hypocotyl length of 7-d-old light-grown seedlings of rsw1-1, CT-2 (SALK_091570), ixr1-1, CT-5 (SALK_023353), prc1-1, ixr2-1,CT-9 (SALK_049129), Col-0, and det2-1 grown on the 1/2 MS medium with or without 10 nM epiBL. Means and standard deviations were calculated from 30–45 seedlings. (B) The relative value of hypocotyl elongation of 4-d-old dark-grown seedlings in the absence or presence of 10 nM epiBL. (C) The fold changes of CESA2 and CESA5 expression in the light, compared to that in the dark. Quantitative real-time PCR was performed using 7-d-old light-grown seedlings and 4-d-old dark-grown seedlings of prc1-1 and the wild type.
To investigate why the hypocotyl elongation of prc1-1 is normal in the light, but dramatically shorter than the wild type in the dark, the gene expression of all CESA genes in prc1-1 and the wild type was measured under both light and dark conditions. Previous studies have revealed that over-expression of CESA2 and CESA5 can partially rescue prc1-1 (Desprez et al., 2007; Persson et al., 2007). In this study, the levels of CESA2 and CESA5 gene expression induced by BRs in the light is about 4.7-fold and 3.5-fold of that in the dark, respectively (Fig. 6C). Together with CESA2 and CESA5 being up-regulated with epiBL treatment (Fig. 2A), it is suggested that BR-induced expression of CESA2 and CESA5 in prc1-1 may promote cellulose synthesis in order to keep the cell elongating in the light.
Discussion
In this study, several lines of evidence have been provided to conclude that BRs can induce the expression of most CESA genes. First, our data clearly demonstrated that BRs regulate the expression of multiple CESA genes in both the short term and the long term. Although most of the CESAs are induced by epiBL in 2 h, the CESA genes related to secondary cell wall synthesis were not induced in long-term treatments in young seedlings (Figs 2C, 3; see Supplementary Fig. S3 at JXB online). Furthermore, CESA9 and CESA10, which are mainly expressed in flowers, also showed the similar induction (Fig. 3; see Supplementary Fig. S3 at JXB online). It is suggested that BRs regulate different sets of CESA expression at different developmental stages. In addition, the BR-related mutants, bri1-301 and det2-1 are both dwarf plants with a similar developmental speed, but the expression of CESAs in the two mutants differ in their response to the epiBL treatment (Fig. 2A), demonstrating that the BR signal is important for inducing CESA expression. When BR signalling was enhanced in the BRI1-GFP over-expression line, the expression of these CESA genes was significantly increased (Fig. 2B). Moreover, the dry weight of aerial parts and the cellulose content in the stems of BR's related materials (Fig. 1) suggested that the BR signal promotes above-ground biomass accumulation.
Our results also indicate that BRs may control both primary and secondary growth through regulating the expression of different sets of CESAs. During primary cell wall synthesis, the CSC, containing CESA1, CESA3, and CESA6, synthesizes cellulose microfibres mainly for cell enlargement and cell elongation. A previous study has shown that the dwarf phenotype of the BR mutants is mainly caused by a reduced cell size, not by cell number (Kauschmann et al., 1996) and our results also proved that the non-allele of cesA6, prc1-1 cannot respond to exogenous BRs. Therefore, the BR-induced CESA genes in young seedlings (Figs 2, 3) may be essential for hypocotyl elongation. Our data also indicate that these CESAs with unknown function, including CESA2, CESA5, and CESA9, also participate in BR-induced cell elongation in young seedlings. Furthermore, BRs also induce the expression of CESA4 and CESA8 involved in the secondary growth in the stem (Fig. 2C). Interestingly, at the early developmental stages, the dry weight above ground per seedling between the BR-related mutants and the wild type was not much different (Fig.1), but after bolting, the height and dry weight of the BR-deficient mutants were dramatically lower than those in the wild type. Thus, at the early stages, BR signalling can regulate the expression of the CESA genes mainly involved in primary growth to promote cell elongation, while at the later developmental stages; BR signalling may affect secondary growth by regulating CESA4 and CESA8 expression.
The BR-activated BES1 can regulate many developmental processes probably by controlling the expression of many structural and regulatory genes. Previous microarray data indicate that the CESA5 (at4g38850) is up-regulated in bzr1-D, a gain-of-function mutation of BZR1 (He et al., 2005). Microarray data also indicate that most CESA genes are induced solely following BR treatment (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi; http://www.arabidopsis.org/portals/expression/microarray/ATGenExpress.jsp). Recently, it was reported that BRs can directly regulate the expression of many key genes involved in pollen and anther development (Ye et al., 2010). In this study, the ChIP assay and genetic studies indicated that CESA genes act downstream in the BR signalling to regulate hypocotyl elongation and plant growth. Recently, a chip on chip study by two groups showed the interesting result that CESA6 is the target of BZR1 (Sun et al., 2010; Yu et al., 2011), However, none of these CESA genes was identified as a BES1 target by Yu et al.’s study (Yu et al., 2011). It suggested that data from the chip on chip assay may miss some important genes.
Like BRs, many other hormones, including auxins, gibberellins, and cytokinins, can also significantly promote plant growth and plant size. For instance, the enhanced expression of AVP, which encodes a vacuolar H+-pyrophosphatase, and auxin-regulated genes, ARGOS and ARF2 (in the auxin signalling pathway), apparently regulate plant size by promoting cell division (Hu et al., 2003; Li et al., 2005; Okushima et al., 2005). In addition, over-expression of GA20ox required for GA biosynthesis can regulate plant size through regulating cell division and expansion (Coles et al., 1999). Recently, a cytokinin binding protein HOG1 has been shown to promote leaf size and seed yield (Godge et al., 2008). However, little is know about how the co-expressed genes related to cell wall expansion, cell wall formation, and the cell cycle, are regulated by various growth-promoting hormones. A large set of microarray data suggest that different hormones may regulate distinct sets of gene families in the same process of plant growth and development (Nemhauser et al., 2006). Interestingly, according to our analysed chip data, it is suggested that most CESA genes can be induced by BRs, but other growth-promoting hormones do not have such broad effect on the expression of CESA genes (https://www.genevestigator.com/gv/user/serveApplet.jsp) (see Supplementary Fig. S6 at JXB online).
Based on our findings and previous observations, a model to illustrate the role of BRs in plant growth in Arabidopsis is proposed (Fig. 7). During primary growth, BR signalling activates the transcription factor BES1 and promotes BES1 to associate with the E-boxes of the CESA1, CESA3, and CESA6 promoters to enhances their expression and cellulose biosynthesis in cell elongation. Following primary growth, the expression of CESA4 and CESA8, which are also promoted by BES1, contributes to plant height and secondary growth. Apparently, almost all CESA genes for primary and secondary cell wall accumulation are regulated by BR signalling in order to provide adequate cellulose to sustain the architecture of enlarged cells. Without CESA6, the CSC cannot generate enough new cellulose for primary cell wall deposition and so the hypocotyl cannot elongate in the dark. In the light, the BR signal can regulate the expression of CESA2, CESA5, or CESA9 and CESA10, which partially substitute for the function of CESA6 and rescue the dwarf phenotype of prc1-1.
Fig. 7.
A model to illustrate the mechanism of BR-regulated CESA expression in plant growth. The BR signalling activates the transcription factor BES1 and promotes its binding to the E-boxes of CESA1, CESA3, and CESA6 promoters to enhance their expression during the primary growth. BES1 can also promote the expression of CESA4 and CESA8 for the secondary growth. These CESAs provide more cellulose to sustain the architecture of enlarged cells in cell elongation. BR signalling can regulate the expression of CESA2, CESA5, or CESA9 and CESA10, which partially substituted for the function of CESA6 and rescued the dwarf phenotype of prc1-1 in the light.
This study on the mechanisms of BR regulating cellulose synthesis provides significant insights into the hormonal regulation of cellulose accumulation in Arabidopsis, but there are still many questions that remain to be studied further. First, many phytohormones can regulate plant size, but what their mechanisms are and how they cross-talk to regulate this process is still unknown. Second, it is known that different cellulose synthase complexes may function at different developmental stages and in different tissues. How cellulose biosynthesis is regulated, especially by BR, in different tissues and at different developmental stages is still poorly understood. Third, the applied epiBL can significantly induce the expression of CESA7, but BES1 cannot bind to the promoter region of CESA7, suggesting that the expression of CESA7 may be regulated by other unknown mechanisms.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. BRs induce hypocotyl elongation.
Supplementary Fig. S2. CESA7, CESA8, and CESA10 were expressed in flowers or stems.
Supplementary Fig. S3. Expression of CESAs was enhanced in seedlings grown on the 1/2 MS medium containing BRs.
Supplementary Fig. S4. BES1 can associate with the upstream elements of CESA genes.
Supplementary Fig. S5. The CESA mutants show an altered BR response.
Supplementary Fig. S6. Microarray data shows the regulation of CESA expression by various plant hormones. (https://www.genevestigator.com/gv/user/serveApplet.jsp).
Acknowledgments
The research was supported by a Grant 08KF02 from Shanghai Key Laboratory of Bio-energy Crop (to X Wang), and Grants 30871330 and 90817004 from the National Natural Science Foundation of China (to X Wang). We thank H Wang, S Zhang, and Z Cai for technical support in vector construction and qPCR. We thank Y Yin (Iowa State University) for kindly providing the anti BES1 antibody and L Li (Iowa State University) for advice on the ChIP experiments. We also thank Y Zhou, L Li, Z Cai, Y Lu, and Y Wei for critically reading the manuscript.
Glossary
Abbreviations
- BR
brassinosteroid
- CESA
cellulose synthase
- CSC
cellulose synthase complex
- epiBL
2, 4-epi-brassinolide
References
- Alexopouloua E, Sharmab N, Papatheoharic Y, Christoua M, Piscionerib I, Panoutsoud C, Pignatellib V. Biomass yields for upland and lowland switch grass varieties grown in the Mediterranean region. Biomass and Bionenergy. 2008;32:926–933. [Google Scholar]
- Aragunde NC, Lombardo C, Lamattina L. Nitric oxide: an active nitrogen molecule that modulates cellulose synthesis in tomato roots. New Phytologist. 2008;179:386–396. doi: 10.1111/j.1469-8137.2008.02466.x. [DOI] [PubMed] [Google Scholar]
- Caderas D, Vogler H, Mandel T, Rose JKC, McQueen-Mason S, Kuhlemeier C, Muster M. Limited correlation between expansin gene expression and elongation growth rate. Plant Physiology. 2000;123:1399–1413. doi: 10.1104/pp.123.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles JP, Phillips AL, Croker SJ, GarcÍ a-Lepe R, Lewis MJ, Hedden P. Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. The Plant Journal. 1999;17:547–556. doi: 10.1046/j.1365-313x.1999.00410.x. [DOI] [PubMed] [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2007;39:15572–15577. doi: 10.1073/pnas.0706569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T, Vernhettes S, Fagard M, Refregier G, Desnos T, Aletti E, Py N, Pelletier S, Höfte H. Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiology. 2002;128:482–490. doi: 10.1104/pp.010822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divi UK, Krishna P. Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. New Biotechnology. 2009;26:131–135. doi: 10.1016/j.nbt.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP. Cellulose biosynthesis in plants: from genes to rosettes. Plant and Cell Physiology. 2002;43:1407–1420. doi: 10.1093/pcp/pcf164. [DOI] [PubMed] [Google Scholar]
- Fujioka S. Characterization of brassinosteroid biosynthesis mutants in. Arabidopsis thaliana. RIKEN Review. 1999;21:5–6. [Google Scholar]
- Godge MR, Kumar D, Kumar PP. Arabidopsis HOG1 gene and its petunia homolog PETCBP act as key regulators of yield parameters. Plant Cell Reports. 2008;27:1497–1507. doi: 10.1007/s00299-008-0576-z. [DOI] [PubMed] [Google Scholar]
- Gonzalez N, Beemster GT, Inzé D. David and Goliath: what can the tiny weed Arabidopsis teach us to improve biomass production in crops? Current Opinion in Plant Biology. 2009;12:157–164. doi: 10.1016/j.pbi.2008.11.003. [DOI] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SSL, Gendron N, Sun CQ, Wang ZY. BZR1 Is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;15:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hématy K, Höfte H. Cellulose and cell elongation. Plant Cell Monograph. 2006;5:33–56. [Google Scholar]
- Hong Z, Tanaka MU, Fujioka S, Takatsuto S, Yoshida S, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. The Plant Cell. 2005;17:2243–2254. doi: 10.1105/tpc.105.030973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Xie Q, Chua NH. The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. The Plant Cell. 2003;15:1951–1961. doi: 10.1105/tpc.013557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurentin A, Edwards CA. A microtiter modification of the anthrone–sulfuric acid colorimetric assay for glucose-based carbohydrates. Analytical Biochemistry. 2003;315:143–145. doi: 10.1016/s0003-2697(02)00704-2. [DOI] [PubMed] [Google Scholar]
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Genetic evidence for an essential role of brassinosteroids in plant development. The Plant Journal. 1996;5:701–713. [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Molecular Biology Reporter. 1987;5:387–405. [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li J, Yang H, Peer WA, Richter G, Blakeslee J, Bandyopadhyay A, Titapiwantakun B, Undurraga S, Khodakovskaya M, Richards EL. Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science. 2005;310:121–125. doi: 10.1126/science.1115711. [DOI] [PubMed] [Google Scholar]
- Li J, Jin H. Regulation of brassinosteroid signaling. Trends in Plant Science. 2006;12:37–41. doi: 10.1016/j.tplants.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Luo M, Xiao YH, Li XB, Lu XF, Deng W, Li DM, Hou L, Hu MY, Li Y, Pei Y. GhDET2, a steroid 5a-reductase, plays an important role in cotton fiber cell initiation and elongation. The Plant Journal. 2007;51:419–430. doi: 10.1111/j.1365-313X.2007.03144.x. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. The Plant Cell. 2005;17:2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozak K, Ohme-Takagi M. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. The Plant Cell. 2007;19:270–280. doi: 10.1105/tpc.106.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Mitina I, Quach HL, Theologis A. AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. The Plant Journal. 2005;43:29–46. doi: 10.1111/j.1365-313X.2005.02426.x. [DOI] [PubMed] [Google Scholar]
- Pauly M, Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. The Plant Journal. 2008;54:559–568. doi: 10.1111/j.1365-313X.2008.03463.x. [DOI] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2007;39:15566–15571. doi: 10.1073/pnas.0706592104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refrégier G, Pelletier S, Jaillard D, Höfte H. Interaction between wall deposition and cell elongation in dark-grown hypocotyl cells in Arabidopsis. Plant Physiology. 2004;135:959–968. doi: 10.1104/pp.104.038711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. The cellulose synthase superfamily. Plant Physiology. 2000;124:495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. Integrative approaches to determining Csl function. Plant Molecular Biology Reporter. 2001;47:131–143. [PubMed] [Google Scholar]
- Robert S, Mouille G, Höfte H. The mechanism and regulation of cellulose synthesis in primary walls: lessons from cellulose-deficient Arabidopsis mutants. Cellulose. 2004;11:351–364. [Google Scholar]
- Sakurai A. Brassinosteroid biosynthesis. Plant Physiology and Biochemistry. 1999;37:351–361. [Google Scholar]
- Salas-Fernandez MG, Becraft PW, Yin YH, Lübberstedt T. From dwarves to giants? Plant height manipulation for biomass yield. Trends in Plant Science. 2009;14:454–461. doi: 10.1016/j.tplants.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Scheible WR, Eshed R, Richmond T, Delmer D, Somerville C. Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proceedings of the National Academy of Sciences, USA. 2001;98:10079–10084. doi: 10.1073/pnas.191361598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Veerabomma S, Abdel-Mageed HA, Fokar M, Asami T, Yoshida S, Allen RD. Brassinosteroid regulates fiber development on cultured cotton ovules. Plant and Cell Physiology. 2005;46:1384–1391. doi: 10.1093/pcp/pci150. [DOI] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Developmental Cell. 2010;5:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Tanabe S, Ashikari M, Fujioka S, et al. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. The Plant Cell. 2005;17:776–790. doi: 10.1105/tpc.104.024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZH. SKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;25:557–260. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao YZ, Zheng J, Xu ZM, Zhang XH, Zhang K, Wang GY. Functional analysis of ZmDWF1, a maize homolog of the Arabidopsis brassinosteroids biosynthetic DWF1/DIM gene. Plant Science. 2004;167:743–751. [Google Scholar]
- Taylor NG. Regulation of cellulose synthesis: another player in the game? New Phytologist. 2008;179:247–249. doi: 10.1111/j.1469-8137.2008.02504.x. [DOI] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vichers K, Turner SR. Interactions among three distinct CesA proteins essential for cellulose synthesis. Proceedings of the National Academy of Sciences, USA. 2003;100:1450–1455. doi: 10.1073/pnas.0337628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. The Plant Cell. 2000;12:2529–2540. doi: 10.1105/tpc.12.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff DM. Semi-micro determination of cellulose in biological materials. Analytical Biochemistry. 1969;32:420–424. doi: 10.1016/s0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
- Vogel J. Unique aspects of the grass cell wall. Current Opinion in Plant Biology. 2008;11:301–307. doi: 10.1016/j.pbi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- Wang X, Li X, Meisenhelder J, Hunter T, Yoshida S, Asami T, Chory J. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Developmental Cell. 2005;8:855–865. doi: 10.1016/j.devcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Setu H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma membrane receptor for plant steroids. Nature. 2001;410:380–382. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- Wightman R, Turner S. Trafficking of the plant cellulose synthase complex. Plant Physiology. 2010;153:427–432. doi: 10.1104/pp.110.154666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Xu ZY, Song J, Conner K, Barrena GV, Wilson ZA. Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. The Plant Cell. 2007;1:534–548. doi: 10.1105/tpc.106.046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Zhu W, Li L, Zhang S, Yin Y, Ma H, Wang X. Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proceedings of the National Academy of Sciences. 2010;13:6100–6105. doi: 10.1073/pnas.0912333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang Z, Garcia SM, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Yuan JS, Tiller KH, Al-Ahmad H, Stewart NR, Stewart CN., Jr. Plants to power: bioenergy to fuel the future. Trends in Plant Science. 2008;13:421–429. doi: 10.1016/j.tplants.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. The Plant Cell. 2011;4:634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou JL, McCarthy RL, Ye ZH. A battery of transcription factors Involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. The Plant Cell. 2008;20:2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Richardson E, Ye ZH. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta. 2007;225:1603–1611. doi: 10.1007/s00425-007-0498-y. [DOI] [PubMed] [Google Scholar]
- Zhou JL, Lee C, Zhong R, Ye ZH. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. The Plant Cell. 2009;21:248–266. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.